Abstract
A major question in hematopoiesis is how the system maintains long-term homeostasis whereby the generation of large numbers of differentiated cells is balanced with the requirement for maintenance of progenitor pools, while remaining sufficiently flexible to respond to periods of perturbed cellular output during infection or stress. We focused on the development of the myeloid lineage and present evidence that promyelocytic leukemia zinc finger (PLZF) provides a novel function that is critical for both normal and stress-induced myelopoiesis. During homeostasis, PLZF restricts proliferation and differentiation of human cord blood-derived myeloid progenitors to maintain a balance between the progenitor and mature cell compartments. Analysis of PLZF promoter-binding sites revealed that it represses transcription factors involved in normal myeloid differentiation, including GFI-1, C/EBPα, and LEF-1, and induces negative regulators DUSP6 and ID2. Loss of ID2 relieves PLZF-mediated repression of differentiation identifying it as a functional target of PLZF in myelopoiesis. Furthermore, induction of ERK1/2 by myeloid cytokines, reflective of a stress response, leads to nuclear export and inactivation of PLZF, which augments mature cell production. Thus, negative regulators of differentiation can serve to maintain developmental systems in a primed state, so that their inactivation by extrinsic signals can induce proliferation and differentiation to rapidly satisfy increased demand for mature cells.
Keywords: Myeloid differentiation, human hematopoiesis, lineage determination, transcription factors
Myeloid cells, including macrophages and neutrophils, are an intrinsic component of the innate immune system. Mature myeloid cells in the periphery rapidly turn over and are replenished by differentiation from myeloid progenitors in the bone marrow. This process is orchestrated by a complex interplay of transcription factors and cytokine signals that govern an ordered activation and repression of lineage-specific genes. Networks of transcription factors that regulate myeloid development have been extensively studied and modeled (Laiosa et al. 2006; Laslo et al. 2006). Macrophage and neutrophil specification require master regulators PU.1 and C/EBPα, which control more “specialized” downstream transcription factors, including GFI1 and EGR2 (Laslo et al. 2006). Since blood production is enormous (∼1012 cells per day in human) and continues for a lifetime, homeostatic mechanisms must be in place to permit ongoing differentiation without depletion of progenitor pools. Although the molecular mechanisms that govern lineage determination are well understood, how myeloid transcription factors maintain the balance between the progenitor and mature cell compartments is less clear. Proper regulation ensures an adequate supply of mature cells, while leukemogenesis and other myeloid malignancies are often driven by inappropriate activity of these transcription factors (Rosenbauer and Tenen 2007). Thus, identification and characterization of novel regulators of myelopoiesis is of interest from the standpoint of both normal and malignant hematopoiesis.
Besides maintaining homeostasis, developmental systems must remain flexible to respond to emergent events. During immune activation, high levels of secreted cytokines stimulate myeloid-mediated innate immunity (Cannistra and Griffin 1988; Oster et al. 1988). Of these, G-CSF, GM-CSF, and IL-3 were initially isolated as colony-stimulating factors owing to their potential to stimulate proliferation, survival, and differentiation of hematopoietic progenitors (Donahue et al. 1988; Lieschke et al. 1994). Following binding to their cognate receptors, cytokines activate parallel JAK/STAT, MAPK, and PI3K signal transduction pathways, which modify expression and activation of downstream transcription factor effectors (Barreda et al. 2004). For instance, STAT3 and C/EBPβ are dispensable for normal, but are required for cytokine-induced, myelopoiesis (Hirai et al. 2006; Panoupolos 2006). Still, the mechanisms by which stress-induced cytokines interact with the network of myeloid transcription factors remain largely unexplored.
Kruppel-like transcription factor promyelocytic leukemia zinc finger (PLZF) is expressed in CD34+ hematopoietic progenitors, but not mature cells, suggesting it may play a role in lineage determination (Reid et al. 1995). PLZF is a negative regulator of cell division in embryogenesis and controls segment patterning through repression of HOX and BMP expression (Barna et al. 2000). In the adult, PLZF augments self-renewal of spermatogonial stem cells and its deletion results in sterility (Buaas et al. 2004; Costoya et al. 2004). In hematopoiesis, PLZF has been implicated in the development of megakaryocytic (Labbaye et al. 2008) and NKT cell lineages (Kovalovsky et al. 2008; Savage et al. 2008). PLZF was identified as a chromosomal fusion partner with RARα in promyelocytic leukemia, a disease marked by an accumulation of undifferentiated myeloid blasts (Chen et al. 1993). Consistent with a role in myelopoiesis, enforced expression of PLZF in myeloid cell lines resulted in inhibition of proliferation and differentiation (Shaknovich et al. 1998; Ward et al. 2001; McConnell et al. 2003). In these distinct systems, PLZF is a sequence-specific transcriptional repressor that recruits nuclear corepressors to establish silenced chromatin structure at target promoters (Hong et al. 1997; Barna et al. 2002). Thus, PLZF can maintain long-term epigenetic repression of target genes, which is an integral aspect of cellular “memory” in fate determination and differentiation (Fisher 2002).
Most studies of mammalian hematopoiesis have used the mouse as a model organism. The improvement of gene transfer methods and “humanized” xenograft mouse models now provide the possibility of carrying out comparable studies with primary human cells, which may be a more relevant model for many aspects of normal human biology and disease. Using this approach, we identify a previously unrecognized mechanism of homeostatic regulation, whereby the balance between maintenance and commitment myeloid progenitors is controlled by cytokine-mediated inhibition of a differentiation-suppressing transcription factor, PLZF.
Results
PLZF is expressed in human hematopoietic stem cells (HSCs) and progenitors
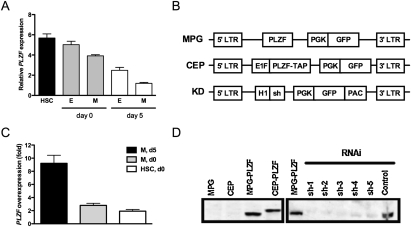
PLZF is expressed in human CD34+ hematopoietic progenitors and its expression declines in differentiated cells (Reid et al. 1995). To refine this analysis, PLZF transcript expression was assessed by quantitative real-time PCR (qPCR) in HSC-enriched (CD34+ CD38−), myeloid progenitor (CD34+ CD38+ CD71−), and erythroid progenitor (CD34+ CD38+ CD71+) fractions, isolated from lineage-depleted (Lin−) umbilical cord blood (CB). Comparable levels of PLZF expression were detected in all three fractions; however, its expression rapidly declined in progenitors isolated from culture under conditions that promote differentiation (Fig. 1A). This pattern of expression suggests that PLZF may play a role in differentiation of hematopoietic progenitors.
Figure 1.
PLZF expression and viral vector design. (A) Expression of PLZF mRNA in human hematopoietic subsets, before and after culture, was quantified by qPCR. Hematopoietic subsets were isolated by FACS-sorting from fresh Lin− CB (day 0) or after 5 d in serum-free culture (day 5) based on the following phenotypes: HSCs, CD34+ CD38−, erythroid progenitors (E), CD34+ CD38+ CD71+, myeloid progenitors (M), and CD34+ CD38+ CD71−. (B) Schematic representation of viral vectors used to overexpress (MPG, retroviral; CEP, lentiviral) or silence human PLZF (shPLZF). (C) Fold overexpression of PLZF in myeloid progenitors transduced with MPG-PLZF and cultured for 5 d, compared with same-day control cells (M, d5), or freshly sorted myeloid progenitors (M, d0) or HSCs (HSC, d0) from the same CB. (D, left panel) Western blot analysis of total protein extracts from 293T cells transduced with control or MPG-PLZF viruses. (Right panel) Silencing vectors (shPLZF1–3) were transduced into 293T cells stably expressing PLZF. Equal total protein was loaded in all lanes and the blots were probed with an anti-PLZF antibody.
