Abstract
RPM1-interacting protein 4 (RIN4), a negative regulator of the basal defense response in plants, is targeted by multiple bacterial virulence effectors. We show that RIN4 degradation is induced by the effector AvrPto from Pseudomonas syringae and that this degradation in Solanaceous plants is dependent on the resistance protein, Pto, a protein kinase, and Prf, a nucleotide binding site–leucine-rich repeat protein. Our data demonstrate overlap between two of the best-characterized pathways for recognition of pathogen virulence effectors in plants. RIN4 interacts with multiple plant signaling components and bacterial effectors in yeast and in planta. AvrPto induces an endogenous proteolytic activity in both tomato (Solanum lycopersicum) and Nicotiana benthamiana that degrades RIN4 and requires the consensus site cleaved by the protease effector AvrRpt2. The interaction between AvrPto and Pto, but not the kinase activity of Pto, is required for proteolysis of RIN4. Analysis of many of the effectors comprising the secretome of P. syringae pv tomato DC3000 led to the identification of two additional sequence-unrelated effectors that can also induce degradation of RIN4. Therefore, multiple bacterial effectors besides AvrRpt2 elicit proteolysis of RIN4 in planta.
INTRODUCTION
Both plants and animals have defense systems comprised of basal defense responses triggered by detection of common components of potential pathogens (pathogen-associated molecular pattern [PAMP]-triggered immunity) as well as more specific defenses induced by recognition of ligands produced by individual pathogens (effector-triggered immunity) (Jones and Dangl, 2006; McDowell and Simon, 2008). While in vertebrates the adaptive immune system has evolved to supplement innate immunity, plants have elaborated their resistance machinery to provide defense responses that are qualitatively and quantitatively more extreme than the PAMP-elicited resistance (Jones and Takemoto, 2004; Jones and Dangl, 2006). In animals, Toll-like receptors and nucleotide binding oligomerization domain (NOD) proteins extracellularly and intracellularly recognize conserved ligands shared by many pathogens and trigger innate immunity (Athman and Philpott, 2004). There are ∼25 NOD-encoding genes in mammals, and their predominant role seems to be in innate immunity (Inohara and Nunez, 2003). In plants, FLAGELLIN SENSITIVE2, a Toll-like receptor-like protein, recognizes flagellin from multiple bacterial pathogens and triggers PAMP-elicited resistance (Gomez-Gomez et al., 2001; Göhre and Robatzek, 2008). The number of genes encoding a nucleotide binding site (NBS) domain, the equivalent of the animal NOD domain, is much greater in plants than animals; however, NBS-encoding genes have so far been shown to be involved in recognition of specific pathogen ligands rather than in PAMP-elicited resistance (Meyers et al., 2003).
To be successful, pathogens must suppress the induction of PAMP-elicited resistance. This has been best characterized for pathogenic Gram-negative bacteria, some of which have been shown to secrete up to 40 effectors into the cytosol of their plant or animal hosts (Chang et al., 2004; Lindeberg et al., 2006; Schechter et al., 2006; Vinatzer et al., 2006). Some of these effectors target components of the defense signaling pathways, and plants have evolved to detect perturbations in these pathways (Mudgett, 2005; Grant et al., 2006; Zhou and Chai, 2008). Specific recognition of a bacterial effector elicits a series of plant defense responses often including callose deposition, the release of active oxygen species, induction of defense gene expression, and the hypersensitive response (HR). These defense responses limit pathogen invasion of the plant (Dangl and Jones, 1998).
In one of the best-characterized plant–pathogen interactions, the effectors AvrRpm1 from Pseudomonas syringae pv maculicola and AvrB from P. syringae pv glycinea cause the phosphorylation of RPM1-interacting protein 4 (RIN4), a negative regulator of innate immunity in Arabidopsis thaliana (Mackey et al., 2002; Kim et al., 2005b), which is then detected by the NBS-leucine-rich repeat (LRR) protein RPM1; this recognition in turn leads to the HR. Elicitation of the HR by RPM1 is abrogated by the proteolytic cleavage of RIN4 by another bacterial effector, AvrRpt2, from P. syringae pv tomato; however, the disappearance of RIN4 can be detected by a second NBS-LRR protein, RPS2, and, thereby, elicit the HR (Axtell et al., 2003; Mackey et al., 2003). Another well-characterized plant–pathogen interaction involves the recognition of the effectors AvrPto and AvrPtoB from P. syringae pv tomato by Pto in tomato (Solanum lycopersicum; Scofield et al., 1996; Tang et al., 1996). Pto is a protein kinase that binds to AvrPto and AvrPtoB and requires Prf, an NBS-LRR protein, to elicit the HR in tomato (Scofield et al., 1996; Tang et al., 1996; Kim et al., 2002). Pto has also been shown to interact with Prf in a multimeric complex in planta and inactivate the E3 ligase activity of AvrPtoB by phosphorylation (Mucyn et al., 2006; Ntoukakis et al., 2009). AvrPto has not been reported to elicit the HR in Arabidopsis; however, transgenic expression of AvrPto in Arabidopsis suppresses basal defense responses (Hauck et al., 2003; Xing et al., 2007; Gohre et al., 2008; Shan et al., 2008; Xiang et al., 2008).
The effect of AvrPto on basal defense responses in Arabidopsis as well as our results from a yeast two-hybrid screen between plant signaling components and bacterial effectors (L. Williams, K. Cavanaugh, J. Greenberg, and R.W. Michelmore, unpublished data) led us to investigate the possible involvement of RIN4 in the Pto-mediated resistance response pathway. In this article, we describe the identification of interactions between RIN4 from both Arabidopsis (At RIN4) and a RIN4 homolog from tomato (Sl RIN4) and unrelated bacterial effectors, including AvrPto and AvrPtoB, using the yeast two-hybrid system. These interactions were recapitulated in planta using coimmunoprecipitation. Furthermore, in planta studies demonstrated that RIN4 degradation was elicited by AvrPto in a Pto- and Prf-dependent manner. Degradation of RIN4 occurred as the result of endogenous proteolytic activity and involved a similar site as the one involved in the cleavage that occurs in response to the bacterial effector AvrRpt2. Multiple experiments demonstrated that the AvrPto-mediated degradation of RIN4 was not a consequence of general proteolysis associated with an HR. Decreased expression of Sl RIN4 in tomato resulted in enhanced resistance to P. syringae pv tomato expressing avrPto. The nonpathogenic wild-type Pseudomonas fluorescens did not cause the disappearance of RIN4 in Nicotiana benthamiana; ectopic expression and secretion of AvrPto by P. fluorescens resulted in proteolysis of RIN4. The disappearance of RIN4 caused by P. syringae pv tomato in tomato is dependent on the type III secretion system (T3SS). Screening of the secretome of P. syringae pv tomato DC3000 identified two additional effectors that elicit degradation of RIN4 in N. benthamiana. Therefore, RIN4 is a common and important host target for pathogen effectors and plays a potential role in AvrPto-Pto–mediated resistance in tomato.
RESULTS
Cloning of Sl RIN4 and Its Enhanced Expression in Plants Challenged by P. syringae pv syringae B728A
A full-length homolog of RIN4 (Sl RIN4) was amplified from tomato cv Rio-Grande 76R (RG-76R) using oligonucleotide primers based on the sequence TC174419 in The Institute for Genomic Research database, cloned, and sequenced. Sl RIN4 was 37% identical to the Arabidopsis RIN4 (At RIN4) at the amino acid level and was the reciprocal best hit to At RIN4 in BLASTP analyses (see Supplemental Figure 1 online). Sl RIN4 contains the cleavage sites for AvrRpt2 at the N and near the C termini (Chisholm et al., 2005).
Sl RIN4 is expressed at a moderate level in unchallenged tomato RG-76R, similar to the housekeeping gene encoding α-tubulin (see Supplemental Figure 2 online). When tomato plants were challenged with the virulent pathogen, P. syringae pv tomato T1, the transcription level of Sl RIN4 was enhanced at least four times, as determined by quantitative real-time RT-PCR (see Supplemental Figure 3 online). Challenge with the avirulent pathogen, P. syringae pv syringae B728A, enhanced the transcription level of Sl RIN4 10 times more than challenge with P. syringae pv tomato T1 (see Supplemental Figure 3 online). Microarray data described in The Arabidopsis Information Resource (http://www.Arabidopsis.org/servlets/TairObject?id=1007966202andtype=expression_set) demonstrated that the transcription level of At RIN4 was similarly enhanced after challenge of Arabidopsis with P. syringae pv phaseolicola and the P. syringae pv tomato DC3000 hrcC mutant. The hrcC mutant contains a nonfunctional T3SS and is therefore nonpathogenic (Collmer et al., 2000).
RIN4 Interacts with Multiple Plant Signaling Components and Bacterial Effectors in Yeast and in Planta
As part of a large yeast two-hybrid screen to investigate interactions between resistance proteins, signaling components, and effector proteins, we identified several interactions between At RIN4, Sl RIN4, plant signaling proteins, and bacterial effectors (Table 1; see Supplemental Figure 4A online). Both At RIN4 and Sl RIN4 interacted with AvrB and the N-terminal 176–amino acid region of RPM1. A weak interaction was detected between Sl RIN4 and Pto. Sl RIN4 but not At RIN4 interacted with AvrPto and full-length AvrPtoB. Both At RIN4 and Sl RIN4 interacted strongly with the N-terminal region of AvrPtoB [AvrPtoB(deltaC), amino acids 1 to 308; Table 1]. This suggested that Sl RIN4 is a potential target of AvrPto and AvrPtoB.
Table 1.
| Yeast Two-Hybrid Assay
|
Coimmunoprecipitation
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Baitc
|
N. benthamiana
|
||||||||||
| AvrB | AvrPto | AvrPtoB | AvrPtoB (deltaC) | AvrRpm1 | RPM1 (N-terminus) | Pto | 3FLAG:SlRIN4 | 3HA:SlRIN4 | |||
| Prey | Sl RIN4 | SI | WI | I | I | WI | SI | WI | AvrPto:3HA | I | |
| At RIN4 | SI | N | N | WI | N | SI | N | AvrPtoB(deltaC) | I | ||
| EV | N | N | N | N | N | N | N | Pto | WI | ||
For the yeast two-hybrid assay, interactions were tested using Sl RIN4, At RIN4 in pSLR4, and AvrPto and the other proteins expressed using the bait vector pSLR3 following standard protocols for the LexA system. Empty vector controls and numerous other resistance signaling components, such as NDR1, NPR1, and multiple bacteria effectors, such as HopPsyE, showed no interactions with either At RIN4 or Sl RIN4 (data not shown).
For coimmunoprecipitations, A. tumefaciens carrying constructs to express 3HA:AvrPtoB(deltaC), AvrPto:3HA, or empty vector (EV) and 3FLAG:SlRIN4 or 3HA:SlRIN4 and Pto were coinfiltrated into N. benthamiana. Protein complexes were immunoprecipitated using anti-HA antibody matrix and immunoblotted using either anti-HA or anti-FLAG antibodies. Empty vector was used as a negative control; those data were not included in this table.
