Abstract
Endoreduplication involves a doubling of chromosomal DNA without corresponding cell division. In plants, many cell types transit from the mitotic cycle to the endoreduplication cycle or endocycle, and this transition is often coupled with the initiation of cell expansion and differentiation. Although a number of cell cycle regulators implicated in endocycle onset have been identified, it is still largely unknown how this transition is developmentally regulated at the whole organ level. Here, we report that a nuclear-localized SUMO E3 ligase, HIGH PLOIDY2 (HPY2), functions as a repressor of endocycle onset in Arabidopsis thaliana meristems. Loss of HPY2 results in a premature transition from the mitotic cycle to the endocycle, leading to severe dwarfism with defective meristems. HPY2 possesses an SP-RING domain characteristic of MMS21-type SUMO E3 ligases, and we show that the conserved residues within this domain are required for the in vivo and in vitro function of HPY2. HPY2 is predominantly expressed in proliferating cells of root meristems and it functions downstream of meristem patterning transcription factors PLETHORA1 (PLT1) and PLT2. These results establish that HPY2-mediated sumoylation modulates the cell cycle progression and meristem development in the PLT-dependent signaling pathway.
INTRODUCTION
Shoot and root apical meristems are the primary sources of newly generated cells in plants. Cells within these meristems actively proliferate through the mitotic cell cycle to support the outgrowth and development of various plant organs. Upon leaving the meristems, many cells start to differentiate and at the same time to increase their cell size through postmitotic cell expansion. Recent studies have shown that the plant growth regulators auxin and cytokinin play crucial roles in determining the balance between cell proliferation and cell expansion/differentiation in Arabidopsis thaliana roots. Auxin concentration gradients, mediated by the PIN-FORMED (PIN) auxin transport facilitators, provide the key instructive signals in this regulation as they promote cell division in the meristem and expansion/differentiation after cells have left the meristem (Blilou et al., 2005). The auxin-inducible PLETHORA (PLT) genes, encoding AP2-domain transcription factors, are thought to act as translators of these auxin gradients because PLT proteins also display similar concentration gradients, and their protein dosages can define the zones of cell proliferation and cell expansion/differentiation in Arabidopsis roots (Galinha et al., 2007). Cytokinin, on the other hand, counteracts auxin to promote cell expansion/differentiation at the root meristem (Dello Ioio et al., 2007; Ruzicka et al., 2009). The antagonistic interaction between auxin and cytokinin appears to rely on their opposing regulations of SHORT HYPOCOTYL2 (SHY2)/INDOLE-3-ACETIC ACID3 (IAA3) protein abundance, which in turn modifies the expression of PIN genes and resulting auxin distributions (Dello Ioio et al., 2008).
The switch from cell proliferation to cell expansion/differentiation at the meristems is often accompanied by the transition from the mitotic cell cycle into the endocycle, an alternative cell cycle in which cells replicate their chromosomal DNA without cell division (Inze and De Veylder, 2006; De Veylder et al., 2007). Successive rounds of endocycling result in a doubling of nuclear DNA content each time, and in Arabidopsis many cell types in leaves, roots, and hypocotyls reach 16C or 32C (C = haploid DNA content) (Galbraith et al., 1991). Cyclins (CYCs) and cyclin-dependent kinases (CDKs) are major drivers of the mitotic cell cycle, and accumulating evidence suggests that one core element of entry into the endocycle is a reduction of mitotic CDK activities (Edgar and Orr-Weaver, 2001; Larkins et al., 2001; Inze and De Veylder, 2006). Plants have two types of CDKs, CDKA and CDKB, that directly regulate the cell cycle. CDKA is a functional homolog of yeast Cdc2/Cdc28p that is present throughout the mitotic cell cycle. CDKBs, which can be further classified into two subtypes, CDKB1 and CDKB2, are plant-specific CDKs that are expressed and active in a cell cycle–dependent manner (De Veylder et al., 2003). CDKB1 is expressed from the late S- to the M-phase, while CDKB2 is expressed from the G2- to the M-phase. In Arabidopsis, CDKA is encoded by a single gene named CDKA;1, whereas there are two homologs for each CDKB, designated as CDKB1;1, CDKB1;2, CDKB2;1, and CDKB2;2. Overexpression of a dominant-negative allele of CDKB1;1 enhances endoreduplication in Arabidopsis leaves, suggesting that CDKB1;1 is one of the key mitosis-inducing/endocycle-repressing factors (Boudolf et al., 2004). The CDK inhibitor KRP2 is also implicated in the regulation of the mitotic-to-endocycle transition. The proposed model suggests that in mitotically dividing cells, CDKB1;1 phosphorylates KRP2 and triggers its proteasomal degradation. By contrast, downregulation of CDKB1;1 is thought to cause an accumulation of KRP2, which in turn specifically inhibits the mitotic CDKA;1 kinase complexes and thus enhances an onset of endoreduplication (Verkest et al., 2005). Premature or delayed transition into endocycling often leads to severe growth phenotypes; thus, the timing of endocycle onset should be regulated by developmental cues. Despite our increasing knowledge on various cell cycle regulators involved in the endocycle onset (Cebolla et al., 1999; Sun et al., 1999; Magyar et al., 2005; Verkest et al., 2005; Vlieghe et al., 2005; Dewitte et al., 2007; Lammens et al., 2008; Larson-Rabin et al., 2009), our understanding of their integration with developmental programs is still very limited.
SMALL UBIQUITIN-RELATED MODIFIER (SUMO) is a posttranslational modifier that is covalently conjugated to Lys residues in a substrate protein. SUMO modification, or sumoylation, can modulate protein activities, localizations, and/or interactions, and recent studies have established that sumoylation influences diverse cellular processes, including transcriptional regulation, maintenance of genome integrity, and signal transduction (Geiss-Friedlander and Melchior, 2007). Like ubiquitination, sumoylation involves SUMO-specific E1 activation, E2 conjugation, and E3 ligation. Several SUMO E3 ligases, including SIZ/PIAS, RanBP2, Pc2, and MMS21, have been characterized in yeasts and mammals, but to date, only one SUMO E3 ligase, SIZ1, has been identified in Arabidopsis (Miura et al., 2005). Previous genetic studies implicate SIZ1 in various stress responses and developmental processes, including phosphate deficiency response, thermotolerance, cold acclimation, innate immunity, abscisic acid signaling, and flowering control (Jin et al., 2008; Lee et al., 2007; Miura et al., 2005, 2007b, 2009; Yoo et al., 2006). Because loss of SUMO, E1, or E2 enzymes leads to embryonic lethality (Saracco et al., 2007), sumoylation is likely to play more profound roles in plant development. Residual SUMO-conjugated protein levels in siz1-3 mutants suggest the existence of additional proteins with SUMO E3 ligase activity in Arabidopsis (Catala et al., 2007), but their molecular identities and potential biological functions remain unknown.
In this study, we have identified HIGH PLOIDY2 (HPY2) as a negative regulator of endocycle onset in Arabidopsis. Loss of HPY2 function results in a premature mitotic-to-endocycle transition, leading to severe dwarfism with compromised meristems. HPY2 possesses an SP-RING (SIZ/PIAS-RING) domain characteristic of MMS21-type SUMO E3 ligases, and we show that HPY2 has SUMO ligase activity both in vivo and in vitro. We demonstrate that HPY2 proteins are found predominantly in the nuclei of root meristems and that meristem-defining transcription factors PLT1 and PLT2 are both necessary and sufficient for HPY2 expression and/or accumulation in the meristem. Furthermore, an activation of PLT2 does not rescue the root meristem defects in hpy2-1, indicating that HPY2 functions downstream of PLT. Based on these findings, we propose that HPY2-mediated sumoylation is part of the regulatory mechanisms linking PLT-dependent developmental signals with the downstream cell cycle progression and meristem development.