PLZF restricts myeloid proliferation and differentiation in vitro
To characterize the requirement for PLZF in human hematopoiesis, we designed lentiviral and retroviral vectors to enforce or silence expression (Fig. 1B). Transduction of 293T cells with retroviral MPG-PLZF or lentiviral TAP-tagged CEP-PLZF vectors yielded protein of expected size (Fig. 1D). Transcript abundance in PLZF-transduced (termed PLZFOX) CD34+ cells was 2.8 ± 0.3-fold and 1.9 ± 0.3-fold compared with Lin− CB myeloid progenitors or HSCs, respectively (Fig. 1C). Finally, infection of PLZFOX 293T cells with short hairpin PLZF knockdown viruses (sh-PLZF) effectively abolished protein expression (Fig. 1D; see also Supplemental Fig. S1).
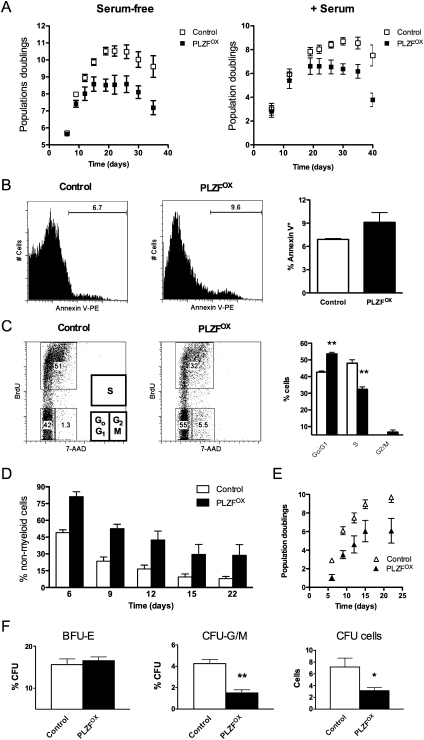
Since PLZF expression is down-regulated rapidly during differentiation, we tested the effect of enforcing its constitutive expression in cultured primary human progenitors. Lin− CB cells were infected with control or MPG-PLZF vectors and GFP+ CD34+ progenitors (which contain myeloid and erythroid progenitors, but negligible HSCs) were assayed in liquid cultures and colony assays. After 3 wk, PLZFOX progenitors displayed 4.0-fold and 5.2-fold lower proliferative capacity in serum-free and serum-supplemented cultures, respectively (Fig. 2A). Annexin V staining revealed no significant differences in the proportion of apoptotic cells in these cultures (Fig. 2B). Notably, BrdU incorporation assays demonstrated that the decrease in proliferation was directly attributable to reduced entry into S phase (Fig. 2C). These observations are consistent with the proposed tumor suppressor function of PLZF (McConnell et al. 2003).
Figure 2.
PLZF restricts myeloid proliferation and differentiation of human progenitors in vitro. (A) Effect of PLZF on progenitor proliferation. Growth of CD34+ progenitors transduced with MPG (control) or MPG-PLZF (PLZFOX) vectors in serum-free and serum-supplemented cultures quantified by total cell counting. (B) Effect of PLZF on apoptosis. Proportion of annexin V+ (7AAD−) apoptotic cells after 7 d in serum-supplemented cultures initiated with control or PLZFOX CD34+ cells; the difference between groups is not significant. (C) Effect of PLZF on cell cycle kinetics. BrdU incorporation of control and PLZFOX CD34+ cells cultured for 7 d in serum-free conditions; representative flow cytometric profiles (left panel) and the proportion of cells in G0/G1, S, and G2/M phases of the cell cycle (right panel) are shown. (D) Effect of PLZF on myeloid differentiation. Proportion of nonmyeloid (CD15− CD14− CD11b−) cells in serum-free cultures initiated with control or PLZFOX CD34+ cells. (E) Production of mature lineage-positive myeloid cells in serum-free cultures initiated with sorted control or PLZFOX CD34+ cells. (F) Effect of PLZF on clonogenic progenitors. Colony-forming capacity (colonies counted as proportion of input; percentage of CFU) of freshly sorted control or PLZFOX CD34+ cells. (BFU-E) Erythroid blast-forming units; (CFU-G/M) granulocyte or monocyte colony-forming units; (CFU cells) total myeloid cells × 105 in a CFU-G/M assay. All data are expressed as mean ± SEM of three independent experiments (CB samples), except F, which has four experiments. (*) P < 0.05; (**) P < 0.005.
To assess the effect of constitutive expression of PLZF on differentiation, we analyzed the frequency of cells that had acquired CD14, CD15, and CD11b mature myeloid lineage markers. As control Lin− cells differentiated, 92.1 ± 2.0% of them acquired these markers after 3 wk in culture. In contrast, 29 ± 9.6% of PLZFOX cells did not express the markers of differentiation at this time point (Fig. 2D). Thus, combined with the 4.0-fold reduction of proliferation, PLZF decreased total output of mature myeloid cells by 12.1-fold over 3 wk (Fig. 2E).
Myeloid development can also be functionally tested using colony assays, which measure the frequency and growth potential of clonogenic progenitors. Freshly sorted (day 0) CD34+ PLZFOX progenitors formed an equal proportion of erythroid colonies (Fig. 2F, BFU-E) as controls, but gave rise to 2.7 ± 0.2-fold fewer granulocyte or macrophage colonies (Fig. 2F, CFU-G/M). Since an equal number of transduced progenitors were seeded, these results represent the effect of PLZF on the clonal growth potential of myeloid progenitors. This conclusion is also supported by the equivalent reduction of the total cell output from PLZFOX progenitors in colony assays (Fig. 2F, CFU cells). Collectively, these findings indicate that PLZF inhibits myeloid proliferation and differentiation in vitro.
Expression of PLZF is quickly lost in culture, as primitive cells differentiate (see Fig. 1B), so further reduction of PLZF levels by knockdown is expected to have no phenotypic consequences. Indeed, three independent sh-PLZF vectors had no effect on the growth of myeloid progenitors when PLZF was not expressed (Supplemental Fig. S1, control) indicating that these hairpins do not have off-target effects that alter cell growth and differentiation. However, all three shRNA vectors partially rescue (72%–74%) colony growth in the context of ectopic PLZF expression providing a functional measure of knockdown efficiency in primary progenitors (Supplemental Fig. S1, PLZFOX). These data demonstrate that sh-PLZF lentiviruses specifically target PLZF and provide a functional rescue of its effects in myeloid progenitors.
PLZF restricts human myelopoiesis in vivo
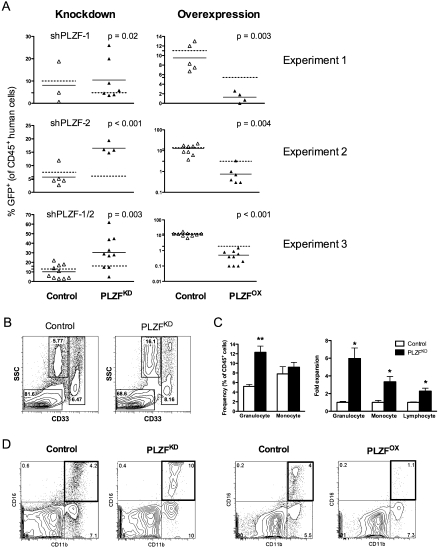
During culture, progenitors are not maintained due to rapid differentiation, making it impossible to study the process of human hematopoiesis over time. In contrast, in vivo xenotransplantion of human cells into immunodeficient mice does allow for maintenance of progenitors, so we used this system to more fully investigate the role of PLZF in human hematopoiesis. Lin− CB cells were transduced with PLZF overexpression and knockdown vectors containing a GFP reporter. The level of gene transfer into CD34+ CD133+ HSCs ranged from 5% to 10% (Fig. 3A, indicated by broken lines), and NOD/SCID mice were transplanted with a mixture of transduced and nontransduced (GFP−) cells. Silencing of PLZF (termed PLZFKD) using two independent sh-PLZF vectors enhanced the capacity of GFP-marked hematopoietic cells to contribute to the repopulation of the NOD/SCID bone marrow with human bone marrow cells (termed BMCs). There was a 2.5 ± 0.4-fold increased proportion of GFP+ PLZFKD BMCs in the human CD45+ graft at 8 wk post-transplant compared with the proportion of GFP+ cells prior to transplantation (three experiments, P < 0.02 for all) (Fig. 3A, Knockdown, cf. solid and broken lines). These data suggest that PLZFKD cells have a modest competitive advantage over their nontransduced counterparts. As expected, the proportion of GFP+ cells after transplantation of empty vector-transduced or irrelevant red fluorescent protein (shRFP) hairpin-transduced cells was unchanged compared with input transduction levels, indicating that these cells had no proliferative advantage over GFP− cells (Fig. 3A). In contrast, the proportion of GFP+ PLZFOX BMCs was decreased in the CD45+ graft by 3.9 ± 0.1-fold (three experiments, P < 0.002) (Fig. 3A, Overexpression).
