Abstract
We determined the role of Phospholipase Dα1 (PLDα1) and its lipid product phosphatidic acid (PA) in abscisic acid (ABA)-induced production of reactive oxygen species (ROS) in Arabidopsis thaliana guard cells. The pldα1 mutant failed to produce ROS in guard cells in response to ABA. ABA stimulated NADPH oxidase activity in wild-type guard cells but not in pldα1 cells, whereas PA stimulated NADPH oxidase activity in both genotypes. PA bound to recombinant Arabidopsis NADPH oxidase RbohD (respiratory burst oxidase homolog D) and RbohF. The PA binding motifs were identified, and mutation of the Arg residues 149, 150, 156, and 157 in RbohD resulted in the loss of PA binding and the loss of PA activation of RbohD. The rbohD mutant expressing non-PA-binding RbohD was compromised in ABA-mediated ROS production and stomatal closure. Furthermore, ABA-induced production of nitric oxide (NO) was impaired in pldα1 guard cells. Disruption of PA binding to ABI1 protein phosphatase 2C did not affect ABA-induced production of ROS or NO, but the PA–ABI1 interaction was required for stomatal closure induced by ABA, H2O2, or NO. Thus, PA is as a central lipid signaling molecule that links different components in the ABA signaling network in guard cells.
INTRODUCTION
The plant hormone abscisic acid (ABA) participates in diverse physiological processes, such as stomatal movement, seed dormancy and germination, vegetative growth, and response to abiotic and biotic stresses (Schroeder et al., 2001; Finkelstein et al., 2002; Assmann, 2003; Xiong and Zhu, 2003; Hirayama and Shinozaki, 2007). Under drought stress, ABA promotes stomatal closing and inhibits stomatal opening, thereby reducing transpiration and water loss. Using forward and reverse genetics and cellular, biochemical, and molecular approaches, a number of components in ABA signal transduction have been characterized, including G proteins, protein kinases/phosphatases, transcriptional factors, receptors, and enzymes involved in producing reactive oxygen species (ROS), such as O2− and H2O2 (Gosti et al., 1999; Merlot et al., 2001; Wang et al., 2001; Himmelbach et al., 2002; Mishra et al., 2006; Shen et al., 2006; Liu et al., 2007; Nilson and Assmann, 2007; Ma et al., 2009; Pandey et al., 2009; Park et al., 2009). In addition, lipid mediators generated by phospholipase D (PLD), phospholipase C (PLC), and sphingosine kinase have been identified as integral parts of the complex signaling cascades in the ABA response (Fan et al., 2004; Zhang et al., 2005; Wang et al., 2006). Recent studies show that PLD and its lipid product phosphatidic acid (PA) interact with a G protein and protein phosphatase to mediate the ABA response in guard cells. Abrogation of Phospholipase Dα1 (PLDα1) renders Arabidopsis thaliana insensitive to ABA in stomatal closure and leads to more water loss than in wild-type plants (Zhang et al., 2004), whereas overexpression of PLDα1 results in less water loss (Sang et al., 2001b). PLDα1-produced PA binds to the ABI1 protein phosphatase 2C, a negative regulator of ABA action. This interaction inhibits the function of ABI1 by inhibiting its phosphatase activity and tethers it to the plasma membrane. Membrane tethering is thought to prevent the interaction of ABI1 with the transcription factor ATHB6 (Zhang et al., 2004). On ABA inhibition of stomatal opening, PLDα1 binds to GPA1 (α-subunit of heterotrimeric G protein) and regulates its function by promoting the conversion of GTP-bound Gα to a GDP-bound Gα, thus producing PA that acts upstream of Gα to inhibit stomatal opening (Mishra et al., 2006).
PLD and PA are both implicated in ROS production. The depletion of PLDα1 in Arabidopsis decreases ROS production in leaves, and addition of PA results in recovery of ROS production in pldα1 mutants (Sang et al., 2001a). In response to an avirulent elicitor in tomato (Solanum lycopersicum), PA induces an oxidative burst (de Jong et al., 2004). PA induces leaf cell death in Arabidopsis by activating small G protein ROP2-mediated pathway of ROS generation (Park et al., 2004). NADPH oxidases are the source of ROS produced in ABA response and other processes, including pathogen recognition and root hair growth (Torres et al., 2002; Foreman et al., 2003; Kwak et al., 2003; Torres and Dangl, 2005). Plant NADPH oxidases, termed respiratory burst oxidase homologs (Rbohs), are homologs of the mammalian NADPH oxidase catalytic subunit gp91phox. Different Rboh genes have been isolated from rice (Oryza sativa), Arabidopsis, tomato (Solanum lycopersicum), tobacco (Nicotiana tabacum), and potato (Solanum tuberosum) (Torres and Dangl, 2005). Arabidopsis has 10 Rboh genes with diversified functions. Arabidopsis RbohC is required for the root hair development (Foreman et al., 2003), whereas RbohD and RbohF are involved in pathogen recognition and ABA-induced stomatal closure (Torres et al., 2002; Kwak et al., 2003; Bright et al., 2006). In Nicotiana benthamiana, RbohA and RbohB participate in ROS accumulation and viral resistance (Yoshioka et al., 2003).
In phagocytic cells, NADPH oxidase contains plasma membrane–bound cytochrome b558 (gp91phox and p22phox) and cytosolic regulatory proteins, including p67phox, p47phox, p40 phox, and small GTP binding protein Rac (Babior, 2004). Activation of the oxidase requires translocation of the cytosolic subunits to membrane-bound gp91phox and p22phox complex, and the p47phox subunit is regarded as a key regulator in the translocation (Karathanassis et al., 2002). During cell stimulation, p47phox is recruited to the membrane via SH3-mediated interaction with p22phox and Phox homology (PX)-mediated binding to phosphoinositides (PtdInsP2) and PA (Karathanassis et al., 2002; Ago et al., 2003). On the other hand, the membrane recruitment of p67phox is mediated via its binding to p47phox (Heyworth et al., 1991; Kami et al., 2002), whereas Rac is independently tethered to the membrane (Heyworth et al., 1994). These cytosolic regulatory proteins translocate to the plasma membrane to form an active enzyme complex capable of producing superoxide.
Homologs of the cytosolic regulator small GTPase Rac of mammalian NADPH oxidase are found in plants (Groom et al., 1996; Keller et al., 1998; Torres et al., 1998; Kawasaki et al., 1999; Wong et al., 2007), but no homologs of other regulatory components of mammalian NADPH oxidase, namely, p22phox, p67phox, p47phox, and p40phox, are found in the Arabidopsis genome (Torres and Dangl, 2005). In contrast with mammalian NADPH oxidases, plant Rboh proteins possess an extended N terminus with two Ca2+ binding EF-hand motifs. Using a heterologous expression system, a Ca2+ increase in the cytosol was found to be necessary for stimulating Arabidopsis RbohD. This stimulation requires a conformational change in the EF region, which is caused by Ca2+ binding (Ogasawara et al., 2008). Ca2+ may also regulate Rboh activity and ROS generation by phosphorylation mediated by calcium-dependent protein kinases (CDPKs) (Kobayashi et al., 2007; Ogasawara et al., 2008). Early genetic evidence pointed to involvement of the small GTPase in the regulation of ROS in plants (Ono et al., 2001; Schultheiss et al., 2002; Suharsono et al., 2002; Morel et al., 2004). The direct binding of rice Rac1 to the N terminus of rice RbohB was characterized recently (Wong et al., 2007). Rac-Rboh binding leads to an increase in ROS, thus inducing cytosolic Ca2+ accumulation, whereas an increase in cytosolic Ca2+ inhibits the Rac–Rboh interaction and reduces ROS production (Wong et al., 2007). Asai et al. (2008) reported that MAPK cascades regulate the expression of N. benthamiana RbohB and oxidative burst. However, compared to mammalian NADPH oxidase, much less is known about the regulatory mechanisms of plant Rboh. Specifically, it is unknown whether membrane factors, such as lipid mediators, regulate Rboh activity.
Arabidopsis RbohD and RbohF are located in the plasma membrane (Torres and Dangl, 2005, and references within), and RbohD and RbohF are expressed in Arabidopsis guard cells (Kwak et al., 2003). pldα1 and rbohD/F mutants show similar phenotypes; they are both insensitive to ABA-induced stomatal closure (Kwak et al., 2003; Zhang et al., 2004). PA stimulates ROS production in Arabidopsis leaves (Sang et al., 2001a). In light of these observations, this study was undertaken to address whether and how PLD and PA regulate ABA-mediated ROS generation in guard cells. The results show that PLDα1 is required for the ABA activation of NADPH oxidase and the production of ROS and that PLDα1-derived PA interacts directly with RbohD. In addition, the data indicate that the production and action of NO are downstream of the PLDα1-mediated production of PA and ROS. The interaction between PA and ABI1 is not required for the ABA-induced production of ROS and NO but is important for mediating the ROS effect on stomatal closure.
RESULTS
Loss of PLDα1 Leads to a Decrease in ABA-Mediated ROS Production in Guard Cells
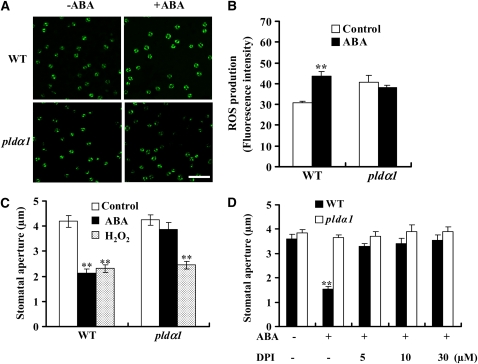
The PLDα1-null Arabidopsis mutant, pldα1, which contains a T-DNA insertion 1027 nucleotides downstream of the initiation codon, was insensitive to ABA-induced stomatal closure (Zhang et al., 2004; Mishra et al., 2006). The ABA response of the mutant was restored to that of the wild type after pldα1 was complemented with the native PLDα1 gene, verifying that the loss of PLDα1 is responsible for the defect in ABA response (Mishra et al., 2006). To further determine the cellular role of PLDα1 in the regulation of stomatal closure, we compared ROS production in pldα1 and wild-type guard cells using the ROS-sensitive, cell-permeable, fluorescent dye, 2′,7′-dichlorofluorescin diacetate (H2DCFDA) (Pei et al., 2000). H2DCFDA diffuses through the cell membrane and is hydrolyzed by intracellular esterases to form nonfluorescent H2DCF, which is oxidized to fluorescent DCF by ROS (Wang and Joseph, 1999). H2DCFDA is an effective dye for measuring ROS in our system because ABA-induced DCF fluorescence was shown to be inhibited by a suicide substrate inhibitor of NADPH oxidases, diphenylene iodonium (DPI). As a positive control, the superoxide donor phenazime methosulfate (5 μM) resulted in an increase in DCF fluorescence (see Supplemental Figure 1 online).
Without ABA treatment, wild-type and pldα1 guard cells exhibited similar basal staining of ROS. Treatment of epidermal strips with 50 μM ABA induced a rapid (within 5 min) increase in ROS levels, as indicated by the change in fluorescent intensity compared with the tissue not exposed to ABA treatment (Figure 1A). Approximately a 38% increase in ROS level occurred in wild-type guard cells after the ABA treatment. However, no increase in ROS was observed in pldα1 guard cells after ABA treatment for up to 10 min (Figures 1A and 1B). ABA was dissolved in ethanol (0.1%, v/v), and this level of ethanol alone had no effect on ROS generation, RAB18 expression, PLDα activity, and stomatal closure (see Supplemental Figure 2 online). The effect of ABA on ROS increase in the wild type and pldα1 was verified by measuring the total H2O2 content in leaves with an Amplex red hydrogen peroxide/peroxidase assay kit (Molecular Probes) (see Supplemental Figure 3A online). The results indicate that PLDα1 is required for ABA-mediated ROS production in guard cells.
Figure 1.
PLDα1 Mediates ABA-Induced ROS Production and Stomatal Closure.
(A) Representative images of ROS production indicated by fluorescent dye DCF. Epidermal peels were loaded with H2DCFDA for 10 min before adding 50 μM ABA. Photomicrographs were taken 5 min after ABA addition. Bar = 100 μm.
(B) Quantification of ROS production in guard cells of the wild type and pldα1 following ABA treatment. Confocal planes were quantified as mean pixel intensities. The data are means ± se from three independent experiments; n = 50 of each genotype per experiment.
(C) Applied H2O2 induces stomatal closure in the wild type and pldα1. The data are means ± se from three independent experiments; n = 60 apertures each genotype per experiment.
(D) Effect of NADPH oxidase inhibitor DPI on ABA-induced stomatal closure. The epidermal peels were pretreated with DPI for 30 min before 50 μM ABA was added. The data are means ± se from three independent experiments; n = 50 apertures each genotype per experiment.
For all stomatal bioassays, at least three separate peels were observed for each treatment per genotype. Data were compared using Student's t test. Two asterisks in (B) to (D) indicate that the mean value is significantly different from that of the control (P < 0.01).
To determine whether the loss of PLDα1 affects the ROS response in guard cells, we tested the effect of H2O2 on stomatal movement in wild-type and pldα1 leaves. H2O2 induced stomatal closure in both the wild type and pldα1 (Figure 1C) in a dosage-dependent manner (see Supplemental Figure 3B online). By contrast, ABA induced stomatal closure in the wild type, but not pldα1 (Figure 1C). Thus, the loss of PLDα1 impairs the production of ROS but does not affect the stomatal response to ROS, suggesting that PLDα1 acts upstream of ROS and may regulate its production in the ABA signaling pathway.
