Abstract
Yeast senses the availability of external energy sources via multiple interconnected signaling networks. One of the central components is SNF1, the homolog of mammalian AMP-activated protein kinase, which in yeast is essential for the expression of glucose-repressed genes. When glucose is available hyperphosphorylated SNF1 is rendered inactive by the type 1 protein phosphatase Glc7. Dephosphorylation requires Reg1, which physically targets Glc7 to SNF1. Here we show that the chaperone Ssb is required to keep SNF1 in the nonphosphorylated state in the presence of glucose. Using a proteome approach we found that the Δssb1Δssb2 strain displays alterations in protein expression and suffers from phenotypic characteristics reminiscent of glucose repression mutants. Microarray analysis revealed a correlation between deregulation on the protein and on the transcript level. Supporting studies uncovered that SSB1 was an effective multicopy suppressor of severe growth defects caused by the Δreg1 mutation. Suppression of Δreg1 by high levels of Ssb was coupled to a reduction of Snf1 hyperphosphorylation back to the wild-type phosphorylation level. The data are consistent with a model in which Ssb is crucial for efficient regulation within the SNF1 signaling network, thereby allowing an appropriate response to changing glucose levels.
Keywords: AMP-activated protein kinase, energy metabolism, glucose starvation, ribosome-associated chaperone, Saccharomyces cerevisiae, translational shutdown
The most abundant Hsp70s in the budding yeast Saccharomyces cerevisiae are the cytosolic subfamilies of Ssa and Ssb. The essential family of Ssa proteins consist of four close homologs, SSA1–SSA4, which are thought to perform the same set of functions, but are regulated differentially on the transcriptional level (Werner-Washburne et al. 1989). Ssa is soluble in the cytosol where it is the central player of the cytosolic protein folding machinery (Kim et al. 1998; Mayer and Bukau 2005). Misfolding and aggregation are more frequent at elevated temperatures. Consistently, SSA1–SSA4 are induced when the temperature is raised (Craig and Jacobsen 1984; Werner-Washburne and Craig 1989) and yeast lacking both SSA1 and SSA2 display a temperature-sensitive phenotype (Craig and Jacobsen 1984). Also, deletions of the Ssa cochaperones YDJ1 or FES1 result in strong temperature-sensitive phenotypes (Atencio and Yaffe 1992; Shomura et al. 2005). Finally, authentic substrates that require Ssa to fold in vivo have been identified (Kim et al. 1998) and, together with its cochaperones, Ssa was shown to refold a denatured model substrate in vitro (Lu and Cyr 1998). In summary, Ssa is a bona fide chaperone, acting as a folding helper.
The second major cytosolic Hsp70 is encoded by two close homologs, SSB1 and SSB2, which are functionally redundant. It is commonly accepted that Ssb, which can interact with nascent polypeptides in the context of the ribosome (Nelson et al. 1992; Pfund et al. 1998; Gautschi et al. 2002) also assists the folding of proteins. The current model suggests that ribosome-bound Ssb functions early during protein biogenesis and Ssa takes over, after synthesis is complete and newly synthesized proteins are released from the ribosome (Craig et al. 2003; Albanese et al. 2006; Dragovic et al. 2006). However, to date, specific proteins that require Ssb as a folding helper have not been reported and, unlike Ssa, Ssb does not bind to peptides mimicking unfolded substrates (Pfund et al. 2001). Quite unexpectedly, recent results revealed that Ssb does not enhance the productive folding of structurally destabilized proteins in vivo (Tomala and Korona 2008). Moreover, the Δssb1Δssb2 strain displays a strong growth defect at low temperature and growth approaches that of wild type when the temperature is raised (Craig and Jacobsen 1985; Nelson et al. 1992). Also, Ssa and Ssb chaperones differ with respect to regulation. In contrast to SSA1–SSA4, the transcription of SSB1–SSB2 is not induced but rather diminished at elevated temperatures (Craig and Jacobsen 1985; Werner-Washburne and Craig 1989). Instead, Ssb levels increase when late log-phase cells are transferred to fresh glucose-containing medium (Brejning and Jespersen 2002) and decrease during late stages of the stationary phase (Werner-Washburne et al. 1989). There is evidence that Ssb might function in processes other than protein folding (Bonner et al. 2000; Dombek et al. 2004; Hurt et al. 2004; Zhang et al. 2004; Bagriantsev et al. 2008). For instance, interaction of Ssb with the heat-shock transcription factor HSF1 was shown to down-regulate the activity of the transcription factor. The current model suggests that Ssb binding sterically interferes with the activation domains of HSF1 (Bonner et al. 2000).
The kinase SNF1 is one of the major components of the yeast glucose-sensing system (for recent reviews on SNF1, see Hardie 2007; Hedbacker and Carlson 2008; Zaman et al. 2008). SNF1 is activated in response to glucose starvation and other stresses, however, not in response to heat shock (Hong and Carlson 2007). Yeast SNF1 consists of the catalytic α subunit Snf1; one of three different β subunits, Sip1, Sip2, or Gal83; and the γ subunit Snf4. When yeast grows in the absence of glucose, SNF1 is activated via phosphorylation of Thr 210 of the Snf1 subunit (McCartney and Schmidt 2001) by any one of three upstream kinases named Sak1, Elm1, and Tos3 (Hong et al. 2003; Sutherland et al. 2003; Elbing et al. 2006). As soon as glucose becomes available the essential type 1 protein phosphatase (PP1) Glc7 dephosphorylates Snf1. In order to recognize Snf1 as a substrate, Glc7 requires targeting via a regulatory subunit. Reg1 is the major regulatory subunit for the Glc7-dependent dephosphorylation of Snf1 (Tu and Carlson 1995; Ludin et al. 1998; Sanz et al. 2000). However, a homolog of Reg1, termed Reg2, also plays a role in targeting of Glc7 within the SNF1 signaling network (Frederick and Tatchell 1996; Jiang et al. 2000). Ssb was found to interact with the Snf1 as well as with the Sip2 subunit of SNF1 (Wiatrowski and Carlson 2003), the SNF1-regulating kinases Sak1 and Elm1 (Elbing et al. 2006), and also the SNF1-regulating phosphatase Glc7/Reg1 (Mayordomo et al. 2003; Dombek et al. 2004). It was suggested that interaction between Ssb and Reg1 may serve as a metabolic sensor to modulate the activity of Glc7/Reg1 (Dombek et al. 2004). However, at present, it remains uncertain whether or not these interactions are functionally important or reflect the general stickiness of the chaperone Ssb.
Here we set out to better understand the major cellular functions of Ssb. Applying an unbiased proteomic approach we uncovered that the lack of Ssb results in the inability of yeast to correctly respond to the presence of glucose. Side-by-side proteome and transcriptome analysis of glucose-grown Δssb1Δssb2 versus wild-type yeast revealed that changes on the protein level were due to transcriptional deregulation rather than to cotranslational or post-translational events. We found that major phenotypic defects of Δssb1Δssb2, some of which had previously escaped detection, are correlated to this defect in glucose sensing. Using biochemical and yeast genetic experiments we show that, on a mechanistic level, Ssb was required to keep SNF1 dephosphorlyated in the presence of glucose. A role of Ssb in SNF1 signaling is supported by the observation that SSB1 was a strong multicopy suppressor of Δreg1, and that suppression was significantly decreased in a Δreg1Δreg2 strain. The data are consistent with a model in that Ssb, most likely in concert with a bona fide Glc7 targeting subunit, affects the dephosphorylation of Snf1.
Results
Protein expression in the absence of Ssb resembled protein expression upon glucose exhaustion
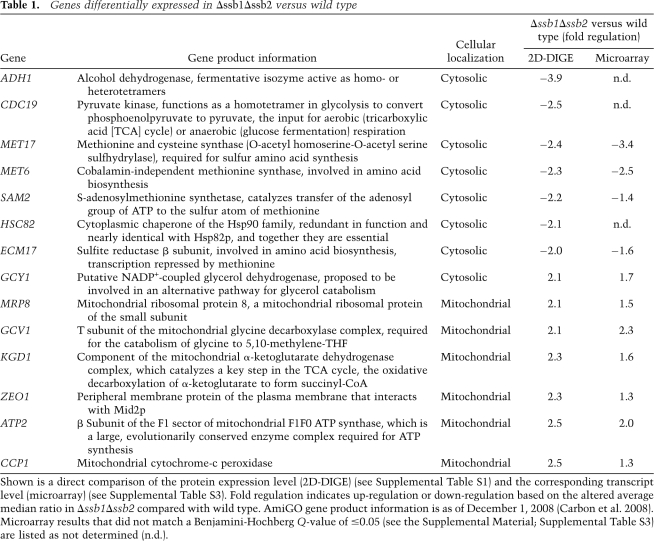
To identify potential substrates of Ssb, we compared the soluble proteome of a Δssb1Δssb2 strain with that of the corresponding wild-type strain using two-dimensional fluorescence difference gel electrophoresis (2D-DIGE). We expected proteins that fail to fold in the absence of Ssb to be reduced in the soluble fraction of the proteome because in vivo misfolded or aggregated proteins are delivered to the proteasome for degradation. In yeast, degradation was previously shown to be dependent on functional Ssa, however, independent of Ssb (Park et al. 2007). In case misfolded proteins in the Δssb1Δssb2 strain would escape degradation and accumulate as insoluble aggregates, these would be depleted from the soluble proteome due to sample preparation. Each three independent protein extracts of wild type and the Δssb1Δssb2 strain were analyzed on a total of 10 2D-DIGE gels (Supplemental Fig. S1). In this data set 995 spots could be matched on at least five of the 2D-DIGE gels. Of the matched spots 44 displayed a significantly (more than two) altered expression level in the Δssb1Δssb2 strain compared with wild type. Thirteen spots were up- and 31 were down-regulated (Supplemental Table S1). To identify differentially regulated proteins in Δssb1Δssb2, spots were excised from the gels and peptides released by trypsin treatment were analyzed by mass spectrometry. As expected, Ssb itself, which runs as a line of seven matched spots (Supplemental Fig. S1A; Supplemental Table S1), was one of the down-regulated proteins. Besides Ssb, 14 proteins were identified, seven of which were expressed at a lower, and seven of which were expressed at a higher level in the Δssb1Δssb2 strain (Table 1).
