Abstract
Vascular injury triggers dedifferentiation and cytoskeletal remodeling of smooth muscle cells (SMCs), culminating in vessel occlusion. Serum response factor (SRF) and its coactivator, myocardin, play a central role in the control of smooth muscle phenotypes by regulating the expression of cytoskeletal genes. We show that SRF and myocardin regulate a cardiovascular-specific microRNA (miRNA) cluster encoding miR-143 and miR-145. To assess the functions of these miRNAs in vivo, we systematically deleted them singly and in combination in mice. Mice lacking both miR-143 and miR-145 are viable and do not display overt abnormalities in smooth muscle differentiation, although they show a significant reduction in blood pressure due to reduced vascular tone. Remarkably, however, neointima formation in response to vascular injury is profoundly impeded in mice lacking these miRNAs, due to disarray of actin stress fibers and diminished migratory activity of SMCs. These abnormalities reflect the regulation of a cadre of modulators of SRF activity and actin dynamics by miR-143 and miR-145. Thus, miR-143 and miR-145 act as integral components of the regulatory network whereby SRF controls cytoskeletal remodeling and phenotypic switching of SMCs during vascular disease.
Keywords: MicroRNA, vascular disease, actin signaling, serum response factor, myocardin
Smooth muscle cells (SMCs) are highly plastic and modulate their phenotype in response to physiological and pathological cues. Differentiated SMCs are quiescent and contractile and contain an organized actin cytoskeleton. In response to vascular injury or growth factor signaling, SMCs dedifferentiate and adopt a proliferative, migratory phenotype that causes vascular occlusion in a variety of disorders, including atherosclerosis, restenosis following angioplasty, and hypertension (Owens 1995).
Serum response factor (SRF) acts as a central regulator of SMC phenotypes by directly activating a battery of genes encoding components of the actin cytoskeleton (Pipes et al. 2006; Miano et al. 2007; Niu et al. 2007). The cardiovascular coactivator myocardin interacts with SRF and stimulates smooth muscle differentiation (Wang et al. 2001, 2003; Chen et al. 2002; Du et al. 2003; Yoshida et al. 2003). Myocardin-related transcription factors (MRTFs) also confer actin sensitivity to SRF. MRTFs are sequestered in the cytoplasm by monomeric G-actin, whereas in response to Rho signaling or other stimuli that promote actin polymerization, MRTFs are released from G-actin and translocate to the nucleus, where they associate with SRF to drive the expression of actin and other cytoskeletal protein genes (Miralles et al. 2003; Medjkane et al. 2009). Thus, SRF functions as a nodal sensor of actin polymerization and as a regulator of cytoskeletal assembly, thereby governing SMC differentiation, proliferation, and migration during development and disease.
Recent studies have revealed key roles for microRNAs (miRNAs) in the control of cardiac and skeletal muscle development and remodeling in response to injury (van Rooij and Olson 2007; Condorelli and Dimmeler 2008; Chen et al. 2009; Cordes and Srivastava 2009). miRNAs act as negative regulators of gene expression by annealing with complementary sequences in the 3′ untranslated regions (UTRs) of mRNAs, causing translational inhibition and mRNA degradation (Bartel 2004). Most studies to date have focused on the functions of miRNAs as determined by overexpression or knockdown experiments in vitro, whereas much remains to be learned about miRNA functions in tissue homeostasis and remodeling in vivo.
Here, we show that SRF and myocardin regulate a bicistronic miRNA gene cluster encoding two cardiovascular-specific miRNAs, miR-143 and miR-145. Smooth muscle differentiation is unperturbed in mice lacking these miRNAs. However, these mutant mice display a unique phenotype in which they fail to develop a neointima in response to vascular injury due to disrupted actin stress fibers and diminished migratory activity. This blockade to SMC phenotypic switching reflects the regulation by miR-143 and miR-145 of a collection of modulators of actin dynamics and cytoskeletal assembly. Our results demonstrate that miR-143 and miR-145 function as components of an elaborate SRF-dependent regulatory network involving multiple positive and negative feedback loops to control SMC phenotypes in response to stress signaling.
Results
Cardiovascular regulation of miR-143 and miR-145 by myocardin and MRTF-A
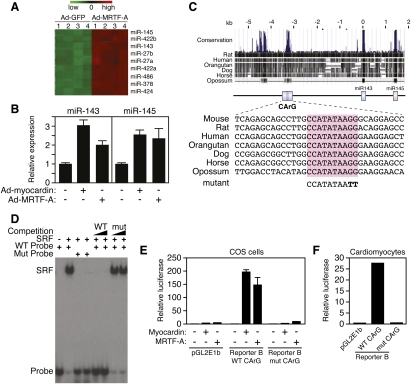
To search for miRNAs potentially regulated by SRF, we compared miRNA expression patterns by microarray analysis of primary rat cardiomyocytes infected with adenovirus expressing MRTF-A, an activator of SRF-dependent transcription, and GFP as a control (Fig. 1A). miR-145 and miR-143 were among the most strongly up-regulated miRNAs in response to MRTF-A, as revealed by microarray and confirmed by real-time PCR (Fig. 1A,B). Myocardin also up-regulated the expression of miR-145 and miR-143 in cardiomyocytes (Fig. 1B). Northern blot analysis of adult mouse tissues showed these miRNAs to be especially abundant in the aorta and bladder, rich sources of SMCs (Supplemental Fig. 1A), consistent with recent studies (Cheng et al. 2009; Cordes et al. 2009).
Figure 1.
Cardiovascular regulation of miR-143/145 by myocardin and MRTF-A. (A) miRNA microarray of rat cardiomyocytes infected with adenovirus expressing GFP or MRTF-A. The most strongly up-regulated miRNAs in response to Ad-MRTF-A are shown. Numbers 1–4 designate independent spots for each miRNA on the arrays. The colored bar at the top of the panel represents changes on a linear scale, where green and red represent minimal and maximal expression, respectively. (B) Up-regulation of miR-143 and miR-145 in rat cardiomyocytes 48 h after infection with adenovirus encoding MRTF-A and myocardin as detected by real-time PCR. Ad-GFP was used as a negative control. (C) Cross-species DNA sequence conservation surrounding the miR-143/145 gene. A conserved island of homology at −3328/−3056 contains a CArG box. A mutation placed in the CArG box is shown at the bottom. (D) Gel mobility shift assay with a labeled probe containing the CArG box sequence shown in C. SRF obtained from cell lysates of SRF-expressing cells bound with high affinity to this probe, and binding was competed by wild-type but not by mutant unlabeled probe. (E) The conserved region between −3.2 and −4.6 kb (Reporter B in Fig. 2A) upstream of the miR-143/145 gene was cloned into the pGL2E1b luciferase expression plasmid and transfected into COS cells with a myocardin expression plasmid. Myocardin potently activated the reporter linked to the miR-143/145 upstream region with the wild-type CArG box, whereas the mutant CArG box abolished responsiveness to myocardin. (F) Reporter B was transfected into rat cardiomyocytes. The reporter with the wild-type CArG box showed high luciferase activity, whereas the reporter with the mutant CArG box showed no activity. The empty pGL2E1b reporter was used as a negative control.
miR-143 and miR-145 are highly conserved across vertebrates but show no homology with each other, indicating that they act on different target sequences. The genomic coding regions for miR-143 and miR-145 are separated by ∼1.4 kb on mouse chromosome 18 (Supplemental Fig. 1B). RT–PCR using primers from the stem–loop sequences of the two miRNAs showed them to be derived from a common bicistronic precursor (Supplemental Fig. 1C), indicating that they are cotranscribed from the same gene.
