Abstract
Centrioles are subcellular organelles composed of a ninefold symmetric microtubule array that perform two important functions: (1) They build centrosomes that organize the microtubule cytoskeleton, and (2) they template cilia, microtubule-based projections with sensory and motile functions. We identified HYLS-1, a widely conserved protein, based on its direct interaction with the core centriolar protein SAS-4. HYLS-1 localization to centrioles requires SAS-4 and, like SAS-4, HYLS-1 is stably incorporated into the outer centriole wall. Unlike SAS-4, HYLS-1 is dispensable for centriole assembly and centrosome function in cell division. Instead, HYLS-1 plays an essential role in cilia formation that is conserved between Caenorhabditis elegans and vertebrates. A single amino acid change in human HYLS1 leads to a perinatal lethal disorder termed hydrolethalus syndrome, and we show that this mutation impairs HYLS-1 function in ciliogenesis. HYLS-1 is required for the apical targeting/anchoring of centrioles at the plasma membrane but not for the intraflagellar transport-dependent extension of the ciliary axoneme. These findings classify hydrolethalus syndrome as a severe human ciliopathy and shed light on the dual functionality of centrioles, defining the first stably incorporated centriolar protein that is not required for centriole assembly but instead confers on centrioles the capacity to initiate ciliogenesis.
Keywords: Centrioles, basal bodies, ciliogenesis, hydrolethalus syndrome, ciliopathy
Centrioles are small cylindrical organelles whose distinguishing feature is an outer wall composed of a ninefold symmetric array of stabilized microtubules. Centrioles perform two distinct functions in eukaryotic cells: (1) They recruit pericentriolar material to form centrosomes that organize the microtubule cytoskeleton, and (2) they template cilia, cellular projections that perform a variety of sensory and motile functions (Marshall 2007). Understanding how centrioles alternate between their two functions, and identifying the structural elements that underlie the distinct roles of centrioles in centrosome assembly and cilia formation, is an important current challenge.
In their capacity as centrosome organizers, centrioles direct the assembly of a filamentous matrix that is often much larger than their own dimensions (for example, during mitosis in the early Caenorhabditis elegans embryo, the volume of the pericentriolar material is ∼1000 times that of the centriole). This matrix, in turn, recruits other pericentriolar material components including the microtubule nucleator γ-tubulin (Moritz et al. 1998; Schnackenberg et al. 1998). Putative components of this matrix have been identified in a number of organisms (Hamill et al. 2002; Lucas and Raff 2007). However, how centrioles direct centrosome formation and the nature of the physical connection between centrioles and the surrounding pericentriolar material remains unknown.
In dividing vertebrate cells, centrioles organize centrosomes and direct the formation of a primary cilium during interphase (Rieder et al. 1979). In other cell types, such as in C. elegans neurons or in multiciliated vertebrate epithelial cells, ciliogenesis is initiated following terminal differentiation in response to expression of specific transcription factors (Chen et al. 1998; Swoboda et al. 2000). Centrioles initiate ciliogenesis by translocating to the cell surface in a step thought to involve vesicle trafficking and the actin cytoskeleton (Dawe et al. 2007; Park et al. 2008). Once anchored at the plasma membrane via a specialized structure termed the transition zone, motor-driven intraflagellar transport (IFT) extends the ciliary axoneme (Scholey 2008). The molecular mechanisms required for centrioles to translocate to the cell surface and serve as a launching pad for IFT to elongate the ciliary axoneme remain largely unknown.
Human cells have multiple types of cilia that differ in structure as well as in their mechanical and signaling properties (Afzelius 2004). A large spectrum of disorders, collectively termed ciliopathies, are associated with ciliary defects. Ciliopathies vary in severity from disorders that affect a single type of cilium and result in a specific pathology—such as progressive blindness, infertility, or polycystic kidney disease—to broad-based disorders such as Bardet-Biedl syndrome, characterized by obesity, retinal degeneration, polydactyly, and mental retardation (Badano et al. 2006). The knockout phenotype for the mouse protein Tg737, a conserved component of the IFT machinery, suggests that severe loss of cilia function in vertebrates results in midgestation lethality accompanied by a spectrum of developmental defects (Murcia et al. 2000).
The nematode C. elegans has emerged as an important model to study centrioles, centrosomes, and cilia. Nonmotile cilia, present on the dendritic endings of 60 of the 302 neurons in the adult C. elegans hermaphrodite, mediate the reception of chemosensory and mechanosensory stimuli. Compromised cilia function leads to defects in easily assayed behaviors, including chemotaxis, foraging, and male mating (Inglis et al. 2007). However, cilia are dispensable for viability and fertility, as C. elegans sperm are amoeboid rather than flagellated. These features have facilitated the study of ciliogenesis and proteins mutated in ciliary disorders (Inglis et al. 2007). Studies in the early C. elegans embryo also led to a major breakthrough in our understanding of centriole assembly by defining a conserved molecular pathway that underlies this process. Mutational analysis and comprehensive genome-wide RNAi-based screens defined a set of four proteins specifically required for centriole assembly: SAS-4, SAS-5, SAS-6, and the kinase ZYG-1/Plk4 (O'Connell et al. 2001; Kirkham et al. 2003; Leidel and Gonczy 2003; Dammermann et al. 2004; Delattre et al. 2004; Leidel et al. 2005). Subsequent ultrastructural work placed these components in a pathway in which ZYG-1/Plk4 triggers centriole assembly by recruiting SAS-5 and SAS-6 to form a cylindrical scaffold called the central tube (Delattre et al. 2006; Pelletier et al. 2006). SAS-4 is recruited to this structure and, together with γ-tubulin, directs the assembly of the stabilized microtubules that comprise the outer centriole wall (Pelletier et al. 2006; Dammermann et al. 2008). As a component of the outer centriolar wall, SAS-4 is a candidate for a scaffold that recruits pericentriolar material proteins to form centrosomes (Kirkham et al. 2003) and/or proteins that enable centrioles to initiate cilia (Basto et al. 2006).
To explore the connection between the outer centriole wall and proteins required for cilia/centrosome formation, we identified proteins that interact with C. elegans SAS-4. This approach yielded a single polypeptide, which we named HYLS-1 based on its homology with a human protein of unknown function mutated in the perinatal lethal disorder hydrolethalus syndrome (Mee et al. 2005). We show that HYLS-1 is a conserved centriolar protein that is stably incorporated into centrioles as they form via a direct interaction with SAS-4. Functional characterization of HYLS-1 in C. elegans and in the ciliated epithelium of the Xenopus embryo revealed that, unlike SAS-4, HYLS-1 is dispensable for centriole assembly and centrosome function during cell division. Instead, HYLS-1 is specifically required for ciliogenesis. HYLS-1 is not essential for the IFT-dependent extension of the ciliary axoneme, but is required for the apical targeting and/or anchoring of centrioles at the plasma membrane. These findings reveal that stable incorporation of HYLS-1 into the outer centriole wall during centriole assembly confers on them the capacity to initiate ciliogenesis and classify hydrolethalus syndrome as one the most severe ciliopathies identified to date.