Table 1.
Genes differentially expressed in Δssb1Δssb2 versus wild type
Shown is a direct comparison of the protein expression level (2D-DIGE) (see Supplemental Table S1) and the corresponding transcript level (microarray) (see Supplemental Table S3). Fold regulation indicates up-regulation or down-regulation based on the altered average median ratio in Δssb1Δssb2 compared with wild type. AmiGO gene product information is as of December 1, 2008 (Carbon et al. 2008). Microarray results that did not match a Benjamini-Hochberg Q-value of ≤0.05 (see the Supplemental Material; Supplemental Table S3) are listed as not determined (n.d.).
Unexpectedly, six of the seven proteins with increased expression localized to mitochondria, suggesting that mitochondrial function might be enhanced in the Δssb1Δssb2 strain (see also below). All proteins that were less abundant localized to the cytosol (Table 1). To discover possible functional relationships between the proteins differentially expressed, we used the Gene Ontology (GO) term enrichment tool (Carbon et al. 2008). The biological process of the down-regulated proteins turned out to be strongly associated with amino acid and sulfur metabolism (Supplemental Table S2).
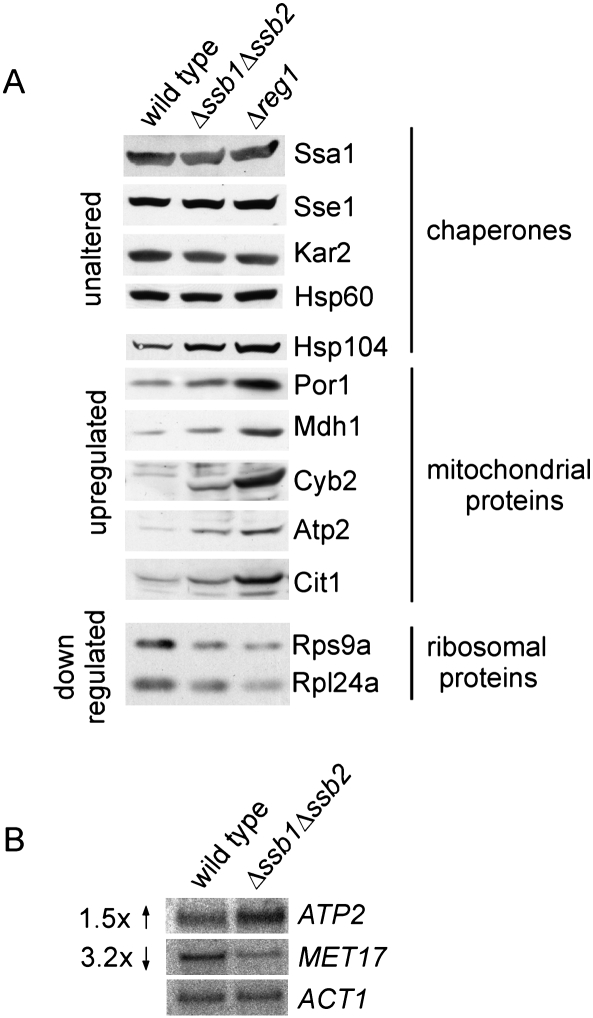
Proteins differentially expressed in the absence of Ssb matched quite closely with those regulated on the transcript level when cells undergo the diauxic shift and progress to the stationary phase (DeRisi et al. 1997; Brauer et al. 2005). Glucose-grown batch cultures generate energy exclusively via fermentation and produce ethanol and CO2. Shortly after glucose is depleted yeast undergoes a major transcriptional reprogramming that now allows it to use ethanol as a substrate for respiration (Brauer et al. 2005). During the diauxic shift, genes involved in oxidative phosphorylation and mitochondrial function are strongly up-regulated, and genes involved in protein biosynthesis, ribosome biogenesis, and amino acid metabolism are down-regulated (DeRisi et al. 1997; Gasch et al. 2000; Brauer et al. 2005). Prompted by the 2D-DIGE data, we compared the expression level of ribosomal proteins and of additional mitochondrial proteins via immunoblotting (Fig. 1A). Indeed, ribosomal proteins were down-regulated and a number of additional mitochondrial proteins, like Por1, Mdh1, Cyb2, Cit1, and Atp2, which was also identified via 2D-DIGE (Table 1), were significantly up-regulated in the Δssb1Δssb2 strain (Fig. 1A). In addition, the expression level of Hsp104 (Fig. 1A) and Hsp26 (Fig. 6A, below) was increased in Δssb1Δssb2. Hsp104 and Hsp26 belong to the group of proteins up-regulated during or soon after the diauxic shift (DeRisi et al. 1997; Gasch et al. 2000; Brauer et al. 2005). The chaperones Ssa, Sse1, Kar2, and Hsp60, however, were expressed at the same level in Δssb1Δssb2 and wild-type strains (see the Discussion).
Figure 1.
The Δssb1Δssb2 strain and the glucose-repression mutant Δreg1 display similar transcript and protein expression levels. (A) Glucose-grown wild type, Δssb1Δssb2, and Δreg1 were harvested during logarithmic growth and were analyzed for the expression of proteins via immunoblotting with antibodies recognizing the proteins indicated. (B) Analysis of ATP2 and MET17 transcript levels via Northern blotting. Total mRNA was extracted from wild type and the Δssb1Δssb2 strain grown as in A. ACT1 served as loading control.
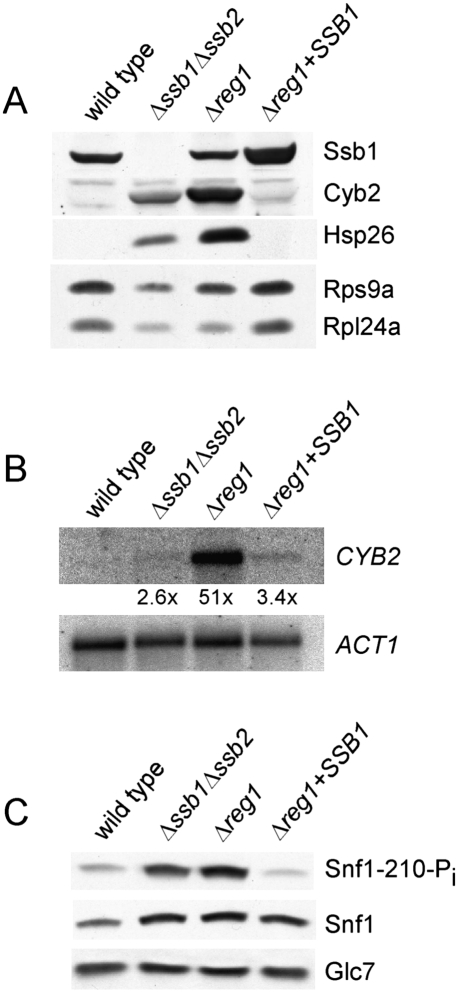
Figure 6.
Complementation of Δreg1 by SSB1 results in dephosphorylation of Snf1 and abolishes deregulation of transcript and protein expression. (A) Total protein extracts of wild type, Δssb1Δssb2, Δreg1, and Δreg1 + SSB1 were generated as in Figure 1A. Aliquots were analyzed via immunoblotting using antibodies specifically recognizing the proteins indicated. (B) Northern blot analysis of CYB2 was performed as described in Figure 1B. ACT1 served as loading control. (C) Snf1 phosphorylation on Thr210 was analyzed using affinity purified antibody PT210, which specifically recognizes the phosphorylated form of Snf1. The expression level of Snf1 and Glc7 was monitored using antibodies specifically recognizing Snf1 and Glc7.
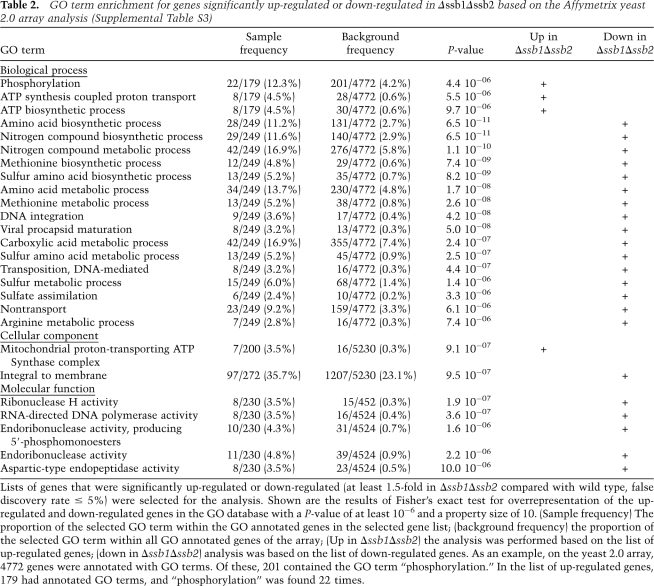
The protein expression pattern of the Δssb1Δssb2 strain suggested a major regulatory defect on the transcriptional level rather than a defect due to a post-translational event. To test this hypothesis, we initially analyzed the transcript level of ATP2 and MET17 (Fig. 1B). The transcript of ATP2 was increased 1.5-fold, while the transcript of MET17 was decreased 3.2-fold in Δssb1Δssb2 cells. Based on this finding we next asked whether the Δssb1Δssb2 strain suffered from general transcriptional deregulation using microarray analysis (for details, see the Supplemental Material). Of the 5814 S. cerevisiae genes analyzed on the array, 219 were up-regulated and 309 were down-regulated ≥1.5-fold in the Δssb1Δssb2 strain with a false discovery rate of 5% (Supplemental Table S3). GO and KEGG pathway analysis revealed that up-regulated transcripts were highly overrepresented among the ones involved in oxidative phosphorylation and the citrate cycle. Down-regulated transcripts were overrepresented among the ones involved in processes related to amino acid and sulfur metabolism (Table 2; Supplemental Table S4). Most genes encoding ribosomal proteins were also down-regulated; only three ribosomal transcripts were enhanced. However, reduction in most cases was <1.5-fold (Supplemental Table S5), and thus ribosomes were not detected in the pathway analysis. While deregulation in the Δssb1Δssb2 strain was only moderate for most transcripts, we found an excellent agreement between deregulation on the level of the transcriptome and proteome (Table 1). The correlation was also evident from a comparison of the GO term analysis. The identified deregulated proteins (Table 1) displayed significant enrichment in eight GO terms of the category “biological process” (Supplemental Table S2). Of these, five GO terms were also indentified in the GO term enrichment based on the microarray results (Table 2).
