Abstract
Development of the germline requires consecutive differentiation events. Regulation of these has been associated with germ cell-specific and pluripotency-associated transcription factors, but the role of general transcription factors (GTFs) remains elusive. TATA-binding protein (TBP) is a GTF involved in transcription by all RNA polymerases. During ovarian folliculogenesis in mice the vertebrate-specific member of the TBP family, TBP2/TRF3, is expressed exclusively in oocytes. To determine TBP2 function in vivo, we generated TBP2-deficient mice. We found that Tbp2−/− mice are viable with no apparent phenotype. However, females lacking TBP2 are sterile due to defective folliculogenesis, altered chromatin organization, and transcriptional misregulation of key oocyte-specific genes. TBP2 binds to promoters of misregulated genes, suggesting that TBP2 directly regulates their expression. In contrast, TBP ablation in the female germline results in normal ovulation and fertilization, indicating that in these cells TBP is dispensable. We demonstrate that TBP2 is essential for the differentiation of female germ cells, and show the mutually exclusive functions of these key core promoter-binding factors, TBP and TBP2, in the mouse.
Keywords: TBPL2, TRF3, TBP, Zp3, RNA Polymerase II, ovary
Regulation of transcription initiation by RNA polymerase II (Pol II) is central to any developmental process. Development of the germline is essential for transmitting information between generations and for sexual reproduction. Female germ cells develop during oogenesis, leading to the formation of a highly differentiated and specialized cell, the oocyte. Oogenesis comprises an orderly number of events, each of which must be regulated. Although some work has been done to understand how specific transcription factors (Rajkovic et al. 2004; Choi and Rajkovic 2006) regulate oocyte-specific gene expression, little is known about the role of general transcription factors (GTFs) in regulation of female germ cell development.
The general RNA Pol II transcription factor TFIID is composed of TATA-binding protein (TBP) and 14 TBP-associated factors (TAFs) (Burley and Roeder 1996). TBP plays a crucial role in preinitiation complex assembly by nucleating the binding of GTFs to the core promoters of genes. TBP also plays a role in Pol I and Pol III transcription. TBP possesses two regions: an N-terminal domain and a highly conserved saddle-like C-terminal DNA-binding domain (Hernandez 1993). The N termini of the different TBP proteins are divergent among species and seem to have a role in regulation of Pol II and Pol III transcription (Muller and Tora 2004). In contrast to the generally accepted idea that TBP is a universal transcription factor, recent studies documented TBP-independent Pol II transcription (Muller and Tora 2004). Indeed, in the absence of TBP, TBP paralogs are able to mediate Pol II and Pol III transcription of developmentally important genes in vertebrate embryos (Takada et al. 2000; Veenstra et al. 2000; Muller et al. 2001; Martianov et al. 2002a; Ferg et al. 2007; Jacobi et al. 2007). TBP-like factor (TLF, also known as TBPL1, TRF2, or TRP) has been found in different metazoan species. TLF shares ∼45% identity with the core domain of TBP and can interact with TFIIA and TFIIB, but cannot bind the TATA box (Muller and Tora 2004). In contrast, another vertebrate-specific TBP-related factor, TBP2 (also known as TBPL2 or TRF3) shows ∼92% conservation in its DNA-binding core domain compared with TBP. Indeed, TBP2 is able to bind the TATA box, interact with TFIIA and TFIIB, and mediate Pol II transcription initiation in vitro (Bartfai et al. 2004; Jallow et al. 2004; Deato et al. 2008). Moreover, a role for TBP2 in directing cell type-specific transcription using an ex vivo model of myotube differentiation has been reported (Deato and Tjian 2007). In mice, the expression of Tbp2 mRNA has been detected exclusively in the ovary, specifically in the oocytes (Bartfai et al. 2004; Xiao et al. 2006). Enrichment of TBP2 in the ovary is conserved in zebrafish and Xenopus, further suggesting a fundamental role for TBP2 in the vertebrate ovary (Bartfai et al. 2004; Jallow et al. 2004).
Most oocytes enter meiosis during the embryonic life or during the early neonatal period. At birth, ovaries contain clusters of oocytes without surrounding granulosa cells that are in transitory stages of prophase (pachytene to early diplotene) or in late diplotene (Pedersen and Peters 1968). From day 3 after birth, some primordial follicles form, undertake a growth phase, and enter folliculogenesis (Matzuk and Lamb 2002). Oocytes within the primary follicle start to increase their size and undergo growth during the preantral follicular stages. Later, in antral, preovulatory follicles, the fully grown germinal vesicle (GV) stage oocyte is ready for ovulation. Maturation follows with the resumption of meiosis induced by ovulatory stimulus and proceeds until the metaphase II, at which stage the oocyte awaits fertilization. Genes specifically expressed in the oocyte are necessary for either growth or communication with follicular cells. These include Zona Pellucida 3 (Zp3) (Bleil and Wassarman 1980), the growth factors Bmp15 (bone morphogenetic protein 15) and Gdf9 (growth differentiation factor 9) (Yan et al. 2001), and transcription factors specific to the oocyte such as Nobox (Rajkovic et al. 2004). Null females for all these genes are sterile (Andreu-Vieyra et al. 2006).
The identification of gonad-specific variants of core promoter-binding factors suggests that, in the gonads of different metazoan organisms, specific transcription initiation mechanisms have evolved (Muller and Tora 2004). The oocyte-specific expression of TBP2 (Xiao et al. 2006; Gazdag et al. 2007) prompted us to determine the role of TBP2 in vivo. We show that Tbp2−/− mice are viable and display no obvious morphological phenotype. However, females lacking TBP2 are sterile due to defective folliculogenesis. Ovaries from Tbp2−/− females show defects in the formation of secondary follicles. Tbp2−/− females lack fully grown GV stage oocytes and do not ovulate. Transcriptome analysis of ovaries from Tbp2-deficient mice revealed a high number of oocyte-specific genes severely deregulated. TBP2 binds to some of their promoters, and this suggests a direct regulation. In contrast to TBP2, we show that TBP is dispensable for oocyte maturation and fertilization. Finally, when TBP2 is misexpressed in mouse embryos, it has a negative effect on cell proliferation, leading to developmental arrest. Thus, TBP2 plays a critical and specialized role in mammalian female germ cell development.
Results
Tbp2−/− mice are viable without obvious abnormalities or apparent anatomical aberration
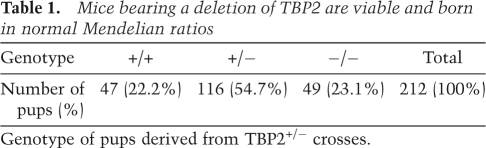
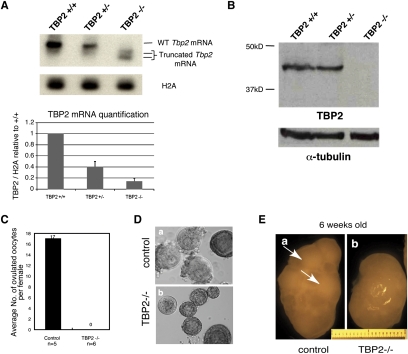
The dynamic nature and specific expression of TBP2 during oogenesis (Xiao et al. 2006; Gazdag et al. 2007) suggests that it may play a role in female germ cell development. To investigate the in vivo role of TBP2, the Tbp2 (Tbpl2) gene was targeted by homologous recombination and a Cre-loxP strategy. Embryonic stem (ES) cells were generated in which exon 4 of Tbp2 was flanked by two loxP sites (Supplemental Fig. S1A). To obtain Tbp2+/− mice, we deleted floxed exon 4 by breeding mice carrying the recombined allele with CMV-Cre mice (White et al. 1997), which led to the generation of the null allele (Supplemental Fig. S1A,B). Deletion of exon 4 is expected to produce a shorter mRNA that, if stable, would give rise to a TBP2 protein truncated at amino acid 206 in which the core domain is absent, thereby eliminating its DNA-binding domain. Tbp2+/− mice were viable and indistinguishable from their wild-type littermates. To generate Tbp2-null mice, we performed Tbp2+/− crosses. Tbp2−/− mice were born at normal Mendelian ratios, indicating no embryonic lethality (Table 1). Expression of TBP2 mRNA was strongly reduced in Tbp2−/− ovaries (Fig. 1A) and TBP2 protein was not detected in extracts from these ovaries, demonstrating that we generated a null allele (Fig. 1B). Tbp2 heterozygotes showed reduced levels of TBP2 mRNA, but protein levels appeared largely unaffected compared with wild-type littermates (Fig. 1A,B). Tbp2−/− mice developed to adulthood normally, were of normal size and weight, and showed no external abnormalities or apparent anatomical aberrations. Hence, TBP2 is not essential for mouse viability.
