Abstract
The mechanisms underlying starch granule initiation remain unknown. We have recently reported that mutation of soluble starch synthase IV (SSIV) in Arabidopsis thaliana results in restriction of the number of starch granules to a single, large, particle per plastid, thereby defining an important component of the starch priming machinery. In this work, we provide further evidence for the function of SSIV in the priming process of starch granule formation and show that SSIV is necessary and sufficient to establish the correct number of starch granules observed in wild-type chloroplasts. The role of SSIV in granule seeding can be replaced, in part, by the phylogenetically related SSIII. Indeed, the simultaneous elimination of both proteins prevents Arabidopsis from synthesizing starch, thus demonstrating that other starch synthases cannot support starch synthesis despite remaining enzymatically active. Herein, we describe the substrate specificity and kinetic properties of SSIV and its subchloroplastic localization in specific regions associated with the edges of starch granules. The data presented in this work point to a complex mechanism for starch granule formation and to the different abilities of SSIV and SSIII to support this process in Arabidopsis leaves.
INTRODUCTION
Starch and glycogen are the most widespread glucose-based reserve polymers in living cells. Both polysaccharides share the common feature of being composed of linear chains of (1→4)-linked α-d-glucose residues, which are interlinked by (1→6) α-d-glucosidic linkages, whereas other characteristics of these polymers are completely different. For example, glycogen is a homogeneous water-soluble polymer whose unit particles are <50 nm in diameter (Manners, 1991), whereas starch accumulates in the form of a huge water-insoluble quaternary structure (from 0.1 to >50 μm in diameter depending on botanical or tissue origins) composed of two structurally distinct polysaccharides. The major component of these starch granules, which comprises ∼75% of the starch dry weight, is amylopectin. Approximately 5% of the sugars of amylopectin have an α(1→6) glycosidic bond, which forms a branch point. These branch points are clustered, allowing the glucan side chains to form double-helical structures and conferring semicrystallinity to amylopectin. The minor component of starch is amylose, an almost unbranched polymer of α(1→4)-linked glucose residues (Buleon et al., 1998).
Glycogen (GS) and starch synthases (SS) are involved in the elongation of the linear chains of glycogen and starch, respectively, by catalyzing the transfer of the glucosyl moiety of the activated glucosyl donor (UDP-glucose or ADP-glucose, depending on the organism in question) to the nonreducing ends of a preexisting α(1→4) glucan primer (for a review, see Ball and Morell, 2003). The enzyme glycogenin participates in the initiation of glycogen primer formation in yeast, fungi, and mammalian cells. Glycogenin uses UDP-glucose to synthesize an α(1→4)-linked glucan covalently bound to one of its own Tyr residues (Alonso et al., 1995; Cao et al., 1995). This glycogenin-linked maltooctasaccharide is then used as a primer by GS for further elongation. In bacteria, the glycogen priming process seems to be controlled by the GS itself, which apparently displays a self-glucosylating activity that results in a GS-linked maltooligosaccharide, which then becomes the substrate for further GS-catalyzed glucan elongation (Ugalde et al., 2003). In plants, by contrast, almost nothing is known about the biochemical and physical processes underlying the initiation of starch granule formation. It has been proposed that debranching enzymes (essentially isoamylases) may play a role in controlling the number of starch granules in barley (Hordeum vulgare) endosperm (Burton et al., 2002) and in potato tuber (Solanum tuberosum; Bustos et al., 2004), although it now seems clear that debranching enzymes are not directly involved in the priming of starch synthesis (Delatte et al., 2005; Wattebled et al., 2005, 2008; Streb et al., 2008). More recently, Satoh et al. (2008) have suggested that starch phosphorylase is involved in starch biosynthesis together with a hypothetical, and as yet unidentified, factor X. However, evidence concerning the role of these proteins in the initiation of starch granule formation is still lacking. Finally, the fact that Arabidopsis thaliana mutants lacking SS class IV (SSIV) are unable to synthesize more than one starch granule per chloroplast has been considered as a clear indication that SSIV is probably involved in the initiation of starch granule formation (Roldán et al., 2007).
Five distinct classes of SS are known in all plants: granule-bound SS (GBSSI), which is responsible for the synthesis of amylose, and SS classes I, II, III, and IV (SSI, SSII, SSIII, and SSIV). These classes are broadly conserved in plant species, thus suggesting that their specific functions have been selected during evolution. Mutational analyses corroborate this hypothesis, as elimination of a specific class results in the alteration of one or more starch features, such as amylopectin structure, amylopectin/amylose ratio, shape, morphology, or number of starch granule (Craig et al., 1998; Delvallé et al., 2005; Zhang et al., 2005, 2008; Roldán et al., 2007). Thus, SSs do not substitute fully one for another.
We have previously characterized Arabidopsis mutant plants lacking SSIV (Roldán et al., 2007). The elimination of SSIV causes chloroplasts to accumulate only one large starch granule in most cases. However, the molecular structure of the residual starch (amylose content, amylopectin chain length distribution, etc.) is not modified when compared with wild-type starch. Taken together, our data showed that SSIV is necessary for the synthesis of the regular number of starch granules observed in wild-type chloroplasts and suggested that SSIV may play a role in the initiation process of starch granule synthesis. The loss of SSIV does not, however, prevent starch granule formation in chloroplasts, thereby suggesting a certain degree of redundancy in the function of SSIV.
In this work, we have analyzed different combinations of SSs mutations in an ssIV mutant background to identify the activity responsible for the formation of starch granules found in ssIV mutant plants and show that only the simultaneous elimination of SSIII and SSIV completely abolishes starch synthesis in Arabidopsis. In addition, by the production and analysis of several combinations of SS mutations, we show that SSIII or SSIV is sufficient to allow the synthesis of starch, although SSIV is mandatory to produce the regular number of starch granule found in wild-type plants. The results presented in this work are therefore consistent with the idea that SSIV participates in the initiation of starch granule formation. This function can only be partially substituted by SSIII.
RESULTS
Starch Synthesis in ssI ssIV, ssII ssIV, and ssIII ssIV Double Mutants
Arabidopsis double mutant plants lacking SSIV activity and each one of the other SS activities were selected by crossing the respective single null, knockout mutants described previously: ssIV-1 (Columbia-0 [Col-0] ecotype) and ssIV-2 (Wassilewskija [Ws] ecotype) (Roldán et al., 2007), ssIII-1 (Col-0 ecotype) (Zhang et al., 2005), ssII-3 (Ws ecotype) (Zhang et al., 2008), and ssI-1 (Ws ecotype) (Delvallé et al., 2005), or selected for this work: ssIII-2 (Ws ecotype; line DSC44, FTS 117H05 from the mutant collection Institut National de la Recherche Agronomique [INRA] of Versailles, France). Homozygous double mutants were identified by PCR (see Supplemental Table 1 online for the primers used) and selected for further studies.
