Abstract
Jasmonates play a number of diverse roles in plant defense and development. CORONATINE INSENSITIVE1 (COI1), an F-box protein essential for all the jasmonate responses, interacts with multiple proteins to form the SCFCOI1 E3 ubiquitin ligase complex and recruits jasmonate ZIM-domain (JAZ) proteins for degradation by the 26S proteasome. To determine which protein directly binds to jasmonoyl-isoleucine (JA-Ile)/coronatine (COR) and serves as a receptor for jasmonate, we built a high-quality structural model of COI1 and performed molecular modeling of COI1–jasmonate interactions. Our results imply that COI1 has the structural traits for binding JA-Ile or COR. The direct binding of these molecules with COI1 was further examined using a combination of molecular and biochemical approaches. First, we used the immobilized jasmonate approach to show that the COI1 protein in crude leaf extracts can bind to the jasmonate moiety of JA-Ile. Second, we employed surface plasmon resonance technology with purified COI1 and JAZ1 protein to reveal the interaction among COI1, JA-Ile, and JAZ1. Finally, we used the photoaffinity labeling technology to show the direct binding of COR with purified insect-expressed COI1. Taken together, these results demonstrate that COI1 directly binds to JA-Ile and COR and serves as a receptor for jasmonate.
INTRODUCTION
Jasmonates (JAs), which are derived from linolenic acid and characterized by a pentacyclic ring structure (Creelman and Mullet, 1997; Feussner and Wasternack, 2002; Wasternack, 2007), are critical for plant defense responses, particularly for defense against insect herbivory and pathogens (McConn et al., 1997; Reymond and Farmer, 1998; Farmer, 2001; Farmer et al., 2003). JAs are also involved in the regulation of many developmental processes, such as root growth, tuberization, tendril coiling, senescence, and fertility (Ueda and Kato, 1980; Staswick et al., 1992; McConn and Browse, 1996; Schommer et al., 2008; Cheng et al., 2009). In addition, JAs play many important roles in the wound response and the production of secondary metabolites (Farmer et al., 1992; Wasternack and Hause, 2002; Lorenzo and Solano, 2005; Shan et al., 2009).
A great deal of knowledge on the action of JAs comes from analysis of Arabidopsis thaliana mutants with alterations in JA biosynthesis and signal transduction. One such mutant is the coronatine insensitive1-1 (coi1-1) mutant, which exhibits male sterility, resistance to JA inhibition of root growth, and defects in the expression of JA-regulated genes (Feys et al., 1994). COI1 is an F-box protein (Xie et al., 1998) that associates with Arabidopsis Skp1-like 1, Arabidopsis Skp1-like 2 (Liu et al., 2004), cullin 1, and ring-box protein 1 to form an E3 ubiquitin ligase known as the SCFCOI1 complex (Xu et al., 2002; Ren et al., 2005). It is well known that E3 ubiquitin ligases in plants are involved in the ubiquitination of target proteins for subsequent degradation by the 26S proteasome (Moon et al., 2004). JAs have been suggested to promote ubiquitination and degradation of negative regulators of JA signaling, which are substrates of the SCFCOI1 complex.
A negative regulator of JA signaling was recently discovered; this protein, JAZ1, belongs to a novel family of transcription regulators named the JA ZIM-domain (JAZ) proteins (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007). JAZ proteins are targeted by SCFCOI1 for degradation via the 26S proteasome in response to JA signaling. The physical interaction of COI1 with the JAZ1 protein is promoted by an Ile-conjugated form of jasmonic acid (JA-Ile), but not by jasmonic acid, jasmonic acid methyl ester (MeJA), or cis(+)-12-oxophytodienoic acid (OPDA) (Thines et al., 2007). Coronatine (COR) is a phytotoxin produced by the plant pathogen Pseudomonas syringae (Mitchell, 1982), which acts as a molecular mimic of JA-Ile and has high biological activity to activate JA signaling (Weiler et al., 1994; Koda et al., 1996; Bender et al., 1999). Indeed, COR is more active than JA-Ile in triggering the COI1–JAZ interaction in vitro (Katsir et al., 2008). COR and JA-Ile bind with a protein complex containing at least COI1 and JAZ1, suggesting that this complex is the site of JA-Ile perception (Katsir et al., 2008). However, the JA binding assays in previous studies were performed with crude plant extracts (Thines et al., 2007; Katsir et al., 2008; Melotto et al., 2008) or with partially purified proteins (Fonseca et al., 2009), where COI1 is undoubtedly associated with other proteins that copurified with it. Coexistence of COI1, JAZ1, and JA-Ile/COR can activate the reporter gene in yeast two-hybrid system (Thines et al., 2007; Melotto et al., 2008; Fonseca et al., 2009), where COI1 might interact with yeast proteins, such as CUL1, SKP1, RBX1, and possibly with other proteins that are highly conserved between Arabidopsis and yeast. It is speculated that COI1 or other proteins, which copurified with COI1, may bind JA-Ile/COR and function as a JA receptor (Fonseca et al., 2009).
In this study, we show that the COI1 protein directly binds to JA-Ile/COR using a combination of the immobilized JA approach, the surface plasmon resonance technology, and the photoaffinity labeling technology. Together with the genetic evidence (Feys et al., 1994; Xie et al., 1998; Xu et al., 2002), we demonstrate that COI1 functions as a receptor for JA-Ile/COR. We also built a high-quality structure model of COI1, performed molecular modeling of COI1–JA interactions, and propose a model to illustrate the mechanism for JA signal perception.
RESULTS
Structural Model of the Arabidopsis COI1 Protein
In plants, the signal transduction pathway of JA is similar to that of auxin, in which an F-box protein, TRANSPORT INHIBITOR RESPONSE1 (TIR1) in the auxin signaling pathway or COI1 in the JA pathway, forms a SCF complex that recruits substrates for ubiquitination and subsequent degradation by the 26S proteasome to regulate plant responses (Dharmasiri et al., 2005; Kepinski and Leyser, 2005; Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Katsir et al., 2008). COI1 displays ∼33% sequence similarity to TIR1, and both contain leucine-rich repeats (LRRs) and an F-box motif (Tan et al., 2007). Based on the striking similarity between COI1 and TIR1, a good structural model of COI1 could be developed adopting the crystal structure data of TIR1.
To build a structural model of the Arabidopsis COI1 protein, we performed homology modeling of COI1 using Accelrys Discovery Studio 1.7 software. We first built an initial structure of COI1 with the MODELER module (Sali et al., 1995) and then performed two optimization steps, including energy minimization and molecular dynamics simulations, to achieve a final structural model of COI1 (Figure 1).
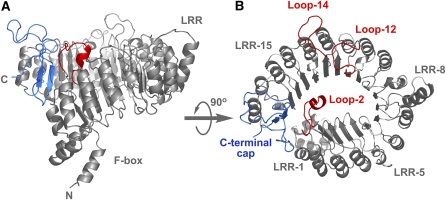
Figure 1.
Structural Model of COI1.
Two views of the structural model of COI1 are shown as ribbon diagrams. The F-box and LRR domains of COI1 are labeled and shown in gray. The C-terminal cap is shown in blue. The 18 LRR motifs are numbered. The three surface loops in LRR-2, LRR-12, and LRR-14 are shown in red and labeled loop-2, loop-12, and loop-14.
To determine the reliability of the final structural model of COI1, we performed comprehensive analysis with multiple model quality assessment programs (Table 1; see Supplemental Figure 1 online). The Ramachandran plot generated by PROCHECK (Laskowski et al., 1993) showed that the ϕφ/ψ angles of the majority of residues were in the favored region, indicating good stereochemical quality. The model received a Z-score of −11.59 in ProSA2003 (Sippl, 1993), which was close to the value of −12.78 obtained for the template. These scores indicate traits of a reasonable native-like fold and good interaction energies for residues in its environment in the model. Verify3D (Luthy et al., 1992), which evaluates the fitness of a protein sequence in a three-dimensional environment, returned an overall score of 245.45 for the model, in which 86% of the residues had a score of above 0.2. The fine packing environment of the residues in the model was evaluated by WHATIF (Vriend, 1990), which returned an overall score of −1.46 compared with −1.12 for the template, indicating high quality of the model. Together, these evaluations indicate that we have built a high-quality structural model of COI1.
Table 1.
Evaluation of the Structural Model of COI1 in Comparison to the Template
| PROCHECK Ramachandran Plot
|
ProSA 2003 Z-Score | Verify 3D
|
||||||
|---|---|---|---|---|---|---|---|---|
| Protein Name | Core (%) | Allowed (%) | Generally Allowed (%) | Disallowed (%) | WHATIF Quality Control Value | Residues with 3D-1D Score > 2.0 (%) | Overall Score | |
| COI1 | 87.4 | 11.4 | 1.2 | 0 | −11.59 | −1.46 | 86.48 | 245.45 |
| TIR1 | 88.8 | 11.0 | 0.2 | 0 | −12.78 | −1.12 | 94.37 | 267.45 |
TIR1, Template structure (PDB: 2p1m, chain B).
The structural model of COI1 consists of two domains: a small N-terminal F-box domain and a large LRR domain (Figure 1A). The LRR domain includes 18 LRRs, which adopt a tandem packed structure of staggered α-helices and β-sheets (Figure 1B). These LRRs assemble into a solenoid fold with a horseshoe-like shape. The characteristics of the LRR domain imply that the integrity of the LRR domain is important to the structural framework of COI1. It is speculated that disruption of the LRR integrity may affect COI1 native structure and result in a reduction in the stability of COI1 in vivo.