Table 1.
Mice bearing a deletion of TBP2 are viable and born in normal Mendelian ratios
Genotype of pups derived from TBP2+/− crosses.
Figure 1.
Characterization of Tbp2-deficient mice. (A) RNA from ovaries derived from females of the indicated genotype was reverse-transcribed and amplified with primers specific for TBP2. PCR products were transferred and hybridized with a radioactive Tbp2-specific probe to confirm the specificity of the products and to allow quantification. Histone H2A was used as internal control. Quantification of TBP2 mRNA levels in Tbp2+/+, Tbp2+/−, and Tbp2−/− ovaries is shown on the graph. Data are normalized to H2A expression and calculated relative to TBP2 mRNA expression in wild-type ovaries. Average ± SEM of two ovaries per genotype are represented. (B) TBP2 protein is absent in Tbp2−/− ovaries. Western blot analysis on total extracts from ovaries of females of the indicated genotype using the α-TBP2 antibody. α-Tubulin was used as a loading control. Shown is a representative experiment of three biological replicates. (C) Tbp2−/− females do not ovulate upon hormonal stimulation. Control and Tbp2−/− 4- to 6-wk-old females were induced for superovulation with intraperitoneal injection of PMSG followed by administration of hCG 48 h later. The number of ovulated oocytes was determined after dissection for each female. The average number of oocytes ovulated per female is indicated. (D) Increased number of immature follicles and abnormal oocytes in Tbp2−/− females. Examples of follicles dissected from control and Tbp2−/− ovaries observed under DIC microscopy. While GV stage (preovulatory) oocytes are easily distinguishable in control females, Tbp2−/− oocytes contain mainly immature follicles or oocytes of abnormal morphology and devoid of zona pellucida. (E) Gross morphology of ovaries from 6-wk-old control and Tbp2−/− females. While control ovaries show clear mature follicles (arrows), such structures are absent in the Tbp2−/− ovaries.
Tbp2−/− females are sterile due to defective folliculogenesis
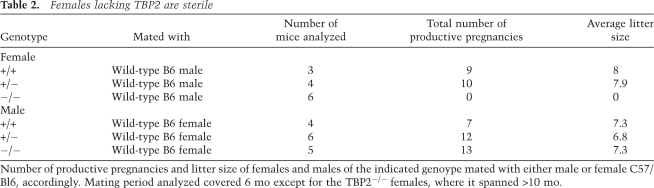
Given the specific expression of TBP2 in the female gonads (Xiao et al. 2006; Gazdag et al. 2007), we next asked whether Tbp2−/− mice exhibited normal reproductive success. Wild-type, heterozygous, and mutant females were caged with wild-type males over a breeding period of 6 mo. While Tbp2+/+ and Tbp2+/− females gave birth repeatedly and with normal litter sizes, Tbp2-null females never gave birth when caged with several fertile males (Table 2). This has been observed for Tbp2−/− females for a continued breeding period of 12 mo, indicating that Tbp2−/− females are sterile. In contrast, Tbp2−/− males were fertile and produced normal litters (Table 2). This female sterility phenotype is consistent with the ovary-restricted expression of TBP2 in the mouse (Bartfai et al. 2004; Xiao et al. 2006; Gazdag et al. 2007), and demonstrates that TBP2 is absolutely required for female fertility.
Table 2.
Females lacking TBP2 are sterile
Number of productive pregnancies and litter size of females and males of the indicated genoype mated with either male or female C57/Bl6, accordingly. Mating period analyzed covered 6 mo except for the TBP2−/− females, where it spanned >10 mo.
In order to get insight into the mechanisms underlying the ovarian failure elicited by TBP2 loss, we first assessed whether the sterility phenotype could be overridden by hormonal stimulation. We superovulated Tbp2+/+ and Tbp2−/− female littermates and determined the number of ovulated oocytes per female. While all control females responded to hormonal stimulation and ovulated mature oocytes (17 oocytes per female, SD = 14, n = 5), we did not find any ovulated oocytes in Tbp2−/− females (Fig. 1C). In agreement with this, superovulated Tbp2−/− females never gave birth (data not shown). Dissection of control and Tbp2−/− ovaries revealed an increased number of immature follicles and abnormal oocytes (Fig. 1D, panel b). In contrast, oocytes dissected from Tbp2+/+ littermates included mature, dictyate GV stage oocytes in antral follicles, indicative of normal ovarian physiology (Fig. 1D, panel a). These observations suggest that TBP2-null females are devoid of mature fully grown oocytes.
To elucidate the defect in oogenesis of Tbp2−/− mice, we performed detailed ovarian morphological analyses. Ovaries from null and wild-type 2-wk-old females (e.g., prepuberty) were of similar size and morphological appearance (data not shown). However, ovaries of 6-wk- old mutant mice were significantly smaller than wild-type ones (Fig. 1E). Moreover, while mature follicles are clearly visible in control ovaries (Fig. 1E, panel a, arrow), ovaries from Tbp2−/− females lack such structures (Fig. 1E, panel b).
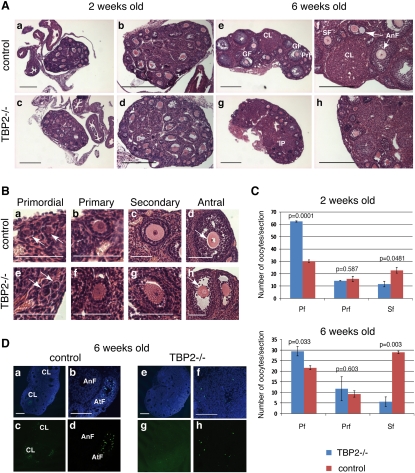
We next analyzed the histology of 2-wk-old and 6-wk-old ovaries in Tbp2−/− mice. In 2-wk-old Tbp2−/− ovaries, follicle development takes place up to the secondary follicle stage, and this appeared to occur normally when compared with wild-type ovaries (Fig. 2A, panels a–d). However, a detailed analysis of the number of oocytes in each stage on individual sections revealed an increased number of oocytes that persisted in the primordial follicle stage and a reduced number of oocytes at the secondary follicle stage (Fig. 2C). This presumably reflects the limited recruitment of follicles for growth and results in reduced numbers of preantral and antral follicles (Fig. 2A,C). Adult ovaries from Tbp2−/− females were remarkably different from the control ovaries (Fig. 2A, panels e–h). At 6 wk of age there was a gradual depletion of follicles in Tbp2−/− ovaries and abnormal oocyte development became pronounced (Fig. 2A, panels e–h). Null ovaries showed greater numbers of small follicles in the cortex that were delayed in their development and that consisted mainly of abnormal secondary follicles (Fig. 2A). The oocytes within the secondary follicles with antrum showed signs of degeneration and irregularly shaped nuclei (Fig. 2A,B). These oocytes lost their symmetrical rounded shape and the zona pellucida was absent (Fig. 2B, panel h, arrow). Phenotypical changes upon TBP2 loss were further evident in the number of small (late primary and secondary) follicles containing irregularly shaped oocytes and the emergence of cyst-containing (empty) structures as well as interstitial cell proliferation. Anovular small, empty follicles with disarranged granulosa cells were also frequent in the mutant compared with the wild type, and no signs of corpus luteum (CL) formation were detected (Fig. 2A,B). Overall, adult Tbp2−/− ovaries showed a strong decrease in secondary follicles (Fig. 2C), suggesting that folliculogenesis is impaired before or during secondary follicle stages. Given the above-described degeneration of oocytes, we next looked for apoptosis in Tbp2−/− ovaries by TUNEL analysis. We did not detect increased apoptosis in ovaries from Tbp2-null females at 2 wk of age (data not shown). In control 6-wk-old ovaries, we observed apoptotic granulosa cells in ovulated follicles that can be distinguishable because of the presence of the CL (Fig. 2D). In contrast, these apoptotic granulosa cells were missing in the Tbp2−/− mice (Fig. 2D, panels e,f), further demonstrating that TBP2-deficient oocytes do not reach a fully grown ovulatory stage. These data indicate that TBP2 is essential for proper oocyte growth and follicular development and its absence leads to infertility.
Figure 2.