Results
Identification of a SAS-4-associated protein that is mutated in hydrolethalus syndrome
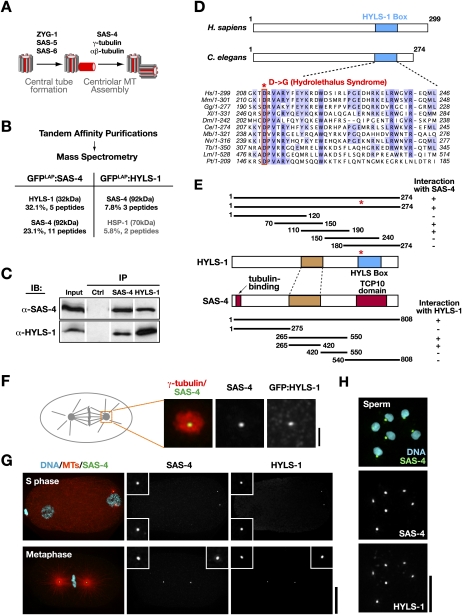
SAS-4 directs a late step in centriole assembly, the formation of the outer centriole wall (Fig. 1A). Depending on the differentiation/cell cycle state, the outer wall, composed of SAS-4 and the stabilized centriolar microtubules, mediates the acquisition of the pericentriolar matrix or directs cilia formation. To elucidate the molecular mechanisms underlying these dual functions, we performed tandem affinity purifications of GFPLAP:SAS-4 from worm extracts and identified copurifying proteins using mass spectrometry (Cheeseman and Desai 2005). This approach identified a single SAS-4-interacting protein encoded by the C05C8.9 predicted gene (Fig. 1B). Primary sequence analysis revealed that C05C8.9 encodes the C. elegans ortholog of HYLS1, a protein of unknown function mutated in the recessively inherited perinatal lethal developmental disorder hydrolethalus syndrome (Mee et al. 2005). We therefore named the C05C8.9 gene hyls-1 and refer to its protein product as HYLS-1. The interaction between HYLS-1 and SAS-4 was confirmed by coimmunoprecipitation using affinity-purified antibodies (Fig. 1C); reciprocal tandem affinity purifications of GFPLAP:HYLS-1 also identified SAS-4 (Fig. 1B). Database searches and sequence alignments indicated that HYLS-1 is a member of a protein family conserved from ciliates to humans defined by an ∼40-amino-acid C-terminal region that we refer to as the “HYLS-1 box” (Fig. 1D). Notably, HYLS-1-like proteins were not found in organisms that lack centrioles, such as fungi and higher plants. Hydrolethalus syndrome in humans is caused by a missense mutation that changes an invariant aspartic acid residue in the HYLS-1 box to glycine (Fig. 1D; Mee et al. 2005).
Figure 1.
HYLS-1 directly associates and colocalizes with the core centriolar structural protein SAS-4. (A) Schematic of centriole assembly pathway in C. elegans: The central tube, a cylindrical intermediate that templates centriole dimensions, assembles in S phase via an SAS-4-independent process. SAS-4 is recruited to the central tube and directs the assembly of the stabilized microtubules that constitute the outer centriole wall during mitotic prophase (Pelletier et al. 2006; Dammermann et al. 2008). (B) Results of mass spectrometry performed on tandem affinity preparations of SAS-4 and HYLS, listing all polypeptides above 5% coverage. (C) Immunoblot showing SAS-4 and HYLS-1 coimmunoprecipitate from embryo extracts. (D) Schematics of human and C. elegans HYLS-1, indicating position of conserved “HYLS-1 box.” Alignment shows HYLS-1 orthologs from vertebrates, Drosophila, C. elegans, and ciliates highlighting invariant aspartic acid mutated in hydrolethalus syndrome. (E) SAS-4 and HYLS-1 directly interact in a yeast two-hybrid assay via their middle portions (brown). For reference, the location of the conserved HYLS-1 box (blue), and the N-terminal tubulin-binding and C-terminal TCP10 motifs in SAS-4 (red), are also shown. The hydrolethalus-associated mutation does not affect interaction with SAS-4. (F) Centrosome from a metaphase embryo stained with antibodies to γ-tubulin as a marker for the pericentriolar material, SAS-4 and GFP. GFP:HYLS-1 colocalizes with SAS-4 at centrioles, which appear as a small focus in the center of the pericentriolar material. (G,H) HYLS-1 colocalizes with SAS-4 to centrioles throughout the embryonic cell cycle (G) and in sperm (H). Bars: F, 1 μm; G, 10 μm; H, 5 μm. Insets in G are magnified 3×.
With the exception of a common contaminant (HSP-1) no other proteins were copurified in the SAS-4 and HYLS-1 tandem affinity preps, suggesting that HYLS-1 interacts directly with SAS-4. Consistent with this, a robust interaction was observed between HYLS-1 and SAS-4 in yeast two-hybrid analysis (Fig. 1E). Deletion studies identified the regions of SAS-4 and HYLS-1 responsible for the interaction (Fig. 1E). The region of HYLS-1 that mediates the interaction with SAS-4 is distinct from the HYLS-1 box, in which the disease-linked mutation is located. Thus, hydrolethalus syndrome is not likely to be a consequence of disrupting the interaction between SAS-4 and HYLS-1.
Consistent with their biochemical association, a GFP:HYLS-1 fusion (Fig. 1F) as well as endogenous HYLS-1 (Fig. 1G,H), colocalized with SAS-4 to centrioles at the center of the pericentriolar material throughout the embryonic cell cycle as well as in mature sperm, where there is no pericentriolar material (Fig. 1F–H). We conclude that HYLS-1, whose human ortholog is linked to hydrolethalus syndrome, directly associates with and colocalizes with the core centriolar structural protein SAS-4.
HYLS-1 is stably incorporated into newly forming centrioles in an SAS-4-dependent step
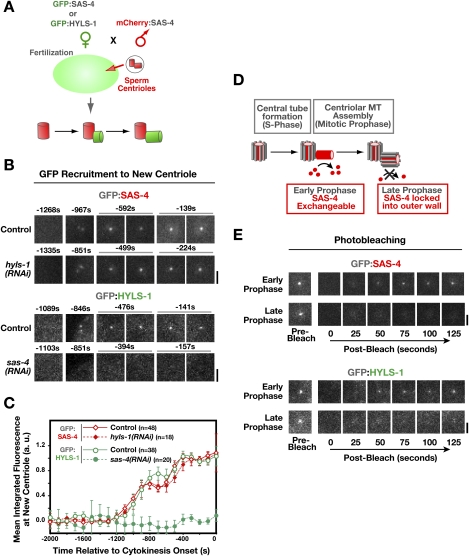
During centriole duplication in the C. elegans embryo, SAS-4 is recruited to the central tube in S phase. In prophase, SAS-4 directs assembly of the microtubules that constitute the outer centriole wall. Centriolar microtubule assembly prevents further exchange of SAS-4 with the cytoplasmic pool, irreversibly locking it into the outer wall of the new centriole (Dammermann et al. 2008). Using a previously developed method (Fig. 2A; Dammermann et al. 2008), we found that GFP:HYLS-1 was recruited to the site of new centriole assembly with timing identical to SAS-4 (Fig. 2B,C). Depletion of SAS-4 by RNAi abolished centriolar recruitment of HYLS-1, but not vice versa, placing HYLS-1 below SAS-4 in the centriole assembly pathway (Fig. 2B,C). Photobleaching experiments revealed that centriolar HYLS-1, like SAS-4, ceases to exchange with the cytoplasmic pool during late prophase (Fig. 2D,E), indicating that HYLS-1 is stably incorporated into the new centriole along with SAS-4 when the outer wall microtubules assemble. These in vivo results validate the interaction between SAS-4 and HYLS-1 observed using biochemistry and two-hybrid analysis and indicate that HYLS-1, like SAS-4, is stably incorporated into centrioles during their assembly, beginning with the first new centrioles formed after fertilization in the one-cell embryo.
Figure 2.
HYLS-1 is stably incorporated into newly forming centrioles in an SAS-4-dependent manner. (A) Schematic of the method used to analyze recruitment of centriolar proteins to site of centriole assembly (Dammermann et al. 2008). Briefly, sperm centrioles labeled with mCherry:SAS-4 were introduced by mating into oocytes expressing a GFP fusion with HYLS-1 or SAS-4. Recruitment of the GFP-labeled protein to newly forming centrioles was quantified by measuring the GFP fluorescence coincident with the mCherry signal at each time point. Measurements from multiple embryos were pooled and averaged to generate each recruitment plot. (B,C) Representative images (B) and quantification (C) of centriolar GFP:SAS-4 or GFP:HYLS-1 recruitment in control embryos and embryos depleted of HYLS-1 or SAS-4 as indicated. HYLS-1 recruitment requires SAS-4 but not vice versa. Times are in seconds relative to cytokinesis onset. Error bars are the 90% confidence interval. (D) SAS-4 is recruited to centrioles during the formation of the central tube in S phase, but remains in dynamic exchange with the cytoplasmic pool of SAS-4 until assembly of the centriolar microtubules in late prophase locks it into the outer centriole wall (Dammermann et al. 2008). (E) GFP:SAS-4 and GFP:HYLS-1 exhibit an identical behavior in a photobleaching-based assay assessing the ability of the centriolar protein to exchange with cytoplasmic pools. Recovery is observed if centrioles are bleached in early prophase (GFP:SAS-4, n = 8 out of 8; GFP:HYLS-1, n = 8 out of 8), but not if centrioles are bleached in late prophase (GFP:SAS-4, n = 1 out of 22; GFP:HYLS-1, n = 0 out of 9). Bars, 2 μm.