PLDα1 Mediates ABA-Induced ROS Production via NADPH Oxidase
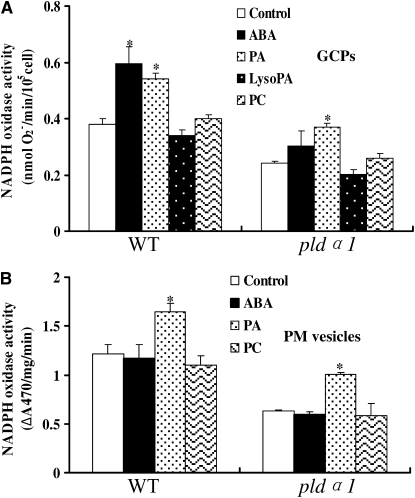
To examine how PLDα1 affects ROS production, we investigated the effect of PLDα1 and PA on NADPH oxidase activity in wild-type and pldα1 plants subjected to ABA treatment. NADPH oxidase has been reported to be an important source of ROS production in ABA signaling in Arabidopsis (Kwak et al., 2003; Suhita et al., 2004). When the epidermal strips were pretreated with the NADPH oxidase inhibitor DPI, the ABA-induced stomatal closure was inhibited in the wild type (Figure 1D), suggesting that activation of NADPH oxidase is required for ABA-induced stomatal closure. The treatment of DPI did not alter the stomatal aperture of pldα1, presumably because NADPH oxidase activity in the mutant guard cells was not activated in response to ABA.
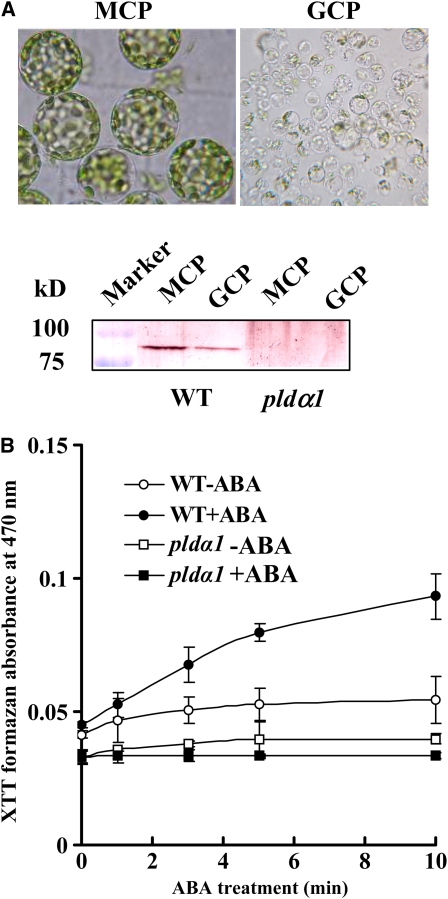
To determine whether PLDα1 is involved in NADPH oxidase activation, we measured NADPH oxidase activity using guard cell protoplasts (GCPs) isolated from leaf epidermal strips. PLDα1 protein was present in both mesophyll cell protoplasts (MCPs) and GCPs of the wild type, but not of pldα1 (Figure 2A). Treatment of GCPs with 10 μM ABA induced an increase in O2− levels, measured as a reduction of tetrazolium dye (XTT) in the wild type compared with the control (0.02% ethanol only, v/v). ABA-induced O2− generation declined sharply in pldα1 GCPs (Figure 2B). These results suggest that PLDα1 is involved in ABA-induced activation of NADPH oxidase and ROS production.
Figure 2.
PLDα1 and PA Mediate ABA-Enhanced NADPH Oxidase Activity and ROS Production.
(A) Isolated MCPs and GCPs (top panel); immunoblotting of PLDα1 (bottom panel). Proteins from the wild type (MCPs, 26 μg; GCPs, 15 μg) and pldα1 (MCPs, 30 μg; GCPs, 22 μg) were separated by 8% SDS-PAGE. PLDα1 was made visible by alkaline phosphatase staining after blotting with PLDα antibody.
(B) ABA-induced O2− production in the wild type and pldα1 GCPs (4 × 104 protoplasts). GCPs were treated with 10 μM (±) ABA dissolved in 0.02% (v/v) ethanol (+ABA) or 0.02% (v/v) ethanol only (−ABA). O2− production was measured by the detection of XTT formazan using spectrophotometric analysis at A470. Data are means ± se of three independent experiments.
ABA Induces Different Changes in PA Species in the Wild Type and pldα1
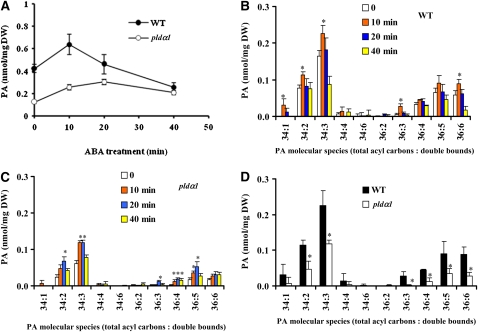
We previously showed that PLDα1 was activated in the ABA response and that phosphatidylcholine (PC) was hydrolyzed to PA in leaf protoplasts labeled with fluorescent PC (Zhang et al., 2004). To further characterize the PA change in response to ABA, we used electrospray ionization–tandem mass spectrometry (ESI-MS/MS) to analyze PA species in leaves. The level of total PA was lower in pldα1 than in wild-type leaves, with or without ABA treatments (Figure 3A). ABA induced a transient increase in PA, with a maximum increase occurring at 10 min after ABA application to wild-type leaves. Total PA in pldα1 leaves also increased after an application of ABA, which may result from activities of other PLDs and/or diacylglycerol kinases (DGKs). However, the amount of PA in the mutant after 10 min of ABA treatment was 2.4-fold lower than that in the wild type.
Figure 3.
ABA-Induced Changes in PA Species in Arabidopsis Leaves.
(A) Change in total PA content in leaves after ABA treatment.
(B) and (C) ABA-induced changes in PA molecular species in the wild type (B) and pldα1 (C).
(D) Comparison of PA molecular species in wild-type and pldα1 leaves treated with ABA for 10 min.
Data in (A) to (D) are means ± se (n = 4 or 5). The asterisk in (B) and (C) indicates that the mean value is significantly higher than that of leaves not subjected to ABA treatment (0 time) (P < 0.05). The asterisk in (D) indicates that the mean value is significantly different from that of the wild type (P < 0.05).
Analysis of PA molecular species revealed further distinguishable changes between wild-type and pldα1 plants after ABA treatment (Figures 3B and 3C). The major molecular species of PAs in the wild type were 34:2 (16:0-18:2), 34:3 (16:0-18:3), 36:4 (18:2-18:2), 36:5 (18:2-18:3), and 36:6 (18:3-18:3) (Devaiah et al., 2006; Figure 3B). Although the basal levels of PA species 34:1 (16:0-18:1) and 36:3 (18:1-18:2) were very low, the level of these PAs dramatically increased 10 min after ABA application. In addition, 34:2, 34:3, and 36:6 PAs were significantly increased in wild-type leaves after ABA treatment for 10 min (Figure 3B). In comparison, in pldα1, the PA species induced by ABA were primarily 34:2, 34:3, 36:4, and 36:5 PAs (Figure 3C). In addition, the magnitude of increase in 34:2 PA at 10 min was lower in pldα1 than in wild-type leaves. After a 10-min ABA treatment, the contents of 34:2, 34:3, 36:3, 36:4, 36:5, and 36:6 PAs in wild-type leaves were significantly higher than those in pldα1 mutant leaves (Figure 3D). The results show that abrogation of PLDα1 leads to changes in the amount, the magnitude, and molecular species of PA in response to ABA.
PA Binds to NADPH Oxidases
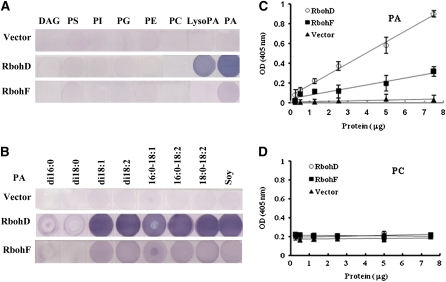
To investigate how PA affects NADPH oxidase function, we examined the potential interaction of PA with RbohD and RbohF, which are the major Rbohs in guard cells and are upregulated by ABA (Kwak et al., 2003). These Rbohs share about 49% identity in predicted amino acid sequences (see Supplemental Figure 4 online). RbohD and RbohF were expressed in Escherichia coli as His-tagged proteins using RbohD and RbohF cDNAs cloned from wild-type Arabidopsis. The isolated proteins were tested for lipid binding using a filter-binding assay. Compared with the empty vector control, both Rboh D and RbohF bound to PA from egg yolk, but not to PC, phosphatidylethanolamine, phosphatidylglycerol, phosphatidylinositol, phosphatylserine, or diacylglycerol (Figure 4A). PA binding to RbohD was stronger than binding to RbohF. RbohD and RbohF also bound to lysoPA, but more weakly than to PA. Phosphatidylinositol 3-phosphate [PtdIns(3)P], which is essential for ROS generation in the salt stress response (Leshem et al., 2007) and stomatal closure induced by ABA (Park et al., 2003), has been demonstrated to bind to mammalian NADPH oxidase subunit p40phox (Kanai et al., 2001; Honbou et al., 2007). Therefore, we further investigated RbohD and RbohF binding to PtdIns(3)P, PtdIns(4)P, and PtdIns(3,4)P2, and no lipid-RbohD (or -RbohF) binding was found (see Supplemental Figure 5 online). To investigate the specific interaction of PA with RbohD (or RbohF), we tested different PA species. Both RbohD and RbohF bound to dioleoyl PA (di18:1 PA), dilinoleoyl PA (di18:2 PA), palmitoyl-oleoyl PA (16:0-18:1 PA), palmitoyl-linoleoyl PA (16:0-18:2 PA), and stearoyl-linoleoyl PA (18:0-18:2 PA), but not to dipalmitoyl PA (di16:0 PA) or distearoyl PA (di18:0 PA). RhohD displayed stronger binding than did RbohF (Figure 4B). Because 16:0-18:2 PA is commercially available and is a PA species that is significantly increased by exposure to ABA in Arabidopsis leaves, we used 16:0-18:2 PA for functional analysis in further experiments.
Figure 4.
PA Binds to RbohD and RbohF Proteins.
(A) Lipid binding specificity of RbohD and RbohF proteins on filters. Lipids (10 μg) were spotted onto nitrocellulose and incubated with purified RbohD and RbohF expressed in E. coli. Equal amounts of RbohD and RbohF protein were used. The protein bound to lipids was visualized by immunoblotting. Lipids from egg yolk were used. DAG, diacylglycerol; PS, phosphatidylserine; PI, phosphatidylinositol; PG, phosphatidylglycerol; PE, phosphatidylethanolamine; PC, phosphatidylcholine; LysoPA, oleoyl lyso PA.
(B) Different PA species bind to RbohD and RbohF proteins on filters. PA species on filters include dipalmitoyl PA (di16:0 PA), distearoyl PA (di18:0 PA), dioleoyl PA (di18:1 PA), dilinoleoyl PA (di18:2 PA), palmitoyl-oleoyl PA (16:0-18:1 PA), palmitoyl-linoleoyl PA (16:0-18:2 PA), stearoyl-linoleoyl PA (18:0-18:2 PA ), and natural PA (from soy).
(C) Binding of RbohD andRbohF proteins to 16:0-18:2 PA by ELISA assay.
(D) Binding of RbohD and RbohF proteins to 16:0-18:2 PC by ELISA assay.
The data in (C) and (D) are means ± se of three replicates.
[See online article for color version of this figure.]
The lipid–RbohD (-RbohF) interaction was further determined using an ELISA-based assay (Ghosh et al., 1996). 16:0-18:2 PA was coated on microtiter plates and incubated with His-tagged RbohD or RbohF. Both RbohD and RbohF bound to PA in a dose-dependent manner, but the binding of RbohF was weaker than that of RbohD (Figure 4C). The results were consistent with those of filter binding experiments (Figure 4B). Increases in RbohD and RbohF concentrations had no effect on PC binding capacity (Figure 4D). These results suggest that RbohD and RbohF specifically bind to PA.
A Specific PA Species Activates NADPH Oxidase
To determine whether PLDα1-derived PA affects NADPH oxidase activity, we tested the effects of 16:0-18:2 PA on ROS generation in GCPs. Treatment of MCPs with 16:0-18:2 PA resulted in the activation of NADPH oxidase activity in the wild type in a dose-dependent manner, reaching a maximum at 1 μM. By contrast, 16:0-18:2 PC had no stimulatory effect (see Supplemental Figure 6 online). We then determined the effects of different lipids on NADPH oxidase activity in wild-type and pldα1 GCPs. 16:0-18:2 PA stimulated NADPH oxidase activity in both the wild type and pldα1, whereas 16:0-18:2 PC or LysoPA did not affect the activity significantly (Figure 5A). Di16:0 PA had no effect on ROS production in GCPs or on stomatal closure (see Supplemental Figures 7A and 7B online).
Figure 5.
PA Activates NADPH Oxidase Activity in GCPs and the Plasma Membrane.
(A) The effect of PA on NADPH oxidase activity in GCPs. GCPs were incubated with XTT dye together with 1 μM 16:0-18:2 PA,16:0-18:2 PC, or LysoPA or 10 μM ABA at 25°C for 10 min. After centrifugation, the supernatant was used for spectrophotometric analysis of XTT formazan production at A470. The NADPH activity calculation is described in Methods.
(B) The effect of PA on NADPH oxidase activity in the plasma membrane. Plasma membrane vesicles (3 to 6 μg protein) were incubated with XTT, together with 1 μM 16:0-18:2 PA,16:0-18:2 PC, or 10 μM ABA at 25°C for 10 min. The reaction solution was used for spectrophotometric analysis of XTT formazan production at A470. NADPH oxidase activity was expressed as ΔA 470 per mg protein per min (ΔA 470 represents the difference of XTT formazan absorbance at 470 nm in the presence and absence of superoxide dismutase [SOD]).
The data are means ± se of three independent experiments. The asterisk in (A) and (B) indicates that the mean value is significantly different from that of the control (P < 0.05).