Alignment of the genomic sequence surrounding the miR-143/145 cluster across species revealed a 272-base-pair (bp) region located between −3328 and −3056 bp upstream of the gene, which was highly conserved from human to opossum (Fig. 1C). This region contained a conserved SRF-binding site [CC(A/T)6GG], known as a CArG box, that bound SRF avidly and specifically in gel mobility shift assays (Fig. 1D). The SRF coactivators MRTF-A and myocardin activated the expression of a luciferase reporter (referred to as Reporter B) containing the conserved CArG box within a 1.4-kb genomic fragment spanning −4.6 to −3.2 kb upstream of the miR143/145 locus. Transcriptional activation was abolished by a mutation in the CArG box of Reporter B that prevented SRF binding (Fig. 1E). Similarly, Reporter B was also strongly expressed in transfected cardiomyocytes, whereas a mutation in the CArG box extinguished expression (Fig. 1F).
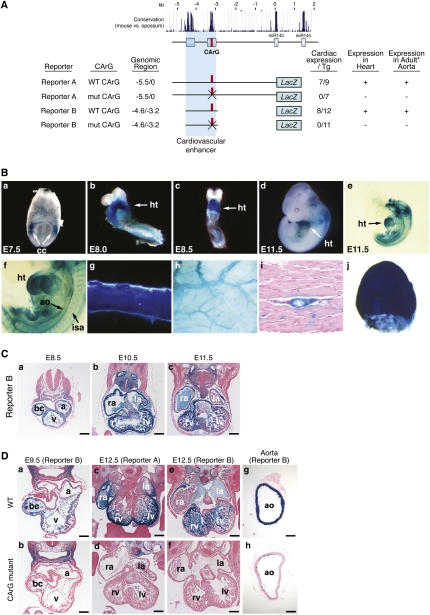
To examine the expression of the miR-143/145 locus during development, we generated transgenic mice harboring 5.5 kb of genomic DNA upstream of the gene linked to a lacZ reporter (Reporter A) (Fig. 2A). Robust expression of the transgene was observed in the cardiac crescent at embryonic day 7.5 (E7.5) (Fig. 2B, panel a) and throughout the developing heart (Fig. 2B, panels b–d) until E16.5 (data not shown) and was down-regulated thereafter. At E11.5, the miR-143/145 enhancer region directed strong expression in the dorsal aorta and developing intersomitic vessels (Fig. 2B, panels e,f). In adult mice, transgene expression was confined to vascular and visceral SMCs (Fig. 2B, panels g–j). The Reporter A and B transgenes were expressed in all vascular SMCs throughout the body, as well as in SMCs lining internal organs such as the bladder, stomach, and intestines. The cardiovascular-specific expression of this transgene mirrors that of the endogenous miR-143/145 miRNAs as revealed by in situ hybridization (Cheng et al. 2009; Cordes et al. 2009).
Figure 2.
An essential CArG box in the miR-143/145 upstream enhancer. (A) Summary of transgenic constructs used to delineate the miR-143/145 enhancer is shown. Evolutionary conservation of the genomic region upstream of pre-miR-143 is shown at the top. Numbers of F0 transgenic embryos showing cardiac expression at E12.5 per total transgenic embryos obtained are indicated in the right column. A single mutation of the CArG in Reporter A abolished expression of the lacZ transgene in the embryonic heart and adult aorta. Embryonic heart expression was assayed in F0 and stable mouse lines, whereas adult aorta expression was assayed in two independent stable mouse lines. The 1.5-kb enhancer construct (Reporter B) contains only the two evolutionarily conserved regions and is sufficient to drive lacZ expression in the embryonic heart and adult aorta. Mutation of the CArG box in Reporter B abolished lacZ expression in the embryonic heart and adult aorta as determined in two stable mouse lines. (B) Transgenic mice were generated with a lacZ reporter linked to 5.5 kb of genomic DNA upstream of the miR-143/145 gene (Reporter A in A). The embryo in panel e was cleared to allow visualization of internal structures. Panel f is a high magnification of the embryo in panel e to show lacZ expression in SMCs lining the dorsal aorta (ao) and intersomitic arteries (isa). (Panel g) Adult aorta. (Panel h) Subdermal vasculature. (Panel i) Transverse section showing lacZ staining in an artery in skeletal muscle. (Panel j) Bladder. (ao) Aorta; (cc) cardiac crescent; (ht) heart. (C) Histological sections of Reporter B transgenic mice at various embryonic stages. (Bars: panel a, 100 μm; panels b,c, 200 μm. (a) Common atrial chamber; (bc) bulbus cordis; (la) left atrium; (lv) left ventricle; (ra) right atrium; (rv) right ventricle; (v) common ventricular chamber. (D) Histological sections of transgenic mice with wild-type genomic regions or the same regions with mutations in the CArG box at the indicated ages. Note that the CArG box mutation abolishes lacZ expression in the heart and vasculature. (a) Common atrial chamber; (ao) aorta; (bc) bulbus cordis; (la) left atrium; (lv) left ventricle; (ra) right atrium; (rv) right ventricle; (v) common ventricular chamber. Bars: panels a,b,g,h, 100 μm; panels c–f, 200 μm.
Transgenic mice containing Reporter B showed strong expression in cardiac and vascular SMCs, comparable with that of the complete 5.5-kb region in Reporter A (Fig. 2A,B,D). Consistent with the results of reporter assays in cultured cells (Fig. 1E), a mutation in the CArG box abolished cardiac and smooth muscle expression of the Reporter A and B transgenes in vivo (Fig. 2D). We conclude that SRF and myocardin/MRTF-A activate miR-143/145 expression through a conserved CArG box within an upstream, cardiovascular-specific enhancer.
These transgenic analyses of the miR-143/145 cis-regulatory elements agree with those of Cordes et al. (2009) with respect to the requisite role of the CArG box in smooth muscle expression in vivo. However, in contrast to our findings demonstrating that the CArG box is required for cardiac expression at multiple developmental time points, Cordes et al. (2009) reported that an evolutionarily conserved binding site for Nkx2.5 located ∼2.57 kb upstream of the miR-143/145 gene was essential for cardiac expression in vivo, whereas our results indicate that only the conserved region between −3.2 and −4.6, which lacks this putative Nkx2.5-binding site, is necessary and sufficient for cardiac expression in vivo. Upon analysis of the Nkx2.5-binding site identified by Cordes et al. (2009), we found that this sequence is not evolutionarily conserved in rats, humans, and other vertebrate genomes. We are uncertain of the basis for these differing results, but based on the strong expression of Reporter B, which lacks the putative Nkx2.5-responsive region, our results do not indicate a role for this region in regulating cardiac expression in vivo. Instead, our results point to the ultraconserved upstream region containing the SRF-binding site as critical for cardiac expression. Since the constructs tested in our study and that of Cordes et al. (2009) were not identical, it is conceivable that DNA sequences that differ between the various reporters contributed to differences in expression.