HYLS-1 is dispensable for centriole duplication, centrosome function in cell division, and embryonic viability
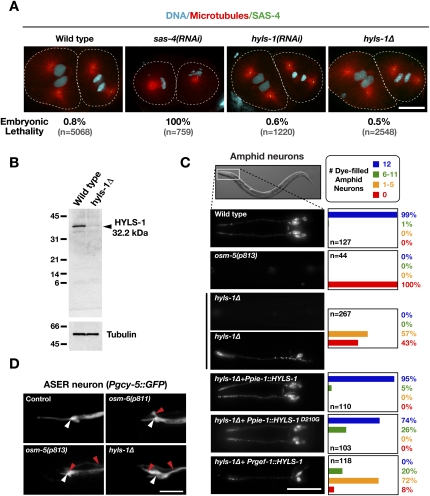
SAS-4 is required for the formation of the microtubule-containing outer centriole wall. Following its depletion in the early embryo, the two sperm-derived centrioles separate but fail to duplicate (Kirkham et al. 2003). Consequently, each daughter cell inherits a single centriole, leading to monopolar spindle formation in the second mitotic division and penetrant embryonic lethality. Surprisingly, despite the direct association of HYLS-1 with SAS-4 and their similar behavior during centriole assembly, HYLS-1-depleted embryos exhibited no defects in centriole duplication and no significant increase in embryonic lethality (Fig. 3A). To confirm this unexpected finding, we obtained a deletion mutant, hyls-1(tm3067) (Supplemental Fig. S1A), hereafter referred to as hyls-1Δ, which has no detectable HYLS-1 expression by immunoblot (Fig. 3B) or immunofluorescence (Supplemental Fig. S1C). In agreement with the RNAi results, hyls-1Δ embryos did not show centriole duplication defects, were viable, and developed into fertile adults (Fig. 3A). Thus, even though HYLS-1 is associated with SAS-4 and stably incorporated into centrioles during their assembly, it is dispensable for centriole duplication and centrosome function during cell division.
Figure 3.
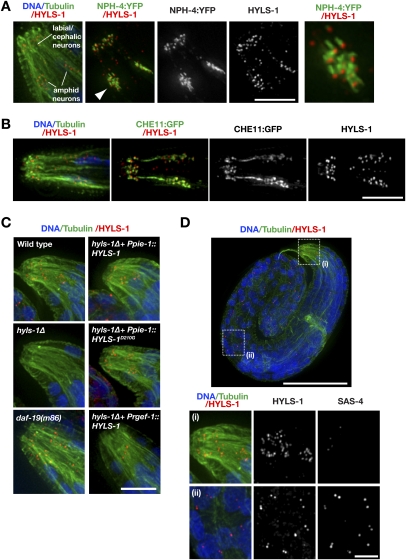
HYLS-1 is dispensable for centriole assembly and centrosome function, but is essential for ciliogenesis. (A) Embryos in second mitotic division stained for DNA, α-tubulin and SAS-4. The percentage embryonic lethality for each condition is indicated. (B) Immunoblot comparing wild-type and hyls-1Δ whole-worm extracts probed with an antibody raised against full-length HYLS-1. Arrowhead indicates wild-type HYLS-1, which runs slightly higher than its predicted molecular weight. A size of 16.7 kDa would be expected for mutant HYLS-1 based on initiation at the first (in-frame) ATG following the deleted region. (C) Representative images and quantification of DiI uptake in amphid neurons of wild-type and mutant animals. hyls-1Δ animals display a dye-fill defect—indicative of a failure of cilia assembly—that is fully rescued by wild-type HYLS1 and partially by HYLS-1D210G when expressed from a germline promoter (Ppie-1). Expression of HYLS-1 from a pan-neuronal promoter (Prgef-1) fails to restore ciliogenesis. (D) Cytoplasmic GFP expressed in the ASER amphid neuron using a cell-specific promoter (Yu et al. 1997). The position of the transition zone is marked (white arrowhead). GFP fills the dendrite (right of white arrowhead) and cilium (left of white arrowhead) of wild-type animals. The ASER neuron lacks ciliary projections in osm-5(p813) and osm-6(p811) IFT mutants as well as in the hyls-1Δ mutant. Red arrowheads indicate aberrant protrusions in cilia-defective mutants. Bars: A,D, 10 μm; C, 50 μm.
HYLS-1 is required for ciliogenesis
In addition to organizing centrosomes, centrioles serve as basal bodies that template cilia. To determine whether HYLS-1 contributes to cilia formation, we monitored ciliogenesis in hyls-1Δ animals using a dye-fill assay. Of the 60 ciliated neurons in C. elegans hermaphrodites, 12 amphid (head) and four phasmid (tail) neurons take up the lipophilic dye DiI through their exposed ciliated endings and accumulate it in their cell bodies (Supplemental Fig. S2A–C; Hedgecock et al. 1985). In hyls-1Δ animals, only ∼10% of the normal number of ciliated neurons were observed to dye-fill (Fig. 3C; Supplemental Fig. S2D). Neuron-specific GFP reporters confirmed normal presence of the amphid and phasmid neurons (Figs. 3D, 5A,B [below]), indicating that the dye-filling defect is not due to absence of the neurons themselves. Albeit infrequent, the low level of dye-filling in hyls-1Δ animals is in contrast to IFT-defective mutants, which completely lack dye-filling neurons. Placing the hyls-1Δ mutant allele over a deficiency did not reduce the number of residual cilia [1.2 ± 0.9 dyf(+) neurons in hyls-1Δ/Δ; 1.6 ± 1.4 hyls-1Δ/deficiency; n ∼ 35 animals per condition], excluding the possibility that the deletion mutant is hypomorphic. Residual cilia were not associated with a particular type of neuron and the dye-fill pattern was not heritable (data not shown). These results establish that HYLS-1 plays an important role in cilia assembly in C. elegans.
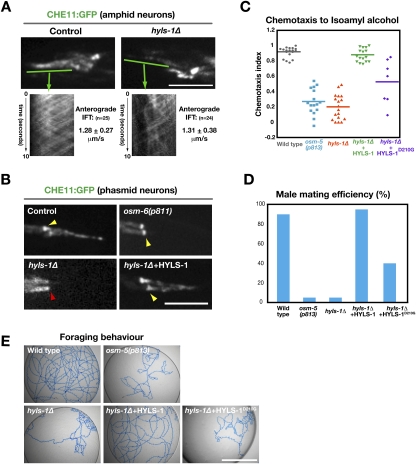
Figure 5.
HYLS-1 acts in an IFT-independent early step in ciliogenesis. (A) IFT is unchanged in residual cilia of hyls-1Δ animals. Stills from time-lapse sequences of amphid neurons expressing CHE-11:GFP (Supplemental Movies S1, S2) and kymographs showing IFT particle movement along ciliary axonemes. IFT rates were determined for particles in the distal segment. Note that there are eight ciliated neurons in each amphid bundle in wild-type, but only a single cilium in the hyls-1Δ mutant, explaining the difference in the number of particles. (B) CHE-11:GFP fails to accumulate at the transition zone of hyls-1Δ animals. Stills from time-lapse sequences of phasmid neurons expressing CHE-11:GFP. Arrowheads indicate position of transition zone. (C) Results of chemotaxis assays performed with wild-type and mutant animals. Each data point represents one assay with >100 animals. (D) Male mating efficiency of wild-type and mutant animals. The ability of individual L4 males to produce cross-progeny with unc-119 mutant hermaphrodites was scored (percentage of males producing crossprogeny; n = 20 per genotype). (E) Foraging behavior of wild-type and mutant animals. Tracks generated by a single animal left for 16 h on an NGM plate seeded with OP50 bacteria are shown. While wild-type animals efficiently explore their environment, cilia-defective mutant animals fail to do so, exhibiting a dwelling phenotype (Fujiwara et al. 2002). Bars: A,B, 5 μm; E, 1 cm.
