Abstract
Plant immune responses depend on dynamic signaling events across the nuclear envelope through nuclear pores. Nuclear accumulation of certain resistance (R) proteins and downstream signal transducers are critical for their functions, but it is not understood how these processes are controlled. Here, we report the identification, cloning, and analysis of Arabidopsis thaliana modifier of snc1,7 (mos7-1), a partial loss-of-function mutation that suppresses immune responses conditioned by the autoactivated R protein snc1 (for suppressor of npr1-1, constitutive 1). mos7-1 single mutant plants exhibit defects in basal and R protein–mediated immunity and in systemic acquired resistance but do not display obvious pleiotropic defects in development, salt tolerance, or plant hormone responses. MOS7 is homologous to human and Drosophila melanogaster nucleoporin Nup88 and resides at the nuclear envelope. In animals, Nup88 attenuates nuclear export of activated NF-κB transcription factors, resulting in nuclear accumulation of NF-κB. Our analysis shows that nuclear accumulation of snc1 and the defense signaling components Enhanced Disease Susceptibility 1 and Nonexpresser of PR genes 1 is significantly reduced in mos7-1 plants, while nuclear retention of other tested proteins is unaffected. The data suggest that specifically modulating the nuclear concentrations of certain defense proteins regulates defense outputs.
INTRODUCTION
Innate immunity in plants against microbial pathogen infection is a dynamic process that requires stimulus-dependent spatial and temporal action of its defense regulatory components. One of the most effective disease resistance mechanisms is mediated by resistance (R) proteins. Upon infection, an R protein recognizes a specific pathogen effector (termed Avirulence [Avr] protein) and mounts a fast and robust response leading to a local hypersensitive response, a form of programmed cell death, to restrict pathogen growth and spread (Jones and Dangl, 2006). Many R genes have been cloned and the majority encodes proteins containing NB-LRR domains in which the NB is a central nucleotide binding site and LRRs are C-terminal leucine-rich repeats. There are two subclasses of NB-LRR proteins, varying according to their N termini (Martin et al., 2003; Belkhadir et al., 2004; McHale et al., 2006). TIR-NB-LRR–type R proteins carry an N-terminal Toll interleukin receptor (TIR) domain, while the CC-type has a predicted coiled-coil domain (also called leucine zipper domain) at its N terminus. These two NB-LRR classes differ in their initial mode of signaling since TIR-NB-LRR proteins activate resistance and cell death through EDS1/PAD4/SAG101 (for Enhanced Disease Susceptibility1/Phytoalexin-Deficient4/Senescence Associated Gene 101) complexes, whereas CC-NB-LRR proteins commonly use NDR1 (for Non Race-Specific Disease Resistance1) (Century et al., 1997; Aarts et al., 1998; Feys et al., 2005; Wiermer et al., 2005). NDR1 associates with the plasma membrane, while EDS1 interacts with PAD4 and SAG101 in distinct complexes in the cytosol and nucleus (Coppinger et al., 2004; Feys et al., 2005). Downstream of EDS1 and NDR1, pathways converge at the synthesis of the defense hormone salicylic acid (SA), a sufficient and necessary signal for systemic acquired resistance (SAR) (Vernooij et al., 1994). SAR represents systemic responses induced throughout the plant to enhance resistance. NPR1 (for Nonexpressor of PR genes 1) is a key positive regulator of SAR whose monomerization and nuclear accumulation is essential for its activity in stimulating defense gene expression (Cao et al., 1997; Mou et al., 2003; Tada et al., 2008).
The detailed biochemical functions of NB-LRR proteins have started to emerge in recent years. They are normally under tight negative control, but upon infection, the release of repression seems to be the driving force for the resistance responses. For example, the Arabidopsis thaliana defense modulator RPM1-interacting protein 4 negatively regulates two different CC-NB-LRR–type R proteins, RPM1 and RPS2 (Mackey et al., 2002; Axtell and Staskawicz, 2003; Mackey et al., 2003; Kim et al., 2005). Although most NB-LRR proteins are predicted to be cytosolic (Jones and Dangl, 2006), the CC-NB-LRR class proteins MLA1 and MLA6 localize partially to and function inside the nucleus (Shen et al., 2007). Upon infection, recognition of its cognate fungal effector induces MLA interaction with repressive WRKY transcription factors, leading to deregulation of downstream defense gene expression. Also, the TIR-type NB-LRR proteins, N in tobacco (Nicotiana tabacum) and RPS4 in Arabidopsis, need to accumulate in nuclei to function (Burch-Smith et al., 2007; Wirthmueller et al., 2007). These recent discoveries suggest there may be a general requirement for nuclear localization of R proteins or their downstream signaling components in R-mediated resistance.
Previous studies of MOS3 (for Modifier of snc1,3; Zhang and Li, 2005), MOS6 (Palma et al., 2005), and RanGAP2 (Sacco et al., 2007; Tameling and Baulcombe, 2007) reveal the importance of two nucleocytoplasmic trafficking pathways in plant innate immunity: mRNA export and nuclear localization signal (NLS)-dependent nuclear protein import. It is not known whether other nucleocytoplasmic trafficking machineries, such as the one governing nuclear export signal (NES)-mediated nuclear protein export, contribute to plant disease resistance. MOS3/NUP96/SAR3 is required for mRNA export (Dong et al., 2006; Parry et al., 2006), and mutations in MOS3 confer enhanced susceptibility to both virulent and avirulent pathogens. Also, mutations in MOS6, an Importin α homolog responsible for importing proteins with an NLS to the nucleus, compromise plant defense against pathogen infection. RanGAP2, another component of the protein nuclear import machinery, interacts with the NB-LRR protein Rx, and silencing of RanGap2 impairs Rx-mediated resistance (Sacco et al., 2007; Tameling and Baulcombe, 2007).
Both MOS3 (Zhang and Li, 2005) and MOS6 (Palma et al., 2005) were identified in a forward genetic screen aimed at finding components that function downstream of R protein activation. In snc1 (for suppressor of npr1-1, constitutive 1), a point mutation resulting in an E-to-K change in the linker region between the NB and LRR of an RPP4 homolog, renders this TIR-type R protein constitutively active without pathogen recognition (Zhang et al., 2003a). As a consequence, snc1 mutant plants are dwarf, accumulate high levels of SA, and exhibit enhanced disease resistance against virulent pathogens (Li et al., 2001; Zhang et al., 2003a). As a TIR-NB-LRR protein, snc1 was accordingly found to be fully dependent on EDS1/PAD4, whose nucleocytoplasmic partitioning and complex formation is probably under tight control (Feys et al., 2005; Wiermer et al., 2005).
In this study, we report the isolation, positional cloning, and detailed functional analysis of MOS7. A partial loss-of-function mutation, mos7-1, suppresses snc1 autoimmune phenotypes, while complete loss of MOS7 in mos7-2 and mos7-3 mutants causes lethality. In the mos7-1 single mutant, basal defense against virulent pathogens, local resistance conditioned by several TIR- and CC-type NB-LRR R proteins, and SAR responses are impaired. MOS7 encodes a protein homologous to the human Nup88 nucleoporin. In Drosophila melanogaster and human, mutations in Nup88 enhance CRM1 (for Chromosomal Region Maintenance 1; also named Exportin 1; XPO1)–dependent nuclear export of activated NF-κB transcription factors (Roth et al., 2003; Xylourgidis et al., 2006). In this study, we establish that MOS7 is required for appropriate nuclear accumulation of the autoactivated R protein snc1, as well as the downstream defense signaling components EDS1 and NPR1.
RESULTS
Identification of the mos7-1 Mutant
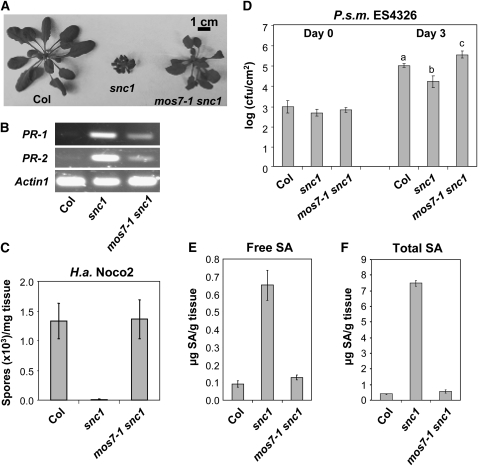
The mos7-1 mutant was identified from a MOS (modifier of snc1) forward genetic screen with fast neutrons, as described earlier (Zhang and Li, 2005). snc1 plants have a stunted stature and curly leaves due to constitutive defense activation (Zhang et al., 2003a). The suppressor screen was designed to search for mutants that resemble wild-type morphology and abolish constitutive pathogen resistance in snc1. mos7-1 snc1 double mutant plants are larger than snc1 plants (Figure 1A). In snc1 plants, several Pathogenesis Related (PR) defense marker genes are constitutively expressed. Analysis using RT-PCR showed that PR-1 and PR-2 expression was suppressed in mos7-1 snc1 compared with snc1 (Figure 1B).
Figure 1.
mos7-1 Suppresses the Autoimmune Responses in snc1.
(A) Morphology of 5-week-old soil-grown plants of Col, snc1, and mos7-1 snc1.
(B) PR gene expression in mos7-1 snc1. RNAs were prepared from 3-week-old plants grown on Murashige and Skoog media and reverse transcribed to obtain total cDNA. The cDNA samples were normalized by real-time PCR using Actin1. PR-1, PR-2, and Actin1 were amplified by 31 cycles of PCR using equal amounts of total cDNA, and the products were analyzed by agarose gel electrophoresis with ethidium bromide staining.
(C) Two-week-old soil-grown seedlings were inoculated with H.a. Noco2 at a concentration of 50,000 conidia per mL of water, and the number of conidia was quantified 7 d after inoculation. Bars represent means of four replicates ± sd.
(D) Five-week-old soil-grown plants were infiltrated with P.s.m. ES4326 (OD600 = 0.0001), and colony-forming units (cfu) were quantified at 0 and 3 d after inoculation, respectively. Bars represent means of six replicates ± sd. All data were analyzed by one-way analysis of variance. Different letters indicate statistically significant differences between genotypes (P < 0.05).