Figure 3.
PLZF negatively regulates myeloid development in vivo as assessed in the NOD/SCID xenotransplant system. (A) Effect of PLZF on CD45+ human cell engraftment. Proportion of GFP+ BMCs in individual mice each denoted with a symbol at 8 wk post-transplant (mean marked as solid lines) compared with mean GFP positivity of input HSCs prior to transplantation (broken lines). Lin− CB cells were transduced with control (clear triangles), PLZF knockdown (PLZFKD), or CEP-PLZF (PLZFOX) vectors (black triangles) and transplanted into NOD/SCID mice. The level of transduction in three independent experiments ranged from 5% to 12%; as a result, mice were transplanted with a mixture of transduced and nontransduced cells. Thus, equivalent means in the values of the pretransplant and post-transplant percentage of GFP in each experiment indicates a lack of competitive advantage of GFP+ cells relative to nontransduced cells, while increased or decreased percentage of GFP+ indicates competitive advantage or disadvantage, respectively, in vivo. (B) Frequency of human myeloid BMCs in mice transplanted with PLZFKD cells (right) and a representative flow analysis (left). The frequency of granulocytes was calculated as the proportion of SSChi CD33lo cells within the GFP+ CD45+ graft and monocytes as SSClo CD33hi cells within the GFP+ CD45+ graft; the remaining cells were SSClo CD33− lymphocytes. To obtain enough cells for accurate flow cytometric analysis, bone marrow was pooled from four to eight mice. Data are expressed as mean ± SEM of five independent experiments. (**) P < 0.01. (C) Expansion of the absolute number of human granulocytes, monocytes, and lymphoid cells in mice transplanted with PLZFKD cells normalized to controls. The absolute number was calculated by multiplying the frequency of each cell type by the number of GFP+ CD45+ cells in both femurs, tibiae, and pelvis. Data are expressed as mean ± SEM of five independent experiments. (*) P < 0.02. (D) Representative analysis of the proportion of CD11b+ CD16+ human neutrophils in the GFP+ fraction of mice engrafted with Lin− CB cells transduced with control, PLZFKD, or PLZFOX viruses. To obtain enough cells for accurate flow cytometric analysis, bone marrow was pooled from four to eight mice.
To determine if the increased mature cell production was specific to a particular lineage, we assessed lineage distribution within the GFP+ graft. PLZFKD BMCs were composed of 77.5 ± 1.6% (control 85.9 ± 1.6%) SSClo CD33− B-lymphocytes, 12.3 ± 1.3% (control 5.2 ± 0.3%) SSChi CD33lo granulocytes, and 9.2 ± 1.0% (control 7.8 ± 1.5%) SSClo CD33hi monocytes (five experiments; granulocytes, P < 0.001) (Fig. 3B). There were not enough cells of other lineages for flow analysis. Taking into account the overall 2.5-fold expansion of PLZFKD BMCs, knockdown of PLZF increased the absolute number of granulocytes by 6.0 ± 1.7-fold, compared with controls (Fig. 3C). Furthermore, increased granulocyte numbers were largely due to augmented production of neutrophils expressing maturation markers CD11b and CD16 (Fig. 3D). These observations suggest that PLZF decreases proliferation and/or differentiation of myeloid cells in vivo.
PLZF regulates the balance between progenitor and mature compartments
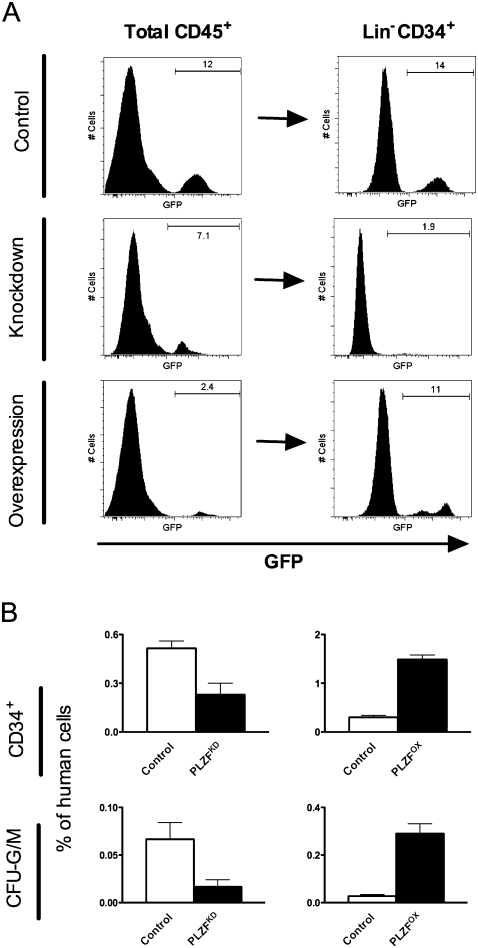
To determine if the increased contribution of PLZFKD cells to the myeloid graft reflects an expansion of immature progenitors, marrow from mice transduced with control or PLZF-bearing viruses was pooled and depleted of murine and mature human cells to enable more precise flow cytometric analysis of the immature engrafted populations (see the Materials and Methods). As expected for empty vector and shRFP controls, the proportion of GFP+ cells in unfractionated CD45+ bone marrow and Lin− CD34+ fractions was equivalent (Fig. 4A, Control). Surprisingly, knockdown of PLZF decreased the contribution of GFP+ cells to the progenitor compartment by 2.6 ± 0.1-fold, compared with total BMCs (Fig. 4A [Knockdown], B). Conversely, the proportion of GFP+ PLZFOX cells in the primitive Lin− CD34+ fraction was increased by 5.4 ± 0.5-fold (Fig. 4A [Overexpression], B). Thus, while negatively regulating the mature myeloid compartment, PLZF also expanded the pool of undifferentiated progenitors.
Figure 4.
PLZF expands the human progenitor compartment in vivo. (A) Proportion of GFP+ cells within the total human CD45+ graft in the bone marrow (left) compared with Lin− CD34+ progenitor fraction (right) in a representative experiment 8 wk post-transplant. Bone marrow was pooled from four to eight mice, depleted of murine and mature human cells, and analyzed for GFP and CD34 expression by flow cytometry before and after lineage depletion. Equal percentage of GFP indicates equal contribution of transduced cells to mature and progenitor compartments, while increased or decreased proportion reflects expansion or depletion of progenitors relative to total cells. (B) Proportion of Lin− CD34+ progenitors and myeloid colony-forming units (CFU-G/M) within the total CD45+ graft in mice transplanted with PLZFKD or PLZFOX cells. Data are expressed as mean ± SEM of two independent experiments, each with Lin− cells isolated from four to eight engrafted mice.