Neuronal expression of HYLS-1 is not sufficient for ciliogenesis
The requirement for HYLS-1 in ciliogenesis but not cell division raises the question as to why HYLS-1 is incorporated into all centrioles, starting with those assembled in the first embryonic division. One possibility is that HYLS-1 must be present on centrioles to perform its function in initiating ciliogenesis. Since centrioles are distributed randomly to daughter cells in C. elegans embryos (our unpublished results), the assembly-coupled incorporation of HYLS-1 into all centrioles would guarantee its presence in the centrioles inherited by ciliated neurons. To test this idea, we generated strains carrying integrated transgenes directing expression of GFP:HYLS-1 under control of the pie-1 promoter, which directs expression in early embryos, and the rgef-1 promoter, which directs expression in all post-mitotic neurons (Altun-Gultekin et al. 2001), and compared their ability to rescue ciliogenesis in the hyls-1Δ mutant. Expression of GFP:HYLS-1 under the control of the pie-1 promoter fully rescued the dye-filling defect in the hyls-1Δ mutant (Fig. 3C; Supplemental Fig. S2D). By contrast, expression under the control of the rgef-1 promoter failed to rescue ciliogenesis in the majority of neurons (Fig. 3C; Supplemental Fig. S2D). These results suggest that the assembly-coupled incorporation of HYLS-1 into centrioles inherited by ciliated neurons is necessary for it to perform its role in ciliogenesis.
HYLS-1 is present at the base of mature cilia
To examine if HYLS-1 protein is present in mature cilia, we performed immunofluorescence of late (threefold stage) embryos and early (L1) larvae with antibodies to HYLS-1. HYLS-1 localized to punctate structures in amphid and labial/cephalic neurons (Fig. 4A–D). This punctate staining was absent in hyls-1Δ worms and was fully restored by pie-1 promoter-regulated HYLS-1 expression in the deletion mutant; by contrast, rgef-1 promoter-regulated neuronal expression of HYLS-1 in the deletion mutant resulted in a few puncta but failed to restore the wild-type pattern (Fig. 4C).
Figure 4.
HYLS-1 is present at the base of mature C. elegans cilia. (A) HYLS-1 localizes to the base of the cilium. Late (threefold)-stage embryo expressing the transition zone marker NPH-4:YFP stained for DNA, tubulin, and HYLS-1. The inset shows 3× magnified view of amphid bundle (arrowhead). (B) L1 larva expressing the IFT protein CHE-11:GFP stained for DNA, tubulin, and HYLS-1. (C) Expression of wild-type or disease mutant HYLS-1 from a germline promoter (Ppie-1) restores localization in hyls-1Δ animals, wheras expression from a pan-neuronal promoter (Prgef-1) fails to do so. Late-stage embryos stained for DNA, tubulin, and HYLS- 1. (D) SAS-4 is not present at the ciliary base (panel i), although it colocalizes with HYLS-1 at centrioles elsewhere in the animal (panel ii). Late-stage embryo stained for DNA, tubulin, HYLS-1, and SAS-4. Bars: A–C, 5 μm; D, top panel, 50 μm; D, panels i,ii, 10 μm.
To determine whether the HYLS-1 puncta are at the base of the cilia, we examined the localization of HYLS-1 relative to NPH-4:YFP, a marker for the ciliary transition zone (Williams et al. 2008). HYLS-1 puncta were closely apposed to the short stretches of NPH-4:YFP (Fig. 4A), indicating that HYLS-1 localizes to a discrete structure at the base of cilia. We confirmed this result using CHE-GFP, an IFT component that localizes to the transition zone and along the ciliary axoneme (Fig. 4B; Ou et al. 2005). Localization to the base of the cilium was also observed in phasmid neurons (Supplemental Fig. S2E). We conclude that HYLS-1 localizes to a discrete structure at the base of the mature cilium, consistent with its function in ciliogenesis.
HYLS-1 localization in ciliated neurons is independent of the master ciliogenesis regulator DAF-19
In C. elegans, ciliogenesis occurs during terminal differentiation in response to expression of the DAF-19 transcription factor, whose targets include the IFT machinery as well as the Bardet-Biedl and Meckel syndrome proteins (Swoboda et al. 2000). In daf-19(m86) mutants, the ciliogenesis program is not activated and cilia assembly fails (Perkins et al. 1986). However, HYLS-1 localization was unaffected (Fig. 4C). This finding is consistent with the observations that HYLS-1 is incorporated into all centrioles during their assembly and that expression of HYLS-1 from a pan-neuronal promoter fails to restore ciliogenesis in hyls-1Δ animals.
C. elegans is unusual in that mature cilia do not terminate in centrioles/basal bodies recognizable by electron microscopy. Instead, structures similar to the “transition zone” found above the basal body in other organisms anchor cilia to the plasma membrane (Inglis et al. 2007). As the initiation and early steps of ciliogenesis, which occur in the late embryo where cells are small and cell density is very high, have not been visualized using either light or electron microscopy, it is unclear when centriolar structure is lost during differentiation. Immunofluorescence of late embryos with antibodies to HYLS-1 and SAS-4 revealed puncta containing both SAS-4 and HYLS-1 throughout the body of the animal, likely corresponding to centrioles. By contrast, in the ciliated neurons of the head, puncta containing HYLS-1, but not SAS-4, were observed (Fig. 4D). Thus, in C. elegans, two interacting outer centriole wall components are uncoupled in mature cilia, with HYLS-1, but not SAS-4, present at the ciliary base. Elucidation of the timing of this uncoupling and the mechanisms underlying loss of centriolar structure will require methods to visualize the early steps of ciliogenesis. We conclude that in C. elegans HYLS-1 localization to the ciliary base is independent of the master ciliogenesis regulator DAF-19 and persists after loss of centriole structure in mature cilia.
HYLS-1 acts in a step functionally distinct from IFT
To determine how HYLS-1 contributes to ciliogenesis, we analyzed a GFP fusion with the IFT component CHE-11 (Fig. 5A,B; Supplemental Movies S1, S2; Ou et al. 2005). In the 10% residual cilia in hyls-1Δ animals, anterograde movement of CHE-11:GFP particles occurred at a similar rate to wild-type, indicating that HYLS-1 is dispensable for IFT (Fig. 5A). In wild-type and IFT-defective osm-6(p811) animals, CHE11:GFP accumulated at a focus, which is the transition zone at the base of the mature C. elegans cilium in the wild-type. By contrast, in hyls-1Δ animals, no CHE-11:GFP accumulation was seen in the majority of neurons (Fig. 5B). Thus, HYLS-1 is required to form a normal transition zone, but not for the IFT-dependent extension of the ciliary axoneme.
Ciliogenesis and cilia-dependent behaviors are compromised by the disease-associated missense mutation in the HYLS-1 box
Cilia, while dispensable for viability and fertility in C. elegans, are required for a variety of behaviors including chemotaxis, foraging, and male mating. Consistent with their severe cilia assembly defect, the performance of hyls-1Δ animals in behavioral assays was similar to that of other ciliary mutants (Fig. 5C–E). Expression of a GFP fusion with wild-type HYLS-1 rescued the dye-fill defect of hyls-1Δ worms as well as all three analyzed cilia-dependent behaviors. To examine the functional consequences of the hydrolethalus syndrome-associated mutation, we generated a GFP fusion with HYLS-1 containing the D210G mutation in the HYLS-1 box. Mutant and wild-type proteins were expressed at similar levels (Supplemental Fig. S1D), and both localized to centrioles (Supplemental Fig. S1C) and the base of cilia (Fig. 4C), consistent with the finding that the region of HYLS-1 that interacts with SAS-4 is distinct from the HYLS-1 box (Fig. 1E). However, mutant HYLS-1 was compromised in its ability to completely restore cilia assembly in the hyls-1 deletion (Fig. 3C; Supplemental Fig. S2D). In behavioral assays, the disease-associated mutation exhibited partial rescue, with the extent of rescue scaling inversely with the complexity of the analyzed behavior (Fig. 5C–E). We conclude that the hydrolethalus syndrome-associated mutation does not alter the interaction of HYLS-1 with SAS-4 or its localization to centrioles but compromises HYLS-1 function in ciliogenesis.