(E) and (F) Free (E) and total (F) SA were extracted from 5-week-old plants and analyzed by HPLC. Bars represent the average of four replicates ± sd.
snc1 plants exhibit enhanced resistance against virulent pathogens, including the bacterium Pseudomonas syringae pv maculicola (P.s.m.) ES4326 and the oomycete pathogen Hyaloperonospora arabidopsidis (H.a.; previously named Peronospora parasitica or Hyaloperonospora parasitica) Noco2 (Zhang et al., 2003a). To determine whether the mos7-1 mutation alters the snc1 autoimmune response, we inoculated mos7-1 snc1 double mutant plants with these pathogens. As shown in Figures 1C and 1D, mos7-1 snc1 double mutants had lost enhanced resistance to both pathogens. Bacterial growth in mos7-1 snc1 was even higher than in wild-type plants (Figure 1D).
SA levels are elevated in the snc1 mutant (Li et al., 2001). To determine whether mos7-1 affects SA accumulation in snc1, SA was extracted and measured from mos7-1 snc1 plants. As shown in Figures 1E and 1F, levels of free and total SA in mos7-1 snc1 were similar to those of wild-type plants and approximately fourfold lower than in snc1. Therefore, mos7-1 fully suppresses all known autoimmune phenotypes of snc1.
When mos7-1 snc1 was backcrossed with snc1, the F1 progeny had snc1 morphology. Of 40 F2 plants, 28 were snc1-like, whereas 12 were wild type–like. The 1:3 wild type to snc1-like ratio (χ2 = 0.53; P > 0.1) together with the F1 phenotype are consistent with mos7-1 being a single, recessive nuclear mutation.
Map-Based Cloning of mos7-1
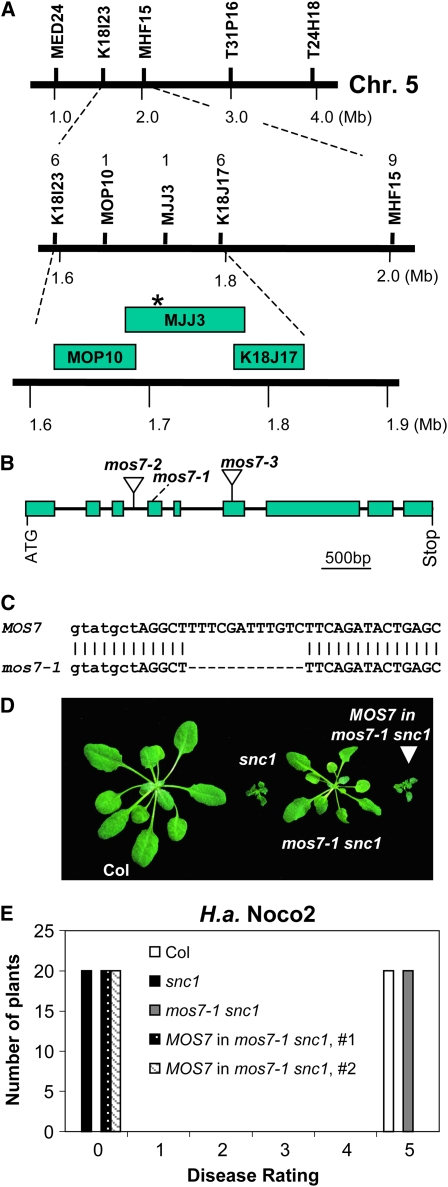
A positional cloning approach was used to identify the mutation in mos7-1 leading to the suppression of snc1. To map mos7-1, mos7-1 snc1 in Columbia (Col) ecotype was crossed with Landsberg erecta (Ler) containing the introgressed snc1 mutation, Ler-snc1 (Zhang and Li, 2005). Linkage analysis was performed on 24 F2 plants that had lost the snc1 morphology. The mos7-1 locus was found to have linkage with markers on the top arm of chromosome 5 that unfortunately was not introgressed from Ler to Ler-snc1. Therefore, a population of 1056 F3 plants was generated for fine mapping from F2 progeny that were homozygous for snc1 and heterozygous for mos7-1 from another cross between mos7-1 snc1 and Ler. The mos7-1 mutation was narrowed to the region between markers MOP10 and MJJ3 on chromosome 5 (Figure 2A). To identify the molecular lesion in mos7-1, a set of overlapping PCR fragments covering coding sequences in this region were amplified from mos7-1 snc1 and sequenced. Comparing sequences from the mutant with the Arabidopsis genome sequence revealed a 12-bp deletion in the fourth exon of At5g05680 (Figure 2C) that leads to an in-frame deletion of four amino acids at the N terminus of MOS7 (see Supplemental Figure 1 online). BLAST analysis showed that MOS7 is related to human Nup88 and Drosophila Mbo (Members only) proteins (see Supplemental Figure 1 online). MOS7 is the only Nup88 homolog in Arabidopsis. The homology between MOS7 and Nup88 and Mbo is throughout the entire length of the protein.
Figure 2.
Map-Based Cloning of mos7-1.
(A) Map position of the mos7-1 locus on chromosome 5. BAC clones and number of recombinants are indicated. Asterisk indicates the physical location of the mos7-1 locus.
(B) Gene structure of At5g05680. The exons are indicated by boxes, and the introns are represented by solid lines. Locations of mos7-1 deletion and T-DNA insertions of mos7-2 (SALK_129301) and mos7-3 (SALK_085349) are indicated.
(C) mos7-1 deletion at the DNA level. Lowercase and uppercase letters indicate intron and exon, respectively.
(D) Morphology of wild-type Col, snc1, mos7-1 snc1, and a representing transgenic line containing MOS7 transgene driven by its native promoter in snc1 mos7-1.
(E) Resistance of Col, snc1, mos7-1 snc1, and a MOS7 complementing line in snc1 mos7-1 against H.a. Noco2. The infection was rated as follows on 20 plants 7 d after infection by counting the number of conidiophores per infected leaf: 0, no conidiophores on the plants; 1, no more than five conidiophores per infected leaf; 2, 6 to 20 conidiophores on a few of the infected leaves; 3, 6 to 20 conidiophores on most of the infected leaves; 4, five or more conidiophores on all infected leaves; 5, 20 or more conidiophores on all infected leaves.
[See online article for color version of this figure.]
To confirm that MOS7 is At5g05680, a wild-type copy of At5g05680 under the control of its own promoter was transformed into mos7-1 snc1. Among 12 T1 transgenic plants obtained, all displayed snc1-like morphology (Figure 2D). Progeny of T1 plants carrying the MOS7 transgene were tested for resistance against H.a. Noco2. Constitutive resistance to H.a. Noco2 was restored in the transgenic plants (Figure 2E), indicating that At5g05680 is able to complement mos7-1 and that MOS7 is At5g05680. Two additional mutant alleles of MOS7 were obtained from the ABRC. mos7-2 (SALK_129301) contains a T-DNA insertion in the third intron and mos7-3 (SALK_085349) has a T-DNA inserted in the sixth exon of MOS7 (Figure 2B). We were unable to identify plants that are homozygous for either mos7-2 or mos7-3 from >200 progeny of plants heterozygous for the mutations, indicating that null mutations of MOS7 are lethal. This is consistent with the lethality phenotype of null mbo alleles in Drosophila (Uv et al., 2000). The viability and recessive nature of mos7-1 suggest that it is a partial loss-of-function allele of MOS7.
To obtain a mos7-1 single mutant, mos7-1 snc1 was crossed with wild-type Col plants. Lines homozygous for mos7-1 and wild type for SNC1 were selected as the mos7-1 single mutant. We tested whether mos7-1 and mos7-2 or mos7-3 are allelic by crossing plants heterozygous for mos7-2 or mos7-3 with mos7-1. mos7-1/mos7-2 plants were identified by PCR and were found to support higher bacterial growth than the wild type (see Supplemental Figure 2 online), indicating that mos7-1 and mos7-2 do not complement each other and therefore carry mutations in the same gene. Similar results were obtained with crosses between mos7-1 and mos7-3 (data not shown).
mos7-1 Single Mutant Plants Exhibit Enhanced Disease Susceptibility
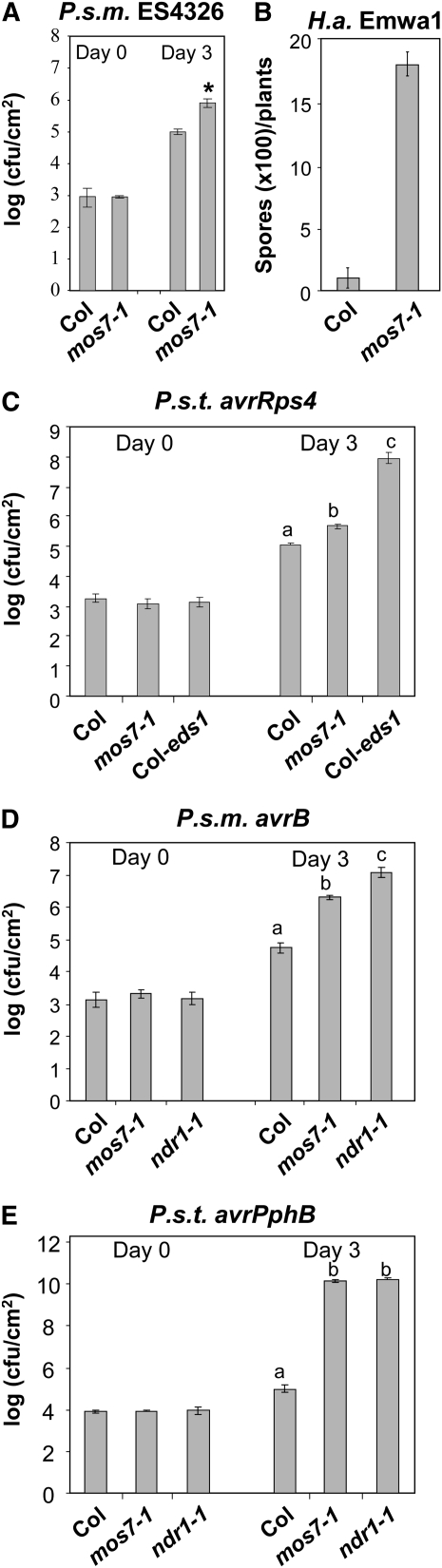
To test whether MOS7 contributes to basal defense against virulent pathogens, mos7-1 plants were challenged with P.s.m. ES4326 at a concentration of OD600 = 0.0001. Wild-type plants usually develop no disease symptoms at this low dose; however, subtle disease symptoms were observed on mos7-1 plants 3 d after inoculation. When bacterial growth was determined, ∼10-fold more bacteria accumulated in mos7-1 than in wild-type leaves (Figure 3A). Therefore, MOS7 contributes to basal resistance.