Explanations of bait construct abbreviations: AvrPtoB(deltaC), the N terminus only (amino acids 1 to 308) of AvrPtoB; N-RPM1(N-terminus), the N-terminal 176–amino acid region of RPM1. The C-terminus of AvrPtoB (amino acids 308 to 543) was unstable in yeast and therefore could not be tested using the yeast two-hybrid assay. SI, strong interaction; WI, weak interaction; I, interaction; N, no interaction detected.
Coimmunoprecipitation confirmed the yeast two-hybrid results. To test the association of Sl RIN4 with the N-terminal region of AvrPtoB or AvrPto, Sl RIN4 with three FLAG tags at its N terminus (3FLAG:SlRIN4) was transiently coexpressed in N. benthamiana with the N-terminal region of AvrPtoB [AvrPtoB(deltaC)] or AvrPto with 3x hemaglutinin (HA) tags at their N [3HA:AvrPtoB(deltaC)] and C (AvrPto:3HA) termini, respectively, using Agrobacterium tumefaciens–mediated transient assays. 3FLAG:SlRIN4 was detected in the pellet following immunoprecipitation of 3HA:AvrPtoB(deltaC) or AvrPto:3HA but not in the pellet of the empty vector control (Table 1; see Supplemental Figures 4B and 4C online). To test the association between Sl RIN4 and Pto, 3HA:SlRIN4 was transiently expressed in a stable transgenic of N. benthamiana overexpressing Pto. Pto was detected in the pellet following immunoprecipitation of 3HA:SlRIN4 but not in the control pellet (Table 1; see Supplemental Figure 4D online). These data indicated that Sl RIN4 can associate with Pto, AvrPto, and the N terminus of AvrPtoB in planta.
RIN4 Modification Is Induced in Planta by Three Bacterial Effectors: AvrRpt2, AvrPto, and AvrPtoB
To explore the biological significance of the protein–protein interactions observed in yeast and plants, we studied the effect of four bacterial effectors on the modification of RIN4 in planta. Both the Sl RIN4 and At RIN4 proteins were degraded in N. benthamiana when coexpressed with AvrRpt2 using Agrobacterium-mediated transient expression (see Supplemental Figure 5 online) as expected due to the conservation of the protease cleavage sites between Sl RIN4 and At RIN4. AvrPto also induced the degradation of At RIN4 and Sl RIN4; however, unlike the situation with AvrRpt2 (Axtell et al., 2003; Mackey et al., 2003; Chisholm et al., 2005; Kim et al., 2005a), we found that proteolytic degradation associated with AvrPto was dependent on the presence of Pto and Prf (Tables 2 and 3, Figure 1A). RIN4 degradation was induced by AvrPto in transgenic N. benthamiana expressing Pto but not in wild-type plants (Table 2; see Supplemental Figure 6A online), which contain a Prf capable of signaling AvrPto recognition (Salmeron et al., 1996). Similarly, AvrPto elicited the degradation of RIN4 in wild-type tomato cv RG-76R but not the isogenic mutants, RG-pto11 and RG-prf3, that have inactivating mutations in Pto and Prf, respectively (Table 3, Figure 1). In addition, the constitutive gain-of-function mutant PtoL205D, which mimics the negative charge conferred by autophosphorylation of Pto and elicits an AvrPto-independent, Prf-dependent HR (Rathjen et al., 1999), also induced degradation of Sl RIN4 in tomato RG-76R but not RG-prf3 (Table 3; see Supplemental Figure 6B online). Therefore, Pto/Prf-dependent degradation of RIN4 appeared to be due to endogenous proteolytic activity rather than a protease activity of AvrPto.
Table 2.
Pto-Dependent Degradation of RIN4 in N. benthamiana
|
N. benthamiana
|
||||
|---|---|---|---|---|
| Wild Type
|
35S:Ptoa
|
|||
| 3HA:AtRIN4 | 3HA:SlRIN4 | 3HA:AtRIN4 | 3HA:SlRIN4 | |
| AvrPto | Pb | P | Dc | D |
| AvrPtoB | P | P | P | P |
| PtoL205D | D | D | – | –d |
N. benthamiana plants constitutively expressing Pto from tomato.
The presence of 3HA:SlRIN4 as detected by immunoblotting.
Degradation of 3HA:SlRIN4.
No experiment was done with this combination.
Table 3.
Pto- and Prf-Dependent Degradation of RIN4 in Tomato
| Tomato
|
||||||
|---|---|---|---|---|---|---|
| RG-76R
|
RG-pto11a
|
RG-prf3b
|
||||
| 3HA:AtRIN4 | 3HA:SlRIN4 | 3HA:AtRIN4 | 3HA:SlRIN4 | 3HA:AtRIN4 | 3HA:SlRIN4 | |
| AvrPto | Dc | D | Pd | P | P | P |
| AvrPtoB | D | D | P | P | P | P |
| PtoL205D | –e | D | – | – | – | P |
Isogenic line of RG-76R with a null allele of Pto.
Isogenic line of RG-76R with a null allele of Prf.
Degradation of 3HA:SlRIN4 or 3HA:AtRIN4 as detected by immunoblotting.
The presence of 3HA:SlRIN4 or 3HA:AtRIN4 as detected by immunoblotting.
No experiment was done with this combination.
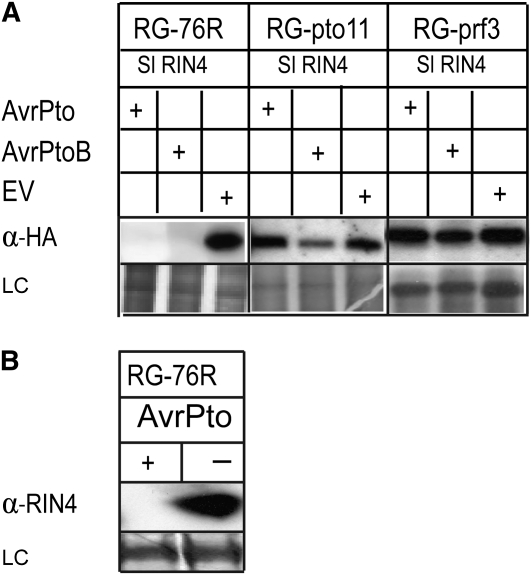
Figure 1.
AvrPto Caused the Proteolysis of Sl RIN4 in Tomato.
(A) AvrPto- and AvrPtoB-triggered degradation of RIN4 was dependent on Pto and Prf in tomato. 3HA:SlRIN4 and AvrPto, AvrPtoB, or empty vector (EV) were transiently expressed using A. tumefaciens in RG-76R tomato plants and RG-76R plants with mutations in Pto (RG-pto11) or Prf (RG-prf3). Twenty-four hours after infiltration, samples were collected and immunoblotted using anti-HA (α-HA) antibody to detect the presence of Sl RIN4. LC, silver staining of the membrane to show equal loading.
(B) AvrPto caused the proteolysis of Sl RIN4 expressed from its native promoter in tomato. AvrPto (+) or empty vector (−) were transiently expressed by A. tumefaciens in RG-76R tomato plants. Twenty-four hours after infiltration, samples were collected and immunoblotted using anti-At RIN4 (α-RIN4) antibody to detect native Sl RIN4. LC, Ponceau S staining of the membrane to show equal loading.
To demonstrate that AvrPto induced the degradation of endogenous Sl RIN4 expressed from Sl RIN4 using its native promoter in tomato (and to exclude the possibility that the degradation of Sl RIN4 was an artifact of transient overexpression of Sl RIN4 from the cauliflower mosaic virus [CaMV] 35S promoter), we transiently expressed AvrPto in RG-76R tomato plants. Samples were immunoblotted using At RIN4 antibody to detect endogenous levels of Sl RIN4. Sl RIN4 had been degraded in leaves expressing AvrPto but was present in samples from leaves infiltrated with the empty vector control (Figure 1B).
RIN4 Degradation Elicited by AvrPto Involves the Proteasomal Pathway
To determine whether the lack of detection of Sl RIN4 was due to the removal of the HA tag or other events, we tested the effect of two inhibitors on the disappearance of Sl RIN4. The general protease inhibitor cocktail developed for inhibition of plant proteases did not block the disappearance of Sl RIN4 elicited by AvrPto (Figure 2A). However, MG132, a proteasomal inhibitor, did prevent the disappearance of Sl RIN4 (Figure 2A). The expression level of AvrPto did not differ between treatments. The experiment was repeated using an artificial substrate with the RIN4 cleavage site (RCS) between the HA tag and green fluorescent protein (GFP; see below). In this case, MG132 did not block the cleavage on the RCS of the artificial substrate (Figure 2B); however, in multiple experiments, the proteolysis of GFP was not complete, possibly indicative of partial inhibition of degradation by MG132. Therefore, the degradation of Sl RIN4 induced by AvrPto seems to involve both a protease activity at the AvrRpt2 cleavage site that is not sensitive to the general protease inhibitor cocktail and the proteosomal degradation pathway. However, the role of the proteosomal pathway does not seem to be simply degradation of the product following protease cleavage. Neither MG132 nor the protease inhibitor cocktail blocked the HR caused by AvrPto (data not shown). Consequently, the degradation of Sl RIN4 appears not to be required for the elicitation of the HR.
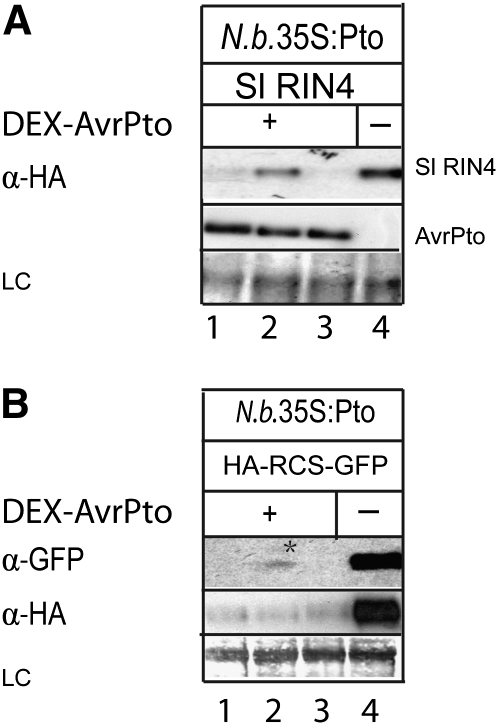
Figure 2.
Degradation of RIN4 Elicited by AvrPto Involves the Proteosomal Pathway.
In both experiments, 1 d after infiltration, leaves were infused with protease inhibitor (lane 1), MG132 proteosomal inhibitor (lane 2), or DMSO (lanes 3 and 4); DEX was applied to induce expression of avrPto 2 h later. Tissues were harvested 4 h after induction with DEX, and immunoblotting was performed using anti-HA antibody (α-HA) or anti-GFP antibody (α-GFP). LC, Ponceau S staining of the membrane to show equal loading.
(A) The proteosomal inhibitor MG132 partially blocked the degradation of RIN4 elicited by AvrPto. A. tumefaciens carrying a gene encoding HA-tagged Sl RIN4 was simultaneously coinfiltrated with either A. tumefaciens carrying HA-tagged avrPto expressed from a DEX-inducible promoter (+) or with A. tumefaciens carrying the empty vector (−) into N. benthamiana expressing Pto.