TBP2 loss results in ovarian failure due to defective folliculogenesis. (A–C) Histological analysis of ovaries from Tbp2−/− mice reveals a block in folliculogenesis around the primary and secondary follicular stage. In A, sections from 2-wk-old and 6-wk-old control (panels a–b,e–f) and Tbp2−/− (panels c–d,g–h) ovaries stained with hematoxylin-eosin are shown at two different magnifications. In control 6-wk-old ovaries, primary (Prf), secondary (SF), and antral follicles (AnF) as well as corpus lutei (CL) are indicated. Tbp2−/− oocytes lack antral secondary follicles and large graafian follicles; instead, intersticial proliferation (IP) is visible in panel g. Data are representative of three biological replicates. Bar, 100 μm. (B) Higher magnifications of oocytes at different stages of follicular development from 6-wk-old control (panels a–d) and Tbp2−/− (panels e–h) ovaries. (Panels a,e) Arrows point to two different primordial follicles, which appear normal when compared with control ovaries. Primary and secondary follicles can be found in null ovaries, albeit in reduced numbers (see C). (Panel h) Following the secondary follicle stage, the few Tbp2−/− oocytes that survive fail to undergo antrum formation and instead show disorganized follicular structure. Note the absence of zona pellucida in these oocytes as seen from the lack of regular white space surrounding the oocyte (arrow). (C) Increased number of underdeveloped oocytes in Tbp2−/− females. Analysis of oocyte counts comparing Tbp2−/− and control 2-wk-old and 6-wk-old ovaries. Oocytes within primordial (PF), primary (PrF), and secondary (SF) follicles were counted on ovarian sections. Numbers represent average of counts of three sequential sections from serially sectioned ovaries. Antral oocytes are only present in the controls and were therefore not included in the counts. P values were calculated using an unpaired t-test for triplicates. (D) TUNEL assay on sections from control and Tbp2−/− ovaries. Apoptotic nuclei were labeled using a modified TUNEL protocol with fluorescein detection. DNA was stained with DAPI. In 6-wk-old control sections, an artetic follicle (AtF) and nuclei within the CL show normal signs of apoptosis. A healthy antral follicle (AnF) is also depicted. Representative merge (panels a,b,e,f) and green (panels c,d,g,h) channel images of lower (panels a,c,e,g) and higher (panels b,d,f,h) magnifications are shown.
TBP2 is essential for transcription and correct chromatin organization in the developing oocytes
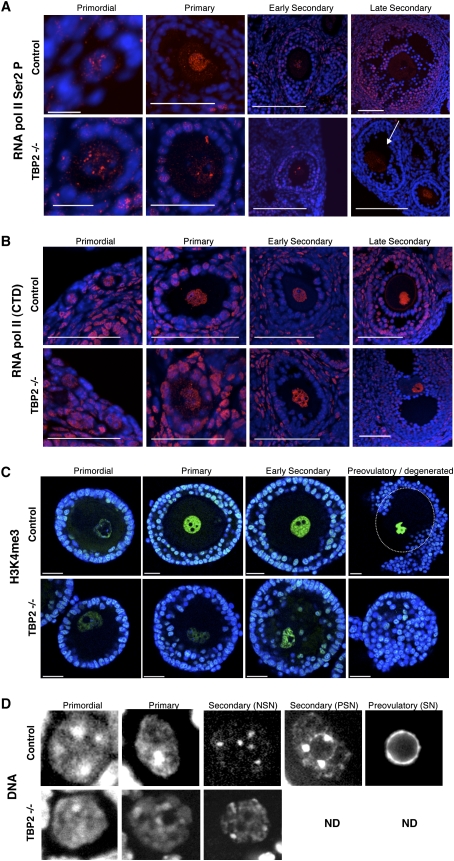
Because TBP2 is able to mediate transcription initiation in vitro (Bartfai et al. 2004; Deato et al. 2008), we asked whether TBP2 can regulate transcription in vivo. To this end, we examined the transcriptional competence of Tbp2−/− oocytes by visualizing the Ser2 phosphorylation state of the C-terminal domain (CTD) of the largest subunit of Pol II, an indicator of Pol II activity (Bellier et al. 1997). Phosphorylation of the CTD is highest at the primary and preantral stages of folliculogenesis. It then decreases gradually until it is almost undetectable at the fully grown preovulatory stage, when the oocyte is transcriptionally silent (Bachvarova 1985). Primordial follicles displayed similar levels of CTD phosphorylation in control and Tbp2−/− mice (Fig. 3A). However, reduction in the Ser2 phosphorylation of the CTD was observed in TBP2-deficient oocytes from 6-wk-old females, mainly at the primary follicle stage (Fig. 3A). We next asked whether the reduction of Ser2 phosphorylation of the Pol II CTD was due to a reduction in the levels of Pol II protein by using an antibody that recognizes all Pol II CTD repeats. No reduction in the levels of Pol II CTD was detected at any of the stages of oocyte development analyzed, when comparing TBP2-deficient and control oocytes (Fig. 3B). Therefore, despite the fact that Pol II levels remain stable, it appears that transcription is perturbed mainly at the primary follicle stage as a result of TBP2 loss, at a stage where wild-type oocytes exhibit extensive transcriptional activity (Bachvarova 1985).
Figure 3.
RNA Pol II activity and chromatin structure are altered in oocytes lacking TBP2. (A,B) Immunofluorescence analysis of control and Tbp2−/− ovarian sections from 6-wk-old females using antibodies recognizing the Ser2-phophorylated CTD of Pol II (A) or all forms of the CTD of Pol II (B). Pol II staining is shown in red. DNA (blue) was counterstained with DAPI. Follicular stages are indicated. (A) The arrow points to the follicle at the stage indicated at the top. Bars: for primordial stages, 10 μm; for later stages, 50 μm. (B) Bar, 50 μm. (C) Tbp2−/− oocytes show reduced H3K4me3 levels from the primary follicular stage. Immunostaining analysis for H3K4me3 in oocytes at different stages of follicular development from 6-wk-old females. DNA (blue) was stained with DAPI. Bar, 10 μm. Confocal acquisition was done using identical parameters to allow comparison. Hence, H3K4me3 levels in preovulatory stage oocyte in the control appear saturated. Note that Tbp2−/− oocytes degenerate after the secondary follicular stage, as seen from the presence of follicular cells within the follicle. The dashed white line demarcates the oocyte membrane. (D) Altered chromatin configuration in Tbp2−/− oocytes. Sections from control and Tbp2−/− mice were stained with DAPI and analyzed under confocal microscopy. (NSN) Nonsurrounded nucleolus; (PSN) partially surrounded nucleolus; (SN) surrounded nucleolus. Representative sections are shown. (ND) Not determined.
As another marker of active transcription, we examined global levels of histone H3K4 trimethylation (H3K4me3) in Tbp2-null ovaries. Consistent with the reduction of levels of Pol II phospho-Ser2, oocytes lacking TBP2 showed diminished levels of H3K4me3 from the primary follicle stage onward (Fig. 3C). This observation further indicates that the transcription-related chromatin organization is impaired in Tbp2−/− oocytes. While control oocytes showed the normal chromatin maturation process from the so-called nonsurrounded nucleolus to a surrounded nucleolus chromatin configuration during folliculogenesis (Zuccotti et al. 2005), Tbp2−/− oocytes displayed a disorganized chromatin (Fig. 3D). Instead of forming one prominent nucleolar-like body, dispersed small DAPI-rich regions were formed (Fig. 3D), indicating that from the primary follicle stage the chromatin organization of the mutant oocytes is abnormal. Hence, these data indicate that TBP2 is required for correct chromatin organization.
TBP2 loss leads to altered transcriptional profile of oocyte-specific genes
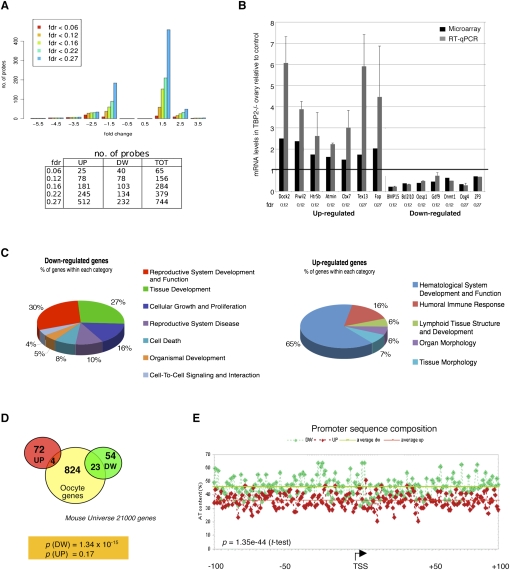
To identify genes misregulated in Tbp2−/− ovaries (see the Discussion), we performed a transcriptome analysis from 2-wk-old females. At the very stringent false discovery rate (FDR) of 0.12 we identified 156 misregulated probes, which correspond to 153 genes, among which half (76) were up-regulated and half (77) were down-regulated upon TBP2 loss (Fig. 4A; Supplemental Tables S1, S2). When a standard FDR value of 0.27 was used, a greater number of misregulated genes was identified, with 744 probes, equivalent to 714 genes (Fig. 4A). Thus, as expected, the number of probes increases as the FDR stringency is lowered. Using the 0.27 FDR, we identified more genes in the up-regulated class as compared with the down-regulated one (Fig. 4A). However, at any given FDR value, we found more genes with high negative fold changes in the down-regulated class than in the up-regulated one (Fig. 4A), suggesting that the down-regulated genes may be direct TBP2 target genes.
