Abstract
The telomere repeat-binding factor 1 (TERF1, referred to hereafter as TRF1) is a component of mammalian telomeres whose role in telomere biology and disease has remained elusive. Here, we report on cells and mice conditionally deleted for TRF1. TRF1-deleted mouse embryonic fibroblasts (MEFs) show rapid induction of senescence, which is concomitant with abundant telomeric γ-H2AX foci and activation of the ATM/ATR downstream checkpoint kinases CHK1 and CHK2. DNA damage foci are rescued by both ATM and ATM/ATR inhibitors, further indicating that both signaling pathways are activated upon TRF1 deletion. Abrogation of the p53 and RB pathways bypasses senescence but leads to chromosomal instability including sister chromatid fusions, chromosome concatenation, and occurrence of multitelomeric signals (MTS). MTS are also elevated in ATR-deficient MEFs or upon treatment with aphidicolin, two conditions known to induce breakage at fragile sites, suggesting that TRF1-depleted telomeres are prone to breakage. To address the impact of these molecular defects in the organism, we deleted TRF1 in stratified epithelia of TRF1Δ/ΔK5-Cre mice. These mice die perinatally and show skin hyperpigmentation and epithelial dysplasia, which are associated with induction of telomere-instigated DNA damage, activation of the p53/p21 and p16 pathways, and cell cycle arrest in vivo. p53 deficiency rescues mouse survival but leads to development of squamous cell carcinomas, demonstrating that TRF1 suppresses tumorigenesis. Together, these results demonstrate that dysfunction of a telomere-binding protein is sufficient to produce severe telomeric damage in the absence of telomere shortening, resulting in premature tissue degeneration and development of neoplastic lesions.
Keywords: Telomeres, TRF1, DNA damage, chromosomal instability, conditional knockout mice, cancer
The telomere repeat-binding factor 1 (TERF1, referred to hereafter as TRF1) is part of the shelterin complex present at mammalian telomeres, formed by TRF1, TRF2, POT1, TPP1, TIN2, and RAP1 (Liu et al. 2004; de Lange 2005). Extensive cell-based in vitro studies using overexpression of TRF1 alleles have suggested a role for TRF1 as a negative regulator of telomere length (van Steensel and de Lange 1997; Smogorzewska et al. 2000; Ancelin et al. 2002). More recently, TRF1 overexpression in the context of transgenic mouse tissues was also shown to lead to telomere shortening mediated by the XPF nuclease (Munoz et al. 2009). Post-transcriptional modification of TRF1 by tankyrases 1 and 2, which poly-ADP-ribosylate TRF1, can regulate its binding to telomeres, thereby influencing telomere length and sister telomere cohesion (Smith et al. 1998; Cook et al. 2002). In particular, tankyrase 1-depleted cells show sister telomere fusions due to excessive telomeric cohesion (Dynek and Smith 2004), which is restored by abrogating the SA1 cohesin subunit or TRF1 and TIN2 (Canudas et al. 2007).
TRF2, a homolog of TRF1 (Bilaud et al. 1997; Broccoli et al. 1997), is also a negative regulator of telomere length in both mice and humans (Smogorzewska et al. 2000; Ancelin et al. 2002; Zhu et al. 2003; Munoz et al. 2005; Blanco et al. 2007). In addition, TRF2 is essential for telomere capping (van Steensel et al. 1998; Celli and de Lange 2005). Conditional deletion of TRF2 in mouse embryonic fibroblasts (MEFs) leads to chromosome end-to-end fusions and severe proliferative defects (Celli and de Lange 2005). TRF2 conditional deletion in the liver, however, did not have an impact on liver regeneration or mouse viability (Lazzerini Denchi et al. 2006), arguing that TRF2 is dispensable for hepatocyte regeneration.
POT1, a TRF1-interacting protein, is proposed to regulate telomere length and telomere protection (Loayza and De Lange 2003). In particular, Pot1a/b deficiency in mice leads to increased DNA damage foci at telomeres (Hockemeyer et al. 2006; Wu et al. 2006), and Pot1b-deficient mice show increased degenerative pathologies when generated in a telomerase-deficient background and crossed for several generations (Hockemeyer et al. 2008; He et al. 2009).
Conventional TRF1 deletion in mice produces very early embryonic lethality at the blastocyst stage; however, no defects are detected at this stage in telomere length or telomere capping (Karlseder et al. 2003). More recently, embryonic stem (ES) cells conditionally deleted for TRF1 were reported to show normal telomere length and increased chromosomal aberrations, including fusions and multitelomeric signals (MTS) (Okamoto et al. 2008), suggesting that TRF1 deficiency leads to telomere aberrations in cultured ES cells. The early embryonic lethality of TRF1-null mice has not allowed the role of TRF1 in differentiated cells to be ascertained.
Here, we address the role of TRF1 in telomere biology and disease in the context of a mammalian organism by generating tissue-specific TRF1 conditionally deleted cells and mice. We deleted TRF1 in MEFs and stratified epithelia in the mouse. TRF1-deleted MEFs show massive induction of telomeric DNA damage foci, activation of ATM/ATR and their downstream checkpoint kinases CHK1 and CHK2, and cell cycle arrest, demonstrating that TRF1 protects telomeres from eliciting a DNA damage response (DDR). TRF1-null cells also show abundant telomere fusions (particularly, sister telomere fusions), as well as the occurrence of MTS. The latter type of telomere aberration was also increased in wild-type cells following inhibition of DNA synthesis by aphidicolin treatment, as well as in ATR-deficient cells recently shown to have increased replicative stress and breakage at fragile sites (Murga et al. 2009), thus suggesting a role for TRF1 in preventing telomere fragility (Durkin and Glover 2007).
In agreement with severe telomere dysfunction, TRF1-null mice die perinatally and show an early onset of pathologies, including epithelial dysplasia, lack of hair follicles, and skin hyperpigmentation. p53 deficiency rescues mouse survival, hair growth defects, and skin hyperpigmentation, but leads to the increased occurrence of spontaneous skin cancer and epithelial abnormalities such as nail distrophy and oral leukoplakia, which are similar to pathologies in human patients with mutations in telomerase components, such as dyskeratosis congenita, aplastic anemia, and some cases of pulmonary fibrosis (Mitchell et al. 1999; Vulliamy et al. 2001; Armanios et al. 2007; Tsakiri et al. 2007). TRF1-deficient mice represent the first mouse model for complete abrogation of a telomere-binding protein that leads to increased cancer and degenerative phenotypes in the absence of telomere shortening and of any generational lag. Together, these results indicate an essential role of TRF1 in telomere protection and stability in vivo, and open a new class of telomere pathologies produced by telomere dysfunction in the absence of telomere shortening.
Results
Rapid induction of p53 and pRb-mediated senescence in TRF1Δ/Δ-Cre MEFs
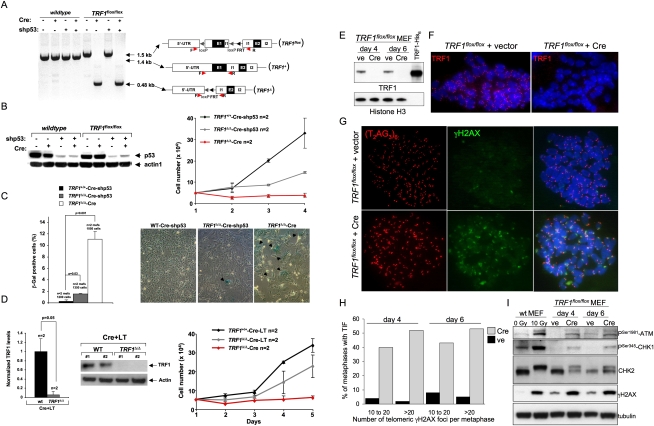
To study the impact of TRF1 deletion in differentiated cells, we generated a conditional TRF1 knockout mouse strain, TRF1flox/flox mice, where TRF1 exon 1 (E1) is flanked by loxP sites (Figs. 1A, 3A [below]. We included FRT sites to remove the neomycin cassette after generation of chimeric mice by crossing with Flp mice (Materials and Methods). To generate TRF1-null MEFs, we infected MEFs derived from wild-type and conditional TRF1flox/flox mice with either a pBabe-Cre retrovirus (TRF1Δ/Δ-Cre MEF) or an empty pBabe vector (Fig. 1A). The conditional TRF1 allele was efficiently excised in TRF1flox/flox MEFs infected with pBabe-Cre (Fig. 1A). The resulting TRF1Δ/Δ-Cre MEFs, however, failed to proliferate due to a rapid induction of senecence, as detected by senescence-associated β-gal staining (Fig. 1B,C). To bypass senescence, we down-regulated the expression of p53, a mediator of senescence (Collado et al. 2005), by using an shRNA against p53, shp53 (Fig. 1B,C). We observed a modest recue of cell proliferation and senescence in TRF1Δ/Δ-Cre-shp53 MEFs compared with TRF1Δ/Δ-Cre MEF (Fig. 1B,C), although these cells ceased proliferation after 4 d in culture (data not shown). Proliferation of TRF1Δ/Δ-Cre MEFs was further augmented by simultaneously canceling the p53 and the pRb pathways upon expression of the SV40 large T (LT) antigen, TRF1Δ/Δ-Cre-LT MEFs (Fig. 1D; Materials and Methods). These findings indicate that TRF1 deletion results in rapid induction of senescence, which is mediated by the p53 and pRb pathways.