(B) MG132 partially blocked the proteolysis of GFP after its being cleaved at the RCS. A. tumefaciens carrying a gene to express an artificial substrate (see Figure 5A) was simultaneously coinfiltrated either with A. tumefaciens carrying avrPto expressed from a DEX-inducible promoter (+) or with A. tumefaciens carrying the empty vector (−) into N. benthamiana expressing Pto. A weak band of GFP was detected as marked by an asterisk.
RIN4 Degradation Is Not Due to General Proteolysis Occurring as Part of the HR
To exclude the possibility that AvrPto-mediated degradation of RIN4 was a secondary consequence of the induction of HR or a component of HR induced by multiple specific resistance reactions, we analyzed degradation of Sl RIN4 in the presence of several elicitors of HR in N. benthamiana. These elicitors of HR included the products of the bacterial avirulence gene avrB or those of the genes encoding the NBS-LRR proteins, RPP8 or RPS2, and an auto-activated mutant of a mitogen-activated protein kinase kinase 2 from tomato (MKK2). None of these elicitors of HR caused the degradation of Sl RIN4 (Figure 3A). Therefore, the initiation of the HR does not necessarily elicit RIN4 degradation. In addition, we transiently coexpressed AvrPto under a dexamethasone (DEX)-inducible promoter, Sl RIN4, and GFP in transgenic N. benthamiana plants that constitutively expressed Pto. Sl RIN4 was undetectable 24 h after induction of AvrPto expression, whereas GFP showed a strong band at this time point (Figure 3B). Consequently, AvrPto-induced degradation of RIN4 is specific rather than the result of general proteolysis.
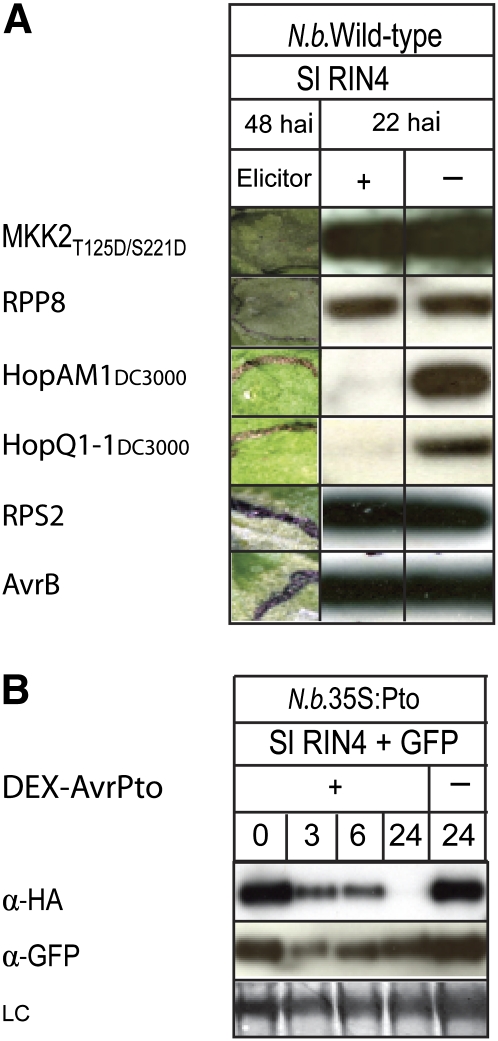
Figure 3.
RIN4 Degradation Is Not Due to General Proteolysis.
(A) The proteolysis of Sl RIN4 is not correlated with the HR phenotype. MKK2T215D/S221D, RPP8, HopAM1DC3000, HopQ1-1DC3000, RPS2, AvrB (+) or empty vector (−), and HA-tagged Sl RIN4 were transiently coexpressed in wild-type N. benthamiana. The leaf tissues were collected 22 h after infiltration (hai), and immunoblotting was performed using anti-HA antibody. The macroscopic necrosis (HR) phenotype was photographed 48 h after infiltration.
(B) Proteolysis of Sl RIN4 elicited by AvrPto was not the result of general proteolysis due to a HR. HA-tagged Sl RIN4 and GFP were coexpressed with AvrPto under a DEX-inducible promoter (DEX-AvrPto) in transgenic N. benthamiana leaves that overexpressed Pto (N.b.35S:Pto). After 24 h, expression of avrPto was induced by applying DEX to the leaves. Plant proteins were extracted at 0, 3, 6, and 24 h after DEX induction and assayed by immunoblotting with anti-HA (α-HA) or anti-GFP (α-GFP) antibodies. No Sl RIN4 was detected 24 h after the application of DEX, but GFP was readily detected.
Two additional effectors, HopQ1-1DC3000 (HopPtoQDC3000) and HopAM1DC3000 (AvrPpiBDC3000) (http://pseudomonas-syringae.org/pst_home.html), that caused the disappearance of RIN4 were identified by screening the secretome of P. syringae pv tomato DC3000 (see below). Neither effector elicited HR in wild-type N. benthamiana 48 h after inoculation (Figure 3A); however, both caused the disappearance of RIN4 (Figure 3A). Therefore, RIN4 degradation occurred independently of HR, confirming that RIN4 degradation was not the result of general proteolysis. HopQ1-1DC3000 was reported to cause HR 48 h after transient expression (Wei et al., 2007). However, we did not observe HR in multiple inoculations; this may have been due to using different vectors to express HopQ1-1DC3000, different experimental conditions, or different genotypes of N. benthamiana.
The Interaction between AvrPto and Pto, but Not the Kinase Activity of Pto, Is Required for Proteolysis of RIN4
To test whether the ability of AvrPto to interact with Pto is required for the proteolysis of RIN4, an AvrPto mutant, AvrPtoI96T, was coexpressed with Sl RIN4 in N. benthamiana. AvrPtoI96T has a polar amino acid substitution in the GINP Ω loop and does not interact with Pto in the yeast two-hybrid assay (Wulf et al., 2004). AvrPtoI96T did not trigger an HR in N. benthamiana expressing Pto (Figure 4A). Coexpression of AvrPtoI96T and Sl RIN4 in N. benthamiana constitutively expressing Pto did not induce the proteolysis of Sl RIN4 (Figure 4B). This indicates that interaction between AvrPto and Pto is required for the AvrPto-elicited proteolysis of RIN4 in planta or that an unknown modified interaction between AvrPtoI96T and RIN4 abrogates proteolysis.
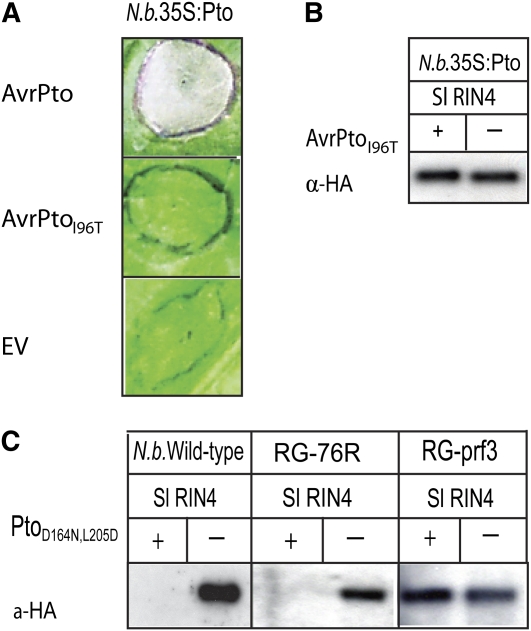
Figure 4.
Pto and AvrPto Interaction but Not Pto Kinase Activity Is Required for RIN4 Proteolysis.
(A) AvrPtoI96T cannot trigger HRs in N. benthamiana expressing Pto. AvrPtoI96T has a polar amino acid substitution in the GINP Ω loop. This mutant does not interact with Pto in the yeast two-hybrid assay. A. tumefaciens carrying genes encoding AvrPtoI96T, AvrPto, or empty vector (EV) were infiltrated into the leaves of N. benthamiana expressing Pto. Photographs of the necrosis phenotype were taken 3 d after infiltration.
(B) Interaction between AvrPto and Pto was required to trigger proteolysis of RIN4 in N. benthamiana expressing Pto. HA-tagged Sl RIN4 and AvrPtoI96T (+) or empty vector (−) were transiently coexpressed in transgenic N. benthamiana that constitutively overexpressed Pto (N.b.35S:Pto), and immunoblotting was performed using anti-HA antibody (α-HA). AvrPtoI96T was unable to elicit RIN4 degradation.
(C) Kinase activity of Pto is not required for the proteolysis of RIN4. The constitutively active but kinase-deficient mutant PtoD164N,L205D and HA-tagged Sl RIN4 were transiently coexpressed in wild-type N. benthamiana, RG-76R tomato plants, or an isogenic mutant RG-prf3. Total protein was extracted 22 h after coinfiltration, and immunoblotting was performed using anti-HA antibody (α-HA). PtoD164N,L205D elicited RIN4 degradation in a Prf-dependent manner.
To determine whether the kinase activity of Pto is required for the degradation of RIN4, we tested a constitutively active, kinase-deficient double mutant, PtoD164N, L205D, which does not interact with AvrPto and elicits a strong HR in N. benthamiana (Wu et al., 2004). Transient expression of PtoD164N, L205D and Sl RIN4 in both N. benthamiana and tomato RG-76R resulted in the degradation of Sl RIN4 (Figure 4C), indicating that the kinase activity of the constitutively active form of Pto is not required for the degradation of RIN4. Coexpression of PtoD164N, L205D and Sl RIN4 in tomato RG-prf3 did not cause an HR as reported previously (Wu et al., 2004) or proteolysis of Sl RIN4 (Figure 4C).
The Site Cleaved by the Effector AvrRpt2 Is Required for Proteolysis of RIN4
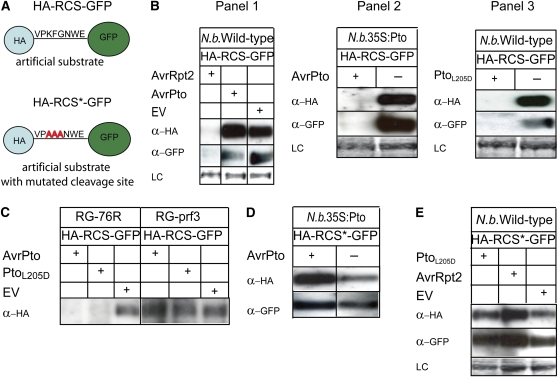
The parallel data from yeast two-hybrid and in planta assays for At RIN4 and Sl RIN4, which have very different sequences except at regions containing the AvrRpt2 cleavage sites (see Supplemental Figure 1 online), the dependency of RIN4 degradation on Pto and Prf, and particularly the degradation induced by PtoL205D in the absence of AvrPto all indicated that AvrPto does not directly degrade RIN4 (in contrast with the situation with the protease effector AvrRpt2) but rather induces the activation of endogenous proteolytic activity. To test the requirement for the site cleaved by AvrRpt2, we generated an artificial substrate containing the RCS used by AvrRpt2 as well as a control substrate comprising a mutated RCS. Several amino acids in the cleavage site, particularly the Phe at position 4, had previously been shown to be important for AvrRpt2-mediated RIN4 cleavage and subsequent degradation (Chisholm et al., 2005; Kim et al., 2005a; Takemoto and Jones, 2005). For this study, the Lys at position 3, Gly at position 5, and the Phe at position 4 were all changed to Ala. The wild-type RCS or the mutated RCS (RCS*), both eight amino acids in length, were then inserted between an HA and the GFP to generate HA-RCS-GFP and HA-RCS*-GFP chimeric proteins, respectively (Figure 5A). When HA-RCS-GFP was coexpressed with AvrPto in wild-type and Pto-expressing N. benthamiana, the artificial substrate was detectable in wild-type N. benthamiana but not in the transgenic plants constitutively expressing Pto (Table 4, Figure 5B, Panels 1 and 2). To determine whether the peptides within the AvrRpt2 cleavage site were sufficient to target GFP to the proteasome, we used GFP antibody to assay whether the GFP persisted after cleavage. However, GFP was not detected, indicating that the cleavage site is sufficient to target the fusion protein to the proteasome (Figure 5B, Panel 2). Similarly, transient expression of AvrPto induced the disappearance of HA-RCS-GFP in RG-76R tomato plants but not in RG-prf3 plants (Table 4, Figure 5C). Coexpressing the constitutive mutant PtoL205D with HA-RCS-GFP resulted in its disappearance in wild-type N. benthamiana and RG-76R tomato, but not in RG-prf3 plants (Table 4, Figures 5B, Panel 3, and 5C). Therefore, the AvrPto-induced disappearance of the artificial substrate required a similar cleavage site as is required for cleavage by AvrRpt2 and was also Pto and Prf dependent. HA-RCS*-GFP, containing mutations in the cleavage site, was not degraded when coexpressed with AvrPto in N. benthamiana expressing Pto (Table 4, Figure 5D). These mutations also blocked the disappearance of HA-RCS*-GFP when it was coexpressed with PtoL205D in wild-type N. benthamiana (Table 4, Figure 5E). These data are consistent with the activation of an endogenous proteolytic activity that requires the same cleavage site as does AvrRpt2.