Figure 3.
Altered Basal and R Protein–Mediated Resistance in mos7-1 Single Mutant Plants.
(A) Enhanced disease susceptibility to P.s.m. ES4326 in mos7-1. The plants were infiltrated with the bacteria at OD600 = 0.0001. Leaf discs within the infiltrated area were taken at days 0 and 3 to measure the bacterial growth. Bars represent means of six replicates ± sd. Susceptibility toward P.s.m. ES4326 in mos7-1 is significantly enhanced compared with Col, as indicated by the asterisk (P < 0.0001, t test).
(B) to (E) mos7-1 mutant plants were challenged with the indicated avirulent pathogens carrying effectors that can trigger the cognate R protein–mediated resistance.
(B) 50,000 conidia/mL was used for H.a. Emwa1 inoculation.
(C) to (E) For bacterial pathogens, an inoculation dose of OD600 = 0.002 was used. Bars represent means of six replicates ± sd. Statistical analyses of the bacterial growth assays were done by one-way analysis of variance provided by StatsDirect statistical software (StatsDirect). Statistical differences among the samples are labeled with different letters (P < 0.05).
MOS7 Is Required for Resistance Mediated by Multiple R Proteins
To determine whether MOS7 is involved in resistance mediated by other TIR-NB-LRR R proteins, we challenged the mos7-1 single mutant with H.a. Emwa1 and P.s. tomato DC3000 carrying avrRps4 that are recognized by RPP4 (van der Biezen et al., 2002) and RPS4 (Hinsch and Staskawicz, 1996), respectively. As shown in Figures 3B and 3C, the mos7-1 mutation markedly reduced resistance mediated by RPP4. By contrast, RPS4 resistance was only slightly compromised.
We then tested whether resistance mediated by CC-NB-LRR–type R proteins is also impaired in mos7-1 by inoculating plants with P.s.m. ES4326 carrying avrB or P.s.t. DC3000 carrying avrPphB that encode effector proteins recognized by RPM1 (Grant et al., 1995) and RPS5 (Simonich and Innes, 1995), respectively. The mos7-1 mutant supported ∼30-fold more bacterial growth compared with wild-type plants when challenged with P.s.m. ES4326 carrying avrB (Figure 3D). Also, mos7-1 was highly susceptible to P.s.t. DC3000 carrying avrPphB, the extent of bacterial growth being the same as in susceptible ndr1 plants (Figure 3E). These data show that mos7-1 compromises resistance mediated by both TIR- and CC-NB-LRR proteins, although the degree of its effect varies depending on the R protein tested.
mos7-1 Is Compromised in SAR
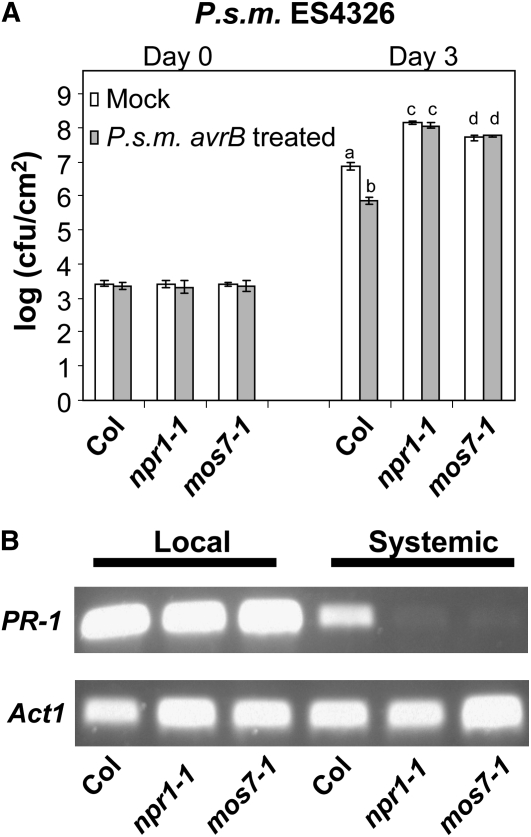
Initiation of local R protein–mediated resistance primes uninfected tissues against subsequent infections by a broad range of pathogens, a process termed SAR. As a result, the entire plant becomes more resistant to secondary infections (Durrant and Dong, 2004). To test whether SAR is affected in mos7-1 plants, we first treated leaves locally with P.s.m. ES4326 expressing avrB at a density of OD600 = 0.2 to trigger a hypersensitive response. A high dose of virulent P.s.m. ES4326 (OD600 = 0.001) was then infiltrated into the distal leaves 24 h after SAR induction. Wild-type plants showed a significant decrease in bacterial growth in avrB pretreated plants compared with mock-inoculated ones. By contrast, leaves of mos7-1 supported bacterial growth in systemic leaves of both avrB pretreated and untreated plants; this bacterial growth was similar to that seen in the SAR-defective npr1-1 mutant (Figure 4A). Therefore, the SAR response is abolished in mos7-1. A defect in SAR was also reflected by the inability of mos7-1 to boost PR-1 expression in systemic tissue after local avrB induction (Figure 4B).
Figure 4.
SAR Is Compromised in mos7-1.
(A) Growth of P.s.m. ES4326 in 5-week-old Col, npr1-1, and mos7-1 plants preinoculated with P.s.m. ES4326 avrB in 10 mM MgCl2 (gray bars) or 10 mM MgCl2 alone (mock; white bars). Bars represent means of four replicates ± sd. Different letters indicate statistically significant differences between genotypes (P < 0.05).
(B) RT-PCR for PR-1 on RNA extracted from local and systemic leaves of the indicated genotypes pretreated with P.s.m. ES4326 avrB. Actin1 was used as control. PCR conditions are as described in Figure 1B.
In SA-mediated defense, npr1-1 and tga2 tga5 tga6 triple mutants were the only genotypes reported to be sensitive to high concentrations of SA (Cao et al., 1997; Zhang et al., 2003b). To determine whether mos7-1 plants also have altered responsiveness to high SA, mos7-1 seeds were plated on Murashige and Skoog medium containing 0.2 mM SA. Similar to npr1-1, mos7-1 plants were highly sensitive to SA at an early stage of development (see Supplemental Figure 3 online). Notably, whereas npr1-1 plants remained bleached on high SA plates, mos7-1 plants slowly recovered after 5 d and eventually restored a wild type–like appearance (see Supplemental Figure 3 online).
mos7-1 Plants Do Not Exhibit Defects in Salt Tolerance, Ethylene, and Auxin Responses
No obvious developmental defects were observed in mos7-1, although mutant plants are slightly smaller than the wild type. Due to the lethality of its null alleles and pleiotropic phenotypes of reported Nup mutants, such as mos3 and nup160 in Arabidopsis (Dong et al., 2006; Parry et al., 2006), we investigated whether mos7-1 has other abiotic response or hormonal defects. As shown in Supplemental Figure 4 online, mos7-1 plants tolerate salt stress to the same level as the wild type. Also, no altered ethylene or auxin responses were observed in mos7-1 seedlings (see Supplemental Figures 5A and 5B online).
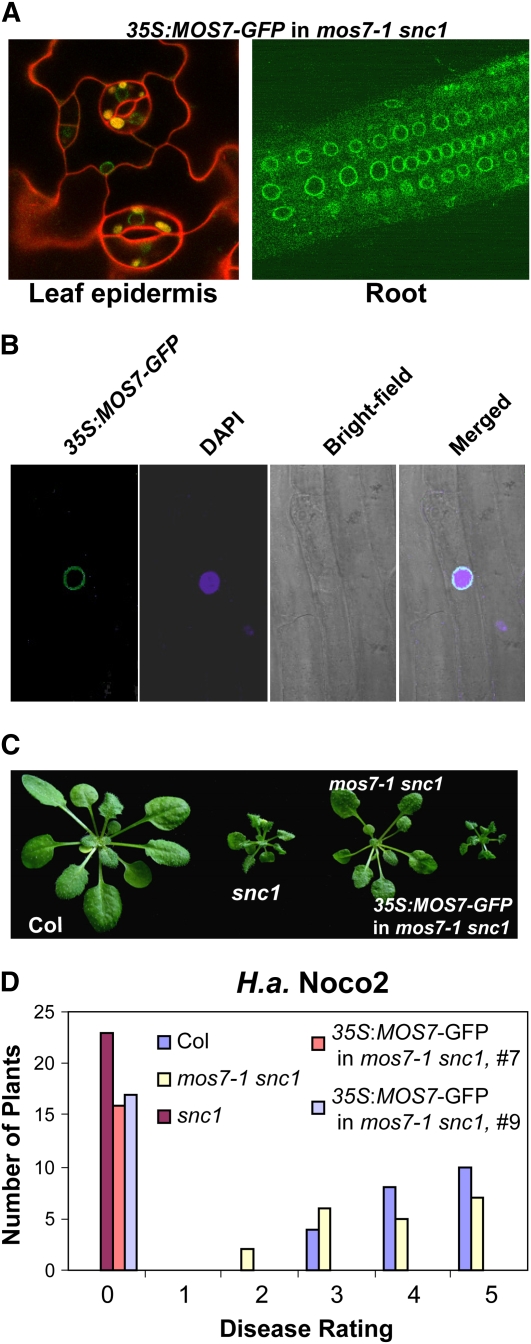
MOS7 Localizes to the Nuclear Rim
To explore the roles of MOS7 in plant innate immunity further, we investigated its subcellular localization. Initially we used the native promoter of MOS7 to drive expression of MOS7 with green fluorescent protein (GFP) fused to its C terminus. Although MOS7-GFP expressed under the native promoter complemented the mos7-1 mutation in regards to all snc1-suppressing phenotypes (data not shown), we did not observe any green fluorescence using confocal fluorescence microscopy, probably due to low abundance of the fusion protein. A construct containing MOS7 fused to a C-terminal GFP tag under the control of the constitutive 35S promoter was then made and transformed into the mos7-1 snc1 mutant. A green fluorescence signal was observed at the nuclear rim (Figures 5A and 5B). The nuclear rim localization was observed in all cell types examined, including root cells (Figures 5A and 5B) and leaf pavement cells (Figure 5A). Concentration of GFP signal at the nuclear envelope is consistent with MOS7 being a member of the nuclear pore complex, as predicted by its similarity to nuclear pore components human Nup88 and Drosophila Mbo. The MOS7-GFP transgene not only complemented the morphological phenotypes of mos7-1 snc1 (Figure 5C) but also restored a constitutive defense response against virulent H.a. Noco2 (Figure 5D), suggesting that the MOS7-GFP fusion protein localizes correctly inside the cell.