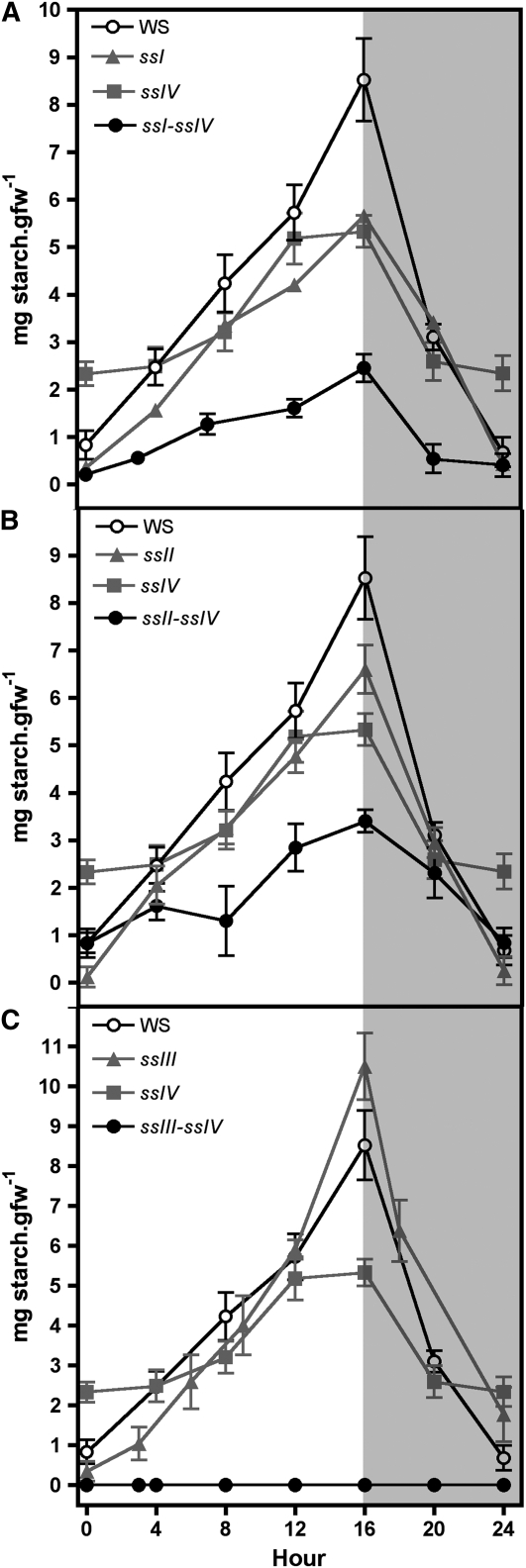
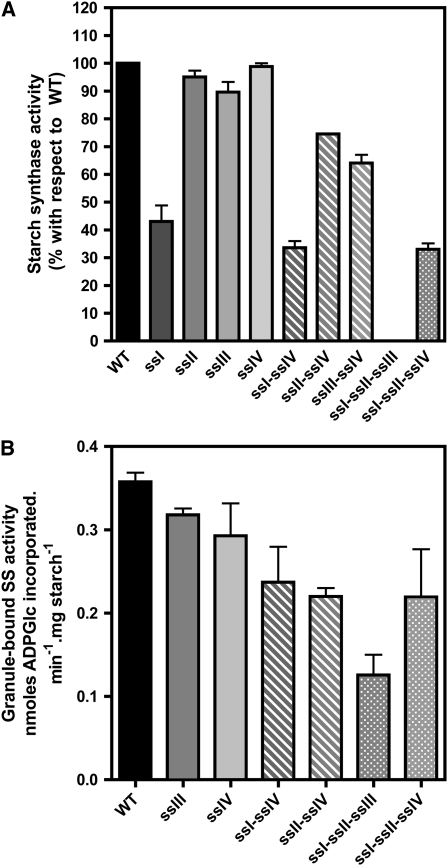
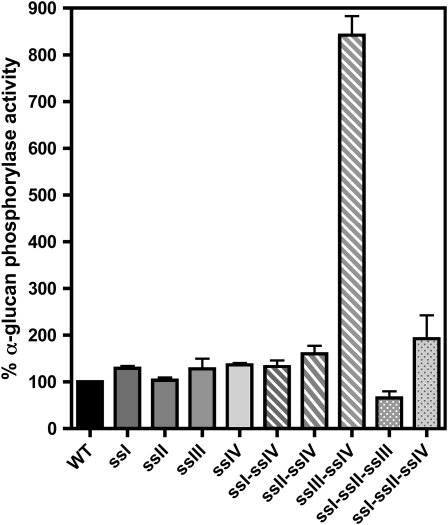
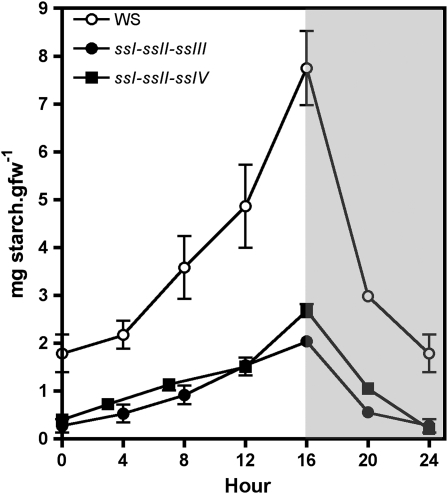
Plants were cultured under 16-h-day/8-h-night photoperiod conditions, and starch levels in leaves were determined in the different double mutants and compared with their respective parental lines and wild-type ecotype plants over a diurnal cycle (Figure 1). The decrease in the levels of starch accumulated in ssI ssIV and ssII ssIV double mutants was found to be equal to the sum of the decreases found in their respective single mutant parental lines (Figures 1A and 1B). A different situation was found for the ssIII ssIV double mutant, where the parental lines accumulated 122% (ssIII) and 62% (ssIV) of starch determined in wild-type plants by the end of the light period, in accordance with previous results described by Zhang et al. (2005) and Roldán et al. (2007). However, the ssIII ssIV double mutant did not accumulate measurable amounts of starch throughout the day/night cycle (Figure 1C). This result was corroborated using two different methods of starch purification as described in Methods. These data suggest that an active class III or class IV SS is mandatory to allow the synthesis of transitory starch in Arabidopsis leaves. This requirement did not lead to the loss of any other activity of the starch biosynthesis pathway. As can be seen in Figure 2A, the decrease of SS activities observed in the ssI, ssII, and ssIII single mutant plants are slightly potentiated in the ssIV mutant background, but SSI and SSII are active in the double ssIII ssIV mutant, accounting for ∼65% of the SS activity of wild-type plants (Figure 2; see Supplemental Figure 1 online). The activity of other starch metabolism enzymes, such as phosphoglucomutase, ADP-glucose pyrophosphorylase, and starch branching, remained unaffected in the different double mutants (Table 1; see Supplemental Figures 1 and 2 online). The SS activity bound to the starch granule decreased slightly in ssI ssIV and ssII ssIV mutants (Figure 2B), whereas an increase in α-glucan phosphorylase activity, which was especially significant in the ssIII ssIV mutants (more than eightfold the activity in wild-type plants), was detected in all the double mutants (Figure 3). An increase in β-amylase activity was also observed in the ssIII ssIV mutant plants (Table 1); this is a common feature of other starchless mutants affected in other steps of the starch biosynthetic pathway (Caspar et al., 1989).
Figure 1.
Starch Accumulation in Leaves of Wild Type, ssI ssIV, ssII ssIV, and ssIII ssIV Double Mutants and Their Respective Parental Lines during a Day/Night Cycle.
Plants were cultured under a 16-h-light/8-h-dark photoperiod for 21 d, and then one leaf was collected from three plants for each line at the indicated time. The starch content in these leaves was determined by enzymatic assay as described in Methods. Each point is the mean ± se from three independent experiments. White and gray areas in the graph correspond to day and night, respectively. ssI ssIV (A), ssII ssIV (B), and ssIII ssIV (C). gfw, gram fresh weight.
Figure 2.
SS Activities of the Different Double and Triple ss Mutants.
Leaves from 3-week-old plants were collected at midday. The material was disrupted using a tissue homogenizer in the presence of 50 mM HEPES, pH 7.6, and proteases inhibitor cocktail. The crude extract was centrifuged for 15 min at 13,000g and 4°C, and the total soluble SS activity (A) and granule-bound SS activity (B) were determined in the supernatant and the purified starch granule, respectively, as described in Methods. Data in (A) are presented as percentage of activity found in wild-type plants. Each point is the mean ± se from three independent experiments.
Table 1.
Activity of Starch-Related Enzymes in the Wild-Type and ssIII ssIV Double Mutant
| Enzyme | Wild Type | ssIII ssIV |
|---|---|---|
| AGPase (biosynthetic assay)a | 44.95 ± 7.3 | 47.88 ± 5.4 |
| Starch branching enzymeb | 168 ± 14.6 | 173 ± 10.7 |
| α-Amylasec | 0.218 ± 0.05 | 0.247 ± 0.03 |
| β-Amylased | 0.90 ± 0.001 | 2.65 ± 0.02 |
Activity defined as nmol of ADP-glucose formed·min−1·mg protein−1.
Activity is measured as the stimulation of the incorporation of carbon-14 from 14C-glucose 1-phosphate into glucan by phosphorylase and is given in nmol·min−1·mg protein−1.
Ceralpha units.
Betamyl units. Values are the mean ± se of three independent determinations.
Figure 3.
α-Glucan Phosphorylase Activity of the Different Double and Triple ss Mutants.
Leaves from 3-week-old plants were collected at midday. The material was disrupted using a tissue homogenizer in the presence of 50 mM HEPES, pH 7.6, and a proteases inhibitor cocktail. The crude extract was centrifuged for 15 min at 13,000g and 4°C, and the total α-glucan phosphorylase activity was determined as described in Methods. Data are presented as percentage of activity found in wild-type plants. Each point is the mean ± se from three independent experiments.
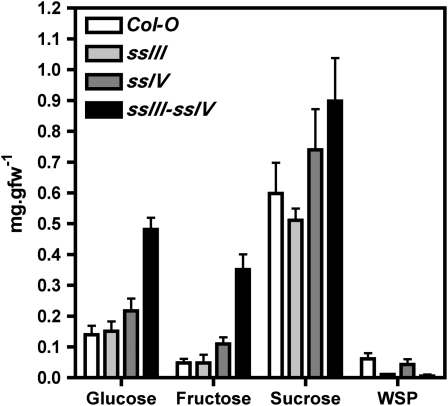
Photosynthetically fixed carbon was not diverted to the synthesis of a soluble glucan in the ssIII ssIV mutant, as no increase in water-soluble polysaccharides (WSPs) was observed in this plant (Figure 4). The starchless phenotype of this mutant plant might be the consequence of an increased starch turnover, which would lead to an increment in the intracellular maltose concentration; however, this idea is not supported by the levels of maltose detected in the ssIII ssIV double mutant, which were not significantly different from those found in wild-type plants (Figure 4). Finally, a clear increase in the intracellular levels of sucrose (1.5 times) and, in particular, glucose (3.5 times) and fructose (7 times) was found (Figure 4), which is characteristic of starch synthesis impairment (Caspar et al., 1985).
Figure 4.
Sugar Content in Leaves of ssIII ssIV Double Mutant, Its Parental Lines, and Wild-Type Plants.
Leaves from plants cultured under a 16-h-light/8-h-dark photoperiod were collected at midday, and their levels of sucrose, glucose, fructose, maltose, and WSPs were determined as described in Methods. Each point is the mean ± se from four independent experiments.
Structural Analysis of Starch in Double Mutants
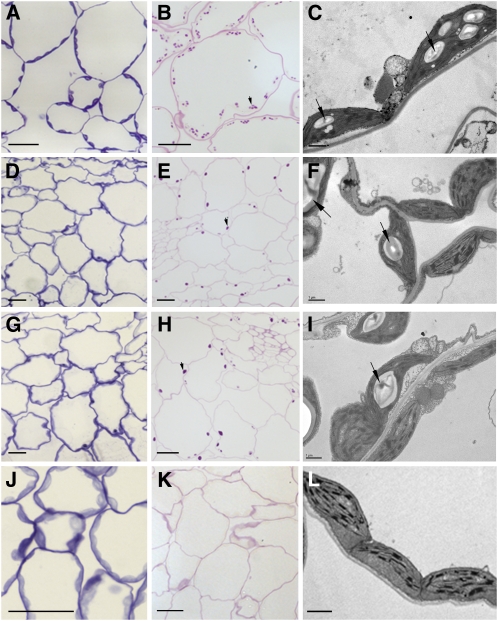
Light and transmission electron microscopy analyses of leaf sections from the different double mutants corroborate the data shown in Figure 1, as no starch granules could be observed in the ssIII ssIV mutant (Figures 5J to 5L). In both ssI ssIV and ssII ssIV mutants, a single large starch granule was found in most of the chloroplasts (Figures 5D to 5I) in a similar way to that described for single ssIV mutants (Roldán et al., 2007). Moreover, and somewhat surprisingly, some chloroplasts seemed to lack starch granules as could be observed by periodic acid-Schiff's (PAS) carbohydrate-specific staining of sections of leaf samples harvested at the end of the photoperiod (Figures 5E and 5H).
Figure 5.
Microscopy of Leaf Sections from the Wild Type (A-B) and ssI ssIV (D-F), ssII ssIV (G-I), and ssIII ssIV (J-L) Mutant Plants.
Light micrographs show tissue stained with toluidine blue (which stains proteins) ([A], [D], [G], and [J]) and with the PAS reaction for carbohydrates ([B], [E], [H], and [K]). Transmission electron microscopy images ([C], [F], [I], and [L]) show the differences in the starch granule sizes of the plants. Samples were processed for microscopy and stained as described in Methods. The wild type ([A] and [B]) and ssI ssIV ([D] to [F]), ssII ssIV ([G] to [I]), and ssIII ssIV ([J] to [L]) mutant plants. Arrows indicate starch granules. Bars = 20 μm in (A), (B), (D), (E), (G), (H), and (J)and 1 μm in (C), (F), (L), and (I).