The role of HYLS1 in ciliogenesis is conserved in vertebrates
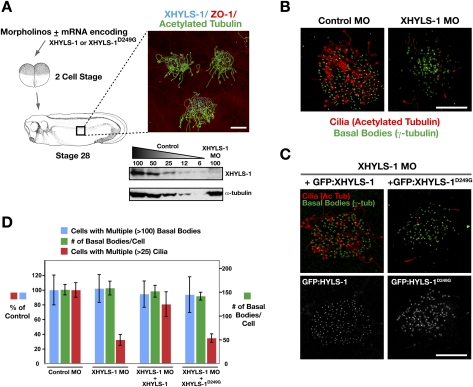
To determine if the function of HYLS-1 in ciliogenesis is conserved in vertebrates, we turned to Xenopus laevis as an experimental model. The epidermis of amphibian larvae is a mucociliary epithelium, with multiciliated cells interspersed among nonciliated mucus-secreting cells. Antisense morpholino oligonucleotides, injected on their own or with morpholino-resistant mRNAs, can be used to examine the requirement of specific factors for cilia assembly or function (Fig. 6A; Mitchell et al. 2007; Park et al. 2008).
Figure 6.
The role of HYLS-1 in ciliogenesis is conserved in vertebrates. (A) Schematic of Xenopus experiments. Inset shows ciliated epithelial cells stained for XHYLS-1, cilia (acetylated tubulin), and apical cell junctions (ZO-1). Immunoblot of control and XHYLS-1 morpholino (MO) tailbud-stage embryo extracts. (B) Basal bodies (γ-tubulin; green) are present, but cilia (acetylated tubulin; red) fail to assemble in XHYLS-1-depleted embryos. (C) Expression of wild-type but not disease mutant (D249G) XHYLS-1 fused to GFP restores ciliogenesis in XHYLS-1-depleted embryos. (D) Quantification of morpholino experiments. n = 10–12 embryos, 30 fields of view per embryo (cells with multiple cilia/basal bodies); 20 cells (basal bodies per cell). Bars, 10 μm. Error bars in D are the 95% confidence interval.
Morpholino-based depletion of XHYLS-1 resulted in a specific loss of cilia, with no effect on the number of cells with multiple basal bodies or the number of basal bodies per cell (Fig. 6B,D). Thus, as in C. elegans, Xenopus HYLS-1 is dispensable for centriole assembly and cell division, but is specifically required for cilia formation. GFP fusions with both wild-type and disease mutant XHYLS-1, expressed using mRNA injections into two-cell embryos, localized to basal bodies in multiciliated cells (Fig. 6C); however, only the wild-type GFP:XHYLS-1 restored ciliogenesis in embryos depleted of endogenous XHYLS-1 (Fig. 6C,D). Thus, HYLS-1 is required for ciliogenesis in vertebrate multiciliated epithelia, indicating that its function is conserved between C. elegans and vertebrates. In addition, the disease-associated mutation has a more severe effect on cilia assembly in multiciliated epithelia, which may reflect a species difference and/or the increased burden associated with building >150 cilia per cell.
HYLS1 is required for the apical organization of basal bodies
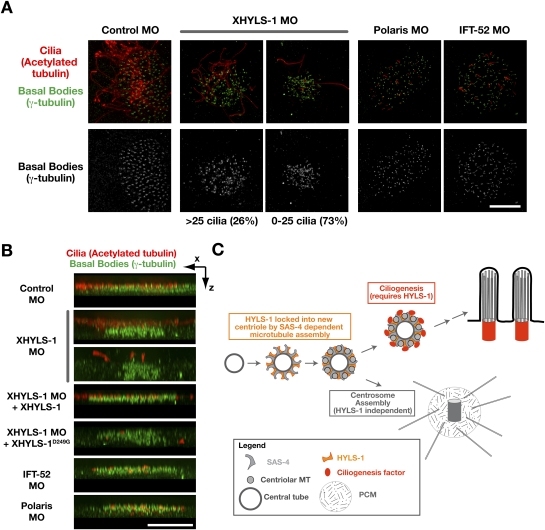
In epithelia, cilia extend from basal bodies that are evenly distributed over the apical cell surface. In XHYLS-1 depletions, basal bodies were clustered and spatially disorganized within the cell, with the most noticeable defect being a lack of apical localization. This defect was most clearly evident in x–z projections of three-dimensional (3D) image stacks (Fig. 7A,B). Apical localization was restored upon expression of wild-type but not disease mutant XHYLS-1 (Fig. 7B). Importantly, the tight junction marker ZO-1 retained its normal apical localization (Supplemental Fig. S3), indicating that depletion of XHYLS-1 did not affect overall apico–basal polarity. To test if the defect in basal body organization reflected a specific function of HYLS-1 in ciliogenesis or was a secondary consequence of the loss of cilia, we depleted the IFT components Polaris and IFT-52. In both IFT component depletions, cilia assembly was inhibited to a similar extent as following XHYLS-1 depletion, but basal bodies remained evenly distributed and in close apposition to the apical cell surface (Fig. 7A,B). We conclude that HYLS-1 is stably incorporated into centrioles during their assembly and acts in an early step in ciliogenesis, likely the apical targeting and/or anchoring of basal bodies, prior to IFT-dependent extension of the ciliary axoneme.
Figure 7.
XHYLS-1 is required for apical targeting/anchoring of basal bodies. (A) Representative images of control embryos and embryos depleted of XHYLS-1, Polaris or IFT-52, stained with antibodies to acetylated tubulin and γ-tubulin to illustrate effect on basal body distribution. Images are x–y projections of deconvolved 3D data stacks. (B) x–z projection of 3D data stacks presented in A and Figure 6C. (C) Model for the role of HYLS-1 in cilia formation. HYLS-1 is recruited to the forming centriole by its association with SAS-4, and like SAS-4 is locked into the centriole during the assembly of centriolar microtubules. Incorporation of HYLS-1 confers on centrioles the capacity to initiate ciliogenesis. Bars, 10 μm.
Discussion
HYLS1 acts as a centriolar scaffold for cilia assembly
It has long been appreciated that a primary role of centrioles, conserved in all species that contain them, is to generate cilia. Our analysis indicates that HYLS-1 is a core centriolar protein that is stably incorporated into the outer wall during centriole assembly specifically to confer on centrioles the capacity to initiate ciliogenesis (Fig. 7C). A defect in this capacity leads to a lethal human developmental disorder, hydrolethalus syndrome. HYLS-1 is conserved in evolution from ciliates to vertebrates, indicating that ciliogenesis in all eukaryotes is likely to require a centriole-intrinsic mechanism involving HYLS-1.
Hydrolethalus syndrome is a cilia-related disorder
The single amino acid substitution in HYLS1 linked to hydrolethalus syndrome impairs its function in ciliogenesis, indicating that this developmental disorder is a previously unrecognized ciliopathy. Despite the severity of hydrolethalus syndrome, our results suggest that the hydrolethalus-associated missense mutation, which changes an invariant aspartic acid in the conserved HYLS-1 box to glycine, is not a null mutation. HYLS-1 containing the hydrolethalus mutation is stable, associates with SAS-4, and targets to centrioles/basal bodies in both C. elegans and Xenopus. In the sensory (primary) cilia of C. elegans, the hydrolethalus mutation only mildly compromised rescue of the ciliary dye-fill phenotype of the hyls-1 deletion, although complex cilia-dependent behaviors were significantly perturbed. In contrast, in the multiciliated epithelium of Xenopus embryos, the mutant protein exhibited little ability to rescue cilia formation. The difference between the consequences of the disease-associated mutation in C. elegans and Xenopus may reflect either a species difference, the nature of the cilia assayed (primary sensory cilia in C. elegans, motile cilia in Xenopus), or the two orders of magnitude difference in the number of cilia assembled per cell (one in C. elegans neurons; ∼150 per ciliated epithelial cell). While future work is needed to discriminate between these possibilities, the strong defect associated with the disease mutation in multiciliated vertebrate epithelia may account for the severity of hydrolethalus syndrome.