Table 2.
GO term enrichment for genes significantly up-regulated or down-regulated in Δssb1Δssb2 based on the Affymetrix yeast 2.0 array analysis (Supplemental Table S3)
Lists of genes that were significantly up-regulated or down-regulated (at least 1.5-fold in Δssb1Δssb2 compared with wild type, false discovery rate ≤ 5%) were selected for the analysis. Shown are the results of Fisher's exact test for overrepresentation of the up-regulated and down-regulated genes in the GO database with a P-value of at least 10−6 and a property size of 10. (Sample frequency) The proportion of the selected GO term within the GO annotated genes in the selected gene list; (background frequency) the proportion of the selected GO term within all GO annotated genes of the array; (Up in Δssb1Δssb2) the analysis was performed based on the list of up-regulated genes; (down in Δssb1Δssb2) analysis was based on the list of down-regulated genes. As an example, on the yeast 2.0 array, 4772 genes were annotated with GO terms. Of these, 201 contained the GO term “phosphorylation.” In the list of up-regulated genes, 179 had annotated GO terms, and “phosphorylation” was found 22 times.
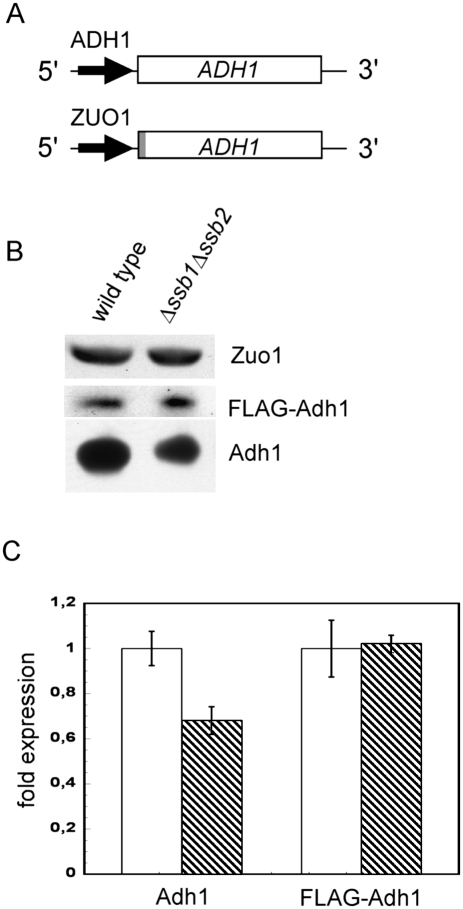
Adh1 was detected as one of the down-regulated proteins via 2D-DIGE analysis (Table 1); however, microarray results for this transcript were ambiguous (Supplemental Table S3), probably because ADH1 microarray expression ratios are unreliable due to cross-hybridization with the ADH2 message as previously observed by others (Young et al. 2003). Yeast Adh1 is the cytosolic isoenzyme of alcohol dehydrogenase that acts during fermentative growth and converts acetaldehyde to ethanol (Piskur et al. 2006). Consistent with its function the enzyme is transcriptionally repressed by growth on nonfermentable carbon sources (Denis et al. 1983). To test whether Adh1 was transcriptionally repressed in Δssb1Δssb2 or was a potential substrate for Ssb-mediated folding, we tested transcriptional regulation of ADH1 expression by a different approach. The coding region of ADH1 was placed under the control of the ZUO1 5′ and 3′ regulatory elements (Fig. 2A). This transcriptional context was chosen because the expression of Zuo1 was unaffected in the Δssb1Δssb2 strain (Fig. 2B; Supplemental Table S3). In order to directly compare expression to endogenous ADH1, an N-terminal Flag tag was fused to ADH1 under control of the ZUO1 promoter (Fig. 2A). Consistent with the 2D-DIGE data (Table 1) Adh1 was reduced in Δssb1Δssb2, however, the expression level of Flag-Adh1 was unaffected (Fig. 2C). We conclude that ADH1 expression was reduced due to an effect on transcriptional regulation. Please note that immunoblotting revealed a less severe reduction of endogenous Adh1 than was suggested by the 2D-DIGE results (Table 1). Most likely the total Adh1 content of the Δssb1Δssb2 strain was underestimated via 2D-DIGE because only one of the multiple Adh1 spots was analyzed.
Figure 2.
The expression level of Adh1 in the glucose-grown Δssb1Δssb2 strain is promoter-dependent. (A) Schematic representation of chromosomal ADH1 under the control of its endogenous regulatory elements and plasmid encoded Flag-tagged ADH1 under the control of the ZUO1 regulatory elements. The N-terminal Flag tag is indicated in gray. (B) Aliquots of total protein extracts of wild-type and Δssb1Δssb2 cells prepared as described in Figure 1A were analyzed via immunoblotting with antibodies recognizing Adh1, the Flag tag, or Zuo1 as indicated. Flag-tagged Adh1 is recognized by the Adh1 antibody as well as by the Flag tag antibody. The Flag-tagged version can be separated from the untagged version on 10% TRIS-Tricine gels and is not shown on the blot developed with the Adh1 antibody. Endogenous Zuo1 was monitored as a control. (C) Quantification of the relative amounts of Adh1 and Flag-Adh1 in wild type and Δssb1Δssb2. Densitometric analysis of immunoblots was performed in the linear range as described in Experimental Procedures. The expression level of endogenous and Flag-tagged Adh1 was determined in three independent clones. Expression in wild type (white bars) was set to 1. (Hatched bars) Expression level in Δssb1Δssb2. Error bars represent the standard error of the mean.
In summary, a direct comparison of proteome and transcriptome data revealed that the Δssb1Δssb2 strain displayed changes on the level of the proteome as a direct consequence of changes on the level of the transcriptome.
The Δssb1Δssb2 strain displayed defects in glucose sensing
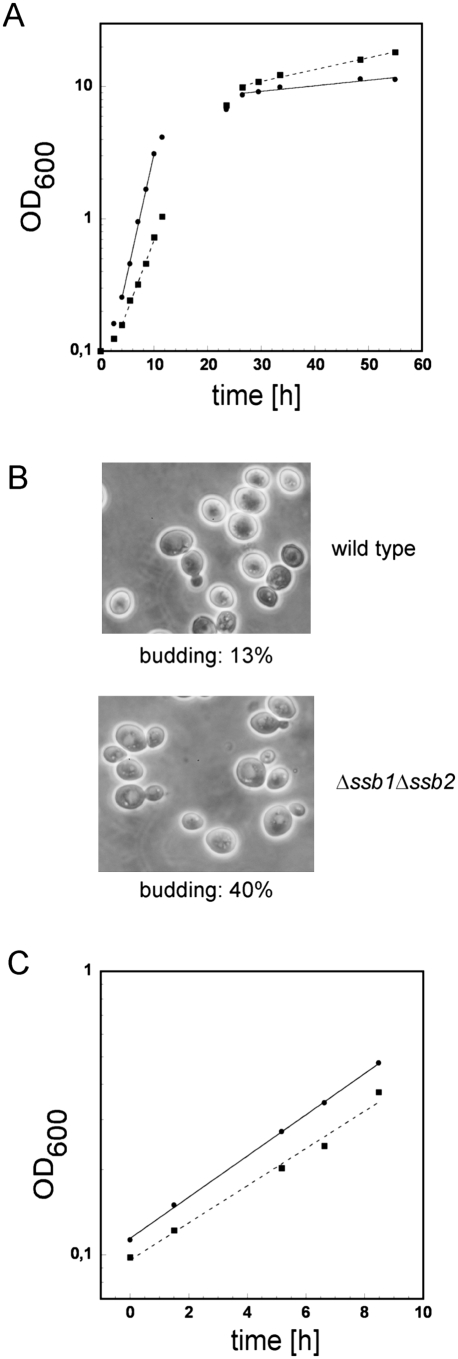
On rich glucose-containing medium at 30°C the Δssb1Δssb2 strain displays slow growth (Craig and Jacobsen 1985). In our strain background the doubling time during early logarithmic growth on glucose was 1.6 h for the wild type and 2.8 h for the Δssb1Δssb2 strain (Fig. 3A). We now noticed that slow growth of the Δssb1Δssb2 strain was confined to the early growth phase. At higher OD600, when cells undergo the diauxic shift and start to grow on ethanol, the doubling time of the wild-type strain was 72 h, while the Δssb1Δssb2 strain grew significantly faster with a doubling time of 33 h and even outgrew the wild-type strain (Fig. 3A). Microscopic inspection revealed that at an OD600 = 7, ∼90% of the wild-type cells were small and round-shaped as expected if cells were to enter the stationary phase; only 13% were still budding (Fig. 3B). At the same OD600 the majority of Δssb1Δssb2 cells was oval shaped, larger in size, and about 40% carried at least one bud (Fig. 3B). To test whether slow growth was indeed related to the carbon source, we determined doubling times also on ethanol (Fig. 3C) or lactate medium (data not shown), which can be used exclusively via respiration. On ethanol the doubling time during logarithmic growth of wild type was 4.2 h, while that of the Δssb1Δssb2 strain was 4.4 h (Fig. 3C). The slow growth phenotype of the Δssb1Δssb2 strain is thus carbon source-dependent.