Figure 5.
Subcellular Localization of MOS7-GFP.
(A) MOS7-GFP fluorescence in leaf pavement and root cells of mos7-1 snc1 transgenic plants expressing MOS7-GFP under the control of 35S promoter. Plant cell walls were stained with 5 mg/mL propidium iodine (red) in the left panel.
(B) MOS7-GFP fluorescence, 4',6-diamidino-2-phenylindole (DAPI) staining of the nucleus, bright-field, and merged fluorescence channels in root cells. Pictures in (A) and (B) were taken on 2-week-old plate-grown plants.
(C) Complementation of mos7-1 by MOS7-GFP expressed by the 35S promoter.
(D) Restoration of enhanced disease resistance in mos7-1 snc1 transformed with MOS7-GFP driven by 35S promoter. The disease ratings are as described in Figure 2E.
Since mos7-1 is a partial loss-of-function allele of MOS7, we investigated whether mos7-1 mutation affected MOS7 expression or protein subcellular localization. No difference in MOS7 gene expression was observed between mos7-1 and the wild type with quantitative RT-PCR (data not shown). mos7-1-GFP localizes to the nuclear rim as does wild-type MOS7 protein (see Supplemental Figure 6 online), indicating that mos7-1 phenotypes are not caused by misexpression or mislocalization of the nucleoporin.
NES-Mediated Nuclear Protein Export Is Enhanced in mos7-1
In fruitfly and human cells, the function of the MOS7 homolog Nup88 is to anchor Nup214 and CRM1 at the nuclear envelop to attenuate NES/CRM-mediated nuclear export (Roth et al., 2003; Bernad et al., 2004; Xylourgidis et al., 2006). Mutations or depletion of Nup88 in animals leads to increased nuclear export of NF-κB transcription factors (Uv et al., 2000; Xylourgidis et al., 2006; Takahashi et al., 2008). In Arabidopsis, CRM1-like transport is mediated by XPO1 (Haasen et al., 1999), a protein with homology to human CRM1.
To test whether mos7-1 also affects NES-mediated nuclear protein export, we took advantage of an established nuclear export/import assay system (Haasen et al., 1999). Here, the cytosolic protein chalcone synthase (CHS) is fused to the NLS from the SV40 large antigen together with the Leu-rich NES from the HIV-1 Rev protein and GFP. Localization of the recombinant protein (GFP-NLS-CHS-NES) can be monitored by microscopy after transfection of protoplasts. When GFP-NLS-CHS-NES was transfected into wild-type Col mesophyll protoplasts, a nucleocytoplasmic localization was observed (see Supplemental Figure 7 online). When the same construct was transformed in mos7-1 protoplasts, minimal nuclear accumulation was observed (see Supplemental Figure 7 online). As a control, we used the same construct in which the NLS was mutated to a nonfunctional form (GFP-NLSmut-CHS-NES). In both the wild type and mos7-1, localization of this protein was mostly cytosolic (see Supplemental Figure 7 online). When only CHS protein fused to GFP was transformed into the same genotypes, weak nuclear and cytosolic localizations were observed (see Supplemental Figure 7 online). These results show that mos7-1 cells fail to retain this NES-containing nuclear protein in the nucleus, whereas a protein that does not have an NES (GFP-CHS) is unaffected (see Supplemental Figure 7 online). The data support an activity of plant Nup88/MOS7 in CRM1/Exportin-mediated nuclear protein export. As its homologs in animal systems, mutation in mos7-1 enhances NES-mediated nuclear export, causing reduced nuclear retention of the chimeric GFP reporter (see Supplemental Figure 7 online).
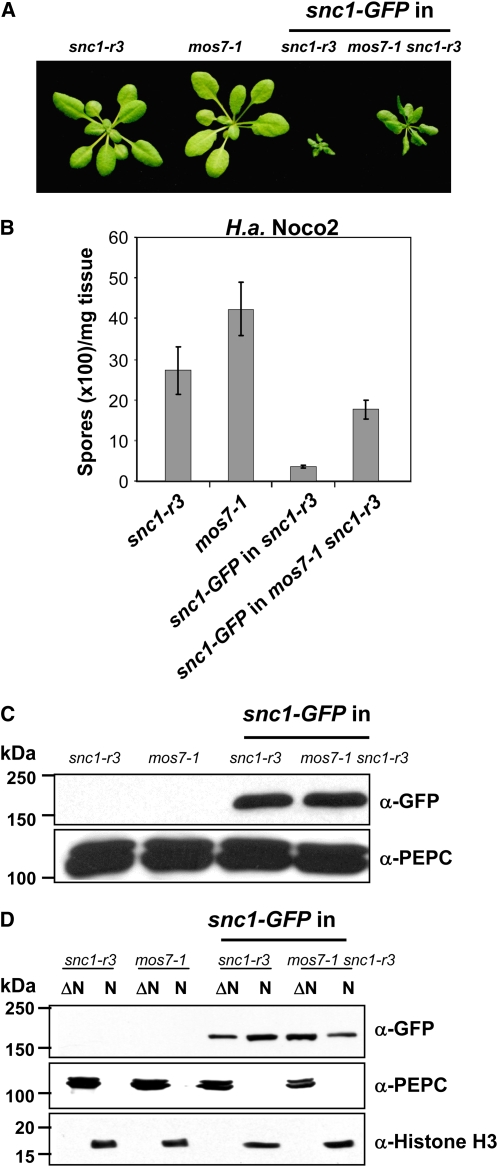
Nuclear Accumulation of snc1 Resistance Protein Is Reduced in mos7-1
Recent studies on tobacco N and Arabidopsis RPS4, two TIR-type NB-LRR proteins, revealed that both are present in the cytoplasm and nucleus and that their nuclear pools are important for triggering immune responses (Burch-Smith et al., 2007; Wirthmueller et al., 2007). However, the mechanism that regulates their partitioning between the cellular compartments remains elusive. The TIR-type R protein SNC1 contains a predicted NLS and two NES motifs. Since mos7-1 was identified as a genetic suppressor of snc1, one obvious candidate protein whose cellular distribution could be affected by mos7-1 is snc1. We therefore made a snc1-GFP fusion gene construct driven by its own promoter and transformed this into snc1-r3 that contains a deletion of the entire RPP4 cluster. snc1-r3 was identified as an snc1 revertant allele from the MOS screen (Zhang et al., 2003a) and was used in this study to avoid potential interference by endogenous SNC1-related proteins. Of many transgenic plants obtained, we were unable to find lines that consistently exhibited green fluorescence, probably due to low levels of the fusion protein. We selected one line with a single insertion site that exhibits an snc1-like morphology and crossed it with mos7-1 to generate a mos7-1 snc1-r3 line expressing the identical snc1-GFP transgene. Homozygous snc1-GFP transgenic plants were much smaller than snc1 plants, probably due to overexpression of the transgene in this particular line. mos7-1 partially suppressed the morphological phenotypes of snc1-GFP in snc1-r3 (Figure 6A) and constitutive defense against virulent H.a. Noco2 (Figure 6B). Immunoblot analysis showed that total snc1-GFP levels were similar in snc1-r3 and mos7-1 snc1-r3 (Figure 6C). Further fractionation revealed that snc1-GFP was present in both the nuclear-depleted and nuclear fraction (Figure 6D). When band intensities were measured by Quantity One 4.6.1 software (Bio-Rad), we estimated from repeated experiments that the majority of snc1 protein accumulates in the cytoplasmic compartment with 5.6 to 11.5% in the nucleus (Figure 6D). In mos7-1, there was a significant increase of snc1-GFP protein in the cytoplasm, whereas the nuclear pool of snc1-GFP was reduced (Figure 6D), ranging between 2.5 and 2.9% of total snc1 protein. We reasoned that the altered cellular distribution of snc1-GFP likely contributes to the snc1-suppressing phenotype of mos7-1.
Figure 6.
Abundance and Cellular Distribution of snc1-GFP in mos7-1 snc1-r3.
(A) Morphology of 3-week-old plants of snc1-r3, mos7-1, snc1-GFP in snc1-r3, and snc1-GFP in mos7-1 snc1-r3.
(B) H.a. Noco2 growth on the same genotypes as (A).
(C) Immunoblot analysis of snc1-GFP expressed under its native promoter in total protein extracts of unchallenged leaf tissues in snc1-r3 and mos7-1 snc1-r3. Equal loading was monitored by probing the membrane with anti-phoshpo-enol-pyruvate carboxylase (PEPC).
(D) Immunoblot analysis of snc1-GFP in nuclei-depleted (ΔN) and nuclear (N) protein extracts of the indicated genotypes. Anti-PEPC was used as a cytosolic marker, and anti-Histone H3 was used as a nuclear marker. Nuclear protein extracts (N) were 20× concentrated compared with nuclei-depleted fractions (ΔN). The experiment was repeated five times; a figure representative of all repetitions is shown. Various exposure times on films were used to make sure that the figure shown was in the linear range.