Generation of miR-143 and miR-145 mutant mice
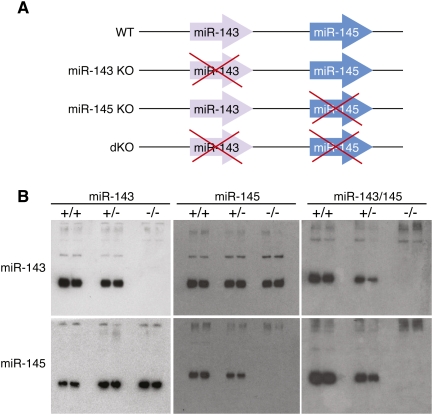
To systematically investigate the functions of miR-143 and miR-145 in vivo, we deleted each gene singly and in combination in mice by introducing loxP sites for Cre-mediated recombination in the regions flanking the pre-miR coding regions of each miRNA through homologous recombination (Fig. 3A; Supplemental Figs. 2–4). The targeting strategy deleted the 70-bp pre-miR stem–loop sequence of each miRNA, or a genomic region encompassing both miRNAs, and replaced them with a neomycin resistance cassette flanked by loxP sites. Chimeric mice obtained by blastocyst injection of targeted embryonic stem (ES) cells transmitted the mutant alleles through the germline, yielding mice heterozygous for miR-143(neo), miR-145(neo), or miR-143/145(neo) alleles. The neomyocin resistance cassette was removed by breeding these mice with mice expressing a ubiquitously expressed CAG-Cre transgene. Breeding of the offspring of these crosses generated homozygous mutants. Deletion of each miRNA was confirmed by Northern blot analysis (Fig. 3B). Expression of either miR-143 or miR-145 did not affect the expression of the other member of the pair (Fig. 3B). Mice homozygous for either single miRNA deletion or the double miRNA deletion (referred to as double knockout [dKO] mice) were viable and were obtained at predicted Mendelian ratios from heterozygous intercrosses. Thus, neither miR-143 nor miR-145 is essential for cardiovascular development in vivo.
Figure 3.
Targeting of the miR-143/145 cluster. (A) Schematic of miR-143/145 mutations. (B) Northern blots of RNA from bladders of duplicate mice of each genotype. Mutant mice used for these experiments had the neomyocin resistance gene removed by Cre-mediated recombination. Deletion of either miR-143 or miR-145 does not affect expression of the other member of the pair.
Smooth muscle abnormalities in miR-143/145 mutant mice
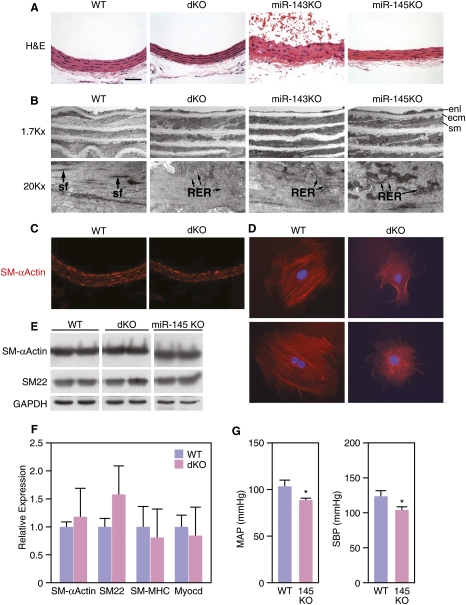
We observed no overt abnormalities in cardiac structure or gene expression in miR-143−/−, miR-145−/−, or dKO mice (data not shown). However, the smooth muscle layers of the aorta and of other arteries of miR-145−/− and dKO mice were noticeably thinner than those of wild type (Fig. 4A), due to a reduction in width of SMCs. The number of elastin layers separating SMCs was comparable in mice of the different genotypes, indicating that a reduction in cell size in miR-145−/− and dKO mice likely accounts for thinning of the vessel wall. Electron microscopy revealed that wild-type SMCs contained prominent actin-based stress fibers throughout the cytoplasm, characteristic of a differentiated phenotype (Fig. 4B). In contrast, the number and prominence of stress fibers were dramatically decreased in SMCs from miR-145−/− and dKO mice, suggesting that these miRNAs modulate actin dynamics and cytoskeletal assembly. Electron microscopy also revealed an increase in rough endoplasmic reticulum in the cytoplasm of aortic SMCs from miR-143−/−, miR-145−/−, and dKO mice, characteristic of a more synthetic state (Fig. 4B). In addition, the vessel walls of miR-145−/− and dKO mice were distinguishable by the presence of thick extracellular matrix (ECM) surrounding mutant SMCs (Fig. 4B).
Figure 4.
Smooth muscle abnormalities in miR-143/145 KO mice. (A) H&E staining of the dorsal aortae of mice of the indicated genotypes. Bar, 40 μm. (B) Electron microscopy of the dorsal aortae of mice of the indicated genotypes. Note the thickened ECM in miR-145 KO and dKO. Prominent actin-based stress fibers (arrows) are apparent in wild-type (WT) and miR-143 KO SMCs, but are absent from miR-145 KO and dKO SMCs. Rough endoplasmic reticulum (RER) is evident in miR-143 KO, miR-145 KO, and dKO SMCs, indicative of a synthetic state, but is not visible in wild-type SMCs. (ecm) Extracellular matrix; (enl) endothelial layer; (sf) stress fiber; (sm) smooth muscle. (C) Immunostaining of sections of aorta of wild-type and dKO mice showing similar staining for SMA. (D) Aortic SMCs were isolated from wild-type and dKO mice, cultured in growth medium, and stained for SMA. Wild-type SMCs display highly organized actin stress fibers, whereas SMCs from dKO mice display only diffuse actin staining. (E) Expression of the smooth muscle differentiation markers SMA and SM22 in aorta of wild-type and dKO mice as detected by Western blot analysis. GAPDH was detected as a loading control. (F) Expression of smooth muscle mRNAs as detected by real-time PCR. (G) Blood pressure measurements in wild-type and miR-145 KO mice. MAP and SBP were measured as described in the Materials and Methods. (*) P < 0.05.
To further examine the role of miR-143 and miR-145 in cytoskeletal assembly, we isolated vascular SMCs from the aortas of wild-type and dKO littermates and examined their properties in culture. SMCs from wild-type mice were larger than those from dKO mice and displayed more intense smooth muscle α-actin (SMA) staining within highly organized stress fibers. Consistent with their appearance in vivo, actin filaments appeared diffuse and stress fibers were not visible in SMCs from dKO mice, suggesting a disruption of actin cytoskeleton dynamics (Fig. 4D).
Despite this less differentiated appearance, mice of all mutant genotypes expressed protein markers of smooth muscle differentiation, such as SMA and SM22, at levels comparable with wild-type littermates (Fig. 4C,E). Similarly, mRNAs encoding smooth muscle structural proteins and myocardin were expressed at comparable levels in SMCs isolated from wild-type and dKO mice (Fig. 4F). We conclude that miR-145 and, to a lesser extent, miR-143 are required for maintenance of actin stress fibers in SMCs in vivo, but neither miR-143 nor miR-145 are required for SMC development or differentiation per se.