Figure 4.
TBP2 ablation results in altered oocyte-specific transcriptional profile in 2-wk-old ovaries. (A) Distribution of the number of probes on the microarray with respect to their FDR and fold change in 2-wk-old the Tbp2−/− ovaries compared with the control. (Top panel) In the histogram, bars of different colors represent the number of probes differentially hybridized at the indicated FDR threshold. (Bottom panel) The exact number of probes is displayed in the table. At FDR = 0.12 (yellow bars), there are as many probes in the down-regulated class (negative fold change) as there are in the up-regulated one (positive fold change). (B) Microarray validation in 2-wk-old Tbp2−/− and Tbp2+/+ ovaries. Up-regulated and down-regulated genes with different FDR and fold change values (ranging from −0.22-fold to 2.5-fold) were randomly selected and their mRNA levels were analyzed by RT-qPCR. mRNA levels are shown relative to 18S RNA and are expressed relative to the control, for which mRNA levels were set at 1 (black line). Microarray (black bars) and RT-qPCR (gray bars) fold changes are average of three independent biological replicates, respectively. Error bars are SD. (C) Most enriched functional categories among the down-regulated and up-regulated genes according to Ingenuity Pathways. (D) Genes down-regulated upon TBP2 loss are enriched in oocyte-specific genes. Comparison of the up-regulated and down-regulated genes with an FDR of 0.12 with genes that are specifically expressed in the oocyte. P values for the intersection of the up-regulated and down-regulated genes are shown at the bottom. Note that only the intersection of down-regulated genes is statistically significant. (E) Sequence composition of the promoters belonging to the two classes of genes according to their nucleotide content. The percentage of AT content of the up-regulated (red) and down-regulated (green) gene promoters within a region from −100 to +100 bp around TSS is shown. The average AT content for the two promoter populations is also indicated. Comparison of the two groups of genes using a Student's t-test indicates that the two populations of promoters differ significantly. The TSS positions follow the current NCBI annotation.
We next validated our microarray analysis by randomly choosing seven up-regulated and seven down-regulated genes that included genes with both a high and a low fold change and an FDR ranging between 0.12 and 0.27 and assessed their expression by RT–qPCR (Fig. 4B). All genes analyzed within this FDR range behaved as predicted by the microarray (Fig. 4B). We next asked whether misregulated genes in TBP2-depleted ovaries are involved in specific functional categories. We used the 153 genes with an FDR of 0.12 for this analysis. Among the down-regulated genes, the most enriched functional categories were reproductive and developmental processes, tissue development, and cellular growth and proliferation (Fig. 4C, left). The significant enrichment of genes (P = 0.002) belonging to the reproductive system category is in full agreement with an oocyte-specific role of TBP2 and the sterility phenotype of Tbp2-null females. The up-regulated class of genes also shows enrichment in various developmental and tissue differentiation pathways, although the statistical significance is lower (P = 0.049). Among these, the hematological system is highly overrepresented, suggesting impaired or delayed vascular development in Tbp2−/− ovaries.
We further compared misregulated genes in Tbp2−/− ovaries with an oocyte-specific set of genes that we built from the Novartis Tissue Expression Atlas (http://expression.gnf.org). This set contained genes expressed in oocytes and genes coding for maternally inherited transcripts in the cytoplasm of the fertilized zygote. We compared this oocyte-specific gene list, which contains 851 genes, to the up-regulated and down-regulated genes identified in Tbp2−/− ovaries (Fig. 4D). Intersection of the down-regulated genes revealed that 29% (23 out of 77) of the genes are specific to the oocyte (P = 1.34e−15), and may thus be direct targets of TBP2 (Supplemental Table S3). In contrast, the up-regulated category did not show a significant enrichment for oocyte-specific genes (P = 0.17) (Fig. 4D).
We next aimed to identify putative common signatures within the regulatory elements of the misregulated genes. We searched for common transcription factor-binding motifs in the regions spanning from −200 to +50 base pairs (bp) relative to their transcription start sites (TSSs) using pScan (Zambelli et al. 2009). This analysis revealed that promoters of the down-regulated genes are significantly enriched in AT-rich binding motifs including the TATA box. This pattern was not found in the up-regulated promoters, for which no significant motif appeared. The nucleotide composition between the two classes of promoters was also significantly different, with promoters of genes positively regulated by TBP2 displaying higher AT content, in particular in the −30, +1, and +60 to +80 regions relative to the TSS (Fig. 4E). Thus, the two groups of genes can be distinguished at the promoter level on the basis of their composition, suggesting different mechanisms of transcriptional regulation for up-regulated and down-regulated genes. Taken together, our data demonstrate that ablation of TBP2 results in major changes in the ovary transcriptome, a significant number of which are genes specifically expressed in the oocyte, substantiating an oocyte-specific transcriptional regulatory function of TBP2.
TBP2 binds promoters of actively transcribed genes in the oocytes
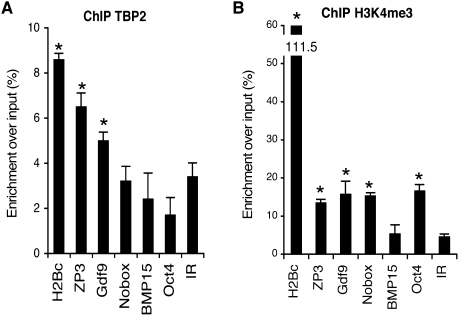
Because TBP2 is required for the correct expression of genes in the oocyte, we next asked whether endogenous TBP2 directly regulates such genes. To this end, we tested whether TBP2 binds to the promoter of transcribed genes in the oocyte. We analyzed the promoters of genes encoding two secreted growth factors (Bmp15 and Gdf9), Zp3, and two transcription factors (Oct4/Pou5f1 and Nobox). These genes are expressed to different extents in the oocyte (Supplemental Fig. S2) and were either down-regulated (Bmp15, Gdf9, and Zp3) or up-regulated (Oct4 and Nobox) in Tbp2−/− ovaries (Supplemental Tables S1, S2). Furthermore, because histone genes are highly transcribed in the oocyte, we also included histone H2B promoter in our analysis. We performed chromatin immunoprecipitation (ChIP) with ovaries from 2-wk-old females, which contain mainly oocytes at early stages of folliculogenesis (Kerr et al. 2006), when TBP2 accumulation is highest (Gazdag et al. 2007). We used primers encompassing the TSS for each gene and two primer pairs for an intergenic region (IR) as negative control. We detected a good and reproducible enrichment of TBP2 at Zp3 and the H2bc promoters (Fig. 5A). The Gdf9 promoter displayed significant, albeit weak, binding of TBP2. ChIP with a H3K4me3 antibody revealed a good enrichment of H3K4me3 at the promoter regions of H2bc, Zp3, Gdf9, Oct4, and Nobox, but not on the Bmp15 promoter, further indicating that these genes are actively transcribed in the oocyte (Fig. 5B). Thus, our data demonstrate that endogenous TBP2 is able to bind to promoters of genes that are actively transcribed in the oocyte; in particular, to those showing a high level of expression and whose expression is severely down-regulated upon TBP2 loss. These data, together with the transcriptome analysis, indicate that TBP2 plays an essential and direct role in mediating oocyte-specific transcription during folliculogenesis.
Figure 5.
TBP2 binds to promoters of active genes in the oocyte. (A–B) ChIP using anti-TBP2 or anti-H3K4me3 antibodies as indicated and promoter regions of oocyte-specific genes were analyzed by real-time qPCR. Data is represented as the enrichment over the input (in percentage). IRs are shown as negative controls. ChIP with a mouse or rabbit IgG antibody showed no enrichment above background levels. Shown are representative results of at least three independent experiments. Asterisk (*) indicates significant enrichment (P ≤ 0.05). Note that although Bmp15 mRNA accumulation in the oocyte is high (Supplemental Fig. S2), H3K4me3 levels on its promoter are not significant, suggesting that H3K4me3 regulation on Bmp15 promoter is different or that Bmp15 mRNA is relatively stable and that actual rate of active transcription of this gene is low.