We next isolated plasma membrane vesicles from leaves. Application of 16:0-18:2 PA (1 μM) to vesicles stimulated plasma membrane–associated NADPH oxidase activity, whereas the same concentration of PC did not stimulate the activity (Figure 5B). Unlike the case in GCPs (Figure 5A), ABA had no stimulation on NADPH oxidase activity associated with the plasma membrane. These results suggest that ABA does not directly stimulate the activity of NADPH oxidase or PLD but acts through other intracellular factors.
Identification of the PA Binding Region and Sites in Rbohs
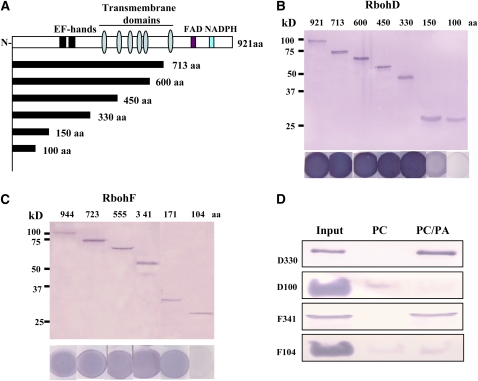
To identify the protein region involved in PA binding, serial deletion mutants of RbohD and RbohF were generated and expressed in E. coli. As shown in Figure 6A, all plant Rbohs have an N-terminal extension with two EF-hand motifs, six transmembrane domains, and flavin adenine dinucleotide and NADPH binding motifs at the C terminus (Torres and Dangl, 2005). Deletion of the amino acid residue 331 to the C terminus did not affect PA binding of RbohD, indicating that the PA binding region resides in the N-terminal 330–amino acid residues. However, the peptide consisting of only the first 100 amino acids displayed no PA binding (Figure 6B), suggesting that the PA binding motif resides in amino acid residues 101 to 330 of RbohD. For RbohF, the PA binding motif was mapped to amino acid residues 104 to 341 (Figure 6C).
Figure 6.
Characterization of the PA Binding Domain of RbohD and RbohF.
(A) Schematic diagram of Rboh protein with the functional domains and deletion structures used in recombinant protein expression and PA binding indicated. Flavin adenine dinucleotide (FAD) and NADPH binding domains, two EF-hands, and six transmembrane domains are shown.
(B) Immunoblotting of His-tagged RbohD N-terminal fragments expressed in E. coli (top panel) and 16:0-18:2 PA binding on filters using the same proteins as in the upper panel (bottom panel).
(C) Immunoblotting of His-tagged RbohF N-terminal fragments (top panel) and 16:0-18:2 PA binding on filters using the same proteins as in the upper panel (bottom panel).
(D) RbohD and RbohF fragments bind to liposomes containing 16:0-18:2 PA. 200 μL of RbohD fragment D330 (D330) and RbohF fragment F341 (F341) (0.5 μg/μL), and 200 μL of D100/F104 (1.25 μg/μL of each protein) were mixed with 200 μL of lipid vesicles comprised of 16:0-18:2 PC:PA (3:1) or PC. The vesicles were pelleted by centrifugation, and the liposomal binding proteins were detected by immunoblotting with anti-His antibody. Twenty percent of the proteins used for liposome binding were loaded as a control (input).
[See online article for color version of this figure.]
The PA–RbohD (or -RbohF) interaction was further examined by a liposome binding assay. Purified N-terminal fragments of RbohD (residues 1 to 100 and 1 to 330) and of RbohF (residues 1 to 104 and 1 to 341) were incubated with lipid vesicles comprised of PC and PA at a ratio of 3:1 or PC only. The vesicles were then pelleted and examined by immunoblotting to determine the amount of RbohD (or RbohF) fragments associated with liposomes. As shown in Figure 6D, RbohD330 (residues 1 to 330) and RbohF341 (residues 1 to 341) bound to the PA-containing vesicles but not to the PC-only vesicles (Figure 6D). When the same amount of each fragment was incubated with PC:PA vesicles, band intensity indicated that more RbohD330 than RbohF was present in the pelleted liposomes (Figure 6D). For RbohD100 and RbohF104, significantly less binding was observed in vesicles comprised of either PC (for D100 and F104) or PC/PA (F104). The results confirm that the PA binding region resides in amino acid residues 101 to 330 (RbohD) and 105 to 341 (RbohF).
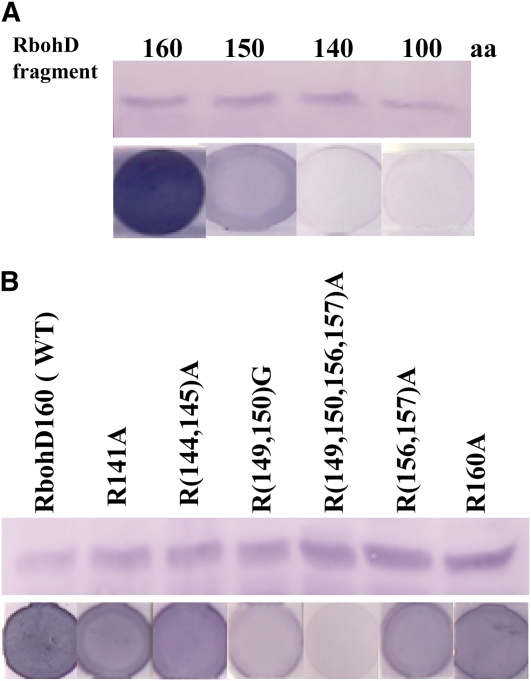
Further deletions were conducted to map the PA binding motif. The RbohD140 fragment (residues 1 to 140) showed no PA binding ability, and RbohD150 had less PA binding activity than did the full-length protein, whereas RbohD160 bound to PA with a similar affinity as did the full-length RbohD (Figures 6B and 7A). The results indicate that amino acid residues 140 to 160 are required for PA binding. The potential PA binding motif of RbohD is rich in the basic residue Arg (Supplemental Figure 4 online). Basic residues are usually involved in PA binding, but a consensus amino acid sequence has yet to be identified as a common PA binding motif in animals and/or plants (Wang et al., 2006). To determine whether the amino acid residues are involved in PA binding, several Arg residues were mutated to Ala or Gly (Figure 7B). Single mutations R141A and R160A did not affect PA binding to RbohD compared with the PA binding fragment RbohD160. The double mutation R(144,145)A had no effect on PA binding (Figure 7B). Another double mutation R(156,157)A slightly decreased PA binding compared to RbohD160. However, the quadruple mutation R(149,150,156,157)A caused RbohD160 to lose the ability to bind PA. The change of R149 and R150 to Gly also resulted in an obvious decrease in RbohD160 binding to PA (Figure 7B). The data suggest that R(149,150,156,157) are important residues in the binding of RbohD to PA.
Figure 7.
Identification of PA Binding Sites in RbohD.
(A) Binding of RbohD fragments to PA. Top panel: immunoblotting of N-terminal fragments (1 to 100, 140, 150, and 160 amino acids, respectively) expressed in E. coli. Bottom panel: binding of protein fragments to PA on filters.
(B) Expression of mutated RbohD fragments and detection of PA binding. Top panel: immunoblotting of wild-type and mutated RbohD160 fragments expressed in E. coli. Bottom panel: binding of RbohD160 fragments to PA on filters using the same protein as in the top panel.
[See online article for color version of this figure.]
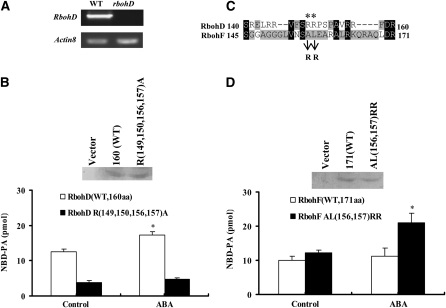
We next determined the effect of these basic amino acid residues on the interaction between PA and RbohD (RbohF) in guard cell protoplasts. We expressed the PA binding fragment RbohD160 (1 to 160 amino acids) tagged with HA in the RbohD-null mutant (SALK-120299, named rbohD; Figure 8A) GCPs. The protein levels of RbohD160 and its quadruple mutant [R(149,150,156,157)A] were similar, as measured by immunoblotting (Figure 8B, top panel). The protoplasts expressing these proteins were labeled with fluorescent 12[(7-nitro-2-1, 3-benzoxadiazol-4-yl) amino] dodecanoyl phosphatidylcholine (NBD-PC; which is converted to NBD-PA by PLD), treated with 10 μM ABA, and tracked by lysis and immunoprecipitation with an HA antibody. In the absence of ABA treatment, the amount of NBD-PA precipitated with wild-type RbohD 160 fragments was about threefold greater than that with the quadruple mutant protein fragment. ABA treatment of GCPs for 10 min resulted in a 45% increase in the amount of PA associated with wild-type RbohD160 but not with the mutant (Figure 8B, bottom panel). When Ala and Leu (residues 156 and 157) in RbohF (corresponding to residues 149 and 150 in Rboh D) were mutated to Arg and Arg (Figure 8C), the amount of PA associated with RbohF increased greatly in the presence of ABA (Figure 8D). The data confirm the PA–Rboh interaction in guard cells and highlight the importance of residues R(149,150) (RbohD) in PA binding.
Figure 8.
Quantification of Fluorescent PA Bound to RbohD and RbohF Fragments Expressed in GCPs.
(A) RT-PCR analysis of RbohD gene expression in the wild type and rbohD mutant. The experiment was repeated three times under the same conditions.
(B) Quantification of fluorescent PA bound to wild-type and non-PA-binding RbohD fragments. Top panel: immunoblotting of HA-tagged RbohD fragments (160 amino acids) of the wild type and non-PA-binding mutant [R(149,150,156,157)A] expressed in rbohD GCPs. Bottom panel: quantification of fluorescent NBD-PA coimmunoprecipated with RbohD fragments in the absence or presence of 10 μM ABA for 10 min.
(C) Amino acid alignment of the PA binding region in RbohD and corresponding region in RbohF. The double mutation [AL(156,157)RR] in the RbohF sequence is indicated.
(D) Quantification of fluorescent PA bound to wild-type and double mutant RbohF [AL(156,157)RR] fragments. Top panel: immunoblotting of RbohF fragments of the wild type and double mutant [AL(156,157)RR]. Bottom panel: quantification of fluorescent NBD-PA coimmunoprecipated with RbohF fragments in the absence or presence of 10 μM ABA for 10 min.
Data in (B) and (D) are means ± se of three independent experiments.
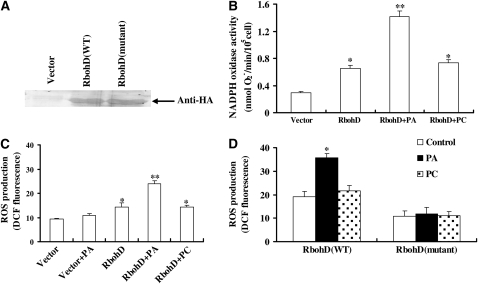
PA Binding Stimulates RbohD NADPH Oxidase Activity
To analyze the effect of the PA–Rboh interaction on NADPH oxidase activity, we expressed HA-tagged full-length RbohD in GCPs that were isolated from rbohD leaves and measured the resulting ROS production. The RbohD protein expressed in the GCPs was detected by immunoblotting with an HA antibody (Figure 9A). We first measured O2− production in GCPs transfected with RbohD-HA and empty vector based on the reduction of XTT. Overexpression of RbohD induced significant increases in O2− production. Addition of 16:0-18:2 PA further stimulated O2− production in the transfected cells. By contrast, 16:0-18:2 PC had no such stimulation on O2− generation (Figure 9B). The PA activation of RbohD-mediated ROS generation was tested with the fluorescent dye H2DCFDA. PA, but not PC, was found to stimulate ROS generation in the GCPs expressing RbohD, whereas PA had a negligible effect on ROS production in GCPs transfected with empty vector (Figure 9C). The results suggest that PA specifically stimulates ROS production in GCPs expressing RbohD.
Figure 9.
PA Stimulates RbohD Activity.
(A) Immunoblotting of full-length proteins of the wild type and the non-PA-binding mutant RbohD expressed in rbohD GCPs. The proteins were separated by SDS-PAGE and probed with anti-HA antibody. The empty vector with HA tag was used as a control.
(B) The effect of PA on RbohD activity. RbohD-HA or empty vector was expressed in rbohD mutant GCPs for 4 h. Reaction buffer and 1 μM 16:0-18:2 PA (or 16:0-18:2 PC) were added to GCPs for 10 min. After centrifugation, the supernatant was used for spectrophotometric analysis of XTT formazan production at A470. NADPH activity calculation is described in Methods. Data are means ± se of 4 × 104 GCPs per treatment per experiment.
(C) The effect of PA on ROS production in GCPs expressing RbohD. RbohD-HA or empty vector was expressed in rbohD mutant GCPs for 4 h. A total of 1 μM 16:0-18:2 PA (or 16:0-18:2 PC) was added to the GCPs for 10 min after the GCPs were stained with H2DCFDA for 10 min. Confocal images were quantified as mean pixel intensities ± se; n = 60 cells per treatment per experiment.
(D) The effect of PA on ROS production by wild-type and mutated RbohD. RbohD-HA or RbohD-HA with R(149,150,156,157)A mutations was expressed in rbohD GCPs for 4 h, followed by DCF fluorescence assaying of ROS production. Confocal images were quantified as mean pixel intensities ± se; n = 55 cells per treatment per experiment.
Data in (B) to (D) are means ± se of three independent experiments. The asterisk in (B) and (C) indicates that the mean value is significantly different from that of the vector (one asterisk for P < 0.05; two asterisks for P < 0.01). An asterisk in (D) indicates that the mean value is significantly different from the control (P < 0.05).
To directly establish if PA-RbohD binding is important for PA-activated generation of ROS in GCPs, we compared the effect of PA on ROS production in rbohD GCPs expressing wild-type RbohD with those expressing the non-PA-binding quadruple mutant. In GCPs expressing the quadruple mutated RbohD protein, PA or PC had no effect on ROS production. However, in GCPs expressing wild-type RbohD, PA, but not PC, increased ROS production significantly (Figure 9D). All of the above data demonstrate that PA-RbohD binding leads to the activation of RbohD NADPH oxidase activity and ROS generation.