RESULTS
HPY2 Is Required for Normal Endocycle Onset and Meristem Development
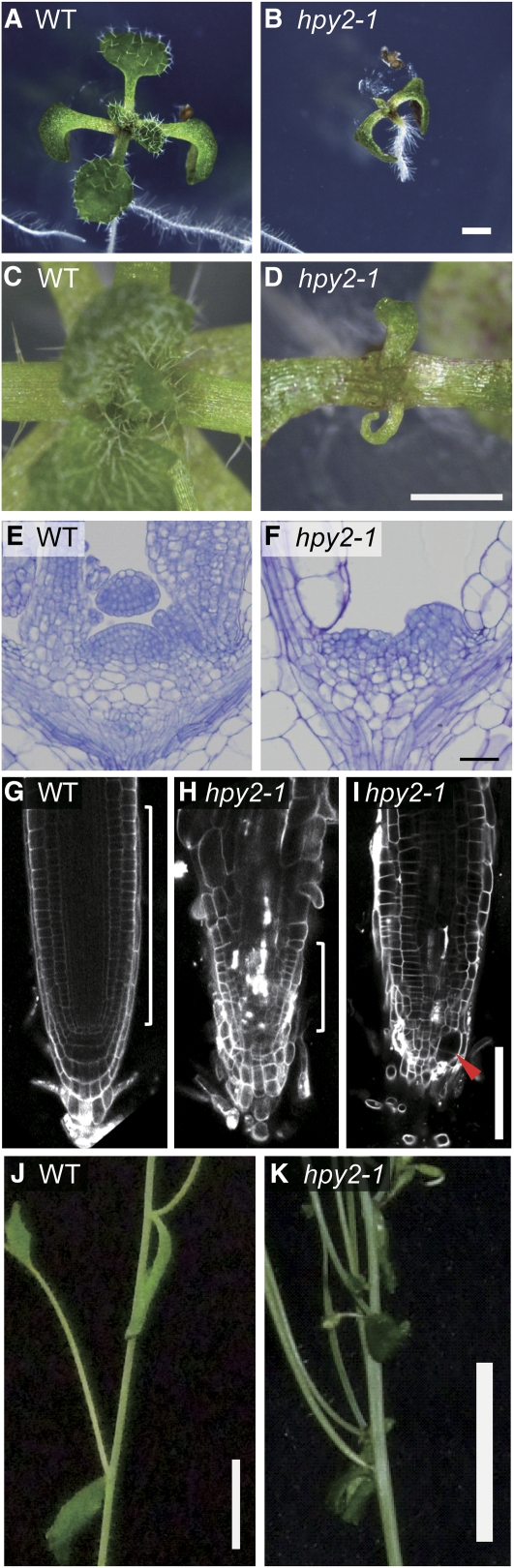
To dissect the molecular mechanisms that govern endocycle onset in Arabidopsis, we performed a genetic screen using ∼3000 independent ethyl methanesulfonate–mutagenized lines. Because endocycle defects are often associated with altered plant organ sizes (Breuer et al., 2007; Sugimoto-Shirasu et al., 2002, 2005), we first searched for mutations that cause larger or smaller organ sizes and subscreened, using flow cytometry, for mutants with ploidy defects. One mutant we identified, high ploidy2-1 (hpy2-1), indeed displays a severe dwarf phenotype (Figures 1A and 1B). The hpy2-1 mutants start to show severe growth retardation soon after germination, and their roots, for instance, reach only ∼10% of the wild-type length by 7 d (Figures 2A and 2B). A closer examination of the shoot apical meristem in 14-d-old seedlings reveals that hpy2-1 has severely compromised meristem structures (Figures 1C to 1F). Similarly, confocal microscopy of the propidium iodide (PI)-stained 6-d-old roots shows strongly reduced meristems in hpy2-1 (Figures 1G and 1H). Unlike wild-type root meristem cells, which have fairly constant sizes, we also find abnormally enlarged cells in hpy2-1 meristems (Figure 1I). Most hpy2-1 seedlings survive for only up to 3 weeks under our standard growth conditions and those that occasionally reach the shoot bolting stage often show fasciation and phyllotaxis defects (Figures 1J and 1K).
Figure 1.
hpy2 Mutants Are Defective in the Meristem Development.
(A) and (B) Ten-day-old seedlings of the wild type (A) and hpy2-1 (B).
(C) and (D) A close-up view of the shoot apices in 14-d-old wild-type (C) and hpy2-1 (D) seedlings.
(E) and (F) Histological sections of 6-d-old wild-type (E) and hpy2-1 (F) shoot apical meristems.
(G) to (I) Confocal microscopy of the PI-stained wild-type (G) and hpy2-1 ([H] and [I]) roots. White brackets in (G) and (H) mark the approximate position of root meristems. An optical section scanning closer to the root surface (I) reveals a swollen epidermal cell, indicated by an arrowhead, near the quiescent center.
(J) and (K) Fasciation and phyllotaxis defects of 30-d-old hpy2-1 plants (K) compared with the wild type (J).
Bars = 1 mm in (A) to (D), 20 μm in (E) and (F), 100 μm in (G) to (I), and 1 cm in (J) and (K).
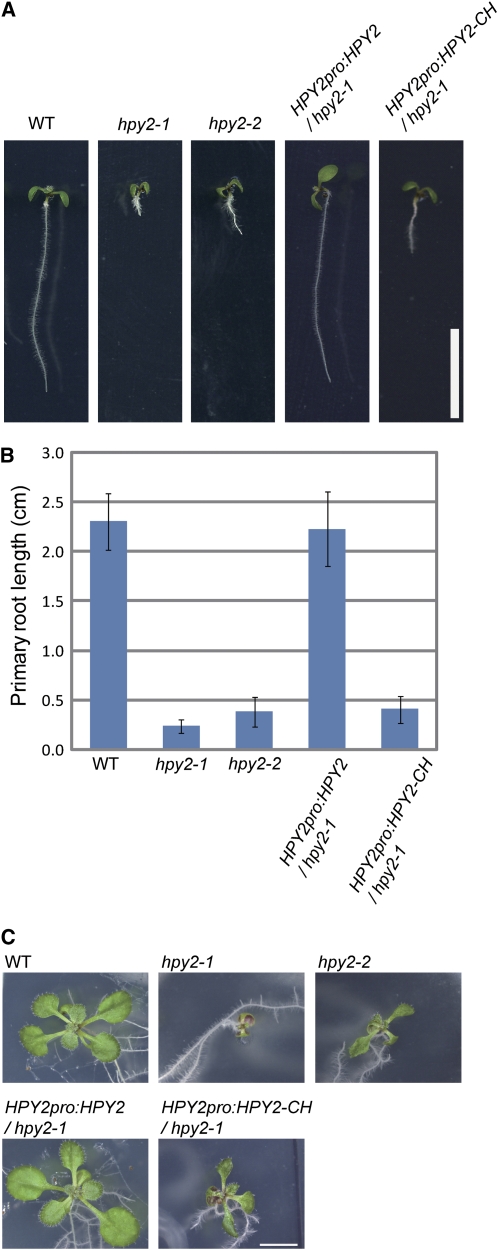
Figure 2.
Quantitative Analysis of hpy2 Phenotypes and Their Complementation with Genomic HPY2.
(A) Seven-day-old seedlings of wild-type, hpy2-1, hpy2-2, hpy2-1 carrying HPY2pro:HPY2, and hpy2-1 carrying HPY2pro:HPY2-CH.
(B) Quantitative analysis of primary root length. The length of 7-d-old roots was measured and shown as averages ± sd (n = 11 to 20).
(C) Fourteen-day-old seedlings of wild-type, hpy2-1, hpy2-2, hpy2-1 carrying HPY2pro:HPY2, and hpy2-1 carrying HPY2pro:HPY2-CH.
Bars = 1 cm in (A) and (C).
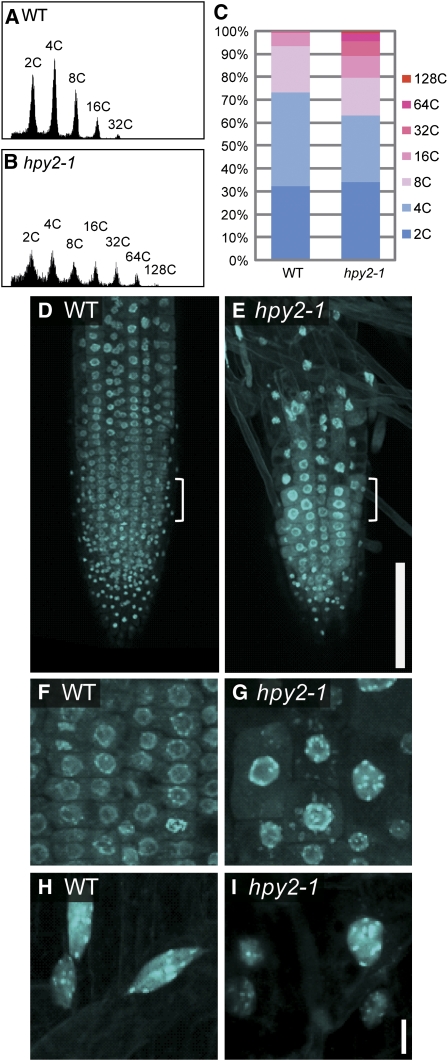
Our flow cytometry analysis demonstrates that ∼75% of nuclei isolated from the aerial tissues of 14-d-old wild-type seedlings give rise to 2C or 4C peaks, indicating that these populations of cells remain within the mitotic cell cycle (Figures 3A and 3C). In addition to these peaks, wild-type plants also possess ∼25% of 8C, 16C, and 32C peaks, representing the cell populations that have entered the endocycle. We found that the proportion of 2C and 4C peaks is reduced to ∼65% in hpy2-1 (Figures 3B and 3C), implying an earlier transition from the mitotic cycle to the endocycle. Furthermore, hpy2-1 has a higher 32C peak and additional 64C and 128C peaks that are not normally present in wild-type plants, suggesting that hpy2-1 cells have undergone additional rounds of endoreduplication. Confocal microscopy of 4',6-diamidino-2-phenylindole (DAPI)-stained nuclei reveals that 6-d-old wild-type root tips contain a large population of meristematic nuclei (Figures 3D and 3F). This nuclear population is mostly replaced with much larger nuclei in hpy2-1 (Figures 3E and 3G), indicating a premature entry into the endocycle in hpy2-1 root meristems. The final nuclei sizes that hpy2-1 root epidermal cells reach appear to be comparable with those found in mature wild-type cells (Figures 3H and 3I), implying that these cell types do not undergo additional endocycling.
Figure 3.
hpy2 Mutants Display an Early Transit into the Endocycle.
(A) and (B) Flow cytometry analysis of 14-d-old wild-type (A) and hpy2-1 (B) aerial tissues.
(C) Quantitative analysis of the flow cytometry data. The values represent averages of three independent biological replicates.
(D) to (I) Confocal microscopy of DAPI-stained roots of the wild type ([D], [F], and [H]) and hpy2-1 ([E], [G], and [I]).
(D) and (E) The size of small dots at the root tip, representing nuclei in root cap cells, is comparable between the wild type and hpy2-1. Further away from these nuclear populations, wild-type roots have large arrays of meristematic nuclei, including those undergoing mitosis. The hpy2-1 roots have very few meristematic nuclei.
(F) and (G) Magnified views of the area marked by white brackets in (D) and (E) show endoreduplicated nuclei in hpy2-1.