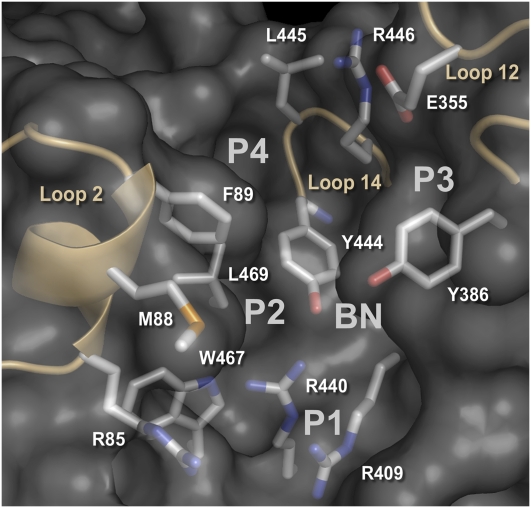
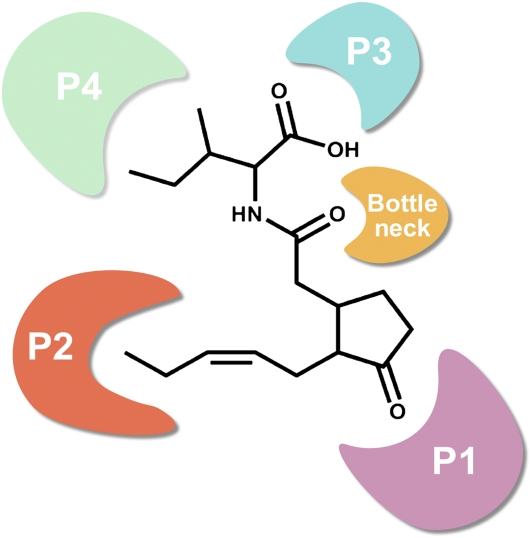
The structural model of COI1 reveals that there is a cavity in the center of the solenoid fold assembled by LRRs. At the top surface of the cavity, a long loop (loop 2), which associates with LRR-2 and contains a 310-helix, together with the other two long loops (loop 12 and loop 14) and the solenoid inner surface, forms a surface pocket (Figure 2). The surface pocket can be divided into four distinct pockets (P1, P2, P3, and P4) that involve different residues and have distinctive surface properties. The P1 pocket contains Arg-409, Arg-440, and Arg-85, which cause the pocket to be highly positively charged. On the opposite side, the P2 pocket is highly hydrophobic because it is encircled by Met-88, Phe-89, and Trp-467. The P1 and P2 pockets form the bottom of the surface pocket. The P3 pocket mainly contains hydrophilic residues (Glu-355 and Arg-446), whereas the P4 pocket contains hydrophobic residues (Leu-445 and Leu-469). The channel that connects the four pockets is occupied by two Tyr residues (Tyr-386 and Tyr-444), which form a bottleneck area in the surface pocket. The characteristics of the surface pocket suggested that it could be a binding site for JAs.
Figure 2.
An Overall View of the Surface Pocket in COI1.
Three long loops (loop 2, loop 12, and loop 14) and the solenoid inner surface of COI1 form a surface pocket that can be divided into four distinct pockets (P1, P2, P3, and P4) and one bottleneck (BN) region based on surface properties. The P1 pocket mainly includes basic residues (Arg-409, Arg-440, and Arg-85), and the P2 pocket is encircled by hydrophobic residues (Met-88, Phe-89, and Trp-467). The P3 pocket mainly contains hydrophilic residues (Glu-355 and Arg-446), while the P4 pocket mainly contains hydrophobic residues (Leu-445 and Leu-469). The channel connecting the four pockets is occupied by the bottleneck region, which contains Tyr-386 and Tyr-444. The residues in the surface pocket are shown as green stick models. Loop 2, loop 12, and loop 14 are shown in yellow.
The structural model of COI1 contains 19 Cys residues, 12 of which are distributed in the LRR domain (Figure 3A). Despite this high frequency of Cys residues, the MODELER module did not return a structure with disulfide bridges. We attempted to construct structural models with disulfide bridges that were manually defined, but these structural models failed to pass the criteria set in the model quality assessment programs. Therefore, our studies indicate that disulfide bridges likely do not exist in the structure of COI1, suggesting that a reducing environment is required for the normal function of COI1.
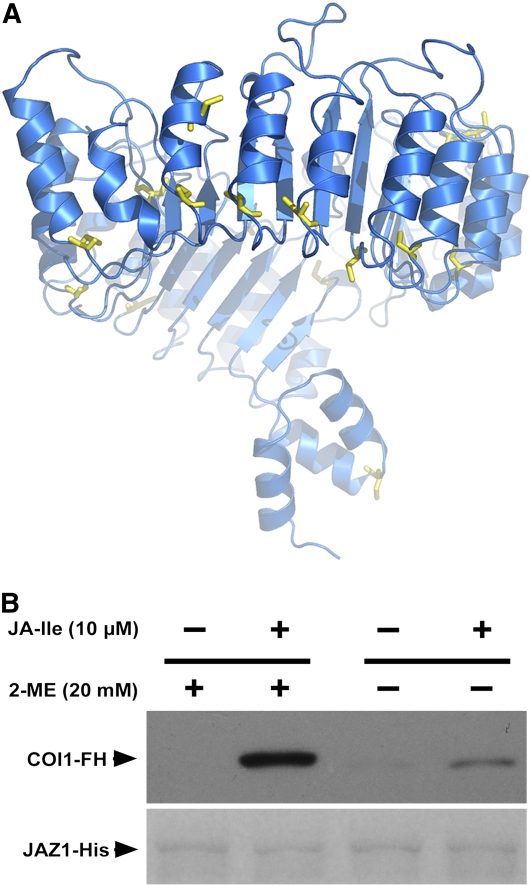
Figure 3.
The Reducing Environment Is Important for the JA-Ile–Induced Interaction between COI1 and JAZ1.
(A) A view of the Cys residues in the COI1 structural model. The structural model of COI1 is shown as a ribbon diagram. The Cys residues of COI1 are shown as yellow stick models. All the sulfhydryl groups of Cys residues are in a reduced form.
(B) Pull-down reactions were performed using recombinant JAZ1-His and total plant extracts prepared from coi1-1 seedlings transgenic for HA-tagged COI1 (COI1-FH). The reactions were performed at 4°C for 1 h with or without 20 mM 2-mercaptoethanol at the indicated JA-Ile concentrations. Protein bound to the Ni-NTA resin was washed, separated on SDS-PAGE, and immunoblotted with an anti-HA antibody (top panel). The polyvinylidene fluoride (PVDF) membrane was stained with Memstain (Applygen Technologies) to visualize the recovery of JAZ1 by the Ni-NTA affinity resin (bottom panel).
The Function of COI1 Is Dependent on a Reducing Environment
Because our modeling studies suggested that a reducing environment is required for the normal function of COI1, we evaluated the effect of a reducing agent (2-mercaptoethanol) on COI1 function through determination of JA-Ile–dependent COI1–JAZ1 interactions using an in vitro pull-down assay. The pull-down reactions were performed in the presence or absence of 2-mercaptoethanol, with recombinant Arabidopsis JAZ1-His and total plant extracts prepared from coi1-1 mutant plants transgenically expressing HA-tagged COI1 protein (COI1-FH). As expected, JAZ1-His was unable to recover the COI1-FH protein in the absence of JA-Ile, regardless of whether 2-mercaptoethanol was added or not. In the presence of both JA-Ile and 2-mercaptoethanol, COI1-FH was recovered by JAZ1-His (Figure 3A). However, the recovery of COI1-FH by JAZ1-His was largely diminished in the absence of 2-mercaptoethanol, although JA-Ile was present (Figure 3B). In support of our predictions from the COI1 structure model, the results suggest that the function of COI1 depends to a great extent on a reducing environment.
The Integrity of the LRR Domain Is Required for COI1 Stability in Vivo
The structural model of COI1 indicates that the integrity of the LRR domain is important to the structural framework of COI1. Mutations within the LRR domain may affect COI1 native structure and result in a reduction in the stability of COI1 in vivo. To examine this hypothesis, we isolated a series of coi1 mutants from ethyl methanesulfonate–mutagenized Arabidopsis seeds, which harbor different amino acid substitutions within the LRR domain of COI1 (Figure 4A). The four amino acid substitutions, G369E (coi1-4), G155E (coi1-7), D452A (coi1-9), and L490A (coi1-10), are present in the α-helix or loops close to the β-sheet of the LRR domain in the structural model of COI1 (Figure 4A). These mutations are located in the important regions of the LRR domain and may disrupt COI1 stability. The G98D substitution in coi1-5 was located at the end of loop 2, and the E543K substitution in coi1-8 was at the end of the LRR domain. G98D and E543K may not affect the LRR domain of COI1 as much as the mutations in the α-helix or β-sheet of the LRR domain and may reduce the stability of COI1 to lesser extents than the other four amino acid substitutions.
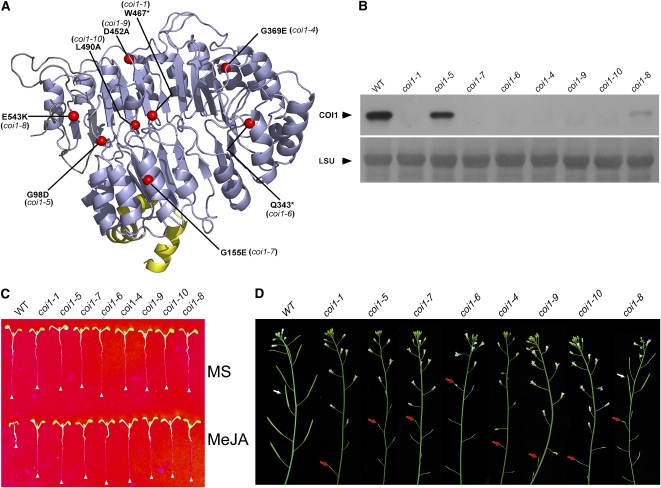
Figure 4.
Mutations in the LRR Domain Disrupt COI1 Stability in Vivo.
(A) A view of the mutant sites in the COI1. The structural model of COI1 is shown as a ribbon diagram. The F-box, LRR domain, and C-terminal domain of COI1 are colored yellow, light blue, and gray, respectively. The mutant sites are labeled.
(B) Determination of COI1 protein levels in coi1 mutant plants. The proteins were extracted from leaves and subjected to immunoblot analyses using COI1 antiserum. The PVDF membrane was stained with Memstain to visualize equal loading. LSU, large subunit of ribulose-1, 5-bisphosphate carboxylase/oxygenase.
(C) Phenotype of 10-d-old seedlings grown on Murashige and Skoog (MS) medium or MS with 25 μM MeJA. The white triangles indicate root tips.
(D) The coi1-8 displays a partial fertility phenotype, while the other mutants, like coi1-1, are male sterile. The red arrows indicate sterile siliques, and the white arrows indicate fertile siliques.