Figure 1.
Activation of a DDR in TRF1-deficient MEF. (A) TRF1 deletion in TRF1flox/flox MEFs upon retroviral infection with Cre recombinase was confirmed by PCR amplification of TRF1 alleles. Alleles are schematically depicted. (B) Knockdown of p53 expression in MEFs infected with a vector containing shRNA-p53 (shp53). (Left) Western blot showing p53 knockdown. (Right) Growth curves of TRF1+/+ and TRFΔ/Δ cells doubly infected with Cre and shp53 and of TRFΔ/Δ cells only infected with Cre. n = independent cultures used per genotype and condition. Error bars indicate standard error. (C, left) Quantification of senescence-associated β-gal-positive cells 4 d post-infection. Error bars indicate standard error. Student's t-test was used for statistical analysis and P-values are indicated. (Right) Representative images of senescence-associated β-gal staining in the indicated genotypes. Black arrows indicate senescent cells. (D, left) Western blot of TRF1 protein and its quantification in the indicated MEFs. Values were normalized to wild-type levels. n = independent MEFs per genotype. Error bars indicate standard error. The Student's t-test was used for statistical analysis and P-values are indicated. (Right) Growth curves of the indicated MEFs. n = independent cultures used per genotype and condition. Error bars indicate standard error. Statistical differences were calculated using the Student's t-test and P-value is indicated. (E) Western blot detection of mouse TRF1 in TRF1Δ/Δ MEFs treated with empty pBabe vector (ve) and H&R Cre (Cre). Extracts from cells collected 4 and 6 d post-selection were immunoblotted as indicated. Recombinant human TRF1-His6, migrating more quickly due the smaller size of the human TRF1 protein, served as a control. Histone H3 was used as loading control. (F) Immunofluorescence detection of mouse TRF1 in TRF1Δ/Δ MEFs treated with empty vector and H&R Cre. Metaphase chromosome spreads were stained with anti-TRF1 rabbit polyclonal antibody (red). DNA was counterstained with DAPI (blue). (G) Immunofluorescence detection of γH2AX (green) combined with FISH staining of the telomeres (red) in TRF1Δ/Δ MEFs treated with empty vector and H&R Cre. (H) The percentage of metaphase nuclei exhibiting 10–20 or >20 γH2AX foci was determined for at least 50 metaphases prepared as in G and collected 4 and 6 d post-selection. (I) Western blot detection of DDR factors in TRF1Δ/Δ MEFs treated with empty pBabe vector (ve) and H&R Cre (Cre). Extracts were collected 4 and 6 d post-selection as indicated. MEFs treated with 0 or 10 Gy of IR served as a control for checkpoint activation. Tubulin was used as loading control.
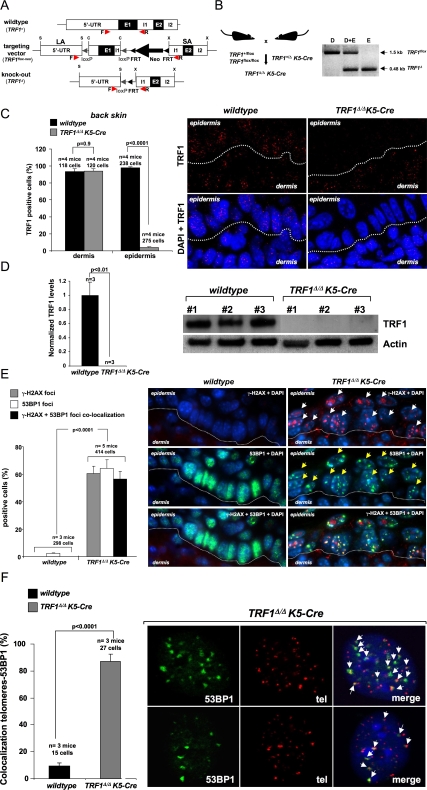
Figure 3.
TRF1 deficiency in mouse tissues leads to massive induction of TIFs. (A) Schematic outline of wild-type (TRF1+), floxed (TRF1flox-neo), and deleted (TRF1Δ) loci. (B, left) Breeding strategy to generate TRF1Δ/ΔK5-Cre mice. (Right) PCR amplification of TRF1 alleles in whole skin (dermis and epidermins, D + E), dermis only (D), and epidermis only (E) of TRF1Δ/ΔK5-Cre mice using the F and R primers depicted in A. (C, left) Quantification of TRF1-positive cells in the dermis and epidermis of the indicated genotypes. n = mice analyzed per genotype. The total number of cells scored per genotype is shown. Error bars indicate standard error. Student's t-test was used for statistical analysis and P-values are indicated. (Right) Representative images of TRF1 immunofluorescence. (D, left) Quantification of TRF1 protein in the epidermis of the indicated genotypes. n = newborn mice used per genotype. Error bars indicate standard error. The Student's t-test was used for statistical analysis and P-values are indicated. (Right) Representative Western blot of TRF1 protein in newborn primary keratinocytes from the indicated genotypes. Actin was used as a loading control. #1, #2, #3 correspond to individual mice. (E, left) Percentage of cells showing γ-H2AX foci, 53BP1 foci, and doubly γ-H2AX–53BP1-positive foci in back skin sections of mice of the indicated genotype. n = mice analyzed per genotype. The total number of cells scored for each genotype is shown. Error bars indicate standard error. Student's t-test was used for statistical analysis and P-values are indicated. (Right) Representative images. Arrows indicate cells with DNA damage foci. (F, left) Percentage of cells with colocalization of 53BP1 foci to telomeres. n = mice analyzed per genotype. The total number of cells scored per genotype is shown. Error bars indicate standard error. Student's t-test was used for statistical analysis and P-values are indicated. (Right) Representative images of 53BP1 foci and of telomeres, and of the combined images. White arrows indicate colocalization of DNA damage foci to telomeres.
TRF1 deficiency results in activation of a DDR at telomeres
To further understand the molecular mechanisms underlying the severe cellular proliferation defects of TRF1-deleted MEFs, we first addressed whether TRF1 abrogation resulted in increased telomeric damage. γH2AX foci have been shown to mark the presence of DNA double-strand breaks (DSBs), including those associated with critically short/dysfunctinal telomeres, also known as TIFs (d'Adda di Fagagna et al. 2003; Takai et al. 2003). To this end, transient expression of Cre recombinase from the self-inactivating Hit-and-Run retrovirus resulted in efficient loss of TRF1 gene expression as monitored by Western blotting at 4 and 6 d after selection (Fig. 1E). Concomitantly, the telomere-associated TRF1 signal on metaphase chromosome spreads became undetectable by imunofluorescence microscopy (Fig. 1F). Most of these cells showed γH2AX foci at telomeres of spread mitotic chromosomes, with at least one telomere per pair of sister chromatids being labeled by γH2AX (Fig. 1G). TRF1Δ/Δ MEFs showed a very robust TIF induction, with 40% of the metaphases carrying 10–20 TIFs and 50% having >20 foci (Fig. 1H). TIF induction following TRF1 deletion was accompanied by phosphorylation of ATM (at Ser 1981) and its downstream checkpoint kinases CHK1 (at Ser 345) and CHK2 (as detected by mobility shift) (Fig. 1I). Also an indication that the checkpoint was activated was the high phosphorylation of H2AX itself (Fig. 1I). As a positive control, we used wild-type MEFs exposed to 10 Gy of ionizing radiation (IR), which show a complete shift of the CHK2 lower band and high levels of ATM and CHK1 phosphorylation. None of these phosphorylation events was observed in control wild-type cells treated with Cre recombinase (data not shown).
To address the relative importance of the ATM and ATR pathways in the DDR induced by TRF1 deficiency, cells were treated with ATM-specific (KU55933) or both ATM/ATR (caffeine) kinase inhibitors and DNA damage foci quantified by using 53BP1 immunofluorescence, a known mediator of the DDR that forms foci in the presence of DNA damage. 53BP1-positive and TIF-positive cells were scored after 6 h of treatment with the inhibitors (Materials and Methods). As shown in Supplemental Figure 1, A–C, 60% of cells deleted for TRF1 were 53BP1-positive and 90% of these cells contained more than four TIFs. Both 53BP1 foci and TIFs induced by TRF1 abrogation were significantly decreased upon ATM inhibition, and an additive effect was observed after ATM/ATR inhibition with caffeine, reaching similar levels to control wild-type cells (Supplemental Fig. 1A–C). These results indicate that TRF1 deletion triggers activation of both ATM and ATR DNA damage signaling pathways.