TBP is dispensable for oocyte maturation, zygotic genome activation, and subsequent development
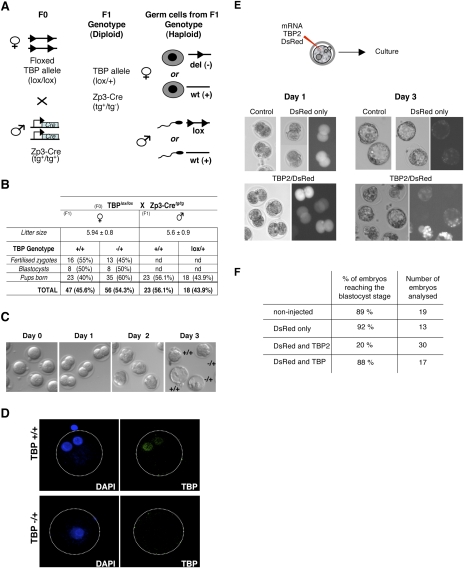
We established that TBP2 is a functional oocyte transcription factor in vivo. The distribution of TBP2 and TBP at the protein level is mutually exclusive during oocyte development, TBP2 being predominant in the oocyte during folliculogenesis (Gazdag et al. 2007). Thus, we next asked whether loss of TBP has an effect on oocyte development and subsequent fertilization. Depletion of TBP in the oocyte was achieved by a genetic approach using a Zp3-Cre transgenic line (Fig. 6A). Males homozygous for the Zp3-Cre transgene (de Vries et al. 2000) were mated with females homozygous for the floxed Tbp allele (Tbplox/lox) (Martianov et al. 2002b). Zp3 promoter-driven Cre expression is restricted to the oocyte, thereby resulting in maternal depletion of floxed Tbp allele. As a result of Cre recombination, 50% of the haploid oocytes ovulated from the females derived from these crosses will inherit the deleted Tbp allele and 50% will inherit the wild-type allele (Fig. 6A). In order to analyze whether inheritance of Tbp in the oocyte is required for ovulation and/or embryonic development, we mated females issued from Tbplox/lox;Zp3-Cretg/tg crosses with wild-type males. As a reciprocal control, littermate males issued from the same Tbplox/lox;ZP3-Cretg/tg crosses were mated with wild-type females (Fig. 6A). We first asked whether Tbp− haploid oocytes could be fertilized in normal ratios: We observed no difference between the oocytes carrying Tbp− or Tbp+ (wild-type) alleles (Fig. 6B). We further assessed whether embryos inheriting the maternal Tbp− allele develop until the blastocyst stage in the same proportions as those inheriting the Tbp+ allele. Both groups of embryos developed equally well and with the same timing (Fig. 6B–C). Thus, developmental progression was unaffected upon loss of the maternal Tbp allele. Finally, we analyzed the number of pups born that carried the maternal Tbp− allele versus those carrying the maternal Tbp+ allele (Fig. 6B). Pups that inherited the Tbp-deleted allele from their mothers were born at 60% frequency, whereas those inheriting the Tbp+ wild-type allele were found at 40%. We verified that TBP protein was efficiently depleted by analyzing zygotes immediately after fertilization by immunostaining using an anti-TBP antibody. As shown in Figure 6D, TBP was undetectable in the pronuclei of 50% of the zygotes analyzed (n = 9). Single embryo genotyping following immunostaining confirmed that zygotes displaying no TBP staining were Tbp−/+, and hence inherited the deleted allele from their mother (Fig. 6D). Collectively, these data suggest that, in contrast to TBP2, maternal TBP is not required for oocyte maturation, fertilization, and subsequent development. More generally, our results show that, in oocytes, TBP2 is the major factor mediating Pol II transcription during female germ cell development, and that TBP is dispensable in this process during oocyte maturation.
Figure 6.
TBP is dispensable for oocyte maturation, ovulation, and subsequent fertilization. (A) Genetic approach used to delete TBP in the maternal germline. Homozygous females for the floxed Tbp allele were mated to homozygous males for the ZP3-Cre transgene. Females express Cre in their oocytes. Haploid genotype of 50% of their oocytes is thus Tbp− (deleted) and the other 50% have a Tbp+ (wild-type) genotype. To determine the competence of these oocytes, females were mated with wild-type males as indicated. Male littermates of the F1 were used as controls. (B) Table summarizing the results obtained from the crosses between transgenic F1 females (Tbplox/+;ZP3-Cretg/+) or males and their respective wild-type partner. (nd) Not determined. (C) Oocytes carrying the maternal Tbp− (deleted) allele can be fertilized in vivo and develop normally until the blastocyst stage. Zygotes derived from F1(Tbplox/+;ZP3-Cretg/+) females mated with wild-type males were collected at fertilization and were cultured until the blastocyst stage. Single embryo genotyping was performed subsequently. (D) Efficient deletion of TBP in oocytes ovulated from F1(Tbplox/+;ZP3-Cretg/+) females upon Cre expression driven from the Zp3 promoter. Zygotes were collected from F1 X wild-type male crosses immediately after fertilization, stained with anti-TBP antibody (3G3), and analyzed using confocal microscopy. Individual embryos were genotyped after acquisition. Shown are representative embryos derived from oocytes inheriting the deleted Tbp allele maternally (Tbp−/+) and those inheriting the Tbp wild-type allele (Tbp+/+). Note that in the Tbp+/− embryo, the two pronuclei mask each other. (E) TBP2 overexpression results in altered embryonic development and in reduced cell proliferation. Experimental design for TBP2 overexpression is shown on the top. Zygotes were microinjected with mRNA for DsRed alone (control) or in combination with mRNA for TBP2 and cultured until control embryos reached the blastocyst stage. (F) Zygotes overexpressing TBP2 cleave to the two-cell stage normally, but show reduced development until the blastocyst stage.
TBP2 overexpression has a negative effect on progression of embryonic development and cell growth
We established that, in primary and secondary follicular stage oocytes, TBP2 is the main TBP-type factor required for Pol II transcription initiation. However, TBP2 protein levels decline upon ovulation to become undetectable after fertilization (Gazdag et al. 2007). The mutually exclusive roles described for TBP2 and TBP in the oocytes are the opposite following fertilization, when only TBP becomes abundant concomitant with zygotic genome activation. Thus, to test whether TBP2 can function in the zygote in parallel with TBP, we used a gain-of-function approach. We microinjected zygotes with mRNA for TBP2 together with mRNA of the fluorescent DsRed protein as a marker for injection. As controls, we used noninjected embryos, embryos injected with mRNA for TBP and DsRed, and embryos injected with DsRed mRNA only (Fig. 6E,F). Injection of TBP and TBP2 mRNA led to efficient protein translation (data not shown). Zygotes overexpressing TBP2 cleaved to the two-cell stage normally and at the same time as the control groups (Fig. 6E, day 1). Noninjected, TBP + DsRed, and DsRed-only embryos reached the blastocyst stage after 3 d. In contrast, TBP2-expressing embryos displayed a low development rate: Only 20% reached the blastocyst stage (Fig. 6E [day 3], F). Among these, 40% ceased development at the two-cell stage, 6.6% ceased development at the two- to four-cell stage, and 33% ceased development between the four- and eight-cell stage (data not shown). Thus, misexpression of TBP2 in the embryo is deleterious to development, further suggesting that its role is restricted to the highly specialized female germ cell transcription program.
Discussion
TBP2 is an oocyte-specific TBP-type factor
Recent data support the idea that an alternative form of transcription initiation machinery has evolved to control the expression of genes required for differentiation and development (Deato and Tjian 2007; Jones 2007; Muller et al. 2007; Reina and Hernandez 2007). Determining the importance of homolog transcription factors in a given species in mediating initiation of transcription by Pol II at different developmental stages and in different cell types is central to understanding how transcription is regulated in a cell-specific context. The two core promoter-binding factors TBP2 and TBP share an almost identical DNA-binding domain and differ in their N termini. Although TBP2 has been shown to interact with TFIIA and TFIIB, to bind a consensus TATA box, and to mediate transcription initiation in vitro, the role of TBP2 in mammalian cells has been only partially addressed. Here we show that (1) TBP2-deficient mice are viable, but females are sterile; (2) TBP2 is the main essential TBP-type transcription factor during follicular female germ cell differentiation; (3) TBP is not required during the later stages of folliculogenesis and subsequent fertilization; (4) TBP2 inhibits cell proliferation when misexpressed in cells that express TBP; and (5) TBP2 loss results in major changes in transcription profile, demonstrating the ability of TBP2 to regulate gene expression globally. Finally we show that in vivo TBP2 is able to bind to promoters of genes that are expressed in the oocyte, suggesting that TBP2 is directly involved in the regulation of oocyte-specific gene expression. Thus, our data indicate for the first time that TBP2 is a central basal transcriptional regulator that drives specialized processes and establishes cell type-specific features in the oocyte.