To examine the stability of COI1, total protein was extracted from these mutants and immunoblotted with the antiserum against COI1 (Xu et al., 2002). The coi1-4, coi1-6, coi1-7, coi1-9, and coi1-10 mutants failed to accumulate detectable amounts of COI1 protein, similar to coi1-1 with the W467* substitution (Figure 4B), and exhibited a complete loss of JA responses, including JA inhibitory root growth (Figure 4C) and plant fertility (Figure 4D). However, the coi1-5 and coi1-8 mutants accumulated COI1 protein, although at low levels compared with the wild type. Consistent with our predictions from the structural model of COI1, these results revealed that the LRR domain is important for the stability of COI1 in vivo.
The Surface Pocket of COI1 Is Important for the JA-Ile–Dependent Interaction between COI1 and JAZ1
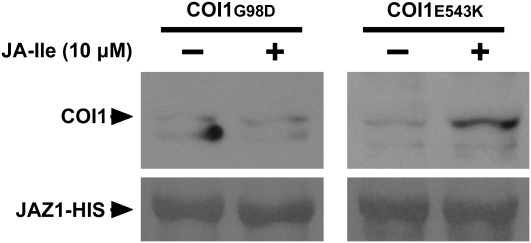
Although both coi1-5 and coi1-8 accumulated detectable amounts of COI1 protein, the coi1-5 mutant was completely male sterile and fully resistant to JA inhibitory root growth (Figures 4C and 4D), whereas coi1-8 exhibited partial fertility (Figure 4D). These results indicated that COI1G98D in coi1-5 had no function, but COI1E543K in the coi1-8 plant still had partial activity.
Through analysis of the mutated sites in the structural model of COI1, we noticed that Gly at position 98 (Gly-98) was located at the end of loop 2, which is an important part of the COI1 surface pocket. The amino acid substitution of Gly-98 with an Asp (G98D) in coi1-5 may influence the flexibility of loop 2, which may affect the conformation of the surface pocket of COI1. By contrast, Glu-543 was present at the end of the LRR domain and far away from the surface pocket of COI1. The amino acid substitution of Glu-543 with Lys (E543K) in coi1-8 may not significantly change the conformation of the surface pocket of COI1.
We further investigated whether the G98D and E543K mutations affect the JA-Ile–dependent interaction between COI1 and JAZ1. The pull-down assay, which was performed with recombinant Arabidopsis JAZ1-His and total plant extracts prepared from coi1-5 and coi1-8 mutant plants, showed that JAZ1-His did not recover COI1G98D in a JA-Ile–dependent manner, whereas COI1E543K still maintained the ability to interact with JAZ1-His in the presence of JA-Ile (Figure 5). These results indicate that the surface pocket of COI1 is important for the JA-Ile–dependent interaction between COI1 and JAZ1,which is consistent with our structure model of COI1. However, complete confirmation of the surface pocket requires determination of COI1's x-ray crystal structure.
Figure 5.
The G98D Mutation in COI1 Disrupts the JA-Ile–Dependent Interaction between COI1 and JAZ1.
Pull-down assays used recombinant JAZ1-His protein and extracts from coi1-5 and coi1-8 mutants. The reactions were performed at 4°C for 1 h in the presence or absence of 10 μM JA-Ile. The recovery of COI1 was detected with an anti-COI1 antiserum (top panel). The PVDF membrane was stained with Memstain to visualize the recovery of JAZ1 by the Ni-NTA affinity resin (bottom panel).
Molecular Modeling of COI1–JA Interactions
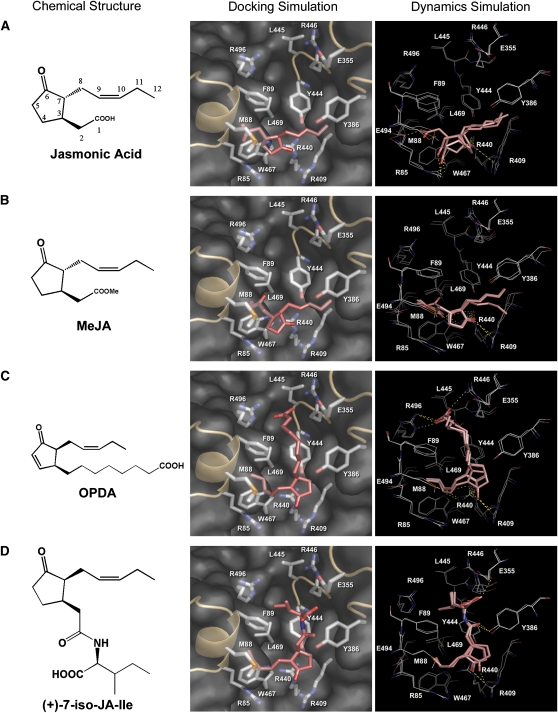
Analysis of the COI1 structural model suggested that the surface pocket in COI1 may be a binding site for JAs. To explore this possibility, we analyzed several common JAs, such as jasmonic acid [(3R,7R)-jasmonic acid], MeJA [(3R,7R)-(−)-methyl JA], OPDA, JA-Ile, and COR, a molecular mimic of JA-Ile, through molecular modeling procedures. We performed molecular docking simulations with GOLD 3.0.1 (Jones et al., 1997) to generate possible poses that were subsequently ranked by the GoldScore scoring function, which rapidly evaluates energy terms (van de Waals, hydrogen bond, and ligand torsional strain energy) by empirical parameters. The pose with the highest GoldScore fitness value was selected for determination of binding stability with a restricted molecular dynamics (MD) simulation. The binding free energies between COI1 and JAs were further estimated by the force field–based molecular mechanics-Poisson–Boltzmann surface area (MM-PBSA) analysis, a more refined way used in calculating protein-ligand binding free energies, which comprehensively evaluates van der Waals, electrostatic, and solvation contributions to the binding process (Kollman et al., 2000).
The molecular docking simulation indicated that both JA and MeJA exhibited low GoldScore fitness values (27.51 for JA and 32.85 for MeJA), denoting unfavorable bindings. In addition, the results of restricted MD simulations showed that both failed to maintain the initial binding conformations during the simulation timescale (Figures 6A and 6B, right panels). These results suggest that JA and MeJA molecules are too small to fit within the pocket of COI1. For OPDA, the binding pose also received a low GoldScore of 34.83, and the subsequent restricted MD simulation showed that OPDA shifted between the keto group of the cyclopentanone ring and Arg-409 as the simulation time elapsed (Figure 6C, right panel), suggesting that OPDA could not form a stable interaction with the surface pocket. Furthermore, the MM-PBSA analysis also returned higher binding free energy values (see Supplemental Table 1 online) for JA (−18.79 kJ mol−1), MeJA (−22.23 kJ mol−1), and OPDA (−18.31 kJ mol−1), which further indicate unfavorable bindings.
Figure 6.
JA-Ile Could Fit within the Surface Pocket of COI1.
Molecular modeling of the interaction between COI1 and JA (A), MeJA (B), OPDA (C), or (+)-7-iso-JA-Ile (D). Left panel: chemical structures of the JAs. Middle panel: the pose with the highest GoldScore fitness value in the molecular docking simulation. The JAs are shown as red sticks. The surface pocket of COI1 is shown in gray. Right panel: superposition of representative frames of the restricted molecular dynamics simulation. Frames at the early, intermediate, and late stages were extracted and superimposed. The JAs are shown as pink sticks, and their interacting residues are shown as white lines. Polar contacts are shown as yellow dotted lines.
By contrast, JA-Ile achieved higher GoldScore [43.06 for (+)-7-iso-JA-Ile and 41.90 for (−)-JA-Ile]. The restricted MD simulation showed that there was no great change in the binding conformation of both JA-Ile isomers during the simulation (Figure 6D; see Supplemental Figure 2 online). These results indicate that both JA-Ile isomers could fit within the surface pocket of COI1. However, the more precise analysis using the MM-PBSA method produced much lower binding free energy values (see Supplemental Table 1 online) for (+)-7-iso-JA-Ile (−47.68 kJ mol−1) compared with that of (−)-JA-Ile (−38.18 kJ mol−1), indicating that (+)-7-iso-JA-Ile may possess higher binding affinity. The docking poses of JA-Ile isomers in the structural model of COI1 showed that the keto group of the cyclopentanone ring in JA-Ile could anchor in the P1 pocket of COI1, the pentenyl moiety of JA-Ile preferred the hydrophobic P2 pocket of COI1, and the oxygen atom in the amide group of JA-Ile could form hydrogen bonds with either Tyr-444 or Tyr-386 in the bottleneck region of the surface pocket of COI1 (Figure 6D; see Supplemental Figure 2 online). These results suggest that JA-Ile probably binds to the surface pocket of COI1 via the keto group of the cyclopentanone ring, the pentenyl side chain, and the oxygen atom of the amide group in the JA moiety.
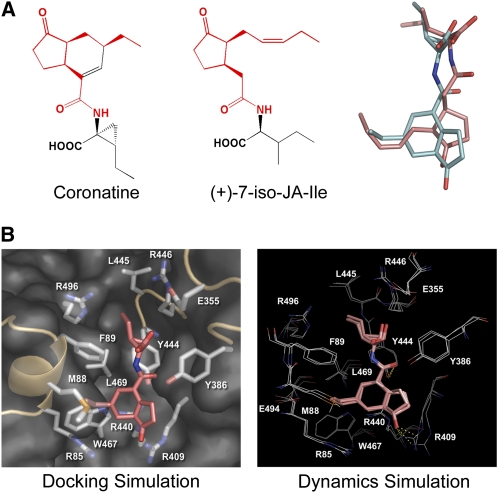
The molecular modeling of COR revealed that the binding mode of COR to COI1 was strikingly similar with that of JA-Ile (Figures 7A and 7B). The highest GoldScore of 51.95 and lowest binding free energy of −78.33 kJ mol−1 (see Supplemental Table 1 online) indicate that COR may have a strong binding affinity to the COI1 protein. The coronafacic acid moiety in COR overlaps well with the JA moiety in JA-Ile, promoting interactions of COR with the P1 and P2 pockets and the bottleneck region of COI1. Moreover, the two coupled ring structures of coronafacic acid moiety in COR could effectively facilitate its binding with COI1, resulting in the high biological activity of COR.