TRF1 protects from telomere fusions and telomere fragility
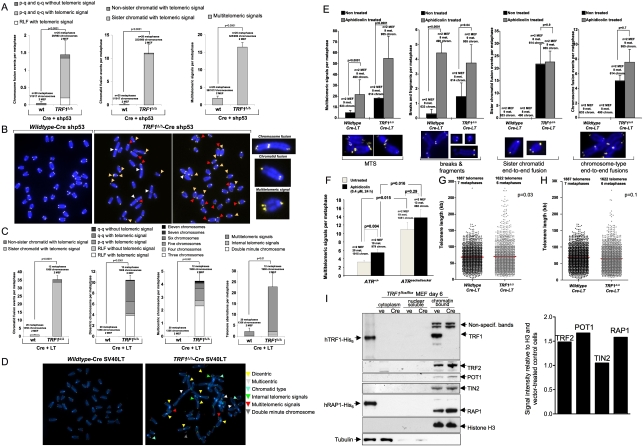
The fact that TRF1 abrogation leads to rapid induction of a DDR and senescence suggests a role for TRF1 in telomere protection. Given that TRF1Δ/Δ-Cre MEFs fail to proliferate, we performed metaphase quantitative telomere and centromere fluorescence in situ hybridization (Q-FISH) analysis on TRF1Δ/Δ-Cre-shp53 MEFs. TRF1Δ/Δ-Cre-shp53 MEFs showed a high frequency of sister telomere fusions (>10 fusions per metaphase), and a lower but significant increase in chromosome end-to-end fusions (up to 1.5 fusions per metaphase) compared with TRF1+/+-Cre-shp53 controls (Fig. 2A,B). In addition, these cells showed a very high increase in MTS (>15 per metaphase) (Fig. 2A,B), a type of telomere aberration described previously in the context of TRF2 overexpression and TRF1 deficiency (Munoz et al. 2005; Blanco et al. 2007; Okamoto et al. 2008). Notably, end-to-end fusions and MTS were further increased after simultaneous abrogation of the p53 and the pRB pathways in TRF1Δ/Δ-Cre-LT MEFs, including now the formation of concatenated chromosomes (up to 11 concatenated chromosomes in a single metaphase) (Fig. 2C,D). MTS could reflect increased breakage at chromosome termini and subsequent repair by homologous recombination mechanisms (Lydeard et al. 2007). In this regard, TRF1-null MEFs show slightly increased recombination events between sister telomeres as determined by chromosome orientation FISH (CO-FISH), although this increase did not reach statistical significance (Supplemental Fig. 2). It had been proposed recently that TRF1-induced MTS reflect telomere fragility during replication (Sfeir et al. 2009). We wanted to extend these observations to our mouse model, and for this we treated wild-type and TRF1-null MEFs with aphidicolin, a drug known to inhibit DNA synthesis and to induce breaks at chromosome fragile sites (Durkin and Glover 2007). MTS were significantly increased in wild-type cells treated with aphidicolin and further increased in aphidicolin-treated TRF1-deficient MEFs (Fig. 2E). Concomintantly, breaks/fragments were also increased in aphidicolin-treated wild-type and TRF1-null MEFs, consistent with induction of breaks at genomic fragile sites (Fig. 2E). In contrast, other types of aberrations produced by TRF1 deficiency, such as sister chromatid and chromosome-type fusions, were not increased by aphidicolin treatment (Fig. 2E), arguing that these aberrations are not the consequence of increased telomere breakage but instead may reflect loss of telomere capping in the absence of TRF1. Next, we studied whether MTS were also induced in a recently generated mouse model for ATR deficiency (ATRSeckel/Seckel mice), which shows increased replicative stress and increased breakage at fragile sites (Murga et al. 2009). As shown in Figure 2F, ATR-deficient MEFs showed increased MTS compared with wild-type controls, reaching similar levels to those of TRF1-null MEFs, further supporting the notion that TRF1-deficient telomeres have characteristics of fragile sites and are prone to breakage. These findings, together with DDR activation in TRF1-null MEFs, confirm and extend the concept that TRF1 deletion leads to increased replication fork stalling at telomeres and subsequent telomere breakage (Sfeir et al. 2009). In summary, TRF1 deletion induces at least two types of telomere defects at the molecular level: (1) telomere uncapping leading to end-to-end fusions (particularly sister telomere fusions), and (2) increased fragility and breakage at telomeres resulting in MTS.
Figure 2.
TRF1 deficiency leads to telomere fusions and increased telomere fragility in the presence of normal binding of other shelterin components. (A,C) Frequency of aberrations in metaphase spreads from the indicated genotypes and conditions. n = metaphases used for the analysis from a total of two MEFs per genotype. Error bars indicate standard error. Statistical comparisons using the χ2 test are shown and P-values are indicated. (B,D) Representative images of metaphases from the indicated genotypes. In B, red arrows indicate MTS, white arrows indicate end-to-end fusions and yellow arrows indicate sister telomere fusions. In D, arrows are described in the figure. (E,F) Chromosomal aberrations in untreated MEFs or upon treatment with aphidicolin of the indicated MEFs. (G,H) Telomere fluorescence (Q-FISH) distribution in metaphase spreads from the indicated genotypes. The fused telomeres were quantified as single telomeres in G or divided by a factor of 2 in H. Mean telomere fluorescence and standard deviation values are shown for each genotype. n = number of telomeres used for the analysis. The Student's t-test was used for statistical calculations and P-values are indicated. (I, left) Subcellular fractionation of TRF1Δ/Δ MEFs treated with empty pBabe vector (ve) and H&R Cre (Cre). Tubulin was used as loading control for the cytoplasmic fraction and histone H3 was used for the chromatin-bound fraction. (Right) Quantification of Western blots probed with TRF2, POT1, TIN2, and RAP1 antibodies is shown. Intensity was calculated relative to the histone H3, which served as a loading control for the chromatin-bound fraction, and was expressed relative to the wild type.
The majority of fusions in TRF1-null MEFs showed detectable telomeric signals at the fusion point, indicating that they occurred in the absence of telomere shortening (Fig. 2A–E). Given the previously proposed role of TRF1 as a negative regulator of telomere length (van Steensel and de Lange 1997; Smogorzewska et al. 2000; Ancelin et al. 2002; Munoz et al. 2009), we addressed the impact of TRF1 deletion on telomere length by performing telomere Q-FISH on metaphases (Materials and Methods). TRF1Δ/Δ-Cre-LT MEF showed increased telomere fluorescence when considering all individual telomere signals, including the fusions (Fig. 2G); however, these apparently longer telomeres were largely eliminated when fusions were corrected (Fig. 2H). These results suggest that the apparently elongated telomeres in TRF1-null metaphase cells correspond to fusions and that TRF1 deletion does not result in significant telomere length changes.
Finally, as some of the telomere defects associated with TRF1 deletion are also observed in cells depleted for TRF2 and Pot1, we determined whether the binding to telomeres of other shelterin components was altered in TRF1-depleted telomeres by using chromatin fractionation assays (Materials and Methods). As shown in Figure 2I, the binding of TRF2, POT1, TIN2, and RAP1 was similar or slightly increased in the absence of TRF1 binding, arguing that the telomere defects triggered by TRF1 deletion are not due to abrogation of other shelterin components from telomeres. These results were also confirmed by confocal immunofluorescence experiments, which showed normal TRF2 and POT1 binding in the absence of TRF1 (Supplemental Fig. 3A,B).
Persistent induction of TIFs in TRF1Δ/ΔK5-Cre mice
To study the in vivo impact of the molecular and chromosomal defects triggered by TRF1 deficiency, we targeted deletion of TRF1 to mouse stratified epithelia. For this, we crossed conditional TRF1flox/flox mice or TRF1+/flox mice with K5-Cre mice that constitutively express the Cre recombinase under the control of the keratin 5 (K5) promoter from day 11.5 of embryonic development (E11.5) onward (Ramirez et al. 1994), thereby generating TRF1Δ/ΔK5-Cre mice (Fig. 3A,B). TRF1Δ/ΔK5-Cre mice show 100% excision of TRF1 exon 1 in the epidermis, while no deletion can be detected in the dermis (Fig. 3B), in agreement with the K5 expression pattern. Accordingly, the TRF1 protein was efficiently abrogated in the epidermis of TRF1Δ/ΔK5-Cre mice as determined by TRF1 immunofluorescence (Fig. 3C) and Western blotting analysis of isolated skin keratinocytes (Fig. 3D; Materials and Methods). Again, TRF1-deleted keratinocytes showed normal levels of TRF2 and POT1 proteins (Supplemental Fig. 3C,D).
We observed a dramatic increase in γH2AX-positive and 53BP1-positive cells in TRF1Δ/ΔK5-Cre epidermis compared with wild-type controls (Fig. 3E). Furthermore, these foci largely colocalized, indicative of strong DDR activation (Fig. 3E). In addition, we found an almost complete colocalization (>80% of the dots colocalized) between 53BP1 and telomeric DNA in TRF1Δ/ΔK5-Cre keratinocytes, indicating DNA damage foci specifically localized to telomeres (Fig. 3F). These results demonstrate that TRF1 abrogation in the epidermis leads to persistent activation of a DDR at telomeres, in agreement with the MEF results (Fig. 1).