The number of progenitors in vivo was determined independently using colony assays. PLZFOX granulocyte or monocyte colony-forming units (CFU-G/M) were overrepresented by 15.7-fold, whereas PLZFKD progenitors were depleted by 4.6-fold (Fig. 4B), providing an independent correlation with the phenotypic data. In contrast to our in vitro experiments, the number of seeded progenitors was not the same between control and PLZF mice. However, since PLZF expanded Lin− CD34+ cells, we infer that the increase in CFU-G/M in vivo is due to an increased number of progenitor cells, rather than their clonogenic potential.
Taken together, these results demonstrate that rather than conferring a general proliferative advantage to human cells in vivo, PLZF specifically regulates the balance between the progenitor and mature compartments. Higher levels of PLZF maintain undifferentiated progenitors and curb their mature output, while silencing causes depletion of progenitors due to an increased rate of proliferation and differentiation.
Cytokines modulate PLZF repression of myeloid development
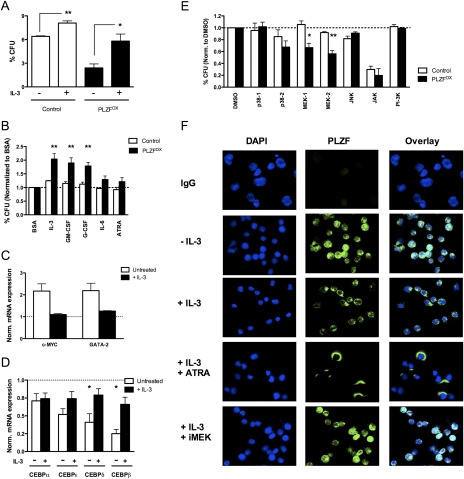
Since our data suggest that PLZF normally functions to negatively regulate entry of progenitors into terminal differentiation, we sought to determine the involvement of PLZF in myelopoiesis stimulated by stress-induced cytokines. Overexpression of PLZF in the absence of cytokine exposure reduced the growth potential of clonogenic progenitors (see Fig. 2F). However, when PLZFOX progenitors were seeded in the presence of IL-3, myeloid colony-forming potential was restored to nearly normal levels, suggesting that IL-3 signaling can alleviate the defect in growth and differentiation (Fig. 5A). As expected, IL-3 also improved the clonogenic potential of control cells, but to a much lesser extent (Fig. 5A). As colony assays are performed in the presence of several cytokines (see the Materials and Methods), we sought to examine the effect of individual cytokines. CD34+ CD71− myeloid progenitors were stimulated with a select cytokine for 24 h in serum-free medium supplemented with BSA, insulin, and transferrin (SFM + BIT) to maintain cellular viability and seeded in methylcellulose. The potential of PLZF to repress colony-forming capacity was significantly alleviated by stimulation with IL-3, GM-CSF, or G-CSF (all P < 0.01), all of which have been implicated as key mediators of stress-induced myelopoiesis (Fig. 5B; Hirai et al. 2006). However, IL-6 or all-trans retinoic acid (ATRA), which have distinct biological roles, did not promote myelopoiesis in the presence of PLZF (Fig. 5B). These results suggest that stress-induced cytokines can act by interfering with the capacity of PLZF to inhibit proliferation and differentiation.
Figure 5.
Cytokines modulate the effects of PLZF on growth and differentiation. (A) Myeloid colony-forming capacity of CD34+ CD71− myeloid progenitors transduced with MPG (control) or MPG-PLZF (PLZFOX) viruses and seeded in methylcellulose ± IL-3. (B) Same as A, except progenitors were stimulated with the indicated cytokines for 24 h in SFM + BIT and seeded in methylcellulose. Data were normalized to BSA-treated samples (dashed line). (C) Expression of MYC and GATA-2 in control or PLZFOX CD34+ CD71− progenitors cultured for 4 d in serum-free media ± IL-3. Expression is normalized to cells infected with a control virus (dashed line). (D) Same as C for C/EBP transcription factors. Expression is normalized to cells infected with a control virus (dashed line). (*) P < 0.05. (E) Same as A, except progenitors were stimulated with IL-3 plus an indicated protein kinase inhibitor or DMSO vehicle for 24 h in SFM + BIT and seeded in methylcellulose. SB203580 inhibits p38-1, SB202190 inhibits p38-2, PD98059 inhibits MEK-1, U0126 inhibits MEK-2, SP600125 inhibits JNK, AG490 inhibits JAK, and LY294002 inhibits PI-3K. Data were normalized to DMSO-treated samples (dashed line). (F) Immunofluorescence staining of PLZF localization (green) in DAPI-stained nuclei or cytoplasm of nontransduced Lin− CD34+ CD71− progenitors cultured in SFM + BIT ± IL-3, ATRA, or PD98059 (iMEK) for 24 h. Magnification, 100×. All data are expressed as mean ± SEM of three independent experiments. (*) P < 0.02; (**) P < 0.006.
To probe the interaction of PLZF and cytokines on the molecular level, we profiled the expression of key indicators of myeloid differentiation in culture by qPCR. Both c-MYC (MYC) and GATA-2 transcription factors are normally expressed in myeloid progenitors and expression declines with differentiation (Johansen et al. 2001; Kobayashi-Osaki et al. 2005). Consistent with their impaired differentiation, PLZFOX myeloid progenitors had higher levels of MYC and GATA-2 transcripts; however, normal levels were restored if the transduced cells were cultured with IL-3 (Fig. 5C). The C/EBP family of transcription factors (C/EBPs) are expressed in an ordered fashion during normal neutrophil differentiation (Bjerregaard et al. 2003). Transcript expression of C/EBPs in CB cells was blocked by PLZF (all P < 0.05, except CEBPA); however, normal levels were restored in the presence of IL-3 (Fig. 5D). Notably, the degree of repression by PLZF correlated with their normal order of expression during differentiation (C/EBPα < ε < δ < β), such that C/EBPs expressed in more mature cells were more strongly repressed (Bjerregaard et al. 2003). The ordered repression of these transcription factors suggests that PLZF establishes a molecular state that impairs differentiation, while the addition of cytokines restores a normal pattern of myeloid development.
To investigate the pathway(s) responsible for cytokine-mediated inhibition of PLZF, sorted CD34+ CD71− myeloid progenitors were cultured in SFM + BIT, treated with IL-3 in the presence of specific protein kinase inhibitors for 24 h, and seeded in methylcellulose. Control and PLZFOX progenitors treated with p38 MAPK (SB203580 and SB202190), JNK (SP600125), or PI-3K (LY294002) inhibitors showed a similar response to IL-3 stimulation (Fig. 5E). Treatment with JAK inhibitor AG490 drastically reduced colony formation of both control and PLZFOX cells, but there was no difference in their response to IL-3. Only specific MEK inhibitors PD98059 and U0126, which impair downstream ERK1/2 (ERK) activation, restored PLZF-mediated repression of myeloid colony formation in the presence of IL-3 (both P < 0.01) (Fig. 5E). Thus, IL-3 interferes with PLZF repression by activating ERK signal transduction.
In CD34+ progenitors and myeloid cell lines, PLZF is localized to distinct nuclear domains, while its loss-of-function mutants exhibit aberrant localization (Reid et al. 1995; Guidez et al. 2005). To examine the effect of cytokines on native cellular localization of PLZF, nontransduced CD34+ CD71− myeloid progenitors were cultured in SFM + BIT with or without IL-3 for 24 h, and distribution of PLZF was visualized by immunofluorescence microscopy. In the absence of cytokines, most cells expressed PLZF protein, which was found in the nucleus and the cytoplasm (Fig. 5F, −IL-3). Stimulation with IL-3 resulted in a redistribution of nuclear PLZF to the cytoplasm (Fig. 5F, +IL-3), and this effect was enhanced by cotreatment with other differentiation-inducing agents, such as ATRA (Fig. 5F, +IL-3 +ATRA). However, nuclear localization was completely restored upon cotreatment with the specific ERK inhibitor PD98059 (Fig. 5F, +IL-3 +iMEK). These results suggest that cytokines mediate inactivation of PLZF by triggering its export from the nucleus in an ERK-dependent manner.