(H) and (I) Magnified views of mature root epidermal cells.
Bars = 100 μm in (D) and (E) and 10 μm in (F) to (I).
CYCB1, CDKB1, and CDKB2 Levels Are Reduced in hpy2-1
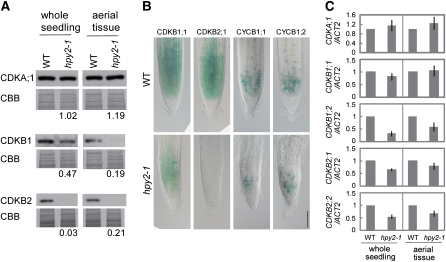
To test whether premature endocycles in hpy2-1 correlate with expression of CDKs, we investigated the transcript and protein levels of CDKA;1, CDKB1, and CDKB2 in the wild type and hpy2-1. Protein levels of CDKB1 and CDKB2 are substantially decreased in both hpy2-1 whole seedlings and aerial tissues compared with the wild type, whereas CDKA;1 levels remain relatively unchanged in hpy2-1 (Figure 4A). Consistently, the β-glucuronidase (GUS) activities deriving from the CDKB1;1pro:CDKB1;1-GUS or CDKB2;1pro:CDKB2;1-GUS translational fusion constructs, in which CDKB promoters drive the expression of a CDKB-GUS fusion protein, are strongly reduced in the hpy2-1 root meristems (Figure 4B), further substantiating low CDKB1 and CDKB2 levels in hpy2-1. The reduction in the CDKB1;1-GUS levels is milder than that of CDKB2;1-GUS but is considerable when compared with the wild type (Figure 4B). In addition, we found that the transcript levels of CDKB1;1, CDKB1;2, CDKB2;1, and CDKB2;2, but not of CDKA;1, are 10 to 60% lower in hpy2-1, although these differences are not as dramatic as those of the protein levels (Figure 4C). These results indicate that the levels of CDKB1 and CDKB2 are modified both transcriptionally and posttranscriptionally in hpy2-1 but that the effects of latter are more profound.
Figure 4.
CYCB1, CDKB1, and CDKB2 Levels Are Reduced in hpy2.
(A) Quantitative immunoblotting of 10-d-old wild-type and hpy2-1 whole seedlings or aerial tissues, using anti-CDKA;1, CDKB1, and CDKB2 antibodies. Coomassie Brilliant Blue (CBB)–stained membranes are shown as loading controls. Representative data from three biological repeats. Values shown below each blot are average protein levels in hpy2-1, relative to those of the wild type (set to 1), calculated from three independent biological trials.
(B) Differential interference contrast microscopy of the root meristems. The numbers of CDKB1;1-GUS, CDKB2;1-GUS, CYCB1;1-GUS, and CYCB1;2-GUS labeled cells are reduced in hpy2-1, and they are confined more tightly to the root meristem. Bar = 100 μm.
(C) Quantitative RT-PCR of CDKA;1, CDKB1;1, CDKB1;2, CDKB2;1, and CDKB2;2 expression in 10-d-old whole seedlings or aerial tissues. Mean values of three repeats are shown with sd.
We also determined the protein levels of mitotic cyclins CYCB1;1 and CYCB1;2 using transgenic lines, harboring CYCB1;1-GUS and CYCB1;2-GUS constructs, in which CYCB1 promoters drive the expression of a fusion protein between the CYCB1 mitotic destruction box and GUS (Colon-Carmona et al., 1999; Donnelly et al., 1999). The expressions of CYCB1;1-GUS and CYCB1;2-GUS are strong in the meristematic regions of 6-d-old wild-type roots, while these GUS signals are significantly reduced in hpy2-1 root meristems (Figure 4B), indicating that the hpy2-1 mutation affects accumulation of CYCB1;1 and CYCB1;2 proteins. These observations strongly suggest that HPY2 is involved in promoting the mitotic cell cycle and/or repressing an entry into the endocycle in Arabidopsis.
HPY2 Encodes SUMO E3 Ligase MMS21
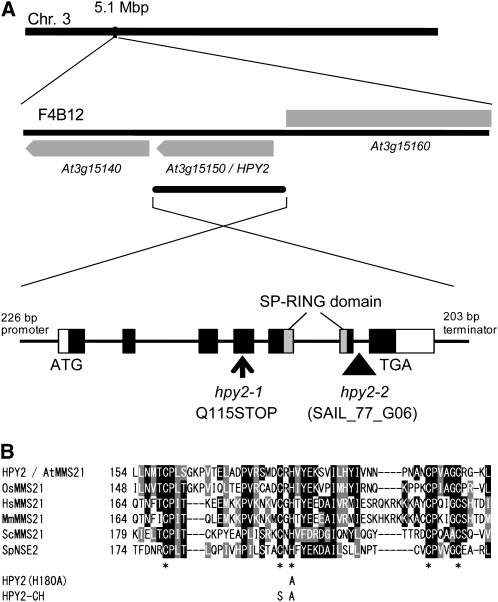
By positional cloning, we found that HPY2 encodes an MMS21 protein (At3g15150) that has significant deduced amino acid sequence similarity to the yeast or mammalian SUMO E3 ligase MMS21 (Figures 5A and 5B). The HPY2 gene comprises seven exons, and the hpy2-1 mutation, a single nucleotide substitution from cytosine to thymine in the fourth exon, results in a premature translational stop codon and a truncation of the C-terminal 135 amino acids (Figure 5A). The hpy2-2 allele, containing a T-DNA insertion in the sixth intron (Figure 5A), shows slightly milder but similar growth phenotypes (Figures 2A to 2C). In addition, the ∼2.6-kb HPY2 genomic fragment (Figure 5A) fully rescues the hpy2-1 phenotypes (Figures 2A to 2C), formally confirming the identity of the HPY2 gene.
Figure 5.
HPY2 Encodes an MMS21 Protein.
(A) Chromosomal position and structure of the HPY2 locus. Gray boxes indicate the coding region of HPY2 (At3g15150) and its neighboring genes in the BAC clone F4B12 of chromosome 3. The HPY2 genomic fragment used for the complementation includes 226 bp upstream sequence, 2.2 kb coding sequence, and 203 bp downstream sequence of the At3g15150 gene. Gene structure of HPY2 illustrates the position of the SP-RING domain in the C-terminal region. The hpy2-1 mutation converts Gln-115 (Q115) to a stop codon. The hpy2-2 allele (SAIL_77_G06) contains a T-DNA insertion in the sixth intron.
(B) Alignment of the SP-RING domain among MMS21 homologs from rice (Oryza sativa; OsMMS21), human (Homo sapiens; HsMMS21), mouse (Mus musculus; MmMMS21), and yeast (Saccharomyces cerevisiae; ScMMS21 and Schizosaccharomyces pombe; SpNSE2). Black boxes indicate conserved residues, and gray boxes indicate similar residues. The core Cys (C) and His (H) residues are marked by asterisks. HPY2 (H180A) shows the His-to-Ala (A) substitution introduced for the in vitro sumoylation assay, and HPY2-CH shows the Cys-to-Ser (S) and His-to-Ala substitutions introduced for the in planta complementation assay.
HPY2 is a 249–amino acid protein that contains an SP-RING domain, a Zn-finger domain required for SUMO E3 ligase activity (Andrews et al., 2005; Miura et al., 2007a) (Figures 5A and 5B). An SP-RING domain has several Cys and His residues conserved across yeasts, mammals, and plants (Andrews et al., 2005; Miura et al., 2007a) (Figure 5B), and these residues facilitate the binding of SUMO E2 to E3 ligases to promote sumoylation. A construct expressing a mutant form of HPY2 (HPY2-CH; Figure 5B), carrying Cys-to-Ser (C178S) and His-to-Ala (H180A) substitutions, under the endogenous HPY2 promoter fails to complement the hpy2-1 phenotypes (Figures 2A to 2C), indicating that these amino acid residues are required for the in vivo function of HPY2.
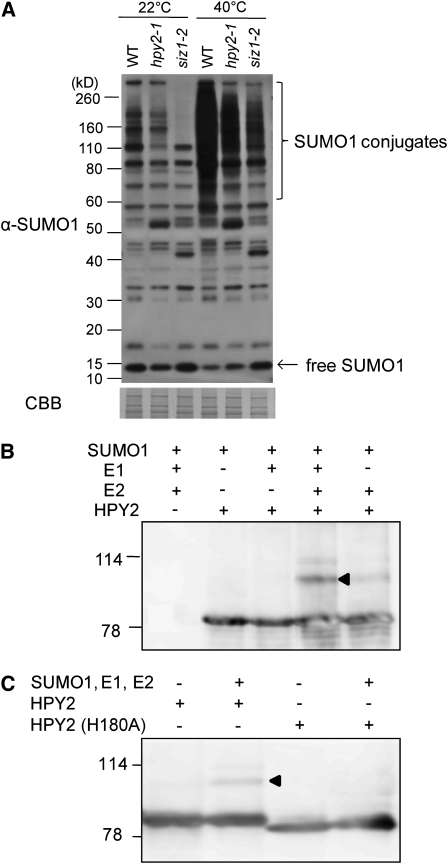
To test whether HPY2 is required for sumoylation in vivo, we first performed an immunoblot experiment using an antiserum raised against Arabidopsis SUMO1 (see Supplemental Figure 1 online). As shown in Figure 6A, the sumoylation patterns of total proteins differ moderately but reproducibly between 10-d-old wild-type and hpy2-1 seedlings grown at 22°C, suggesting that HPY2 is involved in sumoylation in planta. The conjugation of SUMO to target proteins increases dramatically upon various stress responses in Arabidopsis; for example, brief heat shock treatments at 40°C induce rapid and massive sumoylation in wild-type seedlings (Kurepa et al., 2003; Miura et al., 2005, 2007b; Catala et al., 2007; Conti et al., 2008) (Figure 6A). We found that heat shock–induced sumoylation level in hpy2-1 is substantially weaker than that of the wild type (Figure 6A). Interestingly, hpy2-1 and siz1-2 appear to display slightly different sumoylation patterns both at 22 and 40°C, implying that HPY2 and SIZ1 might have some distinct SUMO targets.