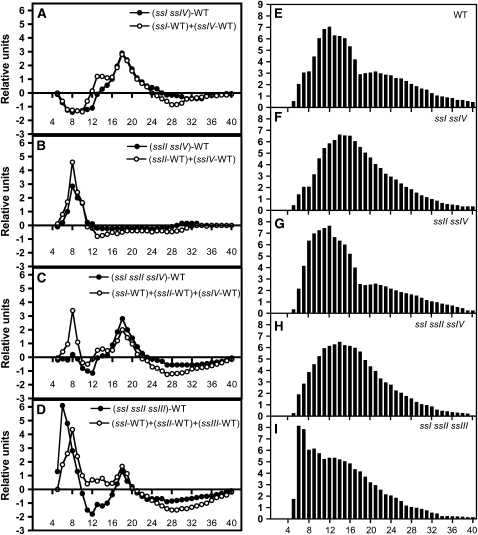
Analyses of chain length (CL) distribution for the amylopectin isolated from ssI ssIV and ssII ssIV mutants showed that the alteration observed in the amylopectin structure was the sum of the alterations caused by the respective single mutations (Figure 6). These modifications were mainly due to the ssI and ssII mutations, as mutations in SSIV gene induce only marginal changes in CL distribution of amylopectin-forming glucans (Roldán et al., 2007). Our data indicate the absence of any synergy between the activity of SSIV and both SSI and SSII as regards the determination of amylopectin structure. This result strengthens the hypothesis that SSIV has no precise role in the determination of amylopectin structure (i.e., in the synthesis of glucan subpopulation of this polymer).
Figure 6.
Amylopectin CL Distribution Profiles for ss Mutants.
Amylopectin was purified on a CL-2B column and subsequently debranched with a mix of isoamylase and pullulanase. The resulting linear glucans were analyzed by HPAEC-PAD.
(A) to (D) The y axes represent mol % difference: for each mutant, the relative proportion for each glucan in the total population, expressed as a percentage of the total number of chains, was calculated and subtracted from the corresponding percentage of that glucan in wild-type amylopectin. The x axes represent the degree of polymerization (dp) of the chains. The closed circles correspond to observed values for each mutant plant, and the open circles correspond to the predicted values resulting from the addition of the subtraction of the amylopectin CL distribution from their respective single mutant parental lines and the wild type.
(E) to (I) The y axes represent the relative proportions for each glucan in the total population expressed as a percentage of the total number of chains. The values are the average of three independent experiments. The sd was less than ±15% of the average values. WT, wild-type plants.
Synthesis of Starch in ssI ssII ssIII and ssI ssII ssIV Triple Mutants
The data obtained from double mutant analyses indicated that starch synthesis requires the presence of an active form of SSIII or SSIV. To investigate whether these enzymes were sufficient to allow the synthesis of starch, we obtained the triple mutants ssI ssII ssIII and ssI ssII ssIV by crossing the double mutants ssI ssII × ssI ssIII and ssI ssII × ssI ssIV, respectively. The soluble SS activity in ssI ssII ssIV mutant plants was 32% ± 3% of the activity determined in wild-type plants, whereas no soluble SS activity could be detected in ssI ssII ssIII mutant plants (Figure 2A) despite using different assay conditions: glycogen, amylopectin, or maltotriose as primer, presence or absence of citrate (an activator for some SS enzymes (Boyer and Preiss, 1979), or reducing agents such as DTT. Immunoblot analysis of ssI ssII ssIII plants showed that the levels of SSIV protein were unaffected, indicating that the absence of measurable SS activity in the ssI ssII ssIII triple mutants was not the consequence of SSIV gene repression (see Supplemental Figure 3 online). Figure 7 shows that both triple mutants were still able to accumulate starch during the day and to mobilize it during the dark period, although the leaf starch content was strongly reduced in comparison to the wild type at the end of the illuminated period (19 and 30% for ssI ssII ssIII and ssI ssII ssIV, respectively).
Figure 7.
Starch Accumulation in Leaves of ssI ssII ssIII and ssI ssII ssIV Triple Mutants and Wild-Type Plants during a Day/Night Cycle.
Plants were cultured under a 16-h-light/8-h-dark photoperiod during 21 d, and then one leaf was collected from three plants for each line at the indicated time. The starch content in the leaves was determined by enzymatic assay as described in Methods. The values are the average of three independent experiments. Vertical bars indicate se. The white and gray areas in the graph correspond to day and night, respectively.
Granule-bound SS activity could be detected in starch granules of both triple mutants although at lower levels than those determined in wild-type starch granules (Figure 2B). An increase in the activity of α-glucan phosphorylase and ADP-glucose pyrophosphorylase was detected in the ssI ssII ssIV mutant (Figure 3 and Supplemental Figure 2 online, respectively), whereas the level of α-glucan phosphorylase activity in ssI ssII ssIII mutant plants was lower than in the wild type. Other starch-metabolizing activities were unaltered in these mutants (see Supplemental Figure 2 online).
Starch Structure in SS Triple Mutants
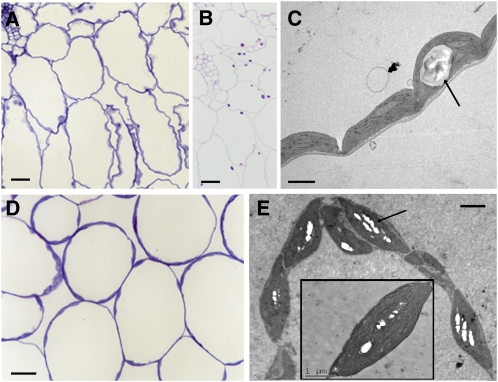
Microscopy analyses of leaves sections from ssI ssII ssIV mutant plants showed the presence of a single, large starch granule per chloroplast similar to that described for ssI ssIV and ssII ssIV mutants (Figures 8A to 8C) and that some chloroplasts lack visible starch granules, as confirmed by serial sections analyses (see Supplemental Figures 4 and 5 online). By contrast, the chloroplasts of triple mutant ssI ssII ssIII leaves displayed the normal number of starch granules observed in wild-type plants, although they were considerably smaller (Figures 8D and 8E).
Figure 8.
Microscopy of Leaf Sections from ssI ssII ssIV and ssI ssII ssIII Mutant Plants.
ssI ssII ssIV ([A] to [C]) and ssI ssII ssIII mutant plants ([D] and [E]). The light micrographs show tissue stained with toluidine blue ([A] and [D]) and with the PAS reaction for carbohydrates (B). The transmission electron microscopy images ([C] and [E]) show the differences in the starch granule sizes of the chloroplasts (indicated by arrows). Bars = 20 μm in (A), (B), and (D) and 2 μm in (C) and (E).
Starch from the ssI ssII ssIV and ssI ssII ssIII lines contained a high proportion of amylose (42 and 41%, respectively), and the CL distribution analysis of the amylopectin-forming glucans showed a strong enrichment of glucans with a degree of polymerization (number of glucose residues [dp]) of 5 to 9, with a maximum at dp 6, for the ssI ssII ssIII mutant (Figures 6D and 6I). A similar strong increase has been described for ssII and in ssII-ssIII Arabidopsis mutants (Zhang et al., 2008) and seems to be a general characteristic of the lack of class II SS (Morell et al., 2003; Zhang et al., 2004), which is amplified when SSI or SSIII, or both, are also absent (although the increase at dp 6 is less pronounced in the latter case than in the double mutant). Conversely an overall depletion of the proportion of dp 11 to 35 glucans was observed in ssI ssII ssIII, except for dp 17 to 20 glucans, which showed a slight enrichment in this triple mutant (this seems to be a general feature of the lack of SSI). This profile looks similar to that calculated from the profile of the corresponding single mutants, although it is not identical (Figure 6D). This may point toward some overlapping functions between isoforms for the synthesis of amylopectin.
In the case of the ssI ssII ssIV triple mutants, the abundance of dp 10 to 13 and dp 16 to 21 amylopectin-forming glucans was lower and higher, respectively, than for wild-type amylopectin. Glucans longer than dp 25 were less abundant in the triple mutant (Figures 6C and 6H). Again, this profile is closely related to that calculated by the addition of the profiles from the respective single mutants with the notable exception of the very short glucans (dp 5 to 9), whose abundance was unmodified in the triple mutant even though they were predicted to be more abundant than in the wild type (Figure 6C). Indeed, the ssI ssII ssIV mutant displays the same profile as the ssI ssII double mutant (N. Szydlowski, unpublished results), again suggesting that SSIV is of low importance for the building of the amylopectin structure. The unmodified number of dp 5 to 9 glucans in both ssI ssII and ssI ssII ssIV amylopectins is likely to be a consequence of the opposite effects observed in both ssI and ssII mutant amylopectins (Delvallé et al., 2005; Zhang et al., 2008), which cancel each other out. However, these effects do not cancel each other in the absence of SSIII, thereby leading to a strong increase in the number of these very short glucans in the ssI ssII ssIII mutant, as is also the case in the ssII ssIII double mutant (Zhang et al., 2008). This behavior may be related to the collapse of a large protein complex (Hennen-Bierwagen et al., 2008), in which SSIII may be an essential component.