Figure 5.
Proteolysis of an Artificial Substrate Induced in N. benthamiana and Tomato.
(A) The artificial substrates comprising either the native or mutated RCS between an HA-tag and GFP. Eight amino acids from the first AvrRpt2 cleavage site in RIN4 were placed between an HA-tag and GFP (HA-RCS-GFP). Similarly, a mutated cleavage site with three Ala substitutions at positions known to be important for cleavage by AvrRpt2 was placed between an HA tag and GFP (HA-RCS*-GFP).
(B) AvrPto induced proteolysis of artificial substrates in a Pto-dependent manner in N. benthamiana. Panel 1: HA-RCS-GFP was transiently coexpressed with AvrRpt2, AvrPto, or empty vector (EV) in wild-type N. benthamiana. Total proteins were extracted before the HR caused by AvrRpt2 or AvrPto was evident, and immunoblotting was performed using anti-HA (α-HA) or anti-GFP (α -GFP) antibody. Panel 2: HA-RCS-GFP was transiently coexpressed with AvrPto (+) or empty vector (−) in transgenic N. benthamiana expressing Pto. Total proteins were extracted and immunoblotted against anti-HA (α-HA) antibody or anti-GFP (α-GFP) antibody. The lack of a detectable HA tag or GFP indicated that the cleavage site in AS is sufficient for AvrPto-induced proteolysis. Panel 3: Constitutively active PtoL205D induced proteolysis in N. benthamiana. HA-RCS-GFP was coexpressed with PtoL205D (+) or empty vector (−) in wild-type N. benthamiana, and immunoblotting was performed using anti-HA (α-HA) or anti-GFP (α-GFP) antibody.
(C) Proteolysis induced by AvrPto or PtoL205D is dependent on Prf in tomato. HA-RCS-GFP was coexpressed in RG-76R tomato plants or an isogenic mutant at the Prf locus (RG-prf3) with AvrPto (+), PtoL205D (+), or empty vector (EV), and immunoblotting was performed using anti-HA (α-HA) antibody.
(D) and (E) Amino acid substitutions in the cleavage site prevented cleavage caused by AvrRpt2, AvrPto, or PtoL205D in N. benthamiana. HA-RCS*-GFP was coexpressed with AvrRpt2, AvrPto, PtoL205D, or empty vector (EV) in wild-type or transgenic N. benthamiana expressing Pto, and immunoblotting was performed using anti-HA (α-HA) or anti-GFP (α-GFP) antibody. LC, Ponceau S staining of the membrane to show equal loading.
Table 4.
Pto- and Prf-Dependent Degradation of Artificial Substrates Required an Intact AvrRpt2 Cleavage Site
|
N. benthamiana
|
Tomato
|
|||||
|---|---|---|---|---|---|---|
| Wild Type
|
35S:Ptoa
|
RG-76R
|
RG-prf3b
|
|||
| HA-RCS-GFP | HA-RCS*-GFP | HA-RCS-GFP | HA-RCS*-GFP | HA-RCS-GFP | HA-RCS-GFP | |
| AvrPto | Pc | P | Dd | P | D | P |
| AvrRpt2 | D | P | –e | – | D | P |
| PtoL205D | D | P | – | – | – | – |
N. benthamiana plants constitutively expressing Pto from tomato.
Isogenic line of RG-76R with a null allele of Prf.
The presence of artificial substrate detected by immunoblotting.
Degradation of artificial substrate as determined by immunoblotting.
No experiment was done with this combination.
Decreased Levels of RIN4 Result in Increased AvrPto-Induced Inhibition of Pathogen Growth
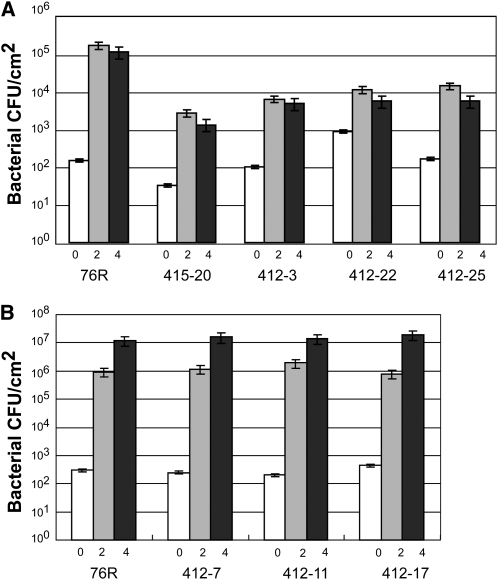
We attempted to generate transgenic plants of tomato cv RG-76R with either elevated or reduced levels of RIN4 to determine if RIN4 degradation is required for AvrPto-Pto–initiated resistance. However, we were able to obtain very few transgenic plants despite multiple transformation attempts. Concurrent transformations with other constructs resulted in normal transformation rates. Therefore, it seems that perturbations of RIN4 levels, either increased or decreased, were deleterious in tomato. Only one plant could be obtained that overexpressed RIN4 from the 35S promoter; this plant had severely abnormal morphology, yielded stunted progeny, and could not be studied further. Four independent transgenics were obtained in which the expression of RIN4 had been reduced by ∼85% using RNA interference (RNAi). One was sterile and only two of the other three provided sufficient numbers of T2 seeds for further analysis. T2 plants were derived from T1 plants 412 and 415 that showed a 97 and 85% reduction in expression of RIN4, respectively, as detected by quantitative PCR. Transmission of the T-DNA to the T2 generation was less than Mendelian expectations in both families, further indicating the deleterious effects of abnormal RIN4 levels in tomato; PCR products representing T-DNA containing the complete RNAi construct were detected in only 8 out of 40 T2 plants in the 412-derived family and 5 out of 20 T2 plants in the 415-derived family. The growth of P. syringae pv tomato T1, expressing AvrPto, was repressed in these silenced T2 plants (Figure 6A). However, the growth of an isogenic P. syringae pv tomato strain T1 carrying only the empty vector and lacking AvrPto was similar on the wild-type and transgenic plants (Figure 6B). These data are consistent with the decreased expression of Sl RIN4 in these transgenic plants specifically enhancing Pto-dependent resistance as opposed to Pto-independent resistance.
Figure 6.
Degradation of RIN4 Is Important for AvrPto-Dependent Inhibition of Bacterial Growth.
(A) Growth of P. syringae pv tomato T1 expressing avrPto was reduced in RIN4 RNAi-silenced T2 tomato plants. T2 plants derived from original (T1) transgenic plants 415 (one plant) and 412 (three plants) were inoculated by infiltrating the pathogen at 103 cfu/mL and assayed after 0 (white), 2 (gray), and 4 (black) d; error bars indicate the sd from three biological replications.
(B) Wild-type P. syringae pv tomato T1 exhibited normal levels of growth in RIN4 RNAi-silenced T2 plants. Three transgenic RNAi T2 plants derived from one RNAi-silenced T1 line (412) with reduced RIN4 expression relative to their isogenic wild-type tomato RG-76R were inoculated by infiltrating with pathogen at 103 cfu/mL and assayed 0, 2, and 4 d after infiltration. Error bars indicate the sd from three measurements.
Additional Effectors Cause Proteolysis of RIN4 in Tomato in a Pto- and Prf-Independent Manner
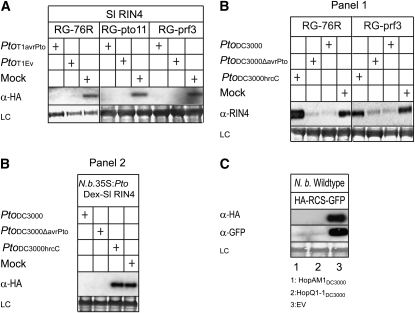
In an attempt to investigate AvrPto-induced degradation of RIN4 when AvrPto is secreted by P. syringae rather than overexpressed in Agrobacterium-mediated transient assays, we studied RIN4 degradation using isogenic strains of P. syringae pv tomato T1, carrying a plasmid with and without avrPto. However, P. syringae pv tomato T1 caused the proteolysis of Sl RIN4 in RG-76R, RG-pto11, and RG-prf3 tomato plants regardless of whether avrPto was present (Figure 7A). Wild-type P. syringae pv tomato strain DC3000, expressing AvrPto and AvrPtoB, caused degradation of Sl RIN4 in stably transformed N. benthamiana plants constitutively expressing Sl Pto and induced to express Sl RIN4 by applying DEX on the leaves; a mutant strain of DC3000 lacking avrPto caused the degradation of Sl RIN4 in these transgenic plants as well (Figure 7B, Panel 2). However, P. syringae pv tomato strain DC3000 hrcC, a nonpathogen because it lacks a functional T3SS (Alfano and Collmer, 1997), did not cause the proteolysis of Sl RIN4, indicating that the T3SS is required for the proteolysis (Figure 7B, Panel 2). To test whether the lack of accumulation of Sl RIN4 in tomato is due to inhibition of agro-transient expression, we inoculated tomato leaves of RG-76R and its isogenic line RG-prf3 with P. syringae pv tomato strain DC3000, DC3000 lacking avrPto, and DC3000 hrcC. Both P. syringae pv tomato strain DC3000 and DC3000 lacking avrPto caused the degradation of Sl RIN4 but DC3000 hrcC did not (Figure 7B, Panel 1). Together, these data suggested that other effectors induced RIN4 degradation independent of Pto and Prf.
Figure 7.
Degradation of RIN4 by Pseudomonas spp.