Figure 6.
HPY2 Acts as a SUMO E3 Ligase Both in Vivo and in Vitro.
(A) HPY2 functions as an E3 SUMO ligase in vivo. Immunoblot analysis using an anti-SUMO1 antiserum shows that the steady state level and heat shock–induced SUMO1 conjugation are reduced in hpy2-1 and siz1-2. Total protein was extracted from 10-d-old wild-type, hpy2-1, and siz1-2 plants with and without heat treatment. A Coomassie (CBB)-stained membrane after immunoblotting is shown as a loading control.
(B) HPY2 is autosumoylated in vitro. Purified maltose binding protein (MBP)-tagged HPY2 proteins were incubated with SUMO reaction mixtures containing Arabidopsis His-SAE1 (E1), His-SAE2 (E1), His-SCE1 (E2), and His-SUMO1. After reaction, MBP-HPY2 was pulled down using amylose resin, followed by SDS-PAGE and immunoblotting with an anti-MBP antibody.
(C) The H180A substitution in the SP-RING domain blocks autosumoylation of HPY2. An arrowhead indicates the MBP-HPY2-SUMO1 conjugates.
Several known SUMO E3 ligases, such as yeast SIZ2 and human MMS21, promote sumoylation of themselves (Takahashi et al., 2003; Potts and Yu, 2005). To test whether HPY2 has a SUMO E3 ligase activity in vitro, we thus performed an in vitro autosumoylation assay. As shown in Figure 6B, HPY2, when incubated with SUMO1, Arabidopsis E1 (SAE1 and SAE2), and E2 (SCE1), efficiently forms high molecular weight conjugates on SDS-PAGE. The removal of SAE1 and SAE2, or SCE1, blocks autosumoylation of HPY2 (Figure 6B). Substitution of His to Ala (H180A; Figure 5B) in the SP-RING domain dramatically reduces the formation of HPY2-SUMO1 conjugates (Figure 6C), indicating that HPY2 can act as a SUMO E3 ligase and not simply as an efficient SUMO substrate for SCE1.
HPY2-GFP Proteins Are Predominantly Found in the Nuclei of Root Meristems
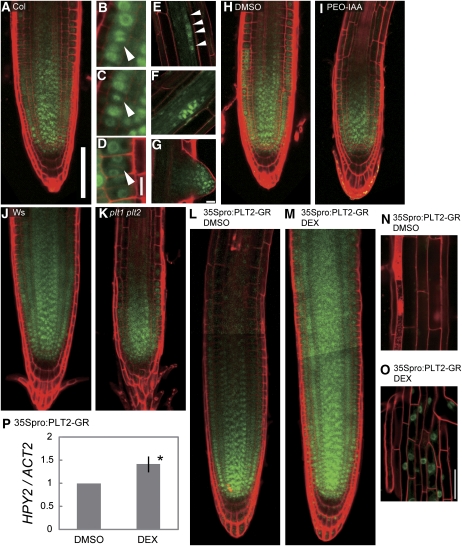
Consistent with the predicted function of HPY2 in the root meristem, the HPY2 promoter-driven HPY2-green fluorescent protein (GFP) expression is found in the meristems of growing roots (Figure 7A). The GFP signal can be found in almost all cell types of primary root meristems, including epidermis, cortex, endodermis, and pericycle, and its expression declines rapidly as cells leave the meristem and start expanding. HPY2-GFP proteins appear to be present throughout the nuclei in most of these cells, but they exhibit localizations similar to those of condensed chromosomes in cells undergoing mitosis (Figures 7B to 7D). The nuclear HPY2-GFP signals can be also detected in cells of developing lateral root primordia and their meristems (Figures 7E to 7G), suggesting an involvement of HPY2 in lateral root formation.
Figure 7.
PLT Family Transcription Factors PLT1 and PLT2 Modulate the HPY2-GFP Accumulation in Arabidopsis Roots.
(A) Confocal microscopy of 6-d-old wild-type Columbia (Col) roots showing the 226-bp HPY2 promoter-driven expression of HPY2-GFP in the root meristematic region.
(B) to (D) HPY2-GFP expression in mitotically dividing cells. Arrowheads indicate cells undergoing mitosis.
(E) to (G) HPY2-GFP expression in the developing lateral root primordia ([E] and [F]) and meristems (G). Arrowheads in (E) indicate four nuclei with HPY2:GFP signals at the very early stage of lateral root initiation.
(H) and (I) HPY2-GFP expression in 6-d-old wild-type roots treated with DMSO (H) and an auxin antagonist PEO-IAA ([I]; 10 μM), blocking TIR1-dependent auxin signaling.
(J) and (K) HPY2-GFP expression in 6-d-old wild-type Wassilewskija (Ws) (J) and plt1-4 plt2-2 (K) roots.
(L) to (O) HPY2-GFP expression in double transgenic plants carrying HPY2pro:HPY2-GFP and 35Spro:PLT2-GR after 1-d treatment of DMSO ([L] and [N]) and 1 μM DEX ([M] and [O]).
(P) Quantitative RT-PCR of HPY2 expression in 6-d-old roots after 1-d exposure to DMSO (left) and 1 μM DEX (right). A mean value of four repeats, relative to that of DMSO-treated samples, is shown with an sd. An asterisk indicates a significant difference between DMSO- and DEX-treated roots (Student's t test, P < 0.05).
Bars = 100 μm in (A) and (H) to (O), 10 μm in (B) to (D), and 20 μm in (E) to (G).
PLT Family Transcription Factors PLT1 and PLT2 Positively Regulate the Expression and/or Accumulation of HPY2 Proteins in the Root Meristem
The plant growth regulator auxin plays a crucial role in the patterning and growth of Arabidopsis roots (Blilou et al., 2005; Dello Ioio et al., 2008). We therefore tested whether the activity of DR5rev:GFP, a reporter of auxin response and distribution (Friml et al., 2003), is modified in hpy2-1. As shown in Supplemental Figures 2A and 2B online, however, the DR5 activity is not markedly different between wild-type and hpy2-1 roots. To explore the possibility that HPY2 acts downstream of auxin signaling, we next examined whether exogenous auxin modifies the expression and/or accumulation of HPY2-GFP proteins. An application of the synthetic auxin α-naphthaleneacetic acid (NAA), at a concentration (10 μM) sufficient to induce lateral root formation (Himanen et al., 2002), strongly induces HPY2-GFP expression in the lateral root primordia (see Supplemental Figure 2C online). Furthermore, an auxin antagonist PEO-IAA (10 μM) that blocks TIR1-dependent auxin signaling (Hayashi et al., 2008) narrows the domain of HPY2-GFP expression in primary roots (Figures 7H and 7I), suggesting that auxin signaling might modulate the expression pattern of HPY2 proteins.
The auxin-inducible AP2-domain transcription factors PLT1 and PLT2 are key regulators of root meristem development in Arabidopsis (Aida et al., 2004). Since the expression pattern of HPY2-GFP proteins largely overlaps with those of PLT1 and PLT2 (Galinha et al., 2007), they may be involved in the induction of HPY2 expression. Meristem size in plt1 plt2 roots is strongly reduced (Galinha et al., 2007) and, as expected, HPY2-GFP expression is also more constrained to the root tip (Figures 7J and 7K). Conversely, activation of PLT2-GR by dexamethasone (DEX) expands the root meristematic region (Galinha et al., 2007), and the expression of HPY2-GFP proteins is also increased dramatically in the DEX-induced double transgenic plants carrying HPY2pro:HPY2-GFP and 35Spro:PLT2-GR constructs (Figures 7L and 7M). Strong nuclear HPY2-GFP signals are also detected in fully differentiated root epidermal cells (Figures 7N and 7O), indicating that the induced accumulation of HPY2 is not simply a downstream consequence of enlarged meristems. Our quantitative RT-PCR, using RNA isolated from 6-d-old roots, further demonstrates that the HPY2 transcript level is significantly increased in DEX-treated PLT2-GR roots (Figure 7P), indicating that at least part of the induced HPY2-GFP expression is due to its transcriptional activation. These results establish that PLT proteins are both necessary and sufficient for the expression and/or accumulation of HPY2 proteins in the root meristem.
HPY2 Functions Downstream of PLT in the Root Meristem
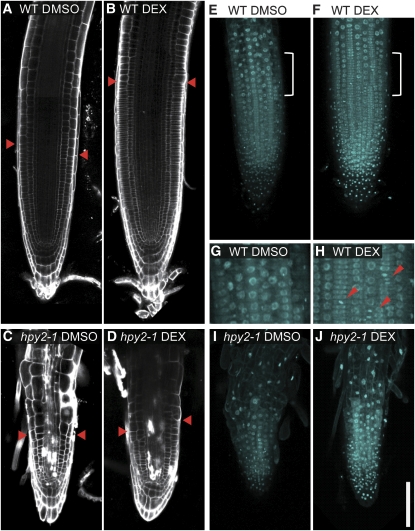
The PLT-dependent HPY2 accumulation in the root meristem suggests that HPY2 may function downstream of PLT to promote meristem development. To test this possibility, we determined whether the DEX-induced root meristem expansion in PLT2-GR plants requires HPY2 proteins. As reported previously (Galinha et al., 2007), our confocal microscopy reveals that 1-d DEX treatment (1 μM) of 6-d-old PLT2-GR plants substantially enlarges their root meristem size as judged by the size of PI-stained epidermal cells (Figures 8A and 8B). In contrast, when PLT2-GR is activated by DEX in hpy2-1, the root meristem size increases only very little, if at all (Figures 8C and 8D), indicating that the PLT-induced meristem enlargement is largely dependent on HPY2 and, thus, that HPY2 acts downstream of PLT to mediate root meristem development.