Enzymatic Characterization of SSIV
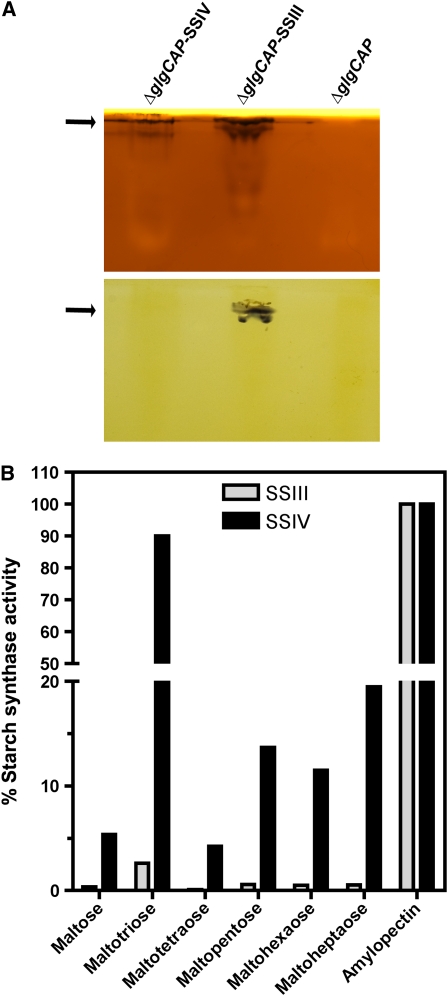
The data shown above indicate that SSIV and SSIII have unique features that differentiate them from other SSs. To identify those features, we proceeded to characterize the enzymatic parameters of expressed and purified mature (without the chloroplast transit peptide) His6-tagged recombinant SSIII and SSIV proteins. To avoid interference by contaminating endogenous bacterial GS, we employed Escherichia coli ΔglgCAP deletion mutants, which lack a large part of the glycogen biosynthetic machinery (Morán-Zorzano et al., 2007). Recombinant SSIII and SSIV displayed activities of 58 ± 2.3 and 31 ± 3.4 nmol·min−1·mg protein−1, respectively, using glycogen as primer and ADP-glucose as glucosyl donor. As shown in Table 2, the His-tagged recombinant SSIV protein could use both glycogen and amylopectin as acceptors of glucose residues, with Km values for ADP-glucose as glucosyl donor of 0.47 mM (amylopectin as primer) or 0.96 mM (glycogen as primer). These values fall within the range of values determined for other SSs (http://www.brenda-enzyme.info/). As shown in Table 2, SSIV is specific for ADP-glucose as glucosyl donor and, unlike GSs from mammalians or fungi, shows no activity when using UDP-glucose as substrate. The glucan-elongating activity of SSIV could also be visualized by native PAGE assay. As shown in Figure 9A, E. coli–expressed SSIV produced an activity band at the same position as that originated by SSIII, although no band corresponding to SSIV activity could be detected in crude leaf extracts using this zymogram assay (see Supplemental Figure 1 online). It is noteworthy that SSIII was able to synthesize linear glucans using ADP-glucose as substrate in the absence of any added primer, whereas SSIV could not, under the assay conditions tested (Figure 9A). Figure 9B shows that SSIV had high activity when using maltotriose as primer, displaying >90% of the activity observed with amylopectin with this maltooligosaccharide (MOS). SSIV could use other MOSs as primers, but with a much lower efficiency (∼15 and 20% of amylopectin-dependent activity for dp5 and dp7, respectively). SSIII also displayed a higher activity when maltotriose was employed as the MOS in the assay, but to a much lower extent than that observed for SSIV (∼3% of the activity detected using amylopectin as primer). The activity of SSIII using other MOSs as substrate was almost negligible (Figure 9B).
Table 2.
Values of Activity and Affinity for ADP-Glucose of SSIV Expressed in E. coli
| Substrate | Activity | Km for ADP-Glucose (mM) |
|---|---|---|
| Rabbit liver glycogen | 31.4 ± 3.4 | 0.96 |
| Potato amylopectin | 25.0 ± 7.5 | 0.47 |
| Rabbit liver glycogen + UDP-glucose | n.d. | n.a. |
Activity is defined as the nmoles of glucose incorporated into α-glucan·min−1·mg protein−1. Values are the mean ± se of three independent determinations. n.d., not detected; n.a., not applicable.
Figure 9.
Activities of SSIII and SSIV with Different Glucans as Primers.
Purified fractions of E. coli–expressed SSIII and SSIV proteins were used to determine the activity of these proteins with different substrates and assay conditions.
(A) Detection of SSIII and SSIV activities in native PAGE containing glycogen (top gel) or without glycogen (bottom gel). After electrophoresis, gels were incubated overnight in a medium containing 2 mM ADP-glucose and then stained with iodine solution to reveal dark bands (indicated by arrows) where SS had elongated linear chains.
(B) In vitro assays of SSIII and SSIV with different maltooligosaccharide and potato amylopectin as substrates. Values are expressed as a percentage of the activity determined using amylopectin as primer and are the mean ± se of three independent experiments.
We could not detect any activity of SSIV protein in plants (mutant ssI ssII ssIII), even though this protein displays SS activity in vitro and can lead the synthesis of glycogen in E. coli, as illustrated by the transformation of ΔglgAP E. coli strain (see Supplemental Figure 6 online). These cells have normal ADP-glucose pyrophosphorylase activity but lack both GlgA and glycogen phosphorylase activities (Morán-Zorzano et al., 2007). As expected, ΔglgAP cells displayed a glycogenless phenotype (see Supplemental Figure 6 online), which was complemented by glgA expression. SSIV expression partially complemented the glycogenless phenotype of ΔglgAP cells, as confirmed by both qualitative and quantitative glycogen content analyses; SSIII expression also complemented the phenotype but to a lower extent (see Supplemental Figure 6 online). SSIV-expressing ΔglgAP ΔgalU cells (which lack the main producer of UDP-glucose in E. coli) accumulated as much glycogen as SSIV-expressing ΔglgAP cells, thus corroborating the observation shown in Table 2 that SSIV uses ADP-glucose specifically as glucosyl donor.
Complementation of the glycogenless phenotype of ΔglgAP cells by SSIV or SSIII raises the question whether these proteins display self-glucosylating activity in a similar way to that described for bacterial GS (Ugalde et al., 2003). Purified fractions of E. coli expressed SSIV and SSIII proteins were incubated with 50 μM ADP-[U-14C]glucose (9.81 GBq/mmol) following the procedure described by de Paula et al. (2005) (see Methods). The incorporation of radioactive glucose into the SSs proteins was analyzed by SDS-PAGE and autoradiography. No radioactive band corresponding to the SSIII or SSIV proteins was detected, thereby indicating that these proteins do not display ADP-glucose dependent self-glucosylating activity under these experimental conditions.
SSIV Localizes in Specific Areas Associated with the Starch Granule
The data shown above indicate that SSIV is required to determine the correct number of starch granules found in chloroplasts of Arabidopsis leaves. To establish whether this feature correlates with a specific localization of SSIV in the chloroplast, we performed confocal fluorescence microscopy analyses of Arabidopsis plants expressing full-length SSIV protein (including its predicted chloroplast transit peptide) fused with green fluorescent protein (GFP). These plants constitutively express the translationally fused SSIV-GFP encoding gene under the control of the cauliflower mosaic virus 35S promoter (see Supplemental Figure 7 online). Plants constitutively expressing potato adenosine diphosphate sugar pyrophosphatase fused with GFP (ASPP-GFP) (Muñoz et al., 2008) and plants expressing Arabidopsis GBSSI (full-length protein including its predicted chloroplast transit peptide) fused with GFP (GBSSI-GFP) (see Supplemental Figure 7 online) were used as controls for stromal and starch granule localizations, respectively.
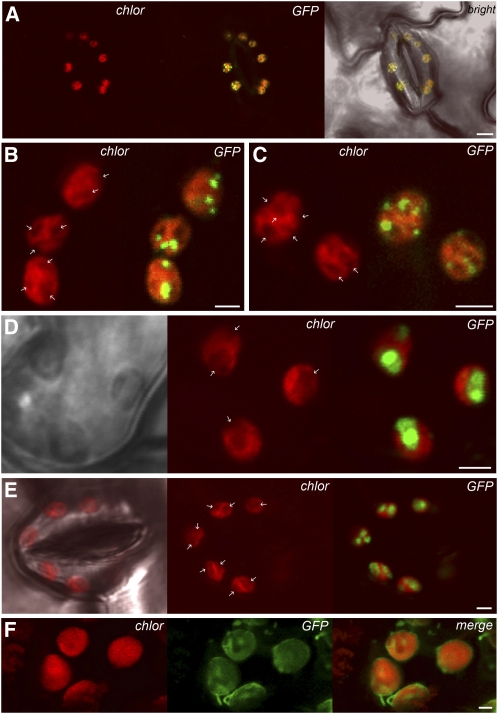
As shown in Figure 10F, the stromal marker ASPP-GFP was present among the grana in the central part of the chloroplast, as well as in the grana-free peripheral part of the chloroplast. In addition, GFP fluorescence labeled long stroma-filled tubular extensions corresponding to plastid stromules. Analyses of SSIV-GFP–expressing plants revealed that SSIV-GFP has a plastidial localization (Figure 10A). However, in contrast with ASPP-GFP plants, the fluorescence in SSIV-GFP–expressing cells was not uniformly distributed within the stroma but was mainly located in specific regions at the boundary of oval structures that were similar in size, number, and distribution to starch granules (Figures 10B and 10C; see Supplemental Figure 8 online). Further analyses of GBSSI-GFP–expressing plants showing uniform GFP fluorescence of these structures confirmed that they were indeed starch granules (Figures 10D and 10E; see Supplemental Figures 8 and 9 online). We must emphasize, however, that in contrast with other starch metabolizing enzymes, such as isoamylases, which uniformly localize to the periphery of the starch granule (Delatte et al., 2006), the GFP fluorescence of SSIV-GFP–expressing plants seemed to be predominantly located in specific areas associated with the edges of the starch granules (Figures 10B and 10C; see Supplemental Figure 8 online). This association seems to be weak, as the SSIV protein was found in the soluble fraction of leaves crude extracts, and no signal was detected by immunoblot analyses of purified starch granules using anti-SSIV under the experimental conditions followed to obtain crude extracts.
Figure 10.
Localization of SSIV-GFP in Arabidopsis Chloroplasts.
(A) Localization of SSIV-GFP in leaf stomata guard cells. Confocal laser scanning microscopy revealed a dot-like pattern of SSIV-GFP distribution in chloroplasts. Chloroplasts are visualized by chlorophyll autofluorescence.
(B)to (E)GFP-positive dots were associated with oval structures ([B]and [C]; indicated by arrows), detected in chloroplast autofluorescence images (chlor), which were confirmed to be starch granules using GBSSI-GFP–expressing plants ([D]and [E]) (see Supplemental Figures 8 and 9 online).
(F) Stromal localization of the ASPP-GFP construct.
Bars = 5 μm in (A) and 2 μm in (B) to (E).