The clinical features of hydrolethalus syndrome are most similar to those of Meckel-Gruber syndrome (Salonen and Herva 1990). Both are characterized by a majority of affected individuals dying in utero, CNS malformations, and polydactyly, although hydrolethalus syndrome does not share the cystic dysplasia of the kidneys or fibrous changes of the liver associated with Meckel-Gruber syndrome. Meckel-Gruber syndrome is related to two less severe ciliopathies, nephronophthisis and Joubert syndrome, in that these three phenotypically distinct disorders are linked to different lesions in the same genetic loci. These loci include MKS3/JBTS6 (Baala et al. 2007b), NPHP1/JBTS4 (Parisi et al. 2004), MKS4/NPHP6/JBTS5 (Sayer et al. 2006; Valente et al. 2006; Baala et al. 2007a), and MKS5/NPHP8/JBTS7 (Arts et al. 2007; Delous et al. 2007). Notably, the genes currently identified for Meckel-Gruber syndrome, nephronophthisis, and Joubert syndrome are only mutated in a small percentage of affected individuals, suggesting that other loci responsible for these disorders remain to be identified (Khaddour et al. 2007; Hildebrandt et al. 2009). It is possible that mutations in HYLS1, distinct from the one linked to hydrolethalus syndrome, are associated with these or other ciliary disorders. Future efforts to sequence the HYLS1 locus in affected individuals should help address this possibility.
HYLS-1 acts in an early step in the ciliogenesis pathway
The apical migration and anchoring of centrioles at the cell surface is a critical initial step in ciliogenesis. Whether the centriole plays an active role in apical migration and anchoring has until now been unclear; our results suggest that HYLS-1 provides a centriole-intrinsic mechanism that interfaces with other processes to organize basal bodies at the cell surface and support elongation of the ciliary axoneme. Perturbations of apico–basal polarity and the cortical actin cytoskeleton have been shown to disrupt apical targeting of basal bodies (Boisvieux-Ulrich et al. 1990; Pan et al. 2007; Park et al. 2008). Early EM studies also suggested potential involvement of vesicle trafficking (Sorokin 1962) and components of the exocyst, a complex involved in tethering vesicles to the plasma membrane, have been localized recently to basal bodies (Park et al. 2008; Zuo et al. 2009).
Elucidating the molecular mechanism by which HYLS-1 contributes to basal body organization at the cell surface and helps initiate cilia formation will require identification of its effectors. The proteins mutated in Meckel syndrome, Joubert syndrome, and Nephronophtisis are good candidates for downstream effectors of HYLS-1 in the ciliogenesis pathway. Homologs of three of the gene products linked to Meckel-Gruber syndrome and nephronophthisis in humans, MKS-1, NPH-1 and NPH-4, have been characterized in C. elegans. MKS-1 contains a B9 domain that is found in two other proteins, called TZA-1 and TZA-2. All three B9 domain proteins as well as the nephrocystins NPH-1 and NPH-4 localize to the transition zone in ciliated neurons. Unlike HYLS-1, expression of the nephrocystins, MKS-1, TZA-1, and TZA-2 is under control of DAF-19 (Winkelbauer et al. 2005; Williams et al. 2008), supporting the idea that HYLS-1 acts upstream of these components. While individual inhibition of MKS-1, TZA-1, TZA-2, NPH-1, or NPH-4 does not result in a significant defect in cilia morphology (Jauregui and Barr 2005; Winkelbauer et al. 2005; Jauregui et al. 2008; Williams et al. 2008; Bialas et al. 2009), simultaneous inhibition of a B9 domain-containing protein and either NPH-1 or NPH-4 results in a dye-filling defect reminiscent of HYLS-1 inhibition (Williams et al. 2008). The reasons for the functional redundancy between these components are currently unclear. Future work is needed to understand the basis for this redundancy and to determine whether there are functional interactions between these ciliogenesis-specific proteins and HYLS-1 in the early stages of the ciliogenesis pathway.
Materials and methods
LAP purifications, immunoprecipitations, and yeast two-hybrid
LAP preps were performed on high-speed supernatants from whole-worm extracts as described (Cheeseman and Desai 2005) and interacting proteins were identified by LC/MS/MS. Immunoprecipitations were performed on embryo extracts as described previously (Desai et al. 2003). Yeast two-hybrid experiments were performed using the Matchmaker two-hybrid system (Clontech).
Sequence analysis
HYLS-1 orthologs were identified by PSI-BLAST using the conserved sequence shown in Figure 1D (which extends from amino acid 205 to 246 in human HYLS-1) as query. The Xenopus ortholog was cloned by PCR from cDNA prepared from tailbud-stage embryos. Sequence alignments were performed with MUSCLE in Jalview (http://www.jalview.org). Accession numbers are as follows: Hs (Homo sapiens) NP_659451; Mm (Mus musculus) AAH35953; Xl (X. laevis) UniGene Xl.13199; Gg (Gallus gallus) XP_001231543; Dm (Drosophila melanogaster) CG42231 (FlyBase); Ce (C. elegans) NP_504840; Mb (Monosiga brevicollis) EDQ89544; Nv (Nematostella vectensis) XP_001640703; Tb (Trypanosoma brucei) XP_803779; Lm (Leishmania major) XP_001687472; Pt (Paramecium tetraurelia) XP_001437786.
C. elegans experiments
Worm strains are listed in Supplemental Table S1. The hyls-1 deletion mutant obtained from the C. elegans knockout consortium (NBP-Japan) was outcrossed six times against N2 wild-type before phenotypic analysis and introduction of GFP markers. Strains were maintained at 16°C (OD19 and OD113) or 22°C (all others). RNAi experiments were performed as described previously (Dammermann et al. 2008) using the dsRNAs listed in Supplemental Table S2. Recruitment curves, high-resolution imaging, and FRAP analyses were performed as described previously (Dammermann et al. 2008). Immunofluorescence experiments on early and late embryos were performed using directly labeled affinity-purified antibodies to α-tubulin (DM1α-FITC, Sigma), SAS-4 (Dammermann et al. 2004), and HYLS-1 (full-length) (this study) as described previously (Oegema et al. 2001). Immunoblots were performed on whole-worm extracts with affinity-purified antibodies to full-length HYLS-1 or GFP, using anti-α-tubulin as a loading control (DM1α, Sigma).
C. elegans ciliary assays
Dye-fill assays were carried out by incubating ∼100 worms in a 1/500 dilution of DiI (Invitrogen) in M9 buffer for 1 h, allowing worms to destain on a seeded NGM plate for ∼30 min before imaging using a 20× 0.75 NA PlanApochromat lens on a Nikon E800 upright microscope equipped with a Hamamatsu Orca ER CCD camera. To monitor IFT and individual neurons, worms were anaesthetized with 10 mM levamisole, mounted on agarose pads and imaged using a 100×, 1.4 NA PlanApochromat lens and 1.5× optivar on a Nikon TE2000-E inverted microscope equipped with a krypton–argon 2.5-W water-cooled laser (Spectra-Physics) and an iXon electron multiplication back-thinned CCD camera (Andor Technology). Kymographs and movies were generated using Metamorph software. Chemotaxis assays were performed as described previously (Bargmann et al. 1993). To assay male mating efficiency of cilia mutant strains, males were generated using him-8 RNAi to induce X-chromosome missegregation in the parental generation.