Figure 8.
HPY2 Functions Downstream of PLT to Promote the Root Meristem Development.
(A) to (D) Confocal microscopy of the PI-stained wild-type ([A] and [B]) and hpy2-1 ([C] and [D]) roots. Six-day-old wild-type or hpy2-1 seedlings carrying 35Spro:PLT2-GR were treated with DMSO ([A] and [C]) and 1 μM DEX ([B] and [D]). Red arrowheads indicate the approximate position where epidermal cells start expanding.
(E) to (J) Confocal microscopy of the DAPI-stained nuclei in wild-type ([E] to [H]) and hpy2-1 ([I] and [J]) roots. Six-day-old wild-type or hpy2-1 seedlings carrying 35Spro:PLT2-GR were treated with DMSO ([E], [G], and [I]) and 1 μM DEX ([F], [H], and [J]).
(G) and (H) Magnified (twofold) views of marked areas in (E) and (F) show the presence of mitotic nuclei further away from the root tip in DEX-treated roots. Red arrowheads indicate mitotic nuclei with condensed chromosomes.
Bar = 100 μm.
We next explored whether these enlargements of root meristems in DEX-activated PLT2-GR plants are accompanied by a delayed transition from the mitotic cycle into the endocycle. Confocal microscopy of DAPI-stained nuclei shows that DEX-induced PLT2-GR roots possess mitotic nuclei with condensed chromosomes much further away from the root tip, compared with the mock-treated PLT2-GR roots (Figures 8E to 8H). These results clearly demonstrate that PLT induction delays an exit from the mitotic cycle. As expected, DEX-induced PLT activities do not delay the mitotic exit in hpy2-1 (Figures 8I and 8J), further substantiating the functional role of HPY2 in the PLT pathway.
DISCUSSION
Role of HPY2 in Cell Cycle Progression and Meristem Development
This study demonstrates that HPY2 functions as a SUMO E3 ligase modulating cell cycle progression and meristem development in Arabidopsis. Loss of HPY2 results in a premature exit from the mitotic cell cycle and an early entry into the endocycle, implicating HPY2 in the repression of the mitotic-to-endocycle transition. The expression of HPY2 proteins in proliferating cells of root meristems suggests that the primary function of HPY2 is to maintain meristematic cells in the mitotic cycle. Since the in vivo targets of HPY2 sumoylation remain unknown, it also might be possible that HPY2 is required to establish and/or maintain the plant meristems through other mechanisms and that the observed endocycle defects are indirect consequences of severely compromised meristems. These two possibilities are not mutually exclusive since HPY2 may target both cell cycle proteins and meristem regulators to promote the meristem development through multiple pathways. We need to point out, however, that some of the phenotypes we observe in hpy2-1 are difficult to explain as merely an effect of reduced meristem sizes. For example, we show that both transcript and protein levels of CDKB1 and CDKB2 are strongly reduced in hpy2-1, while those of CDKA remain largely unchanged. We need to bear in mind that compared with CDKA, the expression of CDKBs is more restricted to the meristems; thus, the lower expression of CDKBs can be explained more readily by a reduced meristem size. However, hpy2-1 mutants still possess small meristems, and we think it is unlikely that almost complete loss of CDKB2 is solely due to the reduced size of meristems. Moreover, reduced meristems in hpy2-1 may account for an early onset of endocycling but not for the higher endocycle levels we find in the aerial tissues of the mutant. Based on these observations, our current hypothesis is that at least part of HPY2's in vivo function is to target cell cycle proteins for sumoylation to modify their activities. Indeed, many core cell cycle proteins and/or their upstream regulators, including CYCs, CDKs, and E2Fs, have putative sumoylation sites and they are all localized to nuclei. Thus, it is plausible that HPY2 directly mediates sumoylation of these proteins, leading to the modulation of CYC and CDK levels through transcriptional and/or posttranscriptional modifications. Relatively strong downregulation of CDKB1 and CDKB2, both of which function during the G2-to-M phase, in hpy2-1 suggests that the preferential inhibition of mitotic CDK activities might prevent cells from entering the M-phase, but not the S-phase, thus leading to the endocycle. In fact, downregulation of CDKB1 and CDKB2 activities is sufficient to induce endocycling in Arabidopsis (Boudolf et al., 2004; Andersen et al., 2008); thus, it would be important in future studies to determine whether the reduced CDKs and/or CYCs are responsible for the premature endocycle onset in hpy2-1. Although CDKs tested in this study are potential targets for HPY2-mediated sumoylation, our immunoblot experiments with anti-CDK antibodies haven't detected bands with altered mobility corresponding to the CDK-SUMO1 conjugates. These data leave two possibilities: one is that CDKs are not the direct SUMO targets, and the other is that CDK-SUMO1 conjugates are below the detection limit, as suggested by previous reports showing that only very small fractions of substrate proteins are sumoylated at a given time (Johnson, 2004).
Our data are also consistent with the idea that HPY2 is required for other processes having downstream consequences for endoreduplication. Yeast and mammalian MMS21 encodes a subunit of the SMC5/6 (structural maintenance of chromosomes) complex and facilitates sumoylation of SMC5/6 (Andrews et al., 2005; Potts and Yu, 2005). The SMC5/6 complex is implicated for DNA damage repair (Potts, 2009) and sister chromatid separation during mitosis (Torres-Rosell et al., 2005); thus, HPY2/MMS21 may facilitate sumoylation of the plant SMC complex, which in turn promotes cell proliferation. The colocalization of HPY2-GFP with condensed chromosomes in mitotically dividing cells suggests an involvement of HPY2 in some basic cell division function, such as chromatin condensation and/or segregation during mitosis. Because endocycle onset is often linked with the initiation of cell expansion and cell differentiation (Inze and De Veylder, 2006), it is also possible that HPY2 plays some regulatory roles in these accompanying processes. It is worth noting in this context that some previously described meristem patterning regulators contain predicted sumoylation sites, suggesting that sumoylation might be involved more generally in the establishment and/or maintenance of plant meristems.
HPY2 Functions in the PLT Pathway
The PLT family of AP2 transcription factors are key regulators of root meristem development (Aida et al., 2004; Galinha et al., 2007), but to date little is established on how they exert their function. In addition to the PIN genes whose expressions are positively regulated by PLT proteins as part of the feedback loops maintaining the active auxin gradients (Blilou et al., 2005; Galinha et al., 2007; Benjamins and Scheres, 2008), only a few genes have been described so far to be regulated by PLTs. We show that in the root meristem, localized HPY2 activities are defined at least partially by the PLT-dependent expression and/or accumulation of HPY2 proteins. In agreement with this, PLT induction in hpy2-1 mutants does not rescue their meristem defects, demonstrating that HPY2 functions downstream of PLT to mediate the root meristem development. Although HPY2 may or may not be a direct target of PLT's transcriptional regulation, these findings represent an important conceptual advance in our understanding of how PLTs define the root meristem. Given that PLT expression is auxin responsive (Aida et al., 2004), it is tempting to speculate that HPY2-mediated sumoylation constitutes part of the regulatory mechanisms connecting auxin- and PLT-dependent developmental signals with downstream cell cycle progression and meristem development.
The severe defects in the shoot apical meristem of hpy2-1 strongly suggest that HPY2 also has vital functions in shoot meristem development. It would be of interest in future studies to explore the regulatory mechanisms that specify HPY2 expression in shoot meristems.
Sumoylation in Plant Growth and Development
An involvement of MMS21 in various cellular processes, such as DNA repair and chromosome segregation, is well documented in yeast and human culture cells (Potts and Yu, 2005), and this study demonstrates that MMS21 proteins also have crucial roles in the growth and development of multicellular organisms. The hpy2 mutants display extended cycles of endoreduplication up to 128C. An increase in ploidy levels is often accompanied by an increase in cell size (Sugimoto-Shirasu and Roberts, 2003), and accordingly we often find abnormally enlarged cells in hpy2 mutants. However, hpy2 mutants exhibit severe dwarf phenotypes, probably because premature endocycling comes at the expense of reduced cell proliferation activities. These observations thus highlight that HPY2-dependent regulation of the balance between mitotic cycles and endocycles is critical for the normal growth of postembryonic plant organs.
In comparison with the siz1 mutants that display general growth retardation phenotypes (Catala et al., 2007), the developmental phenotype of hpy2 mutants is most profound in the meristematic region, suggesting that HPY2 has preferential functions in the meristem. This is in agreement with our observation that compared with SIZ1, HPY2 makes smaller contributions to the overall accumulation of SUMO1 conjugates in planta, implying that HPY2 targets more meristem-related proteins for sumoylation. Furthermore, siz1-2 mutants do not display major ploidy defects or alterations in the expression and/or accumulation of CDKA and CDKB proteins (see Supplemental Figure 3 online). These results together implicate HPY2 and SIZ1 in some distinct regulatory pathways mediating plant organ growth. Our data show that the heat shock–induced sumoylation level is reduced in hpy2, suggesting that like SIZ1, HPY2 might also participate in stress responses.