Figure 7.
The Proposed Mechanism of COR Binding to COI1.
(A) The similarity between (+)-7-iso-JA-Ile and COR. Chemical structures of COR (left) and (+)-7-iso-JA-Ile (middle) are shown. The groups of (+)-7-iso-JA-Ile or COR that contribute to the binding with the surface pocket of COI1 are colored red. The optimized binding conformation of (+)-7-iso-JA-Ile and COR was extracted and superimposed to show the identity of the binding conformations between (+)-7-iso-JA-Ile and COR in the surface pocket of COI1 (right). Carbon atoms in (+)-7-iso-JA-Ile are represented in red, while those in COR are in cyan.
(B) Molecular modeling of the interaction between COI1 and COR. Left panel: the pose with the highest GoldScore fitness value in the molecular docking simulation. COR is shown in red sticks. The surface pocket of COI1 is shown in gray. Right panel: superposition of representative frames of the restricted molecular dynamics simulation. Frames at the early, intermediate, and late stages were extracted and superimposed. COR is shown in pink sticks, and the interacting residues are shown in white lines. Polar contacts are shown as yellow dotted lines.
COI1 Binds to JA Moiety of the Immobilized JA
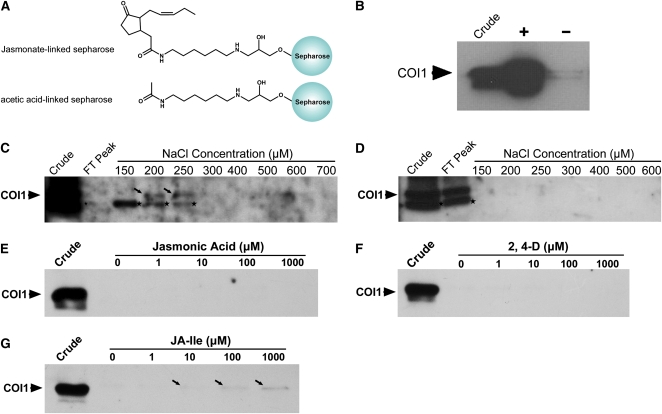
The molecular modeling of interaction between COI1 and JA-Ile suggests that COI1 may directly bind to the JA moiety of JA-Ile. Here, we employed immobilized JA to test this hypothesis. We covalently immobilized the free carboxylic group of JA on EAH-Sepharose 4B to generate JA-linked sepharose. The JA-linked sepharose contains the intact cyclopentanone ring, pentenyl side chain, and amide group (Figure 8A), which are identical to those of the JA moiety of JA-Ile. Therefore, the JA-linked sepharose structurally mimics the JA moiety of JA-Ile. However, JA is different from the JA moiety of JA-linked sepharose due to lack of the amide group and the molecular groups similar to the Ile moiety of JA-Ile or the 11-atom spacer arm of the JA-linked sepharose, which are able to fix the orientation during interaction between active JAs and COI1. As a control, we covalently immobilized acetic acid on the EAH-Sepharose 4B, resulting in acetic acid–linked sepharose without the JA moiety (Figure 8A).
Figure 8.
COI1 Binds Directly to the JA Moiety of JA-Ile.
(A) Structural diagrams of JA-linked sepharose and acetic acid–linked sepharose.
(B) Total wild-type plant protein extracts (Crude) were incubated with JA-linked sepharose (+) and acetic acid-linked sepharose (−). Protein bound to sepharose beads was washed, separated on SDS-PAGE, and immunoblotted with anti-COI1 antiserum.
(C) and (D) Total wild-type plant protein extracts (Crude) were loaded onto JA-linked sepharose (C) and acetic acid–linked sepharose (D) chromatography columns. After washing the columns, the columns were eluted with a stepwise gradient of NaCl from 150 to 900 mM. The flow-through peak (FT peak) and fractions corresponding to the stepwise elutions of NaCl were collected, and COI1 protein levels were detected with anti-COI1 antiserum. The black arrows indicate the COI1 protein band, the smaller band labeled by a star is a nonspecific protein that cross-reacted with the anti-COI1 antiserum.
(E) to (G) Total wild-type plant protein extracts (Crude) were incubated with JA-linked sepharose at 4°C for 3 h. Protein bound to sepharose beads was washed and resuspended in 100 μL extraction buffer for 1 h with different concentrations of JA (E), 2,4-D (F), and JA-Ile (G). The proteins in the supernatant were separated on SDS-PAGE and immunoblotted with anti-COI1 antiserum. The black arrows indicate the COI1 protein band.
[See online article for color version of this figure.]
We incubated total protein extracted from wild-type Arabidopsis plants with the JA- or acetic acid–linked sepharose beads and subsequently immunoblotted with anti-COI1 antiserum (Xu et al., 2002). Our results show that the JA-linked sepharose beads retained COI1, while the acetic acid–linked sepharose did not (Figure 8B). Next, we used column chromatography to further verify the binding between the COI1 protein and the JA-linked sepharose. We loaded the sepharose column with the total protein extracted from wild-type Arabidopsis plants, collected the flow-through peak, and then eluted the column with a stepwise gradient of NaCl from 150 to 900 mM. Using the JA-linked sepharose, we were unable to detect the COI1 protein in the flow-through peak and in the fractions eluted with 150 mM NaCl; however, we found that the COI1 protein was eluted from the JA-linked sepharose column by the washing buffer containing 200 and 250 mM NaCl (Figure 8C). These results confirmed that COI1 in the total protein extract binds the JA-linked sepharose. By contrast, with the acetic acid–linked sepharose column, the COI1 protein was detected in the flow-through fractions but not in the elution fractions with a stepwise gradient of NaCl from 150 to 900 mM (Figure 8D), suggesting that the COI1 protein was not retained by the acetic acid–linked sepharose column. Taken together, these results demonstrate that the COI1 protein effectively binds to the JA-linked sepharose but not the acetic acid–linked sepharose. As the JA-linked sepharose mimics the JA moiety of JA-Ile and exhibits an identical molecular orientation of JA-Ile bound to the surface pocket of COI1, the binding between COI1 and the JA-linked sepharose suggests that COI1 is able to directly bind to the JA moiety of JA-Ile.
The JA-linked sepharose beads retaining the COI1 protein were incubated with extraction buffer containing JA-Ile, JA, or 2,4-D, which is a synthetic auxin that binds directly to TIR1. Both JA and 2,4-D were unable to compete with the JA-linked sepharose for binding to COI1 (Figures 8E and 8F). However, the COI1 protein bound to the JA-linked sepharose beads was obviously eluted by JA-Ile at concentrations of 100 and 1000 μM (Figure 8G). These results demonstrate that the competition of JA-Ile with the JA-linked sepharose for binding to COI1 is specific and that both JA-Ile and the JA-linked sepharose are able to bind to the same site on COI1. Together with the data from molecular modeling of the interaction between JA-Ile and COI1 (Figure 6D), our results indicate that the JA moiety of JA-Ile directly binds to the surface pocket of COI1.
Surface Plasmon Resonance Technology Reveals the Interaction among COI1, JA-Ile, and JAZ1
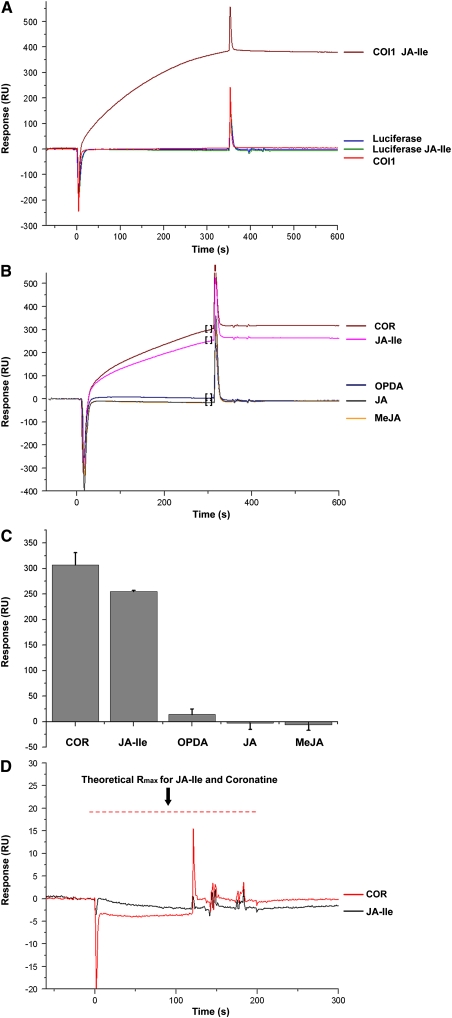
To rule out the possibility that other COI1 copurified proteins may be involved in JA-Ile perception, we used the purified COI1 to investigate the possible interactions among COI1, JAZ1, and JA via surface plasmon resonance (SPR) technology. The SPR technique is an established method to study the effect of ligand binding on protein dimerization (Yue et al., 2005). If the molecule of interest could bind to its interactant that was immobilized on the sensor chip covalently, it would introduce a shift in resonance angle, which is measured in resonance units (RUs) and recorded in real time. Higher RU denotes stronger binding, while quicker rise/fall of RU denotes faster association/dissociate rate.
We expressed His-tagged-COI1 protein (His-COI1) with insect cell lysates in vitro and purified the His-COI1 using Ni-NTA matrices (see Supplemental Figure S3 online). We also purified the insect-expressed His-tagged-luciferase protein (His-LUC) as a negative control. Approximately 1000 RUs of the purified JAZ1-His protein was immobilized onto the chip surface. His-LUC (100 μg/mL) was either directly injected over the sensor chip surface or preincubated with JA-Ile (1 μM) before the injection. As expected, His-LUC, with or without JA-Ile pretreatment, all gave low RU values and did not interact with the JAZ1-His protein (Figure 9A).When His-COI1 (100 μg/mL) was directly injected over the sensor chip surface, it gave low response and made no difference to the negative control (Figure 9A), demonstrating that COI1 alone could not interact with JAZ1. When His-COI1 was preincubated with JA-Ile (1 μM) before injection, the mixture gave significantly high response level (a maximum response around 380 RUs), indicating a significant interaction between the JA-Ile-bound His-COI1 with JAZ1-His (Figure 9A).