Xenopus experiments
Xenopus embryos were obtained by in vitro fertilization according to standard methods and staged as described (Nieuwkoop and Faber 1967). Morpholino antisense oligonucleotides (GeneTools) were designed to inhibit expression of XHYLS-1, IFT52, and Polaris by targeting the initiation codon of the target mRNA. In the case of XHYLS-1, two morpholinos were necessary to target the two forms encoded in the pseudotetraploid Xenopus genome. In the case of IFT52 and Polaris, one morpholino was sufficient. The sequences were as follows: control, 5′-GGATGGTGCAGAGTCTCCATCAGTA-3′; XHYLS-1.1, 5′-GAACTGCCTGTCTCGAAGTGACATG-3′; XHYLS-1.2, 5′-GAACTGCCTGTCTCTCAGTGACATG-3′; IFT52, 5′-AAGCAATCTGTTTGTTGACTCCCAT-3′; Polaris, 5′-GCACGAGATGGACATTTTGCATCAT-3′.
For expression of GFP:XHYLS-1, full-length XHYLS-1 coding sequence was inserted into pCS2+ and fused to GFP at the N terminus. A mutant form corresponding to the single amino acid substitution found in hydrolethalus syndrome (D249G) was generated by site-directed mutagenesis of the wild-type template. To avoid interference with morpholinos targeting the endogenous message, the 5′ end of XHYLS-1 was modified using alternate codons to remove basepair complementarity while preserving amino acid identity. RNA was prepared from the pCS2 template by in vitro transcription using SP6 (Promega). Morpholinos and RNAs were injected separately into each blastomere of two-cell stage embryos using a total of 50–75 ng of morpholino and 1–5 ng RNA per embryo and cultured until stage 28 (tailbud stage).
Xenopus immunofluorescence and immunoblotting
Tailbudstage (stage 28) embryos were fixed in 4% paraformaldehyde in PBS for 2 h, followed by 48 h in Dent's (80% methanol, 20% DMSO) at −20°C. Embryos were rehydrated into PBS and blocked for 1 h in 4% BSA in PBS 0.1% Triton before incubation with primary antibodies overnight at 4°C. Embryos were then washed multiple times before incubation with secondary antibodies overnight. After multiple washes, embryos were mounted for examination. Antibodies used were mouse monoclonal anti-acetylated tubulin (T6793, Sigma), mouse monoclonal anti-γ-tubulin (GTU-88, Sigma) and rabbit anti-ZO-1 (61-7300, Invitrogen). Fluorescent secondary antibodies were purchased from Jackson Laboratories. Goat anti-mouse Fab fragments (Jackson Laboratoires) were used to present γ-tubulin antibody as goat before incubation with acetylated tubulin antibody, permitting double labeling with these two mouse antibodies. 3D wide-field data sets were collected using a 100× 1.3 NA PlanApochromat lens on a DeltaVision microscope and were computationally deconvolved and projected using SoftWorx software (Applied Precision). To determine depletion of XHYLS-1 following morpholino treatment, extracts prepared from tailbud-stage embryos were probed with affinity-purified antibodies to full-length XHYLS-1 (generated as described previously; Oegema et al. 2001), with anti-α-tubulin as a loading control (DM1α, Sigma).
Acknowledgments
We thank members of the Oegema and Desai laboratories for discussions, and Shohei Mitani (Tokyo Women's Medical University, Japan), the Caenorhabditis Genetics Center (University of Minnesota, MN), Maureen Barr (Rutgers University, NY), and Bradley Yoder (University of Alabama at Birmingham, AL) for strains. This work was supported by funding from the Ludwig Institute for Cancer Research to K.O. and A.B.D., and the National Institutes of Health to the Yeast Resource Center, C.K., and K.O. K.O. is a Pew Scholar in the Biomedical Sciences.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1810409.
Supplemental material is available at http://www.genesdev.org.
References
- Afzelius BA. Cilia-related diseases. J Pathol. 2004;204:470–477. doi: 10.1002/path.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altun-Gultekin Z, Andachi Y, Tsalik EL, Pilgrim D, Kohara Y, Hobert O. A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development. 2001;128:1951–1969. doi: 10.1242/dev.128.11.1951. [DOI] [PubMed] [Google Scholar]
- Arts HH, Doherty D, van Beersum SE, Parisi MA, Letteboer SJ, Gorden NT, Peters TA, Marker T, Voesenek K, Kartono A, et al. Mutations in the gene encoding the basal body protein RPGRIP1L, a nephrocystin-4 interactor, cause Joubert syndrome. Nat Genet. 2007;39:882–888. doi: 10.1038/ng2069. [DOI] [PubMed] [Google Scholar]
- Baala L, Audollent S, Martinovic J, Ozilou C, Babron MC, Sivanandamoorthy S, Saunier S, Salomon R, Gonzales M, Rattenberry E, et al. Pleiotropic effects of CEP290 (NPHP6) mutations extend to Meckel syndrome. Am J Hum Genet. 2007a;81:170–179. doi: 10.1086/519494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baala L, Romano S, Khaddour R, Saunier S, Smith UM, Audollent S, Ozilou C, Faivre L, Laurent N, Foliguet B, et al. The Meckel-Gruber syndrome gene, MKS3, is mutated in Joubert syndrome. Am J Hum Genet. 2007b;80:186–194. doi: 10.1086/510499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: An emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Bialas NJ, Inglis PN, Li C, Robinson JF, Parker JD, Healey MP, Davis EE, Inglis CD, Toivonen T, Cottell DC, et al. Functional interactions between the ciliopathy-associated Meckel syndrome 1 (MKS1) protein and two novel MKS1-related (MKSR) proteins. J Cell Sci. 2009;122:611–624. doi: 10.1242/jcs.028621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvieux-Ulrich E, Laine MC, Sandoz D. Cytochalasin D inhibits basal body migration and ciliary elongation in quail oviduct epithelium. Cell Tissue Res. 1990;259:443–454. doi: 10.1007/BF01740770. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci STKE. 2005;2005:pl1. doi: 10.1126/stke.2662005.pl1. [DOI] [PubMed] [Google Scholar]
- Chen J, Knowles HJ, Hebert JL, Hackett BP. Mutation of the mouse hepatocyte nuclear factor/forkhead homologue 4 gene results in an absence of cilia and random left–right asymmetry. J Clin Invest. 1998;102:1077–1082. doi: 10.1172/JCI4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A, Muller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev Cell. 2004;7:815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Dammermann A, Maddox PS, Desai A, Oegema K. SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the γ-tubulin-mediated addition of centriolar microtubules. J Cell Biol. 2008;180:771–785. doi: 10.1083/jcb.200709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe HR, Farr H, Gull K. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J Cell Sci. 2007;120:7–15. doi: 10.1242/jcs.03305. [DOI] [PubMed] [Google Scholar]
- Delattre M, Leidel S, Wani K, Baumer K, Bamat J, Schnabel H, Feichtinger R, Schnabel R, Gonczy P. Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat Cell Biol. 2004;6:656–664. doi: 10.1038/ncb1146. [DOI] [PubMed] [Google Scholar]
- Delattre M, Canard C, Gonczy P. Sequential protein recruitment in C. elegans centriole formation. Curr Biol. 2006;16:1844–1849. doi: 10.1016/j.cub.2006.07.059. [DOI] [PubMed] [Google Scholar]
- Delous M, Baala L, Salomon R, Laclef C, Vierkotten J, Tory K, Golzio C, Lacoste T, Besse L, Ozilou C, et al. The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat Genet. 2007;39:875–881. doi: 10.1038/ng2039. [DOI] [PubMed] [Google Scholar]
- Desai A, Rybina S, Muller-Reichert T, Shevchenko A, Hyman A, Oegema K. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes & Dev. 2003;17:2421–2435. doi: 10.1101/gad.1126303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M, Sengupta P, McIntire SL. Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cGMP-dependent protein kinase. Neuron. 2002;36:1091–1102. doi: 10.1016/s0896-6273(02)01093-0. [DOI] [PubMed] [Google Scholar]