PLZF regulates expression of myeloid transcription factors
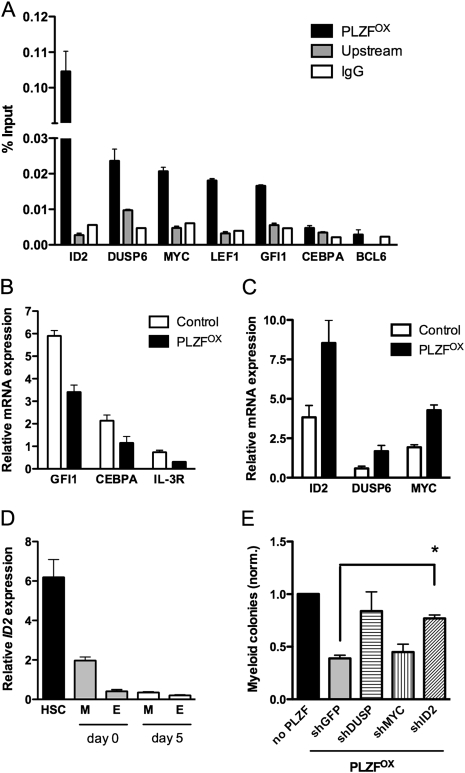
To gain insight into the mechanisms that underlie the differentiation-suppressing function of PLZF, a genome-wide analysis of PLZF promoter binding was performed using chromatin immunoprecipitation microarray (ChIP–chip) in KG1a myeloid cells (K Rice, in prep.). The top candidates were then validated using conventional ChIP (Fig. 6A), and the transcriptional regulation of candidate targets was verified by qPCR in Lin− CB. PLZF bound to promoters of transcription factors that control myeloid differentiation, including LEF1 (TCF1A) and GFI1 and repressed transcription, indicating that these are direct targets of PLZF (Fig. 6A,B). LEF1 is a nuclear cofactor of β-catenin, critical for lymphoid and myeloid development (Skokowa et al. 2006). GFI1 is a proto-oncogene that functions as a neutrophil lineage determinant downstream from C/EBPα (Laslo et al. 2006). Notably, PLZF also repressed CEBPA transcription but did not bind to its promoter, indicating indirect regulation (Fig. 6A,B).
Figure 6.
Transcriptional regulation of myeloid target genes by PLZF. (A) ChIP analysis of target promoter occupancy by PLZF in KG1a myeloid cells. ChIP was performed using an anti-PLZF antibody (PLZF) or control IgG (IgG). Binding is represented as fold enrichment over input determined by qPCR for promoter sequence containing consensus PLZF-binding sites, and an internal control sequence 6 kb upstream of transcriptional start site (Upstream). Binding to an intronic sequence of the BCL6 gene was used as a negative control. (B) Expression levels of GFI1, CEBPA, and IL-3R in control or PLZFOX CD34+ CD71− myeloid progenitors 24 h after transduction. (C) Expression levels of ID2, DUSP6, and MYC in control or PLZFOX CD34+ CD71− myeloid progenitors 24 h after transduction. (D) Expression levels of ID2 in human CD34+ CD38− HSCs, CD34+ CD38+ CD71+ erythroid (E), and CD34+ CD38+ CD71− myeloid (M) progenitors sorted from fresh Lin− CB (day 0) or after 5 d culture in serum-free media (day 5). (E) Myeloid colony-forming capacity of CD34+ CD71− progenitors cotransduced with control or PLZFOX vectors and knockdown viruses for GFP, ID2, DUSP6, and MYC. All data are expressed as mean ± SEM of three independent experiments, except E, which includes four experiments. (*) P < 0.005.
Classically, PLZF has been regarded as a transcriptional repressor that recruits nuclear corepressor complexes to silence promoter activation (Hong et al. 1997). However, PLZF also bound to promoter sites of negative regulators of signaling and differentiation, including MYC, DUSP6, and ID2, and activated their transcription (Fig. 6A,C). MYC is a proto-oncogene whose down-regulation is critical for myeloid differentiation (Johansen et al. 2001). DUSP6 is an ERK-specific phosphatase that acts as part of a negative feedback loop in FGF signaling (Li et al. 2007). ID2 is a member of the basic helix–loop–helix (bHLH) transcription factor family and a negative regulator of differentiation (Ji et al. 2008). These data suggest that PLZF can recruit either corepressors or coactivators to target promoters to regulate gene expression.
Whereas MYC is expressed ubiquitously in hematopoiesis, expression of DUSP6 and ID2 is highest in HSCs and declines with differentiation (Fig. 6D; K Takenaka, in prep.). This observation suggests that DUSP6 and ID2 may act as cooperating factors downstream from PLZF. To test this hypothesis, CD34+ CD71− myeloid progenitors were first transduced with the control (MPG) or PLZFOX vectors to induce ectopic PLZF expression. Cells were then cotransduced with shRNA vectors for PLZF target genes (MYC, ID2, and DUSP6) and plated in colony assays. Since each shRNA was introduced side-by-side into control and PLZF-overexpressing cells, any putative off-target effects would be equal in the presence or absence of ectopic PLZF. By normalizing the number of colonies formed by PLZFOX + shRNA-transduced progenitors to those transduced with MPG + shRNA, we selectively measured the effects of gene silencing that are specific to PLZF-overexpressing cells. PLZFOX myeloid progenitors transduced with shGFP or shMYC gave rise to fewer colonies compared with shGFP or shMYC alone (0.41 ± 0.03-fold and 0.46 ± 0.06-fold, respectively) indicating that PLZF retained its inhibitory effect on myeloid growth (Fig. 6E). However, shDUSP6 or shID2 restored the colony-forming potential of PLZFOX myeloid progenitors to 0.84 ± 0.18-fold and 0.78 ± 0.03-fold of the knockdown alone, although only the shID2 effect was significant (P < 0.005) (Fig. 6E). These data identify ID2 as a necessary downstream effector of the differentiation-suppressing function of PLZF.
Discussion
In this study, we present evidence that transcription factor networks that control hematopoietic specification also incorporate factors that inhibit normal differentiation, thereby playing an essential role in maintaining the steady-state output of mature cells and priming the system to be able to respond to conditions of immune activation. Although several transcription factors that suppress normal differentiation have been described, their biological role has remained unclear (Khanna-Gupta et al. 2001; Gery et al. 2004). In our model, the significance of this regulatory mechanism is readily apparent in PLZF-deficient cells, which generate increased numbers of mature progeny in a competitive transplant setting, but suffer from a depletion of the progenitor pool. Thus, the lack of negative regulation destines progenitors to eventual exhaustion as a result of excessive differentiation. At the other extreme, overexpression of PLZF is equally disruptive, shutting off the production of mature cells and causing a buildup of undifferentiated progenitors in vitro and in vivo. Thus, negative control of differentiation is essential for setting the balance between proliferation and differentiation, such that the loss of myeloid progenitors by differentiation is offset by their regeneration from primitive precursors including HSCs.
Modulation of PLZF by stress-induced myeloid cytokines
In addition to its role during homeostasis, negative control of proliferation and differentiation by PLZF is involved in priming the system for increased production of mature cells in response to cytokines. Cytokine stimulation induces progenitor cycling and causes a rapid differentiative response that depletes the progenitor pool (Hirai et al. 2006). Accordingly, sufficient numbers of progenitors must be made available in the event of emergencies; our data indicate that this is dependent on PLZF function. When the production of mature cells must be rapidly ramped up, this restriction on proliferation and differentiation is lifted via cytokine-induced signal transduction pathways, most notably p38 and ERK MAPKs. These pathways directly augment differentiation by activating positive regulators of differentiation, such as C/EBPβ and C/EBPε (Hu et al. 2001; Williamson et al. 2005). We show that activation of ERK by stress-induced cytokines, including IL-3, GM-CSF, and G-CSF, also interferes with PLZF-mediated repression of growth and differentiation. Thus, our data reveal greater complexity in the cytokine response, whereby the inactivation of anti-differentiation factors can function as a switch that shifts the balance in favor of differentiation.