[See online article for color version of this figure.]
In Drosophila and human, mutations in Nup88 enhance NES-mediated nuclear export (Roth et al., 2003; Xylourgidis et al., 2006). We further tested whether adding an NES to snc1-GFP would affect snc1-mediated resistance. When a construct expressing snc1-GFP-NES driven by its native promoter was transformed into wild-type Col, none of the T1 transgenic plants showed snc1-like morphology, while 61% of the transgenic plants carrying the control snc1-GFP transgene in Col showed snc1-like morphology. The numbers here support that snc1 nuclear localization might be critical for its autoimmunity. Enhancing nuclear export of snc1-GFP results in reduced autoimmunity, a similar effect as observed in mos7-1.
Nuclear Accumulation of NPR1 Is Reduced in mos7-1
In Drosophila, the MOS7 homolog DNup88/Mbo affects an immune response against bacterial infection through nuclear retention of master immune regulators of the NF-κB family (Uv et al., 2000; Xylourgidis et al., 2006). Upon infection, the I-κB homolog Cactus becomes degraded, allowing the Rel/NF-κB proteins Dorsal and Dif to translocate to the nucleus and activate gene expression. During this process, DNup88 attenuates CRM1-mediated nuclear export of Dorsal and Dif, leading to their nuclear accumulation, whereas nup88 mutant larvae exhibit enhanced nuclear export of the Rel/NF-κB proteins and fail to activate an immune response (Uv et al., 2000; Roth et al., 2003; Xylourgidis et al., 2006).
The plant defense regulator NPR1 controls basal resistance and SAR downstream of the defense hormone SA and displays a somewhat analogous pattern of activation as NF-κB. Using 35S promoter-driven NPR1-GFP, it was shown that under noninducing conditions, NPR1 is sequestered in the cytoplasm as an oligomeric complex (Mou et al., 2003; Tada et al., 2008). Upon SA application, increased SA levels result in a change of the cellular redox state that in turn leads to monomerization of NPR1, allowing it to translocate from the cytosol to the nucleus to regulate downstream PR gene expression. The SAR defect observed in mos7-1 mutant plants (Figures 4A and 4B; Supplemental Figure 3 online) prompted us to investigate the contribution of MOS7 to NPR1 nuclear accumulation.
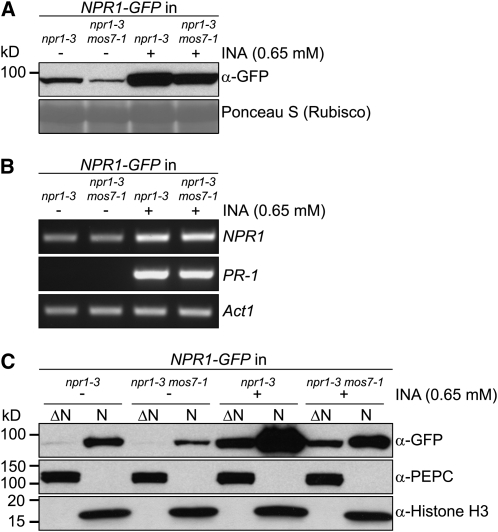
To avoid artifacts that can result from overexpression from the 35S promoter, we constructed an NPR1-GFP fusion gene driven by its native promoter. When the NPR1-GFP transgene was transformed into the null npr1-3 mutant, none of the transgenic lines consistently exhibited green fluorescence even upon SAR induction, suggesting that levels of the fusion protein in the transgenic plants are very low. We selected one line that carried a single transgene insertion and complemented fully all npr1 phenotypes (see Supplemental Figure 8 online), suggesting that NPR1-GFP expressed by its own promoter functions similarly to wild-type NPR1 protein. The NPR1-GFP transgenic line was then crossed with mos7-1 to create a mos7-1 npr1-3 double mutant expressing the same NPR1-GFP protein. As shown in Figure 7A, NPR1 total protein levels increased markedly in both the wild type and the mos7-1 mutant upon SAR induction by spraying plants with the SA analog 2,6-dichloroisonicotinic acid (INA). Lower amounts of NPR1 protein accumulated in mos7-1 before and after SAR induction compared with the wild type. No differences in NPR1 transcript abundance were observed between uninduced npr1-3 and mos7-1 npr1-3 tissues determined by RT-PCR (Figure 7B), suggesting that reduced NPR1 accumulation in mos7-1 likely results from decreased protein synthesis or stability rather than reduced transcription.
Figure 7.
NPR1 Protein Abundance and Subcellular Localization in mos7-1.
(A) NPR1-GFP expressed by its native promoter in mos7-1. Immunoblot analysis of NPR1-GFP in total protein extracts of unchallenged leaf tissues (−) and leaf tissues harvested 24 h after spraying plants with 0.65 mM INA (+). Equal loading was monitored by staining the membrane with Ponceau S.
(B) RT-PCR for NPR1 and PR-1 on RNA extracted from 4-week-old plants of the indicated genotypes treated with (+) or without (−) INA. Actin1 expression was used as control. NPR1 and Actin1 were amplified by 29 cycles of PCR and PR-1 by 30 cycles of PCR using equal amounts of cDNA. PCR products were analyzed by agarose gel electrophoresis and ethidium bromide staining.
(C) Immunoblot analysis of NPR1-GFP in nuclei-depleted (ΔN) and nuclear (N) protein extracts of unchallenged (−) and INA treated (+) tissues of the indicated genotypes. Anti-PEPC was used as a cytosolic marker, and anti-Histone H3 was used as a nuclear marker. Nuclear protein extracts (N) were 35× concentrated compared with nuclei-depleted fractions (ΔN). The experiment was repeated twice with similar results.
We further investigated the cellular distribution of NPR1-GFP in mos7-1 before and after INA induction by comparing NPR1-GFP levels in nuclear and nuclei-depleted protein extracts of untreated and INA-induced tissues. As shown in Figure 7C, lower levels of NPR1-GFP accumulated in nuclei of both healthy and INA-induced mos7-1 plants than in the wild type. In contrast with the preferential depletion of nuclear snc1 in mos7-1 tissues, lower amounts of NPR1-GFP were also observed in nuclei-depleted (cytosolic) extracts of INA-treated mos7-1 compared with the wild type (Figure 7C). Since mos7-1 plants exhibit a SAR defect and nuclear accumulation of NPR1 is required for SAR induction, these data suggest that NPR1 may not be able to attain sufficient abundance in the nucleus for activation of SAR in mos7-1.
It is notable that NPR1-GFP expressed under its native promoter was detected in the nucleus of uninduced tissues (Figure 7C); this pattern was observed in multiple independent NPR1-GFP transgenic lines. This partitioning contrasts with data derived from NPR1-GFP expressed under control of the 35S constitutive promoter that showed a cytoplasmic localization of NPR1 without SAR induction (Mou et al., 2003). To rule out the possibility that unchallenged plants were already stressed, causing increased nuclear translocation of NPR1-GFP, we analyzed the expression of the SAR marker gene PR-1 in the same uninduced tissues from which nuclear extracts were generated. No PR-1 transcripts were detected in uninduced tissues, whereas strong expression was detected after INA induction (Figure 7B), suggesting that the observed nuclear pool of NPR1 represents its uninduced state. These data suggest that NPR1 is present in the nucleus of both uninduced and INA-induced tissues.
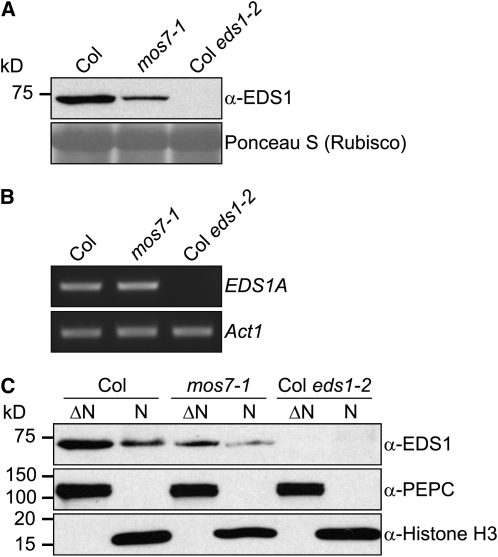
EDS1 Nuclear Accumulation Is Reduced in mos7-1
Another key plant immune regulator known to localize to both cytosol and nucleus is EDS1 (Feys et al., 2005). We investigated whether accumulation and cellular distribution of native EDS1 is affected by mos7-1. As with NPR1, EDS1 total protein was reduced in mos7-1 mutant plants compared with Col wild type (Figure 8A), although the wild type and mos7-1 have comparable EDS1 transcript levels (Figure 8B). In mos7-1, the ratio of EDS1 distributing in the cytosol and nucleus was not strongly affected. However, overall lower accumulation of EDS1 resulted in only very low levels being detected in mos7-1 nuclei (Figure 8C). We conclude that MOS7 is also necessary for EDS1 protein accumulation in the nucleus. The effect of mos7-1 on EDS1 nuclear accumulation may also contribute to the ability of mos7-1 to suppress snc1.
Figure 8.
EDS1 Protein Abundance and Subcellular Localization in mos7-1.
(A) EDS1 in mos7-1. Immunoblot analysis of EDS1 in total protein extracts of unchallenged leaf tissues. Equal loading was monitored by staining the membrane with Ponceau S.
(B) RT-PCR for EDS1A on RNA extracted from 4-week-old unchallenged plants of the indicated genotypes. Actin1 expression was used as control. EDS1 and Actin1 were amplified by 29 cycles of PCR using equal amounts of cDNA. PCR products were analyzed by agarose gel electrophoresis and ethidium bromide staining.
(C) Immunoblot analysis of EDS1 in nuclei-depleted (ΔN) and nuclear (N) protein extracts of unchallenged leaf tissues. Anti-PEPC was used as a cytosolic marker, and anti-Histone H3 was used as a nuclear marker. Nuclear protein extracts (N) were 35× concentrated compared with nuclei-depleted fractions (ΔN). The experiment was repeated twice with similar results.