The thinning of blood vessels walls in the mutant mice prompted us to investigate whether these mice might be hypotensive due to a reduction in vascular tone. Wild-type mice had normative steady-state mean arterial pressures (MAPs) and systolic blood pressures (SBPs) of 103.5 ± 6.5 mmHg and 124.3 ± 7.9 mmHg, respectively (Fig. 4G). In contrast, the miR-145−/− mice had statistically significant arterial hypotension under steady-state conditions, with MAPs of 90.2 ± 3.6 mmHg (P = 0.05) and 104.2 ± 3.9 mmHg (P = 0.021), respectively. Systemic hemodynamic alterations were associated with statistically significant reductions in ventricular mass that likely reflect chronic unloading of the heart (Supplemental Fig. 5). Wild-type mice had a heart weight index (HWI) of 8.66 ± 0.38 mg/mm and a left ventricular index (LVI) of 6.21 ± 0.28 mg/mm, while miR-145−/− mice had an HWI of 7.7 ± 0.31 mg/mm (P = 0.035) and an LVI of 5.56 ± 0.24 mg/mm (P = 0.049). Thus, arterial hypotension was associated with reductions in cardiac and LV mass.
miR-143 and miR-145 are required for vascular remodeling in response to injury
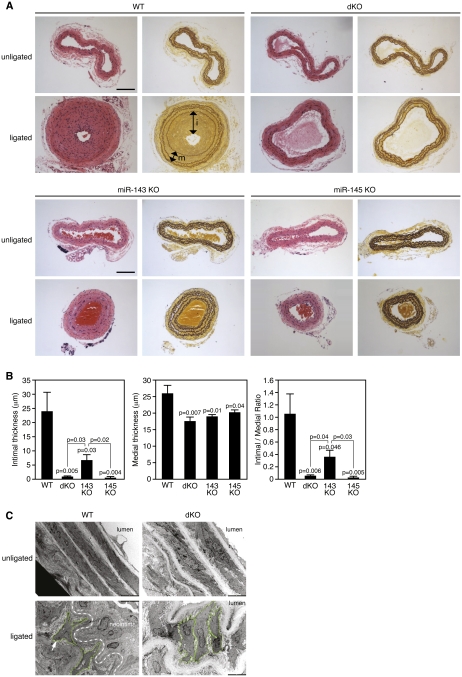
To test whether miR-143 or miR-145 might participate in vascular remodeling in response to injury, we compared the responses of wild-type and mutant mice following carotid artery ligation, a well-established model of vascular injury that results in neointima formation due to dedifferentiation, proliferation, and migration of SMCs within the vessel wall. In wild-type mice, ligation of the carotid artery promoted the generation of a thick neointima, which almost completely occluded the lumen of the vessel (Fig. 5A). Surprisingly, this response was nearly abolished in miR-145−/− and dKO mice. Instead, the architecture of the vessel wall in these animals was preserved following carotid artery ligation (Fig. 5A). In miR-143−/− mice, neointima formation was also reduced relative to wild type, but to a lesser extent than in miR-145−/− or dKO animals (Fig. 5A). Neointimal formation in mice of the different genotypes was quantified by measuring the width of the intimal layer (the distance between the inner elastin layer and the lumen) and the medial layer (the distance between the inner and outer elastin layers) (Fig. 5B). miR-145−/− and dKO mice formed virtually no neointimal layer, and neointima formation in miR-143−/− was reduced by at least 60%. The medial layer was also significantly thinner in the mutants, suggesting a more relaxed, less contractile state (Fig. 5B).
Figure 5.
Reduced neointima formation in miR-143/145 mutant mice following carotid artery ligation. (A) Mice of the indicated genotypes were subjected to carotid artery ligation for 28 d, after which histological sections were obtained from the ligated artery and the unligated artery as a control 1900 μm proximal to the ligature and stained with H&E (left side) or for elastin (right side). The intimal (i) and medial (m) layers are designated with arrows in the ligated wild-type (WT) artery stained for elastin. Bar, 100 μm. (B) The intimal thickness in mice of each genotype 28 d after carotid artery ligation was determined by measuring the distance between the inner elastin layer and the lumen in at least five mice of each genotype. Medial thickness was measured as the distance between the inner and outer elastin layers. (C) Electron microscopy of carotid arteries of wild-type and dKO mice 14 d after ligation. SMCs from the wild-type artery adopt a migratory phenotype in response to ligation, giving rise to a neointima. In contrast, SMCs from the dKO artery retain a more organized arrangement and fail to form a neointima. The boundaries of representative SMCs are outlined in green. The dashed line in the bottom left panel indicates the internal boundary of the neointima.
Electron microscopy revealed that wild-type SMCs in the ligated carotid artery became dedifferentiated, displaying a loss of spindle-like cell morphology as they became migratory and proliferative (Fig. 5C). In contrast, individual SMCs in the ligated carotid artery from dKO animals showed little morphological change in response to injury and appeared to be ensheathed in a thickened ECM (Fig. 5C).
We also observed lethality in ∼50% of miR-145−/− and dKO mice that underwent carotid ligation, whereas <10% of wild-type or miR-143−/− littermates died after the surgery. Lethality of miR-145−/− and dKO mice was preceded by severe neurological abnormalities—including circling, loss of coordination, and seizures—that likely reflect an inability of the blood vessel to respond appropriately to injury due to a failure in SMC phenotypic modulation. Histological sections revealed severe necrosis in the left hemisphere of the brains of miR-145−/− and dKO mice, downstream from the ligated carotid artery (Supplemental Fig. 6). These findings suggest a failure of vasoconstriction of the circle of Willis and failure of shunting of blood to the left hemisphere in response to carotid artery ligation in miR-145−/− and dKO mice. These results indicate miR-145 plays an unexpected role in the response of vascular SMCs to vessel injury.
miR-143 and miR-145 target multiple regulators of actin dynamics
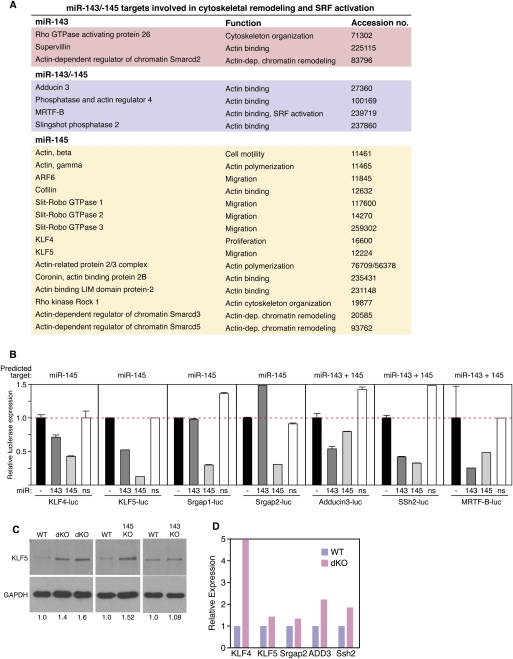
To identify mRNA targets that might contribute to the observed aberrations in SMC phenotypic switching and actin stress fiber formation in miR-143 and miR-145 mutant mice, we used TargetScan (Grimson et al. 2007) and Pic Tar (Krek et al. 2005) prediction algorithms to search for evolutionarily conserved targets for these miRNAs. Intriguingly, despite the lack of sequence homology between miR-143 and miR-145, both miRNAs were predicted to target a disproportionate number of the same genes involved in the regulation of SRF activity and actin dynamics (Fig. 6A; Supplemental Figs. 7–13). Among these predicted targets are the zinc finger proteins; Kruppel-like factor 4 (KLF4) and KLF5, which repress SRF activity and either promote or inhibit SMC phenotypic switching (Nagai et al. 2005; C Wang et al. 2008; Yoshida et al. 2008; Suzuki et al. 2009); Slit–Robo GTPase-activating protein 1 (Srgap1) and Srgap2, which function as negative regulators of actin polymerization (Wong et al. 2001); Adducin-3 (Add3), an F-actin capping protein involved in cell migration (Barkalow et al. 2003); Sling-shot 2 (Ssh2) phosphatase, which stimulates the activity of the actin depolymerizing factor cofilin (San Martin et al. 2008; Eiseler et al. 2009); and MRTF-B, an actin-regulated coactivator of SRF (Kuwahara et al. 2005; Hinson et al. 2007; Parmacek 2007).
Figure 6.