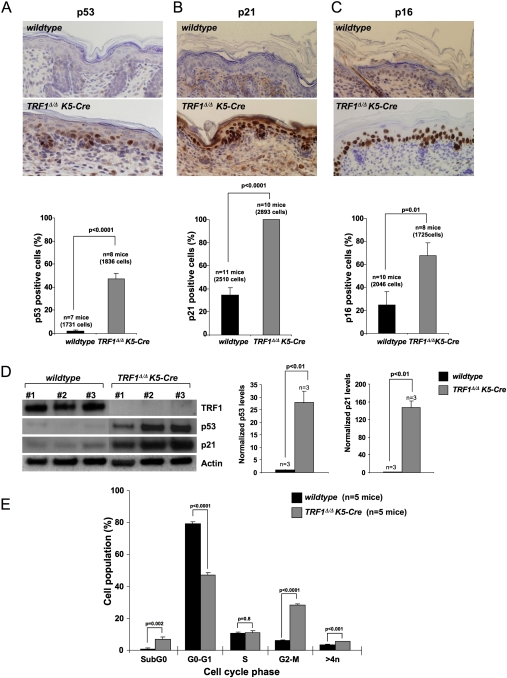
TRF1Δ/ΔK5-Cre mice show increased p53, p21, and p16 cell cycle inhibitors and G2/M cell cycle arrest
Persistent DNA damage at telomeres as the consequence of critical telomere shortening induces a senescence-like cycle arrest mediated by the p21 cell cycle inhibitor, which is thought to restrict tissue regeneration and prevent tumor development in the context of telomerase-deficient mice (Choudhury et al. 2007; Feldser and Greider 2007). To address whether TRF1 deficiency leads to similar proliferative defects in vivo, we studied expression of p53, p21, and p16 cell cycle inhibitors known to be up-regulated in association with cellular senescence in vivo (Collado et al. 2005). TRF1Δ/ΔK5-Cre epidermis showed a dramatic increase in cell cycle inhibitors compared with the controls (Fig. 4A–C), which was confirmed by Western blot analysis of freshly isolated newborn keratinocytes (Fig. 4D). Concomitantly, we detected decreased proliferation (lower percentage of Ki67-positive and BrdU-positive cells) in TRF1Δ/ΔK5-Cre epidermis compared with wild-type skin, while no significant differences in apoptosis were detected (caspase-3-positive cells) (Supplemental Fig. 4A–C). Flow cytometry analysis (FACS) of isolated skin keratinocytes further demonstrated that a significant percentage of TRF1Δ/ΔK5-Cre epidermal cells were arrested in G2/M, indicating failure to undergo mitosis upon TRF1 deletion (Fig. 4E; Materials and Methods). Together, these results indicate that TRF1 deletion in the epidermis leads to cell cycle arrest concomitant with up-regulation of both the p53/p21 and p16 pathways, in agreement with the rapid induction of p53/Rb-dependent senescence observed in TRF1-abrogated MEFs (Fig. 1B–D).
Figure 4.
TRF1 deficiency leads to increased p53, p21, and p16 levels and severe proliferative defects in vivo. (A–C) Percentage of p53-positive cells (A), p21-positive cells (B), and p16-positive cells (C) in back skin sections from mice of the indicated genotypes. Representative images are also shown. n = mice analyzed per genotype. The total number of cells scored per genotype is shown in brackets. Error bars indicate standard error. Student's t-test was used for statistical analysis and P-values are indicated. (D, left) Representative Western blot showing TRF1, p53, and p21 protein levels in primary keratinocytes isolated the indicated genotypes. Actin was used as a loading control. #1, #2, #3 correspond to individual mice. (Right) Quantification of p53 and p21 protein levels after normalization to wild-type levels. n = independent mice used per genotype. Error bars indicate standard error. The Student's t-test was used for statistical analysis and P-values are indicated. (E) Quantification of percentage of cells at different phases of the cell cycle by FACS analysis of newborn keratinocytes. Cells were stained with propidium iodide. n = independent newborn mice used per genotype. Error bars indicate standard error. Student's t-test was used for statistical analysis and P-values are indicated.
Of interest, severe proliferative defects in TRF1Δ/ΔK5-Cre epidermis occurred in the absence of any detectable telomere shortening and in the presence of normal telomerase activity (Supplemental Fig. 5A–F). In particular, TRF1Δ/ΔK5-Cre skin showed higher telomere fluorescence than wild-type skin when measuring individual telomere dots, as well as when considering average telomere fluorescence per nuclei (Supplemental Fig. 5A,C,E). However, the fact that centromere fluorescence was also increased in TRF1Δ/ΔK5-Cre skin (Supplemental Fig. 5B,D,E) suggested that the higher telomere and centromere signals associated with TRF1 deletion are a consequence of increased fusions (Fig. 2A–D) and/or increased poliploidy (Fig. 4E). In this regard, we did not observe differences in telomere length between wild-type and TRF1Δ/ΔK5-Cre keratinocytes when using telomere restriction fragment (TRF) analysis, a technique based on Southern blotting (Supplemental Fig. 6A). TRF analysis also indicated that TRF1 deletion was accompanied by a slight decrease in G-strand overhang signal compared with wild-type mice (Supplemental Fig. 6A,B). Together, these findings indicate that persistent telomere damage signaling in TRF1-deleted epidermis occurs in the absence of detectable telomere shortening, in contrast to mouse models for telomerase and other telomere-binding proteins (Munoz et al. 2005; Blanco et al. 2007; Hockemeyer et al. 2008; He et al. 2009).
TRF1 deficiency leads to severe skin atrophy and skin hyperpigmentation
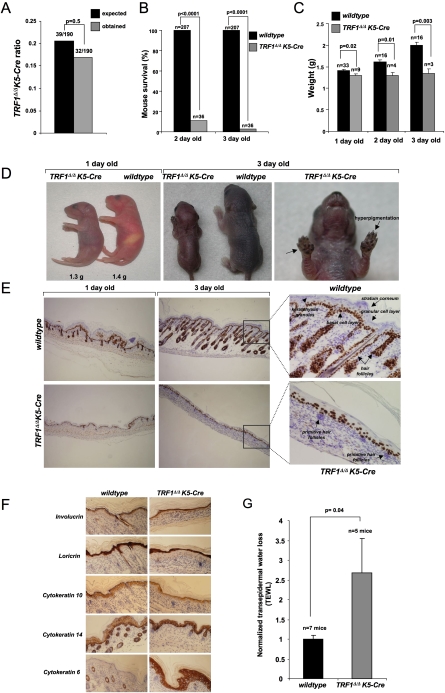
TRF1Δ/ΔK5-Cre mice were born at the expected Mendelian ratios, indicating an absence of embryonic lethality associated with TRF1 abrogation in stratified epithelia from E11.5 onward (Fig. 5A; Ramirez et al. 1994); however, they showed a severe perinatal mortality, with only 8% of the TRF1Δ/ΔK5-Cre mice reaching day 3 post-partum (P3) (Fig. 5B). Moreover, while wild-type mice progressively gained weight after birth, no increase in body weight was observed in the TRF1Δ/ΔK5-Cre cohorts (Fig. 5C,D).
Figure 5.
TRF1 deficiency results in perinatal mortality and severe epithelial abnormalities. (A) TRF1Δ/ΔK5-Cre mice were born at the expected Mendelian ratios. (B) Early perinatal lethality of TRF1Δ/ΔK5-Cre mice. The χ2 test was used to determine statistical significance. P-values are indicated. n = number of mice. (C) TRF1Δ/ΔK5-Cre mice are born with low body weight and do not gain weight after birth. The Student's t-test was used for statistical calculations, and standard error and P-values are shown.(D, left panel) Note the lack of milk in the stomach of a 1-d-old TRF1Δ/ΔK5-Cre mouse compared with an age-matched wild type. (Middle and right panels) Note that the skin of the 1-d-old TRF1Δ/ΔK5-Cre mouse shows a shiny appearance and it is hyperpigmented, a phenotype that is aggravated at day 3. The mean weight of each genotype is indicated. Arrows indicate severe hyperpigmentation of the paws. (E,F) Representative images of back skin sections stained for p63 at P1 and P3 (E), as well as stained for involucrin, loricrin, cytokeratin 10, cytokeratin 14, and cytokeratin 6 (F). (G) Transepidermal water loss (TEWL) normalized to wild-type levels (set to 1). n = mice analyzed per genotype. Error bars indicate standard error. Student's t-test was used for statistical analysis and P-values are indicated.