(A) P. syringae pv tomato T1 with or without AvrPto caused proteolysis of Sl RIN4 in tomato in a Pto- and Prf-independent manner. P. syringae pv tomato strain T1 (PtoT1) with and without avrPto (avrPto and EV, respectively) were coinoculated with A. tumefaciens strain C58C1 with a binary vector that provided expression of HA-tagged Sl RIN4 in planta. The negative control (Mock) was infiltrated with 10 mM MgCl2. Coinoculations were made with A. tumefaciens at OD600 = 0.5 and P. syringae pv tomato at OD600 = 10−2 into wild-type RG-76R, RG-pto11, and RG-prf3 tomato leaves, and total proteins were extracted 24 h after infiltration and the presence of RIN4 assayed using anti-HA antibody (α-HA). LC, Ponceau S staining of the membrane to show equal loading.
(B) P. syringae pv tomato DC3000 and its derivative lacking AvrPto caused T3SS-dependent proteolysis of RIN4 in tomato and in N. benthamiana. Wild-type P. syringae pv tomato DC3000 (PtoDC3000), a knockout derivative lacking AvrPto (PtoDC3000ΔavrPto), or a T3SS-deficient mutant (PtoDC3000hrcC) were individually inoculated at OD600 = 10−2 into leaves of tomato RG-76R, RG-prf3, or transgenic N. benthamiana constitutively expressing Pto and expressing HA-tagged Sl RIN4 under the DEX-inducible promoter. In tomato, total protein was extracted and detected using anti-At RIN4 (α-RIN4) antibody (Panel 1). In N. benthamiana, 24 h after inoculation, Sl RIN4 expression was induced by applying DEX onto the leaves. Total plant protein was extracted 24 h after induction and the presence of RIN4 assayed using anti-HA antibody (α-HA) (Panel 2). LC, Ponceau S staining of the membrane to show equal loading.
(C) Effectors HopAM1 and HopQ1-1 from PtoDC3000 cleaved the artificial substrate (HA-RCS-GFP) containing the native RCS independent of Pto. HA-RCS-GFP and HopAM1DC3000 (lane 1), HopQ1-1DC3000 (lane 2), or empty vector (lane 3) were coexpressed in wild-type N. benthamiana leaves, and immunoblotting was performed using anti-HA antibody (α-HA) or anti-GFP antibody (α-GFP). LC, Ponceau S staining of the membrane to show equal loading.
To determine which other bacterial effectors induced the degradation of RIN4, we coexpressed Sl RIN4 with each of 30 different effectors from P. syringe pv tomato DC3000 in N. benthamiana and assayed for degradation of Sl RIN4. We identified two additional effectors, HopQ1-1DC3000 (HopPtoQDC3000) and HopAM1DC3000 (AvrPpiBDC3000) (http://pseudomonas-syringae.org/pst_home.html), that induced the proteolysis of Sl RIN4 in wild-type N. benthamiana (Figure 3A). Homologs of HopQ1 are present in P. syringae pv tomato T1 (http://staff.vbi.vt.edu/jcslab/pseudomonas/), and homologs of HopQ1-1 and HopAM1 exist in many strains of P. syringae (Lindeberg et al., 2005; Sarkar et al., 2006). Therefore, it is likely that the Pto- and Prf-independent Sl RIN4 degradation observed with P. syringae pv tomato strain T1, as well as that observed with P. syringae pv tomato strain DC3000, is due to the direct or indirect action of other effectors. Both effectors also induced degradation of the artificial RCS substrate in wild-type N. benthamiana (Figure 7C), indicating that the RCS is necessary and sufficient for effectors to elicit RIN4 degradation in a Pto-independent manner. HopAM1DC3000 caused an HR in tomato but not in N. benthamiana, while HopQ1-1DC3000 did not cause an HR in tomato and elicited chlorosis in N. benthamiana (http://charge.ucdavis.edu/). These results provide further evidence that the induction of RIN4 degradation by bacterial effectors was not a consequence of the general proteolysis that occurs as part of the HR. The biochemical, particularly proteolytic, activities of these two effectors are not known and are currently under investigation.
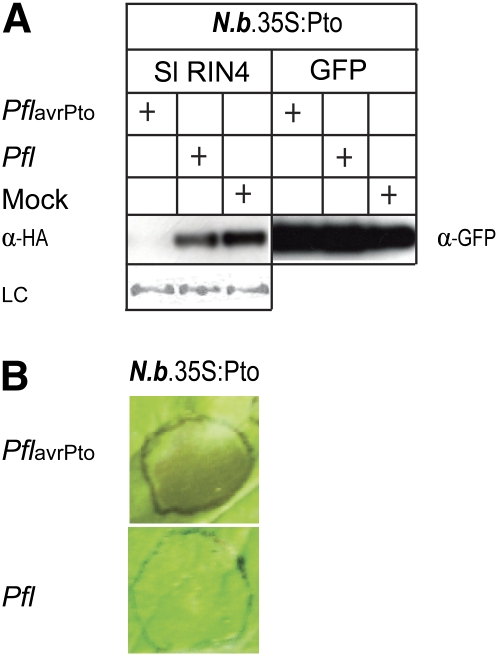
RIN4 Is Degraded When AvrPto Is Secreted by P. fluorescens
To further investigate AvrPto-induced degradation of RIN4 when secreted rather than overexpressed in Agrobacterium-mediated transient assays, we used isogenic strains of transgenic P. fluorescens, one carrying a plasmid with and one without avrPto. P. fluorescens strain 55 is a nonpathogen. It carries pHIR11, which expresses a portion of the pathogenicity island from P. syringae pv syringae and enables secretion of effectors from P. fluorescens (Collmer et al., 2000). Wild-type P. fluorescens barely grows in N. benthamiana; however, expression of AvrPto at low inoculum concentrations resulted in two- to threefold increased growth of P. fluorescens consistent with a virulence phenotype of AvrPto; however, as with P. syringae pv tomato DC3000 at this concentration, no degradation of RIN4 was observed (data not shown). On the other hand, inoculation of P. fluorescens 55 (pHIR11), expressing or not expressing AvrPto, into N. benthamiana at a higher concentration (colony-forming units [cfu] = 108/mL), resulted in the AvrPto-specific degradation of Sl RIN4 (Figure 8A). GFP was not degraded in the control inoculations with either strain of P. fluorescens (Figure 8A), providing further evidence that RIN4 degradation is specific to the activity of AvrPto rather than a consequence of HR induction. AvrPto-specific induction of HR after 24 h was not observed unless inoculations were made with 109 cfu/mL (Figure 8B).
Figure 8.
AvrPto-Specific Degradation of Sl RIN4 by P. fluorescens in N. benthamiana.
(A) AvrPto-specific degradation of Sl RIN4 by P. fluorescens. Strains of P. fluorescens 55 (pHIR11) (OD600 = 0.1) expressing (PflavrPto) or not expressing (Pfl) avrPto were coinoculated with A. tumefaciens carrying Sl RIN4 (OD600 = 1.0) or GFP (as a control) into leaves of transgenic N. benthamiana that expressed Pto. After 24 h, leaf samples were harvested and immunoblotted using anti-HA (α-HA) or anti-GFP (α-GFP) antibodies. The mock treatment (Mock) was infiltration with 10 mM MgCl2 solution. LC, Ponceau S staining of the membrane to show equal loading.
(B) P. fluorescens at a high inoculum concentration elicited necrosis in an avrPto-dependent manner. Strains of P. fluorescens 55 (pHIR11) (OD600 = 1.0) expressing (PflavrPto) or not expressing (Pfl) avrPto were inoculated individually into leaves of transgenic N. benthamiana that expressed Pto. Only P. fluorescens expressing avrPto elicited macroscopic necrosis indicative of a HR 48 h after the inoculation.
DISCUSSION
Proteolysis of RIN4 seems to be an important aspect of plant–bacteria interactions that can be achieved by a variety of mechanisms, including direct enzymatic activity of a bacterial protein elicitor or indirectly through induction of the endogenous proteolytic pathway. Sl RIN4 seems to be targeted directly or indirectly by multiple effectors from P. syringae. At least seven effectors, AvrRpt2, AvrPto, AvrPtoB, HopQ1-1DC3000 (HopPtoQ DC3000), HopAM1DC3000 (AvrPpiB DC3000), AvrB, and AvrRpm1, cause various modifications of Sl RIN4. Yeast two-hybrid and coimmunoprecipitation data indicate that AvrPto and RIN4 can directly interact; the importance of this interaction to the elicitation of RIN4 degradation is unclear because degradation can occur in the absence of AvrPto. The experiment with the artificial substrate demonstrated that the site of RIN4 cleavage by AvrRpt2 is required for Pto-AvrPto–mediated degradation and is sufficient to target GFP with the cleavage site at its N terminus to the proteasome (Figure 5B). However, the mechanisms of RIN4 degradation by AvrRpt2 and AvrPto are different. AvrRpt2 is known to function as a protease (Axtell et al., 2003). By contrast, AvrPto is not thought to be a protease, and the constitutive gain-of-function PtoL205D mutant caused proteolysis of Sl RIN4 in the absence of AvrPto, indicating that proteolysis of RIN4 is not dependent on a proteolytic activity of AvrPto. Also, unlike AvrRpt2-induced cleavage, AvrPto-induced degradation of RIN4 is Pto and Prf dependent, and the interaction between AvrPto and Pto is required for AvrPto-induced cleavage of RIN4.
The role of RIN4 degradation in plant–bacterial interactions is unclear. Multiple lines of evidence demonstrated that the proteolysis of RIN4 elicited by AvrPto is not due to general protein degradation as part of the HR. Even when there was extensive necrosis 48 h after ectopic expression of several HR inducers, RIN4 degradation was not apparent (Figure 3A). Conversely, RIN4 was degraded well before necrosis in multiple experiments, including when AvrPto was secreted by P. fluorescens (Figure 8B). RIN4 has been reported to be a negative regulator of basal resistance in Arabidopsis (Kim et al., 2005b). However, our data indicate that the situation is considerably more complex than currently described. Pto and Prf may be part of one or more macromolecular complexes (Mucyn et al., 2006) with multiple negative regulators, including RIN4. One possible role of the Prf-dependent proteolysis of RIN4 is to alleviate basal defense suppression by RIN4 or other related proteins. This scenario is similar to the derepression of WRKY-mediated suppression of basal defenses upon activation of MLA10 (Shen et al., 2007). However, the dynamics, cellular localization, and coincidence of such complexes and their roles in resistance are not currently known. Clearly, AvrPto interacts with RIN4 in the absence of Pto, but this does not trigger RIN4 degradation or resistance. Also, Pto interacts with RIN4 in the absence of AvrPto; however, binding of AvrPto to Pto results a Prf-dependent activation of an endogenous proteolytic pathway that degrades RIN4.