DISCUSSION
SSIII or SSIV Is Necessary for the Synthesis of Starch in Arabidopsis Leaves
In this study, we have found that SSIII is responsible for the synthesis of the one or two starch granules per chloroplast observed in single ssIV mutant plants (Roldán et al., 2007). Double mutant ssIII ssIV plants do not accumulate starch or any other soluble or insoluble α-linked glucans, thereby indicating that the synthesis of such polymers requires the participation of an active form of either a class III or class IV SS. This redundancy of the functions of SSIV and SSIII does not occur with other SSs. The phenotypes of both ssI ssIV and ssII ssIV double mutants have been found to be the addition of the phenotypes of their respective single mutant lines, as illustrated by the accumulation of starch along the diurnal cycle (Figure 1) and the length distribution of the amylopectin-forming glucans (Figure 6). However, both double mutants display a more severe phenotype with respect to the number of starch granules per chloroplasts than that observed in the parental lines. Thus, whereas single ssI and ssII mutants do not show any alteration in the number of starch granules per chloroplast (Delvallé et al., 2005; Zhang et al., 2008) and ssIV contains only one (sometimes two) granule per chloroplast (Roldán et al., 2007), the double mutants ssI ssIV and ssII ssIV have two types of chloroplasts, one of which contains a huge single starch granule, whereas the other has no starch granule or similar structure visible by electron microscopy (Figure 5). No correlation between the distribution of chloroplasts with or without starch and the different types of leaf tissues is observed, thus ruling out the possibility that the presence of starch is related to the metabolic characteristics of the cell. There is an increasing body of evidence illustrating the interaction between proteins of the starch biosynthetic pathway, including SSI, SSII, and SSIII (Hennen-Bierwagen et al., 2008; Tetlow et al., 2008). Therefore, a possible explanation for those results is that the priming of starch granule synthesis by SSIII in the absence of SSIV is enhanced by the presence of the other soluble SSs, SSI and SSII in a large protein complex. The subsequent elimination of one or both of these (in addition to SSIV) would decrease the starch granule initiating activity of SSIII and lead to some granule-free chloroplasts.
SSIII and SSIV Are Sufficient to Promote the Synthesis of Starch
The analyses of ssI-ssII ssIII and ssI ssII-ssIV triple mutant plants indicate that either SSIII or SSIV is sufficient to promote the synthesis of starch granules in the absence of the other soluble SSs. We must emphasize, however, that GBSSI is still present in these mutants (Figure 2B) and may well be responsible for the high amylose content of the starch found in these plants; indeed, the low residual levels of soluble SS activity found in these mutants may lead to an increase of the intraplastidial concentration of ADP-glucose and thereby an increase in the activity of GBSSI, which shows a lower affinity for ADP-glucose than the soluble SSs. However, the redundancy in the function of these two proteins is not complete, and the presence of an active form of SSIV appears to be mandatory for the synthesis of the normal number of starch granules found in the wild type or other ss mutants. As in the case of ssI ssIV and ssII ssIV, some starch-free chloroplasts could be found in ssI ssII ssIV plants (Figure 7; see Supplemental Figures 4 and 5 online), which supports the idea that SSIII alone can promote the initiation of starch granule synthesis, although, as already discussed above, this capability is greatly reduced when other SSs are absent. The presence of several small granules in the ssI ssII ssIII plants shows that the granule initiation mechanism is not altered in this mutant even though the granule enlargement capacities are strongly impaired in the absence of SSI, SSII, and SSIII.
Distinct Roles of SSIII and SSIV in the Initiation of the Starch Granule
An understanding of the mechanisms underlying the initiation of the starch granule has remained elusive to date. It has been suggested recently that α-glucan phosphorylase could be involved in this process in rice (Oryza sativa) endosperm (Satoh et al., 2008). We have previously shown the induction of the two genes encoding for both cytosolic and chloroplastic α-glucan phosphorylases in an ssIV single mutant (Roldán et al., 2007), probably as a result of the nutritional starvation experienced by the plant (Zeeman et al., 2004). Here, we have shown an increment of α-glucan phosphorylase activity in all mutants lacking the SSIV protein. This increment is especially dramatic in the case of ssIII ssIV double mutant, which displays up to a ninefold higher α-glucan phosphorylase activity than that detected in wild-type plants. In addition, the growth rate of this mutant is severely arrested, thus indicating an intense nutritional starvation. Despite the elevated levels of phosphorylase activity in the ssIII ssIV double mutant, however, this plant cannot synthesize starch, thus suggesting distinct roles for α-glucan phosphorylases in photosynthetic and long-term storage organs.
The genetic analyses of the ss mutant plants presented in this work indicate that SSIII and SSIV are directly involved in starch granule initiation, although these enzymes play different roles in this mechanism. Phylogenetic analyses have provided evidence that SSIII and SSIV are closely related, whereas SSI, SSII, and GBSSI clustered together (Ball and Morell, 2003; Patron and Keeling, 2005). These data could explain the partially overlapping function of SSIII and SSIV described in our work. Nevertheless, both proteins display distinct features, which suggest that they play unique roles in the synthesis of the starch granule. Thus, the N-terminal half of SSIII contains three repeated starch binding domains that modulate the affinity of the protein for amylopectin (Valdez et al., 2008). These domains are relevant for the activity of SSIII, as their elimination increases the affinity of the enzyme for ADP-glucose 18-fold (Valdez et al., 2008). These changes could explain why a truncated version of SSIII containing just the C-terminal part of the protein, homologous to the GS, can fully complement the glycogenless phenotype of a glgA− Agrobacterium tumefaciens strain (Busi et al., 2008), whereas the glycogenless E. coli strain transformed with the full-length mature SSIII protein only accumulated ∼5% of the glycogen detected in the glycogen-accumulating parental line (see Supplemental Figure 6 online). These starch binding domains are absent in the N-terminal part of SSIV, which contains two long coiled-coil domains, extending from amino acids 194 to 407 and from amino acids 438 to 465 in the Arabidopsis amino acids sequence of SSIV (Rose et al., 2004). Long coiled-coil structural motifs, such as those located in SSIV, are found in a number of functionally distinct proteins and are often involved in attaching functional protein complexes to larger cellular structures and, in general, as anchors for the regulation of protein positioning in the cell (Rose et al., 2004). These motifs are highly conserved in SSIV proteins from other sources, such as Vigna unguiculata (accession number AJ006752), O. sativa (accession numbers AY373257 and AY373258), or Triticum aestivum (accession number AY044844), thereby suggesting that they play a role in the function of this class of SS.
Analysis of the activity levels and substrate specificity of SSIII and SSIV also indicates that both enzymes may play different roles in vivo. Thus, SSIV displays a greater affinity for ADP-glucose (Km = 0.96 mM; Table 2) than SSIII (Km = 4.28 mM) (Valdez et al., 2008) and shows a specifically high activity and affinity for maltotriose that is not observed for other SSs (Figure 9) and whose physiological meaning is still not clear. Figure 9 illustrates another feature that differentiates both enzymes, namely that SSIII can synthesize linear glucans (long enough to be stained by iodine) in the absence of a primer in a gel activity assay, whereas E. coli–expressed SSIV seems to lack this capability. Analysis of the SS activity levels in the different mutants also indicates a difference between these two proteins. Thus, the decrease of SS activity in the ssIII single mutant and the activity detected in the ssI ssII ssIV triple mutant indicate that SSIII accounts for 15 to 30% of the total soluble SS activity in Arabidopsis leaves. By contrast, we could not detect any SS activity in the triple ssI ssII ssIII mutant or a decrease in SS activity in the ssIV mutant (Figure 2). This could indicate either that the in vivo activity of SSIV is below the detection limit of the enzymatic assay used or that SSIV is inactive in the plant because of posttranslational modifications or physical interaction with other proteins yet to be determined. Further studies will be necessary to clarify this point.
The different functions of SSIII and SSIV could also be the consequence of distinct patterns of gene-expression at both temporal and spatial levels, rather than of different specificities of these enzymes. The expression of SSIII and SSIV genes seems not to be controlled by the circadian clock in Arabidopsis leaves (Smith et al., 2004) as opposed to the GBSSI gene (Tenorio et al., 2003). Analogously, the expression of SSIV-1 and SSIV-2 genes has been shown to be relatively constant during the rice grain filling (Hirose and Terao, 2004). The activity of SSIII and SSIV could, however, be regulated during the diurnal cycle, in the case of transitory starch, or during the starch-accumulation process in long-term storage organs such as cereals endosperm or tubers, and could therefore account for some of the different functions of SSIII and SSIV proteins.
SSIII can synthesize a glucan long enough to be stained by iodine in the absence of primer and using ADP-glucose as substrate (Figure 9A). However, if the early synthesis of small α-glucans were sufficient to prime starch synthesis by itself, then the activity of SSIII alone would be enough to prime the synthesis of the correct number of starch granules in the plastid despite the absence of SSIV. The role of SSIV in the process of starch initiation is therefore probably not limited to the synthesis of the primer molecule itself. It also appears to be required for the seeding of the starch granule (i.e., the formation of a center of nucleation required to allow the correct three-dimensional expansion of the granule). This idea is supported by the subchloroplastic localization of SSIV, which is not uniformly distributed in the stroma or associated throughout the surface of the granule, as would be expected for an enzyme involved in the elongation of the amylopectin-forming glucans. By contrast, SSIV localization is restricted to some specific areas associated with the edges of starch granules (Figures 10A to 10C; see Supplemental Figure 8 online). The architecture of SSIV, probably the coiled-coils domains located in its long and unique N-terminal extension, could be required to adopt (alone or in combination with other as yet unidentified proteins) a quaternary structure that would account for the spatial localization of SSIV observed in Figure 10. This complex would allow the formation of a specific glucan, which would serve as this nucleation center. Glycogen-like structures are known to facilitate the priming of insoluble starch-like molecules (Putaux et al., 2006). The presence of SSIV may facilitate the synthesis and/or the preservation of such glycogen-like molecule and, thus, the seeding of a starch granule. This may explain why starch granules in the ssIV mutants display anomalous organization in their hilum as reported by Roldán et al. (2007).
SSIII may be unable to allow the formation of such a nucleation center, or it could form at such a low rate (at random frequency) that, on average, only one starch granule can be synthesized when SSIV is missing. The initiating capacities of SSIII would be further reduced in the absence of other classes of soluble SSs, thus leading to some starch-free chloroplasts in the ssI ssIV, ssII ssIV, and ssI ssII ssIV mutants. Indeed, the initial conditions that would lead to the formation of a starch granule are probably not very common, even in the wild-type genetic background, since only a few starch granules are generally found in plastids. In this respect, it is worth noting that a reduction in isoamylase activity leads to the accumulation of large numbers of tiny starch granules in transgenic potato tubers and mutant barley seeds (Burton et al., 2002; Bustos et al., 2004). Therefore, we cannot rule out the possibility that the number of nucleation centers that would originate a new starch granule results from balanced counteracting activities of formation (SSIV, SSIII, and possibly other yet unidentified elements) and elimination (such as isoamylases). Further investigations will be required to decipher the precise mechanism of starch granule seeding and to definitely determine which factors are involved in this process.