TBP2 regulates chromatin organization and oocyte-specific gene expression pathways
We found that TBP2 is able to bind directly to certain genes transcribed in the oocyte. Consistent with a positive regulatory function of TBP2 in transcription initiation, oocyte development does not proceed beyond the secondary follicle stage in females lacking TBP2. Indeed, the TBP2-dependent genes we identified begin to be transcribed in primary follicles (Supplemental Fig. S3). This suggests that in oocytes at the primary and early secondary stages, TBP2-dependent Pol II transcription is essential to reach the transcriptionally quiescent preovulatory follicle stage (Bachvarova 1985) in which the chromatin has reached a maximum degree of condensation (Zuccotti et al. 2005). Indeed, we found that TBP2 loss results in disorganization of the chromatin in nuclei of oocytes and that in the TBP2−/− oocytes chromatin never reaches a fully condensed status. These observations together suggest that TBP2 affects both transcription of specific genes and global chromatin organization. In this sense, note that mouse TLF, which is essential for spermatogenesis, has been shown to participate in the organization of the chromocenter during spermiogenesis and is therefore conceived as a factor with a dual role: as a classical transcription factor and as a structural factor (Martianov et al. 2002a). Whether TBP2 directly regulates these chromatin condensation steps or acts on a particular set of genes involved in chromatin remodeling remains to be investigated. However, it is remarkable that genes important for proper chromatin configuration such as the oocyte-specific linker histone H1, H1foo, and the DNA methyltransferase Dnmt1 are down-regulated in Tbp2−/− ovaries.
The identified set of misregulated genes upon TBP2 loss is in full agreement with the sterility phenotype that we report. Among these there are several genes (i.e., Gdf9, Zp3, Bmp15, Rfpl4, H1foo, Zar1, and Dnmt1) (see Supplemental Table S1) that are necessary for oocyte development and function. This group of misregulated genes in the Tbp2−/− ovary is strikingly similar to those identified in mice lacking other oocyte-specific transcription factors such as NOBOX or FIGLA. Mice lacking either of these genes display a complete female sterility phenotype and reduced expression of Gdf9, Bmp15, Rfpl4, H1foo, Zar1, and Dnmt1o (Rajkovic et al. 2004; Joshi et al. 2007). Within the ovary, the oocyte has a central role in regulating granulosa cell metabolism (Su et al. 2009). The full complement of GDF9 and BMP15 is necessary for follicular function and female fertility (Yan et al. 2001; Su et al. 2004). We observed reduced expression of Gdf9 and Bmp15 upon TBP2 loss, which suggests that oocyte–granulosa cell communication is altered in TBP2-deficient ovaries (Yan et al. 2001; Su et al. 2004). Indeed, we observed down-regulation of Kif23, which is a Gdf9 downstream target and is involved in the formation of the intercellular bridges in the ovary (Greenbaum et al. 2009), further suggesting that GDF9 pathway is indeed seriously affected.
We performed the microarray analysis on whole ovaries because we sought to identify any possible effect of TBP2 loss on the communication between the oocyte and the follicular cells. However, this type of analysis may mask higher fold changes of gene expression in the oocytes, specialy if a gene is expressed also in other cells of the ovary. Nevertheless, the robustness of our microarray analysis is supported by the fact that we identified many oocyte-specific genes and also genes involved in the oocyte/granulosa cell communication. Moreover, we also observe that (1) the down-regulated class of genes is enriched in both oocyte-specific genes (tissue specificity analysis) and functions related to developmental pathways (ontology analysis); (2) the promoters of the two classes of genes differ significantly in terms of their AT percentage content (promoter analysis), which is noticeably higher in the down-regulated gene pool; and (3) some of these down-regulated genes turned out to be TBP2 direct targets as determined by ChIP. Thus, our results together indicate that the folliculogenesis program of the oocyte is severely disregulated in the absence of TBP2.
TBP2, but not TBP, is an essential transcription factor in the developing oocyte
We showed previously that TBP is not expressed during oocyte development from the primary follicular stage (Gazdag et al. 2007). In agreement, we show that deletion of the Tbp gene in oocytes, where TBP is not detected, does not affect oocyte maturation and subsequent fertilization. Why is it important to replace TBP function with that of TBP2 in the developing oocytes? While TBP has a positive role in cell proliferation in somatic cells (Johnson et al. 2003), we found that TBP2 has a negative effect on proliferation in the early embryo. Thus, it is possible that TBP2, in contrast to TBP, has acquired a highly specialized role in promoting cell growth, but not cell proliferation. This is one of the main features of the oocytes, which are among the largest cells with a single nucleus in vertebrates, and therefore require a long growing period without cell division. TBP2 could then be able to block (directly or indirectly) the cell division program, but at the same time promote the transcription of genes whose products accumulate in the growing oocyte.
TBP2(TRF3) has been suggested to play a role in orchestrating muscle differentiation in an ex vivo model (Deato and Tjian 2007). However, we find that, in vivo, TBP2 expression is undetectable in tissues other than the ovary or in other cell differentiation models (Supplemental Fig. S4A,B). Consistent with this restricted pattern of TBP2 expression, skeletal muscle in Tbp2−/− mice is undistinguishable from that of control animals, and the expression of key myogenic factors involved in muscle differentiation and integrity is unaffected (Supplemental Fig. S4C,D).
Because of the high identity between the core domain of TBP and TBP2, it is conceivable that TBP2 can potentially assemble with the same TAFs as TBP. The high divergence on the N terminus between TBP2 and TBP raises the interesting question of whether and how the N-terminal domain of TBP2 can diversify its function from that of TBP. One possibility would be that the N-terminal domain of TBP2 could determine the association of TBP2 with TAFs or TAF-like proteins. Similar to Drosophila germ cells (Hiller et al. 2004; Chen et al. 2005), tissue-specific TAF homologs may be expressed in mouse oocytes, thereby allowing association of TBP2 with a TFIID-like complex.
Interestingly, the genes that require TBP2 for their transcription in oocytes have a high frequency of AT nucleotides around their TSSs. The high homology between the DNA-binding core domains of TBP2 and TBP and our results showing that TBP2 can bind to oocyte-specific genes further suggest that TBP2 could recognize AT-rich sequences that may resemble the canonical TATA box.
Groups of genes clustered near the telomeres and specifically expressed in the oocyte have been identified recently (Hamatani et al. 2008). Importantly, our microarray analysis revealed an overrepresentation of genes within such clusters (e.g., Obox1, Obox2, Obox5, Obox3, Nlrp9c, Nlrp9b, Nlrp4a, and Nlrp4c) among the down-regulated genes in Tbp2−/− ovaries (Supplemental Table S1). Analysis of the core promoter sequences of the oocyte-specific homeobox cluster Obox revealed a consensus for a TATA box, further suggesting that TBP2 can bind directly to TATA boxes and regulate these genes.
Specialized TBP-type factors as mediators of germline-specific transcription
Our results indicate that TBP2 is essential for female germ cell differentiation in mice. In a mirroring situation, differentiation of male germ cells in the mouse requires another TBP-type factor: TLF. Indeed, absence of TLF results in increased germ cell apoptosis and male infertility (Martianov et al. 2001). Why have different TBP-type factors such as TBP2 and TLF acquired such a specialized germ cell-specific function? The beginning of TBP2 accumulation in the oocyte nuclei or TLF accumulation in male germ cell nuclei coincides with the phase of meiosis I. The abundance of TBP2 in oocytes during meiosis I therefore suggests that it may be involved in the control of expression of genes in a meiotic context. Interestingly, the murine TLF was not detected in mitotically dividing spermatogonia. Thus, contrary to TBP2 in oocytes, and to TLF in forming spermatocytes, TBP expression is largely restricted to mitotically dividing cells and drops below the detection limit in primary germ cells that are not proliferating. Interestingly, loss of TBP2 resulted in the induction of expression of several genes that are usually only expressed in the male gonad (Tex11, Tex13, Tex14, Tex19, Piwil2, and Tekt2) (Wang et al. 2001). This suggests that TBP2 may have a role in repressing the spermatogenesis transcriptional program in the female germline. These findings together further lead to the concept that in the germ cells of opposite sexes transcription initiation, with either TBP2 in oocytes or with TLF in spermatocytes, has evolved to carry out specialized functions, and that these respective functions cannot be performed by other TBP-type factors.
Finally, it will be important to determine whether TBP2 is linked to cases of female infertility in humans. Increased infertility in humans has attracted considerable attention in recent years, with ∼20% of couples of reproductive age being infertile. Thus, the identification of new animal models and molecules involved in sterility phenotypes, such as TBP2, will aid in tackling the genetic and cellular base of human infertility.