The PA–Rboh Interaction Regulates ABA-Mediated ROS Generation and Stomatal Closure
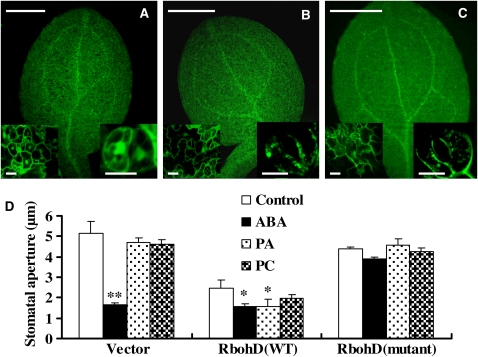
We then asked whether the PA–Rboh interaction is essential for PA- and ABA-mediated ROS generation and stomatal closure. To address this question, we expressed full-length RbohD tagged with green fluorescent protein (GFP) in rbohD using an in planta transient transformation assay (Marion et al., 2008). As shown in Figures 10A to 10C, GFP expression was observed in epidermal cells, including guard cells. Wild-type RbohD and non-PA-binding RbohD [quadruple mutation R(149,150,156,157)A] proteins had similar patterns of expression in cotyledons, according to the GFP fluorescent density (Figures 10A to 10C, inset).
Figure 10.
Abrogation of PA-RbohD Binding Impairs ABA-Induced Stomatal Closure.
(A) to (C) Transient expression of wild-type and non-PA-binding RbohD fused to GFP in rbohD seedlings. RbohD-GFP was visualized in whole cotyledons, in epidermal cells (left bottom), and in guard cells (right bottom). Cotyledons transformed with GFP alone (A), RbohD-GFP (B), and non-PA-binding RbohD-GFP (C). Bars = 0.5 mm for cotyledons and 10 μm for insets.
(D) Changes in stomatal aperture in response to ABA, PA, and PC. rbohD seedlings expressing GFP vector, RbohD-GFP, or non-PA-binding RbohD-GFP were incubated with 2 μM ABA or 50 μM 16:0-18:2 PA (or 16:0-18:2 PC) for 2 h, and stomatal apertures were measured. Data are means ± se of three independent experiments; n = 65 apertures per treatment per experiment. The asterisk indicates that the mean value is significantly different from that of the control (one asterisk for P < 0.05; two asterisks for P < 0.01).
As compared with the GFP vector control, overexpression of wild-type RbohD protein resulted in significant stomatal closure in the absence of ABA, whereas overexpression of the non-PA-binding RbohD led to slight stomatal closure (Figure 10D). Cotyledons became yellow in seedlings transiently expressing wild-type RbohD (see Supplemental Figure 8 online). When 2 μM ABA was applied to the cotyledons, the stomata closed in plants carrying the vector or wild-type RbohD protein. By contrast, ABA-induced stomatal closure was impaired in plants expressing the non-PA-binding RbohD protein (Figure 10D). PA induced stomatal closure in plants expressing wild-type RbohD but not in those carrying the vector or non-PA-binding RbohD. PC had no effects on stomatal closure in any tested plants (Figure 10D). ROS production in the cotyledons of these plants was analyzed by 3,3′-diaminobenzidine staining. The results showed that ABA induced ROS production in guard cells of the cotyledons transformed with wild-type RbohD but that ROS production was seriously impaired in the guard cells of cotyledons transformed with non-PA-binding RbohD (see Supplemental Figure 9 online). Taken together, these results demonstrate that the PA–RbohD interaction is necessary for ABA-mediated ROS generation and stomatal closure.
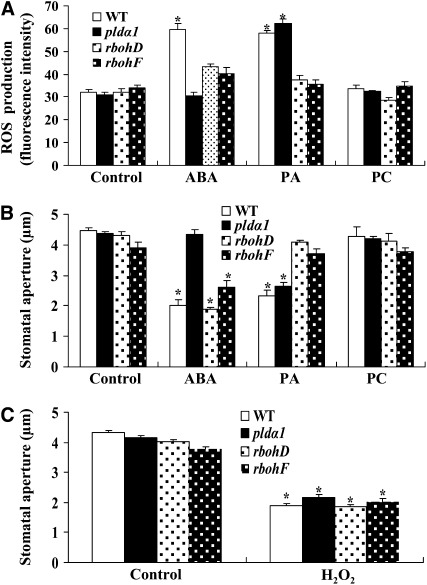
We next compared the effects of PA and of ABA on ROS production in guard cells and stomatal closure in pldα1, rbohD, and rbohF mutants. The ABA-induced ROS increase in guard cells was abolished in pldα1 but was considerably reduced in rbohD and rbohF. PA increased ROS generation in the wild type and pldα1 but not in rbohD and rbohF. PC and LysoPA did not stimulate ROS production in any of the four genotypes (Figure 11A; see Supplemental Figure 10 online).
Figure 11.
Abrogation of RbohD and RbohF Alters ABA- and PA-Induced ROS Production and Stomatal Closure.
(A) ROS production in guard cells in response to ABA or lipids. Epidermal peels preloaded with H2DCFDA for 10 min were treated with 50 μM ABA for 5 min or 50 μM 16:0-18:2 PA (or 16:0-18:2 PC) for 8 min. Confocal images were quantified as mean pixel intensities. Values are means ± se; n = 50 per genotype per experiment.
(B) Changes in stomatal aperture after ABA or lipid treatment. Stomatal apertures were measured after 2.5 h of treatment. Values are means ± se; n = 65 apertures each genotype per experiment.
(C) Addition of H2O2 induces stomatal closure. Stomatal apertures were measured after 2 h of treatment with H2O2. Values are means ± se; n = 80 apertures per genotype per experiment.
Data in (A) to (C) are from three independent experiments. The asterisk in (A) to (C) indicates that the mean value is significantly different from that of the corresponding control (P < 0.05).
pldα1 plants were insensitive to ABA promotion of stomatal closure. ABA-induced stomatal closure was partially inhibited in the rbohF mutant, but rbohD exhibited the same response to ABA-induced stomatal closure as did the wild type (Figure 11B). This result was consistent with that reported by Kwak et al. (2003), who monitored ABA-induced stomatal closure in RbohD knockout plants carrying a single dSpm transposon insertion. However, wild-type and pldα1 plants were sensitive to PA-promoted stomatal closure, whereas rbohD and rbohF were not (Figure 11B). PC had no effect on stomatal closure in any tested genotype (Figure 11B). By contrast, the application of H2O2 induced stomatal closure in the wild type, pldα1, rbohD, and rbohF (Figure 11C). These results indicate that PLDα1, PA, RbohD, and RbohF all act upstream of ROS.
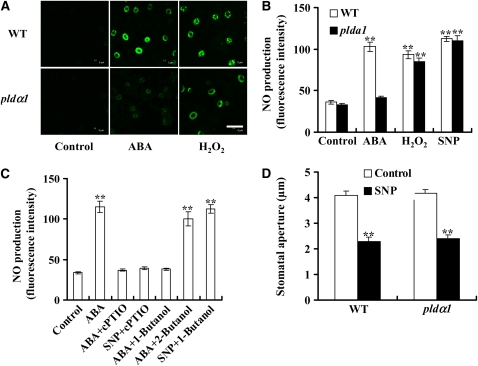
NO Acts Downstream of PLDα1, PA, and ROS
In addition to ROS, nitric oxide (NO) is also involved in ABA-induced stomatal closure (Desikan et al., 2002, 2004). ABA-induced ROS production is reported to mediate NO synthesis (Bright et al., 2006). To investigate the role of PLDα1 and PA in NO production and the relationship between PA, ROS, and NO, we measured NO production in wild-type and pldα1 guard cells. Treatment with ABA increased NO production in the wild type, but the increase was greatly reduced in pldα1 guard cells (Figures 12A and 12B). However, application of H2O2 increased NO generation in both wild-type and pldα1 guard cells (Figures 12A and 12B), and the magnitude of increase was similar in these genotypes (Figure 12B). When the NO scavenger 2-phenyl-4,4,5,5-tetremethylimidazolinone-1-oxy 3-oxide (PTIO) was present, the ABA-induced NO increase was greatly reduced. Such decrease in NO production was also found when 1-butanol was used to decrease PA production (Figure 12C). 1-Butanol decreases PLD-mediated formation of PA because PLD can transfer the phosphatidyl group to primary alcohols to produce phosphatidylalcohol at the expense of PA. As a control, 2-butanol, which is not a substrate of PLD, was used and was found not to reduce NO formation (Figure 12C). When a NO donor, sodium nitroprusside (SNP), was used, an increase in NO occurred in both wild-type and pldα1 guard cells (Figure 9B). 1-Butanol did not affect SNP-produced NO increase (Figure 12C). As with H2O2, NO (SNP) induced stomatal closure in both wild-type and pldα1 plants (Figure 12D). These data indicate that PLDα1 and PA are involved in promoting ABA-induced NO production and that NO functions downstream of ROS.
Figure 12.
PLDα1 and PA Mediate ABA-Induced NO Production.
(A) Representative images of NO production indicated by the fluorescent dye DAF-2DA. Epidermal peels were loaded with 10 μM DAF-2DA for 10 min before the addition of ABA (50 μM) or H2O2 (100 μM) for another 10 min. Bar = 50 μm.
(B) NO production in guard cells induced by ABA and H2O2. The NO donor, SNP (100 μM), was used as a positive control for NO production. Data are means ± se; n = 70 per genotype per experiment.
(C) Inhibition of ABA-induced NO production by 1-butanol. Epidermal peels were preincubated with 200 μM PTIO for 30 min before ABA (50 μM) or SNP (100 μM) treatment; 0.1% (v/v) 1- or 2-butanol was added together with ABA, and 1-butanol was added together with SNP. Data are means ± se; n = 65 per treatment per experiment.
(D) NO induced stomatal closure in wild-type and pldα1 plants. The epidermal peels were treated with 100 μM SNP. Data are means ± se; n = 80 per genotype per experiment.
Data in (B) to (D) are from three independent experiments. Two asterisks indicate that the value is significantly different from that of the control (P < 0.01).
The PA and ABI1 Interaction Is Important for ROS and NO Signal Transduction
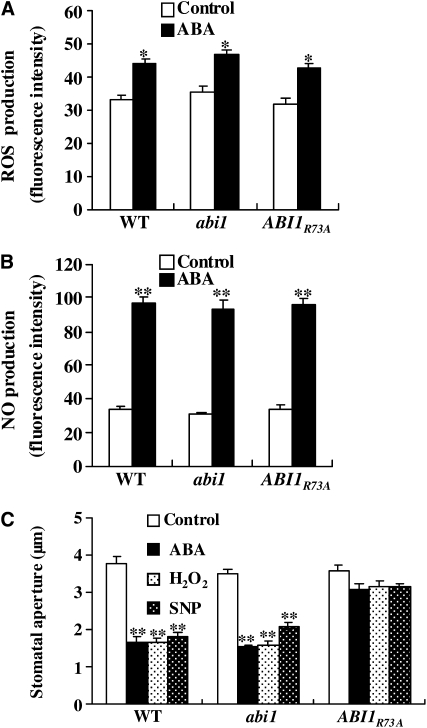
Previous studies showed that one mechanism by which PLDα1 mediates ABA-promoted stomatal closure is through PA binding to ABI1 to inhibit its function (Zhang et al., 2004). This raises intriguing questions as to whether PA and ABI1 interaction is essential for the production of ROS and NO and/or their signal transduction in guard cells. To address these questions, we compared ROS production in guard cells of wild-type plants and in those of the non-PA-binding mutant (ABI1R73A). This mutant was generated by expressing ABI1R73A, driven by the ABI1 native promoter, in an ABI1-knockout (abi1) background (Mishra et al., 2006). ABI1R73A has normal phosphatase activity but cannot bind to PA (Zhang et al., 2004). The ABI1R73A transgenic plants exhibited the ABA-induced increases in ROS levels similar to wild-type cells (Figure 13A). Likewise, ABA-induced NO generation was also similar in wild-type and ABI1R73A plants (Figure 13B).
Figure 13.
Disruption of PA-ABI1 Binding Affects ROS or NO Promotion of Stomatal Closure but Not ROS or NO Production.
(A) ROS production in guard cells as monitored by H2DCFDA. The epidermal peels were treated with 50 μM ABA for 5 min. Data are means ± se; n = 60 per genotype per experiment.
(B) NO production in guard cells as monitored by DAF-2DA. The epidermal peels were treated with 50 μM ABA for 10 min. Data are means ± se; n = 65 per genotype per experiment.
(C) Changes in stomatal aperture in response to ABA, H2O2, and NO. The epidermal peels with fully opened stomata were incubated with 50 μM ABA, 100 μM H2O2, or 100 μM SNP. Stomatal apertures were measured after 2.5 h of treatment. Data are means ± se; n = 80 apertures per genotype per experiment.
Values in (A) to (C) are means ± se of three experiments. The asterisk indicates that the mean value is significantly different from that of the corresponding control (one asterisk for P < 0.05; two asterisks for P < 0.01).
However, ABI1R73A plants were insensitive to the promotion of stomatal closure by ABA, ROS, or NO, whereas wild-type plants were sensitive (Figure 13C). These data indicate that the PA–ABI1 interaction is not required for ROS or NO production but that the PA–ABI1 interaction is involved in mediating the effect of ROS and NO on stomatal closure. The abi1 plants produced similar amounts of ROS and NO in response to ABA treatment as the wild-type plants and had similar degrees of stomatal closure following ABA, H2O2, or NO treatment as the wild type (Figure 13). These similarities are consistent with the notion that ABI1 is a negative effector in the ABA response.