METHODS
Plant Material and Growth Conditions
Mutant lines of Arabidopsis thaliana were obtained from the T-DNA mutant collections generated at INRA, Versailles (Bechtold et al., 1993; Bouchez et al., 1993), Syngenta (Sessions et al., 2002), the Salk Institute Genomic Analysis Laboratory, and the GABI-KAT mutant Collection (Rosso et al., 2003). Information on all mutants used is provided in Supplemental Table 1 online. Construction of SSIV-GFP and GBSSI-GFP is described in Supplemental Figure 7 online. Transgenic Arabidopsis plant expressing SSIV-GFP and GBSSI-GFP fused proteins were obtained by Agrobacterium tumefaciens–mediated transformation of Col-0 wild-type plants using the floral dip method described by Clough and Bent (1998). Wild-type ecotypes (Ws and Col-0) and mutant lines were grown in growth cabinets under a 16-h-light/8-h-dark photoregime at 23°C (day)/20°C (night), 70% humidity, and a light intensity at the plant levels of 120 μE m−2 s−1 supplied by white fluorescent lamps. Seeds were sown in soil and irrigated with 0.5× Murashige and Skoog medium (Murashige and Skoog, 1962).
Bacterial Strains, Plasmids, and Culture Media
The bacterial strains and plasmids used in this work and their characteristics are summarized in Supplemental Table 2 online. Escherichia coli BL21(DE3) cells were used to producte glgCAP, glgAP, and double glgAP galU deletion mutants (ΔglgCAP, ΔglgAP, and ΔglgAP ΔgalU, respectively), as well as for glgA, SSIII, and SSIV expression experiments. Cells simultaneously expressing SSIII and SSIV were transformed with the two compatible pACYCDuet-SS3 and pET45-SS4 plasmids. DNA manipulations were conducted following the procedures indicated by Ausubel et al. (2001). For glycogen measurement experiments, cells were grown with rapid gyratory shaking at 37°C in Kornberg liquid medium (1.1% K2HPO4, 0.85% KH2PO4, and 0.6% yeast extract from Duchefa), liquid media supplemented with 50 mM glucose, and the appropriate selection antibiotic. Cells from cultures entering the stationary phase were centrifuged at 4400g for 15 min, rinsed with fresh Kornberg medium, resuspended in 40 mM Tris/HCl, pH 7.5, and disrupted by sonication. Solid Kornberg medium was prepared by addition of 1.5% bacteriological agar to the liquid medium.
glgCAP, glgAP, and galU Disruptions
ΔglgCAP, ΔglgAP, and ΔglgAP ΔgalU cells were produced essentially as described by Datsenko and Wanner (2000). A selectable antibiotic resistance gene was generated by PCR from a freshly isolated colony of E. coli MC4100, using 80-nucleotide-long primer pairs that included 60-nucleotide homology extensions for the targeted locus and 20-nucleotide priming sequences for the resistance gene (see Supplemental Table 3 online). Mutants were confirmed by both DNA gel blot hybridization and PCR using specific primers.
Extraction and Determination of Starch and Glycogen
For analysis of the structure and composition of starch, Arabidopsis leaves were harvested at the end of the light period. Approximately 10 g of fresh material was homogenized using a Tissue Tearor (Biospec Products) in 30 mL of the following buffer: 100 mM MOPS, pH 7.2, 5 mM EDTA, and 10% (v/v) ethanediol. The homogenate was filtered through two layers of Miracloth and centrifuged for 15 min at 4°C and 4000 g. The pellet was resuspended in 30 mL Percoll 90% (v/v) and centrifuged for 40 min at 4°C and 10,000g. The starch pellet was washed six times with sterile distilled water (10 min at 4°C and 10,000g between each wash). Starch was finally stored at 4°C in 20% ethanol. This method was scaled down for the analysis of starch content in leaves along the diurnal cycle, and three leaves (∼300 mg) from three different plants were used at each point. The material was frozen with liquid nitrogen, homogenized with a mortar and pestle, and resuspended in 1 mL of 50 mM HEPES, pH 7.6, 1% Triton X-100 buffer. The homogenate was filtered through one layer of 100 μm Nylon mesh and centrifuged for 15 min at 4°C and 4000g. The pellet was resuspended in 1 mL Percoll 90% (v/v) and centrifuged for 40 min at 4°C and 10,000g. The pellet obtained was washed three times with 1 mL 80% ethanol and finally air-dried. The pellet was resuspended in 1 mL of 0.2 n KOH, boiled for 30 min, and centrifuged for 10 min at 14,000g at 4°C. Finally, the supernatant was adjusted to pH 5.5 with 1 n acetic acid.
An alternative starch isolation method, which omits the Nylon mesh filtering and the Percoll centrifugation, was employed to confirm the levels of starch determined in the different mutants. This method is based in the method described by Lin et al. (1988) with some modifications: leaves (300 to 500 mg) were cut into strips and extracted three times with 10 mL 80% ethanol in a boiling water bath for 5 min. The leaves were then resuspended in 2 mL of 0.2 n KOH, ground with a mortar and pestle, and heated at 100°C for 30 min. After cooling, the mixture was centrifuged at 17,000g for 10 min, and the supernatant was adjusted to pH 5.5 with 1 n acetic acid. In both cases, an aliquot of 200 μL of the pH 5.5 starch solution was used to determine the starch amount using the enzymatic method described by Lin et al. (1988).
For bacterial glycogen measurement experiments, cells were grown with rapid gyratory shaking at 37°C in Kornberg liquid medium supplemented with 50 mM glucose and the appropriate selection antibiotic. Cells from cultures entering the stationary phase were centrifuged at 4400g for 15 min, rinsed with fresh Kornberg medium, resuspended in 40 mM Tris/HCl, pH 7.5, and disrupted by sonication. Glycogen content was then determined as described by Morán-Zorzano et al. (2007) using an amyloglucosidase/hexokinase/glucose-6P dehydrogenase-based test kit from Sigma-Aldrich.
Extraction and Determination of Sugars
Leaf tissue (∼0.5 g) was harvested and frozen in liquid N2. The material was powdered and extracted with 1.5 mL of 0.7 M perchloric acid as described by Critchley et al. (2001). The fructose, glucose, and sucrose content in the buffered extract was determined by enzymatic analysis as described by Stitt et al. (1989). Maltose levels were determined by enzymatic analysis as described by Shirokane et al. (2000).
Determination of WSP Contents
Water-soluble glucan contents in leaves were determined as described by Zeeman et al. (1998).
Separation of Starch Polysaccharides by Size-Exclusion Chromatography
Starch (1.5 to 2.0 mg) was dissolved in 200 μL of 100% DMSO and boiled for 10 min in a water bath. After the addition of 800 μL of 100% ethanol, the glucans were precipitated overnight at −20°C. After centrifugation for 10 min at 5000g and 4°C, the glucan pellet was dissolved in 500 μL of 10 mM NaOH and subsequently applied to a Sepharose CL-2B column (0.5 cm i.d. × 65 cm), which was equilibrated and eluted with 10 mM NaOH. Fractions of 300 μL were collected at a rate of one fraction per 1.5 min. The glucans in these fractions were detected by their reaction with iodine, and the levels of amylopectin and amylose were determined by amyloglucosidase assays (after pooling of the corresponding fractions).
Amylopectin CL Distribution
Amylopectin CL distribution was established by HPAEC-PAD (Dionex) equipped with a CarboPac PA200 column (4 mm i.d. x 250 mm length) after complete enzymatic debranching as follows: after purification on a Sepharose CL-2B column, 500 μg of amylopectin was dialyzed against distilled water and subsequently lyophilized. The amylopectin pellet was solubilized in 500 μL of 55 mM sodium acetate, pH 3.5, and incubated overnight at 42°C with 20 units of isoamylase (Megazyme International). Prior to injection, salts were removed by running the sample through an Extract Clean Carbograph column (Alltech).
Zymograms Techniques
A complete description of these techniques has been described by Delvallé et al. (2005).
In Vitro Assays of Starch Synthesis Enzymes
ADP-glucose pyrophosphorylase was assayed in the synthesis direction according to the procedure described by Crevillén et al. (2003). Soluble SS activity was assayed using amylopectin, glycogen, or different maltooligosaccharides as primers. Samples were incubated at 30°C for 30 min in 100 μL of the following buffer: 100 mM Tricine, pH 8.0; 25 mM potassium acetate; 5 mM EDTA; 0.5 M sodium citrate; 0.5 mg/mL BSA; 1 mM ADP-[U-14C]glucose (3.7 GBq/mol). MOS (10 mg/mL; DP2 to DP7) glycogen or potato amylopectin were added to the buffer depending on the tested substrate. The reaction was stopped by boiling for 10 min, and the glucans were elongated by incubation overnight at 30°C with 7.5 units of phosphorylase “a” from rabbit muscle (Sigma-Aldrich) in the presence of 50 mM of Glc-1-P (final concentration). The reaction was stopped by addition of 3.5 mL of 75% methanol, 1% KCl solution, and the samples were further treated as described previously (Delvallé et al., 2005) before counting. Elongation with phosphorylase was omitted when amylopectin was used as primer. Granule-bound SS activity was determined as described by Tenorio et al. (2003). α-Glucan phosphorylase and branching enzymes activities were determined according to the procedure described by Zeeman et al. (1998).
Production of cDNAs Coding for Mature SSIII and SIV
Total RNA was isolated as described by Prescott and Martin (1986). First-strand cDNA was synthesized from 10 μg of total RNA using Moloney murine leukemia virus reverse transcriptase and oligo(dT)12–18 primer, according to the manufacturer's instructions (GE Healthcare). The reaction mixture was incubated at 37°C for 2 h and stopped by adding 1 mL of nuclease-free MilliQwater. cDNA fragments encoding for the mature (without the chloroplast transient peptides) SSIII and SSIV proteins were obtained by PCR amplification using high-fidelity iProof Polymerase (Bio-Rad), first-strand cDNA previously synthesized as template, and specific oligonucleotides for both genes: SS3F, 5′-ctgcagGGAAGTGCTCAGA AAAGAACTCAGA-3′ (PstI site in italics), and SS3R, 5′-ctcgagTTACTTGCGTGCAGAGTGATA-3′ (XhoI site in italics), for the SSIII gene and SS4F, 5′-ggatccgTGTAAGATGCGACAACAACGT-3′ (BamHI site in italics), and SS4R, 5′-ctcgagTCACGTGCGATTAGGAACAGC-3′ (XhoI site in italics) for the SSIV gene. Amplified fragments were cloned in the pGEM-T Easy Vector (Promega) and sequenced.