Modulation of actin dynamics by miR-143/145. (A) miR-143/145 targets involved in cytoskeletal remodeling and SRF activation. (B) The 3′ UTRs for the indicated mRNA targets of miR-143 and miR-145 were linked to luciferase and tested for repression by miR-143 and miR-145 expression plasmids in transfected COS cells. A miR-126 expression plasmid was used as a nonspecific (ns) control. The black bar (−) indicates the level of expression of the luciferase reporter without a cotransfected miRNA expression plasmid. miRNAs predicted to target each 3′ UTR are indicated at the top of each graph. (C) Aortae from mice of each genotype were isolated and protein lysates were analyzed for expression of KLF5 protein by Western blot analysis. GAPDH was included as a loading control. Numbers below each lane indicate relative expression of KLF5 normalized to GAPDH. (D) Carotid arteries were obtained from 10 wild-type and 10 dKO mice, pooled from each group, and expression of the indicated miR-143/145 targets was detected by real-time PCR.
We tested each of the above predicted targets for repression by miR-143 and miR-145 by placing their 3′ UTRs downstream from a cytomegalovirus (CMV)-driven luciferase reporter and performing reporter assays in COS cells transfected with expression plasmids for miR-143, miR-145, or a control plasmid encoding an unrelated miRNA. As shown in Figure 6B, each of these reporters was repressed to varying degrees by miR-143 and miR-145 and, in general, repression correlated with the presence of predicted target sites for these miRNAs. Consistent with these findings, expression of KLF5 protein was up-regulated in SMCs from miR-145−/− and dKO mice (Fig. 6C). Moreover, the above targets of miR-143 and miR-145 were up-regulated in carotid arteries isolated from dKO mice compared with wild-type littermates (Fig. 6D), in agreement with the destabilizing influence of miRNAs on their targets.
Discussion
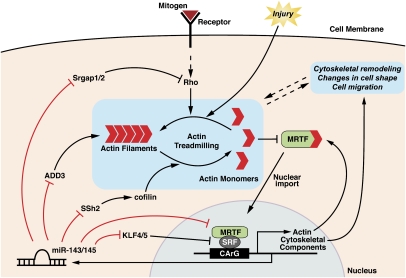
The results of this study reveal that SRF controls the expression of miR-143 and miR-145 in SMCs, and these miRNAs, in turn, feed back to control the expression and function of multiple components of the cytoskeleton and the SRF regulatory network (Fig. 7). In response to injury, differentiated SMCs adopt a migratory, synthetic phenotype, and SRF plays a central role in such phenotypic switching (Wang et al. 2004; Pipes et al. 2006; Miano et al. 2007). In mice lacking miR-143 and miR-145, the formation of normal actin stress fibers is perturbed and SMCs do not form a neointima in response to injury.
Figure 7.
Model for the role of miR-143 and miR-145 in the control of actin remodeling. MRTFs are sequestered in the cytoplasm by monomeric actin. Upon release from actin, MRTF translocates to the nucleus and interacts with SRF to activate the transcription of genes encoding actin and other cytoskeletal components, as well as miR-143 and miR-145. These miRNAs repress the expression of a collection of regulators of actin dynamics and MRTF/SRF activity, thereby creating a complex set of feedback loops to modulate cytoskeletal assembly and dynamics. (Adapted from Pipes et al. 2006.)
Control of actin dynamics by miR-143 and miR-145
Although miR-143 and miR-145 are nonhomologous, they both share a disproportionate number of common targets involved in actin dynamics, cytoskeletal function, and phenotypic switching of SMCs. Among these is the actin-dependent SRF coactivator MRTF-B (Miralles et al. 2003; Kuwahara et al. 2005) and Add3, which caps the barbed ends of actin filaments and acts as a bridge between the membrane and actin cytoskeleton (Gardner and Bennett 1987; Barkalow et al. 2003). Rho kinase-dependent phosphorylation of Adducin results in enhanced F-actin binding and cell motility (Fukata et al. 1999). Ssh2, another target of miR-143 and miR-145, promotes cell motility and enhances F-actin reorganization by dephosphorylating and activating cofilin, an actin depolymerizing factor (Eiseler et al. 2009). Ssh1 is activated by platelet-derived growth factor (PDGF) and promotes SMC migration, whereas knockdown of Ssh1 with antisense RNA inhibits migration (San Martin et al. 2008). miR-145 selectively targets KLF4 and KLF5, which repress SRF activity, and can either inhibit or promote SMC differentiation or proliferation, depending on the context (Suzuki et al. 2009). Srgap1 and Srgap2, also targeted by miR-145, modulate Slit–Robo-dependent repulsive cues and cell migration by inactivating the small GTPase Cdc42 and inhibiting actin polymerization (Wong et al. 2001). The preferential targeting of these modulators of cytoskeletal function by miR-145 may account, at least in part, for the stronger phenotype resulting from miR-145 versus miR-143 deletion. Other predicted targets of miR-145 and/or miR-143 with roles in actin dynamics include actin, cofilin, Rho GTPase-activating protein, phosphatase and actin regulator 4, and others (Fig. 6A). Given the plethora of targets of miR-143 and miR-145 within the actin–SRF regulatory network, we suggest that the absence of these miRNAs creates an imbalance among the various feedback loops required for cytoskeletal homeostasis, thereby perturbing stress fiber formation and actin remodeling, which are required for sensing of mechanical stress and phenotypic switching (Fig. 7). Dysregulation of additional targets also likely contributes to the phenotypic abnormalities of miR-143/145 mutant mice.
Control of neointimal growth by miR-143 and miR-145
A recent study reported that adenoviral overexpression of miR-145 inhibits neointimal growth of the rat carotid artery in response to balloon injury (Cheng et al. 2009). Forced expression of miR-145 in cultured SMCs has also been reported to increase the expression of smooth muscle differentiation markers, and, conversely, inhibition of miR-145 expression with an antisense oligonucleotide interferes with smooth muscle differentiation in vitro (Cordes et al. 2009; Xu et al. 2009). Thus, the block to restenosis in miR-143/145 mutant mice following carotid artery ligation seems paradoxical. How might this be explained? We propose that disparities between results obtained from transient overexpression or knockdown experiments in vitro and genetic loss-of-function studies in vivo reflect the relative simplicity of in vitro assays, as well as the existence of adaptive mechanisms in vivo that do not exist in vitro. In addition, in our studies of mutant mice, SMCs are already aberrant at the time they receive the stress stimulus, whereas in overexpression or knockdown experiments, the cells are normal at the onset of injury. Finally, there is a precedent in other systems for overexpression and knockdown of miRNAs having similar consequences, which likely reflects the importance of miRNAs in stabilizing intracellular processes by buffering fluctuations of gene expression such that tipping the balance one way or the other perturbs that process (Choi et al. 2007). These findings highlight the importance of genetic model systems for the complete understanding of the complex functions of miRNAs in disease settings.
It is also noteworthy that miR-145 mutant mice displayed a significant reduction in blood pressure, which likely reflects reduced vascular tone in these animals. Thus, although the SMCs differentiate normally in these animals, their lack of a mature actin cytoskeleton may diminish vascular rigidity. It is conceivable that reduced vascular tone also contributes to the lack of responsiveness of the vessel wall to carotid artery ligation.