DISCUSSION
Recent work in nerve cells has revealed functions for lipids in protein trafficking, cellular communication, and the production of the hundreds of molecules that carry information both within and across cells (Piomelli et al., 2007). Lipids can regulate ion channels, receptors, and other signal transduction proteins, not only by influencing the curvature or supramolecular organization of the membrane, but also by directly binding to the proteins (Hancock, 2007; Piomelli et al., 2007). There is no doubt that lipids can act as signal messengers in plant cells (Wang et al., 2006; Munnik and Testerink, 2009; and references within). Plant PLD and derived PA are involved in ABA-mediated stomatal movement by interaction with the α-subunit of G protein, through PA regulation of the inwardly rectifying K+ channel, and/or ABI1 PP2C localization and activity (Jacob et al., 1999; Zhang et al., 2004; Mishra et al., 2006). Other lipids, including phosphoinositides and sphingolipids, are also important regulators of ABA-mediated stomatal movement (Jacob et al., 1999; Coursol et al., 2003; Park et al., 2003; Perera et al., 2008). However, much less is known about lipid functions in cell signaling and their mechanisms in plant cells than in animal cells. In this study, we have identified the transmembrane protein Rboh as a direct target of the lipid messenger PA, which is derived from ABA-activated PLDα1 and showed that PA–RbohD interaction is essential to ABA-mediated ROS production and stomatal closure.
Rbohs Are Intermediates of PLD Signaling in ABA-Mediated Stomatal Closure
Both PLDs and NADPH oxidases play important roles in producing signals that mediate the cellular response to environmental stress by activation of downstream targets (Bargmann and Munnik, 2005; Torres and Dangl, 2005; Wang, 2005; and references within). We showed here that PLDα1 and PA were required for the production of ROS in guard cells in response to ABA and that PA directly interacts with Rboh. The conclusion is supported by several lines of evidence. (1) ABA-induced ROS production was impaired in pldα1 guard cells. When H2O2 was added, stomata closed in pldα1, as well as in wild-type plants. (2) The ABA-stimulated increase in NADPH oxidase activity was reduced in pldα1 GCPs, whereas addition of PA restored NADPH oxidase activity. The data suggest that PLDα1-derived PA is an activator of NADPH oxidase. (3) PA species 34:1(16:0-18:1), 34:2(16:0-18:2), and 36:3(18:1-18:2) were significantly increased 10 min after ABA application, producing potential PA sources for binding to the NADPH oxidases (RbohD and RbohF) because PA species with 18:1 or 18:2 fatty acids were bound to the Rbohs. (4) 16:0-18:2 PA, one of the PA species increased most by ABA, was found to bind to RbohD and RbohF in vitro and in vivo. (5) The disruption of PA-Rboh (RbohD) binding compromised ABA-mediated ROS production and stomatal closure.
The 34:2 (16:0-18:2) PA and other PA species were also increased in the pldα1 mutant treated with ABA, although the magnitude and absolute amount of the increase was lower than in the wild type. The results suggest that there are other enzymes that respond to ABA to produce PA. In Arabidopsis, multiple PLDs have diverse functions in response to environmental and developmental stimuli (Wang, 2005), and some of them are probably activated by ABA treatment. In addition, DGK may phosphorylate diacylglycerol that is produced from PLC, which is known to be activated by ABA; the coupled PLC/DGK is another route that produces signaling PA (Testerink and Munnik, 2005; Wang et al., 2006). DGKs play a role in plant development (Gómez-Merino et al., 2005), but whether they are involved in the ABA response remains to be investigated.
The difference in magnitude in ABA-induced PA increase between the wild type and pldα1 mutant is probably one of reasons for lower NADPH oxidase activity and ROS generation in response to ABA in the mutant guard cells, as the amount of PA-bound Rbohs increased with increasing PA concentrations (see Supplemental Figure 11 online). PA binding to NADPH oxidase is essential for Rboh(D) activation. Even in the control condition, NADPH oxidase activity in the pldα1 mutant was lower than in the wild type, which may contribute to the greater water loss from mutant plants during the 3-d growth period (see Supplemental Figure 12 online). This is consistent with results obtained from PLDα-antisense suppressed plants that had an earlier drought phenotype after withholding irrigation than did the wild type. In addition, applied ABA could not alleviate the drought phenotype in the mutant, but it enhanced the tolerance to drought in the wild-type plants (Sang et al., 2001b).
Although it showed a lower level of NADPH oxidase activity and ROS generation than the wild type, pldα1 had a background level of ROS before ABA treatment. Besides NADPH oxidase, there are intracellular and extracellular routes for ROS production in plants. Inside the plant cell, ROS can be produced as byproducts of photosynthesis and respiration. In the apoplast, ROS can be generated by cell wall–bound peroxidases, oxalate oxidases, and amino oxidases (Torres and Dangl, 2005; An et al., 2008). These pathways together with ROS scavengers may contribute to basal ROS homeostasis. Despite this, our data from the plasma membrane, guard cells, and plants show that PLDα1 and PA are important regulators of plasma membrane–associated NADPH oxidase activity in response to ABA in stomatal closure.
PA–Rbohs Interaction Suggests a Novel Regulatory Mechanism for Plant NADPH Oxidase
In mammalian systems, PA activates NADPH oxidase via interaction with the regulatory subunit p47phox. The mammalian NADPH oxidase component p47phox has a PX domain with two distinct lipid binding sites. Simultaneous occupancy of the PA and PtdInsP2 binding sites leads to a synergistic increase in membrane affinity of the protein (Karathanassis et al., 2002). However, the Arabidopsis genome has no ortholog of p47phox, and Rboh proteins do not contain a PX or PH domain (Torres and Dangl, 2005). The PA binding region of RbohD shares little sequence similarity with that of p47phox (Karathanassis et al., 2002) but more sequence similarity with the ABI1 PA binding motif in plants (Zhang et al., 2004; 20% versus 30%). The amino acid sequence of the PA binding region (140 to 160 amino acids for RbohD and 145 to 171 amino acids for RbohF) is not highly conserved between RbohD and RbohF (Figure 8C). The major PA binding basic residues (Arg-149 and -150) in RbohD are replaced with nonpolar residues Ala-156 and Leu-157 in RbohF. Simultaneous mutation of Ala-156 and Leu-157 to Arg in RbohF resulted in an increase in PA binding after ABA treatment. The results indicate that the difference between the two major PA binding residues of RbohF (Ala-156 and Leu-157) and RbohD (Arg-149 and Arg-150) may contribute to lower PA biding in RbohF. We do not rule out the possibility that other mediators of RbohF regulation, such as Rac (Wong et al., 2007), Ca2+ (Ogasawara et al., 2008), and CDPK (Kobayashi et al., 2007), have a role in the ABA response.
Knockout of RbohF results in less sensitivity to ABA-induced stomatal closure than the wild type and rbohD (Kwak et al., 2003; Figure 11). It seems that RbohF plays a more important role than RbohD in ABA-mediated stomatal closure. The rbohD/F double mutant was more impaired in ABA-induced stomatal closure than rbohF or the rbohD/E double mutant (Kwak et al., 2003). Conversely, overexpression of wild-type RbohD led to stomatal closure (Figure 10D) as well as ROS increase even in the absence of ABA (Figure 9C; see Supplemental Figure 9 online). The results suggest that both RbohD and RbohF are major enzymes in ROS generation during ABA response. These proteins probably have overlapping functions or interactions during ABA-induced ROS generation in guard cells and in the pathogen response (Kwak et al., 2003; Torres and Dangl, 2005). Furthermore, the overexpression of the non-PA-binding Rboh protein in rbohD mutant impaired ABA-induced stomatal closure. It seems that the introduction of the non-PA-binding protein has a dominant-negative effect, which could result from the interference of the overexpressed protein with other Rbohs and/or with cellular effectors of Rbohs.
Knockout of RbohD and RbohF both impaired the increase of ROS production in guard cells exposed to ABA. PA could not induce ROS production in guard cells of these mutants. Therefore, the PA–Rboh interaction is essential to ABA-mediated ROS production, regardless of RbohD or RbohF, although their PA binding abilities are different. Using a transient expression system, we have demonstrated directly the importance of PA-RbohD binding in ABA-induced stomatal closure in planta. However, an interesting question still remains as to why ROS generation in both rbohD and rbohF is impaired, but ABA-induced stomatal closure is more impaired in rbohF than in atbohD. In fact, when expression of the ABA-responsive gene RAB18 was assayed in these mutants, we found that RAB18 expression was enhanced in rbohD but was abolished in atrbohF (see Supplemental Figure 13 online). These results suggest that RbohD and RbohF have different functions in mediating the ABA response.
The mechanism by which PA regulates the NADPH oxidases is different from that of other lipids, such as PtdIns(3)P, which participates in ABA-induced ROS generation and stomatal closure (Park et al., 2003). PtdIns(3)P is synthesized by phosphatidylinositol 3-kinase. Both PtdIns(3)P and phosphatidylinositol 3-kinase are involved in ROS production in root hairs (Lee et al., 2008) or roots exposed to salt stress treatment (Leshem et al., 2007). In this work, we did not detect binding of PtdIns(3)P or PtdIns(4)P to RbohD or RbohF (see Supplemental Figure 5 online). In root systems, phosphatidylinositol signaling is thought to regulate the intracellular assembly of the active NADPH oxidase complex by endocytotic membrane trafficking (Leshem et al., 2007); PA binding may not be involved in this pathway.
The PA binding site in RbohD was located in the cytosolic region between the two EF-hands and N terminus (Figure 14). Recently, Kobayashi et al. (2007) reported CDPK-mediated phosphorylation of StrbohB at Ser-82 and Ser-97 in potato. RbohD and StrbohB have a high degree of amino acid identity, and both have the conserved phosphorylation motif B-X-X-S (where B is a basic residue, S is a Ser residue, and X is any residue). It is worth noting that the major PA binding residues (Arg-149 and -150) are in the vicinity of the B-X-X-S motif (Kobayashi et al., 2007; see Supplemental Figure 4 online). Phosphorylation was proposed to induce conformational changes in RbohB, leading to the interaction with the small GTPase Rac in the vicinity of the EF-hand motifs (Wong et al., 2007). However, the phosphorylation of Rboh is not sufficient for full activation of Rboh (Kobayashi et al., 2007; Nuhse et al., 2007), implying that there may be additional determinants that can regulate the activity. As discussed above, the binding of Rbohs by the lipid messenger PA identified in this study may reveal a novel regulatory mechanism for Rboh activation. As PA production is induced by a wide spectrum of stimuli (Wang et al., 2006), this mechanism is likely to have a central role in the coupling of extracellular signals to Rboh activation.
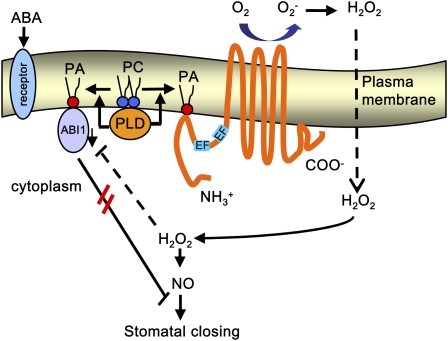
Figure 14.
Working Model of PA Activation of Rboh in ABA-Induced Stomatal Closure in Arabidopsis.
ABA stimulates PLD activity probably through a putative ABA receptor. The lipid messenger PA produced by ABA activation of PLD binds to Rboh in the cytolosolic region of the N terminus. The activation of Rboh by PA binding results in ROS production (including H2O2). The PA–ABI1 interaction impairs ABI1 inhibition of H2O2 and NO signaling. Possible interactions between the identified components and other intermediates are discussed in the Discussion. The double red lines between ABI1 and stomatal closing indicate that the recruitment of ABI1 to the plasma membrane impairs ABI1 inhibition of stomatal closing.
A Complex Signaling Network Involving PA, ROS, NO, and ABI1 in ABA-Mediated Signaling during Stomatal Closure
Several targets downstream of ROS have been reported to respond to ABA treatment in guard cells. These include protein kinases, protein phosphatases, ion channels, and NO (Li et al., 2006; Neill et al., 2008; and references within). Pharmacological and genetic evidence reveals that NO accumulation induced by ABA in guard cells is dependent on ROS (Bright et al., 2006). NO selectively regulates K+ and Cl− channels in Vicia faba guard cells by enhancing cytosolic Ca2+ release from intracellular Ca2+ stores, which leads to solute loss and stomatal closure (Garcia-Mata et al., 2003). Using the dominant-negative abi1-1 and abi2-1 mutants, ABI1 and ABI2 protein phosphatases have been placed downstream of NO in the ABA signal transduction cascade in guard cells (Desikan et al., 2002). We previously showed that PA interacts directly with ABI1, which leads to membrane tethering of ABI1 and a decrease in its PP2C activity (Zhang et al., 2004). ABI1 is a negative regulator of ABA action. The recruitment of ABI1 to the plasma membrane and the inhibition of its activity are essential for preventing its interaction with the transcription factor ATHB6 and, therefore, for transducing the ABA signal (Zhang et al., 2004; Mishra et al., 2006). This could explain why the disruption of PA-ABI1 did not affect ABA-induced ROS and NO production but inhibited H2O2- and NO-induced stomatal closure. These results suggest that PA–ABI1 interaction is downstream of ROS and NO signaling and that the interaction is necessary for both ROS- and NO-mediated stomatal closure.
ROS and NO may also act independently in ABA-evoked stomatal closure. For example, H2O2 elevates the concentration of Ca2+ in the cytosol by regulating Ca2+ gating in the plasma membrane in guard cells (Pei et al., 2000), instead of drawing on the intracellular Ca2+ store via NO (Garcia-Mata et al., 2003). A very low level of NO has no influence on outward-rectifying K+ channels, while a moderate elevation of NO results in a decrease in K+ channel activity, which is reversed by a reducing reagent (Garcia-Mata et al., 2003; Sokolovski and Blatt, 2004). H2O2 suppresses both inward- and outward-rectifying K+ channels (Köhler et al., 2003), but the residual activity of outward-rectifying K+ channels may be sufficient for K+ efflux after H2O2 application (Kwak et al., 2006, and references within). Therefore, it is reasonable to hypothesize that ROS and NO may function harmoniously and/or in parallel to regulate K+ channels in the ABA response.