Figure 9.
Evaluation on Interactions of His-COI1 and/or JAs with JAZ1-His by Surface Plasmon Resonance.
(A) SPR sensorgrams for His-COI1 and His-LUC to interact with the sensor chip surface immobilized with JAZ1-His (1000 RUs). The samples at a concentration of 100 μg/mL were injected to the instrument without preincubation with JA-Ile or pretreated with 1 μM JA-Ile on ice for 2 h before the injection. Three replications were made.
(B) The His-COI1 protein at a concentration of 60 μg/mL was incubated with 1 μM COR, JA-Ile, OPDA, JA, and MeJA and injected over the JAZ1-His (1000 RUs) sensor chip surface.
(C) The statistical analysis of three replications made for each analyte in (B). The response for each measurement was determined by linear averaging in a 20-s time span starting at 30 s before the injection stop (data points in the square brackets in [B]). Bars represent the mean of three replicates and the error bars represent the sd.
(D) Sensorgrams of 100 μM of COR or JA-Ile running over the highly immobilized JAZ-His (5000 RUs) sensor chip surface. The theoretical maximum response (Rmax) for the COR and JA-Ile was calculated and shown in red dotted lines. Neither COR nor JA-Ile interacts with JAZ1-His. Three replications were made.
We also preincubated His-COI1 (60 μg/mL) with JA, MeJA, OPDA, and COR before flowing over the sensor chip surface. Only the COR-pretreated His-COI1 gave significant response level, even with higher RU values than that of JA-Ile–pretreated His-COI1 (Figures 9B and 9C). These results demonstrated that COR is more active than JA-Ile in triggering the COI1–JAZ interaction, which is consistent with our molecular modeling adopting GoldScore scoring function [51.95 for COR and 43.06 for (+)-7-iso-JA-Ile] and MM-PBSA binding free energy analysis [−78.33kJ mol−1 for COR and −47.68 kJ mol−1 for (+)-7-iso-JA-Ile].
We further made the CM5 sensor chip with a high immobilization level (5000 RUs) of JAZ1-His, which will increase the binding capacity for small molecules, to test whether JAZ1 could bind JA-Ile/COR. The theoretical maximum binding signal for JA-Ile/COR is ∼20 RUs (Figure 9D), which can be easily detected. Nonetheless, though a high concentration (100 μM) of JA-Ile/COR was used, the response level was around the baseline level (Figure 9D). Therefore, the JA-Ile/COR cannot bind with JAZ1, consistent with the previous data (Katsir et al., 2008).
To gain direct evidence that COI1 binds JA-Ile/COR, we made enormous efforts to immobilize His-COI1 to the chip surface. However, this attempt did not turn out to be successful, as His-COI1 lost its activity during the coupling procedures. To compensate this, we used photoaffinity labeling technology to validate the direct interaction of COI1 with COR.
The Photoaffinity Labeling Technology Reveals the Direct Binding of Coronatine to COI1
To test if COI1 directly interacts with JA-Ile/COR, we adopted a photoaffinity labeling technology, which serves as a very useful tool to identify a specific receptor for a small-molecule ligand and to localize the ligand binding site within the receptor (Qiu et al., 2007). The photoaffinity labeling technology has been used to successfully demonstrate the direct binding of brassinosteroids to the receptor BRI1 (Kinoshita et al., 2005).
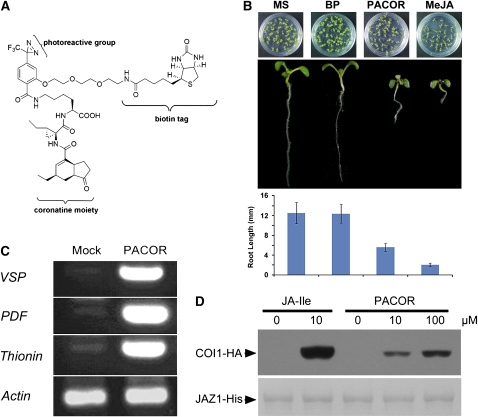
We have designed and synthesized a biotin-tagged photoaffinity probe for coronatine (PACOR) based on molecular modeling of the COI1–COR interaction and according to the careful analysis of structure-activity relationship of COR. PACOR is an analog of COR and comprises a coronatine moiety, a photoreactive group, and a biotin tag (Figure 10A). The coronatine moiety ferries PACOR to the binding site of the receptor; upon UV irradiation, the photoreactive group covalently binds to the receptor at the ligand binding region; and the biotin tag enables subsequent detection of PACOR-bound protein using an antibiotin antibody.
Figure 10.
Biotin-Tagged PACOR Is a Biologically Active JA Mimic.
(A) The structure of PACOR. The PACOR probe comprises a COR moiety, a photoreactive group, and a biotin tag. The COR moiety takes the probe to the binding site of the receptor. Upon UV irradiation, the photoreactive group covalently binds to the receptor. The biotin tag enables subsequent detection using an anti-biotin antibody.
(B) The growth of Arabidopsis seedlings was inhibited by MeJA and PACOR but not by BP. The top panel shows the phenotype of 10-d-old seedlings grown on MS medium or MS with 100 μM of BP, PACOR, or MeJA. The middle panel shows the phenotype of a single seedling from the top panel. The bottom panel is the statistical results of root length of the seedlings from the top panel. Error bars represent sd (n > 30). BP is a compound comprising a photoreactive group and a biotin tag and served as a negative control.
(C) The JA-inducible genes, vegetative storage protein gene 1 (VSP), PDF1.2 (PDF), and Thionin2.1 (Thionin), were induced by PACOR but not by its solvent (mock). The actin transcript was detected by RT-PCR and served as control.
(D) Interaction between COI1 and JAZ1 is promoted by PACOR at the concentrations indicated. Pull-down reactions were performed using recombinant JAZ1-His (bottom panel) and total plant extracts prepared from coi1-1 seedlings with transgenic expressed COI1-FH. The reactions were performed at 4°C for 1 h with PACOR or JA-Ile. Proteins bound to the Ni-NTA resin were washed, separated on SDS-PAGE, and immunoblotted with an anti-HA antibody (top panel). The PVDF membrane was stained with Memstain to visualize the recovery of JAZ1 by the Ni-NTA affinity resin (bottom panel).
Comprehensive physiological and biochemical studies showed that the PACOR probe conserved good biological activities (Figure 10). PACOR showed a clear inhibitory effect on the Arabidopsis seedling growth (Figure 10B) and exhibited significant induction on JA-inducible genes, including VSP, Thionin2.1, and PDF transcript (Figure 10C). Moreover, PACOR can trigger the interaction between COI1 and JAZ1 (Figure 10D).
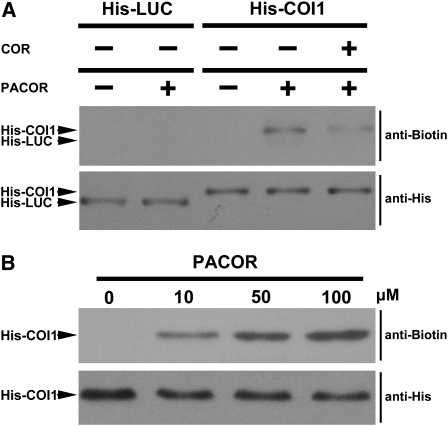
PACOR was incubated with the purified His-COI1 or His-LUC protein and subsequently exposed to UV irradiation. The reaction mixture was then separated on SDS-PAGE and immunoblotted with an antibiotin antibody. As expected, PACOR is unable to bind with the negative control His-LUC (Figure 11A). Figure 11 showed that the antibiotin antibody could clearly detect PACOR-bound His-COI1 and that the binding of PACOR to His-COI1 could be competed by the parent active compound COR (Figure 11A). These results showed that PACOR physically binds with the purified insect-expressed His-COI1. Together with genetic evidence (Figure 4; Feys et al., 1994; Xie et al., 1998; Xu et al., 2002), we demonstrated that COI1 specifically binds JA-Ile/COR and functions as a receptor for JA-Ile/COR.
Figure 11.
Biotin-Tagged PACOR Specifically Binds to His-COI1.
(A) The purified insect-expressed His-COI1 and His-LUC were incubated with (+) or without (−) PACOR (50 μM) in the presence (+) or absence (−) of coronatine (500 μM) at 4°C for 1 h and then photolabeled by exposure to UV. The PACOR-labeled proteins were separated on SDS-PAGE and detected with an antibiotin antibody (top panel). His-COI1 and His-LUC were detected with an anti-His antibody (bottom panel). The protein position was indicated in the left side of each panel.
(B) His-COI1 was incubated with various concentrations of PACOR and then photolabeled by exposure to UV. The PACOR-labeled His-COI1 was detected with an antibiotin antibody (top panel). His-COI1 was detected with an anti-His antibody (bottom panel).
DISCUSSION
The F-box protein COI1, which is able to interact with several proteins, including JAZs (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007), ASK1, ASK2, Cullin1, and Rbx1 (Xu et al., 2002; Wang et al., 2005), was found to be a component of the complex that binds to JAs. The JA binding assays in previous studies were performed with crude plant extracts (Thines et al., 2007; Katsir et al., 2008; Melotto et al., 2008) or partially purified proteins (Fonseca et al., 2009), where COI1 undoubtedly associates with other proteins that copurified with it. In this study, we demonstrated that COI1 directly interacts with JA-Ile/COR and functions as a receptor for JA.
We built a high-quality structure model of COI1 (Figure 1) and tested its ability of binding to JAs through molecular modeling (Figures 6 and 7). We found that COI1 harbors a surface pocket (Figure 2), which could be a potential binding site for JA-Ile (Figure 6D) or COR (Figure 7B). The direct binding of COI1 with JA-Ile or COR was further investigated by immobilized JA approach, SPR technology, and photoaffinity labeling technology.
First, we prepared JA-linked sepharose that mimics the JA moiety of JA-Ile based on our molecular modeling of COI1-JA-Ile interaction (Figure 6D). We found that the JA-linked sepharose retained COI1 in crude plant extracts and that JA-Ile was able to specifically compete with the JA-linked sepharose for binding to COI1 (Figure 8). These results imply that COI1 is likely to bind with the JA moiety of JA-Ile. However, our immobilized JA binding assay was performed with crude plant extracts, which cannot exclude the possibility that other COI1-interacting proteins may contribute to this interaction.