Heterologous Expression and Purification of SSIII and SSIV
cDNAs coding for the complete mature SSIII and SSIV proteins were inserted as a translational fusion into the pET45b prokaryote expression vector (Novagen) at the BamHI-XhoI (SSIV) and PstI-XhoI (SSIII) sites. Recombinant clones were transformed into ΔglgCAP BL21 (DE3) E. coli cells lacking the whole glycogen biosynthetic machinery (Morán-Zorzano et al., 2007). Expression of SS polypeptides was induced by the addition of isopropyl-β-d-thiogalactopyranoside to the culture medium, and cells were harvested by centrifugation, resuspended in Buffer A (20 mM sodium phosphate, pH 7.0, 0.5 M NaCl, and 20 mM imidazole), and disrupted using a French press at 1200 p.s.i. in the presence of 1 mM PMSF and protease inhibitor cocktail (Sigma-Aldrich). Extracts were centrifuged for 30 min at 20,000g and 4°C, and the supernatant constituted the crude extracts.
For the purification of both SSIV and SSIII proteins, the crude extract was applied onto a 5-mL HisTrap HP column (GE Healthcare), which was washed with 100 mL Buffer A, and SSIV or SSIII proteins were eluted using a 0.02 to 0.5 M imidazole lineal gradient in Buffer A. Samples with SS activity were analyzed by SDS-PAGE and by activity in native polyacrylamide gel and selected for further studies.
Self-Glucosylation Assay
The self-glucosylation reaction was performed using purified fractions of E. coli–expressed SSIV and SSIII proteins (3 μg) in 50 mM HEPES, pH 7.6, 5 mM MnSO4, 2 mM DTT, and 50 μM ADP-[U-14C]glucose (9.81 GBq/mmol) at 30°C for 20 min (de Paula et al., 2005). The reactions were stopped by addition of 4× SDS-PAGE sample buffer, boiled, and subjected to SDS-PAGE. The gel was autoradiographed using a Storage Phosphor System Cyclone Plus (Perkin-Elmer). An alternative assay was performed following the same procedure described above except the samples boiling step, which was omitted to discard any possible removal of glucose incorporated to the proteins by the boiling process.
Light and Transmission Electron Microscopy
Fully expanded leaves from plants cultured under a 16-h-light/8-h-dark photoregime were collected at the midpoint of the light period. Small pieces (2 mm2) of leaves were immediately fixed by submersion in a solution of 3% glutaraldehyde (v/v) in 0.05 M sodium cacodylate buffer, pH 7.4 (3 h at 4°C, under vacuum). After fixing, the specimens were washed in a cacodylate buffer (0.05 M sodium cacodylate and 1% sucrose), three times for 30 min each at 4°C, and postfixed with a solution of 1% osmium tetroxide in the same cacodylate buffer (overnight, 4°C). After two washes, 30 min each, at 4°C with the same cacodylate buffer, the samples were dehydrated in a series of increasing concentrations of ethanol in water (30, 50, and 70%) for 10 min each, at the same temperature. Dehydration was continued at 4°C with 1% uranyl acetate in 70% ethanol for 16 h, 90% ethanol for 10 min, 96% ethanol for 30 min, and 100% ethanol for 2 h with one change after the first hour. Infiltration in London Resin White and polymerization at 60°C was performed as described by González-Melendi et al. (2008).
Semi- (1 μm) and ultrathin (70 to 90 nm) sections were stained and observed as described previously by Roldán et al. (2007). Alternatively, the PAS reaction for carbohydrate staining was performed by placing the slices with semithin sections in a 0.5% periodic acid solution in distilled water (30 min at room temperature) and in Schiff's reagent for 40 min as described by Shvaleva et al. (2009).
Confocal Microscopy
Subcellular localization of GFP-tagged proteins was performed using a D-Eclipse C1 confocal microscope (Nikon) equipped with standard argon 488-nm laser excitation, a BA515/30 filter for green emission, a BA650LP filter for red emission, and transmitted light detector for bright-field images.
Analytical Procedures
Bacterial growth was followed spectrophotometrically by measuring the absorbance at 600 nm. Protein content was measured by the Bradford method using a Bio-Rad prepared reagent. Iodine staining of colonies on solid Kornberg medium was performed following the method of Eydallin et al. (2007) according to which, in the presence of iodine vapors, glycogen-excess clones stain darker than their brownish parent cells, whereas glycogen-deficient clones stain yellow.
Accession Numbers
The accession numbers for the Arabidopsis SS genes used in this study can be found in Supplemental Table 1 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Starch Synthase and Phosphoglucomutase Activity of the ssIII ssIV Double Mutant Using Native PAGE
Supplemental Figure 2. Analysis of the Activities of Starch Metabolism Enzymes in Different ss Mutant Plants.
Supplemental Figure 3. Immunoblot Analysis of Wild-Type, ssIV, and ssI ssII ssIII Plants Using Antibody against SSIV Protein.
Supplemental Figure 4. Serial Sections of ssI ssII ssIV Triple Mutant Leaves Stained with the Periodic Acid-Schiff's Reaction for Carbohydrates.
Supplemental Figure 5. Serial Sections of ssI ssII ssIV Triple Mutant Leaves Stained with the PAS Reaction for Carbohydrates and Toluidine Blue.
Supplemental Figure 6. SSIV and SSIII Expression Complements the Glycogenless Phenotype of ΔglgAP E. coli Cells.
Supplemental Figure 7. Cloning Schemes of SSIV-GFP and GBSSI-GFP Constructs.
Supplemental Figure 8. Serial Analyses of 0.8-μm Optical Sections of the z-Axis Capturing SSIV-GFP and GBSSI-GFP Fluorescence in Arabidopsis Chloroplasts.
Supplemental Figure 9. Further Details Regarding Chloroplastic Localization of GBSSI-GFP in Arabidopsis.
Supplemental Table 1. Starch Synthase Genes, Mutant Lines, and Primers Used to Select Mutant Alleles.
Supplemental Table 2 E. coli Strains and Plasmids Used in This Study.
Supplemental Table 3. Primer Pairs for glgCAP, glgAP, and galU Disruptions.
Supplementary Material
Acknowledgments
We thank Steven G. Ball for fruitful scientific discussions. This work was supported financially by the Ministerio de Ciencia e Innovacion and the European Union-FEDER (Grant BIO2006-00359 to A.M.), Junta de Andalucia (Grant P07-CVI-02795 to A.M.), Agence Nationale de la Recherche Génoplante (Grant GPLA0611G to C.D. and N.S.), the European Union-FEDER and the Région Nord Pas de Calais (Grant ARCir PlantTEQ to C.D. and N.S.), the Ministerio de Ciencia e Innovacion and the European Union-FEDER (Grant BIO2007-63915 to J.P.-R.), and by Iden Biotechnology. We thank Fernando Pinto, César N. Morcillo, Susana Fajardo, and Emilie Perrin for their technical assistance in the microscopy analysis and Maria Teresa Morán-Zorzano for expert technical support.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ángel Mérida (angel@ibvf.csic.es).
Online version contains Web-only data.
References
- Alonso, M.D., Lomako, J., Lomako, W.M., and Whelan, W.J. (1995). A new look at the biogenesis of glycogen. FASEB J. 9: 1126–1137. [DOI] [PubMed] [Google Scholar]
- Ausubel, F., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.G., and Struhl, K. (2001). Current Protocols in Molecular Biology 2. (New York: Wiley Interscience).
- Ball, S., and Morell, M. (2003). From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 54: 207–233. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316: 1194–1199. [Google Scholar]
- Bouchez, D., Camilleri, C., and Caboche, M. (1993). A binary vector based on BASTA resistance for in planta transformation of Arabidopsis thaliana. C. R. Acad. Sci. Paris Life Sci. 316: 1188–1193. [Google Scholar]
- Boyer, C.D., and Preiss, J. (1979). Properties of citrate-stimulated starch synthesis catalyzed by starch synthase I of developing maize kernels. Plant Physiol. 64: 1039–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buleon, A., Colonna, P., Planchot, V., and Ball, S. (1998). Starch granules: Structure and biosynthesis. Int. J. Biol. Macromol. 23: 85–112. [DOI] [PubMed] [Google Scholar]
- Burton, R.A., Jenner, H., Carrangis, L., Fahy, B., Fincher, G.B., Hylton, C., Laurie, D.A., Parker, M., Waite, D., van Wegen, S., Verhoeven, T., and Denyer, K. (2002). Starch granule initiation and growth are altered in barley mutants that lack isoamylase activity. Plant J. 31: 97–112. [DOI] [PubMed] [Google Scholar]
- Busi, M.V., Palopoli, N., Valdez, H.A., Fornasari, M.S., Wayllace, N.Z., Gomez-Casati, D.F., Parisi, G., and Ugalde, R.A. (2008). Functional and structural characterization of the catalytic domain of the starch synthase III from Arabidopsis thaliana. Proteins 70: 31–40. [DOI] [PubMed] [Google Scholar]
- Bustos, R., Fahy, B., Hylton, C., Seale, R., Nebane, N.M., Edwards, A., Martin, C., and Smith, A.M. (2004). Starch granule initiation is controlled by a heteromultimeric isoamylase in potato tubers. Proc. Natl. Acad. Sci. USA 101: 2215–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Y., Steinrauf, L.K., and Roach, P.J. (1995). Mechanism of glycogenin self-glucosylation. Arch. Biochem. Biophys. 319: 293–298. [DOI] [PubMed] [Google Scholar]
- Caspar, T., Huber, S.C., and Somerville, C. (1985). Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol. 79: 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar, T., Lin, T.P., Monroe, J., Bernhard, W., Spilatro, S., Preiss, J., and Somerville, C. (1989). Altered regulation of beta-amylase activity in mutants of Arabidopsis with lesions in starch metabolism. Proc. Natl. Acad. Sci. USA 86: 5830–5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Craig, J., Lloyd, J.R., Tomlinson, K., Barber, L., Edwards, A., Wang, T.L., Martin, C., Hedley, C.L., and Smith, A.M. (1998). Mutations in the gene encoding starch synthase II profoundly alter amylopectin structure in pea embryos. Plant Cell 10: 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crevillén, P., Ballicora, M.A., Mérida, A., Preiss, J., and Romero, J.M. (2003). The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct kinetic and regulatory properties to the heterotetrameric enzyme. J. Biol. Chem. 278: 28508–28515. [DOI] [PubMed] [Google Scholar]