Several protein modifications are required for full transcriptional activity of PLZF; notably, its acetylation by p300 (Guidez et al. 2005). The acetylated form localizes to discrete nuclear subdomains, while the deacetylated protein has a diffuse nuclear staining (Guidez et al. 2005). In another example, the proteolytic fragment of the heparin-binding epidermal growth factor inactivates PLZF by interacting with it in the nucleus and targeting it for export (Nanba et al. 2003). In both cases, its activation state is linked with altered cellular localization in the cytoplasm. Consistent with the previous reports (Reid et al. 1995), our data show that in CB-derived progenitors, PLZF is localized primarily in the nucleus. Following stimulation with IL-3, PLZF is redistributed to the cytoplasm, a localization that can be reversed by MEK inhibitors. This observation illustrates a potential mechanism by which cytokines interfere with PLZF function to quickly ramp up proliferation and differentiation. It remains to be shown whether active ERK inhibits PLZF directly targeting it for nuclear export, or if inhibition involves other factors. It was shown previously that activating FLT3 mutations antagonize PLZF by impairing its interaction with corepressors (Takahashi et al. 2004). A similar mechanism may be involved in PLZF inactivation by ERK, with reduced corepressor interaction targeting it for nuclear export.
Molecular mechanisms of differentiation downstream from PLZF
Our evidence indicates that PLZF shifts the balance against differentiation by concertedly repressing expression of myeloid transcription factors GFI1, LEF1, and C/EBPα, and activating MYC and ID2. Normal granulocytic differentiation is orchestrated by the C/EBPα–GFI1 regulatory circuit (Hock et al. 2003; Laslo et al. 2006). Consistent with this, our results showed that granulocyte numbers were most significantly increased in mice transplanted with PLZF-deficient cells, compared with other lineages. Interestingly, mutations in GFI1 and the loss of LEF1 expression are associated with severe congenital neutropenia, a disorder marked by susceptibility to opportunistic infections due to low neutrophil counts (Person et al. 2003; Skokowa et al. 2006). Our study therefore provides the rationale to investigate the role of PLZF in this disorder.
ID2 may establish a molecular state refractory to differentiation by repressing tissue-specific transcription factors, such as E2A and SCL (Lasorella et al. 2001). In hematopoiesis, ID2 functions as a negative regulator of B-lymphoid and myeloid commitment owing to its interaction with E2A and PU.1 (Ji et al. 2008). We demonstrated that ID2 mRNA is expressed at the highest levels in HSCs and declines in progenitors. Furthermore, our results establish that PLZF up-regulates ID2 expression in myeloid progenitors and that ID2 is required to maintain the repressive effect of PLZF on differentiation. Thus, a major mechanism by which PLZF regulates myeloid development is by inducing ectopic ID2 expression, which antagonizes other lineage-specific transcription factors.
In conclusion, our findings point to a central role of differentiation-suppressing mechanisms in establishing developmental homeostasis by regulating the balance between the progenitor pool and the production of mature cells. More broadly, our study demonstrates the practicality of taking a genetic approach to unraveling the developmental program in primary human hematopoietic cells. Such an approach is important, as surrogate models may not entirely recapitulate the intrinsic complexity of the human developmental processes.
Materials and methods
Sample collection and purification
CB samples were obtained according to procedures approved by the institutional review boards of the University Health Network and Trillium Hospital, and were collected, processed, and stored as described (Mazurier et al. 2004). Briefly, Lin− CB cells were purified by negative selection using the StemSep Human Progenitor Cell Enrichment Kit according to the manufacturer's protocol (StemCell Technologies).
Viral constructs
Full-length PLZF cDNA was cloned into a two-promoter MSCV-PGK-EGFP (MPG) retroviral vector and a cppt-PGK-EGFP-WPRE-derived lentiviral vector (Mazurier et al. 2004) with a truncated E1Fα promoter. shRNAs against PLZF were designed using the Dharmacon algorithm (Dharmacon, Inc.; see the Supplemental Material for sequences), synthesized as complimentary 5′-P oligonucleotides, annealed, and cloned into the modified cppt-PGK-EGFP-WPRE vector containing the H1 promoter and an ires-PAC selection cassette. Viral particles pseudotyped with VSV-G were generated by transient transfection as described (Mazurier et al. 2004).
Viral transduction
Lin− CB cells were prestimulated and transduced in X-VIVO 10 (BioWhittaker) medium with 1% BSA, 2 mM L-glutamine, 100 U/mL penicillin–streptomycin, plus SCF (100 ng/mL), FLT3L (100 ng/mL), TPO (50 ng/mL), and IL-6 (20 ng/mL) (all Amgen) for 4 h (lenti) or 24 h (retro). For lentiviral infections, cells were transduced at multiplicity of infection (MOI) 50–100 for 24 h. For retroviral infections, ∼1 × 106 cells were transduced with four changes of 1 × 107 to 5 × 107 viral particles each for 48 h; wells were precoated with CH-296 fibronectin (4 μg/cm2; Retronectin, Takara Bio, Inc.).
Cell sorting
Retrovirus-transduced cells were cultured in transduction medium for 24 h after gene transfer. Cells were stained with antibodies against CD34 and CD71 (BD Pharmingen). GFP+ CD34+ (CD71−) progenitors were sorted on BD FACSAria cytometer operating in the “Purity” mode, consistently yielding >98% purity.
Flow cytometry
Routine flow cytometry was performed using BD FACSCalibur or BD LSRII cytometers on fresh cells using monoclonal mouse anti-human antibodies (see the Supplemental Material). Data were analyzed with FlowJo software (Tree Star, Inc.).
Liquid cultures
Sorted cells (2 × 104 to 5 × 104) were seeded into 1 mL IMDM + 15% FCS plus SCF (20 ng/mL) and IL-3 (2 ng/mL) or IMDM + 20% BIT (StemCell Technologies), 2 mM L-glutamine, 0.1 mM β-mercaptoethanol, plus SCF (100 ng/mL), FLT3L (100 ng/mL), IL-6 (20 ng/mL), G-CSF (20 ng/mL), and GM-CSF (20 ng/mL) (all cytokines; Amgen). Cultures were maintained at a density of ∼1 × 106 cells per milliliter and 1 × 105 cells were used for analysis of lineage markers with antibodies against CD11b, CD14, and CD15 (Beckman Coulter), and CD33 (BD Pharmingen).
Colony assays
Sorted progenitors (1 × 103) were seeded into 3 mL of MethoCult medium (StemCell Technologies) with 20% FCS, 5% human plasma, 5% CB plasma, 0.1 mM β-mercaptoethanol, plus “basic” cytokines SCF (10 ng/mL), FLT3L (10 ng/mL), TPO (50 ng/mL), IL-6 (10 ng/mL), GM-CSF (50 ng/mL), and EPO (4 U/mL). Duplicate 1-mL aliquots were plated into 35-mm dishes (Nunc) and hematopoietic colonies of erythroid, myeloid, or mixed lineage were scored after 14 d.
BrdU and annexin assays
BrdU was added to day 7 liquid cultures at 10 μM final concentration. After 3 h of incubation, 1 × 105 cells were removed, stained with anti-CD11b-PE (Beckman Coulter), and assayed with BrdU Flow Kit and Annexin V-PE Apoptosis Detection Kit (both BD Pharmingen) according to the manufacturer's protocols.