In vitro analyses indicate that H2O2 could in turn inactivate the enzymatic activities of ABI1 and ABI2 PP2Cs (Meinhard and Grill, 2001; Merlot et al., 2001; Meinhard et al., 2002). ROS produced by PA–Rboh interaction may inhibit ABI1 activity, which is likely another mechanism by which PA regulates stomatal closure. On the other hand, recent evidence shows that NO triggers PA accumulation during auxin-induced adventitious root formation (Lanteri et al., 2008) and elicitor response (Laxalt et al., 2007). Whether such a feedback regulation loop exists in ABA-mediated stomatal closure needs to be tested.
In conclusion, this study demonstrates a novel role for PLD/PA-mediated ABA signaling pathway in stomatal closure and for the Rboh regulation in plants. Activation of PLD by ABA results in PA production. PA binds to the cytosolic region in the N-terminus of Rboh(D), resulting in the stimulation of NADPH oxidase activity and ROS production in guard cells. PA–Rboh interaction is essential for ABA-mediated ROS production and stomatal closure. Other regulators, such as Rac, Ca2+, CDPKs, and MAPK cascades are probably involved in the processes. ROS regulates channel activity through NO or independently, leading to solute loss and stomatal closure. On the other hand, the PA–ABI1 interaction results in membrane recruitment of ABI1 and a decrease in PP2C activity, thereby antagonizing ABI1's inhibition of H2O2 and NO signaling (Figure 14). The interactions of PLD and PA with different effector proteins suggest that the lipid messengers play an important role in the regulation of membrane-associated signaling complexes in plant response to ABA and stresses.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana wild-type and mutant plants were from the Columbia ecotype. Seeds were germinated in soil and kept at 4°C for 2 d. Plants were grown in a growth room at a light intensity of 160 μmol m−2 s−1 and 12-h/12-h (23°C/18°C) day/night regimes. Four- to six-week-old plants were used to prepare protoplasts and epidermal peels.
Chemicals
Most chemicals were from Sigma-Aldrich. PtdIns were purchased from Echelon Biociences, and other lipids were purchased from Avanti Polar Lipids.
Genotyping of Mutants
The identification and verification of T-DNA-insertion mutants of PLDα1 (pldα1), ABI1 (abi1), and abi1 complemented with ABI1WT (ABI1WT) or ABI1R73A (ABI1R73A) were reported previously (Zhang et al., 2004; Mishra et al., 2006). The T-DNA insertion RbohD mutant (rbohD, SALK_120299) was obtained from ABRC at Ohio State University (www.arabidopsis.org/abrc). To identify homozygous lines and to confirm the exact insertion position of the T-DNA in the rbohD mutant, a PCR-based approach was used. Primers were designed using the SALK T-DNA verification primer design program (signal.SALK.edu/tdnaprimers): RbohD-SALK-F/RbohD-SALK-R and LB3.1/RbohD-SALK-R. Actin 8 was used as an internal control, with primers Actin8-F/Actin8-R (see Supplemental Table 1 for all primer sequences used in this article). rbohD and rbohF mutants containing dSpm transposon insertions were generous gifts from J.L. Dangl and J.M. Kwak.
RbohD and RbohF Cloning and Construction of Expression Vectors
The cDNA sequences of Arabidopsis Rboh genes RbohD and RbohF were amplified from leaf cDNA using the following primer combinations: RbohD-C-F/RbohD-C-R for RbohD and RbohF-C-F/RbohF-C-R for RbohF. cDNA fragments for nine C-terminal deletion mutants of RbohD were amplified by PCR using RbohD cDNA as a template. A common forward primer, RbohD921F, was paired with the following nine reverse primers: RbohD921R for the full-length RbohD921 (1 to 921 amino acids), RbohD713R for RbohD713 (1 to 713 amino acids), RbohD600R for RbohD600 (1 to 600 amino acids), RbohD450R for RbohD450 (1 to 450 amino acids), RbohD330R for RbohD330 (1 to 330 amino acids), RbohD160R for RbohD160 (1 to 160 amino acids), RbohD150R for RbohD150 (1 to 150 amino acids), RbohD140R for RbohD140 (1 to 140 amino acids), and RbohD100R for RbohD100 (1 to 100 amino acids). All of these fragments were cloned into a pET28a vector.
cDNAs for the full-length and five C-terminal deletion mutants of RbohF were amplified by PCR using RbohF cDNA as a template. The same forward primer, RbohF944F, was paired with the following reverse primers: RbohF944R for RhohF944 (1 to 944 amino acids), RbohF723R for RhohF723 (1 to 723 amino acids), RhohF555R for RhohF555 (1 to 555 amino acids), RhohF341R for RhohF341 (1 to 341 amino acids), RhohF171R for RhohF171 (1 to 171 amino acids), and RhohF104R for RhohF104 (1 to 104 amino acids). All these fragments were cloned into a pET28a vector.
For PA binding site characterization in RbohD and RbohF, the Quickchange site-directed mutagenesis kit (Stratagene) was used to generate the site-directed mutants. Using RbohD160 cDNA as template, the primers used to produce the single mutant R141A and R160A were R141A-F and R141A-R, and R160A-F and R160A-R, respectively. The primers used to generate the double mutants R(144,145)A, R(149,150)G, and R(156,157)A were R(144,145)A-F and R(144,145)A-R, R(149,150)G-F and R(149,150)G-R, and R(156,157)A-F and R(156,157)A-R, respectively. The primers for the quadruple mutant R(149,150,156,157)A were R(149,150,156,157)A-F and R(149,150,156,157)A-R.
For protein expression in Escherichia coli, RbohD and RbohF cDNAs were cloned into a pET28a vector (Novagen) at EcoRI/NotI and BamHI/XhoI sites, respectively. Fusion proteins were expressed in E. coli strain BL21 (DE3; Promega) according to the manufacturer's instructions.
For hemagglutinin (HA)-tagged protein expression in guard cell protoplasts, site-specific mutants were generated using the Quickchange site-directed mutagenesis kit. The cDNAs for native and quadruple mutated HA-RbohD160 were amplified by PCR using cDNAs of the RbohD160 and RbohD160 mutant with R(149,150,156,157)A mutation as templates, respectively. A common forward primer D-160-HA-F was paired with the reverse primer D-160-HA-R (for native HA-RbohD160) and D-R(149,150,156,157)A-HA-R (for mutated HA-RbohD160). cDNA for the C-terminal deletion HA-RbohF171 was amplified using RbohF cDNA as the template with the RbohF171-HA-F and RbohF171-HA-R primers. The cDNA for HA-RbohF171 (AL156-157RR) was generated using native HA-RbohF171 cDNA as the template with forward primer F171-AL(156,157)RR–F and reverse primer F-AL(156,157)RR-R. cDNA for full-length native HA-RbohD was amplified using RbohD cDNA as the template with the D-160-HA-F and D-921-HA-R primer pair. cDNA for full-length HA-RbohD with the quadruple mutation was generated using native HA-RbohD cDNA as the template with forward primer R(149,150,156,157)A-F and reverse primer R(149,150,156,157)A-R.
For the RbohD activity assay in protoplasts, RbohD cDNA was placed under the control of the 35S promoter and a HA tag was fused to the C terminus (Zhang et al., 2004).
The full-length native RbohD-GFP and RbohD-GFP with the quadruple mutation were amplified using cDNAs of RbohD and HA-RbohD with quadruple mutation as the template with gene-specific primers D-GFP-F and D-GFP-R.
All amplified target sequences were confirmed by DNA sequencing.
RNA Isolation and RT-PCR
Total RNA was extracted from expanded leaves of 5-week-old Arabidopsis plants using Trizol reagent (Invitrogen) according to the procedure described by the manufacturer. cDNA was synthesized from 1 μg of total RNA using an oligo(dT) primer and AMV reverse transcriptase (Promega). The RbohD transcript was amplified using the gene-specific primers RbohD-RT-F and RbohD-RT-R. PCR conditions were as follows: 94°C for 4 min; 30 cycles of 94°C for 30 s, 56°C for 45 s, and 72°C for 3 min; and a final extension at 72°C for 10 min. Amplicons were visualized on a 1% agarose gel under UV light. The agarose gel was stained with ethidium bromide.
Expression and Purification of RbohD and RbohF Proteins in E. coli
Recombinant plasmids carrying RbohD and RbohF with six histidine residues were transformed into E. coli BL21(DE3). The proteins were expressed at 25°C for 4 to 5 h. Bacterial cells were harvested and resuspended in lysis buffer (25 mM HEPES, 300 mM NaCl, 0.5% Triton X-100, 10 mM imidazole, and 1 mM PMSF, pH 8.0). After incubation on ice for 30 min, samples were sonicated and centrifuged at 10,000g for 20 min. The resulting supernatant was used for protein purification with Ni-affinity agarose (Qiagen) according to the manufacturer's instructions.
Purified proteins were used for immunoblotting and the analysis of protein–lipid interaction. For immunoblotting, proteins were separated on an 8% SDS-PAGE gel and immunoblotted with anti-His antibody (Sigma-Aldrich), followed by the secondary antibody conjugated with alkaline phosphatase. Proteins were quantified with the Bradford method using the reagent from Bio-Rad.
Analysis of Protein-Lipid Binding
Protein–lipid interaction was assayed using filter, liposome binding, and ELISA approaches. The filter binding was performed as described (Zhang et al., 2004). Briefly, lipids were immobilized on a nitrocellulose filter, followed by incubation with RbohD, RbohF, or their fragments. The filter was then incubated with anti-His antibody, followed by incubation with a second antibody conjugated with alkaline phosphatase. RbohD and RbohF proteins (or their fragments) bound to lipids on filters were visualized by staining alkaline phosphatase activity.
Liposomes were prepared according to references (Avantilipids.com; Hayes et al., 2004) with some modifications. Briefly, 16:0-18:2 PC (3.2 μmol) and mixtures of 16:0-18:2 PC and 16:0-18:2 PA in a 3:1 molar ratio (2.4 μmol and 0.8 μmol, respectively) were dried under gas nitrogen. Lipid films were rehydrated in 1 mL of extrusion buffer (25 mM HEPES, pH 7.5, and 250 mM raffinose) at 42°C for 1 h and then processed by vortexing for six 30-s cycles, freezing under ethanol-dry ice mixture, and thawing under 42°C. The lipid suspension was then extruded 11 times through a polycarbonate membrane (pore size 0.2 μm; Avanti Polar Lipids) to produce an optically clear suspension. The suspension was diluted with 3 volumes of binding buffer (25 mM HEPES, pH 7.5, 50 mM KCl, 1 mM CaCl2, and 1 mM MgCl2) and centrifuged at 100,000g at 20°C for 20 min. The pellet was washed once with binding buffer and then gently resuspended in 1 mL of binding buffer to obtain liposomes of 3.2 mM phospholipid. For liposome binding, the supernatants of expressed RbohD330 and RbohF341 were diluted to 0.5 μg/μL, while supernatants of RbohD100 and RbohF104 were diluted to 1.25 μg/μL with binding buffer. Two hundred microliters of liposome and 200 μL diluted protein extracts were mixed and incubated at 4°C for 1 h by gentle shaking. The mixture was then centrifuged at 14,000g for 20 min to obtain a pellet. The liposome pellet was washed three times with binding buffer. The bound protein was separated on a 12% SDS-PAGE gel, transferred onto a polyvinylidene difluoride membrane, and immunoblotted with anti-His antibody conjugated with an alkaline phosphatase. The protein was made visible by staining for alkaline phosphatase activity.
The ELISA-based assay was carried out according to Ghosh et al. (1996). Phospholipids were coated on the wells of a 96-well titer plates and incubated with His-tagged RbohD or RbohF, followed by incubation with anti-His antibody and spectrometric measurements.
Protoplast Isolation, Transient Expression of Proteins, and Assaying ROS Production and NADPH Oxidase Activity
Mesophyll cell protoplasts were isolated from Arabidopsis leaves as described previously (Zhang et al., 2004). GCPs were isolated according to a procedure described by Pandey et al. (2002) with the following modifications. The basic solution contained 5 mM MES-Tris, pH 5.5, 0.5 mM CaCl2, 0.5 mM MgCl2, 10 μM KH2PO4, 0.5 mM ascorbic acid (AsA), and 0.45 M sorbitol. Enzyme 1 consisted of 1% cellulase R10, 0.1% (w/v) PVP-40, 0.25% BSA, and 0.5 mM AsA, and Enzyme 2 consisted of 2.5% cellulase R10, 1.2% macerozyme R10, 0.25% BSA, and 0.5 mM AsA. After digestion, GCPs were purified using Histopaque (Sigma-Aldrich) as described by Pandey et al. (2002). The purity of GCPs was >96% with contamination from mesophyll cell protoplasts and few chloroplasts. The GCPs were kept in basic solution on ice for 30 min before the transfection according to a procedure described previously (Zhang et al., 2004). Ten to twenty micrograms of cDNA of wild-type RbohD, non-PA-binding RbohD, and RbohD160 and RbohF171 (including their mutants) with HA tag was added into 100 μL GCPs and mixed gently but well. PEG/Ca2+(110 μL; 40% PEG, v/v, 0.2 M sorbitol, and 0.1 M CaCl2) was added to the mixture, which was mixed well, and incubated at 23°C for 5 to 30 min. After transfection, the mixture was washed twice with the basic solution and incubated at 23°C for 3 to 5 h to allow protein expression.
The protein from protoplasts was isolated according to the method of Zhang et al. (2003). Equal amounts of GCP proteins were separated by 12% SDS-PAGE gel and transferred to polyvinylidene difluoride filters. The filters were blotted with anti-HA antibody, followed by incubation with a second antibody conjugated to an alkaline phosphatase. Proteins were observed by staining for phosphatase activity.
ROS production in GCPs (or MCPs in supplemental data) was detected by DCF fluorescence (Pei et al., 2000) using a fluorescence microscope (Nikon Eclipse 800; excitation 488 nm, emission 525 nm). The fluorescence intensity was measured using Imaris 5.0.3 (Bitplane).