- Hamill DR, Severson AF, Carter JC, Bowerman B. Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev Cell. 2002;3:673–684. doi: 10.1016/s1534-5807(02)00327-1. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Culotti JG, Thomson JN, Perkins LA. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev Biol. 1985;111:158–170. doi: 10.1016/0012-1606(85)90443-9. [DOI] [PubMed] [Google Scholar]
- Hildebrandt F, Attanasio M, Otto E. Nephronophthisis: Disease mechanisms of a ciliopathy. J Am Soc Nephrol. 2009;20:23–35. doi: 10.1681/ASN.2008050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis PN, Ou G, Leroux MR, Scholey JM. The sensory cilia of Caenorhabditis elegans. WormBook. 2007;8:1–22. doi: 10.1895/wormbook.1.126.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauregui AR, Barr MM. Functional characterization of the C. elegans nephrocystins NPHP-1 and NPHP-4 and their role in cilia and male sensory behaviors. Exp Cell Res. 2005;305:333–342. doi: 10.1016/j.yexcr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Jauregui AR, Nguyen KC, Hall DH, Barr MM. The Caenorhabditis elegans nephrocystins act as global modifiers of cilium structure. J Cell Biol. 2008;180:973–988. doi: 10.1083/jcb.200707090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaddour R, Smith U, Baala L, Martinovic J, Clavering D, Shaffiq R, Ozilou C, Cullinane A, Kyttala M, Shalev S, et al. Spectrum of MKS1 and MKS3 mutations in Meckel syndrome: A genotype–phenotype correlation. Mutation in brief #960. Online. Hum Mutat. 2007;28:523–524. doi: 10.1002/humu.9489. [DOI] [PubMed] [Google Scholar]
- Kirkham M, Muller-Reichert T, Oegema K, Grill S, Hyman AA. SAS-4 Is a C. elegans centriolar protein that controls centrosome size. Cell. 2003;112:575–587. doi: 10.1016/s0092-8674(03)00117-x. [DOI] [PubMed] [Google Scholar]
- Leidel S, Gonczy P. SAS-4 is essential for centrosome duplication in C. elegans and is recruited to daughter centrioles once per cell cycle. Dev Cell. 2003;4:431–439. doi: 10.1016/s1534-5807(03)00062-5. [DOI] [PubMed] [Google Scholar]
- Leidel S, Delattre M, Cerutti L, Baumer K, Gonczy P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat Cell Biol. 2005;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- Lucas EP, Raff JW. Maintaining the proper connection between the centrioles and the pericentriolar matrix requires Drosophila centrosomin. J Cell Biol. 2007;178:725–732. doi: 10.1083/jcb.200704081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF. What is the function of centrioles? J Cell Biochem. 2007;100:916–922. doi: 10.1002/jcb.21117. [DOI] [PubMed] [Google Scholar]
- Mee L, Honkala H, Kopra O, Vesa J, Finnila S, Visapaa I, Sang TK, Jackson GR, Salonen R, Kestila M, et al. Hydrolethalus syndrome is caused by a missense mutation in a novel gene HYLS1. Hum Mol Genet. 2005;14:1475–1488. doi: 10.1093/hmg/ddi157. [DOI] [PubMed] [Google Scholar]
- Mitchell B, Jacobs R, Li J, Chien S, Kintner C. A positive feedback mechanism governs the polarity and motion of motile cilia. Nature. 2007;447:97–101. doi: 10.1038/nature05771. [DOI] [PubMed] [Google Scholar]
- Moritz M, Zheng Y, Alberts BM, Oegema K. Recruitment of the γ-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. J Cell Biol. 1998;142:775–786. doi: 10.1083/jcb.142.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcia NS, Richards WG, Yoder BK, Mucenski ML, Dunlap JR, Woychik RP. The Oak Ridge Polycystic Kidney (orpk) disease gene is required for left–right axis determination. Development. 2000;127:2347–2355. doi: 10.1242/dev.127.11.2347. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. North-Holland, Amsterdam: 1967. Normal table of Xenopus laevis. [Google Scholar]
- O'Connell KF, Caron C, Kopish KR, Hurd DD, Kemphues KJ, Li Y, White JG. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 2001;105:547–558. doi: 10.1016/s0092-8674(01)00338-5. [DOI] [PubMed] [Google Scholar]
- Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G, Blacque OE, Snow JJ, Leroux MR, Scholey JM. Functional coordination of intraflagellar transport motors. Nature. 2005;436:583–587. doi: 10.1038/nature03818. [DOI] [PubMed] [Google Scholar]
- Pan J, You Y, Huang T, Brody SL. RhoA-mediated apical actin enrichment is required for ciliogenesis and promoted by Foxj1. J Cell Sci. 2007;120:1868–1876. doi: 10.1242/jcs.005306. [DOI] [PubMed] [Google Scholar]
- Parisi MA, Bennett CL, Eckert ML, Dobyns WB, Gleeson JG, Shaw DW, McDonald R, Eddy A, Chance PF, Glass IA. The NPHP1 gene deletion associated with juvenile nephronophthisis is present in a subset of individuals with Joubert syndrome. Am J Hum Genet. 2004;75:82–91. doi: 10.1086/421846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TJ, Mitchell BJ, Abitua PB, Kintner C, Wallingford JB. Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat Genet. 2008;40:871–879. doi: 10.1038/ng.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L, O'Toole E, Schwager A, Hyman AA, Muller-Reichert T. Centriole assembly in Caenorhabditis elegans. Nature. 2006;444:619–623. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Jensen CG, Jensen LC. The resorption of primary cilia during mitosis in a vertebrate (PtK1) cell line. J Ultrastruct Res. 1979;68:173–185. doi: 10.1016/s0022-5320(79)90152-7. [DOI] [PubMed] [Google Scholar]
- Salonen R, Herva R. Hydrolethalus syndrome. J Med Genet. 1990;27:756–759. doi: 10.1136/jmg.27.12.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayer JA, Otto EA, O'Toole JF, Nurnberg G, Kennedy MA, Becker C, Hennies HC, Helou J, Attanasio M, Fausett BV, et al. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat Genet. 2006;38:674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- Schnackenberg BJ, Khodjakov A, Rieder CL, Palazzo RE. The disassembly and reassembly of functional centrosomes in vitro. Proc Natl Acad Sci. 1998;95:9295–9300. doi: 10.1073/pnas.95.16.9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey JM. Intraflagellar transport motors in cilia: Moving along the cell's antenna. J Cell Biol. 2008;180:23–29. doi: 10.1083/jcb.200709133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin S. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J Cell Biol. 1962;15:363–377. doi: 10.1083/jcb.15.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda P, Adler HT, Thomas JH. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol Cell. 2000;5:411–421. doi: 10.1016/s1097-2765(00)80436-0. [DOI] [PubMed] [Google Scholar]
- Valente EM, Silhavy JL, Brancati F, Barrano G, Krishnaswami SR, Castori M, Lancaster MA, Boltshauser E, Boccone L, Al-Gazali L, et al. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat Genet. 2006;38:623–625. doi: 10.1038/ng1805. [DOI] [PubMed] [Google Scholar]
- Williams CL, Winkelbauer ME, Schafer JC, Michaud EJ, Yoder BK. Functional redundancy of the B9 proteins and nephrocystins in Caenorhabditis elegans ciliogenesis. Mol Biol Cell. 2008;19:2154–2168. doi: 10.1091/mbc.E07-10-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelbauer ME, Schafer JC, Haycraft CJ, Swoboda P, Yoder BK. The C. elegans homologs of nephrocystin-1 and nephrocystin-4 are cilia transition zone proteins involved in chemosensory perception. J Cell Sci. 2005;118:5575–5587. doi: 10.1242/jcs.02665. [DOI] [PubMed] [Google Scholar]
- Yu S, Avery L, Baude E, Garbers DL. Guanylyl cyclase expression in specific sensory neurons: A new family of chemosensory receptors. Proc Natl Acad Sci. 1997;94:3384–3387. doi: 10.1073/pnas.94.7.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Guo W, Lipschutz JH. The exocyst protein Sec10 is necessary for primary ciliogenesis and cystogenesis in vitro. Mol Biol Cell. 2009;20:2522–2529. doi: 10.1091/mbc.E08-07-0772. [DOI] [PMC free article] [PubMed] [Google Scholar]