Nuclear Accumulation of HDA19, CDC5, and TGA2 Is Not Affected in mos7-1
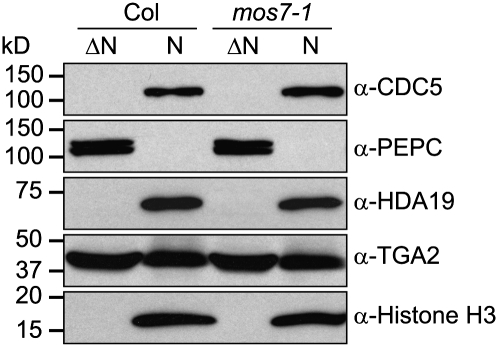
One enigma to mos7-1 is its specificity. Lethality of its null alleles indicates that wild-type MOS7 is probably required for general nuclear export. While mos7-1 appeared not to exhibit pleiotropic phenotypes, we could not rule out a global effect on protein export. To test this, we fractionated proteins from mos7-1 and the wild type and examined the localization and relative protein abundance of the known nuclear proteins CDC5, HDA19, and TGA2 using respective antibodies. CDC5 is a myb-like transcription factor, containing a strong NES and belonging to a nuclear MOS4-associated complex (Palma et al., 2007). HDA19 is a histone deacetylase, and TGA2 is a transcription factor that interacts with NPR1 (Zhang et al., 1999). Neither HDA19 nor TGA2 contains a strong NES. As shown in Figure 9, nuclear accumulation of these proteins (as well as histone H3) is unaffected in mos7-1. These data suggest that the defects we observe in mos7-1 in nuclear retention of snc1, NPR1, and EDS1 are rather selective.
Figure 9.
CDC5, PEPC, HDA19, TGA2, and Histone H3 Protein Abundance and Subcellular Localization Is Unaltered in mos7-1.
Immunoblot analysis of CDC5, PEPC, HDA19, TGA2, and Histone H3 in nuclei-depleted (ΔN) and nuclear (N) protein extracts of unchallenged leaf tissues. Anti-PEPC was used as a cytosolic marker, and anti-Histone H3 was used as a nuclear marker. Nuclear protein extracts (N) were 35× concentrated compared with nuclei-depleted fractions (ΔN).
DISCUSSION
Several recent lines of evidence suggest that nucleocytoplasmic trafficking pathways play important roles in plant innate immunity through dynamic partitioning of signaling regulators between the nucleus and cytosol. Studies of MOS3/SAR3 indicate that mRNA export, regulated by nucleoporins of the Nup107-160 complex, is required for both basal defense and R protein–mediated resistance (Zhang and Li, 2005; Dong et al., 2006; Parry et al., 2006). Also, a requirement for MOS6 and RanGAP2 in R protein–triggered resistance points to import to the nucleus of protein regulators that have NLSs as an important process in plant immunity (Palma et al., 2005; Sacco et al., 2007; Tameling and Baulcombe, 2007). However, contributions of other nucleocytoplasmic trafficking pathways or components controlling nuclear protein export and retention are unclear. In this study, we identified mos7-1 as a genetic suppressor of the autoimmune mutant snc1. We isolated the MOS7 gene and found it to encode a plant homolog of human and Drosophila Nup88. Our analysis reveals that MOS7 contributes to several aspects of plant immunity. Importantly, resistance defects of mos7-1 mutants correlate specifically with reduced nuclear accumulation of the autoactivated TIR-NB-LRR immune receptor, snc1. Nuclear pools of the downstream signaling components NPR1 and EDS1 are also strongly depleted in mos7-1, although unlike snc1, reduced abundance in nuclei appears to reflect lower total steady state levels of these defense regulators.
While only limited studies have been performed on nucleoporins in plants, nuclear pore complexes seem to be conserved among eukaryotes, in both structure and functionality (Terry et al., 2007). For example, Nup107-160 complex components in yeast and animals are critical for mRNA export (Dockendorff et al., 1997; Emtage et al., 1997). In Arabidopsis, mutations in homologs of Nup160 and Nup96, both belonging to the corresponding Nup107-160 complex, similarly affect mRNA export (Dong et al., 2006; Parry et al., 2006). Mutations in Arabidopsis MOS3/Nup96/SAR3 partially disable innate immunity (Zhang and Li, 2005). In mice, although a complete knockout of Nup96 causes lethality, knockdown of Nup96 exhibits defects in both innate and adaptive immunity (Faria et al., 2006). Our studies on MOS7 suggest that the function of Nup88 is also conserved between plants and animals since Nup88s in Drosophila and human are critical for the regulation of innate immunity. Consistent with this idea, MOS7-GFP fusion protein resides mainly at the nuclear rim (Figures 5A and 5B). In animals, Nup88 interacts directly with CAN/Nup214 and Nup358/RanBP2. The major function of Nup88 is to anchor Nup214 and CRM1 on the nuclear envelope to attenuate NES-mediated nuclear export (Roth et al., 2003; Bernad et al., 2004; Xylourgidis et al., 2006). The lethality phenotype of null mos7 mutations agrees with its potential function in general NES-mediated nuclear export. Partial loss-of-function mutations in Drosophila mbo or depletion of mammalian Nup88 lead to increased nuclear export, resulting in reduced retention of Drosophila Rel protein Dorsal and NF-κB, respectively (Uv et al., 2000; Xylourgidis et al., 2006; Takahashi et al., 2008). Our analysis suggests that nuclear accumulation of snc1, NPR1, and EDS1 is controlled in part by MOS7. mos7-1 affects nuclear levels of all three proteins as well as those of a synthetic GFP-NLS-CHS-NES but did not affect the NES-containing CDC5, indicating that the partial loss-of-function mutation in mos7-1 enhances NES-mediated nuclear export in plants with some degree of specificity.
Previous studies revealed nuclear pools of the plant NB-LRR proteins MLA, N, and RPS4 and showed that their nuclear accumulation is required for disease resistance (Burch-Smith et al., 2007; Shen et al., 2007; Wirthmueller et al., 2007). Future in-depth nuclear partitioning analysis of wild-type SNC1 should resolve whether SNC1 has nuclear activity, and if so whether the nuclear activity of SNC1 is essential for activation of defense responses. We find that snc1 (an autoactivated NB-LRR protein) localizes to both the cytosol and the nucleus. Nuclear accumulation of snc1 is disproportionately reduced in mos7-1, and adding an NES to snc1-GFP abolishes its autoimmunity, suggesting that MOS7/Nup88 promoting retention of snc1 in the nucleus is a critical process in defense activation. It is notable that resistance mediated both by CC (RPM1 and RPS5) and TIR-NB-LRR (RPP4) proteins was strongly compromised in mos7-1, suggesting that some R proteins may require MOS7 for nuclear retention. Whether these tested NB-LRR receptors have a nuclear activity remains to be characterized. On the other hand, RPS4 resistance was only marginally compromised in mos7-1, and no detectable change in RPS4 nuclear accumulation was observed in mos7-1 (see Supplemental Figure 9 online). Further analysis should establish whether regulation of nucleocytoplasmic partitioning of NB-LRR proteins by MOS7 is a general phenomenon or whether MOS7 is selective for certain R proteins depending on their intracellular abundance, spatial dynamics, and activities.
One striking phenotype of mos7-1 is its defect in SAR. Similar to the SAR-deficient npr1 mutants, mos7-1 plants had reduced PR gene expression and systemic resistance after SAR induction and exhibited reduced tolerance to high levels of SA. Since NPR1 nuclear accumulation is essential for SAR and lower NPR1 levels were observed in nuclei of mos7-1 plants after SAR induction, the SAR defects in mos7-1 are likely caused by the reduced nuclear NPR1 pool. Nuclear accumulation of EDS1 was also reduced in mos7-1. Reducing the nuclear EDS1 pool partially disables plant defenses (A.V. García and J.E. Parker, unpublished data); therefore, lower amounts of nuclear EDS1 probably contribute to the enhanced disease susceptibility phenotype of mos7-1. For both EDS1 and NPR1, there is a general decrease in protein accumulation in both nonnuclear and nuclear compartments of the mos7-1 mutant. Thus, the influence of mos7-1 on these proteins is different to its effect on snc1. The data support selective retention of snc1 inside the nucleus by MOS7. By contrast, MOS7 may act more indirectly on EDS1 and NPR1. One scenario is that lower cellular accumulation of EDS1 and NPR1 reflects an indirect influence of MOS7 by lowering basal resistance and, thus, the flux through various positive feedback loops (Feys et al., 2001; Shah, 2003). However, mos7-1 did not affect expression of NPR1 or EDS1 mRNAs, suggesting that such feedback mechanisms are not operating at the transcriptional level. Alternatively, MOS7 may act directly on these proteins, but they are subject to increased degradation after being exported from the nucleus, thereby reducing the amount in a cytosolic pool available for nuclear import.
Plant defense responses rely on dynamic translocation of signaling components across the nuclear envelope, and nucleocytoplasmic trafficking might constitute a central regulatory node for the integration of distinct signaling pathways. Although mos7-1 exhibits strong immunity defects, overexpression of MOS7 did not lead to enhanced disease resistance (see Supplemental Figure 10 online), indicating that MOS7 itself is probably not a rate-limiting defense signaling component. The identification of MOS7 reveals how the nuclear protein export pathway contributes to cellular innate immune responses and provides us with a system to test the relevance of different cellular compartments in plant pathogen recognition and defense activation.