Histopathological analysis of newborn TRF1Δ/ΔK5-Cre mice revealed severe skin hyperpigmentation (Fig. 5D) and reduced skin development, including a complete absence of mature skin hair follicles and sebaceous glands in TRF1Δ/ΔK5-Cre mice (cf. 1-d-old and 3-d-old in Fig. 5E). In addition, all TRF1Δ/ΔK5-Cre mice examined presented areas of dysplastic skin (Supplemental Table 1; Supplemental Fig. 7). These observations indicate that TRF1 abrogation in the epidermis results in severe skin morphogenesis defects, which are acompanied by focal development of preneoplastic lesions (hyperplastic and dysplastic lesions). Interestingly, while we observed a relatively normal expression of various differentiation markers in TRF1Δ/ΔK5-Cre epidermis, including involucrin, loricrin, and cytokeratins K10 and K14 (Fig. 5F), TRF1Δ/ΔK5-Cre epidermis showed an abnormally high and ubiquitous expression of cytokeratin K6, which is restricted to the hair follicles in wild-type epidermis (Fig. 5F). Of interest, up-regulated K6 expression is associated with skin diseases such as psoriasis and skin cancer (Stoler et al. 1989), suggesting that increased K6 expression in TRF1Δ/ΔK5-Cre epidermis could be related to the appearance of preneoplastic lesions in all mice studied. Finally, these severe skin defects in TRF1Δ/ΔK5-Cre mice were accompanied by a defective epithelial barrier function (Fig. 5G).
Other stratified epithelia in TRF1Δ/ΔK5-Cre mice (tongue, palate, esophagus, and nongrandular stomach), also showed severe hyperkeratosis and dysplasia, which in the case of the oral epithelium could result in lack of nourishment (Supplemental Table 1; Supplemental Fig. 7), and together with the defective epithelial barrier function of these mice may contribute to their fully penetrant perinatal mortality (McGavin and Zachary 2007).
Finally, persistent DDR activation triggered by TRF1 deletion in the epidermis (Fig. 3) could explain the severe skin hyperpigmentation present in these mice, a phenotype also described in mice with severe telomere dysfunction due to critically short telomeres (Munoz et al. 2005; Blanco et al. 2007; Hockemeyer et al. 2008; He et al. 2009).
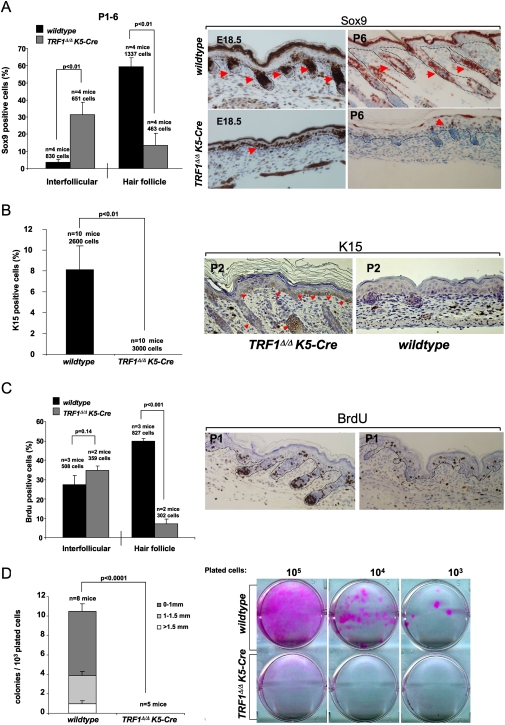
Severe epidermal stem cell defects in TRF1Δ/ΔK5-Cre mice
The lack of mature hair follicles and of sebaceous glands in TRF1Δ/ΔK5-Cre skin suggests a defect in the proliferative capacity of epidermal stem cells. Epidermal stem cells are particularly sensitive to telomere dysfunction, as shown in the context of telomerase-deficient mice, by restricting their capacity to regenerate the hair and the skin (Flores et al. 2005). Here, we examined the impact of TRF1 abrogation on epidermal stem cell functionality by studying the expression of Sox9, a stem cell gene that establishes epidermal stem cell compartments in mice during embryonic and early postnatal development (Vidal et al. 2005). Sox9 expression begins early in skin morphogenesis and is essential to establish the stem cell niche, so that in the absence of Sox9 the early hair bulge stem cell population never forms and all epithelial lineages (hair bulge and sebaceous glands) are affected (Vidal et al. 2005; Nowak et al. 2008). In agreement with the normal Sox9 pattern of expression, wild-type mice showed intense Sox9 expression in the early hair primordia at E18.5 and in the hair bulge area at P6 (Fig. 6A, arrows). In contrast, TRF1-deleted epidermis showed Sox9 expression restricted to the basal layer and absent from the hair primordia from E18.5 to P1–P6 (Fig. 6A, arrows). The abnormal Sox9 expression is in accordance with the absence of mature hair follicles and sebaceous glands in TRF1Δ/ΔK5-Cre skin. We confirmed the absence of epidermal stem cell compartments in TRF1-deficient epidermis by undetectable expresion of K15, a bona fide marker of the hair bulge stem cell niche (Fig. 6B, arrows; Liu et al. 2003). Together, these results suggest that TRF1 deletion has a profoud negative impact on the establishment of the mature hair bulge stem cell compartment. Interestingly, we observed reduced proliferation specifically at the hair primordia of P1 TRF1Δ/ΔK5-Cre epidermis compared with age-matched wild-type epidermis, while this defect was not so apparent in the interfollicular epidermis (Fig. 6C; Supplemental Fig. 4B,C), indicating that epidermal stem cell compartments are particularly sensitive to persistent damage induced by TRF1 deletion. We did not detect increased apoptosis in the hair primordia of TRF1Δ/ΔK5-Cre epidermis (Supplemental Fig. 4A).
Figure 6.
TRF1 deficiency leads to severe epidermal stem cell defects. (A) Percentage of Sox9-positive cells at interfollicular epidermis and at hair follicles of back skin sections from mice of the indicated genotypes. (Right) Representative images of Sox9 staining in E18.5 embryos and P6 mice. Arrows indicate Sox9 expression. (B) Percentage of cytokeratine 15 (K15)-positive cells in back skin sections from mice of the indicated genotypes. (Right) Representative images of K15-positive staining in the skin of P2 mice. Arrows indicate K15 expression. (C) Percentage of BrdU-positive cells in the interfollicular epidermis and in the hair follicles of back skin sections from mice of the indicated genotypes. (Right) Representative images of BrdU-positive staining in the skin of P1 mice. (D) Quantification of size and number of clones obtained. (Right) Representative examples of clones obtained from newborn mice (P1–P2) at the indicated dilutions. Clones are visualized by rhodamine stainning. n = mice analyzed per genotype. The total number of cells scored per genotype is indicated. Error bars indicate standard error. Students's t-test was used for statistical analysis and P-values are shown.
Next, we addressed whether the impact of TRF1 deletion on epidermal stem cell proliferation was a cell-autonomous effect by assessing the proliferative potential of epidermal stem cells ex vivo using clonogenic assays (Materials and Methods). The number and size of the colonies in this assay are considered to reflect, respectively, the number of epidermal stem cells and their ability to proliferate in vitro (Barrandon and Green 1987). TRF1Δ/ΔK5-Cre keratinocytes did not form any detectable colonies (Fig. 6D) and showed a reduced plating efficiency (data not shown). These ex vivo defects, together with abnormal Sox9 and K15 expression in the epidermis, may explain the severe atrophies present in stratified epithelia of these mice.
p53 deficiency rescues degenerative pathologies but leads to increased cancer in TRF1Δ/ΔK5-Cre p53−/− mice
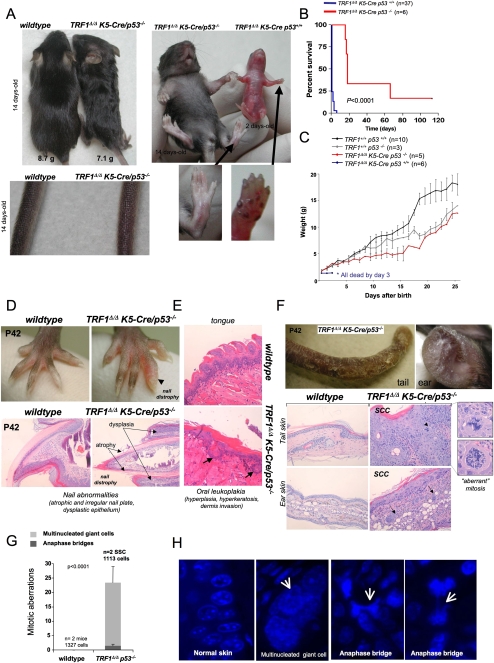
Analysis of TRF1-deleted cells and mice suggests that p53 is an important mediator of cell cycle arrest produced by TRF1 deficiency (Figs. 1B–D, 4A,D). We addressed this by using mouse genetics and generating TRF1Δ/ΔK5-Cre p53−/− mice. Notably, TRF1Δ/ΔK5-Cre p53−/− mice showed normal hair growth and a lack of skin hyperpigmentation (Fig. 7A), as well as a significant rescue of mouse survival and body weight (Fig. 7B,C), suggesting that p53 mediates the severe developmental defects associated with TRF1 deficiency.