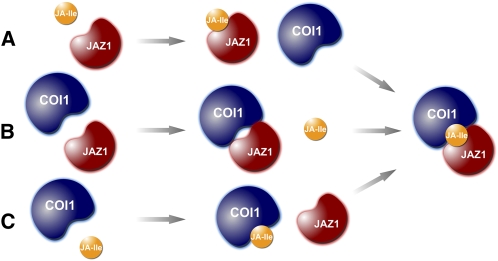
Second, we employed SPR technology with purified COI1 and JAZ1 protein. We found that the three components, COI1, JA-Ile, and JAZ1, are sufficient to form a complex (Figures 9A and 9B), which excludes the possibility that other COI1-interacting proteins may contribute to JA-Ile perception. We also used SPR to verify that JAZ1 alone could neither bind to JA-Ile/COR (Figure 9D) nor COI1 (Figure 9A), which excludes the possibilities that JAZ1 alone binds JA-Ile/COR (Figure 12A; Katsir et al., 2008) and that the JAZ1 and COI1 assemble a complex subsequently to bind with JA-Ile/COR (Figure 12B). Furthermore, we proved that the preincubation of COI1 with JA-Ile or COR is essential for COI1 to interact with JAZ1, which suggests the possibility that COI1 directly binds to JA-Ile/COR and then interacts with JAZ1 (Figure 12C). To determine the direct interaction between His-COI1 and JA-Ile/COR using the SPR technology, we have made enormous efforts to immobilize His-COI1 on the sensor chip. However, His-COI1 lost its activity upon covalently binding to the sensor chip surface, which may be caused by the harsh pH/ironic strength conditions or immobilization of protein residues during the coupling procedures.
Figure 12.
Three Possibilities for Assembly of the COI1-JA-Ile-JAZ1 complex.
(A) JAZ1 binds JA-Ile and subsequently binds to COI1.
(B) JAZ1 and COI1 form a complex that subsequently binds to JA-Ile.
(C) COI1 binds JA-Ile and subsequently binds to JAZ1.
[See online article for color version of this figure.]
Finally, we used the photoaffinity labeling technology to gain the direct evidence that COI1 binds JA-Ile/COR. The photoaffinity labeling technology has the advantage to form a permanent covalent bond between the probe and its receptor through photolysis, which allows us to efficiently and directly detect the probe-labeled receptor (Qiu et al., 2007). We designed and synthesized PACOR (Figure 10A) based on molecular modeling of COI1–COR interaction (Figure 7B) and the careful analysis of coronatine structure-activity relationship. We found that the PACOR probe, which conserved reasonable biological activities (Figure 10), could bind to purified insect-expressed His-COI1 (Figure 11). Application of the photoaffinity labeling technology mainly relies on the successful synthesis of probes with biological activities. The photoaffinity labeling probes may bind to various proteins; however, specific binding of probes with their receptors can be competed by their parent molecules while nonspecific binding with other proteins cannot. We found that the PACOR probe could not bind to the negative control His-LUC (Figure 11A); however, we could not exclude the possibility that PACOR, like other photoaffinity labeling probes, may also nonspecifically bind to other proteins. To test whether PACOR could specifically bind to His-COI1, we further showed that the binding of PACOR to His-COI1 could be competed by the parent active compound COR (Figure 11A). In summary, we successfully synthesized PACOR (Figure 10A) and verified the specific binding of PACOR to His-COI1 protein (Figure 11) in an activity-based manner (Figure 10). Taken together with the genetic evidence (Figure 4; Feys et al., 1994; Xie et al., 1998; Xu et al., 2002), we demonstrated that COI1 directly binds to JA-Ile/COR and functions as a receptor for JA-Ile/COR.
Based on the structure model of COI1, molecular modeling of COI1-JA interactions, and the evidence that COI1 directly binds JA-Ile/COR, we propose a model for JA signal perception. As illustrated in Figure 13, JA-Ile binds to the COI1 surface pocket probably via three important functional groups in the JA moiety of JA-Ile. The keto group of the cyclopentanone ring of JA-Ile may interact with two COI1 Arg residues (Figure 6D) from the P1 pocket by forming stable hydrogen bonds. Meanwhile, the pentenyl side chain of JA-Ile may stack on the P2 region of the COI1 surface pocket through hydrophobic interactions and van der Waals contacts. The oxygen atom of the amide group of JA-Ile may interact with two COI1 Tyr residues (Figure 6D) in the bottleneck region via hydrogen bonds. Together, these three interactions may anchor JA-Ile into COI1 (Figure 13) and form a new interface, which comprises the Ile moiety of JA-Ile, the P3 and P4 pockets of the surface pocket on CO11 (Figure 13), for interaction with JAZs. The P3 pocket mainly contains hydrophilic residues (Figure 2), whereas the P4 pocket contains hydrophobic residues (Figure 2). These features of the P3 and P4 pockets may imply that both hydrophobic and electrostatic interactions are involved in the interaction with JAZs, which is different from the auxin perception mode in which only hydrophobic interaction was involved (Tan et al., 2007). Elucidation of a more detailed mechanism for JA perception will be the subject of future studies.
Figure 13.
Model of JA-Ile Binding to COI1.
JA-Ile binds to the COI1 pocket via three important functional groups in the JA moiety of JA-Ile: the keto group of the cyclopentanone ring, the pentenyl side chain, and the oxygen atom of the amide group. The keto group of the cyclopentanone ring interacts with the P1 pocket by forming hydrogen bonds. The pentenyl side chain of JA-Ile stacks in the P2 pocket through hydrophobic interactions and van der Waals contacts. In the bottleneck region of the surface pocket, the oxygen atom of the amide group interacts with two Tyr residues via hydrogen bonds. Together, the three interactions anchor JA-Ile to the COI1 protein. The Ile moiety of JA-Ile may fix the molecular orientation during interaction between JA-Ile and COI1. The Ile moiety of JA-Ile with the P3 and P4 pockets may form a new interface for interaction with JAZs.
[See online article for color version of this figure.]
METHODS
Plant Materials and Growth Conditions
Seedlings were grown on MS plates containing 2% (w/v) sucrose and used for experiments 10 d after sowing. The wild-type Arabidopsis thaliana Columbia (Col-0), coi1-1, and other mutant plants were grown in soil under a light/dark cycle of 16/8 h at 22°C.
Transgenic Lines
We made the COI1-FH construct by a PCR amplification method with the HA epitope sequence (YPYDVPDYA) fused in frame before the COI1 stop codon and with the FLAG epitope sequence (DYKDDDDK) fused in frame after the COI1 start codon. The full-length COI1-FH fragment was cloned into pROK2 under control of the cauliflower mosaic virus 35S promoter. We transformed the COI1-FH construct into coi1-1 plants and found that COI1-FH fully complemented the coi1-1 mutation.
Homology Modeling
All procedures in the homology modeling of COI1 were performed in the Accelrys Discovery Studio 1.7 package (Accelrys). After sequence alignment by the Align123 program, the correct alignment was delivered to the MODELER module. Overhangs in the N terminus (14 residues) and C terminus (two residues) of COI1 were truncated. The Arabidopsis TIR1 (PDB ID: 2P1M, chain B) was used as the template. To sufficiently sample the side chain orientations, 15 models were generated and the loop regions were optimized. For further refinement, residues with improper parameters were picked out and energy minimized by the Dreiding force field (Mayo et al., 1990). Hydrogen atoms were added to the model, and the protonation states of amino acid side chains were set to their typical pKa values to ensure the neutrality of the protein. CHARMm force field (Brooks et al., 1983) parameters were assigned to every atom. Several rounds of energy minimization runs were performed using steepest descent and conjugated gradient algorithms, from fixing the heavy atoms to gradually releasing the whole structure (the sequence was loop side chain, core side chain, and backbone). In the MD simulations, the structure was immersed in a cubic water box (TIP3 water model) with explicit periodic boundaries. The assembled molecules were energy minimized by steepest descent and conjugated gradient algorithms and underwent a dynamics cascade, including heating, equilibration, and production stages for 100, 100, and 200 ps, respectively. The temperature of the system was set to 300K, and the NVT ensemble was adopted in the production stage. During the MD simulations, fixed atom constraints were applied to backbone atoms. At the end of the run, the side chains were further minimized with 3000 steps of the steepest descent method to remove bad contacts. Verify3D, WHATIF, PROCHECK, and ProSA2003 were used to evaluate the quality of the acquired structure.
Molecular Modeling of Interactions between JAs and COI1
Automated docking simulations were performed with the genetic algorithm-based program GOLD 3.0.1 (Cambridge Crystallographic Data Centre). An area with a radius of 12 Å centered at residue Tyr-386 was defined as the binding site origin, and the detect cavity algorithm was used (Hendlich et al., 1997). Each JA underwent 100 docking runs, and the GoldScore fitness function was adopted. The parameters for the genetic algorithms included the following: population size 100, selection pressure 1.1, number of operations 105, number of islands 5, niche size 2, migrate 10, mutate 95, and crossover 95. The best result was selected by conformational clustering analysis and the scoring function. To refine the docked solution, in situ ligand minimization was performed using the CHARMm force field in Discovery Studio 1.7. The COI1-JA complex was subjected to restricted MD simulations. The system was solvated using a sphere boundary condition, with SHAKE constraints applied. The system was then minimized using 1500 steps of the conjugated gradient method, heated to 300K, and equilibrated for 50 ps. The production phase of the restricted MD simulations lasted for 1 ns, with a time step of 2 fs. During this MD simulation, the CHARMm force field was applied to every atom in the system, and only residues with at least one atom within a 7-Å radius of the ligand were free to move. Extraction and analysis of the resulting trajectories were performed, and PyMOL 0.98 package (DeLano Scientific) was used to perform ray tracing and graphic representation.