Figure 3.
The Δssb1Δssb2 strain displays glucose-specific growth defects and does not normally enter stationary phase. (A) Growth curves of wild type (circles, solid line) and the Δssb1Δssb2 strain (squares, dotted line) on glucose-containing medium prior and after the diauxic shift in batch culture. Doubling times (see the Results) were calculated independently for the early exponential (between 4 and 10 h) and the early stationary phase (between 26.5 and 55 h). (B) Samples taken after 24 h of growth (OD600 7.2–7.3) were analyzed microscopically. The ratio of budded cells versus unbudded cells was determined using a Neubauer improved counting chamber. (C) Growth curves of wild type (circles, solid line) and the Δssb1Δssb2 strain (squares, dotted line) logarithmically growing on ethanol-containing medium.
Deregulation of protein expression in the Δssb1Δssb2 strain resembled that of a Δreg1 strain
In a Δreg1 strain, Snf1 remains constitutively phosphorylated, and glucose-repressed genes are derepressed in the presence of glucose (McCartney and Schmidt 2001; Dombek et al. 2004 for details refer to the Introduction). Thus, Δreg1 can serve as a paradigm for a mutant with a defect in glucose sensing. We directly compared the expression level of proteins deregulated in the Δssb1Δssb2 and Δreg1 strains. Both mutants expressed mitochondrial proteins and the chaperones Hsp104 and Hsp26 at higher levels than the corresponding wild-type strain. Up-regulation was more pronounced in the Δreg1 compared with the Δssb1Δssb2 strain. Ribosomal proteins were down-regulated in the Δreg1 and Δssb1Δssb2 strains to a similar extend (Figs. 1A, 6A [below]).
Both the Δssb1Δssb2 and Δreg1 strain failed to respond to glucose removal with a transient translational shutdown
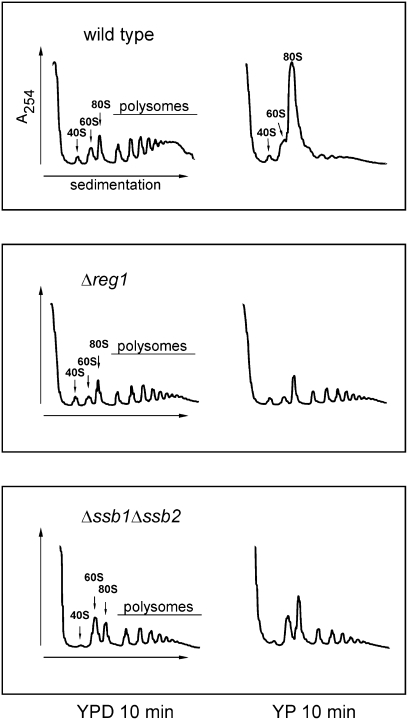
When yeast is suddenly depleted from glucose it responds within minutes by a complete shutdown of translation. This transient and reversible effect of carbon source removal is specific for glucose, and does not occur when cells have used other carbon sources (Ashe et al. 2000). A set of mutants that display defects in glucose sensing, fail to perform this rapid translational shutdown; one of these mutants is Δreg1 (Ashe et al. 2000). We tested whether glucose removal affects translational shutdown also in the Δssb1Δssb2 strain. To that end, logarithmically growing yeast cultures of Δssb1Δssb2, Δreg1, and wild-type strains were depleted of glucose for 10 min prior to harvest and extracts were analyzed via ribosome profiles (Fig. 4). A direct comparison of the ribosome profiles of glucose-grown Δreg1 and Δssb1Δssb2 showed that cells have ∼30% less ribosomes than the wild-type strain (Fig. 1A; Supplemental Fig. S6; Supplemental Table S5), but show a rather normal relative polysome content. In the case of the Δssb1Δssb2, but not of the Δreg1 mutant, the relative amount of free 60S ribosomes was increased compared with wild type (Fig. 4). As expected, glucose removal in wild type resulted in a dramatic increase in nontranslating 80S ribosomes, indicative for the inhibition of translation initiation (Ashe et al. 2000). In contrast, in the Δssb1Δssb2 and Δreg1 strains polysomes were significantly more stable after glucose depletion (Fig. 4, YP 10 min). Both strains failed to respond to glucose removal with a transient translational shutdown.
Figure 4.
The Δssb1Δssb2 mutant and the glucose repression mutant Δreg1 display a defect in the rapid inhibition of translation initiation upon glucose depletion. Glucose-grown wild-type, Δreg1, and Δssb1Δssb2 strains were collected at an OD600 ≈ 0.4. Cells were immediately resuspended in YPD or YP lacking glucose and were incubated on a shaker for 10 min. Ribosome profiles were generated as described in the Materials and Methods. Peaks representing the small ribosomal subunit (40S), the large ribosomal subunit (60S), monosomes (80S), and polysomes are indicated.
Overexpression of Ssb1 rescued growth defects of Δreg1
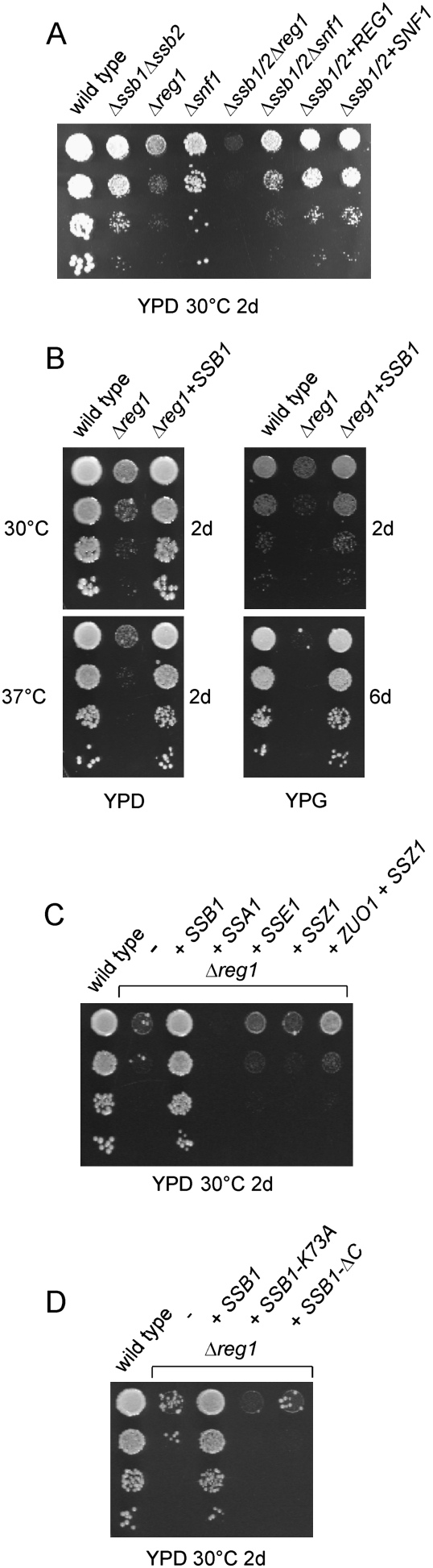
The Δsnf1 mutation causes slow growth on glucose and a complete failure to grow on respiratory media (Fig. 5A; Thompson-Jaeger et al. 1991; data not shown). A Δreg1 strain displays even more severe growth defects on all carbon sources and is unable to grow at 20°C as well as 37°C (Fig. 5; data not shown). To test for synthetic interaction between SSB1/SSB2, REG1, and SNF1, we generated the triple deletion strains Δssb1Δssb2Δreg1 and Δssb1Δssb2Δsnf1. Growth defects of these mutants were slightly enhanced compared with the strains lacking either SSB1/SSB2, REG1, or SNF1 as expected if growth defects of already compromised strains were additive. Overexpression of REG1 or SNF1 did not affect growth of the Δssb1Δssb2 strain (Fig. 5A). However, when SSB1 was expressed on a 2μ plasmid in a Δreg1 background, defects on glucose as well as on glycerol were complemented effectively (Fig. 5B). Even at 37°C SSB1 was an efficient multicopy suppressor of Δreg1-related growth defects (Fig. 5B). The effect was specific, as only high levels of SSB1 but not of the other cytosolic Hsp70 homologs, SSA1, SSZ1, or SSE1 were able to complement growth of the Δreg1 strain (Fig. 5C). Weak suppression, however, was also observed when Zuo1 and Ssz1, the subunits of the Ssb cochaperone ribosome-associated complex (RAC) (Gautschi et al. 2002; Huang et al. 2005), where simultaneously overexpressed in the Δreg1 strain (Fig. 5C). The ability of SSB1 to suppress Δreg1 was dependent on its ability to hydrolyze ATP as well as on the presence of its peptide-binding domain: SSB1-K73A, a mutant that is unable to hydrolyze ATP, and SSB1-ΔC, a mutant lacking the peptide-binding domain (Conz et al. 2007), failed to suppress the phenotype of the Δreg1 strain (Fig. 5D).
Figure 5.
SSB1 is an efficient high-copy suppressor of growth defects displayed by the glucose repression mutant Δreg1. Serial dilutions of logarithmically growing wild-type and mutant cells were analyzed on glucose-containing (YPD) or glycerol-containing (YPG) plates. Temperatures and incubation times are indicated. (A) Growth defects of the triple deletion strains Δssb1Δssb2Δreg1, Δssb1Δssb2Δsnf1, and of Δssb1Δssb2 overexpressing either REG1 or SNF1. (B) Complementation of Δreg1-related growth defects by high levels of SSB1 at 30°C and at 37°C. (C) Overexpression of different cytosolic chaperones in the Δreg1 strain. Expression of SSB1, SSA1, SSE1, SSZ1, and ZUO1 in the different strain backgrounds was from 2μ plasmids under the control of the respective endogenous promoter. Growth medium, temperatures, and incubation times are indicated. (D) Overexpression of mutant versions of SSB1 in Δreg1. Expression of SSB1, SSB1-K73A (ATPase-deficient mutant), and SSB1-ΔC (mutant lacking the peptide-binding domain) were overexpressed from 2μ plasmids under the control of the SSB1 promoter.