Figure 7.
p53 deficiency rescues proliferative defects and hyperpigmentation of TRF1Δ/Δ K5-Cre mice but results in increased skin cancer. (A, left) TRF1Δ/ΔK5-Cre p53−/− mice grow hair. (Right) TRF1Δ/ΔK5-Cre p53−/− mice do not show skin hyperpigmentation. (B) Kaplan-Meyer survival curves of the different mouse cohorts. n = mice of each genotype included in the analysis. P-value for statistical comparisons between genotypes was calculated using log rank test. (C) Weight gain of the different mouse cohorts. n = mice of each genotype used for the analysis. (D) TRF1Δ/ΔK5-Cre p53−/− mice develop nail atrophy and dysplastic epithelium at the proximal and ventral nail fold. (E) TRF1Δ/ΔK5-Cre p53−/− mice develop oral leukoplakia characterized by severe epithelia hyperplasia, hyperkeratosis, and dermis invasion by basal keratinocytes (arrows). (F) SCCs in tail and ear skin of TRF1Δ/ΔK5-Cre p53−/− mice. (Top panel) Macroscopic appearance of tail and ear skin in a TRF1Δ/ΔK5-Cre p53−/− P42 mouse. (Bottom panel) Histopathological analysis of the SCC shows high mitotic index, pleomorphic cells with bizarre nuclei, and aberrant mitosis (arrowhead). Nest of epithelial tumoral cells invade the ear dermis (arrows). (G) Multinucleated giant cells and anaphase bridges are observed in SCC. n = mice or SCC analyzed. The total number of scored cells is indicated. The P-value is calculated by the Student's t-test. (H) Representative images of multinucleated giant cells and anaphase bridges.
Strikingly, longer-lived TRF1Δ/ΔK5-Cre p53−/− mice developed a number of epithelial pathologies, which are considered hallmarks of human telomere-related pathologies, such as occurrence of nail dystrophy (Fig. 7D) and oral leukoplakia (Fig. 7E). In addition, longer-lived TRF1Δ/ΔK5-Cre p53−/− mice spontaneously developed squamous cell carcinomas (SCCs) in the tail and ear skin of adult (P42) TRF1Δ/ΔK5-Cre p53−/− mice (Fig. 7F). Furthermore, the SCCs invaded the dermis (Fig. 7F) and were genetically unstable as indicated by the presence of abundant multinucleated giant cells and anaphases bridges (Fig. 7F–H). As single p53 mutant mice never develop SCCs at these early ages, these results indicate that TRF1 suppresses skin tumorigenesis by preventing telomere-induced chromosomal instability.
Discussion
TRF1 has been attributed roles in telomere length regulation, telomere cohesion, and telomere silencing, among others (van Steensel and de Lange 1997; Koering et al. 2002; Dynek and Smith 2004; Schoeftner and Blasco 2008; Munoz et al. 2009). Most of these studies have been based on TRF1 overexpression experiments in both cells and mice. However, the impact of TRF1 abrogation in the context of the adult organism has not been studied so far. Here, we generated both TRF1-null cells and mice, thus allowing us to connect molecular and cellular defects associated with TRF1 deficiency with cancer and degenerative pathologies in mice.
The data presented here indicate that TRF1 acts as a suppressor of the DDR at mammalian telomeres and plays an essential role in telomere integrity by preventing generation of telomere fusions (particularly, sister telomere fusions) and of chromosome termini with multiple telomeric signals or MTS, a type of telomere aberration that results from increased telomere fragility (Sfeir et al. 2009; this study). In particular, conditional deletion of TRF1 in MEFs leads to massive TIF induction, which we visualize at mitotic telomeres. This is concomitant with a robust activation of ATM/ATR and the downstream CHK1 and CHK2 checkpoint kinases, and formation of the above-described telomere aberrations. The level of binding of other shelterin components to the telomere is not changed in our TRF1-deleted MEFs compared with controls, suggesting that the molecular and chromosomal phenotypes described here are inherent to TRF1 deletion. Of note, the fact that the majority of telomere fusions in TRF1-deleted MEFs involved sister chromatid telomeres may suggest an enhanced telomere cohesion in the absence of TRF1, analogous to that described previously for tankyrase 1 depletion in human cells (Smith et al. 1998; Dynek and Smith 2004; Canudas et al. 2007; Muramatsu et al. 2007). Interestingly, increased telomere breakage and activation of the DDR as a consequence of TRF1 deficiency is in agreement with recently published data suggesting that replication stalling at terminal forks leads to DDR activation and generation of abnormal terminal structures (Doksani et al. 2009). Supporting the notion that MTS result from increased telomere fragility (Sfeir et al. 2009; this study), we show here that induction of fragile sites upon aphidicolin treatment or in ATR-deficient cells also leads to MTS. Together, these findings indicate that TRF1 deletion induces at least two types of telomere defects at the molecular level: (1) telomere uncapping leading to end-to-end fusions, as well as (2) increased telomere fragility and breakage resulting in MTS. Finally, MEFs conditionally deleted for TRF1 show a rapid induction of cellular senescence, which is largely mediated by the p53 and pRB pathways.
Importantly, we extend the molecular and cellular defects resulting from TRF1 deficiency in the context of in vitro cultured MEFs to the adult organism by generating mice deficient for TRF1 in stratified epithelia, TRF1Δ/ΔK5-Cre mice. The epidermis of these mice shows a dramatic induction of DNA damage and cell arrest responses as indicated by the induction of DNA damage foci at most telomeres, as well as induction of the p53, p21, and p16 cell cycle inhibitors and G2/M cell cycle arrest. Furthermore, by using mouse genetics, we demonstrate rescue of mouse survival and skin pathologies by p53 abrogation, confirming that TRF1-induced cell cycle arrest is partially mediated by p53.
Extensive TRF1 overexpression data have established a model in which aberrantly overexpressed TRF1 acts as a negative regulator of telomere length (de Lange 2005; Munoz et al. 2009). Here we show, however, that TRF1 abrogation does not result in telomere length changes in both MEFs and the context of mouse tissues. Our results are in agreement with those obtained previously with TRF1-deleted ES cells (Karlseder et al. 2003), which also reported normal-length telomeres in the absence of TRF1. On the other hand, TRF1 overexpression in mice leads to telomere shortening mediated by the XPF nuclease, suggesting a model in which TRF1 titrates XPF levels at telomeres (Munoz et al. 2009). Although TRF1 deletion could conceivably lead to decreased XPF activity at telomeres, this is unlikely to have any effect on telomere length, as we and others have shown previously that XPF-deficient mice have normal-length telomeres (Zhu et al. 2003; Munoz et al. 2005, 2009).
Finally, we show here that TRF1 deletion has a profound impact on cancer and degenerative pathologies in mice. In particular, while TRF1 deletion in stratified epithelia led to epithelial atrophies and hyperpigmentation of the skin, longer-lived TRF1/p53 double-null mice developed additional epithelial abnormalities such as oral leukoplakia and nail dystrophy, which are characteristic of human diseases produced by mutations in telomerase-related genes and the presence of short telomeres, such as dyskeratosis congenita, aplastic anemia, and idiopathic pulmonary fibrosis (Mitchell et al. 1999; Vulliamy et al. 2001; Armanios et al. 2007; Tsakiri et al. 2007). Concomitant with severe atrophies, all stratified epithelia in TRF1-deficient mice showed development of preneoplastic lesions (dysplasia and hyperkeratosis) as early as 1–6 d after birth. Longer-lived TRF1/p53 double-null mice further developed spontaneous SCCs, which were invasive and genetically unstable as indicated by the occurrence of numerous mitotic abnormalities. These results suggest that TRF1 normally acts as a tumor supressor in the context of the organism by preventing telomere-induced genetic instability.
In conclusion, the TRF1 conditional knockout mice described here represent the first mouse model in which full abrogation of a telomere-binding protein per se, without having to simultaneously abrogate telomerase expression, leads to induction of severe telomere damage; in vivo up-regulation of the p53, p21, and p16 cell cycle inhibitors; and subsequent induction of cellular senescence and premature tissue degeneration, in the absence of telomere shortening (Blasco et al. 1997; Hockemeyer et al. 2006, 2008; Wu et al. 2006; He et al. 2009). Degenerative phenotypes in TRF1-deficient mice are accompanied by a higher incidence of neoplastic lesions, most likely as the consequence of increased chromosomal instability, thus linking aging and cancer at the organismal level through altering a telomere-binding protein in the absence of telomere shortening. The conditional TRF1 mouse model described here is likely to become a valuable tool in the study of the molecular mechanisms underlying human telomere diseases.