Total NADPH oxidase activity in protoplasts was determined using a modified assay based on the reduction of the tetrazolium dye, XTT, by superoxide radicals, O2− (Able et al., 1998). Lipids (indicated in the legends for specificity and concentration) or 10 μM (±) ABA from a stock of 50 mM in 95% ethanol (final ethanol, 0.02% [v/v]) was added to GCPs (in supplemental data, MCPs). XTT (0.5 mM) was added, followed by 50 μM NADPH to activate the reaction. After the desired time of treatment, the reaction solution was centrifuged at 200g for 30 s to pellet protoplasts. Supernatants were collected for spectrophotometric analysis of XTT formazan production at A470. The quantity of O2− was determined using the molar extinction coefficient 2.16 × 104 M−1 cm−1 for XTT at 470 nm (Sutherland and Learmonth, 1997). NADPH activity was calculated according to O2− production per minute per 105 cells with or without 100 units of SOD.
Isolation of the Plasma Membrane and Determination of NADPH Oxidase Activity in the Plasma Membrane
Plasma membrane vesicles were isolated from Arabidopsis leaves using two-phase partitioning according to a procedure described previously (Qiu et al., 2002). The membrane vesicles were resuspended in a 50 mM Tris-HCl buffer, pH 7.5, and used immediately for NADPH oxidase activity assays. A total of 1 μM 16:0-18:2 PA, 16:0-18:2 PC, or lysoPA or 10 μM (±) ABA (final ethanol, 0.02% [v/v]) was applied to the membrane vesicles (3 to 6 μg), respectively, and incubated in the reaction buffer (50 mM Tris-HCl buffer, pH 7.5, and 0.5 mm XTT). NADPH (50 μM) was used to initiate the reaction. After 10 min at 25°C, the reaction solution was used for spectrophotometric analysis of XTT formazan production at A470. NADPH activity was expressed as ΔA470 per milligram protein per minute. ΔA470 represents the difference in XTT formazan absorbance at 470 nm in the presence and absence of 100 units of SOD.
Immunoblotting of PLDα Protein in Protoplasts
Total proteins from mesophyll cell and guard cell protoplasts were isolated according to the method of Zhang et al. (2003). PLDα immunoblotting was carried out according to previous procedures (Sang et al., 2001b).
RbohD-GFP Expression
The PCR fragments of native RbohD-GFP and mutated RbohD-GFP with the quadruple mutation R(149,150,156,157)A were cloned into the EcoRI and KpnI restriction sites of the pCAMBIA1205 vector. Both binary plasmids were transformed into Agrobacterium tumefaciens (C58C1, GV3101) by electroporation. The suspension of Agrobacterium was then infiltrated into 4-d-old Arabidopsis seedlings according to a procedure described by Marion et al. (2008). The fluorescence in cotyledons was observed under a confocal microscope (Leica) after 3 d. Approximately 70 to 85% of seedlings were transformed. The cotyledons were incubated with 2 μM (±) ABA, 50 μM PA, or 50 μM PC for 2 h before stomatal aperture measurements.
ROS and NO Detection in Guard Cells of Epidermal Peels
ROS production in guard cells in epidermal peels was detected using DCF according to the method described by Pei et al. (2000). Epidermal peels were floated in incubation buffer (10 mM MES-KOH, pH 6.15, 10 mM KCl, 0.2 mM CaCl2, and 0.1 mM EGTA) for 2 h under cool white light (150 μmol m−2 s−1) at 22°C to induce stomatal opening and then loaded with 50 μM H2DCFDA for 10 min and washed for 20 min in an incubation buffer. ABA was added at the desired time of the treatment. Ethanol (0.1%) was added as a control experiment. Epidermal peels were observed for fluorescence using a confocal microscope (Leica), with excitation at 488 nm and emission at 525 nm.
NO level in guard cells was analyzed using an NO-sensitive dye, 4, 5-diaminofluorescein diacetate (DAF-2DA) (Sigma-Aldrich). Epidermal peels were floated in an incubation buffer under light for 2 h to induce stomatal opening and loaded with 10 μM DAF-2DA for 10 min before washing in incubation buffer for 20 min. The epidermal peels were incubated for an additional 10 min in the presence of various compounds (as indicated in the legends) before being viewed under a confocal microscope. For PTIO treatment, PTIO was applied for 30 min prior to ABA or SNP treatment. The images were observed under a confocal microscope with excitation at 488 nm and emission at 535 nm.
DCF and DAF fluorescence intensity was measured by Leica confocal software (version 2.5). Data were quantified as mean pixel intensities.
Stomatal Aperture Measurement
Stomatal aperture was measured according to the procedure described previously (Zhang et al., 2004). Briefly, epidermal peels were floated in an incubation buffer (10 mM KCl, 0.2 mM CaCl2, 0.1 mM EGTA, and 10 mM MES-KOH, pH 6.15). After incubation for 2.5 h under a cool white light at 23°C to induce stomatal opening, 50 μM (±) ABA from a stock of 50 mM in 95% ethanol (final ethanol, 0.1% [v/v]), 100 μM H2O2, or 100 μM SNP was applied separately. For DPI treatment, DPI was added to an incubation buffer for 30 min before treatment. Stomatal aperture was recorded under a microscope and measured using Image J (National Institutes of Health).
ESI-MS/MS Analysis of Lipid Molecular Species
For lipid profiling, expanded leaves of 4- to 5-week-old plants were sprayed with 100 μM (±) ABA. The excised leaves were immersed in 3 mL of isopropanol with 0.01% butylated hydroxytoluene (75°C) immediately after sampling to terminate lipolytic activities. Lipid extraction, ESI-MS/MS analysis, and quantification were done as described previously (Devaiah et al., 2006). Five replicates of each treatment were carried out and analyzed.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: PLDα1, At3g15730; ABI1, At4g26080; RbohD, At5g47910; RbohF, At1g64060; and Actin8, At1g49240.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. ROS Assay with NADPH Oxidase Inhibitor and Superoxide Donor.
Supplemental Figure 2. Vehicle Effects on ROS Production, Stomatal Closure, RAB 18 Expression, and PLDα Activity.
Supplemental Figure 3. H2O2 Content in Wild-Type and pldα1 Leaves, and Effect of Applied H2O2 on Stomatal Closure.
Supplemental Figure 4. Sequence Alignment of RbohD and RbohF Proteins.
Supplemental Figure 5. Detection of Potential PtdInsPs Binding to Rbohs on Filter.
Supplemental Figure 6. PA Activation of NADPH Oxidase Activity in a Dose-Dependent Manner.
Supplemental Figure 7. Effect of di16:0 PA on ROS Production and Stomatal Closure.
Supplemental Figure 8. Phenotypes of Seedlings Transiently Expressing RbohD.
Supplemental Figure 9. DAB Staining for ROS in Cotyledons Transiently Expressing RbohD.
Supplemental Figure 10. Lipid Specificity for ROS Generation in GCPs of pldα1, rbohD, and rbohF.
Supplemental Figure 11. Detecting PA Concentration-Dependent Rboh Binding by the ELISA Assay.
Supplemental Figure 12. Water Loss from Wild-Type and pldα1 Plants.
Supplemental Figure 13. RAB18 Expression in the Wild Type, pldα1, rbohD, and rbohF.
Supplemental Table 1. PCR Primers Used in This Article.
Supplemental Methods.
Supplemental References.
Supplementary Material
Acknowledgments
We thank June M. Kwak (University of Maryland) and Jeffery L. Dangl (University of North Carolina) for rboh mutants and Yan Guo (National Institute of Biology Science, China) for GFP vector. We also thank Jiangzhe Zhao, Yakang Jin, Mary R. Roth, and Ye Lu for assistance in the experiments. The work was supported by the Chinese National Key Basic Research Project (2006CB100100), by grants from National Science Foundation of China (30625027 and 90817014) and Ministry of Education of China (B07030, 20060307019, and C-0705) to W.Z., and by grants from National Science Foundation (IOS-0818740) and the USDA (2007-35318-18393) to X.W. Lipidomic analysis development and instrument acquisition at Kansas Lipidomics Research Center was supported by the National Science Foundation (EPS 0236913, MCB 0455318, and DBI 0521587), Kansas Technology Enterprise Corporation, K-IDeA Networks of Biomedical Research Excellence of National Institutes of Health (P20RR16475), and Kansas State University.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Wenhua Zhang (whzhang@njau.edu.cn).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Able, A.J., Guest, D.I., and Sutherland, M.W. (1998). Use of a new tetrazolium-based assay to study the production of superoxide radicals by tobacco cell cultures challenged with avirulent zoospores of Phytophthora parasitica var nicotianae. Plant Physiol. 117: 491–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ago, T., Kuiibayashi, F., Hiroaki, H., Takeya, R., Ito, T., and Kohda, D. (2003). Phosphorylation of p47phox directs phox homology domain from SH3 domain toward phosphoinositides, leading to phagocyte NADPH oxide activation. Proc. Natl. Acad. Sci. USA 100: 4474–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, Z., Jing, W., Liu, Y., and Zhang, W. (2008). Hydrogen peroxide generated by copper amine oxidase is involved in abscisic acid-induced stomatal closure in Vicia faba. J. Exp. Bot. 59: 815–825. [DOI] [PubMed] [Google Scholar]
- Asai, S., Ohta, K., and Yoshioka, H. (2008). MAPK signaling regulates nitric oxide and NADPH oxidase-dependent oxidative bursts in Nicotiana benthamiana. Plant Cell 20: 1390–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann, S.M. (2003). OPEN STOMATA1 opens the door to ABA signaling in Arabidopsis guard cells. Trends Plant Sci. 8: 151–153. [DOI] [PubMed] [Google Scholar]
- Babior, B.M. (2004). NADPH oxidase. Curr. Opin. Immunol. 16: 42–47. [DOI] [PubMed] [Google Scholar]
- Bargmann, B.O., and Munnik, T. (2005). The role phospholipase D in plant stress responses. Curr. Opin. Plant Biol. 9: 515–522. [DOI] [PubMed] [Google Scholar]
- Bright, J., Desikan, R., Hancock, J.T., Weir, I.S., and Neill, S.J. (2006). ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 45: 113–122. [DOI] [PubMed] [Google Scholar]
- Coursol, S., Fan, L.-M., Le Stunff, H., Splegel, S., Gilroy, S., and Assmann, S.M. (2003). Sphingolipid signaling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature 423: 651–654. [DOI] [PubMed] [Google Scholar]
- de Jong, C.F., Laxalt, A.M., Bargmann, B.O.R., De Wit, P.J.G.M., Joosten, M.H.A.J., and Munnik, T. (2004). Phosphatidic acid accumulation is an early response in the Cf-4/Avr4 interaction. Plant J. 39: 1–12. [DOI] [PubMed] [Google Scholar]
- Desikan, R., Cheung, M.-K., Bright, J., Henson, D., Hancock, J.T., and Neill, S.J. (2004). ABA, hydrogen peroxide and nitric oxide signaling in stomatal guard cells. J. Exp. Bot. 55: 205–212. [DOI] [PubMed] [Google Scholar]
- Desikan, R., Griffiths, R., Hancock, J., and Neill, S.J. (2002). A new role for old enzyme: Nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 99: 16314–16318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah, S.P., Roth, M.R., Baughman, E., Li, M., Tamura, P., Jeannotte, R., Welti, R., and Wang, X. (2006). Quantitative profiling of polar glycerolipid species and the role of phospholipase Dα1 in defining the lipid species in Arabidopsis tissues. Phytochemistry 67: 1907–1924. [DOI] [PubMed] [Google Scholar]
- Fan, L.-M., Zhao, Z., and Assmann, S.M. (2004). Guard cells: A dynamic signaling model. Curr. Opin. Plant Biol. 7: 537–546. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R., Gampala, S.S., and Rock, C.D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14: S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman, J., Demidchik, V., Bothwell, J.H.F., Mylona, P., Miedema, H., Torres, M.A., Linstead, P., Costa, S., Brownlee, C., Jones, J.D.G., Davies, J.M., and Dolan, L. (2003). Reactive oxygen species produced by NADPH oxidase regulated plant cell growth. Nature 422: 442–446. [DOI] [PubMed] [Google Scholar]
- Garcia-Mata, C., Gay, R., Sokolovski, S., Hills, A., Lamattina, L., and Blatt, M.R. (2003). Nitric oxide regulates K+ and Cl− channels in guard cells through a subset of abscisic acid-evoked signaling pathways. Proc. Natl. Acad. Sci. USA 100: 11116–11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, S., Strum, J.C., Sciorra, V.A., Danniel, L., and Bell, R.M. (1996). Raf-1 kinase posseeees distinct binding domains for phosphatidylserine and phosphatidic acid. J. Biol. Chem. 271: 8472–8480. [DOI] [PubMed] [Google Scholar]
- Gómez-Merino, F.C., Arana-Ceballos, F.A., Trejo-Téllez, L.I., Skirycz, A., Brearley, C.A., Dörmann, P., and Mueller-Roeber, B. (2005). Arabidopsis AtDGK7, the smallest member of plant diacylglycerol kinases (DGKs), displays unique biochemical features and saturates at low substrate concentration. J. Biol. Chem. 280: 34888–34899. [DOI] [PubMed] [Google Scholar]
- Gosti, F., Beaudoin, N., Serizet, C., Webb, A.A.R., Vartanian, N., and Giraudat, J. (1999). ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11: 1897–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groom, Q.J., Torres, M.A., Fordham-Skelton, A.P., Hammond-Kosack, K.E., Robinson, N.J., and Jones, J.D.G. (1996). rbohA, a rice homologue of the mammalian gp91phox respiratory burst oxidase gene. Plant J. 10: 515–522. [DOI] [PubMed] [Google Scholar]
- Hancock, J.F. (2007). PA promoted to manager. Nat. Cell Biol. 9: 615–617. [DOI] [PubMed] [Google Scholar]
- Hayes, M.J., Merrifield, C.J., Shao, D., Ayala-Sanmartin, J., Schorey, C.D., Levine, T.P., Proust, J., Curran, J., Bailly, M., and Moss, S.E. (2004). Annexin 2 binding to phosphatidylinositol 4,5-bisphosphate on endocytic vesicles is regulated by the stress response pathway. J. Biol. Chem. 279: 14157–14164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyworth, P.G., Bohl, B.P., Bokoch, G.M., and Curnutte, J.T. (1994). Rac translocates independently of the neutrophil NADPH oxidase components p47phox and p67phox. Evidence for its interaction with flavocytochrome b558. J. Biol. Chem. 269: 30749–30752. [PubMed] [Google Scholar]