Roles for miR-143 and miR-145 in SMC differentiation
The apparently normal differentiation of SMCs in miR-143/145-null mice further highlights the differences in functions of these miRNAs as determined by in vitro overexpression or knockdown studies and gene deletion analyses in vivo. Recently, miR-145 was shown to be necessary for reprogramming of fibroblasts to an SMC fate by myocardin and sufficient to direct SMC fate in neural crest cells (Cordes et al. 2009). Our results suggest that the functions of myocardin are less reliant on miR-145 in vivo than in vitro, since we observed little diminution of smooth muscle differentiation markers in miR-145 mutant mice. If miR-145 were indeed required for myocardin activity in vivo, miR-145-null mice might be expected to resemble myocardin-null mice, which die during early embryogenesis from a failure in smooth muscle differentiation (Li et al. 2003). Perhaps overexpression and knockdown experiments in vitro do not fully reflect the functions of miR-145 when expressed at physiological levels in vivo. Alternatively, there may be compensatory mechanisms in vivo that can override the dependency of myocardin on miR-145. In this regard, we also isolated embryonic fibroblasts from miR-143/145-null mice and expressed myocardin in them through adenoviral delivery. In these experiments, smooth muscle markers were activated to comparable levels as in wild-type fibroblasts, again indicating that the genetic deletion of these miRNAs does not strongly diminish myocardin activity (M Xin, L Sutherland, and EN Olson, unpubl.).
miRNA effectors of SRF action
Our results demonstrate that SRF is an obligate activator of miR-143/145 transcription in the developing cardiovascular system, as well as in SMCs of the adult aorta, and underscore an emerging theme with respect to SRF–miRNA regulatory circuits. Similar to the regulation of miR-143/145, SRF directly regulates the expression of the miR-1-1/133a-2 and miR-1-2/133a-1 clusters in developing skeletal and cardiac muscle, and these miRNAs feed back to modulate the actions of SRF in these types of muscle cells through precisely titrated regulatory loops (Chen et al. 2006; Liu et al. 2008). Numerous other miRNA genes are coupled to one or more CArG boxes that have not yet been functionally analyzed (Niu et al. 2007). Together, these findings suggest that SRF regulation of miRNAs represents a common form of regulation that likely reflects the key role of SRF as a sensor of extracellular signals, and the need to precisely modulate its activity so as to control cell proliferation and differentiation.
Therapeutic implications
The results of this study further underscore the pivotal roles of miRNAs in mediating changes in gene expression and cell functions in response to injury and stress. Such functions of miRNAs are especially prominent in the cardiovascular system, in which miRNAs have been shown to mediate numerous pathological processes (van Rooij and Olson 2007; Barringhaus and Zamore 2009), including cardiac hypertrophy (van Rooij et al. 2006), cardiac fibrosis and remodeling in response to myocardial infarction (van Rooij et al. 2008), arrhythmias (Yang et al. 2007), and angiogenesis (S Wang et al. 2008; Bonauer et al. 2009). Depending on whether a miRNA is pathogenic or protective in a particular disease setting, it should be possible, in principle, to inhibit or enhance its function through the use of anti-miRNAs or miR mimics, respectively. The discovery of miRNAs as modulators of disease processes can also provide unanticipated insights into disease mechanisms through the identification of the cellular effectors of miRNA action. Thus, manipulation of disease-associated miRNAs offers interesting therapeutic opportunities for the future.
Materials and methods
Generation of miR-143/145 mutant mice
All targeting vectors for generating null alleles of miR-143, miR-145, and miR-143/145 compound deletion mutations were constructed using the pGKNEO-F2DTA vector, which contains a neomycin resistance gene driven by the pGK promoter, flanked by loxP sites, and a diphtheria toxin gene cassette. Both miR-143 and miR-145 targeting strategies were designed to replace the pre-miR sequences with the neomycin resistance cassette flanked by loxP sites. For the targeting vector of miR-143, a 3.5-kb fragment upstream of pre-miR-143 and a 4.5-kb fragment immediately downstream from pre-miR-143 were generated as the 5′ and 3′ arms, respectively, by TAKARA Taq LA PCR amplification (TAKARA) of 129SvEv genomic DNA. For the targeting vector of miR-145, a 3.5-kb fragment upstream of pre-miR-145 and a 5-kb fragment downstream were generated as the 5′ and 3′ arms, respectively. For the targeting vector of the miR-143/145 double mutant, a 3.5-kb fragment upstream of pre-miR-143 and a 4.5-kb fragment immediately downstream from pre-miR-145 were generated as the 5′ and 3′ arms, respectively. A KO arm was generated by inserting a 1.5-kb genomic DNA sequence containing pre-miR-143, pre-miR-145, and the 1.4-kb DNA between the two pre-miRs into the targeting vector between the two loxP sites. The targeting vectors were then linearized and electroporated into 129SvEv-derived ES cells. Five-hundred ES cell clones for each targeting vector were isolated and analyzed for homologous recombination by Southern blotting. Three clones with a properly targeted miR-143, miR-145, or miR-143/145 allele were injected into 3.5-d C57BL/6 blastocysts, and high-percentage chimeric male mice were crossed to C57BL/6 females to achieve germline transmission of the targeted alleles. Heterozygous miR-143neo/+ or miR-145neo/+ mice were intercrossed with Cre-transgenic mice under the control of the CAG promoter to remove the neo cassette for miR-143 and miR-145 KOs, and to remove both miR-143 and miR-145 together with the neo cassette for dKO mice. All the miR-145 KO mice in the study were products of intercrossing miR-145neo/+ mice.
All experimental procedures involving animals in this study were reviewed and approved by the Institutional Animal Care and Research Advisory Committee at the University of Texas Southwestern Medical Center.
Northern blot analysis
Ten micrograms of total RNA from different tissues were electrophoresed on a 20% polyacrylamide (7.6 M urea) gel in 1× TBE. RNA was then transferred onto a Zetaprobe GT membrane (Bio-Rad) in 0.5× TBE buffer at 80 V for 1 h. Hybridization was performed at 39°C according to a standard protocol. 32P-labeled Star-Fire oligonucleotide probes (IDT) against mature miR-143, miR-145, and U6 were used in the hybridization.
Histology and immunohistochemistry
Tissues were fixed in 4% paraformaldehyde, embedded in paraffin (Moller and Moller 1994), and sectioned at 5-μm intervals. Hematoxilin and eosin (H&E) and elastin stains were performed using standard procedures. Immunohistochemisty was carried out on paraffin-embedded sections as described previously (Xin et al. 2006). Cy3-conjugated SMA antibody was from Sigma (Clone 1A4; 1:200). Slides were mounted with Vectashield mounting medium with DAPI (Vector Laboratories).
Cell culture
Mouse SMCs were isolated from postnatal day 2 aortas. After removing the fat and connective tissues surrounding the aorta, the tissue was sliced into small pieces and placed on a 12-well dish. The explant was kept in DMEM/FM-12 medium containing 20% FBS, streptomycin, and penicillin. After SMCs migrated out of the explant, the cells were maintained in 10% FBS containing DMEM/F-12 medium. Cells between passages 2 and 5 were used in the experiments.
Reporter assays
A DNA fragment containing full-length 3′ UTRs for putative miR-143 and miR-145 targets were cloned into pMIR-REPORT vector (Ambion). Cell culture and luciferase assays were performed as described previously (van Rooij et al. 2007). Briefly, COS cells were transfected using Fugene 6 reagent according to the manufacturer's instructions (Roche). pCMV-lacZ was used as transfection control, and the total plasmid amount was kept constant with empty pcDNA3.1. Forty-eight hours after transfection, cell extracts were assayed for luciferase expression using the luciferase assay kit (Promega). Relative reporter activities are expressed as luminescence units normalized for β-galactosidase expression in the cell extracts.