Inhibiting degradation of a negative regulator, such as RIN4, could be anticipated to contribute to virulence. Therefore, the proteolysis of RIN4, directly or indirectly induced by bacterial effectors, is seemingly counterintuitive if this results in increased induction of the resistance response. However, the effector-elicited resistance response is much stronger than the PAMP-induced basal resistance response (Jones and Dangl, 2006). Consistent with AvrPto-elicited RIN4 degradation increasing resistance, downregulation of RIN4 expression in tomato using RNAi suppressed growth of P. syringae pv tomato specifically expressing AvrPto (Figure 6A). The benefit of proteolysis of RIN4, or of other potential defense regulators that contain the same cleavage site (Chisholm et al., 2005; Takemoto and Jones, 2005), caused directly by AvrRpt2 and indirectly by AvrPto may lie in the abrogation of the detection of RIN4 modification by effectors such as AvrRpm1 and AvrB as exemplified by AvrRpt2 blocking the HR induced by AvrB or AvrRpm1 in the presence of RPM1 in Arabidopsis (Ritter and Dangl, 1996). Additional effectors may act by other mechanisms to minimize the effects of removing the negative-regulatory effects of RIN4 on the induction of basal resistance. AvrPto is known to have other Pto- and Prf-independent effects of virulence (Shan et al., 2008; Zipfel and Rathjen, 2008). The effect of AvrPto-mediated RIN4 proteolysis on virulence would only be detected when effectors that modify RIN4 and resistance genes capable of detecting such modifications are present.
Our data provide further evidence for the overlap and interplay of PAMP-triggered and effector-triggered resistance in plants and are consistent with roles for AvrPto and AvrPtoB in modulating PAMP-elicited resistance. Effector-triggered and PAMP-triggered resistance share signaling components in common, including RIN4, and similar sets of response genes are induced (Jones and Dangl, 2006). Effectors that repress PAMP-triggered resistance in one species may elicit an HR in another (Lahaye and Bonas, 2001). RIN4 is both a negative regulator of basal resistance as well as a trigger for HR (Mackey et al., 2003; Kim et al., 2005b). AvrPto and AvrPtoB suppress PAMP-triggered resistance in compatible interactions but trigger an HR in incompatible ones in a Prf- and Pto-dependent manner (Hauck et al., 2003; He et al., 2006; Hann and Rathjen, 2007). Overexpression of Pto and Prf leads to constitutive, ligand-independent induction of the resistance response or an HR (Salmeron et al., 1996; Tang et al., 1999; Tobias et al., 1999; Xiao et al., 2003). Tomato mutants lacking Prf were more susceptible to virulent strains of P. syringae pv tomato; one of several explanations of this phenotype is a role for Prf in PAMP-triggered resistance (Chang et al., 2001). It is also possible that Prf is involved in the recognition of other effectors from the virulent strain. This leads to the consideration of what extent Pto/Prf-mediated resistance is similar to or different from other effector-triggered resistance systems and whether Pto and Prf are primarily involved in PAMP- or effector-triggered resistance. In the Pto/Prf pathway, the kinase gene component is multicopy and exhibits intraspecific variation in sequence and function (Riely and Martin, 2001; Rose et al., 2005, 2007). In other specific resistances, the genes encoding the NBS-LRR proteins are the ones that have been duplicated and are the more variable component (McHale et al., 2006). This may reflect the involvement of Pto and Prf in PAMP-triggered resistance that is thought to predate the evolution of specific induction of an HR in plants (Alfano and Collmer, 2004; Jones and Dangl, 2006). The first targets of bacterial effectors were, therefore, likely to have been components of PAMP-elicited resistance. Consequently, RIN4 and Pto homologs may have been early targets for bacterial effectors, and selection on host populations to detect interference with these components could have subsequently resulted in the evolution of specific resistance proteins. The recognition of AvrPto and AvrPtoB and elicitation of HR in tomato is likely, therefore, the more highly evolved situation relative to that in N. benthamiana. Consistent with this, coexpression of tomato Prf and Pto in N. benthamiana resulted in an HR induced by AvrPtoB (Mucyn et al., 2006).
Several proteins from Arabidopsis contain the same proteolytic cleavage site as RIN4 and have been shown to be cleaved by AvrRpt2 (Hirano et al., 1999; Chisholm et al., 2005; Takemoto and Jones, 2005). It will be interesting to determine which and how many of these other proteins are also targeted by the virulence effectors we identified as inducing RIN4 degradation. Some may be functional proteins; others, including RIN4 itself, may be decoys or sentinels for the presence of proteolytic effectors (van der Hoorn and Kamoun, 2008). The presence of multiple proteins targeted by many functionally redundant effectors leads to several possible evolutionary questions and scenarios. The lack of sequence similarity between the multiple effectors that induce RIN4 degradation suggests convergent evolution to modify the same plant targets. Alternatively, RIN4 may function primarily as a sentinel for proteolytic activities from the pathogen or host. Such a role would allow the protection of a large number of proteins and explain the conservation of the cleavage site in diverse species.
Our data suggest several avenues for future research. It will be interesting to determine whether the Pto/Prf pathway is primarily involved in PAMP-triggered resistance or in resistance resulting from the detection of specific effectors. Further characterization of the proteolytic pathway leading to degradation of RIN4 by the proteasome is a high priority. Also, it will be intriguing to determine whether RIN4 is targeted by effectors from other types of pathogens, such as fungi and oomycetes. In addition, it will be important to test whether other plant resistance-signaling components are also points of vulnerability that are targeted by multiple effectors.
METHODS
Plasmids and Cloning Procedures
The RIN4 homologs from tomato (Solanum lycopersicum; Sl RIN4) and Arabidopsis thaliana (At RIN4) were amplified from cDNA by RT-PCR (Advantage 2 RT-PCR kit; Clontech) using the primers described in Supplemental Table 1 online and cloned into Gateway entry vector pDONR207 (Invitrogen). The primers for At RIN4 were based on At3G25070. The primers for Sl RIN4 were based on the Tomato Gene Index (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=tomato) SlGI TC174419; this was the only RIN4 homolog available in the databases at the time the experiments were conducted. The RIN4 homologs were transferred into the Gateway-compatible Agrobacterium tumefaciens binary vector pGWB15 to provide expression in planta with an N-terminal HA tag (courtesy of T. Nakagawa). Similar HA-tagged versions of At RIN4 had previously been shown to be functional (Day et al., 2005). Recently, the sequence of a second RIN4 homolog (TC18672) became available from tomato. The two tomato sequences are equally similar to At RIN4. This second sequence has the conserved cleavage sites but has not been studied further. The bacterial effector genes, avrRpt2, avrPtoB, avrPto, and avrB, were cloned into Gateway binary vector pCB302-3 (Xiang et al., 1999) and transformed into A. tumefaciens strain C58C1. For yeast two-hybrid assays, effectors and plant genes were transferred into Gateway-compatible yeast two-hybrid vector pSLR4 as bait and pSLR3 as prey. To generate 3HA:AvrPtoB(deltaC) and AvrPto:3HA, AvrPtoB(deltaC) and AvrPto were cloned into Gateway-compatible binary vectors pGWB15 and pGWB14 with three HA tags at their N or C termini, respectively. To generate 3FLAG:SlRIN4, three FLAG tags were fused onto Sl RIN4 using primer 3FLAG+SlRIN4F (see Supplemental Table 1 online). To generate HA-RCS-GFP, the cleavage site from RIN4 was first introduced next to GFP using the primers Cyscleave+GFPF and GFPR and then the recombination sites for the Gateway system were added using CyscleaveF and GFPR (see Supplemental Table 1 online for primer sequences). The codons encoding the eight amino acids of the cleavage site are underlined in Supplemental Table 1 online. HA-RCS*-GFP was generated using the same strategy but with altered forward primers that encoded a mutated cleavage site (mutated codons are indicated in bold in Supplemental Table 1 online). All the clones that were used in this article are listed in Supplemental Table 3 online.
RNA Isolation and Real-Time PCR
Leaf tissues were collected from tomato plants at 0, 6, 12, and 24 h after challenge with either Pseudomonas syringae pv tomato T1 or P. syringae pv syringae B728A. Total RNA was extracted using Plant RNeasy mini kits (Qiagen) and treated with RNase-free DNase (Sigma-Aldrich). First-strand cDNA was synthesized using 1 μg of total RNA, oligo(dT) primer, and MMLV reverse transcriptase (Clontech). Real-time PCR was performed using SyBr-Green master mix (Sigma-Aldrich) on a DNA Engine Opticon 2 quantitative real-time PCR machine (Bio-Rad). The results were analyzed by Opticon software provided by Bio-Rad. The data were normalized using the gene encoding α-tubulin (TC170178) as the reference gene. The oligonucleotide primers used are listed in Supplemental Table 2 online.
A. tumefaciens–Mediated Transient Assays
A. tumefaciens strain C58C1 carrying RIN4 homologs or virulence effectors in the binary vector were grown overnight in Luria-Bertani liquid medium containing 50 μg/mL kanamycin and 5 μg/mL tetracycline. Cells were resuspended in infiltration medium (10 mM MES, pH 5.6, 10 mM MgCl2, and 150 μM acetosyringone) to a total concentration of OD600 of 1.0 and infiltrated into leaves of Nicotiana benthamiana as previously described (Wroblewski et al., 2005). For transient assays of tomato, a total concentration of OD600 of 0.5 was used to avoid a necrotic reaction to A. tumefaciens C58C1 (Wroblewski et al., 2005). Infiltrated plants were kept in the laboratory until assayed.
RG-76R, RG-pto11, and RG-prf3 tomato leaves were coinoculated with P. syringae pv tomato strain T1 at OD600 = 10−3 and A. tumefaciens strain C58C1 carrying Sl RIN4 at OD600 = 0.5. Leaves of transgenic N. benthamiana expressing Sl Pto were coinoculated with P. syringae pv tomato DC3000, a knockout derivative of P. syringae pv tomato DC3000 lacking avrPto, or a T3SS-deficient mutant of the same strain (at OD600 = 10−3) and A. tumefaciens strain C58C1 (at OD600 = 1.0) carrying Sl RIN4 expressed from the DEX-inducible promoter (Aoyama and Chua, 1997). After 24 h, expression of Sl RIN4 was induced by applying 10 mM DEX (Sigma-Aldrich) on the plant leaves.
Leaves of transgenic N. benthamiana that expressed Pto were coinoculated with a strain of Pseuodmonas fluorescens 55 (pHIR11) (OD600 = 0.1) expressing or not expressing avrPto and A. tumefaciens (OD600 = 1.0) carrying Sl RIN4 or GFP (as a control). After 24 h, leaf samples were harvested and immunoblotted using anti-HA or anti-GFP antibodies (see below). The mock treatment was infiltration with 10 mM MgCl2 solution.
For testing the HR of P. fluorescens in N. benthamiana expressing Pto, leaves of the transgenic N. benthamiana plants were inoculated with individual strains (OD600 = 1.0) of P. fluorescens 55 (pHIR11) expressing or not expressing avrPto. Only P. fluorescens expressing avrPto elicited a HR 48 h after the inoculation.
For proteasome and protease inhibitor treatments, leaves of transgenic N. benthamiana expressing Pto were coinfiltrated with A. tumefaciens C58C1 carrying pTA7002-avrPto and Sl RIN4. One day after inoculation, aqueous solutions of 100 μM MG132 or 50 μM protease inhibitor cocktail (Sigma-Aldrich) were infiltrated into the leaves. After an additional 2 h, expression of AvrPto was induced by spraying the leaves with 30 μM DEX solution. Four hours after induction, the samples were harvested for immunoblotting as described below.