Ssb reversed the aberrant protein expression caused by Δreg1 and affected the phosphorylation state of Snf1
Based on the strong multicopy suppressor effect of SSB1 in Δreg1 we next asked whether aberrant protein expression was affected. Indeed, Hsp26, Cyb2, Rps9, and Rpl24 were back to the wild-type expression level when SSB1 was overexpressed in the Δreg1 strain (Fig. 6A). SSB1 already exerted its effect on the level of transcription, as the transcript of CYB2, highly abundant in glucose-grown Δreg1 cells, was significantly reduced when SSB1 was overexpressed (Fig. 6B). A major reason for deregulation in a Δreg1 strain is hyperphosphorylation of Snf1 on Thr210 (Tu and Carlson 1995; McCartney and Schmidt 2001). Our data suggested that Ssb affected the phosphorylation status of Snf1 in a Δreg1 background. With the aid of an antibody specifically recognizing phosphorylated Thr210 we found that Snf1 was not only hyperphosphorylated in the Δreg1 strain (McCartney and Schmidt 2001) but also in the Δssb1Δssb2 strain (Fig. 6C). Strikingly, hyperphosphorylation of Snf1 was abolished in the Δreg1 background when SSB1 was overexpressed (Fig. 6C). This was not due to changes in expression or stability of Snf1 or Glc7, as both proteins were expressed at similar levels in wild-type and the different mutant strains (Fig. 6C). We conclude that high-level expression of Ssb can overcome Snf1 hyperphosphorylation caused by the Δreg1 mutation.
Ssb-mediated suppression of Δreg1 correlated with the dephosphorylation of Snf1 and is affected by Reg2
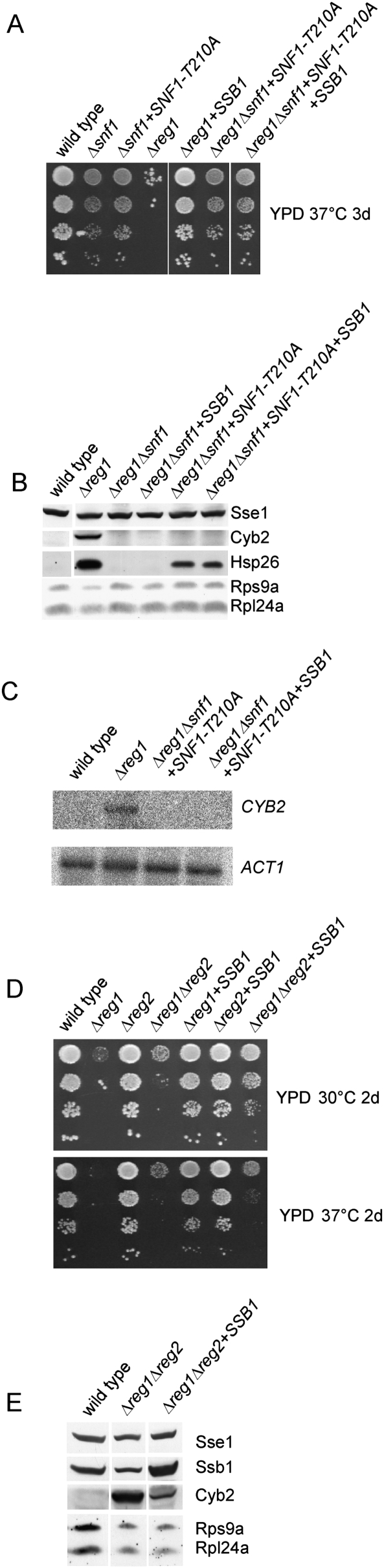
Snf1-T210A is a mutant that mimics constitutively dephosphorylated and thus inactive Snf1 (McCartney and Schmidt 2001). Consistently, Δsnf1 and Δsnf1 expressing Snf1-T210A both grew slowly on glucose and failed to grow on respiratory media (Fig. 7A; data not shown). Snf1-T210A cannot be hyperphosphorylated in a Δreg1 strain; thus, growth defects of Δreg1 are partly suppressed when Snf1 is replaced by Snf1-T210A (Fig. 7A). Please note that suppression of Δreg1 by SNF1-T210A was less effective than suppression by high levels of SSB1 (Fig. 7A). Importantly, overexpression of SSB1 in the Δreg1Δsnf1 + Snf1-T210A strain did not further complement growth defects, indicating that the mechanism of Ssb-mediated suppression involves the dephosphorylation of Snf1.
Figure 7.
Complementation of Δreg1 by SSB1 is dependent on phosphorylatable Snf1, and is significantly reduced in Δreg1Δreg2. (A) Serial dilutions of logarithmically growing cultures were analyzed as in Figure 5. The SNF1-T210A mutant and SSB1 were expressed from a 2μ plasmid. (B) Total protein extracts were generated as in Figure 1A. Aliquots were analyzed via immunoblotting using antibodies specifically recognizing the proteins indicated. (C) Northern blot analysis of CYB2 was performed as described in Figure 1B. ACT1 served as loading control. (D) Analysis was as in (A). SSB1 was expressed from a 2μ plasmid. (E) Total protein extracts were generated as in Figure 1A. Aliquots were analyzed via immunoblotting using antibodies specifically recognizing the proteins indicated.
The Δsnf1Δreg1 and Δsnf1Δreg1 + Snf1-T210A strains on the whole exhibit the glucose-repressed phentoype of Δsnf1 rather than the derepressed phenotype of a Δreg1 strain (Fig. 7A–C; see also Neigeborn and Carlson 1987; Tung and Hopper 1995; Ashe et al. 2000). However, in respect to HSP26 expression, glucose repression was fully restored in Δreg1Δsnf1 but not in Δreg1Δsnf1 + Snf1-T210A (Fig. 7B). The finding suggested that nonphosphorylated Snf1 was not completely inactive, an observation previously made by others in respect to specific functions of the kinase (Portillo et al. 2005). The result enabled us to test the effect of SSB1 overexpresssion in the Δreg1Δsnf1 + Snf1-T210A strain. Interestingly, Hsp26, which was below the detection level in the Δreg1 + SSB1 strain (Fig. 6A), was not reduced when SSB1 was overexpressed in the Δreg1Δsnf1 + Snf1-T210A + SSB1 strain (Fig. 7B). The finding supports the notion that suppression of the Δreg1 phenotype by SSB1 is connected to the phosphorylation state of Snf1.
A number of different regulatory subunits act in concert with Glc7 (Nigavekar et al. 2002; Ceulemans and Bollen 2004). However, besides Reg1, its homolog Reg2 is the only other regulatory subunit that was suggested to target Glc7 to Snf1 and/or to Snf1 substrates (Frederick and Tatchell 1996; Jiang et al. 2000). The Δreg2 mutation causes no significant growth defects (Fig. 7D; Frederick and Tatchell 1996). The Δreg1Δreg2 double deletion in our strain background resulted in a slightly less severe phenotype compared with the Δreg1 single deletion (Fig. 7D; data not shown). Importantly, a direct comparison between the Δreg1 + SSB1 and Δreg1Δreg2 + SSB1 strains revealed that SSB1 was a less efficient multicopy suppressor of the Δreg1Δreg2 double mutant (Fig. 7D). Consistently, overexpression of SSB1 in the Δreg1Δreg2 strain did not fully restore normal protein expression levels. In the Δreg1Δreg2 + SSB1 strain Cyb2 was reduced, but still detectable, and expression of ribosomal proteins was not enhanced compared with the Δreg1Δreg2 strain (Fig. 7E). The data are consistent with a model in that Ssb, in concert with a bona fide phosphatase regulatory subunit, enhances Glc7-mediated dephosphorylation of Snf1.
Discussion
The SNF1 signaling network is essential for the regulation of carbohydrate metabolism. Yeast uses its preferred carbon source glucose via fermentation even under aerobic conditions. When glucose is present, enzymes required for the use of other carbon sources and respiration are repressed (Hedbacker and Carlson 2008). A fast and sensitive response to changes in the glucose level is mediated via several interconnected signaling networks, one of which is the SNF1 network (Hedbacker and Carlson 2008; Zaman et al. 2008). Activated SNF1 relieves glucose repression, both by rapid effects via direct phosphorylation of metabolic enzymes and transporters, and by longer-term effects via regulation of transcription. Yeast SNF1 is the evolutionary conserved homolog of mammalian AMP-activated protein kinase (AMPK), which is required to maintain the balance between ATP production and consumption. In humans, AMPK modulates multiple metabolic pathways, and an impairment of its function is associated with the metabolic syndrome, insulin resistance, and cardiovascular disorders. The importance of AMPK is emphasized by the fact that commonly administered drugs in the treatment of type 2 diabetes—such as, e.g., metformin—exert their effects through the activation of AMPK (Hardie 2007, 2008). The SNF1/AMPK signaling network is remarkably conserved, and yeast has been exploited in the past to identify and characterize a number of the mammalian components (Hedbacker and Carlson 2008). Our data indicating that the yeast Hsp70 homolog Ssb is part of this SNF1 network raise the interesting question of whether one or more chaperones of the Hsp70 family take part also in the mammalian AMPK system.
Ssb is involved in regulation within the SNF1 signaling pathway
Three findings strongly suggest that Ssb is involved in regulating the phosphorylation status of Snf1. First, SSB1 overexpression reduced Snf1 hyperphosphorylation in a Δreg1 strain (Fig. 6C), second, suppression of the effects caused by the Δreg1 mutation by overexpression of SSB1 was dependent on phosphorylatable Snf1 (Fig. 7A). Third, overexpression of SSB1 abolished up-regulation of HSP26 in a Δreg1 mutant, while it did not affect up-regulation of HSP26 in the Δreg1Δsnf1 + SNF1-T210A background (Fig. 7B). The latter observation has also interesting implications for Snf1 function. It has been reported that upon glucose starvation HSF1 is phosphorylated by SNF1. This SNF1-dependent phosphorylation of HSF1 leads to the expression of only a subgroup of target genes, including HSP26 and HSP104 (Hahn and Thiele 2004). Consistent with a direct role of SNF1 in the regulation of HSP26 in the Δreg1 strain, up-regulation was abolished in Δreg1Δsnf1. However, we find that HSP26 expression was only partly dependent on activation of Snf1 via phosphorylation (Fig. 7B).