MM-PBSA Analysis
Snapshot structures of the COI1 protein, JA, and the COI1-JA complex were taken from the MD trajectory of the COI1-JA complex. The binding free energies are averages from 50 snapshots. The binding free energy (ΔGbind) is the sum of average molecular mechanical gas-phase energies (ΔEMM), salvation-free energies (ΔΔGsolv), and entropy contributions (-TΔS) of the binding reaction (ΔGbind =ΔEMM + ΔΔGsolv – TΔS). The ΔEMM term was calculated using the CHARMm force field, while the solvation free energy (ΔGsolv) was estimated as the sum of electrostatic solvation free energy (ΔGpb) and nonpolar solvation free energy (ΔGnp). ΔGpb was calculated by the PBEQ program in CHARMm, and ΔGnp was calculated from the solvent accessible surface area (SASA) using the equation (ΔGnp = γSASA + β), where γ = 0.0227 kJ mol−1 Å−2, and β = 3.85 kJ mol−1. The entropy term (–TΔS) was not included in our calculation because it does not contribute much to the relative binding free energies of the JAs to the COI1 protein.
Pull-Down Assay
The procedure for protein pull-down experiments was as described previously (Thines et al., 2007). Total protein was extracted from seedlings in a buffer containing 50 mM Tris-Cl, pH 7.8, 100 mM NaCl, 25 mM imidazole, 10% (v/v) glycerol, 0.1% (v/v) Tween 20, 20 mM 2-mercaptoethanol, 10 mM MG132, and the EDTA-free complete miniprotease inhibitor cocktail according to the manufacturer's instructions (Roche). Cell debris was removed by centrifugation at 12,000g for 15 min. The total protein concentration was estimated by the Bradford assay (Bio-Rad). Recombinant JAZ1-His was expressed in Escherichia coli and purified by Ni affinity chromatography according to manufacturer's instructions (Qiagen). Fifty micrograms of JAZ1-His was incubated with 5 mg of total plant extract, with gentle rocking, for 1 h at 4°C in the absence or presence of 10 μM JA-Ile, which was ordered from BioDuro Beijing and used as a mixture of isomers. Ni-NTA resin was recovered by a brief centrifugation and washed four times with 1 mL of washing buffer (50 mM Tris-Cl, pH 7.8, 100 mM NaCl, 25 mM imidazole, 10% [v/v] glycerol, 0.1% [v/v] Tween 20, and 20 mM 2-mercaptoethanol). Pull-down mixtures were separated on SDS-PAGE, transferred to PVDF membrane, and detected with an anti-HA antibody (Sigma-Aldrich) or anti-COI1 antiserum (Xu et al., 2002).
Preparation of JA-linked Sepharose
JA-linked sepharose was prepared according to the manufacturer's protocol (GE Healthcare). (±)-JA (500 mg; Sigma-Aldrich) was coupled to prewashed EAH-Sepharose 4B (10 mL) with 5 g of 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (Sigma-Aldrich) as the condensing reagent. The reaction was performed with shaking for 20 h at 4°C in the dark. The pH was occasionally adjusted with NaOH to 4.5.
The amount of JA that coupled to the EAH-Sepharose 4B was determined by assaying incorporation of tritiated jasmonic acid. Five milligrams of (±)-JA was coupled to prewashed EAH-Sepharose 4B (0.1 mL) in the presence of 100 nM tritiated JA in 50% (v/v) dioxane (1 mL). After the coupling reaction, the radioactivity of the sepharose was quantified by scintillation counting.
JA-Linked Sepharose Binding Assay
Total protein from 5 g of seedlings was extracted with 15 mL extraction buffer (50 mM Tris-Cl, pH 7.4, 100 mM NaCl, 5 mM KCl, 1 mM DTT, 10 mM MG132, and the EDTA-free complete miniprotease inhibitor cocktail according to the manufacturer's instructions [Roche]). The extract was centrifuged at 12,000g for 15 min. The total protein concentration was estimated by the Bradford assay (Bio-Rad). Two milligrams of total plant extracts were incubated with prewashed JA-linked sepharose (50 μL) with gentle rocking for 3 h at 4°C. JA-linked sepharose beads were recovered by a brief centrifugation and washed five times with 1 mL of extraction buffer. The beads were resuspended in 100 μL of 1× sample buffer. The bound COI1 was detected by immunoblot assay.
The competitive elution experiment was performed with the JA-linked sepharose beads retaining the COI1 protein. The JA-linked sepharose beads were recovered after incubation, washed, and resuspended in 100 μL extraction buffer in the absence or presence of JA, 2,4-D, or JA-Ile. The mixtures were incubated for 1 h at 4°C, and the COI1 protein in the supernatant was detected by immunoblot assay. All experiments were repeated three times using total protein from independent extraction.
The affinity chromatography procedure was performed at 4°C. The total wild-type plant protein extracts were prepared with the extraction buffer (50 mM Tris-Cl, pH 7.4, 100 mM NaCl, 5 mM KCl, 1 mM DTT, 10 mM MG132, and the EDTA-free complete miniprotease inhibitor cocktail according to the manufacturer's instructions [Roche]) and loaded onto the JA-linked sepharose column or the acetic acid–linked sepharose column that were preequilibrated with a buffer containing 50 mM Tris-Cl, pH 7.4, 100 mM NaCl, 5 mM KCl, and 1 mM DTT. After the flow-through peak was collected, the column was washed with a stepwise gradient of NaCl from 150 to 900 mM in buffer containing 50 mM Tris-Cl, pH 7.4, 100 mM NaCl, 5 mM KCl, and 1 mM DTT. Eluted fractions were dialyzed against 50 mM Tris-Cl, pH 7.4, and detected by immunoblot assay.
In Vitro Synthesis of 6×His-COI1 in Insect Cell Lysates
6×His-tagged COI1 (His-COI1) was synthesized in vitro using insect cell lysates (Qiagen) in accordance with the manufacturer's instructions. The COI1 DNA template for in vitro synthesis was generated using the EasyXpress linear template kit (Qiagen). The COI1 coding sequence was amplified with forward primer 5′-ACCCACGCGCATGTCGTAAAAAGCACCCAAGAGGATCCTGATATCAAGA-3′ and reverse primer 5′-CTTGGTTAGTTAGTTATTATATTGGCTCCTTCAGGAC-3′. The His-COI1 was purified by two runs of Ni affinity chromatography. 6×His tagged luciferase (His-LUC) DNA template for in vitro synthesis was supplied by the insect cell lysates (Qiagen). The purified 6×His-tagged COI1 protein was verified with anti-His antibody (Sigma-Aldrich) and the antiserum against COI1 (Xu et al., 2002).
SPR Assays
The binding affinities of His-COI1 with JAZ1-His were evaluated by a Biacore X100 instrument. Immobilization of the proteins on the sensor chip CM5 was performed using the primary amine coupling kit provided by Biacore. During the immobilization procedures, JAZ1-His protein was solvated in 10 mM sodium acetate buffer, pH 4.0, to a final concentration of 60 μg/mL before being covalently bound to the sensor chip surface. The running buffer used in our experiment was modified HBS buffer (10 mM HEPES, 100 mM NaCl, 5 mM KCl, and 0.005% [v/v] surfactant P20, pH 7.4). All the data were collected at 25°C with a running buffer flow rate of 5 μL/s. Before injection, sample protein was diluted to the corresponding concentration using the running buffer. For regeneration runs, 5 mM of Glycine-HCl buffer at pH 2.0 was injected to wash the remaining proteins on the sensor chip surface. For pretreatment of His-COI1 protein, 1 μM of JA, MeJA, OPDA, JA-Ile, and COR were added to the protein sample and incubated on ice for 2 h before injection.
Photoaffinity Labeling
Biotin-tagged PACOR was synthesized from COR (see Supplemental Figure 4 online). Proton NMR spectra were recorded at 300 MHz in CDCl3 as follows: δ 0.93(m,6H), 1.24(m,6H), 1.41(m,6H), 1.61(m,7H), 1.79(m,5H), 2.13(m,3H), 2.36(m,3H), 2.67(m,1H), 2.88(m,1H), 3.13(m,2H), 3.41(m,4H), 3.53(m,2H), 3.63(m,2H), 3.68 (m,2H), 3.89 (t,2H), 4.29 (m,3H), 4.48 (m,2H), 4.88 (s,1H), 6.13 (s,1H), 6.42 (s,1H), 6.62 (s,1H), 6.69 (s,1H), 6.89 (d,1H), 7.45 (d,1H), 7.62 (s,1H), 8.02 (t,1H), 8.16 (d,1H). Electrospray ionization-mass spectrometry 1031.3 [M-H]-.
For root length measurement experiments, seedlings were grown on MS medium supplemented with 100 μM of PACOR for 10 d before measurement. To test the inducible ability on the accumulation of JA-dependent gene expression, seedlings were grown on MS medium with 2% sucrose for 2 to 3 weeks and then treated with 100 μM of PACOR or water for 8 h. The total RNAs extracted from these materials were used in reverse transcription by M-MLV (Invitrogen). The first-strand cDNA was used as the template for the subsequent RT-PCR. RT-PCR was performed to amplify the Vegetative Storage Gene 1 (using primers 5′-GAACTCGGGATTGAACCCAT-3′ and 5′-GCTTAAAAACCCTTCCAGGAGTA-3′), PDF1.2 (5′-TTTGCTGCTTTCGACGCAC-3′ and 5′-TAACATGGGACGTAACAGATA-3′), and Thionin2.1 (5′-CAAGTTCAAGTAGAAGCAAAGA-3′ and 5′-CCGACGCACCATTCACAA -3′) transcripts with 25 PCR cycles. Actin (5′-CACCGCTTAACCCGAA-3′ and 5′-GTGAGGTCACGACCAG-3′) was also amplified as a control.
The labeling reaction was initiated by incubation of purified His-COI1 or His-LUC with a combination of PACOR and COR, as indicated in Figure 11, in buffer (50 mM Tris-Cl, pH 7.8, 100 mM NaCl, 25 mM imidazole, 10% [v/v] glycerol, 0.1% [v/v] Tween 20, and 20 mM 2-mercaptoethanol) at 4°C for 1 h and then exposed to UV at 365 nm (220 v, 6 w, 365 nm) at a distance of 3 cm for 30 min. After UV irradiation, the labeling mixtures were separated on SDS-PAGE, transferred to PVDF membrane, and detected with an anti-Biotin antibody (Jackson ImmunoResearch) or anti-His antibody (Sigma-Aldrich).