Electron microscopy
Eight-week-old miR-143/145 mutant mice and control littermate mice were anesthetized and transcardially perfused with 0.1 M phosphate buffer (pH 7.3), followed by 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M sodium cacodylate buffer. Descending dorsal aortae were removed and fixed overnight at 4°C. Samples were processed by the University of Texas Southwestern Medical Center Electron Microscopy Core Facility. Sections were imaged on Tecnai TEM at the indicated magnifications. The left carotid ligated and the right carotid control arteries from wild-type and dKO mice were harvested after 14 d of ligation and prepared for electron microscopic observation as described (Davis 1993).
Carotid artery ligation
Carotid artery ligation was performed as described previously (Kumar and Lindner 1997). For histological analysis, the aortic trunk including the left (ligated) and right (control) carotid arteries were harvested on post-operative day 28, fixed in 4% paraformaldehyde overnight at 4°C, and embedded transversely in paraffin. Histological sections were obtained beginning at the ligature and collected every 100 μm over the next 2000 μm. All data were obtained using histological sections taken at 1900 μm proximal to the ligature.
Hemodynamic and cardiac morphologic measurements
To measure systemic hemodynamics, mice were anesthetized via isoflurane inhalation and transferred to a heat-controlled pad to maintain a constant body temperature. At a stable plane of anesthesia (1.5% isoflurane) the right carotid artery was exposed via direct midline neck incision, the distal end was ligated, and the artery was catheterized (1.4F Millar, Millar Instruments). After allowing the animal a minimum of 20 min for stabilization, a minimum of 15 min of steady-state data were obtained. Following completion of hemodynamic evaluation, the mice were euthanized and the heart was excised for morphological analysis. Under a stereomicroscope, hearts were cleaned of extraneous tissue and total heart and LV (free wall + septum) weights were recorded. Tibias were isolated from each animal and used to index whole heart and LV mass.
RNA purification and real-time PCR
Total RNA was isolated with Trizol reagent (Invitrogen) according to the manufacturer's protocol. After treatment with DNase I (RNase-free; Ambion), random hexamer-primed single-strand cDNAs were synthesized from 2 μg of total RNA using SuperScriptIII (Invitrogen). miRNA microarray analysis was performed using the mammalian miRNA probe set (LC Sciences). miRNA levels were confirmed using Northern analysis and TaqMan miRNA real-time probes (Applied Biosystems). Real-time PCR was performed using TaqMan probes on an ABI-PE Prism 7000 sequence detection system (Applied Biosystems) according to the protocol provided by the manufacturer. The relative quantities of mRNA were determined using comparative CT method and were normalized against GAPDH for mRNA or RNU6 for miRNA.
Microarray analysis
Primary neonatal rat ventricular cardiac myocytes were prepared as described (Molkentin et al. 1998). Twenty-four hours after plating, cardiac myocytes were infected with 10 MOI (multiplicity of infection) of adenovirus directing the expression of GFP or Flag epitope-tagged MRTF-A. Forty-eight hours after infection, total RNA was purified from cardiac myocytes using Trizol reagent (Invitrogen). miRNA microarray analysis was performed using the mammalian miRNA probe set (LC Sciences).
Western blot analysis
Ten micrograms to 50 μg of total protein extracts from descending aorta of adult mice were loaded on SDS-PAGE gels for Western blotting. Western blotting was performed by a standard protocol. Antibodies against KLF5 (Abcam), SM22 (Abcam), SMA (Sigma, A5228), and GAPDH (Chemicon) were used as described previously. Quantification of Western blots was performed by densitometry using the Storm 820 PhosphorImager and NIH ImageJ software.
Gel shift assays
Gel shift assays were performed using double-stranded oligonucleotides corresponding to the miR-143/145 CArG sequence. The oligonucleotide probe sequences used were as follows (with binding site in bold and mutations underlined): miR-143/145 CArG, 5′-GCCTTGCCATATAAGGGCAGGA-3′; miR-143/145 mut CArG, 5′-GCCTTGCCATATAATTGCAGGA-3′.
Protein lysates were incubated with 32P-labeled oligonucleotide probes and 1 μL of poly dIdC (1.0 μg/μL). Nondenaturing electrophoresis was carried out essentially as described previously using Flag-SRF or control plasmid containing COS cell extract (McFadden et al. 2000).
Transgenic mice
Transgenic mice were generated by pronuclear injection according to standard procedures. A 5.5-kb genomic sequence (Reporter A) upstream of the pre-miR-143 coding region, containing the two conserved regions, and a series of deletion constructs (including Reporter B) were cloned upstream of the hsp68 basal promoter and a lacZ reporter gene. The reporter construct was digested with SalI to remove vector sequence and was injected into fertilized oocytes from B6C3F1 female mice and implanted into pseudopregnant ICR mice. Embryos and adult tissues were collected at indicated stages and stained for β-galactosidase activity (Naya et al. 1999), and mice carrying lacZ transgenes were identified by PCR analysis. The primer sequences used to generate the 5.5-kb upstream and enhancer constructs were as follows: Reporter A, forward primer, 5′-CTCACCGACAAGTCCTGCAGTG-3′; Reporter A, reverse primer, 5′-GCGGCTTTCTTCTTGGGCAGGA-3′; Reporter B, forward primer, 5′-CAGAGGAAGGACTGGAGGTGAG-3′; Reporter B, reverse primer, 5′-CAACAGCTTGAAGGCCTGCTG-3′.
Acknowledgments
We thank Caroline Song for assistance with transfection experiments; Hiromi Yanagisawa and Shelby Chapman for assistance with carotid artery ligations; Kunhua Song, Ning Liu, and Eva van Rooij for discussions; Jose Cabrera for graphics; and J. Brown for editorial assistance. Knockout mice were generated by the University of Texax Southwestern Transgenic Core Facility. Electron microscopy was performed by the University of Texas Southwestern Electron Microscopy Core Facility. Work in the laboratory of E.N.O. was supported by grants from the NIH, the Robert A. Welch Foundation, the American Heart Association, the Donald W. Reynolds Center for Clinical Cardiovascular Research, and the Foundation Leducq TransAtlantic Network of Excellence Program.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1842409.
Supplemental material is available at http://www.genesdev.org.