Materials and methods
Generation of TBP2 mice
The Tbp2/Trf3 conditional mutant mouse line was generated at the Mouse Clinical Institute/Institut Clinique de la Souris (http://www-mci.u-strasbg.fr). The targeting vector was constructed to obtain a floxed fragment comprising exon 4, and was verified by sequencing. After electroporation into 129S2/SvPas ES cells, recombined clones were analyzed by Southern blot with Neo and 5′ and 3′ external probes. A positive ES clone was microinjected into C57BL/6J blastocysts, and male chimaeras with germline transmission were obtained. Genotyping was performed with tail DNA. To verify TBP2 mRNA levels in Tbp2−/− and Tbp2+/− mice, total RNA was extracted from ovaries of 2-wk-old females with Trizol (Invitrogen), reverse-transcribed, and analyzed by PCR using specific primers for TBP2. PCR products were transferred onto an Amersham Hybond-XL membrane and probed with a radioactive probe specific for TBP2. Autoradiography was quantified using SynGene. ZP3-Cre mice were obtained from the Jackson laboratory. TBPlox/lox mice have been described previously (Martianov et al. 2002b).
Embryo culture and microinjection
Zygotes were microinjected with 1–2 pL of a mixture containing 300 ng/μL mRNA for mouse TBP2 or TBP and 200 ng/μL mRNA for DsRed, which were capped and transcribed in vitro from the pRN3P plasmid. Embryos were cultured in KSOM medium under a 5% CO2 atmosphere at 37°C and their development was monitored daily.
Immunostaining of cells and embryos
Oocytes or embryos were washed and fixed as described (Torres-Padilla et al. 2006), and incubated with the TBP (1:800) (Gazdag et al. 2007) or the H3K4me3 (Abcam) antibodies and the corresponding Alexa488-coupled secondary antibody (Molecular Probes). Embryos were mounted in Vectashield with DAPI (Vector Laboratories) and analyzed under confocal microscopy on a Leica SP2 AOBS confocal microscope.
Western blot
Ovaries from Tbp2+/+, Tbp2+/−, or Tbp2−/− 2-wk-old females were homogenized in 20 μL of loading blue containing SDS and separated by 8% PAGE. Western blotting was performed using the α-TBP2 2B12 monoclonal antibody, and α-tubulin was used as loading control.
Histology and immunohistochemistry
Ovaries from Tbp2−/− or control 2-wk-old and 6-wk-old females were fixed in 4% paraformaldehyde, dehydrated in ethanol, and embedded in paraffin. Sections (5 μm thick) were analyzed using the following antibodies: mouse monoclonal anti-TBP2 2B12 antibody (Gazdag et al. 2007), anti-CTD antibody 7G5 (kind gift from M. Vigneron), and H5 clone recognizing RNA Pol II phosphorylated at Ser2 (Covance). Sections were mounted in Vectashield with DAPI (Vector Laboratories) and analyzed using confocal microscopy.
ChIP and real-time PCR
Ovaries from 12- to 15-d-old CD1 females were fixed in 1% formaldehyde for 15 min and subsequently homogenized in 50 mM sodium phosphate buffer (pH 7.5), 10% glycerol, 10 mM β-mercaptoethanol, 0.1 mM phenylmethylsulfonyl fluoride, and protease inhibitors with a pestle. Fifteen micrograms of DNA were used for each immunoprecipitation with the mouse anti-TBP2 2B12 or the H3K4me3 antibodies (Abcam). Immunoprecipitated chromatin samples were washed and eluted from the antibody/beads complex by adding 0.1% SDS and 0.1 M NaHCO3. DNA was purified after cross-linking reversal, and the enrichment of specific DNA sequences in the immunoprecipitated samples (bound fractions) compared with the input samples (input fractions) was quantified by qPCR using SYBR Green JumpStart Taq ReadyMix (Sigma). Primer sequences are available upon request. The enrichment of each sequence is given by the ratio of the bound to input content (B/I).
Microarray analysis and RT-qPCR
RNA extraction from ovaries of 2-wk-old females was performed with the PicoPure RNA Kit (Arcturus). Total mRNA was amplified in two rounds, and three independent biological replicates were hybridized with Cy3 or Cy5 dyes onto the Affymetrix GeneChip Mouse Gene 1.0 ST microarrays (http://www-microarrays.u-strasbg.fr). Misregulated genes were analyzed using RMA (robust multiarray) normalization. Statistical analysis was performed with the Limma package within R-Bioconductor and the empirical Bayes method. Microarray data were validated in at least two independent biological replicates by real-time qPCR using SYBR Green JumpStart Taq ReadyMix (Sigma). PCR reactions were carried out in triplicate. To ensure proper biological cross-comparison, expression levels were compared between littermates.
Acknowledgments
We are grateful to I. Davidson and I. Martianov for the Tbplox/lox mice; to M. Boeglin and D. Hensch for advice; to M. Vigneron for antibodies; to A. Fadloun, F. Müller, and B. Murphy for critical reading of the manuscript; and to A. Buj-Bello for advice. The TBP2 mutant mice were generated at the Mouse Clinical Institute (MCI/ICS) in the Targeted Mutagenesis and Transgenesis Department. E.G. was supported by a fellowship from the European Community (HPRN-CT-00504228). This work was supported by funds from CNRS, INSERM, Universite de Strasbourg, FRM, ANR (05-BLAN-0396-01; Regulome), and European Community (HPRN-CT 00504228 and STREP LSHG-CT-2004-502950; EUTRACC LSHG-CT-2007-037445) to L.T., and PNRRE/INSERM number P006427 and AVENIR to M.E.T-P.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.535209.
Supplemental material is available at http://www.genesdev.org.
References
- Andreu-Vieyra C, Lin YN, Matzuk MM. Mining the oocyte transcriptome. Trends Endocrinol Metab. 2006;17:136–143. doi: 10.1016/j.tem.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Bachvarova R. Gene expression during oogenesis and oocyte development in mammals. Dev Biol. 1985;1:453–524. doi: 10.1007/978-1-4615-6814-8_11. [DOI] [PubMed] [Google Scholar]
- Bartfai R, Balduf C, Hilton T, Rathmann Y, Hadzhiev Y, Tora L, Orban L, Muller F. TBP2, a vertebrate-specific member of the TBP family, is required in embryonic development of zebrafish. Curr Biol. 2004;14:593–598. doi: 10.1016/j.cub.2004.03.034. [DOI] [PubMed] [Google Scholar]
- Bellier S, Chastant S, Adenot P, Vincent M, Renard JP, Bensaude O. Nuclear translocation and carboxyl-terminal domain phosphorylation of RNA polymerase II delineate the two phases of zygotic gene activation in mammalian embryos. EMBO J. 1997;16:6250–6262. doi: 10.1093/emboj/16.20.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleil JD, Wassarman PM. Structure and function of the zona pellucida: Identification and characterization of the proteins of the mouse oocyte's zona pellucida. Dev Biol. 1980;76:185–202. doi: 10.1016/0012-1606(80)90371-1. [DOI] [PubMed] [Google Scholar]
- Burley SK, Roeder RG. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- Chen X, Hiller M, Sancak Y, Fuller MT. Tissue-specific TAFs counteract Polycomb to turn on terminal differentiation. Science. 2005;310:869–872. doi: 10.1126/science.1118101. [DOI] [PubMed] [Google Scholar]
- Choi Y, Rajkovic A. Characterization of NOBOX DNA binding specificity and its regulation of Gdf9 and Pou5f1 promoters. J Biol Chem. 2006;281:35747–35756. doi: 10.1074/jbc.M604008200. [DOI] [PubMed] [Google Scholar]
- Deato MD, Tjian R. Switching of the core transcription machinery during myogenesis. Genes & Dev. 2007;21:2137–2149. doi: 10.1101/gad.1583407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deato MD, Marr MT, Sottero T, Inouye C, Hu P, Tjian R. MyoD targets TAF3/TRF3 to activate myogenin transcription. Mol Cell. 2008;32:96–105. doi: 10.1016/j.molcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries WN, Binns LT, Fancher KS, Dean J, Moore R, Kemler R, Knowles BB. Expression of Cre recombinase in mouse oocytes: A means to study maternal effect genes. Genesis. 2000;26:110–112. [PubMed] [Google Scholar]