On a mechanistic level different roles for Ssb in the regulation of Snf1 may be envisaged. Ssb might be involved in mediating interactions between components of the SNF1 signaling network. Such a role in regulated complex assembly would not be unique, but has been put forward for other chaperones, including Hsp70 homologs (Lan et al. 2004; Shaner et al. 2008). The best-studied example of a chaperone devoid of classical folding activity is Hsp90. Together with its cochaperones, Hsp90 is thought to function via promoting subtle changes in the conformations or domain arrangements of otherwise folded client proteins. These clients include kinases or transcription factors that rely on Hsp90 for proper signaling activity (Wandinger et al. 2008). As outlined above, several studies have detected interaction between Ssb and components of the SNF1 signaling pathway. Whether these reflect functional interactions remains to be established. However, even if Ssb is directly involved in the Glc7/Reg1-dependent dephosphorylation of Snf1 it is surprising how well SSB1 can suppress the defects of a Δreg1 strain. Ssb seems to fully overcome the Reg1 targeting function and direct Glc7 to Snf1. On the other hand, copurification of Ssb and Reg1 combined with mild derepression of ADH2 in a Δssb1Δssb2 strain suggests that Ssb plays a Reg1-dependent role in glucose repression (Mayordomo et al. 2003; Dombek et al. 2004). How then is Ssb able to exert an effect on Snf1 when Reg1 is absent? A possible explanation for this paradox comes from experiments on the role of Reg2 in Ssb1-mediated suppression. Reg2, a protein of 38.7 kDa, is a distant homolog of the 112.6 kDa protein Reg1 and is the only targeting subunit of Glc7, which was shown to display partial functional redundancy with Reg1 (Frederick and Tatchell 1996; Jiang et al. 2000). Overexpression of REG2 suppresses growth defects caused by Δreg1; however, does not directly suppress defects in glucose repression (Frederick and Tatchell 1996). SSB1-mediated suppression of the phenotype as well as abnormal protein expression was significantly less efficient in the Δreg1Δreg2 compared with the Δreg1 strain (Fig. 7D,E). The mechanism thus seems to involve the Glc7 targeting subunit Reg2. Possibly, when Ssb concentrations are high in a Δreg1 mutant dephosphorylation of Snf1 by Glc7/Reg2 becomes a more likely event. However, other, more indirect functions of Ssb in SNF1 signaling are possible. Most importantly, Ssb might affect folding or stability of one or more intrinsically labile or aggregation prone components of the SNF1 signaling pathway. These would then be present in higher concentrations when Ssb is overexpressed. Alternatively, a Δreg1 strain may suffer from folding defects, which can be suppressed by high levels of SSB1. As suppression by SSB1 is affected by REG2, we regard this as a less likely scenario. Future experiments will have to analyze and define interactions between Ssb and the SNF1 signaling machinery in more detail.
Hyperphosphorylation of Snf1 is not the only cause for growth defects of the Δssb1Δssb2 strain
Snf1 was hyperphosphorylated when the Δssb1Δssb2 strain grew on glucose and the strain suffered from growth defects similar to that of the Δreg1 strain. The results of the microarray analysis are consistent with a transcriptional defect that at least in part is mediated via hyperphosphorylated Snf1 (Table 2; Supplemental Table S4). However, hyperphosphorylation of Snf1 does not fully account for the growth defects of the Δssb1Δssb2 strain. Unlike defects caused by the Δreg1 mutation, the Δssb1Δssb2 mutation was not rescued by simultaneous deletion of SNF1 or by replacing wild-type SNF1 with the SNF1-T210A mutant (Fig. 5A; data not shown). Also, deregulation of protein expression in the Δssb1Δssb2Δsnf1 strain was not reversed to wild-type level (data not shown). The finding is not very surprising; chaperones are versatile devices and serve multiple functions. This holds true also for Ssb, which is expected to play a role in de novo protein folding (Craig et al. 2003; Albanese et al. 2006; Dragovic et al. 2006).
In respect to the folding function of Ssb, a question arises: What are its client proteins and why did we not detect any via 2D-DIGE? Our data strongly suggest that the reduced steady-state levels of the proteins identified (Table 1) was not directly related to a folding defect displayed by the Δssb1Δssb2 strain. However, the 2D-DIGE approach only detects abundant cellular proteins. If Ssb was essential for the folding of, e.g., a transcription factor, or other cellular proteins expressed at only low concentrations, these would not have come up in the analysis. Also, in the context of this study, we focused on the analysis of the soluble proteome. Based on the finding that the Δssb1Δssb2 strain is sensitive to a variety of cations and cation uptake is affected, a specific role for the biogenesis of membrane proteins has been suggested (Jones et al. 2003; Kim and Craig 2005). The identification of Ssb folding clients will be one of the future goals to better understand the function of Ssb. Here we show that the chaperone affects a pivotal post-translational protein modification. The chaperone is required to keep SNF1 in a state of low phosphorylation. Since dephosphorylation reversibly switches off the activity of SNF1, Ssb is crucial for the regulation of the energy metabolism of yeast.
Materials and methods
Yeast strains and plasmids
MH272-3f a/α (ura3/ura3, leu2/leu2, his3/his3, trp1/trp1, ade2/ade2) is the parental wild-type strain of all haploid derivatives used in this study (Gautschi et al. 2002). Deletion strains lacking SSB1 and SSB2 (Δssb1Δssb2) have been described previously (Rakwalska and Rospert 2004). Δsnf1 was constructed by replacing SNF1 with the snf1∷kanMX4 deletion cassette amplified by PCR from strain Y14311 (Euroscarf). Δreg1 (reg1∷LEU2) was constructed by replacing a 755-base-pair (bp) Cla1/Nde1 fragment in the REG1 coding region with the LEU2 ±300 bp upstream and downstream. We also constructed a Δreg1 strain using the reg1∷kanMX4 cassette from strain Y03967 (Euroscarf). The effect of overexpression of SSB1 in reg1∷kanMX4 was the same as in reg1∷LEU2 confirming that SSB1 was a strong multicopy suppressor of Δreg1 (data not shown). Both strains, reg1∷LEU2 and reg1∷kanMX4, had a strong tendency to spontaneously form suppressor mutations. These reg1 suppressor mutants, which are visible in the lowest dilution of Δreg1 platings (Figs. 5, 7), displayed a less severe phenotype than the original Δreg1 strain; however, they still failed to grow on respiratory media at 37°C (data not shown). Δreg2 was constructed by replacing REG2 with the reg2∷kanMX4 deletion cassette amplified by PCR from strain Y03187 (Euroscarf). The double deletion Δreg1Δreg2 was generated by replacing REG2 in a Δreg1 strain expressing REG1 on pYEplac195 (2μ, URA3). Δreg1Δreg2 was then generated by growth on 5-fluoroorotic acid plates, resulting in loss of the URA3 plasmid. For a list of strains see Supplemental Table S6.
REG1, SNF1, SSB1, SSA1, SSZ1, and SSE1 ±300 bp upstream of and downstream from the respective orf were amplified from genomic DNA and were cloned into the multicopy plasmid pYEPlac195 (2μ, URA3) (Gietz and Sugino 1988). ZUO1 ±300 bp upstream of and downstream from the coding region was expressed from pYEPlac555, a derivative of pYEPlac181 in which the LEU2 marker gene was replaced with the ADE2 marker gene (Conz et al. 2007). The resulting plasmids are termed pYEPlac195-REG1, pYEPlac195-SNF1, pYEPlac195-SSB1 (Gautschi et al. 2002), pYEPlac195-SSA1, pYEPlac195-SSZ1 (Gautschi et al. 2002), pYEPlac195-SSE1, and pYEPlac555-ZUO1 (Conz et al. 2007). Plasmids encoding SSB1-K73A and SSB1-ΔC are described (Conz et al. 2007). pYEPlac195-SNF1-T210A was generated using pYEPlac195-SNF1 as a template by exchanging A at position 628 of the SNF1 orf for G via QuickChange (Stratagene). The single base pair exchange results in the replacement of Thr for Ala at position 210 on the amino acid level. The mutation leads to a constitutively unphosphorylated and therefore inactive version of Snf1 (Estruch et al. 1992). Flag-ADH1 was expressed from the low-copy plasmid pYCPlac33 (cen, URA3) (Gietz and Sugino 1988) under control of the ZUO1 regulatory elements. The construct is based on pYCPlac33 containing ZUO1 ±300 bp upstream of and downstream from the ZUO1 coding region. An Nco1 site was introduced at the site of the ZUO1 start codon. The ADH1 coding region was then amplified from genomic DNA of a Δadh2 strain (Euroscarf) with primers introducing an N-terminal Flag tag coding for DYKDDDDK. N-terminally Flag-tagged ADH1 version was cloned into the Nco1/PshA1 site of pYCPlac33-ZUO1 replacing the ZUO1 orf.
Culture conditions
Mutants and transformants were selected on glucose minimal medium containing the appropriate supplements. Growth of strains was analyzed on rich media containing glucose (YPD: 1% yeast extract, 2% peptone, 2% dextrose), ethanol (YPEtOH: 1% yeast extract, 2% peptone, 2% ethanol), glycerol (YPG: 1% yeast extract, 2% peptone, 3% glycerol), or galactose (YPGal: 1% yeast extract, 2% peptone, 2% galactose) as a carbon source. Growth defects were analyzed either in liquid cultures or on solid media. Doubling times were determined for 100-mL batch cultures inoculated to an OD600 ≈ 0.1 from a preculture logarithmically growing on the same medium. Liquid cultures were incubated at 30°C on a shaker at 200 rpm. To avoid anaerobiosis, samples for the determination of OD600 were withdrawn without interrupting agitation of the cultures. Cells were harvested for 3 min at 16,000g and were resuspended to an appropriate density for determination of OD600 in water. Growth defects at different temperatures were also determined on plates. To that end, 10-fold serial dilutions of early log-phase cultures containing the same number of cells were spotted onto solid YPD or YPG. Incubation time and temperature is indicated in the figure legends.