Protein Assays
Plant leaves were harvested at the point of incipient chlorosis, prior to any macroscopic necrosis. This was typically 24 h after inoculation or infiltration but varied from 16 to 24 h after inoculation or infiltration depending on the genes being expressed, the age of the plant, and the temperature. Samples were ground into powder in liquid nitrogen. The ground tissues were resuspended in extraction buffer (100 mM Na-phosphate, pH 7.0, 5 mM DTT, and 1% SDS) and boiled for 10 min. Samples were centrifuged at 14,000 rpm for 15 min. The protein concentration in the supernatant was measured using the BCA assay (Pierce). Eight micrograms of total protein was loaded into a mini SDS-PAGE gel (Invitrogen) and transferred to Immobilon-P membrane (Millipore) according to the manufacturer's protocols. Each membrane was hybridized with HA-tag antibody (Convance) or GFP antibody (Convance) to detect the presence of RIN4 protein or GFP protein, respectively. Dilutions of HA-tag primary and secondary antibodies were 1:1000 and 1:10,000, respectively. Dilutions of GFP primary and secondary antibodies were 1:2000 and 1:5000, respectively.
Yeast Two-Hybrid Assays
Yeast two-hybrid assays used yeast strains EGY48 and YM4721 as described previously (Finley and Brent, 1994). Interactions were tested using Sl RIN4, At RIN4, and AvrPto expressed from the LexA bait vector pSLR3 and other components expressed from the prey vector pSLR4 following standard protocols for the LexA system (Clontech).
Immunoprecipitation
N. benthamiana tissues for immunoprecipitation were harvested at least 22 h after infiltration of Agrobacteria solutions for A. tumefaciens–mediated transient expression of the target protein. Harvested tissues were ground to a powder in liquid nitrogen. Ground samples were resuspended in IP buffer (50 mM HEPES, 150 mM NaCl, 10 mM EDTA, 1% Triton X-100, 5 mM DTT, and 1× Complete Protease Inhibitor [Roche]). The crude lysates were centrifuged at 14,000 rpm for 20 min at 4°C. The protein concentration in each sample was measured by the BCA assay, and equal amounts of each supernatant were used for immunoprecipitation. Samples were incubated with end-over-end agitation with 25 μL anti-HA beads (Roche) for 3 h at 4°C in a spin column (Sigma-Aldrich). Immunocomplexes were washed three times in washing buffer (50 mM HEPES, 150 mM NaCl, 10 mM EDTA, and 1% Triton X-100) and then resuspended in 25 μL of sample buffer (Invitrogen), boiled for 5 min, pelleted, and loaded onto SDS-PAGE gels.
Pathogenicity Assays
P. syringae pv tomato strain T1, carrying avrPto or the empty vector, was assayed for growth in both tomato RG-76R and a transformant of RG-76R with reduced levels of RIN4 due to RNAi. After inoculation, the plants were kept in 80% humidity and at 28°C in a growth chamber. Three leaf discs from five plants of each genotype were removed 4 h after vacuum-inoculation (day 0) using a 2-cm-diameter cork borer and ground in 10 mM MgCl2; three leaf discs were removed from each plant at day 2 and at day 4 after inoculation and treated similarly. Serial dilutions were plated on KB agar plates (20 g/L DIFCO protease peptone No.3 [BD], 1.5 g/L K2HPO4, 1.6 g/L MgSO4x7H2O, and 10 mL/L glycerol, pH 7.2) containing 25 μg/mL rifampicin and 25 μg/mL kanamycin. Bacterial colonies were incubated at 28°C for 2 d and then colonies were counted.
For measuring the transcription level of RIN4 in response to host or nonhost pathogen, plants were inoculated with P. syringae pv tomato T1 or P. syringae pv syringae B728A at concentrations of OD600 = 1.0. Leaf tissues were collected at different time points and prepared for real-time PCR (see above).
Nonspecific Elicitation of HR
To elicit the HR independently of the Pto pathway, we transiently overexpressed the Arabidopsis resistance gene RPP8 from the 35S promoter. In A. tumefaciens–mediated transient expression assays, RPP8 expression elicited an HR similar in magnitude and timing to that elicited by the Pto/AvrPto interaction. After 48 h, total protein was extracted from the infiltrated tissue and hybridized to the HA-tag antibody.
Generation of Stable Transgenic Plants
We attempted to generate transgenic tomato plants that either overexpressed RIN4 in the sense orientation or expressed an RNAi construct containing a fragment of RIN4. The overexpressing transgenic plants were generated using vector pGWB15 (T. Nakagawa, Research Institute of Molecular Genetics, Shimane University, Japan). RNAi constructs were made by cloning a fragment of Sl RIN4 (1 to 500 bp from the initial ATG) into pGollum (T. Wroblewski, unpublished data) behind the CaMV 35S promoter. This vector results in transcription of an inverted repeat of the target gene in concert with a fragment of the β-glucuronidase (GUS) reporter gene. This allows the degree of silencing to be monitored using transient expression with A. tumefaciens carrying a CaMV 35S:GUS gene (Wroblewski et al., 2005). Cocultivation of tomato with A. tumefaciens to generate transgenic plants was performed at the UC Davis Plant Transformation Facility (http://ucdptf.ucdavis.edu/index.shtml). Only one transgenic plant that overexpressed RIN4 using pGWB15 was obtained despite multiple attempts in which other genes were simultaneously successfully introduced. The frequency of obtaining transgenic plants containing the RIN4 RNAi construct was also markedly reduced compared with other constructs introduced at the same time, and only 10 independent transgenics were obtained. Silencing in T1 and T2 plants was assayed by assessing the level of A. tumefaciens–mediated transient expression of GUS (Wroblewski et al., 2005). The 10 T1 plants varied in their levels of expression of RIN4 as measured by quantitative real-time PCR; some had nearly wild-type levels; in others, levels were variously reduced to up to 90% less than wild type. T1 plants were self-pollinated to generate sufficient T2 material for analysis. Four T2 lines from two original T1 plants that exhibited at least 85% suppression of RIN4 expression were available for pathogenicity assays of P. syringae pv tomato T1 with AvrPto. Three T2 lines from one transgenic line were available for pathogenicity assays of P. syringae pv tomato T1 with empty vector.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AT3G25070 (At RIN4), TC174419 (Sl RIN4), L20425 (AvrPto), AY074795 (AvrPtoB), AE016858 (HopQ1-1), AY208298 (HopAm1), TC170178 (Sl α-tubulin), AAF76306 (Sl Pto), and AF220602 (Sl Prf).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Amino Acid Sequence Alignment of At RIN4 and Sl RIN4.
Supplemental Figure 2. Real-Time PCR Analysis Showing the Similar Expression Level of Sl RIN4 and the Housekeeping Gene Encoding α-Tubulin.
Supplemental Figure 3. Enhanced Transcription of Sl RIN4 after Challenge with Virulent or Avirulent Strains.
Supplemental Figure 4. Protein–Protein Interactions in Yeast and in Planta.
Supplemental Figure 5. Degradation of RIN4 by AvrRpt2 in N. benthamiana.
Supplemental Figure 6. Degradation of RIN4 Was Triggered by Two Bacteria Effectors, AvrPto and AvrPtoB, and an Autoactive Pto Mutant and Was Dependent on Pto and Prf.
Supplemental Table 1. Oligonucleotide Primers Used for Molecular Constructions.
Supplemental Table 2. Primers Used for Quantitative PCR Analysis of Sl RIN4 Expression in Transgenic Plants.
Supplemental Table 3. The List of Constructs in This Article.
Supplementary Material
Acknowledgments
This study was supported by an award from the National Science Foundation Plant Genome Program (NSF-DBI-02-11923). We thank M.-J. Truco for bioinformatics support, J. Greenberg for critical reading of the manuscript, G. Coaker for generously providing anti-At RIN4 serum, K. Cavanaugh for technical assistance, B. Martineau for editing the manuscript, A. Collmer for P. syringae pv tomato strain DC3000 hrcC and P. fluorescens 55 pHIR11, as well as T. Nakagawa for vectors pGWB15 and pGWB14.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Richard W. Michelmore (rwmichelmore@ucdavis.edu).