Taken together, this study establishes that a SUMO E3 ligase, HPY2, mediates cell cycle progression and meristem development in Arabidopsis. HPY2 is likely to target multiple proteins for sumoylation; thus, identification and functional analyses of its target proteins should provide further mechanistic insights into how HPY2-dependent sumoylation participates in postembryonic plant development.
METHODS
Plant Materials and Growth Conditions
The hpy2-1 mutant was isolated from a screen using ∼3000 M2 populations independently established after ethyl methanesulfonate mutagenesis. Each M2 line was first screened for altered organ sizes, and up to 30 M2 lines that display altered organ sizes were subscreened for ploidy phenotypes using flow cytometry. All other mutants and transgenic lines used in this study were provided by Peter Dooner (CYCB1;1-GUS; Colon-Carmona et al., 1999), Goro Horiguchi (CYCB1;2-GUS; Donnelly et al., 1999), Iris Meier (nua-4 [Xu et al., 2007] and esd4-2 [Murtas et al., 2003]), Eva Benkova (DR5rev:GFP; Friml et al., 2003), Ben Scheres (plt1-4 plt2-2 [Aida et al., 2004] and 35Spro:PLT2-GR [Galinha et al., 2007]), and the ABRC (hpy2-2 [SAIL_77_G06] and siz1-2 [Miura et al., 2005]). The plt1-4 plt2-2 mutants were in the Wassilewskija background, and all other lines were in Col background. Seeds were surface sterilized in 70% ethanol for 1 min and then in 20% (v/v) sodium hypochlorite for 5 min and rinsed three times in sterile water. Sterilized seeds were plated on Murashige and Skoog (MS) media supplemented with 1% (w/v) sucrose and 0.5% Gelrite. After cold treatment in the dark for 2 d, seeds were incubated under continuous light at 22°C.
For chemical treatments, PEO-IAA, DEX, and NAA were dissolved in DMSO as a stock solution of 100, 1, and 10 mM, respectively, and diluted into MS media. For the treatment with PEO-IAA, seeds were germinated on MS media containing 10 μM PEO-IAA. To induce transient expression of PLT2, transgenic plants harboring the 35Spro:PLT2-GR construct were incubated for 5 d after germination and then transferred to MS media containing 1 μM DEX. To induce lateral root formation, seedlings were incubated for 5 d after germination and then transferred to MS media containing 10 μM NAA. For heat shock experiments, 10-d-old plants were heat-treated at 40°C for 1 h.
Ploidy Measurement
Ploidy levels were measured using the ploidy analyzer PA-I (Partec) as described previously (Sugimoto-Shirasu et al., 2002). At least 7000 nuclei isolated from aerial tissues of 14-d-old seedlings were used for each ploidy measurement.
Microscopy
Histological analyses of shoot apical meristems were performed as described (Hibara et al., 2006). Six-day-old seedlings were fixed in a mixture of 1.8% formaldehyde, 5% acetic acid, and 90% ethanol overnight at 4°C, embedded in Technovit 7100 (Heraeus Kulzer), and sectioned at 6-μm thickness with a microtome (Leica). Sections were briefly stained with Toluidine Blue and observed using an Olympus BX51 fluorescence microscope.
Root meristem organization was visualized using seedlings stained with 5 mg/mL PI. For the visualization of nuclei in roots, 6-d-old seedlings were fixed in the PEMT buffer described by Sugimoto et al. (2000). Fixed seedlings were washed with PEMT buffer three times followed by washing with PBS buffer three times. Seedlings were stained for 10 min with DAPI using 25% CyStain B solution (Partec) in PBS buffer, washed with PBS buffer three times, and then mounted in PBS buffer. PI, DAPI, and GFP signals were visualized by fluorescence microscopy using a Carl Zeiss LSM510 META confocal laser microscope.
Histochemical GUS staining of roots were performed based on the protocol described by Malamy and Benfey (1997). Seedlings were fixed in 90% acetone overnight, stained in the GUS buffer with 0.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronide for 1 h, mounted in chloral hydrate solution (8 g chloral hydrate, 2 mL water, and 1 mL glycerol), and observed using an Olympus BX51 fluorescence microscope.
Positional Cloning of the HPY2 Gene and Its Amino Acid Sequence Analysis
To map the HPY2 locus, the hpy2-1 mutant (Col background) was crossed with the Landsberg erecta line, and the F1 plants were self-pollinated to produce the F2 mapping populations. In the F2 population, genomic DNA was purified from seedlings showing the hpy2-1 dwarf phenotypes, and the mutation was mapped using the Cereon polymorphism database (http://www.Arabidopsis.org/). The genomic sequence of At3g15150 in hpy2-1 was amplified by PCR (using primers gHPY2-5′F, 5′-CACCAAACCCCCTAGACGGGCTATGGATTG-3′ [TOPO site underlined], and gHPY2-3′R, 5′-GGGAAGCTAAAAGCCTTTAGTTTG-3′) and sequenced with ABI PRISM 3100 sequencer (Applied Biosystems). Sequenced data were compared with the wild-type sequence, based on the TAIR9 data set, by the BLAST program (http://www.Arabidopsis.org/). Amino acid sequences of MMS21 from rice (Oryza sativa), human, mouse, and yeast were collected and aligned as previously described (Miura et al., 2007a).
Construction of HPY2 Genomic Vectors
The 2.6-kb HPY2 genomic fragments were PCR amplified with the primers gHPY2-5′F (5′-CACCAAACCCCCTAGACGGGCTATGGATTG-3′) and gHPY2-3′R (5′-GGGAAGCTAAAAGCCTTTAGTTTG-3′) and cloned into entry vectors with the pENTR-D-TOPO cloning kit (Invitrogen) following the manufacturer's instructions. To generate the HPY2-CH construct, these entry vectors were modified by site-directed mutagenesis with primers gHPY2-C178S/H180A-F (5′-CCAGGGCCGTCTATGAAAAATCTGTAATCC-3′) and gHPY2-C178S/H180A-R (5′-CATAGACGGCCCTGGAATCCATACTGCAAC-3′). To generate the translational fusion construct HPY2pro:HPY2-GFP, a SmaI restriction site was introduced to the C-terminal end of the HPY2 genomic sequence by site-directed mutagenesis with primers gHPY2-C-SmaI-F (5′-GATCCCGGGTAGAGATCAGACATTTATTGG-3′) and gHPY2-C-SmaI-R (5′-CTACCCGGGATCTTCATCCACATCTTCTG-3′). The GFP fragments were amplified by PCR (with primers GFP-Sma1-F, 5′-GGGGGTGGCATGGTGAGCAAGGGCGAGGAG-3′, and GFP-Sma1-R, 5′-GGGTCCACCTCCCTTGTACAGCTCGTCC-3′ [3XGly linker underlined]) using the pGWB6 vector (Nakagawa et al., 2007) as DNA templates. After inserting the GFP fragments with DNA Ligation Kit Mighty Mix (TaKaRa), resulting entry vectors were transferred into the pGWB1 destination vectors (Nakagawa et al., 2007) using LR clonase II (Invitrogen).
Construction of CDKB-GUS Markers
The 3.9-kb CDKB1;1 genomic fragments, including the 2.5-kb promoter region, and the 4.0-kb CDKB2;1 genomic fragments, including the 2.2-kb promoter region, were PCR amplified with primers CDKB1;1 genomic-F (5′-AAAAAGCAGGCTAGAATGAGTCCTTTGTTCC-3′) and CDKB1;1 genomic-R (5′-AGAAAGCTGGGTAGAACTGAGACTTGTCAAG-3′) for CDKB1;1 and CDKB2;1 genomic-F (5′-AAAAAGCAGGCTGTCTTTTGCTTGTTCCCTAAC-3′) and CDKB2;1 genomic-R (5′-AGAAAGCTGGGTAGAGAGAGGACTTTTCTGGCAG-3′) for CDKB2;1. These fragments were reamplified and cloned into pDONR221 with the PCR-mediated Gateway directional BP cloning system (Invitrogen). The CDKBpro:CDKB fragments were transferred into the pGWB3 destination vectors (Nakagawa et al., 2007) with LR clonase II (Invitrogen).
RNA Extraction and Gene Expression Analyses
Total RNA was extracted using the Qiagen RNeasy plant mini kit with DNase I treatment (Qiagen) following the manufacturer's instructions, and subsequent reverse transcription was performed using the SuperScript III first-strand synthesis system (Invitrogen). The SYBR Green real-time PCR master mix (Toyobo) was used for quantitative real-time PCR with following primers: CDKA;1F (5′-AGAGGATCCGTCGGTGTGCT-3′) and CDKA;1R (5′-TCTGTGGCTTCAGATCACGATGGA-3′) for CDKA;1, CDKB1;1F (5′-GTTATCAACATCGATCTATGTTGTTCG-3′) and CDKB1;1R (5′-TCTCATGCGTATAAGACTTAAGAGG-3′) for CDKB1;1, CDKB1;2F (5′-TGGATGCATCTTTGCCGAGA-3′) and CDKB1;2R (5′-TCTCTCACTCTCGCTGAACAA-3′) for CDKB1;2, CDKB2;1F (5′-AGAGCCTTCACTCTGCCAATGAA-3′) and CDKB2;1R (5′-CATAGAGGGGGAAGCAGCACACT-3′) for CDKB2;1, CDKB2;2F (5′-TCACAATCTCTTGATGGACCGGAA-3′) and CDKB2;2R (5′-CTCAGAGAGAGGACTTGTCAGGCA-3′) for CDKB2;2, and qRT-HPY2-1F (5′-TGTCTCCGATAACAGTTCCACG-3′) and qRT-HPY2-1R (5′-TCAAGGTCCTTAACCTTGTCCG-3′) for HPY2. In all experiments, expression levels were quantified using the Mx3000P QPCR system (Stratagene) and normalized using a standard Microsoft Excel package against those of the ACTIN2 (ACT2) gene amplified using primers ACT2F (5′-ACATTGTGCTCAGTGGTGGA-3′) and ACT2R (5′-GAGATCCACATCTGCTGGAAT-3′).