- Heyworth, P.G., Curnutte, J.T., Nauseef, W.M., Volpp, B.D., Pearson, D.W., Rosen, H., and Clark, R.A. (1991). Neutrophil nicotinamide adenine dinucleotide phosphate oxidase assembly. Translocation of p47phox and p67phox requires interaction between p47phox and cytochrome b558. J. Clin. Invest. 87: 352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach, A., Hoffmann, T., Leube, M., Höhener, B., and Grill, E. (2002). Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 21: 3029–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama, T., and Shinozaki, K. (2007). Perception and transduction of abscisis acid signals: Keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 12: 343–351. [DOI] [PubMed] [Google Scholar]
- Honbou, K., Minakami, R., Yuzawa, S., Takeya, R., Suzuki, N.N., Kamakura, S., Sumimoto, H., and Inagaki, F. (2007). Full-length p40phox structure suggests a basis for regulation mechanism of its membrane binding. EMBO J. 26: 1176–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob, T., Ritchie, S., Assmann, S.M., and Gilroy, S. (1999). Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc. Natl. Acad. Sci. USA 96: 12192–12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami, K., Takeya, R., Sumimoto, H., and Kohda, D. (2002). Diverse recognition of non-PxxP peptide ligands by the SH3 domains from p67phox, Grb2 and Pex13p. EMBO J. 21: 4268–4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai, F., Liu, H., Field, S.J., Akbary, H., Matsuo, T., Brown, G.E., Cantley, L.C., and Yaffe, M.B. (2001). The PX domains of p47phox and p40phox bind to lipid products of PI(3). Nat. Cell Biol. 3: 675–678. [DOI] [PubMed] [Google Scholar]
- Karathanassis, D., Stahelin, R.V., Bravo, J., Perisic, O., Pacold, C.M., Cho, W., and Williams, R.L. (2002). Binding of the PX domain of p47phox to phosphatidylinositol 3,4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction. EMBO J. 21: 5057–5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki, T., Henmi, K., Ono, E., Hatakeyama, S., Iwano, M., Satoh, H., and Shimamoto, K. (1999). The small GTP-binding protein rac is a regulator of cell death in plants. Proc. Natl. Acad. Sci. USA 96: 10922–10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, T., Damude, H.G., Wener, D., Doerner, P., Dixon, R.A., and Lamb, C. (1998). A plant homolog of the neutrophil NADPH oxidase gp91phox subunit gene encodes a plasma membrane protein with Ca2+ binding motifs. Plant Cell 10: 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, M., Ohura, I., Kawakita, K., Yokota, N., Fujiwara, M., Shimamoto, K., Doke, N., and Yoshioka, H. (2007). Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 19: 1065–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler, B., Hills, A., and Blatt, M.R. (2003). Control of guard cell ion channels by hydrogen peroxide and abscisic acid indicates their action through alternate signaling pathways. Plant Physiol. 131: 385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak, J.M., Mori, I.C., Pei, Z.-M., Leonhardt, N., Torres, M.A., Dangl, J.L., Bloom, R.E., Boddle, S., Jones, J.D.G., and Schroeder, J.I. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22: 2623–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak, J.M., Nguyen, V., and Schroeder, J.I. (2006). The role of reactive oxygen species in hormonal responses. Plant Physiol. 141: 323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanteri, M.L., Laxalt, A.M., and Lamattina, L. (2008). Nitric oxide triggers phosphatidic acid accumulation via phospholipase D during auxin-induced adventitious root formation in cucumber. Plant Physiol. 147: 188–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxalt, A.M., Raho, N., ten Have, A., and Lamattina, L. (2007). Nitric oxide is critical for inducing phosphatidic acid accumulation in xylanase-elicited tomato cells. J. Biol. Chem. 282: 21160–21168. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Bak, G., Chuang, W.-I., Cho, H.-T., and Lee, Y. (2008). Roles of phosphatidyinositol 3-kinase in root hair growth. Plant Physiol. 147: 624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshem, Y., Seri, L., and Levine, A. (2007). Induction of phosphatidyinositol 3-kinase-mediated endocytosis by salt leads to intracellular production of reactive oxygen species and salt tolerance. Plant J. 51: 185–197. [DOI] [PubMed] [Google Scholar]
- Li, S., Assmann, S.M., and Albert, R. (2006). Predicting essential components of signal transduction network: A dynamic model of guard cell abscisic acid signaling. PLoS Biol. 4: 1732–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X.G., Yue, Y.L., Li, B., Nie, Y.L., Li, W., Wu, W.H., and Ma, L.G. (2007). A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science 315: 1712–1716. [DOI] [PubMed] [Google Scholar]
- Ma, Y., Szostkiewicz, I., Krote, A., Moes, D., Yang, Y., Christmann, A., and Grill, E. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068. [DOI] [PubMed] [Google Scholar]
- Marion, J., Bach, L., Bellec, Y., Meyer, C., Gissot, L., and Faure, J.D. (2008). Systematic analysis of protein subcellular localization and interaction using high-throughput transient transformation of Arabidopsis seedlings. Plant J. 56: 169–179. [DOI] [PubMed] [Google Scholar]
- Meinhard, M., and Grill, E. (2001). Hydrogen peroxide is a regulator of ABI1, a protein phosphatase 2C from Arabidopsis. FEBS Lett. 508: 443–446. [DOI] [PubMed] [Google Scholar]
- Meinhard, M., Rodriguez, P.L., and Grill, E. (2002). The sensitivity of ABI2 to hydrogen peroxide links the abscisic acid-response regulator to redox signalling. Planta 214: 775–782. [DOI] [PubMed] [Google Scholar]
- Merlot, S., Gosti, F., Guerrier, D., Vavasseur, A., and Giraudat, J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25: 295–303. [DOI] [PubMed] [Google Scholar]
- Mishra, G., Zhang, W., Deng, F., and Wang, X. (2006). A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science 312: 264–266. [DOI] [PubMed] [Google Scholar]
- Morel, J., Fromentin, J., Blein, J.P., Simon-Plas, F., and Elmayan, T. (2004). Rac regulation of NtrbohD, the oxidase responsible for the oxidative burst in elicited tobacco cell. Plant J. 37: 282–293. [DOI] [PubMed] [Google Scholar]
- Munnik, T., and Testerink, C. (2009). Plant phodpholipid signaling – In a nutshell. J. Lipid Res. 50: S260–S265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill, S., Barros, R., Bright, J., Desikan, R., Hancock, J., Harrison, J., Morris, P., Ribeiro, D., and Wilson, I. (2008). Nitric oxide, stomatal closure, and abiotic stress. J. Exp. Bot. 59: 165–176. [DOI] [PubMed] [Google Scholar]
- Nilson, S.E., and Assmann, S.M. (2007). The control of transpiration. Insights from Arabidopsis. Plant Physiol. 143: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuhse, T.S., Bottrill, A.R., Jones, A.M., and Peck, S.C. (2007). Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 51: 931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara, Y., et al. (2008). Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by Ca2+ and phosphorylation. J. Biol. Chem. 283: 8885–8892. [DOI] [PubMed] [Google Scholar]
- Ono, E., Wong, H.L., Kawasaki, T., Hasegawa, M., Kodama, O., and Shimamoto, K. (2001). Essential role of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA 98: 759–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, S., Nelson, D.C., and Assmann, S.M. (2009). Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136: 136–148. [DOI] [PubMed] [Google Scholar]
- Pandey, S., Wang, X.-Q., Coursol, S.A., and Assmann, S.M. (2002). Preparation and applications of Arabidopsis thaliana guard cell protoplasts. New Phytol. 153: 517–526. [DOI] [PubMed] [Google Scholar]
- Park, J., Gu, Y., Lee, Y., Yang, Z., and Lee, Y. (2004). Phosphatidic acid induces leaf cell death in Arabidopsis by activating the Rho-related small G protein GTPase-mediated pathway of reactive oxygen species generation. Plant Physiol. 134: 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, K.-Y., Jung, J.-Y., Park, J., Hwang, J.-U., Kim, Y.-W., Hwang, I., and Lee, Y.A. (2003). Role for phosphatidylinositol 3-phosphate in abscisic acid-induced reactive oxygen species generation in guard cells. Plant Physiol. 132: 92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.-Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PRY/PYL family of START proteins. Science 324: 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, Z.-M., Murata, Y., Benning, G., Thomine, S., Klüsener, B., Allen, G.J., Grill, E., and Schroeder, J.I. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406: 731–734. [DOI] [PubMed] [Google Scholar]
- Perera, I.Y., Hung, C.-Y., Moore, C.D., Stevenson-Paulik, J., and Boss, W.J. (2008). Transgenic Arabidopsis plants expressing the type 1 inositol 5-pPhosphatase exhibit increased drought tolerance and altered abscisic acid signaling. Plant Cell 20: 2876–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piomelli, D., Astarita, G., and Rapaka, R. (2007). A neuroscientist's guide to lipidomics. Nat. Rev. Neurosci. 8: 743–754. [DOI] [PubMed] [Google Scholar]
- Qiu, Q.-S., Guo, Y., Dietrich, M.A., Schumaker, K.S., and Zhu, J.-K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 99: 8436–8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang, Y., Cui, D., and Wang, X. (2001. a). Phospholipase D and phosphatidic acid-mediated generation of superoxide in Arabidopsis. Plant Physiol. 126: 1449–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang, Y., Zheng, S., Li, W., Huang, B., and Wang, X. (2001. b). Regulation of plant water loss by manipulating the expression of phospholipase Dα. Plant J. 28: 135–144. [DOI] [PubMed] [Google Scholar]
- Schroeder, J.I., Kwak, J.M., and Allen, G.J. (2001). Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410: 327–330. [DOI] [PubMed] [Google Scholar]
- Schultheiss, H., Dechert, C., Kogel, K.H., and Huckelhoven, R. (2002). A small GTP-binding host protein is required for entry of powdery mildew fungus into epidermal cells of barley. Plant Physiol. 128: 1447–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Y.Y., et al. (2006). The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443: 823–826. [DOI] [PubMed] [Google Scholar]
- Sokolovski, S., and Blatt, M.R. (2004). Nitric oxide block of outward-rectifying K+ channels indicates direct control by protein nitrosylation in guard cells. Plant Physiol. 136: 4275–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suharsono, U., Fujisawa, Y., Kawasaki, T., Iwasaki, Y., Satoh, H., and Shimamoto, K. (2002). The heterotrimeric G protein α subunit acts upstream of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA 99: 13307–13312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhita, D., Raghavendra, A.S., Kwak, J.M., and Vavasseur, A. (2004). Cytoplasmic alkalization precedes reactive oxygen species production during methyl jasmonate- and abscisic acid- induced stomatal closure. Plant Physiol. 134: 1536–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland, M.W., and Learmonth, B.A. (1997). The tetrazolium dyes MTS and XTT provide new quantitative assays for superoxide and superoxide dismutase. Free Radic. Res. 27: 283–289. [DOI] [PubMed] [Google Scholar]
- Testerink, C., and Munnik, T. (2005). Phosphatidic acid: A multifunctional stress signaling lipid in plants. Trends Plant Sci. 10: 368–375. [DOI] [PubMed] [Google Scholar]
- Torres, M.A., and Dangl, J.L. (2005). Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 8: 397–403. [DOI] [PubMed] [Google Scholar]
- Torres, M.A., Dangl, J.L., and Jones, J.D.G. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99: 523–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, M.A., Onouchi, H., Hamada, S., Machida, C., Hammond-Kosack, K.E., and Jones, J.D.G. (1998). Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91phox). Plant J. 14: 365–370. [DOI] [PubMed] [Google Scholar]
- Wang, H., and Joseph, J.A. (1999). Quantifying cellular oxidative stress by dichlorofluorescein assay using microplate reader. Free Radic. Biol. Med. 27: 612–616. [DOI] [PubMed] [Google Scholar]
- Wang, X. (2005). Regulatory functions of phospholipase D and phosphatidic acid in plant growth, development, and stress responses. Plant Physiol. 139: 566–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Devaiah, S.P., Zhang, W., and Welti, R. (2006). Signaling functions of phosphatidic acid. Prog. Lipid Res. 45: 250–278. [DOI] [PubMed] [Google Scholar]
- Wang, X.-Q., Ullah, H., Jones, A.M., and Assmann, S.M. (2001). G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cell. Science 292: 2070–2072. [DOI] [PubMed] [Google Scholar]
- Wong, H.L., Pinontoan, R., Hayashi, K., Tabata, R., Yaeno, T., Hasegawa, K., Kojima, C., Yoshioka, H., Iba, K., Kawasaki, T., and Shimamoto, K. (2007). Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 19: 4022–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., and Zhu, J.-K. (2003). Regulation of abscisic acid biosynthesis. Plant Physiol. 133: 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka, H., Numata, N., Nakajima, K., Katou, S., Kawakita, K., Rowland, O., Jones, J.D., and Doke, N. (2003). Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15: 706–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., Qin, C., Zhao, J., and Wang, X. (2004). Phospholipase Dα1-derived phosphatidic acid interacts with ABI1 phophatase 2C and regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA 101: 9508–9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., Wang, C., Qin, C., Wood, T., Olafsdottir, G., Welti, R., and Wang, X. (2003). The oleate-stimulated phospholipase D, PLDδ, and phosphatidic acid decrease H2O2-induced cell death in Arabidopsis. Plant Cell 15: 2285–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., Yu, L., Zhang, Y., and Wang, X. (2005). Phospholipase D in the signaling networks of plant response to abscisic acid and reactive oxygen species. Biochim. Biophys. Acta 1736: 1–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.