Online version contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Alfano, J.R., and Collmer, A. (1997). The type III (Hrp) secretion pathway of plant pathogenic bacteria: Trafficking harpins, Avr proteins, and death. J. Bacteriol. 179: 5655–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano, J.R., and Collmer, A. (2004). Type III secretion system effector proteins: Double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42: 385–414. [DOI] [PubMed] [Google Scholar]
- Aoyama, T., and Chua, N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612. [DOI] [PubMed] [Google Scholar]
- Athman, R., and Philpott, D. (2004). Innate immunity via Toll-like receptors and Nod proteins. Curr. Opin. Microbiol. 7: 25–32. [DOI] [PubMed] [Google Scholar]
- Axtell, M.J., Chisholm, S.T., Dahlbeck, D., and Staskawicz, B.J. (2003). Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol. Microbiol. 49: 1537–1546. [DOI] [PubMed] [Google Scholar]
- Chang, J.H., Tobias, C.M., Staskawicz, B.J., and Michelmore, R.W. (2001). Functional studies of the bacterial avirulence protein AvrPto by mutational analysis. Mol. Plant Microbe Interact. 14: 451–459. [DOI] [PubMed] [Google Scholar]
- Chang, J.H., Urbach, J.M., Law, T.F., Arnold, L.W., Hu, A., Gombar, S., Grant, S.R., Ausubel, F.M., and Dangl, J.L. (2004). A high-throughput, near-saturating screen for type III effector genes from Pseudomonas syringae. Proc. Natl. Acad. Sci. USA 102: 2549–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, S.T., Dahlbeck, D., Krishnamurthy, N., Day, B., Sjolander, K., and Staskawicz, B.J. (2005). Molecular characterization of proteolytic cleavage sites of the Pseudomonas syringae effector AvrRpt2. Proc. Natl. Acad. Sci. USA 102: 2087–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collmer, A., Badel, J.L., Charkowski, A.O., Deng, W.L., Fouts, D.E., Ramos, A.R., Rehm, A.H., Anderson, D.M., Schneewind, O., van Dijk, K., and Alfano, J.R. (2000). Pseudomonas syringae Hrp type III secretion system and effector proteins. Proc. Natl. Acad. Sci. USA 97: 8770–8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J., and Jones, J.D. (1998). Plant-microbe interactions. Affairs of the plant: Colonization, intolerance, exploitation and co-operation in plant-microbe interactions. Curr. Opin. Plant Biol. 1: 285–287. [DOI] [PubMed] [Google Scholar]
- Day, B., Dahlbeck, D., Huang, J., Chisholm, S.T., Li, D., and Staskawicz, B.J. (2005). Molecular basis for the RIN4 negative regulation of RPS2 disease resistance. Plant Cell 17: 1292–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley, R.L., Jr., and Brent, R. (1994). Interaction mating reveals binary and ternary connections between Drosophila cell cycle regulators. Proc. Natl. Acad. Sci. USA 91: 12980–12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göhre, V., and Robatzek, S. (2008). Breaking the barriers: Microbial effector molecules subvert plant immunity. Annu. Rev. Phytopathol. 46: 189–215. [DOI] [PubMed] [Google Scholar]
- Göhre, V., Spallek, T., Haweker, H., Mersmann, S., Mentzel, T., Boller, T., de Torres, M., Mansfield, J.W., and Robatzek, S. (2008). Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 18: 1824–1832. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez, L., Bauer, Z., and Boller, T. (2001). Both the extracellular leucine-rich repeat domain and the kinase activity of FSL2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell 13: 1155–1163. [PMC free article] [PubMed] [Google Scholar]
- Grant, S.R., Fisher, E.J., Chang, J.H., Mole, B.M., and Dangl, J.L. (2006). Subterfuge and manipulation: Type III effector proteins of phytopathogenic bacteria. Annu. Rev. Microbiol. 60: 425–449. [DOI] [PubMed] [Google Scholar]
- Hann, D.R., and Rathjen, J.P. (2007). Early events in the pathogenicity of Pseudomonas syringae on Nicotiana benthamiana. Plant J. 49: 607–618. [DOI] [PubMed] [Google Scholar]
- Hauck, P., Thilmony, R., and He, S.Y. (2003). A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. USA 100: 8577–8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, P., Shan, L., Lin, N.C., Martin, G.B., Kemmerling, B., Nurnberger, T., and Sheen, J. (2006). Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125: 563–575. [DOI] [PubMed] [Google Scholar]
- Hirano, S.S., Charkowski, A.O., Collmer, A., Willis, D.K., and Upper, C.D. (1999). Role of the Hrp type III protein secretion system in growth of Pseudomonas syringae pv. syringae B728a on host plants in the field. Proc. Natl. Acad. Sci. USA 96: 9851–9856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara, N., and Nunez, G. (2003). NODs: Intracellular proteins involved in inflammation and apoptosis. Nat. Rev. Immunol. 3: 371–382. [DOI] [PubMed] [Google Scholar]
- Jones, D.A., and Takemoto, D. (2004). Plant innate immunity - Direct and indirect recognition of general and specific pathogen-associated molecules. Curr. Opin. Immunol. 16: 48–62. [DOI] [PubMed] [Google Scholar]
- Jones, J.D., and Dangl, J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Kim, H.S., Desveaux, D., Singer, A.U., Patel, P., Sondek, J., and Dangl, J.L. (2005. a). The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc. Natl. Acad. Sci. USA 102: 6496–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M.G., da Cunha, L., McFall, A.J., Belkhadir, Y., DebRoy, S., Dangl, J.L., and Mackey, D. (2005. b). Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 121: 749–759. [DOI] [PubMed] [Google Scholar]
- Kim, Y.J., Lin, N.C., and Martin, G.B. (2002). Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109: 589–598. [DOI] [PubMed] [Google Scholar]
- Lahaye, T., and Bonas, U. (2001). Molecular secrets of bacterial type III effector proteins. Trends Plant Sci. 6: 479–485. [DOI] [PubMed] [Google Scholar]
- Lindeberg, M., Cartinhour, S., Myers, C.R., Schechter, L.M., Schneider, D.J., and Collmer, A. (2006). Closing the circle on the discovery of genes encoding Hrp regulon members and type III secretion system effectors in the genomes of three model Pseudomonas syringae strains. Mol. Plant Microbe Interact. 19: 1151–1158. [DOI] [PubMed] [Google Scholar]
- Lindeberg, M., Stavrinides, J., Chang, J.H., Alfano, J.R., Collmer, A., Dangl, J.L., Greenberg, J.T., Mansfield, J.W., and Guttman, D.S. (2005). Proposed guidelines for a unified nomenclature and phylogenetic analysis of type III Hop effector proteins in the plant pathogen Pseudomonas syringae. Mol. Plant Microbe Interact. 18: 275–282. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Belkhadir, Y., Alonso, J.M., Ecker, J.R., and Dangl, J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Holt III, B.F., Wiig, A., and Dangl, J.L. (2002). RIN4 interacts with Pseudomonas syringae Type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754. [DOI] [PubMed] [Google Scholar]
- McDowell, J.M., and Simon, S.A. (2008). Molecular diversity at the plant-pathogen interface. Dev. Comp. Immunol. 32: 736–744. [DOI] [PubMed] [Google Scholar]
- McHale, L., Tan, X., Koehl, P., and Michelmore, R.W. (2006). Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 7: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., Kozik, A., Griego, A., Kuang, H., and Michelmore, R.W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucyn, T.S., Clemente, A., Andriotis, V.M., Balmuth, A.L., Oldroyd, G.E., Staskawicz, B.J., and Rathjen, J.P. (2006). The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell 18: 2792–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgett, M.B. (2005). New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu. Rev. Plant Biol. 56: 509–531. [DOI] [PubMed] [Google Scholar]
- Ntoukakis, V., Mucyn, T.S., Gimenez-Ibanez, S., Chapman, H.C., Gutierrez, J.R., Balmuth, A.L., Jones, A.M., and Rathjen, J.P. (2009). Host inhibition of a bacterial virulence effector triggers immunity to infection. Science 324: 784–787. [DOI] [PubMed] [Google Scholar]
- Rathjen, J.P., Chang, J.H., Staskawicz, B.J., and Michelmore, R.W. (1999). Constitutively active Pto induces a Prf-dependent hypersensitive response in the absence of avrPto. EMBO J. 18: 3232–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riely, B.K., and Martin, G.B. (2001). Ancient origin of pathogen recognition specificity conferred by the tomato disease resistance gene Pto. Proc. Natl. Acad. Sci. USA 98: 2059–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter, C., and Dangl, J.L. (1996). Interference between two specific pathogen recognition events mediated by distinct plant disease resistance genes. Plant Cell 8: 251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, L.E., Langley, C.H., Bernal, A.J., and Michelmore, R.W. (2005). Natural variation in the Pto pathogen resistance gene within species of wild tomato (Lycopersicon). I. Functional analysis of Pto alleles. Genetics 171: 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, L.E., Michelmore, R.W., and Langley, C.H. (2007). Natural variation in the Pto disease resistance gene within species of wild tomato (Lycopersicon) II. Population genetics of Pto. Genetics 175: 1307–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron, J.M., Oldroyd, G.E., Rommens, C.M., Scofield, S.R., Kim, H.S., Lavelle, D.T., Dahlbeck, D., and Staskawicz, B.J. (1996). Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell 86: 123–133. [DOI] [PubMed] [Google Scholar]
- Sarkar, S.F., Gordon, J.S., Martin, G.B., and Guttman, D.S. (2006). Comparative genomics of host-specific virulence in Pseudomonas syringae. Genetics 174: 1041–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter, L.M., Vencato, M., Jordan, K.L., Schneider, S.E., Schneider, D.J., and Collmer, A. (2006). Multiple approaches to a complete inventory of Pseudomonas syringae pv. tomato DC3000 type III secretion system effector proteins. Mol. Plant Microbe Interact. 19: 1180–1192. [DOI] [PubMed] [Google Scholar]
- Scofield, S.R., Tobias, C.M., Rathjen, J.P., Chang, J.H., Lavelle, D.T., Michelmore, R.W., and Staskawicz, B.J. (1996). Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science 274: 2063–2065. [DOI] [PubMed] [Google Scholar]
- Shan, L., He, P., Li, J., Heese, A., Peck, S.C., Nurnberger, T., Martin, G., and Sheen, J. (2008). Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4: 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Q.H., Saijo, Y., Mauch, S., Biskup, C., Bieri, S., Keller, B., Seki, H., Ulker, B., Somssich, I.E., and Schulze-Lefert, P. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103. [DOI] [PubMed] [Google Scholar]
- Takemoto, D., and Jones, D.A. (2005). Membrane release and destabilization of Arabidopsis RIN4 following cleavage by Pseudomonas syringae AvrRpt2. Mol. Plant Microbe Interact. 18: 1258–1268. [DOI] [PubMed] [Google Scholar]
- Tang, X., Frederick, R.D., Zhou, J., Halterman, D.A., Jia, Y., and Martin, G.B. (1996). Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274: 2060–2063. [DOI] [PubMed] [Google Scholar]
- Tang, X., Xie, M., Kim, Y.J., Zhou, J., Klessig, D.F., and Martin, G.B. (1999). Overexpression of Pto activates defense responses and confers broad resistance. Plant Cell 11: 15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias, C.M., Oldroyd, G.E., Chang, J.H., and Staskawicz, B.J. (1999). Plants expressing the Pto disease resistance gene confer resistance to recombinant PVX containing the avirulence gene AvrPto. Plant J. 17: 41–50. [DOI] [PubMed] [Google Scholar]
- van der Hoorn, R.A., and Kamoun, S. (2008). From guard to decoy: A new model for perception of plant pathogen effectors. Plant Cell 20: 2009–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinatzer, B.A., Teitzel, G.M., Lee, M.W., Jelenska, J., Hotton, S., Fairfax, K., Jenrette, J., and Greenberg, J.T. (2006). The type III effector repertoire of Pseudomonas syringae pv. syringae B728a and its role in survival and disease on host and non-host plants. Mol. Microbiol. 62: 26–44. [DOI] [PubMed] [Google Scholar]
- Wei, C.F., Kvitko, B.H., Shimizu, R., Crabill, E., Alfano, J.R., Lin, N.C., Martin, G.B., Huang, H.C., and Collmer, A. (2007). A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J. 51: 32–46. [DOI] [PubMed] [Google Scholar]
- Wroblewski, T., Tomczak, A., and Michelmore, R. (2005). Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol. J. 3: 259–273. [DOI] [PubMed] [Google Scholar]
- Wu, A.J., Andriotis, V.M., Durrant, M.C., and Rathjen, J.P. (2004). A patch of surface-exposed residues mediates negative regulation of immune signaling by tomato Pto kinase. Plant Cell 16: 2809–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulf, J., Pascuzzi, P.E., Fahmy, A., Martin, G.B., and Nicholson, L.K. (2004). The solution structure of type III effector protein AvrPto reveals conformational and dynamic features important for plant pathogenesis. Structure 12: 1257–1268. [DOI] [PubMed] [Google Scholar]
- Xiang, C., Han, P., Lutziger, I., Wang, K., and Oliver, D.J. (1999). A mini binary vector series for plant transformation. Plant Mol. Biol. 40: 711–717. [DOI] [PubMed] [Google Scholar]
- Xiang, T., Zong, N., Zou, Y., Wu, Y., Zhang, J., Xing, W., Li, Y., Tang, X., Zhu, L., Chai, J., and Zhou, J.M. (2008). Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 18: 74–80. [DOI] [PubMed] [Google Scholar]
- Xing, W., Zou, Y., Liu, Q., Liu, J., Luo, X., Huang, Q., Chen, S., Zhu, L., Bi, R., Hao, Q., Wu, J.-W., and Chai, J. (2007). The structural basis for activation of plant immunity by bacterial effector protein AvrPto. Nature 449: 243–247. [DOI] [PubMed] [Google Scholar]
- Xiao, F., Lu, M., Li, J., Zhao, T., Yi, S.Y., Thara, V.K., Tang, X., and Zhou, J.M. (2003). Pto mutants differentially activate Prf-dependent, avrPto-independent resistance and gene-for-gene resistance. Plant Physiol. 131: 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J.M., and Chai, J. (2008). Plant pathogenic bacterial type III effectors subdue host responses. Curr. Opin. Microbiol. 11: 179–185. [DOI] [PubMed] [Google Scholar]
- Zipfel, C., and Rathjen, J.P. (2008). Plant immunity: AvrPto targets the frontline. Curr. Biol. 18: R218–R220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.