Accession Numbers
The Arabidopsis Genome Initiative accession numbers for the genes and gene products mentioned in this article are as follows: COI1 (At2g39940), JAZ1 (At1g19180), VSP (AT5G24780), PDF1.2 (AT5G44420), Thionin2.1 (AT1G72260), Actin (AT2G37620), and TIR1 (At3g62980). The Protein Data Bank accession number for TIR1 is 2p1m.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Evaluation of the COI1 Structural Model.
Supplemental Figure 2. Molecular Modeling of the Interaction between COI1 and (−)-JA-Ile.
Supplemental Figure 3. Expression and Purification of His-COI1 in Insect Cell-Free Expression System.
Supplemental Figure 4. The Synthesis of Biotin-Tagged Photoaffinity Probe for Coronatine (PACOR).
Supplemental Table 1. Calculated Average Binding Free Energies (in kJ mol-1) for Jasmonates.
Supplementary Material
Acknowledgments
We thank Lianhui Wang for helpful suggestion in preparation of the JA-linked sepharose. We also thank Zhaohu Li for providing COR for the synthesis PACOR. We also thank Zhishu Huang for technical assistance in the molecular modeling. This work was financially supported by the National Science Foundation of China, The National Basic Research 973 Program of China, and the Tsinghua-Yue-Yuen Medical Sciences Fund and Grant from Chinese Academy of Sciences.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Daoxin Xie (daoxinlab@tsinghua.edu.cn).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Bender, C.L., Alarcon-Chaidez, F., and Gross, D.C. (1999). Pseudomonas syringae phytotoxins: Mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63: 266–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, B.R., Bruccoleri, R.E., Olafson, B.D., States, D.J., Swaminathan, S., and Karplus, M. (1983). CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4: 187–217. [Google Scholar]
- Cheng, H., Song, S.S., Xiao, L.T., Soo, H.M., Cheng, Z.W., Xie, D.X., and Peng, J.R. (2009). Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 5: e1000440. [DOI] [PMC free article] [PubMed]
- Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J.M., Lorenzo, O., Garcia-Casado, G., Lopez-Vidriero, I., Lozano, F.M., Ponce, M.R., Micol, J.L., and Solano, R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. [DOI] [PubMed] [Google Scholar]
- Creelman, R.A., and Mullet, J.E. (1997). Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 355–381. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, N., Dharmasiri, S., and Estelle, M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. [DOI] [PubMed] [Google Scholar]
- Farmer, E.E. (2001). Surface-to-air signals. Nature 411: 854–856. [DOI] [PubMed] [Google Scholar]
- Farmer, E.E., Almeras, E., and Krishnamurthy, V. (2003). Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Curr. Opin. Plant Biol. 6: 372–378. [DOI] [PubMed] [Google Scholar]
- Farmer, E.E., Johnson, R.R., and Ryan, C.A. (1992). Regulation of expression of proteinase inhibitor genes by methyl jasmonate and jasmonic acid. Plant Physiol. 98: 995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feussner, I., and Wasternack, C. (2002). The lipoxygenase pathway. Annu. Rev. Plant Biol. 53: 275–297. [DOI] [PubMed] [Google Scholar]
- Feys, B.F., Benedetti, C.E., Penfold, C.N., and Turner, J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male-sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca, S., Chini, A., Hamberg, M., Adie, B., Porzel, A., Kramell, R., Miersch, O., Wasternack, O., and Solano, R. (2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5: 344–350. [DOI] [PubMed] [Google Scholar]
- Hendlich, M., Rippmann, F., and Barnickel, G. (1997). LIGSITE: Automatic and efficient detection of potential small molecule-binding sites in proteins. J. Mol. Graph. Model. 15: 359–363. [DOI] [PubMed] [Google Scholar]
- Jones, G., Willett, P., Glen, R.C., Leach, A.R., and Taylor, R. (1997). Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 267: 727–748. [DOI] [PubMed] [Google Scholar]
- Katsir, L., Schilmiller, A.L., Staswick, P.E., He, S.Y., and Howe, G.A. (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105: 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski, S., and Leyser, O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Cano-Delgado, A., Seto, H., Hiranuma, S., Fujioka, S., Yoshida, S., and Chory, J. (2005). Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433: 167–171. [DOI] [PubMed] [Google Scholar]
- Koda, Y., Takahashi, K., Kikuta, Y., Greulich, F., Toshima, H., and Ichihara, A. (1996). Similarities of the biological activities of coronatine and coronafacic acid to those of jasmonic acid. Phytochemistry 41: 93–96. [Google Scholar]
- Kollman, P.A., et al. (2000). Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 33: 889–897. [DOI] [PubMed] [Google Scholar]
- Laskowski, R., MacArthur, M., Moss, D., and Thornton, J. (1993). PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26: 283–291. [Google Scholar]
- Liu, F., Ni, W., Griffith, M., Huang, Z., Chang, C., Peng, W., Ma, H., and Xie, D. (2004). The ASK1 and ASK2 genes are essential for Arabidopsis early development. Plant Cell 16: 5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo, O., and Solano, R. (2005). Molecular players regulating the jasmonate signalling network. Curr. Opin. Plant Biol. 8: 532–540. [DOI] [PubMed] [Google Scholar]
- Luthy, R., Bowie, J.U., and Eisenberg, D. (1992). Assessment of protein models with three-dimensional profiles. Nature 356: 83–85. [DOI] [PubMed] [Google Scholar]
- Mayo, S.L., Olafson, B.D., and Goddard, W.A. (1990). DREIDING: A generic force field for molecular simulations. J. Phys. Chem. 94: 8897–8909. [Google Scholar]
- McConn, M., and Browse, J. (1996). The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8: 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn, M., Creelman, R.A., Bell, E., Mullet, J.E., and Browse, J. (1997). Jasmonate is essential for insect defense Arabidopsis. Proc. Natl. Acad. Sci. USA 94: 5473–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto, M., Mecey, C., Niu, Y.J., Chung, H.S., Katsir, L., Yao, J., Zeng, W.Q., Thines, B., Staswick, P., Browse, J., Howe, G.A., and He, S.Y. (2008). A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 55: 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, R.E. (1982). Coronatine production by some phytopathogenic pseudomonads. Physiol. Plant Pathol. 20: 83–89. [Google Scholar]
- Moon, J., Parry, G., and Estelle, M. (2004). The ubiquitin-proteasome pathway and plant development. Plant Cell 16: 3181–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, W.W., Xu, J., Li, J.Y., Li, J., and Nan, F.J. (2007). Activity-based protein profiling for type I methionine aminopeptidase by using photo-affinity trimodular probes. ChemBioChem 8: 1351–1358. [DOI] [PubMed] [Google Scholar]
- Ren, C., Pan, J., Peng, W., Genschik, P., Hobbie, L., Hellmann, H., Estelle, M., Gao, B., Peng, J., Sun, C., and Xie, D. (2005). Point mutations in Arabidopsis Cullin1 reveal its essential role in jasmonate response. Plant J. 42: 514–524. [DOI] [PubMed] [Google Scholar]
- Reymond, P., and Farmer, E.E. (1998). Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1: 404–411. [DOI] [PubMed] [Google Scholar]
- Sali, A., Potterton, L., Yuan, F., Vanvlijmen, H., and Karplus, M. (1995). Evaluation of comparative protein modeling by modeler. Proteins 23: 318–326. [DOI] [PubMed] [Google Scholar]
- Schommer, C., Palatnik, J.F., Aggarwal, P., Chetelat, A., Cubas, P., Farmer, E.E., Nath, U., and Weigel, D. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6: 1991–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, X., Zhang, Y., Peng, W., Wang, Z., and Xie, D. (July 12, 2009). Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J. Exp. Bot., http://dx.doi.org/10.1093/jxb/erp223. [DOI] [PubMed]
- Sippl, M.J. (1993). Recognition of errors in three-dimensional structure of proteins. Proteins 17: 355–362. [DOI] [PubMed] [Google Scholar]
- Staswick, P.E., Su, W., and Howell, S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89: 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, X., Calderon-Villalobos, L.A., Sharon, M., Zheng, C.X., Robinson, C.V., Estelle, M., and Zheng, N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645. [DOI] [PubMed] [Google Scholar]
- Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G.H., Nomura, K., He, S.Y., Howe, G.A., and Browse, J. (2007). JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448: 661–665. [DOI] [PubMed] [Google Scholar]
- Ueda, J., and Kato, J. (1980). Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium L.). Plant Physiol. 66: 246–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriend, G. (1990). WHAT IF: A molecular modeling and drug design program. J. Mol. Graph. 8: 52–56. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Dai, L., Jiang, Z., Peng, W., Zhang, L., Wang, G., and Xie, D. (2005). GmCOI1, a soybean F-box protein gene, shows ability to mediate jasmonate-regulated plant defense and fertility in Arabidopsis. Mol. Plant Microbe Interact. 18: 1285–1295. [DOI] [PubMed] [Google Scholar]
- Wasternack, C. (2007). Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. (Lond.) 100: 681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C., and Hause, B. (2002). Jasmonates and octadecanoids: Signals in plant stress responses and development. Prog. Nucleic Acid Res. Mol. Biol. 72: 165–221. [DOI] [PubMed] [Google Scholar]
- Weiler, E.W., Kutchan, T.M., Gorba, T., Brodschelm, W., Niesel, U., and Bublitz, F. (1994). The Pseudomonas phytotoxin coronatine mimics octadecanoid signalling molecules of higher plants. FEBS Lett. 345: 9–13. [DOI] [PubMed] [Google Scholar]
- Xie, D., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xu, L., Liu, F., Lechner, E., Genschik, P., Crosby, W.L., Ma, H., Peng, W., Huang, D., and Xie, D. (2002). The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Y.X., Stolz, S., Chetelat, A., Reymond, P., Pagni, M., Dubugnon, L., and Farmer, E.E. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, L.D., Ye, F., Gui, C.S., Luo, H.B., Cai, J.H., Shen, J.H., Chen, K.X., Shen, X., and Jiang, H.L. (2005). Ligand-binding regulation of LXR/RXR and LXR/PPAR heterodimerizations: SPR technology-based kinetic analysis correlated with molecular dynamics simulation. Protein Sci. 14: 812–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.