References
- Barkalow KL, Italiano JE, Jr, Chou DE, Matsuoka Y, Bennett V, Hartwig JH. α-Adducin dissociates from F-actin and spectrin during platelet activation. J Cell Biol. 2003;161:557–570. doi: 10.1083/jcb.200211122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barringhaus KG, Zamore PD. MicroRNAs: Regulating a change of heart. Circulation. 2009;119:2217–2224. doi: 10.1161/CIRCULATIONAHA.107.715839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- Chen J, Kitchen CM, Streb JW, Miano JM. Myocardin: A component of a molecular switch for smooth muscle differentiation. J Mol Cell Cardiol. 2002;34:1345–1356. doi: 10.1006/jmcc.2002.2086. [DOI] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Callis TE, Wang DZ. microRNAs and muscle disorders. J Cell Sci. 2009;122:13–20. doi: 10.1242/jcs.041723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Liu X, Yang J, Lin Y, Xu DZ, Lu Q, Deitch EA, Huo Y, Delphin ES, Zhang C. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ Res. 2009;105:158–166. doi: 10.1161/CIRCRESAHA.109.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- Condorelli G, Dimmeler S. MicroRNAs: Components of an integrated system controlling cardiac development, physiology, and disease pathogenesis. Cardiovasc Res. 2008;79:551–552. doi: 10.1093/cvr/cvn189. [DOI] [PubMed] [Google Scholar]
- Cordes KR, Srivastava D. MicroRNA regulation of cardiovascular development. Circ Res. 2009;104:724–732. doi: 10.1161/CIRCRESAHA.108.192872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis EC. Smooth muscle cell to elastic lamina connections in developing mouse aorta. Role in aortic medial organization. Lab Invest. 1993;68:89–99. [PubMed] [Google Scholar]
- Du KL, Ip HS, Li J, Chen M, Dandre F, Yu W, Lu MM, Owens GK, Parmacek MS. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol Cell Biol. 2003;23:2425–2437. doi: 10.1128/MCB.23.7.2425-2437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiseler T, Doppler H, Yan IK, Kitatani K, Mizuno K, Storz P. Protein kinase D1 regulates cofilin-mediated F-actin reorganization and cell motility through slingshot. Nat Cell Biol. 2009;11:545–556. doi: 10.1038/ncb1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata Y, Oshiro N, Kinoshita N, Kawano Y, Matsuoka Y, Bennett V, Matsuura Y, Kaibuchi K. Phosphorylation of adducin by Rho-kinase plays a crucial role in cell motility. J Cell Biol. 1999;145:347–361. doi: 10.1083/jcb.145.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner K, Bennett V. Modulation of spectrin-actin assembly by erythrocyte adducin. Nature. 1987;328:359–362. doi: 10.1038/328359a0. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinson JS, Medlin MD, Lockman K, Taylor JM, Mack CP. Smooth muscle cell-specific transcription is regulated by nuclear localization of the myocardin-related transcription factors. Am J Physiol Heart Circ Physiol. 2007;292:H1170–H1180. doi: 10.1152/ajpheart.00864.2006. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Kumar A, Lindner V. Remodeling with neointima formation in the mouse carotid artery after cessation of blood flow. Arterioscler Thromb Vasc Biol. 1997;17:2238–2244. doi: 10.1161/01.atv.17.10.2238. [DOI] [PubMed] [Google Scholar]
- Kuwahara K, Barrientos T, Pipes GC, Li S, Olson EN. Muscle-specific signaling mechanism that links actin dynamics to serum response factor. Mol Cell Biol. 2005;25:3173–3181. doi: 10.1128/MCB.25.8.3173-3181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang DZ, Wang Z, Richardson JA, Olson EN. The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc Natl Acad Sci. 2003;100:9366–9370. doi: 10.1073/pnas.1233635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes & Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden DG, Charite J, Richardson JA, Srivastava D, Firulli AB, Olson EN. A GATA-dependent right ventricular enhancer controls dHAND transcription in the developing heart. Development. 2000;127:5331–5341. doi: 10.1242/dev.127.24.5331. [DOI] [PubMed] [Google Scholar]
- Medjkane S, Perez-Sanchez C, Gaggioli C, Sahai E, Treisman R. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat Cell Biol. 2009;11:257–268. doi: 10.1038/ncb1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miano JM, Long X, Fujiwara K. Serum response factor: Master regulator of the actin cytoskeleton and contractile apparatus. Am J Physiol Cell Physiol. 2007;292:C70–C81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, Grant SR, Olson EN. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller W, Moller G. Chemical dehydration for rapid paraffin embedding. Biotech Histochem. 1994;69:289–290. doi: 10.3109/10520299409106304. [DOI] [PubMed] [Google Scholar]
- Nagai R, Suzuki T, Aizawa K, Shindo T, Manabe I. Significance of the transcription factor KLF5 in cardiovascular remodeling. J Thromb Haemost. 2005;3:1569–1576. doi: 10.1111/j.1538-7836.2005.01366.x. [DOI] [PubMed] [Google Scholar]
- Naya FJ, Wu C, Richardson JA, Overbeek P, Olson EN. Transcriptional activity of MEF2 during mouse embryogenesis monitored with a MEF2-dependent transgene. Development. 1999;126:2045–2052. doi: 10.1242/dev.126.10.2045. [DOI] [PubMed] [Google Scholar]
- Niu Z, Li A, Zhang SX, Schwartz RJ. Serum response factor micromanaging cardiogenesis. Curr Opin Cell Biol. 2007;19:618–627. doi: 10.1016/j.ceb.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens GK. Regulation of differentiation of vascular smooth muscle cells. Physiol Rev. 1995;75:487–517. doi: 10.1152/physrev.1995.75.3.487. [DOI] [PubMed] [Google Scholar]
- Parmacek MS. Myocardin-related transcription factors: Critical coactivators regulating cardiovascular development and adaptation. Circ Res. 2007;100:633–644. doi: 10.1161/01.RES.0000259563.61091.e8. [DOI] [PubMed] [Google Scholar]
- Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: Versatile regulators of cell growth, migration, and myogenesis. Genes & Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- San Martin A, Lee MY, Williams HC, Mizuno K, Lassegue B, Griendling KK. Dual regulation of cofilin activity by LIM kinase and Slingshot-1L phosphatase controls platelet-derived growth factor-induced migration of human aortic smooth muscle cells. Circ Res. 2008;102:432–438. doi: 10.1161/CIRCRESAHA.107.158923. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Sawaki D, Aizawa K, Munemasa Y, Matsumura T, Ishida J, Nagai R. Kruppel-like factor 5 shows proliferation-specific roles in vascular remodeling, direct stimulation of cell growth, and inhibition of apoptosis. J Biol Chem. 2009;284:9549–9557. doi: 10.1074/jbc.M806230200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Olson EN. MicroRNAs: Powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007;117:2369–2376. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang DZ, Pipes GC, Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proc Natl Acad Sci. 2003;100:7129–7134. doi: 10.1073/pnas.1232341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wang DZ, Hockemeyer D, McAnally J, Nordheim A, Olson EN. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- Wang C, Han M, Zhao XM, Wen JK. Kruppel-like factor 4 is required for the expression of vascular smooth muscle cell differentiation marker genes induced by all-trans retinoic acid. J Biochem. 2008;144:313–321. doi: 10.1093/jb/mvn068. [DOI] [PubMed] [Google Scholar]
- Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K, Ren XR, Huang YZ, Xie Y, Liu G, Saito H, Tang H, Wen L, Brady-Kalnay SM, Mei L, et al. Signal transduction in neuronal migration: Roles of GTPase activating proteins and the small GTPase Cdc42 in the Slit–Robo pathway. Cell. 2001;107:209–221. doi: 10.1016/s0092-8674(01)00530-x. [DOI] [PubMed] [Google Scholar]
- Xin M, Davis CA, Molkentin JD, Lien CL, Duncan SA, Richardson JA, Olson EN. A threshold of GATA4 and GATA6 expression is required for cardiovascular development. Proc Natl Acad Sci. 2006;103:11189–11194. doi: 10.1073/pnas.0604604103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, Wang H, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13:486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Sinha S, Dandre F, Wamhoff BR, Hoofnagle MH, Kremer BE, Wang DZ, Olson EN, Owens GK. Myocardin is a key regulator of CArG-dependent transcription of multiple smooth muscle marker genes. Circ Res. 2003;92:856–864. doi: 10.1161/01.RES.0000068405.49081.09. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Kaestner KH, Owens GK. Conditional deletion of Kruppel-like factor 4 delays downregulation of smooth muscle cell differentiation markers but accelerates neointimal formation following vascular injury. Circ Res. 2008;102:1548–1557. doi: 10.1161/CIRCRESAHA.108.176974. [DOI] [PMC free article] [PubMed] [Google Scholar]