Ribosome profiles
Ribosome profiles were generated as described (Ashe et al. 2000; Raue et al. 2007). Cultures (250 mL) grown on YPD to an OD600 ≈ 0.4 were harvested at 8000g for 10 min at room temperature, and were resuspended in 200 mL of YPD or YP (1% yeast extract, 2% peptone). After 10 min on a shaker at 30°C cultures were transferred to precooled 200-mL centrifuge bottles containing 2 mL of 10 mg/mL cycloheximide. All subsequent steps were performed at 4°C. Cells were collected at 8000g for 10 min, and were washed with 50 mL of lysis buffer (20 mM Hepes-KOH at pH 7.4, 2 mM MgAc2, 100 mM KAc, 100 μg/mL cycloheximide, 0.5 mM DTT). Cell extracts were prepared using glass beads as described (Ashe et al. 2000). Ten A260 units were loaded onto an 11-mL 15%–55% linear sucrose gradient. Gradients were centrifuged for 2.5 h at 200,000g in a TH-641 rotor (Sorvall, Thermoscientific) and were subsequently fractionated from top to bottom with a density gradient fractionator (Teledyne Isco, Inc.) monitoring A254.
Immunoblotting
Total yeast extract for immunoblot analysis was prepared as described (Raue et al. 2007). Because wild-type and Δssb1Δssb2 cells differ in size, loading according to the OD600 reproducibly resulted in lower overall protein content of the Δssb1Δssb2 sample. We thus normalized loading according to the expression level of Ssa, the expression of which is known to be unaffected in the Δssb1Δssb2 strain (Chernoff et al. 1999). Comparative immunoblot analysis was performed at least in triplicate. In order to make sure that the signal on the immunoblot was within the linear range, x and 2x of each sample were analyzed on a single gel and doubling of the intensity of the band was confirmed via quantification using the AIDA ImageAnalyzer (Raytest) (data not shown). For simplicity, the figures show a gel with only one representative sample per total extract analyzed. Phosphorylation of Snf1 on Thr210 was analyzed as described (McCartney and Schmidt 2001). In brief, cultures (12 mL, OD600 = 0.4–0.8) were directly supplemented with NaOH to a final concentration of 0.1 M, the mixture was incubated for 5 min at room temperature, and subsequently harvested by centrifugation. Immunoblots were developed using ECL as described in Raue et al. (2007). Antibodies are described in the Supplemental Material.
Northern blotting
Steady-state levels of mRNA in wild type and the Δssb1Δssb2 strain were analyzed in YPD-grown cultures at an OD600 of 0.4–0.8. Total RNA was extracted according to the manufacturer's instructions (RNeasy, Qiagen) after cell disruption with glass beads. For detection of the CYB2 transcript, mRNA was purified from total RNA according to the manufacturer's instructions (Oligotex, Qiagen). A total of 10 μg of RNA was loaded onto 1.2% agarose gels. Blotting and hybridization was performed as described (Sambrook and Russel 2001). Labeling of the probes was carried out using purified DNA and the Rediprime II Random Prime Labeling System (GE Healthcare BioSciences). A radiolabeled 1500-bp PCR product containing the entire coding region of ATP2 was used as a probe for the ATP2 transcript and a 705-bp EcoRI/HindIII fragment of MET17 was used for detection of the MET17 transcript. The probe for ACT1 was prepared as described (Carvin and Kladde 2004). For detection of the CYB2 transcript hybridization was carried out using a labeled 1200-bp BamHI/BglII fragment of the CYB2 orf (Lodi and Guiard 1991).
Acknowledgments
We thank Martin Schmidt and Rhonda McCartney for the PT210 antibody, Andreas Mayer for the Glc7 antibody, and Chris Meisinger for the Por1 antibody. We also thank Dietmar Martin and Mike Hall for helpful discussion on target of rapamycin (TOR) and protein kinase A (PKA) pathways and for kindly providing the plasmid coding for the Ras2val19 mutant; Dr. Yong Li for excellent technical support; and Agnieszka Chacinska, Klaus Pfanner, Yves Dubaquie, and members of the laboratory for discussion and critical reading of the manuscript. This work was supported by SFB 746, Forschergruppe 967, by the Excellence Initiative of the German Federal and State Governments (EXC 294) (to S.R.), and by the Wilhelm Gottfried Leibniz association (WGL) (to A.S.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.529409.
Supplemental material is available at http://www.genesdev.org.
References
- Albanese V, Yam AY, Baughman J, Parnot C, Frydman J. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 2006;124:75–88. doi: 10.1016/j.cell.2005.11.039. [DOI] [PubMed] [Google Scholar]
- Ashe MP, De Long SK, Sachs AB. Glucose depletion rapidly inhibits translation initiation in yeast. Mol Biol Cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atencio DP, Yaffe MP. MAS5, a yeast homolog of dnaJ involved in mitochondrial protein import. Mol Cell Biol. 1992;12:283–291. doi: 10.1128/mcb.12.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagriantsev SN, Gracheva EO, Richmond JE, Liebman SW. Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition. Mol Biol Cell. 2008;19:2433–2443. doi: 10.1091/mbc.E08-01-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner JJ, Carlson T, Fackenthal DL, Paddock D, Storey K, Lea K. Complex regulation of the yeast heat shock transcription factor. Mol Biol Cell. 2000;11:1739–1751. doi: 10.1091/mbc.11.5.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Saldanha AJ, Dolinski K, Botstein D. Homeostatic adjustment and metabolic remodeling in glucose-limited yeast cultures. Mol Biol Cell. 2005;16:2503–2517. doi: 10.1091/mbc.E04-11-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brejning J, Jespersen L. Protein expression during lag phase and growth initiation in Saccharomyces cerevisiae. Int J Food Microbiol. 2002;75:27–38. doi: 10.1016/s0168-1605(01)00726-7. [DOI] [PubMed] [Google Scholar]
- Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. AmiGO: Online access to ontology and annotation data. Bioinformatics. 2008;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvin CD, Kladde MP. Effectors of lysine 4 methylation of histone H3 in Saccharomyces cerevisiae are negative regulators of PHO5 and GAL1-10. J Biol Chem. 2004;279:33057–33062. doi: 10.1074/jbc.M405033200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceulemans H, Bollen M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol Rev. 2004;84:1–39. doi: 10.1152/physrev.00013.2003. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Newnam GP, Kumar J, Allen K, Zink AD. Evidence for a protein mutator in yeast: Role of the Hsp70-related chaperone ssb in formation, stability, and toxicity of the [PSI] prion. Mol Cell Biol. 1999;19:8103–8112. doi: 10.1128/mcb.19.12.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conz C, Otto H, Peisker K, Gautschi M, Wölfle T, Mayer MP, Rospert S. Functional characterization of the atypical Hsp70 subunit of yeast ribosome-associated complex. J Biol Chem. 2007;282:33977–33984. doi: 10.1074/jbc.M706737200. [DOI] [PubMed] [Google Scholar]
- Craig EA, Jacobsen K. Mutations of the heat inducible 70 kilodalton genes of yeast confer temperature sensitive growth. Cell. 1984;38:841–849. doi: 10.1016/0092-8674(84)90279-4. [DOI] [PubMed] [Google Scholar]
- Craig EA, Jacobsen K. Mutations in cognate genes of Saccharomyces cerevisiae hsp70 result in reduced growth rates at low temperatures. Mol Cell Biol. 1985;5:3517–3524. doi: 10.1128/mcb.5.12.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig EA, Eisenman HC, Hundley HA. Ribosome-tethered molecular chaperones: The first line of defense against protein misfolding? Curr Opin Microbiol. 2003;6:157–162. doi: 10.1016/s1369-5274(03)00030-4. [DOI] [PubMed] [Google Scholar]
- Denis CL, Ferguson J, Young ET. mRNA levels for the fermentative alcohol dehydrogenase of Saccharomyces cerevisiae decrease upon growth on a nonfermentable carbon source. J Biol Chem. 1983;258:1165–1171. [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Dombek KM, Kacherovsky N, Young ET. The Reg1-interacting proteins, Bmh1, Bmh2, Ssb1, and Ssb2, have roles in maintaining glucose repression in Saccharomyces cerevisiae. J Biol Chem. 2004;279:39165–39174. doi: 10.1074/jbc.M400433200. [DOI] [PubMed] [Google Scholar]
- Dragovic Z, Shomura Y, Tzvetkov N, Hartl FU, Bracher A. Fes1p acts as a nucleotide exchange factor for the ribosome-associated molecular chaperone Ssb1p. Biol Chem. 2006;387:1593–1600. doi: 10.1515/BC.2006.198. [DOI] [PubMed] [Google Scholar]
- Elbing K, McCartney RR, Schmidt MC. Purification and characterization of the three Snf1-activating kinases of Saccharomyces cerevisiae. Biochem J. 2006;393:797–805. doi: 10.1042/BJ20051213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch F, Treitel MA, Yang X, Carlson M. N-terminal mutations modulate yeast SNF1 protein kinase function. Genetics. 1992;132:639–650. doi: 10.1093/genetics/132.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick DL, Tatchell K. The REG2 gene of Saccharomyces cerevisiae encodes a type 1 protein phosphatase-binding protein that functions with Reg1p and the Snf1 protein kinase to regulate growth. Mol Cell Biol. 1996;16:2922–2931. doi: 10.1128/mcb.16.6.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi M, Mun A, Ross S, Rospert S. A functional chaperone triad on the yeast ribosome. Proc Natl Acad Sci. 2002;99:4209–4214. doi: 10.1073/pnas.062048599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Hahn JS, Thiele DJ. Activation of the Saccharomyces cerevisiae heat shock transcription factor under glucose starvation conditions by Snf1 protein kinase. J Biol Chem. 2004;279:5169–5176. doi: 10.1074/jbc.M311005200. [DOI] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated/SNF1 protein kinases: Conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- Hardie DG. Role of AMP-activated protein kinase in the metabolic syndrome and in heart disease. FEBS Lett. 2008;582:81–89. doi: 10.1016/j.febslet.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Hedbacker K, Carlson M. SNF1/AMPK pathways in yeast. Front Biosci. 2008;13:2408–2420. doi: 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SP, Carlson M. Regulation of snf1 protein kinase in response to environmental stress. J Biol Chem. 2007;282:16838–16845. doi: 10.1074/jbc.M700146200. [DOI] [PubMed] [Google Scholar]