NOD/SCID transplantation
NOD_LtSz-scid_scid (NOD/SCID) mice were sublethally irradiated (300 cGy) and injected i.p. with 200 μg of anti-CD122 monoclonal antibody 24 h prior to transplantation (McKenzie et al. 2005). Transduced cells were washed, resuspended at 7.5 × 104 cells in 20 μL of PBS + 1% FCS, and injected intrafemorally into anesthetized mice. Mice were sacrificed after 8 wk, BMCs were isolated by flushing with 2 mL of IMDM, and 50 μL were stained for surface markers (see the Supplemental Material for antibodies). Remaining cells were pooled from all mice within a group (n = 4–8), red blood cell lysed, and used for surface antigen staining and lineage-depletion (see below).
Lineage depletion
Human progenitors were isolated from pooled bone marrow using the Mouse/Human Chimera Enrichment Kit (StemCell Technologies) according to the manufacturer's protocol, with the addition of 100 μL/mL StemSep Human Hematopoietic Progenitor Enrichment Cocktail (StemCell Technologies) and the anti-biotin antibody. Fresh column-purified cells were used for flow cytometry and seeded in colony assays at 1 × 103 cells per plate, and GFP+ colonies were scored using an inverted fluorescent microscope.
Cytokine stimulation
Sorted GFP+ CD34+ CD71− progenitors were either directly seeded into methylcellulose with basic cytokines, without EPO, ±50 ng/mL IL-3, or 1 × 104 cells were stimulated in IMDM + 20% BIT + 2 mM L-glutamine ± 20 ng/mL IL-3, G-CSF, GM-CSF, IL-6, or 10−7 M ATRA (Sigma Aldrich) for 24 h, after which 1.5 × 103 cells were seeded into methylcellulose with basic cytokines. For inhibitor studies, cells were stimulated as above with the addition of 10 μM SB203580, 50 μM SB202190, 10 μM PD98059, 10 μM U0126, 25 μM SP600125, 25 μM AG490, 50 μM LY294002, or DMSO vehicle (all Tocris Bioscience). All inhibitors were used at previously determined concentrations (see the Supplemental Material for references).
Immunofluorescence microscopy
Nontransduced CD34+ CD71− progenitors were cultured in IMDM + 20% BIT + 2 mM L-glutamine ± 50 ng/mL IL-3, 10−7 M ATRA or 10 μM PD98059 for 24 h. Cells were fixed in 2% paraformaldehyde and 5 × 104 cells were cytospun onto microscope slides. See the Supplemental Material for a complete staining protocol.
Quantitative RT–PCR
More than 5 × 104 sorted GFP+ CD34+ CD71− progenitors were cultured in IMDM + 20% BIT + 2 mM L-glutamine ± 20 ng/mL IL-3 for 24 h. RNA was extracted with the Trizol reagent (Invitrogen), DNase I-treated (Qiagen), and reverse-transcribed using SuperScript II (Invitrogen). Real-time PCR reactions were prepared using SYBR Green PCR Master Mix (Applied Biosystems), with 200 nM each primer and >20 ng of cDNA per reaction (all 60°C annealing T; see the Supplemental Material for the primer list). Reactions were performed in triplicate on Applied Biosystems 7900HT instruments. Absolute gene expression was quantified using SDS software (Applied Biosystems) based on the standard curve method and presented as transcript expression (picograms)/GAPDH (picograms) × 100.
ChIP
See the Supplemental Material for the complete protocol.
Statistical analysis
Unless otherwise stated, significance of differences among groups was determined by two-tailed unpaired Student's t-test. Standard errors of the products or norms of two independent data sets were calculated using the MSTAT software (McArdle Laboratory for Cancer Research).
Acknowledgments
We thank J.E.D. laboratory members for critical reading and discussion of the manuscript, UHN/SickKids Flow Cytometry Facility staff (P. Pentilla and S. Zhao) for flow cytometry assistance, and Jason Moffat (University of Toronto) for providing lentiviral shRNA vectors. This work was supported by a Canadian Institute of Health Research (CIHR)-University of Toronto Collaborative Graduate Training Program in Molecular Medicine studentship (to S.D.), s CIHR Doctoral Research award (to F.N.), grants from the CIHR and the Ontario Institute for Cancer Research, and a Summit Award both with funds from the Province of Ontario, Genome Canada through the Ontario Genomics Institute, a Canada Research Chair, the Leukemia and Lymphoma Society, the Canadian Cancer Society, and the Terry Fox Foundation (to J.E.D.). J.D.L and A.Z. were sponsored by NIH grant CA 59936-JDL.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1788109.
Supplemental material is available at http://www.genesdev.org.
References
- Barna M, Hawe N, Niswander L, Pandolfi PP. Plzf regulates limb and axial skeletal patterning. Nat Genet. 2000;25:166–172. doi: 10.1038/76014. [DOI] [PubMed] [Google Scholar]
- Barna M, Merghoub T, Costoya JA, Ruggero D, Branford M, Bergia A, Samori B, Pandolfi PP. Plzf mediates transcriptional repression of HoxD gene expression through chromatin remodeling. Dev Cell. 2002;3:499–510. doi: 10.1016/s1534-5807(02)00289-7. [DOI] [PubMed] [Google Scholar]
- Barreda DR, Hanington PC, Belosevic M. Regulation of myeloid development and function by colony stimulating factors. Dev Comp Immunol. 2004;28:509–554. doi: 10.1016/j.dci.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Bjerregaard MD, Jurlander J, Klausen P, Borregaard N, Cowland JB. The in vivo profile of transcription factors during neutrophil differentiation in human bone marrow. Blood. 2003;101:4322–4332. doi: 10.1182/blood-2002-03-0835. [DOI] [PubMed] [Google Scholar]
- Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, de Rooij DG, Braun RE. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet. 2004;36:647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- Cannistra SA, Griffin JD. Regulation of the production and function of granulocytes and monocytes. Semin Hematol. 1988;25:173–188. [PubMed] [Google Scholar]
- Chen Z, Brand NJ, Chen A, Chen SJ, Tong JH, Wang ZY, Waxman S, Zelent A. Fusion between a novel Kruppel-like zinc finger gene and the retinoic acid receptor-α locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. EMBO J. 1993;12:1161–1167. doi: 10.1002/j.1460-2075.1993.tb05757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36:653–659. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- Donahue RE, Seehra J, Metzger M, Lefebvre D, Rock B, Carbone S, Nathan DG, Garnick M, Sehgal PK, Laston D, et al. Human IL-3 and GM-CSF act synergistically in stimulating hematopoiesis in primates. Science. 1988;241:1820–1823. doi: 10.1126/science.3051378. [DOI] [PubMed] [Google Scholar]
- Fisher AG. Cellular identity and lineage choice. Nat Rev Immunol. 2002;2:977–982. doi: 10.1038/nri958. [DOI] [PubMed] [Google Scholar]
- Gery S, Park DJ, Vuong PT, Chih DY, Lemp N, Koeffler HP. Retinoic acid regulates C/EBP homologous protein expression (CHOP), which negatively regulates myeloid target genes. Blood. 2004;104:3911–3917. doi: 10.1182/blood-2003-10-3688. [DOI] [PubMed] [Google Scholar]
- Guidez F, Howell L, Isalan M, Cebrat M, Alani RM, Ivins S, Hormaeche I, McConnell MJ, Pierce S, Cole PA, et al. Histone acetyltransferase activity of p300 is required for transcriptional repression by the promyelocytic leukemia zinc finger protein. Mol Cell Biol. 2005;25:5552–5566. doi: 10.1128/MCB.25.13.5552-5566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Zhang P, Dayaram T, Hetherington CJ, Mizuno S, Imanishi J, Akashi K, Tenen DG. C/EBPβ is required for ‘emergency’ granulopoiesis. Nat Immunol. 2006;7:732–739. doi: 10.1038/ni1354. [DOI] [PubMed] [Google Scholar]