Materials and methods
Construction of targeting vectors
A 0.992-kb genomic fragment containing part of the upstream regulatory region, Exon I (E1), and part of intron 1(I1) was amplified with primers F-ClaClKO (5′-CCATCGATTCCAGGGTCAGCACCATC-3′) and R2-ClaClKO (5′-GGATCGATACAGGAAGCGGCCGGTTTC-3′), which introduce a ClaI restriction site. The ClaI-restricted E1-containing fragment was cloned into the unique ClaI site between two LoxP repeats in the pDELBOY vector (Rossi et al. 2001), generating the pDELBOY-E1 vector. A 4.471-kb fragment containing the upstream TRF1 region (5′-untranslated region [UTR]) was amplified using primers SalIFClKl (5′-ACGCGTCGACTGAGGAGGATTTAGGTCAGC-3′) and SalIRClKO (5′-GCGTGTCGACAAGTCTAGTCAGTTCCCCATGA-3′), which introduce a SalI restriction site. The SalI-restricted 5′-UTR genomic region was cloned into the unique SalI site of pDELBOY-E1, generating the long arm (LA)-containing pDELBOY-E1-LA vector. The 1.865-kb short arm (SA) of the targeting vector that contains part of intron 1 (I1), Exon 2 (E2), and part of intron 2 (I2) was amplified with primers XScaFClKO (5′-CCGCTCGAGTACTGCTGGAAGTCCTGGCTCC-3′) and XhoRCLKO (5′-CGGCTCGAGAGTTTGAGAGTCACTGTTCTAG-3′), which introduce a XhoI restriction site. The XhoI SA fragment was cloned into the unique XhoI site of pDELBOY-E1-LA, generating the complete targeting vector pDELBOY-E1-LA-SA (Figs. 1A, 3A). This vector harbors the Neo marker flanked by two FRT repeats in between the E1 and SA and a TK marker downstream from the SA arm that serve as a positive and negative selection, respectively, upon microinjection into ES cells (Figs. 1A, 3A).
Cell fractionation and immunoblotting
Cell fractionation was performed as described before (Mendez and Stillman 2000). For whole-cell extracts, cells were harvested by trypsinization, washed once with cold HBSS, resuspended in SDS-PAGE loading buffer, and sonicated. Equal amounts of protein (50–100 μg) were analyzed by gel electrophoresis followed by Western blotting. NuPAGE-Novex 10% Bis-Tris gels (Invitrogen) were run according to the manufacturer's instructions. Anti-tubulin, anti-histone H3, or anti-histone H2AX antibodies were used as loading controls. For details on the antibodies used, see the Supplemental Material.
IF-FISH
Mitotic cells were collected by mitotic shake-off, swollen in hypotonic buffer (10 mM Tris-HCl at pH 7.5, 10 mM NaCl, 5 mM MgCl2) for 10 min at 37°C, and spun onto coverslips using a cytospin apparatus (Cytospin 4, Fisher). Metaphase spreads were then fixed and subjected to immunofluorescence staining as described (Tarsounas et al. 2003). After washing, samples were fixed again in 4% formaldehyde in PBS, and FISH was performed as described previously (Samper et al. 2000). For details on antibodies used, see the Supplemental Material.
Generation of conditional TRF1 knockout mice
R1 ES cells were electroporated with 30 μg of NotI-linearized targeting vector. ES clones showing homologous recombination at the TRF1 locus were identified by PCR. The results of PCR screening were confirmed by Southern blot analysis of BamHI- and ScaI-restricted genomic DNA using 5′ and 3′ external probes, respectively (data not shown). Chimeric mice were generated by microinjection of two independently targeted ES clones into C57BL/6J host blastocysts, which were then implanted into pseudopregnant CD1 foster females. The resulting offspring showed a high level of chimerism as shown by coat color, and were mated to C57BL/6J mice to assess germline transmission. The resulting heterozygous TRF1+/flox-Neo mice were then bred to transgenic mice expressing the Flp recombinase (Rodriguez et al. 2000) to induce excision of the Neo marker. The TRF1+/flox heterozygous mice were then intercrossed to generate TRF1flox/flox and TRF1+/flox mice. Homozygous TRF1flox/flox mice were crossed with transgenic mice expressing the Cre recombinase under the control of the keratine 5 promoter (Fig. 3B; Tarutani et al. 1997). Heterozygous TRF1+/Δ K5-Cre were crossed either to TRF1flox/flox or TRF1+/flox to generate TRF1Δ/ΔK5-Cre (Fig. 3B). The removal of exon I by Cre-mediated recombination was confirmed by PCR analysis using the following primers: F: E1-popout (5′-ATAGTGATCAAAATGTGGTCCTGGG-3′), and R: SA1 (5′-GCTTGCCAAATTGGGTTGG-3′) (Figs. 1A, 3A,B). Amplification of the wild-type, flox, and knockout alleles rendered 1.4-kb, 1.5-kb, and 0.48-kb fragments, respectively.
To generate TRF1Δ/Δp53−/−K5-Cre mice, TRF1+/+p53−/− mice were first crossed to TRF1flox/floxp53−/− mice. The resultant double heterozygote TRF1+/floxp53+/− mice were then crossed to TRF1+/ΔK5-Cre mice.
All mice were generated and maintained at the Spanish National Cancer Center under specific pathogen-free conditions in accordance with the recommendation of the Federation of European Laboratory Animal Science Associations.
Retroviral infections
Retroviral supernatants were produced in 293T cells (5 × 105 cells per 100-mm diameter dish) transfected with the ecotropic packaging plasmid pCL-Eco and either pBabe-Cre, pRS-p53sh (Brummelkamp et al. 2002), or pZip-Neo-sv40-LT (Brown et al. 1986) as described. Two days later, retroviral supernatant (10 mL) was collected. MEFs were seeded the previous day (8 × 105 cells per 100-mm diameter dish) and received 2 mL of each corresponding retroviral supernatant. This procedure was repeated three times at 12-h intervals. Twelve hours after infection, puromycin was added to the cultures (2 μg/mL). Deletion of TRF1 was confirmed by PCR with primers E1-popout-F and SAR1 (Figs. 1A, 3A) and by immunofluorescence on cytospin chromosomes (Fig. 1F). p53 expression was analyzed by Western blotting (Fig. 1B).
Transepidermal water loss (TEWL)
TEWL was measured with a Tewameter TM300 (CK Electronic).
FACS
Keratinocytes and MEFs (1 × 106) were washed twice with PBS and fixed/permeabilized with ice-cold 70% ethanol. Fixed cells were washed with PBS and resuspended in 1 mL of PBS containing 0.2 μg of propidium iodide and 100 μg of RNase. The samples were incubated for 30 min at 37°C and the FACS analysis was performed in a FACSCANTO II flow cytometry system (BD Biosciences). The data were analyzed with FacsDiva version 5.1.1.
Telomere and centromere fluorescence analyses on skin sections
Q-FISH directly on skin sections was performed as described previously (Munoz et al. 2005). For the centromeric Q-FISH, a probe against the major satellite was used (Flores et al. 2008).
Telomere length and cytogenetic analysis using telomere Q-FISH on metaphases
Primary MEFs were incubated with 0.1 μg/mL colcemide (Gibco) for 4 h at 37°C and then fixed in methanol:acetic acid (3:1). FISH was performed as described (Samper et al. 2000). When indicated, primary MEFs were treated with aphidicolin at 0.5 μM for 24 h. For analysis of chromosomal aberrations, metaphases were analyzed by superimposing the telomere image on the DAPI image using TFL-telo.
Isolation of keratinocytes from newborn mouse skin and epidermal stem cell clonogenic assays
Primary keratinocyte from newborn mice (0- to 2-d-old) were obtained as described previously (Munoz et al. 2005) and clonogenic assays were performed as described previously (Flores et al. 2005).
Western blots
Nuclear extracts were prepared from the indicated cells. Protein concentration was determined using the Bio-Rad DC Protein Assay (Bio-Rad). Twenty micrograms of each extract were separated in 4%–20% gradient SDS–polyacrylamide gels by electrophoresis. After transfer, the membranes were incubated with the indicated antibodies (Supplemental Material). Antibody binding was detected after incubation with a secondary antibody coupled to horseradish peroxidase using enhanced chemiluminescence.
Acknowledgments
We thank R. Serrano for mouse care and the Comparative Pathology Unit at CNIO for technical assistance. P.M. is a “Ramon y Cajal” senior scientist. M.A.B.'s laboratory is funded by the Spanish Ministry of Innovation and Science, the Consolider-Ingenio 2010 Programme, the European Union (FP7-Genica,) the European Reseach Council (ERC Advance Grants), the Spanish Association Against Cancer (AECC), and the Körber European Science Award to M.A.B. M.T.'s laboratory is funded by Cancer Research UK. Travel related to this collaboration was funded by a Joint International Project from The Royal Society. M.A.B. conceived the original idea. M.A.B. and P.M. designed experiments and M.A.B. and P.M. wrote the manuscript. P.M. performed most of the experiments. P.M.U. constructed the knockout allele and did the stem cell screen. M.T., C.L., and M.T.A. designed and performed the experiments shown in Figure 1, E–I, and Figure 2I. C.M. and O.F.C. contributed to the results shown in Figure 2F. A.T. measured telomerase activity and G-strand signals. J.M.F. performed the pathology analysis.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.543509.
Supplemental material is available at http://www.genesdev.org.