Production of SUMO1 Antiserum
The SUMO1 cDNA fragments were amplified with primers SUM1-NdeI-F (5′-ATAATTAACATATGTCTGCAAACCAGGAGGAAGAC-3′) and SUM1-XhoI-R (5′-TAATATACTCGAGTCAGCCACCAGTCTGATGGAGCATCGC-3′) and cloned into the expression vector pET-16b (Stratagene). His-SUMO1 proteins heterologously expressed in the Escherichia coli strain BL21 (DE3) pLys (Novagen) were purified by Ni-NTA agarose (Qiagen) via the N-terminal 10xHis-tag according to the manufacturer's instructions and used to raise antiserum (Biogate). The same protein samples were used to affinity purify the raised antibodies (Biogate).
Protein Extraction and Immunoblot Analyses
Protein extracts were prepared from 10-d-old seedlings. Seedlings were ground in liquid nitrogen, resuspended in extraction buffer (100 mM Tris·Cl, pH 7.5, 1 mM EDTA, 0.5% Nonidet P-40, 2 mM NaF, 2 mM Na3VO4, 1 mM DTT, 1 × Protease Inhibitor Cocktail [Sigma-Aldrich], 5 mM N-ethylmaleimide [Thermo Scientific], and 50 μM MG132 [Sigma-Aldrich]) and clarified by centrifugation at 16,000g (15 min at 4°C). Supernatant was resuspended in 1× LDS loading buffer (Invitrogen) and 1× Sample Reducing Agent (Invitrogen) and incubated at 93°C for 1 min. Fifty to one hundred micrograms of protein was fractionated on NuPAGE Novex 4-12% Bis-Tris gels (Invitrogen). Protein was transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad). The membrane was blocked in 1× TBST (10 mM Tris·Cl, pH 8.0, 150 mM NaCl, and 0.05% Tween 20) containing 5% nonfat dried milk at room temperature for 1 h, incubated with the appropriate primary antibodies (1:2000 dilution; α-CDKA;1 [Adachi et al., 2006], α-CDKB1 [Takatsuka et al., 2009], α-CDKB2 [Kono et al., 2003], and affinity-purified α-SUMO1 antiserum) for 2 h at room temperature or overnight at 4°C. The membrane was developed using the peroxidase-conjugated secondary antibody (1:15,000 dilution; GE Healthcare) by ECL plus or ECL advance (GE Healthcare). The membranes after protein gel blot analyses were stained by Coomassie Brilliant Blue. The signal intensities were quantified by ImageJ (NIH).
In Vitro SUMO Conjugation Assay
The HPY2 cDNA fragments were PCR amplified with primers MMS21-F-BamHI (5′-GGATCCATGGCGTCGGCGTCCTCGTC-3′) and MMS21-R-BamHI (5′-GATGGATCCCTAATCTTCATCCAC-3′) and inserted in-frame into the pMal-c2x vector (New England Biolabs). Substitution of His to Ala was performed by site-directed mutagenesis with primers MMS21-H180A-F (5′-GGATTGCAGGGCCGTCTATG-3′) and MMS21-H180A-R (5′-CATAGACGGCCCTGCAATCC-3′). The resulting plasmids were designated as pMBP-HPY2 and pMBP-HPY2(H180A). Recombinant proteins expressed from pMBP-HPY2 and pMBP-HPY2(H180A) were purified with Amylose Resin (New England Biolabs) according to the manufacturer's instructions. Expression of other recombinant proteins from the expression vectors (kindly provided by HP Stuible) and the in vitro sumoylation assay were performed as described (Colby et al., 2006; Miura et al., 2007b, 2009) with modification. Eight micrograms of SUMO1, 0.5 μg of E1 enzyme, 0.5 μg of E2 enzyme (instead of 2 μg of E2), and 4 μg of MBP-HPY2 were added to the reaction mixture (50 mM Tris-HCl, pH 7.8, 100 mM NaCl, 15% glycerol, 5 mM ATP, and 10 mM MgCl2). Reactions were performed in a total volume of 50 μL at 25°C for 2-4 h, then at 30°C for 2 to 4 h. After reaction, MBP-HPY2 was purified with Amylose Resin (New England Biolabs) and separated by SDS-PAGE. Immunoblot analyses were performed with anti-MBP antibody (1:4000 dilution; New England Biolabs) and peroxidase-conjugated anti-mouse IgG antibody (1:3000 dilution; GE Healthcare). The membrane was developed by ECL plus (GE Healthcare).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ACT2 (AT3G18780), CDKA;1 (AT3G48750), CDKB1;1 (AT3G54180), CDKB1;2 (AT2G38620), CDKB2;1 (AT1G76540), CDKB2;2 (AT1G20930), CYCB1;1 (AT4G37490), CYCB1;2 (AT5G06150), ESD4 (AT4G15880), HPY2 (AT3G15150), NUA (AT1G79280), PLT1 (AT3G20840), PLT2 (AT1G51190), SAE1 (AT3G57870), SAE2 (AT2G21470), SCE1 (AT3G57870), SIZ1 (AT5G60410), SUMO1 (AT4G26840), OsMMS21 (NM_001062862), HsMMS21 (NP_775956), MmMMS21 (NP_081022), ScMMS21 (NP_010896), and SpNSE2 (XP_001713077).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. An Antiserum Raised against SUMO1 Recognizes Both Free SUMO1 and Its Conjugates.
Supplemental Figure 2. The hpy2-1 Mutation Does Not Modify the Overall Expression Pattern of DR5rev:GFP.
Supplemental Figure 3. The siz1-2 Mutants Do Not Display Major Defects in Cell Cycle Progression.
Supplementary Material
Acknowledgments
We thank Peter Doener for CYCB1;1-GUS lines, Goro Horiguchi for CYCB1;2-GUS lines, Iris Meier for nua-4 and esd4-2 lines, Eva Benkova for DR5rev:GFP lines, Ben Scheres for plt1-4 plt2-2 and 35Spro:PLT2-GR lines, ABRC for SAIL_77_G06 (CS803668) and siz1-2 lines, Hans-Peter Stuible for His-SUMO1, His-SAE1, His-SAE2, and His-SCE1 vectors, and Kenichiro Hayashi for PEO-IAA. We thank Taeko Kawada and Mariko Mori for their technical assistance, Masao Tasaka for his advice on the histological analysis of shoot meristems, and Keith Roberts and members of the Sugimoto Lab for their critical comments on the manuscript. This work was supported by a UK Biotechnology and Biological Science Research Council David Phillips Fellowship (K.S.), Japan Society for the Promotion of Science grants (T.I., K.S., and K.S.), RIKEN grants (T.I. and S.F.), and a Japanese Ministry of Education, Culture, Sports, Science, and Technology grant (K.M.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Keiko Sugimoto (sugimoto@psc.riken.jp).