- Hock H, Hamblen MJ, Rooke HM, Traver D, Bronson RT, Cameron S, Orkin SH. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity. 2003;18:109–120. doi: 10.1016/s1074-7613(02)00501-0. [DOI] [PubMed] [Google Scholar]
- Hong SH, David G, Wong CW, Dejean A, Privalsky ML. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor α (RARα) and PLZF-RARα oncoproteins associated with acute promyelocytic leukemia. Proc Natl Acad Sci. 1997;94:9028–9033. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Roy SK, Shapiro PS, Rodig SR, Reddy SP, Platanias LC, Schreiber RD, Kalvakolanu DV. ERK1 and ERK2 activate CCAAAT/enhancer-binding protein-β-dependent gene transcription in response to interferon-γ. J Biol Chem. 2001;276:287–297. doi: 10.1074/jbc.M004885200. [DOI] [PubMed] [Google Scholar]
- Ji M, Li H, Suh HC, Klarmann KD, Yokota Y, Keller JR. Id2 intrinsically regulates lymphoid and erythroid development via interaction with different target proteins. Blood. 2008;112:1068–1077. doi: 10.1182/blood-2008-01-133504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen LM, Iwama A, Lodie TA, Sasaki K, Felsher DW, Golub TR, Tenen DG. c-Myc is a critical target for c/EBPα in granulopoiesis. Mol Cell Biol. 2001;21:3789–3806. doi: 10.1128/MCB.21.11.3789-3806.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna-Gupta A, Zibello T, Sun H, Lekstrom-Himes J, Berliner N. C/EBP ε mediates myeloid differentiation and is regulated by the CCAAT displacement protein (CDP/cut) Proc Natl Acad Sci. 2001;98:8000–8005. doi: 10.1073/pnas.141229598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi-Osaki M, Ohneda O, Suzuki N, Minegishi N, Yokomizo T, Takahashi S, Lim KC, Engel JD, Yamamoto M. GATA motifs regulate early hematopoietic lineage-specific expression of the Gata2 gene. Mol Cell Biol. 2005;25:7005–7020. doi: 10.1128/MCB.25.16.7005-7020.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbaye C, Spinello I, Quaranta MT, Pelosi E, Pasquini L, Petrucci E, Biffoni M, Nuzzolo ER, Billi M, Foa R, et al. A three-step pathway comprising PLZF/miR-146a/CXCR4 controls megakaryopoiesis. Nat Cell Biol. 2008;10:788–801. doi: 10.1038/ncb1741. [DOI] [PubMed] [Google Scholar]
- Laiosa CV, Stadtfeld M, Graf T. Determinants of lymphoid-myeloid lineage diversification. Annu Rev Immunol. 2006;24:705–738. doi: 10.1146/annurev.immunol.24.021605.090742. [DOI] [PubMed] [Google Scholar]
- Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, Dinner AR, Singh H. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- Lasorella A, Uo T, Iavarone A. Id proteins at the cross-road of development and cancer. Oncogene. 2001;20:8326–8333. doi: 10.1038/sj.onc.1205093. [DOI] [PubMed] [Google Scholar]
- Li C, Scott DA, Hatch E, Tian X, Mansour SL. Dusp6 (Mkp3) is a negative feedback regulator of FGF-stimulated ERK signaling during mouse development. Development. 2007;134:167–176. doi: 10.1242/dev.02701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieschke GJ, Grail D, Hodgson G, Metcalf D, Stanley E, Cheers C, Fowler KJ, Basu S, Zhan YF, Dunn AR. Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood. 1994;84:1737–1746. [PubMed] [Google Scholar]
- Mazurier F, Gan O, McKenzie J, Doedens M, Dick J. Lentivector-mediated clonal tracking reveals intrinsic heterogeneity in the human hematopoietic stem cell compartment and culture-induced stem cell impairment. Blood. 2004;103:545–552. doi: 10.1182/blood-2003-05-1558. [DOI] [PubMed] [Google Scholar]
- McConnell MJ, Chevallier N, Berkofsky-Fessler W, Giltnane JM, Malani RB, Staudt LM, Licht JD. Growth suppression by acute promyelocytic leukemia-associated protein PLZF is mediated by repression of c-myc expression. Mol Cell Biol. 2003;23:9375–9388. doi: 10.1128/MCB.23.24.9375-9388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie JL, Gan OI, Doedens M, Dick JE. Human short-term repopulating stem cells are efficiently detected following intrafemoral transplantation into NOD/SCID recipients depleted of CD122+ cells. Blood. 2005;106:1259–1261. doi: 10.1182/blood-2005-03-1081. [DOI] [PubMed] [Google Scholar]
- Nanba D, Mammoto A, Hashimoto K, Higashiyama S. Proteolytic release of the carboxy-terminal fragment of proHB-EGF causes nuclear export of PLZF. J Cell Biol. 2003;163:489–502. doi: 10.1083/jcb.200303017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster W, Lindemann A, Mertelsmann R, Herrmann F. Regulation of gene expression of M-, G-, GM-, and multi-CSF in normal and malignant hematopoietic cells. Blood Cells. 1988;14:443–462. [PubMed] [Google Scholar]
- Panopoulos AD, Zhang L, Snow JW, Jones DM, Smith AM, El Kasmi KC, Liu F, Goldsmith MA, Link DC, Murray PJ, et al. STAT3 governs distinct pathways in emergency granulopoiesis and mature neutrophils. Blood. 2006;108:3682–3690. doi: 10.1182/blood-2006-02-003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Person RE, Li FQ, Duan Z, Benson KF, Wechsler J, Papadaki HA, Eliopoulos G, Kaufman C, Bertolone SJ, Nakamoto B, et al. Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat Genet. 2003;34:308–312. doi: 10.1038/ng1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid A, Gould A, Brand N, Cook M, Strutt P, Li J, Licht J, Waxman S, Krumlauf R, Zelent A. Leukemia translocation gene, PLZF, is expressed with a speckled nuclear pattern in early hematopoietic progenitors. Blood. 1995;86:4544–4552. [PubMed] [Google Scholar]
- Rosenbauer F, Tenen DG. Transcription factors in myeloid development: Balancing differentiation with transformation. Nat Rev Immunol. 2007;7:105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaknovich R, Yeyati PL, Ivins S, Melnick A, Lempert C, Waxman S, Zelent A, Licht JD. The promyelocytic leukemia zinc finger protein affects myeloid cell growth, differentiation, and apoptosis. Mol Cell Biol. 1998;18:5533–5545. doi: 10.1128/mcb.18.9.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skokowa J, Cario G, Uenalan M, Schambach A, Germeshausen M, Battmer K, Zeidler C, Lehmann U, Eder M, Baum C, et al. LEF-1 is crucial for neutrophil granulocytopoiesis and its expression is severely reduced in congenital neutropenia. Nat Med. 2006;12:1191–1197. doi: 10.1038/nm1474. [DOI] [PubMed] [Google Scholar]
- Takahashi S, McConnell MJ, Harigae H, Kaku M, Sasaki T, Melnick AM, Licht JD. The Flt3 internal tandem duplication mutant inhibits the function of transcriptional repressors by blocking interactions with SMRT. Blood. 2004;103:4650–4658. doi: 10.1182/blood-2003-08-2759. [DOI] [PubMed] [Google Scholar]
- Ward JO, McConnell MJ, Carlile GW, Pandolfi PP, Licht JD, Freedman LP. The acute promyelocytic leukemia-associated protein, promyelocytic leukemia zinc finger, regulates 1,25-dihydroxyvitamin D(3)-induced monocytic differentiation of U937 cells through a physical interaction with vitamin D(3) receptor. Blood. 2001;98:3290–3300. doi: 10.1182/blood.v98.12.3290. [DOI] [PubMed] [Google Scholar]
- Williamson EA, Williamson IK, Chumakov AM, Friedman AD, Koeffler HP. CCAAT/enhancer binding protein ε: Changes in function upon phosphorylation by p38 MAP kinase. Blood. 2005;105:3841–3847. doi: 10.1182/blood-2004-09-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]