- Ferg M, Sanges R, Gehrig J, Kiss J, Bauer M, Lovas A, Szabo M, Yang L, Straehle U, Pankratz MJ, et al. The TATA-binding protein regulates maternal mRNA degradation and differential zygotic transcription in zebrafish. EMBO J. 2007;26:3945–3956. doi: 10.1038/sj.emboj.7601821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdag E, Rajkovic A, Torres-Padilla ME, Tora L. Analysis of TATA-binding protein 2 (TBP2) and TBP expression suggests different roles for the two proteins in regulation of gene expression during oogenesis and early mouse development. Reproduction. 2007;134:51–62. doi: 10.1530/REP-06-0337. [DOI] [PubMed] [Google Scholar]
- Greenbaum MP, Iwamori N, Agno JE, Matzuk MM. Mouse TEX14 is required for embryonic germ cell intercellular bridges but not female fertility. Biol Reprod. 2009;80:449–457. doi: 10.1095/biolreprod.108.070649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamatani T, Yamada M, Akutsu H, Kuji N, Mochimaru Y, Takano M, Toyoda M, Miyado K, Umezawa A, Yoshimura Y. What can we learn from gene expression profiling of mouse oocytes? Reproduction. 2008;135:581–592. doi: 10.1530/REP-07-0430. [DOI] [PubMed] [Google Scholar]
- Hernandez N. TBP, a universal eukaryotic transcription factor? Genes & Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- Hiller M, Chen X, Pringle MJ, Suchorolski M, Sancak Y, Viswanathan S, Bolival B, Lin TY, Marino S, Fuller MT. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development. 2004;131:5297–5308. doi: 10.1242/dev.01314. [DOI] [PubMed] [Google Scholar]
- Jacobi UG, Akkers RC, Pierson ES, Weeks DL, Dagle JM, Veenstra GJ. TBP paralogs accommodate metazoan- and vertebrate-specific developmental gene regulation. EMBO J. 2007;26:3900–3909. doi: 10.1038/sj.emboj.7601822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallow Z, Jacobi UG, Weeks DL, Dawid IB, Veenstra GJ. Specialized and redundant roles of TBP and a vertebrate-specific TBP paralog in embryonic gene regulation in Xenopus. Proc Natl Acad Sci. 2004;101:13525–13530. doi: 10.1073/pnas.0405536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Dubeau L, Kawalek M, Dervan A, Schonthal AH, Dang CV, Johnson DL. Increased expression of TATA-binding protein, the central transcription factor, can contribute to oncogenesis. Mol Cell Biol. 2003;23:3043–3051. doi: 10.1128/MCB.23.9.3043-3051.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA. Transcription strategies in terminally differentiated cells: Shaken to the core. Genes & Dev. 2007;21:2113–2117. doi: 10.1101/gad.1598007. [DOI] [PubMed] [Google Scholar]
- Joshi S, Davies H, Sims LP, Levy SE, Dean J. Ovarian gene expression in the absence of FIGLA, an oocyte-specific transcription factor. BMC Dev Biol. 2007;7:67. doi: 10.1186/1471-213X-7-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JB, Duckett R, Myers M, Britt KL, Mladenovska T, Findlay JK. Quantification of healthy follicles in the neonatal and adult mouse ovary: Evidence for maintenance of primordial follicle supply. Reproduction. 2006;132:95–109. doi: 10.1530/rep.1.01128. [DOI] [PubMed] [Google Scholar]
- Martianov I, Fimia GM, Dierich A, Parvinen M, Sassone-Corsi P, Davidson I. Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol Cell. 2001;7:509–515. doi: 10.1016/s1097-2765(01)00198-8. [DOI] [PubMed] [Google Scholar]
- Martianov I, Brancorsini S, Gansmuller A, Parvinen M, Davidson I, Sassone-Corsi P. Distinct functions of TBP and TLF/TRF2 during spermatogenesis: Requirement of TLF for heterochromatic chromocenter formation in haploid round spermatids. Development. 2002a;129:945–955. doi: 10.1242/dev.129.4.945. [DOI] [PubMed] [Google Scholar]
- Martianov I, Viville S, Davidson I. RNA polymerase II transcription in murine cells lacking the TATA binding protein. Science. 2002b;298:1036–1039. doi: 10.1126/science.1076327. [DOI] [PubMed] [Google Scholar]
- Matzuk MM, Lamb DJ. Genetic dissection of mammalian fertility pathways. Nat Cell Biol. 2002;4:S41–S49. doi: 10.1038/ncb-nm-fertilitys41. [DOI] [PubMed] [Google Scholar]
- Muller F, Tora L. The multicoloured world of promoter recognition complexes. EMBO J. 2004;23:2–8. doi: 10.1038/sj.emboj.7600027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F, Lakatos L, Dantonel J, Strahle U, Tora L. TBP is not universally required for zygotic RNA polymerase II transcription in zebrafish. Curr Biol. 2001;11:282–287. doi: 10.1016/s0960-9822(01)00076-8. [DOI] [PubMed] [Google Scholar]
- Muller F, Demeny MA, Tora L. New problems in RNA polymerase II transcription initiation: matching the diversity of core promoters with a variety of promoter recognition factors. J Biol Chem. 2007;282:14685–14689. doi: 10.1074/jbc.R700012200. [DOI] [PubMed] [Google Scholar]
- Pedersen T, Peters H. Proposal for a classification of oocytes and follicles in the mouse ovary. J Reprod Fertil. 1968;17:555–557. doi: 10.1530/jrf.0.0170555. [DOI] [PubMed] [Google Scholar]
- Rajkovic A, Pangas SA, Ballow D, Suzumori N, Matzuk MM. NOBOX deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science. 2004;305:1157–1159. doi: 10.1126/science.1099755. [DOI] [PubMed] [Google Scholar]
- Reina JH, Hernandez N. On a roll for new TRF targets. Genes & Dev. 2007;21:2855–2860. doi: 10.1101/gad.1623207. [DOI] [PubMed] [Google Scholar]
- Su YQ, Wu X, O'Brien MJ, Pendola FL, Denegre JN, Matzuk MM, Eppig JJ. Synergistic roles of BMP15 and GDF9 in the development and function of the oocyte–cumulus cell complex in mice: Genetic evidence for an oocyte-granulosa cell regulatory loop. Dev Biol. 2004;276:64–73. doi: 10.1016/j.ydbio.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Su YQ, Sugiura K, Eppig JJ. Mouse oocyte control of granulosa cell development and function: Paracrine regulation of cumulus cell metabolism. Semin Reprod Med. 2009;27:32–42. doi: 10.1055/s-0028-1108008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Lis JT, Zhou S, Tjian R. A TRF1:BRF complex directs Drosophila RNA polymerase III transcription. Cell. 2000;101:459–469. doi: 10.1016/s0092-8674(00)80857-0. [DOI] [PubMed] [Google Scholar]
- Torres-Padilla ME, Bannister AJ, Hurd PJ, Kouzarides T, Zernicka-Goetz M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int J Dev Biol. 2006;50:455–461. doi: 10.1387/ijdb.052073mt. [DOI] [PubMed] [Google Scholar]
- Veenstra GJ, Weeks DL, Wolffe AP. Distinct roles for TBP and TBP-like factor in early embryonic gene transcription in Xenopus. Science. 2000;290:2312–2315. doi: 10.1126/science.290.5500.2312. [DOI] [PubMed] [Google Scholar]
- Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- White JK, Auerbach W, Duyao MP, Vonsattel JP, Gusella JF, Joyner AL, MacDonald ME. Huntingtin is required for neurogenesis and is not impaired by the Huntington's disease CAG expansion. Nat Genet. 1997;17:404–410. doi: 10.1038/ng1297-404. [DOI] [PubMed] [Google Scholar]
- Xiao L, Kim M, DeJong J. Developmental and cell type-specific regulation of core promoter transcription factors in germ cells of frogs and mice. Gene Expr Patterns. 2006;6:409–419. doi: 10.1016/j.modgep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Yan C, Wang P, DeMayo J, DeMayo FJ, Elvin JA, Carino C, Prasad SV, Skinner SS, Dunbar BS, Dube JL, et al. Synergistic roles of bone morphogenetic protein 15 and growth differentiation factor 9 in ovarian function. Mol Endocrinol. 2001;15:854–866. doi: 10.1210/mend.15.6.0662. [DOI] [PubMed] [Google Scholar]
- Zambelli F, Pesole G, Pavesi G. Pscan: Finding over-represented transcription factor binding sites motifs in sequences of co-regulated and co-expressed genes. Nucleic Acids Res. 2009;37:W247–W252. doi: 10.1093/nar/gkp464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccotti M, Garagna S, Merico V, Monti M, Alberto Redi C. Chromatin organisation and nuclear architecture in growing mouse oocytes. Mol Cell Endocrinol. 2005;234:11–17. doi: 10.1016/j.mce.2004.08.014. [DOI] [PubMed] [Google Scholar]