Online version contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Adachi, S., Uchimiya, H., and Umeda, M. (2006). Expression of B2-type cyclin-dependent kinase is controlled by protein degradation in Arabidopsis thaliana. Plant Cell Physiol. 47: 1683–1686. [DOI] [PubMed] [Google Scholar]
- Aida, M., Beis, D., Heidstra, R., Willemsen, V., Blilou, I., Galinha, C., Nussaume, L., Noh, Y.S., Amasino, R., and Scheres, B. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120. [DOI] [PubMed] [Google Scholar]
- Andersen, S.U., Buechel, S., Zhao, Z., Ljung, K., Novak, O., Busch, W., Schuster, C., and Lohmann, J.U. (2008). Requirement of B2-type cyclin-dependent kinases for meristem integrity in Arabidopsis thaliana. Plant Cell 20: 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, E.A., Palecek, J., Sergeant, J., Taylor, E., Lehmann, A.R., and Watts, F.Z. (2005). Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage. Mol. Cell. Biol. 25: 185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins, R., and Scheres, B. (2008). Auxin: The looping star in plant development. Annu. Rev. Plant Biol. 59: 443–465. [DOI] [PubMed] [Google Scholar]
- Blilou, I., Xu, J., Wildwater, M., Willemsen, V., Paponov, I., Friml, J., Heidstra, R., Aida, M., Palme, K., and Scheres, B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. [DOI] [PubMed] [Google Scholar]
- Boudolf, V., Vlieghe, K., Beemster, G.T., Magyar, Z., Torres Acosta, J.A., Maes, S., Van Der Schueren, E., Inze, D., and De Veylder, L. (2004). The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer, C., Stacey, N.J., West, C.E., Zhao, Y., Chory, J., Tsukaya, H., Azumi, Y., Maxwell, A., Roberts, K., and Sugimoto-Shirasu, K. (2007). BIN4, a novel component of the plant DNA topoisomerase VI complex, is required for endoreduplication in Arabidopsis. Plant Cell 19: 3655–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala, R., Ouyang, J., Abreu, I.A., Hu, Y., Seo, H., Zhang, X., and Chua, N.H. (2007). The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19: 2952–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebolla, A., Vinardell, J.M., Kiss, E., Olah, B., Roudier, F., Kondorosi, A., and Kondorosi, E. (1999). The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J. 18: 4476–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby, T., Matthai, A., Boeckelmann, A., and Stuible, H.P. (2006). SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol. 142: 318–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colon-Carmona, A., You, R., Haimovitch-Gal, T., and Doerner, P. (1999). Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20: 503–508. [DOI] [PubMed] [Google Scholar]
- Conti, L., Price, G., O'Donnell, E., Schwessinger, B., Dominy, P., and Sadanandom, A. (2008). Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell 20: 2894–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio, R., Linhares, F.S., Scacchi, E., Casamitjana-Martinez, E., Heidstra, R., Costantino, P., and Sabatini, S. (2007). Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17: 678–682. [DOI] [PubMed] [Google Scholar]
- Dello Ioio, R., Nakamura, K., Moubayidin, L., Perilli, S., Taniguchi, M., Morita, M.T., Aoyama, T., Costantino, P., and Sabatini, S. (2008). A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384. [DOI] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., and Inze, D. (2007). The ins and outs of the plant cell cycle. Nat. Rev. Mol. Cell Biol. 8: 655–665. [DOI] [PubMed] [Google Scholar]
- De Veylder, L., Joubes, J., and Inze, D. (2003). Plant cell cycle transitions. Curr. Opin. Plant Biol. 6: 536–543. [DOI] [PubMed] [Google Scholar]
- Dewitte, W., Scofield, S., Alcasabas, A.A., Maughan, S.C., Menges, M., Braun, N., Collins, C., Nieuwland, J., Prinsen, E., Sundaresan, V., and Murray, J.A. (2007). Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl. Acad. Sci. USA 104: 14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly, P.M., Bonetta, D., Tsukaya, H., Dengler, R.E., and Dengler, N.G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215: 407–419. [DOI] [PubMed] [Google Scholar]
- Edgar, B.A., and Orr-Weaver, T.L. (2001). Endoreplication cell cycles: More for less. Cell 105: 297–306. [DOI] [PubMed] [Google Scholar]
- Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., Offringa, R., and Jurgens, G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153. [DOI] [PubMed] [Google Scholar]
- Galbraith, D.W., Harkins, K.R., and Knapp, S. (1991). Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol. 96: 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinha, C., Hofhuis, H., Luijten, M., Willemsen, V., Blilou, I., Heidstra, R., and Scheres, B. (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057. [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander, R., and Melchior, F. (2007). Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8: 947–956. [DOI] [PubMed] [Google Scholar]
- Hayashi, K., Hatate, T., Kepinski, S., and Nozaki, H. (2008). Design and synthesis of auxin probes specific to TIR1, auxin receptor. Regul. Plant Growth Dev. 43 (suppl.), 95. [Google Scholar]
- Hibara, K., Karim, M.R., Takada, S., Taoka, K., Furutani, M., Aida, M., and Tasaka, M. (2006). Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18: 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen, K., Boucheron, E., Vanneste, S., de Almeida Engler, J., Inze, D., and Beeckman, T. (2002). Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14: 2339–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inze, D., and De Veylder, L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40: 77–105. [DOI] [PubMed] [Google Scholar]
- Jin, J.B., et al. (2008). The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J. 53: 530–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E.S. (2004). Protein modification by SUMO. Annu. Rev. Biochem. 73: 355–382. [DOI] [PubMed] [Google Scholar]
- Kono, A., Umeda-Hara, C., Lee, J., Ito, M., Uchimiya, H., and Umeda, M. (2003). Arabidopsis D-type cyclin CYCD4;1 is a novel cyclin partner of B2-type cyclin-dependent kinase. Plant Physiol. 132: 1315–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa, J., Walker, J.M., Smalle, J., Gosink, M.M., Davis, S.J., Durham, T.L., Sung, D.Y., and Vierstra, R.D. (2003). The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J. Biol. Chem. 278: 6862–6872. [DOI] [PubMed] [Google Scholar]
- Lammens, T., Boudolf, V., Kheibarshekan, L., Zalmas, L.P., Gaamouche, T., Maes, S., Vanstraelen, M., Kondorosi, E., La Thangue, N.B., Govaerts, W., Inzé, D., and De Veylder, L. (2008). Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc. Natl. Acad. Sci. USA 105: 14721–14726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkins, B.A., Dilkes, B.P., Dante, R.A., Coelho, C.M., Woo, Y.M., and Liu, Y. (2001). Investigating the hows and whys of DNA endoreduplication. J. Exp. Bot. 52: 183–192. [PubMed] [Google Scholar]
- Larson-Rabin, Z., Li, Z., Masson, P.H., and Day, C.D. (2009). FZR2/CCS52A1 expression is a determinant of endoreduplication and cell expansion in Arabidopsis. Plant Physiol. 149: 874–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J., et al. (2007). Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 49: 79–90. [DOI] [PubMed] [Google Scholar]
- Magyar, Z., De Veylder, L., Atanassova, A., Bako, L., Inze, D., and Bogre, L. (2005). The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell 17: 2527–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy, J.E., and Benfey, P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44. [DOI] [PubMed] [Google Scholar]
- Miura, K., Jin, J.B., and Hasegawa, P.M. (2007. a). Sumoylation, a post-translational regulatory process in plants. Curr. Opin. Plant Biol. 10: 495–502. [DOI] [PubMed] [Google Scholar]
- Miura, K., Jin, J.B., Lee, J., Yoo, C.Y., Stirm, V., Miura, T., Ashworth, E.N., Bressan, R.A., Yun, D.J., and Hasegawa, P.M. (2007. b). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19: 1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, K., Lee, J., Jin, J.B., Yoo, C.Y., Miura, T., and Hasegawa, P.M. (2009). Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA 106: 5418–5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, K., Rus, A., Sharkhuu, A., Yokoi, S., Karthikeyan, A.S., Raghothama, K.G., Baek, D., Koo, Y.D., Jin, J.B., Bressan, R.A., Yun, D.J., and Hasegawa, P.M. (2005). The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. USA 102: 7760–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtas, G., Reeves, P.H., Fu, Y.F., Bancroft, I., Dean, C., and Coupland, G. (2003). A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. Plant Cell 15: 2308–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., Toyooka, K., Matsuoka, K., Jinbo, T., and Kimura, T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Potts, P.R. (2009). The Yin and Yang of the MMS21-SMC5/6 SUMO ligase complex in homologous recombination. DNA Repair (Amst.) 8: 499–506. [DOI] [PubMed] [Google Scholar]
- Potts, P.R., and Yu, H. (2005). Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol. Cell. Biol. 25: 7021–7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka, K., Simaskova, M., Duclercq, J., Petrasek, J., Zazimalova, E., Simon, S., Friml, J., Van Montagu, M.C., and Benkova, E. (2009). Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc. Natl. Acad. Sci. USA 106: 4284–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracco, S.A., Miller, M.J., Kurepa, J., and Vierstra, R.D. (2007). Genetic analysis of SUMOylation in Arabidopsis: Conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 145: 119–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto-Shirasu, K., Roberts, G.R., Stacey, N.J., McCann, M.C., Maxwell, A., and Roberts, K. (2005). RHL1 is an essential component of the plant DNA topoisomerase VI complex and is required for ploidy-dependent cell growth. Proc. Natl. Acad. Sci. USA 102: 18736–18741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto-Shirasu, K., and Roberts, K. (2003). “Big it up”: Endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6: 544–553. [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu, K., Stacey, N.J., Corsar, J., Roberts, K., and McCann, M.C. (2002). DNA topoisomerase VI is essential for endoreduplication in Arabidopsis. Curr. Biol. 12: 1782–1786. [DOI] [PubMed] [Google Scholar]
- Sugimoto, K., Williamson, R.E., and Wasteneys, G.O. (2000). New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol. 124: 1493–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y., Dilkes, B.P., Zhang, C., Dante, R.A., Carneiro, N.P., Lowe, K.S., Jung, R., Gordon-Kamm, W.J., and Larkins, B.A. (1999). Characterization of maize (Zea mays L.) Wee1 and its activity in developing endosperm. Proc. Natl. Acad. Sci. USA 96: 4180–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y., Toh, E.A., and Kikuchi, Y. (2003). Comparative analysis of yeast PIAS-type SUMO ligases in vivo and in vitro. J. Biochem. 133: 415–422. [DOI] [PubMed] [Google Scholar]
- Takatsuka, H., Ohno, R., and Umeda, M. (2009). The Arabidopsis cyclin-dependent kinase-activating kinase CDKF;1 is a major regulator of cell proliferation and cell expansion but is dispensable for CDKA activation. Plant J. (in press). [DOI] [PubMed]
- Torres-Rosell, J., Machin, F., and Aragon, L. (2005). Smc5-Smc6 complex preserves nucleolar integrity in S. cerevisiae. Cell Cycle 4: 868–872. [DOI] [PubMed] [Google Scholar]
- Verkest, A., Manes, C.L., Vercruysse, S., Maes, S., Van Der Schueren, E., Beeckman, T., Genschik, P., Kuiper, M., Inze, D., and De Veylder, L. (2005). The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 17: 1723–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe, K., Boudolf, V., Beemster, G.T., Maes, S., Magyar, Z., Atanassova, A., de Almeida Engler, J., De Groodt, R., Inze, D., and De Veylder, L. (2005). The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr. Biol. 15: 59–63. [DOI] [PubMed] [Google Scholar]
- Xu, X.M., Rose, A., Muthuswamy, S., Jeong, S.Y., Venkatakrishnan, S., Zhao, Q., and Meier, I. (2007). NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell 19: 1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, C.Y., Miura, K., Jin, J.B., Lee, J., Park, H.C., Salt, D.E., Yun, D.J., Bressan, R.A., and Hasegawa, P.M. (2006). SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol. 142: 1548–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.