- Hong SP, Leiper FC, Woods A, Carling D, Carlson M. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc Natl Acad Sci. 2003;100:8839–8843. doi: 10.1073/pnas.1533136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Gautschi M, Walter W, Rospert S, Craig EA. The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat Struct Mol Biol. 2005;12:497–504. doi: 10.1038/nsmb942. [DOI] [PubMed] [Google Scholar]
- Hurt E, Luo MJ, Rother S, Reed R, Strasser K. Cotranscriptional recruitment of the serine–arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc Natl Acad Sci. 2004;101:1858–1862. doi: 10.1073/pnas.0308663100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Tatchell K, Liu S, Michels CA. Protein phosphatase type-1 regulatory subunits Reg1p and Reg2p act as signal transducers in the glucose-induced inactivation of maltose permease in Saccharomyces cerevisiae. Mol Gen Genet. 2000;263:411–422. doi: 10.1007/s004380051185. [DOI] [PubMed] [Google Scholar]
- Jones GW, Song Y, Masison DC. Deletion of the Hsp70 chaperone gene SSB causes hypersensitivity to guanidine toxicity and curing of the [PSI+] prion by increasing guanidine uptake in yeast. Mol Genet Genomics. 2003;269:304–311. doi: 10.1007/s00438-003-0838-y. [DOI] [PubMed] [Google Scholar]
- Kim SY, Craig EA. Broad sensitivity of Saccharomyces cerevisiae lacking ribosome-associated chaperone Ssb or Zuo1 to cations, including aminoglycosides. Eukaryot Cell. 2005;4:82–89. doi: 10.1128/EC.4.1.82-89.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Schilke B, Craig EA, Horwich AL. Folding in vivo of a newly translated yeast cytosolic enzyme is mediated by the SSA class of cytosolic yeast Hsp70 proteins. Proc Natl Acad Sci. 1998;95:12860–12865. doi: 10.1073/pnas.95.22.12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan C, Lee HC, Tang S, Zhang L. A novel mode of chaperone action: heme activation of Hap1 by enhanced association of Hsp90 with the repressed Hsp70–Hap1 complex. J Biol Chem. 2004;279:27607–27612. doi: 10.1074/jbc.M402777200. [DOI] [PubMed] [Google Scholar]
- Lodi T, Guiard B. Complex transcriptional regulation of the Saccharomyces cerevisiae CYB2 gene encoding cytochrome b2: CYP1(HAP1) activator binds to the CYB2 upstream activation site UAS1-B2. Mol Cell Biol. 1991;11:3762–3772. doi: 10.1128/mcb.11.7.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Cyr DM. Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J Biol Chem. 1998;273:27824–27830. doi: 10.1074/jbc.273.43.27824. [DOI] [PubMed] [Google Scholar]
- Ludin K, Jiang R, Carlson M. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1998;95:6245–6250. doi: 10.1073/pnas.95.11.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayordomo I, Regelmann J, Horak J, Sanz P. Saccharomyces cerevisiae 14–3–3 proteins Bmh1 and Bmh2 participate in the process of catabolite inactivation of maltose permease. FEBS Lett. 2003;544:160–164. doi: 10.1016/s0014-5793(03)00498-8. [DOI] [PubMed] [Google Scholar]
- McCartney RR, Schmidt MC. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J Biol Chem. 2001;276:36460–36466. doi: 10.1074/jbc.M104418200. [DOI] [PubMed] [Google Scholar]
- Neigeborn L, Carlson M. Mutations causing constitutive invertase synthesis in yeast: Genetic interactions with snf mutations. Genetics. 1987;115:247–253. doi: 10.1093/genetics/115.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- Nigavekar SS, Tan YS, Cannon JF. Glc8 is a glucose-repressible activator of Glc7 protein phosphatase-1. Arch Biochem Biophys. 2002;404:71–79. doi: 10.1016/s0003-9861(02)00231-x. [DOI] [PubMed] [Google Scholar]
- Park SH, Bolender N, Eisele F, Kostova Z, Takeuchi J, Coffino P, Wolf DH. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin–proteasome system. Mol Biol Cell. 2007;18:153–165. doi: 10.1091/mbc.E06-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfund C, Lopez-Hoyo N, Ziegelhoffer T, Schilke BA, Lopez-Buesa P, Walter WA, Wiedmann M, Craig EA. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J. 1998;17:3981–3989. doi: 10.1093/emboj/17.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfund C, Huang P, Lopez-Hoyo N, Craig EA. Divergent functional properties of the ribosome-associated molecular chaperone Ssb compared with other Hsp70s. Mol Biol Cell. 2001;12:3773–3782. doi: 10.1091/mbc.12.12.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskur J, Rozpedowska E, Polakova S, Merico A, Compagno C. How did Saccharomyces evolve to become a good brewer? Trends Genet. 2006;22:183–186. doi: 10.1016/j.tig.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Portillo F, Mulet JM, Serrano R. A role for the non-phosphorylated form of yeast Snf1: Tolerance to toxic cations and activation of potassium transport. FEBS Lett. 2005;579:512–516. doi: 10.1016/j.febslet.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Rakwalska M, Rospert S. The ribosome-bound chaperones RAC and Ssb1/2p are required for accurate translation in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:9186–9197. doi: 10.1128/MCB.24.20.9186-9197.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raue U, Oellerer S, Rospert S. Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. J Biol Chem. 2007;282:7809–7816. doi: 10.1074/jbc.M611436200. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. Extraction, purification, and analysis of mRNA from eukaryotic cells. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Sanz P, Alms GR, Haystead TA, Carlson M. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol Cell Biol. 2000;20:1321–1328. doi: 10.1128/mcb.20.4.1321-1328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner L, Gibney PA, Morano KA. The Hsp110 protein chaperone Sse1 is required for yeast cell wall integrity and morphogenesis. Curr Genet. 2008;54:1–11. doi: 10.1007/s00294-008-0193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomura Y, Dragovic Z, Chang HC, Tzvetkov N, Young JC, Brodsky JL, Guerriero V, Hartl FU, Bracher A. Regulation of Hsp70 function by HspBP1: Structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol Cell. 2005;17:367–379. doi: 10.1016/j.molcel.2004.12.023. [DOI] [PubMed] [Google Scholar]
- Sutherland CM, Hawley SA, McCartney RR, Leech A, Stark MJ, Schmidt MC, Hardie DG. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr Biol. 2003;13:1299–1305. doi: 10.1016/s0960-9822(03)00459-7. [DOI] [PubMed] [Google Scholar]
- Thompson-Jaeger S, Francois J, Gaughran JP, Tatchell K. Deletion of SNF1 affects the nutrient response of yeast and resembles mutations which activate the adenylate cyclase pathway. Genetics. 1991;129:697–706. doi: 10.1093/genetics/129.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomala K, Korona R. Alleviation of deleterious effects of protein mutation through inactivation of molecular chaperones. Mol Genet Genomics. 2008;280:409–417. doi: 10.1007/s00438-008-0374-x. [DOI] [PubMed] [Google Scholar]
- Tu J, Carlson M. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 1995;14:5939–5946. doi: 10.1002/j.1460-2075.1995.tb00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung KS, Hopper AK. The glucose repression and RAS-cAMP signal transduction pathways of Saccharomyces cerevisiae each affect RNA processing and the synthesis of a reporter protein. Mol Gen Genet. 1995;247:48–54. doi: 10.1007/BF00425820. [DOI] [PubMed] [Google Scholar]
- Wandinger SK, Richter K, Buchner J. The Hsp90 chaperone machinery. J Biol Chem. 2008;283:18473–18477. doi: 10.1074/jbc.R800007200. [DOI] [PubMed] [Google Scholar]
- Werner-Washburne M, Craig EA. Expression of members of the Saccharomyces cerevisiae hsp70 multigene family. Genome. 1989;31:684–689. doi: 10.1139/g89-125. [DOI] [PubMed] [Google Scholar]
- Werner-Washburne M, Becker J, Kosic-Smithers J, Craig EA. Yeast Hsp70 RNA levels vary in response to the physiological status of the cell. J Bacteriol. 1989;171:2680–2688. doi: 10.1128/jb.171.5.2680-2688.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiatrowski HA, Carlson M. Yap1 accumulates in the nucleus in response to carbon stress in Saccharomyces cerevisiae. Eukaryot Cell. 2003;2:19–26. doi: 10.1128/EC.2.1.19-26.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ET, Dombek KM, Tachibana C, Ideker T. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J Biol Chem. 2003;278:26146–26158. doi: 10.1074/jbc.M301981200. [DOI] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
- Zhang H, Richardson DO, Roberts DN, Utley R, Erdjument-Bromage H, Tempst P, Cote J, Cairns BR. The Yaf9 component of the SWR1 and NuA4 complexes is required for proper gene expression, histone H4 acetylation, and Htz1 replacement near telomeres. Mol Cell Biol. 2004;24:9424–9436. doi: 10.1128/MCB.24.21.9424-9436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]