METHODS
Plant Growth Conditions, Gene Expression Analysis, and Mutant Phenotypic Characterization
All plants were grown at 22°C under 16-h-light/8-h-night or 10-h-light/14-h-night cycles. The snc1 suppressor screen was described previously (Zhang and Li, 2005). Gene expression analysis was done by extracting RNA from 3-week-old plate-grown or 4-week-old soil-grown plants using the Totally RNA kit (Ambion). The extracted RNA was then reverse transcribed using the RT-for-PCR kit (Clontech) or SuperScript II reverse transcriptase (Invitrogen). Expression analysis for PR-1, PR-2, and Actin1 was as previously described by Zhang et al. (2003a) with cDNA samples being normalized by real-time PCR using Actin1 and the QuantiTect SYBR Green PCR kit (Qiagen). For EDS1 and NPR1 expression analysis, 0.5 μg of total RNA were reverse transcribed using SuperScript II reverse transcriptase and 0.5 μg of oligo(dT)18 primer at 42°C in a 20-μL reaction volume. Aliquots of 1 μL RT reaction products were subsequently used for PCR analysis with PCR conditions as follows: 94°C for 3 min and 29 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 45 s. Single-band PCR products were analyzed by agarose gel electrophoresis and ethidium bromide staining. Actin1 expression was used to standardize transcript levels in each sample. Gene-specific primers for RT-PCR analyses used in this study are as follows: PR-1F, 5′-GTAGGTGCTCTTGTTCTTCCC-3′, and PR-1R, 5′-CACATAATTCCCACGAGGATC-3′; PR-2F, 5′-GCTTCCTTCTTCAACCACACAGC-3′, and PR-2R 5′-CGTTGATGTACCGGAATCTGAC-3′; Actin1-F, 5′-CGATGAAGCTCAATCCAAACGA-3′, and Actin1-R, 5′-CAGAGTCGAGCACAATACCG-3′; Actin7-F, 5′-GGTGTCATGGTTGGTATGGGTC-3′, and Actin7-R, 5′-CCTCTGTGAGTAGAACTGGGTGC-3′; EDS1A-F, 5′-ATCATCATAGCTATGAGGAACTGG-3′, and EDS1A-R, 5′-CAGCTCTCTTGACGTTTGC-3′; and NPR1-F, 5′-AGAAGACAAACGAGAACAAATTCC-3′, and NPR1-R, 5′-TCAGCAGTGTCGTCTTCTCC-3′. Infection experiments with Pseudomonas syringae and Hyaloperonospora arabidopsidis (previously Hyaloperonospora parasitica) were performed as described (Li et al., 2001; Feys et al., 2005). Endogenous SA levels were determined as described by Li et al. (1999).
Map-Based Cloning of mos7-1
Positional cloning of mos7-1 was performed according to procedures described by Zhang and Li (2005). The markers used to map mos7-1 were derived from insertion-deletion polymorphisms (see Supplemental Table 1 online) identified from the genomic sequences of Col and Ler ecotypes provided by Monsanto on The Arabidopsis Information Resource homepage (Jander et al., 2002).
Construction of Plasmids
The construct used to complement the mos7-1 mutation was generated by PCR amplifying a genomic fragment containing the MOS7 coding region and its promoter 1.1 kb upstream of the ATG start codon. The primers MOS7 pro-F (5′-ccaatacacaaaatactctggc-3′) and MOS7-3′ (5′-cgcGGATCCtgcgtccctgttacagtga-3′) were used for PCR, and the fragment was subsequently cloned into pGreen0229 (Hellens et al., 2000) to obtain pG229-MOS7 for complementation analysis. To generate 35S-driven MOS7-GFP construct, full-length At5g05680 cDNA lacking a stop codon was cloned into the pBS-GFP5 vector with GFP in frame at the C terminus (Haseloff et al., 1997). The resulting MOS7-GFP fusion construct was subsequently excised and cloned into pBI1.4 containing the 35S promoter to obtain pBI-MOS7-GFP. To localize mos7-1, a 35S-driven mos7-1-GFP construct was generated by cloning full-length At5g05680 cDNA lacking a stop codon from mos7-1 (MOS7-CDS-F, 5′-CGGGGTACCATGAAATTTAACTTTAACGAGAC-3′, and MOS7-CDS-R, 5′-CGCGGATCCCATGAAACTGCTTTCTTGC-3′) into the binary vector pCAMBIA 1300 (http://www.cambia.org.au) with GFP in frame at the C terminus. NPR1-GFP construct was generated by PCR amplifying NPR1 cDNA without a stop codon and a 1.6-kb genomic region upstream of the NPR1 ATG start codon. These two fragments were subsequently cloned into a modified pGreen0229 vector that has a sequence encoding GFP in frame at the C terminus. To generate promSNC1-snc1-GFP, pG229-snc1 (Zhang et al., 2003a) lacking a stop codon was used as the template for PCR amplification. The amplified fragment was subsequently cloned into pGreen0229-GFP vector with GFP in frame at the C terminus. All constructs were sequenced to ensure accuracy in PCR and cloning. All constructs were transformed into designated genotypes using the floral dip method (Clough and Bent, 1998) to generate transgenic lines for subsequent analysis.
SAR Experiments
The infection experiment used to test SAR was performed as described by Cao et al. (1994) with minor modifications. In brief, two leaves of each 5-week-old soil-grown plant were infiltrated with P.s.m. ES4326 expressing avrB at an OD600 = 0.2 in 10 mM MgCl2 to induce SAR or 10 mM MgCl2 without bacteria (mock). Twenty-four hours after inoculation, the upper uninoculated leaves were challenged with P.s.m. ES4326 at OD600 = 0.001. Leaf discs within the infiltrated systemic area were taken immediately (day 0) and 3 d after inoculation (day 3) to measure the bacterial growth in those leaves.
Cellular Distribution of snc1-GFP
Three-week-old plants (0.5 g) were harvested and ground to a fine powder in liquid nitrogen and mixed with 2 volumes of lysis buffer (20 mM Tris-HCl, pH 7.4, 25% glycerol, 20 mM KCl, 2 mM EDTA, 2.5 mM MgCl2, 250 mM sucrose, and 1 mM PMSF). The homogenate was filtered through a 95- and 37-μm nylon netting successively. The flow-through was spun at 1500g for 10 min, and the supernatant consisting of the cytosolic fraction was collected and mixed with 5× Laemmli loading buffer and heated at 95°C for 5 min. The pellet was washed four times with 5 mL of nuclear resuspension buffer NRBT consisting of 20 mM Tris-HCl, pH 7.4, 25% glycerol, 2.5 mM MgCl2, and 0.2% Triton X-100. The final pellet was mixed with 50 μL of 1× Laemmli buffer and heated at 95°C for 5 min. Fifty microliters of each fraction was loaded on an 8% SDS-PAGE gel for protein separation. Antibodies used for immunoblot analyses were as described: anti-Histone H3 (Feys et al., 2005), anti-GFP (Wirthmueller et al., 2007), and anti-PEPC (Noël et al., 2007). Band intensities were measured by Quantity One 4.6.1 software (Bio-Rad). The experiment was repeated five times; a figure representative of all repetitions is shown. Various exposure times on films were used to make sure that the exposure used was in the linear range.
NPR1 and EDS1 Protein Expression and Localization Analyses
For NPR1 protein extraction from INA-induced tissues, 4-week-old plants were sprayed to imminent runoff with an aqueous solution of 0.65 mM INA with 0.01% Silwett L-77 surfactant. Plants were harvested 24 h after being sprayed for the first time and 3 h after being treated a second time with INA. Total protein extracts and preparation of nuclear/nuclei-depleted protein extracts were described previously (Feys et al., 2005). Anti-EDS1 antibody used for immunoblot analysis was described earlier (Feys et al., 2005).
Nuclear Protein Export Assay in Protoplast
Protoplast were prepared and transformed from 3- to 4-week-old Arabidopsis plants as previously described (Sheen, 2001; Yoo et al., 2007). Plasmid construct encoding NES, NLS variant of chalcone synthase were kindly provided by Thomas Merckle (Universitat Bielefeld, Germany; Haasen et al., 1999). Transformed protoplasts were kept in the dark overnight and observed using a confocal microscope the following day.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: At5g05680, NP_196187 (MOS7); NP_002523 (hNup88); NP_524330 (DNup88/Mbo); At4g16890 (SNC1); At2g14610 (PR-1); At3g57260 (PR-2); At2g37620 (Actin1); At5g09810 (Actin7); At3g48090 (EDS1A); and At1g64280 (NPR1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Amino Acid Alignment of MOS7, Human Nup88, and Drosophila Nup88.
Supplemental Figure 2. Allelism Test between mos7-1 and mos7-2 Using P.s.m. ES4326.
Supplemental Figure 3. Tolerance of Col, npr1-1, and mos7-1 Plants to High Concentrations of SA.
Supplemental Figure 4. Tolerance of Col and mos7-1 Plants to High Concentrations of Sodium Chloride.
Supplemental Figure 5. Hormonal Assays on mos7-1.
Supplemental Figure 6. Subcellular Localization of mos7-1-GFP Fusion Proteins in Leaf Pavement Cells.
Supplemental Figure 7. In Vivo Nuclear Transport Assay of Chalcone Synthase Fused to GFP.
Supplemental Figure 8. Complementation of npr1-3 by NPR1-GFP Expressed under the Control of Its Native Promoter.
Supplemental Figure 9. Nuclear RPS4 in mos7-1.
Supplemental Figure 10. Overexpression of MOS7 Leads to No Enhanced Disease Resistance.
Supplemental Table 1. Molecular Markers Used for Map-Based Cloning of mos7-1.
Supplemental Data Set 1. Text File of Alignment Corresponding to Supplemental Figure 1.