- Critchley, J.H., Zeeman, S.C., Takaha, T., Smith, A.M., and Smith, S.M. (2001). A critical role for disproportionating enzyme in starch breakdown is revealed by a knock-out mutation in Arabidopsis. Plant J. 26: 89–100. [DOI] [PubMed] [Google Scholar]
- Datsenko, K.A., and Wanner, B.L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte, T., Trevisan, M., Parker, M.L., and Zeeman, S.C. (2005). Arabidopsis mutants Atisa1 and Atisa2 have identical phenotypes and lack the same multimeric isoamylase, which influences the branch point distribution of amylopectin during starch synthesis. Plant J. 41: 815–830. [DOI] [PubMed] [Google Scholar]
- Delatte, T., Umhang, M., Trevisan, M., Eicke, S., Thorneycroft, D., Smith, S.M., and Zeeman, S.C. (2006). Evidence for distinct mechanisms of starch granule breakdown in plants. J. Biol. Chem. 281: 12050–12059. [DOI] [PubMed] [Google Scholar]
- Delvallé, D., Dumez, S., Wattebled, F., Roldán, I., Planchot, V., Berbezy, P., Colonna, P., Vyas, D., Chatterjee, M., Ball, S., Mérida, Á., and D'Hulst, C. (2005). Soluble starch synthase I: A major determinant for the synthesis of amylopectin in Arabidopsis thaliana leaves. Plant J. 43: 398–412. [DOI] [PubMed] [Google Scholar]
- de Paula, R.M., Wilson, W.A., Roach, P.J., Terenzi, H.F., and Bertolini, M.C. (2005). Biochemical characterization of Neurospora crassa glycogenin (GNN), the self-glucosylating initiator of glycogen synthesis. FEBS Lett. 579: 2208–2214. [DOI] [PubMed] [Google Scholar]
- Eydallin, G., Viale, A.M., Morán-Zorzano, M.T., Muñoz, F.J., Montero, M., Baroja-Fernández, E., and Pozueta-Romero, J. (2007). Genome-wide screening of genes affecting glycogen metabolism in Escherichia coli K-12. FEBS Lett. 581: 2947–2953. [DOI] [PubMed] [Google Scholar]
- González-Melendi, P., Uyttewaal, M., Morcillo, C.N., Hernández Mora, J.R., Fajardo, S., Budar, F., and Lucas, M.M. (2008). A light and electron microscopy analysis of the events leading to male sterility in Ogu-INRA CMS of rapeseed (Brassica napus). J. Exp. Bot. 59: 827–838. [DOI] [PubMed] [Google Scholar]
- Hennen-Bierwagen, T.A., Liu, F., Marsh, R.S., Kim, S., Gan, Q., Tetlow, I.J., Emes, M.J., James, M.G., and Myers, A.M. (2008). Starch biosynthetic enzymes from developing maize endosperm associate in multisubunit complexes. Plant Physiol. 146: 1892–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose, T., and Terao, T. (2004). A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.). Planta 220: 9–16. [DOI] [PubMed] [Google Scholar]
- Lin, T.P., Caspar, T., Somerville, C., and Preiss, J. (1988). Isolation and characterization of a starchless mutant of Arabidopsis thaliana (L.) Heynh lacking ADP-glucose pyrophosphorylase activity. Plant Physiol. 86: 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manners, D.J. (1991). Recent developments in our understanding of glycogen structure. Carbohydr. Polym. 16: 37–82. [Google Scholar]
- Morán-Zorzano, M.T., Alonso-Casajus, N., Muñoz, F.J., Viale, A.M., Baroja-Fernández, E., Eydallin, G., and Pozueta-Romero, J. (2007). Occurrence of more than one important source of ADPglucose linked to glycogen biosynthesis in Escherichia coli and Salmonella. FEBS Lett. 581: 4423–4429. [DOI] [PubMed] [Google Scholar]
- Morell, M.K., Kosar-Hashemi, B., Cmiel, M., Samuel, M.S., Chandler, P., Rahman, S., Buleon, A., Batey, I.L., and Li, Z. (2003). Barley sex6 mutants lack starch synthase IIa activity and contain a starch with novel properties. Plant J. 34: 173–185. [DOI] [PubMed] [Google Scholar]
- Muñoz, F.J., Baroja-Fernandez, E., Ovecka, M., Li, J., Mitsui, T., Sesma, M.T., Montero, M., Bahaji, A., Ezquer, I., and Pozueta-Romero, J. (2008). Plastidial localization of a potato “Nudix” hydrolase of ADPglucose linked to starch biosynthesis. Plant Cell Physiol. 49: 1734–1746. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiol. 15: 473–497. [Google Scholar]
- Patron, N.J., and Keeling, P. (2005). Common evolutionary origin of starch biosynthetic enzymes in green and red algae. J. Phycol. 41: 1131–1141. [Google Scholar]
- Prescott, A., and Martin, C. (1986). A rapid method for the quantitative assessment of levels of specific mRNAs in plants. Plant Mol. Biol. Rep. 4: 219–224. [Google Scholar]
- Putaux, J.L., Potocki-Veronese, G., Remaud-Simeon, M., and Buleon, A. (2006). Alpha-D-glucan-based dendritic nanoparticles prepared by in vitro enzymatic chain extension of glycogen. Biomacromolecules 7: 1720–1728. [DOI] [PubMed] [Google Scholar]
- Roldán, I., Lucas, M.M., Delvalle, D., Planchot, V., Jimenez, S., Perez, R., Ball, S., D'Hulst, C., and Mérida, A. (2007). The phenotype of soluble starch synthase IV defective mutants of Arabidopsis thaliana suggests a novel function of elongation enzymes in the control of starch granule formation. Plant J. 49: 492–504. [DOI] [PubMed] [Google Scholar]
- Rose, A., Manikantan, S., Schraegle, S.J., Maloy, M.A., Stahlberg, E.A., and Meier, I. (2004). Genome-wide identification of Arabidopsis coiled-coil proteins and establishment of the ARABI-COIL database. Plant Physiol. 134: 927–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259. [DOI] [PubMed] [Google Scholar]
- Satoh, H., et al. (2008). Mutation of the plastidial alpha-glucan phosphorylase gene in rice affects the synthesis and structure of starch in the endosperm. Plant Cell 20: 1833–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirokane, Y., Ichikawa, K., and Suzuki, M. (2000). A novel enzymic determination of maltose. Carbohydr. Res. 329: 699–702. [DOI] [PubMed] [Google Scholar]
- Shvaleva, A., Coba de la Peña, T., Rincón, A., Morcillo, C.N., Lucas, M.M., and Pueyo, J.J. (April 8, 2009). Flavodoxin overexpression reduces cadmiun-induced damage in alfalfa root nodules. Plant Soil http://dx.doi.org/.
- Smith, S.M., Fulton, D.C., Chia, T., Thorneycroft, D., Chapple, A., Dunstan, H., Hylton, C., Zeeman, S.C., and Smith, A.M. (2004). Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol. 136: 2687–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt, M., Mc, C., Lilley, R., Gerhardt, R., and Heldt, H.W. (1989). Determination of metabolite levels in specific cells and subcellular compartments of plants leaves. Methods Enzymol. 174: 518–552. [Google Scholar]
- Streb, S., Delatte, T., Umhang, M., Eicke, S., Schorderet, M., Reinhardt, D., and Zeeman, S.C. (2008). Starch granule biosynthesis in Arabidopsis is abolished by removal of all debranching enzymes but restored by the subsequent removal of an endoamylase. Plant Cell 20: 3448–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenorio, G., Orea, A., Romero, J.M., and Mérida, A. (2003). Oscillation of mRNA level and activity of granule-bound starch synthase I in Arabidopsis leaves during the day/night cycle. Plant Mol. Biol. 51: 949–958. [DOI] [PubMed] [Google Scholar]
- Tetlow, I.J., Beisel, K.G., Cameron, S., Makhmoudova, A., Liu, F., Bresolin, N.S., Wait, R., Morell, M.K., and Emes, M.J. (2008). Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes. Plant Physiol. 146: 1878–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugalde, J.E., Parodi, A.J., and Ugalde, R.A. (2003). De novo synthesis of bacterial glycogen: Agrobacterium tumefaciens glycogen synthase is involved in glucan initiation and elongation. Proc. Natl. Acad. Sci. USA 100: 10659–10663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez, H.A., Busi, M.V., Wayllace, N.Z., Parisi, G., Ugalde, R.A., and Gomez-Casati, D.F. (2008). Role of the N-terminal starch-binding domains in the kinetic properties of starch synthase III from Arabidopsis thaliana. Biochemistry 47: 3026–3032. [DOI] [PubMed] [Google Scholar]
- Wattebled, F., Dong, Y., Dumez, S., Delvallé, D., Planchot, V., Berbezy, P., Vyas, D., Colonna, P., Chatterjee, M., Ball, S., and D'Hulst, C. (2005). Mutants of Arabidopsis lacking a chloroplastic isoamylase accumulate phytoglycogen and an abnormal form of amylopectin. Plant Physiol. 138: 184–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattebled, F., Planchot, V., Dong, Y., Szydlowski, N., Pontoire, B., Devin, A., Ball, S., and D'Hulst, C. (2008). Further evidence for the mandatory nature of polysaccharide debranching for the aggregation of semicrystalline starch and for overlapping functions of debranching enzymes in Arabidopsis leaves. Plant Physiol. 148: 1309–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman, S.C., Northrop, F., Smith, A.M., and Ap Rees, T. (1998). A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch-hydrolysing enzyme. Plant J. 15: 357–385. [DOI] [PubMed] [Google Scholar]
- Zeeman, S.C., Thorneycroft, D., Schupp, N., Chapple, A., Weck, M., Dunstan, H., Haldimann, P., Bechtold, N., Smith, A.M., and Smith, S.M. (2004). Plastidial α-glucan phosphorylase is not required for starch degradation in Arabidopsis leaves but has a role in the tolerance of abiotic stress. Plant Physiol. 135: 849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Colleoni, C., Ratushna, V., Sirghie-colleoni, M., James, M., and Myers, A. (2004). Molecular characterization demonstrates that the Zea mays gene sugary2 codes for the starch synthase isoform SSIIa. Plant Mol. Biol. 54: 865–879. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Myers, A.M., and James, M.G. (2005). Mutations affecting starch synthase III in Arabidopsis alter leaf starch structure and increase the rate of starch synthesis. Plant Physiol. 138: 663–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Szydlowski, N., Delvalle, D., D'Hulst, C., James, M.G., and Myers, A.M. (2008). Overlapping functions of the starch synthases SSII and SSIII in amylopectin biosynthesis in Arabidopsis. BMC Plant Biol. 8: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.