References
- Ancelin K, Brunori M, Bauwens S, Koering CE, Brun C, Ricoul M, Pommier JP, Sabatier L, Gilson E. Targeting assay to study the cis functions of human telomeric proteins: Evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol Cell Biol. 2002;22:3474–3487. doi: 10.1128/MCB.22.10.3474-3487.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios MY, Chen JJ, Cogan JD, Alder JK, Ingersoll RG, Markin C, Lawson WE, Xie M, Vulto I, Phillips JA, 3rd, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N Engl J Med. 2007;356:1317–1326. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- Barrandon Y, Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci. 1987;84:2302–2306. doi: 10.1073/pnas.84.8.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud T, Brun C, Ancelin K, Koering CE, Laroche T, Gilson E. Telomeric localization of TRF2, a novel human telobox protein. Nat Genet. 1997;117:236–239. doi: 10.1038/ng1097-236. [DOI] [PubMed] [Google Scholar]
- Blanco R, Munoz P, Flores JM, Klatt P, Blasco MA. Telomerase abrogation dramatically accelerates TRF2-induced epithelial carcinogenesis. Genes & Dev. 2007;21:206–220. doi: 10.1101/gad.406207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Broccoli D, Smogorzewska A, Chong L, de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- Brown M, McCormack M, Zinn KG, Farrell MP, Bikel I, Livingston DM. A recombinant murine retrovirus for simian virus 40 large T cDNA transforms mouse fibroblasts to anchorage-independent growth. J Virol. 1986;60:290–293. doi: 10.1128/jvi.60.1.290-293.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Canudas S, Houghtaling BR, Kim JY, Dynek JN, Chang WG, Smith S. Protein requirements for sister telomere association in human cells. EMBO J. 2007;26:4867–4878. doi: 10.1038/sj.emboj.7601903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- Choudhury AR, Ju Z, Djojosubroto MW, Schienke A, Lechel A, Schaetzlein S, Jiang H, Stepczynska A, Wang C, Buer J, et al. Cdkn1a deletion improves stem cell function and lifespan of mice with dysfunctional telomeres without accelerating cancer formation. Nat Genet. 2007;39:99–105. doi: 10.1038/ng1937. [DOI] [PubMed] [Google Scholar]
- Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguria A, Zaballos A, Flores JM, Barbacid M, et al. Tumour biology: Senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- Cook BD, Dynek JN, Chang W, Shostak G, Smith S. Role for the related poly(ADP-Ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol Cell Biol. 2002;22:332–342. doi: 10.1128/MCB.22.1.332-342.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- de Lange T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes & Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Doksani Y, Bermejo R, Fiorani S, Haber JE, Foiani M. Replicon dynamics, dormant origin firing, and terminal fork integrity after double-strand break formation. Cell. 2009;137:247–258. doi: 10.1016/j.cell.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–172. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- Dynek JN, Smith S. Resolution of sister telomere association is required for progression through mitosis. Science. 2004;304:97–100. doi: 10.1126/science.1094754. [DOI] [PubMed] [Google Scholar]
- Feldser DM, Greider CW. Short telomeres limit tumor progression in vivo by inducing senescence. Cancer Cell. 2007;11:461–469. doi: 10.1016/j.ccr.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores I, Cayuela ML, Blasco MA. Effects of telomerase and telomere length on epidermal stem cell behavior. Science. 2005;309:1253–1256. doi: 10.1126/science.1115025. [DOI] [PubMed] [Google Scholar]
- Flores I, Canela A, Vera E, Tejera A, Cotsarelis G, Blasco MA. The longest telomeres: A general signature of adult stem cell compartments. Genes & Dev. 2008;22:654–667. doi: 10.1101/gad.451008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Wang Y, Guo X, Ramchandani S, Ma J, Shen MF, Garcia DA, Deng Y, Multani AS, You MJ, et al. Pot1b deletion and telomerase haploinsufficiency in mice initiate an ATR-dependent DNA damage response and elicit phenotypes resembling dyskeratosis congenita. Mol Cell Biol. 2009;29:229–240. doi: 10.1128/MCB.01400-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Palm W, Wang RC, Couto SS, de Lange T. Engineered telomere degradation models dyskeratosis congenita. Genes & Dev. 2008;22:1773–1785. doi: 10.1101/gad.1679208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlseder J, Kachatrian L, Takai H, Mercer K, Hingorani S, Jacks T, de Lange T. Targeted deletion reveals an essential function for the telomere length regulator Trf1. Mol Cell Biol. 2003;23:6533–6541. doi: 10.1128/MCB.23.18.6533-6541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koering CE, Pollice A, Zibella MP, Bauwens S, Puisieux A, Brunori M, Brun C, Martins L, Sabatier L, Pulitzer JF, et al. Human telomeric position effect is determined by chromosomal context and telomeric chromatin integrity. EMBO Rep. 2002;3:1055–1061. doi: 10.1093/embo-reports/kvf215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzerini Denchi E, Celli G, de Lange T. Hepatocytes with extensive telomere deprotection and fusion remain viable and regenerate liver mass through endoreduplication. Genes & Dev. 2006;20:2648–2653. doi: 10.1101/gad.1453606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Lyle S, Yang Z, Cotsarelis G. Keratin 15 promoter targets putative epithelial stem cells in the hair follicle bulge. J Invest Dermatol. 2003;121:963–968. doi: 10.1046/j.1523-1747.2003.12600.x. [DOI] [PubMed] [Google Scholar]
- Liu D, O'Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem. 2004;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- Loayza D, De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- McGavin M, Zachary JF. Pathologic basis of veterinary disease. Mosby Elsevier; St Louis, MO: 2007. [Google Scholar]
- Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: Assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- Munoz P, Blanco R, Flores JM, Blasco MA. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat Genet. 2005;37:1063–1071. doi: 10.1038/ng1633. [DOI] [PubMed] [Google Scholar]
- Munoz P, Blanco R, de Carcer G, Schoeftner S, Benetti R, Flores JM, Malumbres M, Blasco MA. TRF1 controls telomere length and mitotic fidelity in epithelial homeostasis. Mol Cell Biol. 2009;29:1608–1625. doi: 10.1128/MCB.01339-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu Y, Ohishi T, Sakamoto M, Tsuruo T, Seimiya H. Cross-species difference in telomeric function of tankyrase 1. Cancer Sci. 2007;98:850–857. doi: 10.1111/j.1349-7006.2007.00462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga M, Bunting S, Montaña M, Soria R, Mulero F, Cañamero M, Lee Y, McKinnon PJ, Nussenzweig A, Fernandez-Capetillo O. A mouse model of the ATR-Seckel syndrome reveals that replicative stress during embryogenesis limits mammalian lifespan. Nat Genet. 2009 doi: 10.1038/ng.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak JA, Polak L, Pasolli HA, Fuchs E. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell. 2008;3:33–43. doi: 10.1016/j.stem.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Iwano T, Tachibana M, Shinkai Y. Distinct roles of TRF1 in the regulation of telomere structure and lengthening. J Biol Chem. 2008;283:23981–23988. doi: 10.1074/jbc.M802395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez A, Bravo A, Jorcano JL, Vidal M. Sequences 5′ of the bovine keratin 5 gene direct tissue- and cell-type-specific expression of a lacZ gene in the adult and during development. Differentiation. 1994;58:53–64. doi: 10.1046/j.1432-0436.1994.5810053.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Rossi DJ, Londesborough A, Korsisaari N, Pihlak A, Lehtonen E, Henkemeyer M, Makela TP. Inability to enter S phase and defective RNA polymerase II CTD phosphorylation in mice lacking Mat1. EMBO J. 2001;20:2844–2856. doi: 10.1093/emboj/20.11.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samper E, Goytisolo FA, Slijepcevic P, van Buul PP, Blasco MA. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 2000;1:244–252. doi: 10.1093/embo-reports/kvd051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T. Control of human telomere length by TRF1 and TRF2. Mol Cell Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler A, Duvic M, Fuchs E. Unusual patterns of keratin expression in the overlying epidermis of patients with dermatofibromas: Biochemical alterations in the epidermis as a consequence of dermal tumors. J Invest Dermatol. 1989;93:728–738. doi: 10.1111/1523-1747.ep12284397. [DOI] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- Tarsounas M, Davies D, West SC. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene. 2003;22:1115–1123. doi: 10.1038/sj.onc.1206263. [DOI] [PubMed] [Google Scholar]
- Tarutani M, Itami S, Okabe M, Ikawa M, Tezuka T, Yoshikawa K, Kinoshita T, Takeda J. Tissue-specific knockout of the mouse Pig-a gene reveals important roles for GPI-anchored proteins in skin development. Proc Natl Acad Sci. 1997;94:7400–7405. doi: 10.1073/pnas.94.14.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakiri KD, Cronkhite JT, Kuan PJ, Xing C, Raghu G, Weissler JC, Rosenblatt RL, Shay JW, Garcia CK. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc Natl Acad Sci. 2007;104:7552–7557. doi: 10.1073/pnas.0701009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Vidal VP, Chaboissier MC, Lutzkendorf S, Cotsarelis G, Mill P, Hui CC, Ortonne N, Ortonne JP, Schedl A. Sox9 is essential for outer root sheath differentiation and the formation of the hair stem cell compartment. Curr Biol. 2005;15:1340–1351. doi: 10.1016/j.cub.2005.06.064. [DOI] [PubMed] [Google Scholar]
- Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason PJ, Dokal I. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol Cell. 2003;12:1489–1498. doi: 10.1016/s1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]