Supplementary Material
Acknowledgments
We thank the ABRC for seeds of mos7-2 and mos7-3 and Roger Innes for P.s.t. DC3000 carrying avrPphB. We also thank Fred Sack and EunKyoung Lee for their help with confocal microscopy. The research is supported by funds to X.L. from the Natural Sciences and Engineering Research Council of Canada, the Canadian Foundation for Innovation, the British Columbia Knowledge Development Fund, the University of British Columbia Blusson Fund, and the University of British Columbia Michael Smith Laboratories. We are thankful for a Feodor Lynen research fellowship of the Alexander von Humboldt Foundation to M.W., and Le Fonds Québécois de la Recherche sur la Nature et les Technologies to H.G. J.E.P. is grateful for an International Max Planck Research School PhD fellowship supporting A.V.G. and Deutsche Forschungsgemeinschaft SFB 670 funding of L.W.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Xin Li (xinli@interchange.ubc.ca).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Aarts, N., Metz, M., Holub, E., Staskawicz, B.J., Daniels, M.J., and Parker, J.E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 10306–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell, M.J., and Staskawicz, B.J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377. [DOI] [PubMed] [Google Scholar]
- Belkhadir, Y., Subramaniam, R., and Dangl, J.L. (2004). Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr. Opin. Plant Biol. 7: 391–399. [DOI] [PubMed] [Google Scholar]
- Bernad, R., van der Velde, H., Fornerod, M., and Pickersgill, H. (2004). Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export. Mol. Cell. Biol. 24: 2373–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith, T.M., Schiff, M., Caplan, J.L., Tsao, J., Czymmek, K., and Dinesh-Kumar, S.P. (2007). A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 5: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, A.S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63. [DOI] [PubMed] [Google Scholar]
- Century, K.S., Shapiro, A.D., Repetti, P.P., Dahlbeck, D., Holub, E., and Staskawicz, B.J. (1997). NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278: 1963–1965. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Coppinger, P., Repetti, P.P., Day, B., Dahlbeck, D., Mehlert, A., and Staskawicz, B.J. (2004). Overexpression of the plasma membrane-localized NDR1 protein results in enhanced bacterial disease resistance in Arabidopsis thaliana. Plant J. 40: 225–237. [DOI] [PubMed] [Google Scholar]
- Dockendorff, T.C., Heath, C.V., Goldstein, A.L., Snay, C.A., and Cole, C.N. (1997). C-terminal truncations of the yeast nucleoporin Nup145p produce a rapid temperature-conditional mRNA export defect and alterations to nuclear structure. Mol. Cell. Biol. 17: 906–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, C.H., Hu, X., Tang, W., Zheng, X., Kim, Y.S., Lee, B.H., and Zhu, J.K. (2006). A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol. Cell. Biol. 26: 9533–9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant, W.E., and Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42: 185–209. [DOI] [PubMed] [Google Scholar]
- Emtage, J.L., Bucci, M., Watkins, J.L., and Wente, S.R. (1997). Defining the essential functional regions of the nucleoporin Nup145p. J. Cell Sci. 110: 911–925. [DOI] [PubMed] [Google Scholar]
- Faria, A.M., Levay, A., Wang, Y., Kamphorst, A.O., Rosa, M.L., Nussenzveig, D.R., Balkan, W., Chook, Y.M., Levy, D.E., and Fontoura, B.M. (2006). The nucleoporin Nup96 is required for proper expression of interferon-regulated proteins and functions. Immunity 24: 295–304. [DOI] [PubMed] [Google Scholar]
- Feys, B.J., Moisan, L.J., Newman, M.A., and Parker, J.E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20: 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J., Wiermer, M., Bhat, R.A., Moisan, L.J., Medina-Escobar, N., Neu, C., Cabral, A., and Parker, J.E. (2005). Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17: 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, M.R., Godiard, L., Straube, E., Ashfield, T., Lewald, J., Sattler, A., Innes, R.W., and Dangl, J.L. (1995). Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269: 843–846. [DOI] [PubMed] [Google Scholar]
- Haasen, D., Kohler, C., Neuhaus, G., and Merkle, T. (1999). Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana. Plant J. 20: 695–705. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., Siemering, K.R., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94: 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832. [DOI] [PubMed] [Google Scholar]
- Hinsch, M., and Staskawicz, B. (1996). Identification of a new Arabidopsis disease resistance locus, RPS4, and cloning of the corresponding avirulence gene, avrRps4, from Pseudomonas syringae pv. pisi. Mol. Plant Microbe Interact. 9: 55–61. [DOI] [PubMed] [Google Scholar]
- Jander, G., Norris, S.R., Rounsley, S.D., Bush, D.F., Levin, I.M., and Last, R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129: 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D., and Dangl, J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Kim M.G., da Cunha L., McFall A.J., Belkhadir Y., DebRoy, S., Dangl, J.L., and Mackey, D. (2005). Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 121: 749–759. [DOI] [PubMed] [Google Scholar]
- Li, X., Clarke, J.D., Zhang, Y., and Dong, X. (2001). Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol. Plant Microbe Interact. 14: 1131–1139. [DOI] [PubMed] [Google Scholar]
- Li, X., Zhang, Y., Clarke, J.D., Li, Y., and Dong, X. (1999). Identification and cloning of a negative regulator of systemic acquired resistance, SNlI1, through a screen for suppressors of npr1–1. Cell 98: 329–339. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Belkhadir, Y., Alonso, J.M., Ecker, J.R., and Dangl, J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Holt, B.F., Wiig, A., and Dangl, J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754. [DOI] [PubMed] [Google Scholar]
- Martin, G.B., Bogdanove, A.J., and Sessa, G. (2003). Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54: 23–61. [DOI] [PubMed] [Google Scholar]
- McHale, L., Tan, X., Koehl, P., and Michelmore, R.W. (2006). Plant NBS-LRR proteins: adaptable guards. Genome Biol. 7: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou, Z., Fan, W.H., and Dong, X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113: 935–944. [DOI] [PubMed] [Google Scholar]
- Noël, L.D., Cagna, G., Stuttmann, J., Wirthmuller, L., Betsuyaku, S., Witte, C.P., Bhat, R., Pochon, N., Colby, T., and Parker, J.E. (2007). Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. Plant Cell 19: 4061–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma, K., Zhang, Y., and Li, X. (2005). An importin α homolog, MOS6, plays an important role in plant innate immunity. Curr. Biol. 15: 1129–1135. [DOI] [PubMed] [Google Scholar]
- Palma, K., Zhao, Q., Cheng, Y.T., Bi, D., Monaghan, J., Cheng, W., Zhang, Y., and Li, X. (2007). Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes Dev. 21: 1484–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry, G., Ward, S., Cernac, A., Dharmasiri, S., and Estelle, M. (2006). The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell 18: 1590–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, P., Xylourgidis, N., Sabri, N., Uv, A., Fornerod, M., and Samakovlis, C. (2003). The Drosophila nucleoporin DNup88 localizes DNup214 and CRM1 on the nuclear envelope and attenuates NES-mediated nuclear export. J. Cell Biol. 163: 701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco, M.A., Mansoor, S., and Moffett, P. (2007). A RanGAP protein physically interacts with the NB-LRR protein Rx, and is required for Rx-mediated viral resistance. Plant J. 52: 82–93. [DOI] [PubMed] [Google Scholar]
- Shah, J. (2003). The salicylic acid loop in plant defense. Curr. Opin. Plant Biol. 6: 365–371. [DOI] [PubMed] [Google Scholar]
- Sheen, J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127: 1466–1475. [PMC free article] [PubMed] [Google Scholar]
- Shen, Q.H., Saijo, Y., Mauch, S., Biskup, C., Bieri, S., Keller, B., Seki, H., Ülker, B., Somssich, I.E., and Schulze-Lefert, P. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103. [DOI] [PubMed] [Google Scholar]
- Simonich, M.T., and Innes, R.W. (1995). A disease resistance gene in Arabidopsis with specificity for the avrPph3 gene of Pseudomonas syringae pv. phaseolicola. Mol. Plant Microbe Interact. 8: 637–640. [DOI] [PubMed] [Google Scholar]
- Tada, Y., Spoel, S.H., Pajerowska-Mukhtar, K., Mou, Z., Song, J., and Dong, X. (2008). Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 321: 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, N., van Kilsdonk, J.W., Ostendorf, B., Smeets, R., Bruggeman, S.W., Alonso, A., van de Loo, F., Schneider, M., van den Berg, W.B., and Swart, G.W. (2008). Tumor marker nucleoporin 88 kDa regulates nucleocytoplasmic transport of NF-κB. Biochem. Biophys. Res. Commun. 374: 424–430. [DOI] [PubMed] [Google Scholar]
- Tameling, W.I., and Baulcombe, D.C. (2007). Physical association of the NB-LRR resistance protein Rx with a Ran GTPase-activating protein is required for extreme resistance to Potato virus X. Plant Cell 19: 1682–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry, L.J., Shows, E.B., and Wente, S.R. (2007). Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science 318: 1412–1416. [DOI] [PubMed] [Google Scholar]
- Uv, A.E., Roth, P., Xylourgidis, N., Wickberg, A., Cantera, R., and Samakovlis, C. (2000). members only encodes a Drosophila nucleoporin required for Rel protein import and immune response activation. Genes Dev. 14: 1945–1957. [PMC free article] [PubMed] [Google Scholar]
- van der Biezen, E.A., Freddie, C.T., Kahn, K., Parker, J.E., and Jones, J.D. (2002). Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 29: 439–451. [DOI] [PubMed] [Google Scholar]
- Vernooij, B., Friedrich, L., Morse, A., Reist, R., Kolditz-Jawhar, R., Ward, E., Uknes, S., Kessmann, H., and Ryals, J. (1994). Salicylic acid is not the translocated signal responsible for inducing systemic acquired resistance but is required in signal transduction. Plant Cell 6: 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer, M., Feys, B.J., and Parker, J.E. (2005). Plant immunity: The EDS1 regulatory node. Curr. Opin. Plant Biol. 8: 383–389. [DOI] [PubMed] [Google Scholar]
- Wirthmueller, L., Zhang, Y., Jones, J.D., and Parker, J.E. (2007). Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr. Biol. 17: 2023–2029. [DOI] [PubMed] [Google Scholar]
- Xylourgidis, N., Roth, P., Sabri, N., Tsarouhas, V., and Samakovlis, C. (2006). The nucleoporin Nup214 sequesters CRM1 at the nuclear rim and modulates NFκB activation in Drosophila. J. Cell Sci. 119: 4409–4419. [DOI] [PubMed] [Google Scholar]
- Yoo, S.D., Cho, Y.H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Fan, W., Kinkema, M., Li, X., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 96: 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Goritschnig, S., Dong, X., and Li, X. (2003. a). A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15: 2636–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., and Li, X. (2005). A putative nucleoporin 96 is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1, constitutive 1. Plant Cell 17: 1306–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Tessaro, M.J., Lassner, M., and Li, X. (2003. b). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15: 2647–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.