Abstract
Metastatic disease is a primary cause of cancer-related death, and factors governing tumor cell metastasis have not been fully elucidated. Here, we address this question by using tumor cell lines derived from mice that develop metastatic lung adenocarcinoma owing to expression of mutant K-ras and p53. Despite having widespread somatic genetic alterations, the metastasis-prone tumor cells retained a marked plasticity. They transited reversibly between epithelial and mesenchymal states, forming highly polarized epithelial spheres in three-dimensional culture that underwent epithelial-to-mesenchymal transition (EMT) following treatment with transforming growth factor-β or injection into syngeneic mice. This transition was entirely dependent on the microRNA (miR)-200 family, which decreased during EMT. Forced expression of miR-200 abrogated the capacity of these tumor cells to undergo EMT, invade, and metastasize, and conferred transcriptional features of metastasis-incompetent tumor cells. We conclude that tumor cell metastasis is regulated by miR-200 expression, which changes in response to contextual extracellular cues.
Keywords: Lung cancer, EMT, microRNA-200, mouse model, metastasis
Lung cancer is the leading cause of cancer-related death in Western countries, and metastasis is the most common cause of death in patients with lung cancer. Approximately two-thirds of patients are diagnosed at an advanced stage, and of the remaining patients who undergo surgery, 30%–50% develop recurrence with metastatic disease. The lack of curative treatment options emphasizes the need for a better understanding of the biologic processes that drive metastasis. Toward that goal, genetic mouse models have been generated that develop lung adenocarcinoma, the most common histologic subtype of lung cancer, with differing propensities to invade and metastasize (Liu et al. 2000; Fisher et al. 2001; Jackson et al. 2001, 2005; Johnson et al. 2001; Guerra et al. 2003; Lang et al. 2004; Olive et al. 2004; Ji et al. 2006; Zheng et al. 2007). Work by our group identified a mutant p53 allele (p53R172HΔG) that confers metastatic potential to lung adenocarcinomas arising in mice due to a latent, somatically activated KrasG12D allele (KrasLA1) (Zheng et al. 2007). Messenger RNA (mRNA) expression profiling of metastatic versus matched lung tumors from this model revealed a signature of differentially expressed genes, which we used to mine publicly available databases and show that this signature identified a subset of lung cancer patients who have a poor prognosis (Gibbons et al. 2009). These findings provide evidence that the KrasG12D;p53R172HΔG mice recapitulate features of patients with metastasis-prone lung cancer and are a useful platform on which to study the basis for lung cancer metastasis.
One model of metastasis proposes that a subset of tumor cells within epithelial malignancies acquires the ability to invade and disseminate by undergoing an epithelial–mesenchymal transition (EMT), characterized by the loss of cell–cell attachments, loss of apical–basal polarity, and appearance of mesenchymal differentiation properties (Huber et al. 2005). Recent evidence from several groups (Mani et al. 2008; Morel et al. 2008) has also demonstrated that induction of EMT in breast cancer cells results in the acquisition of stem cell-like properties, including the ability to form mammospheres in culture and to resist the anti-tumor effects of cytotoxic chemotherapy, a common finding in patients with advanced malignancy. The process of EMT is regulated by several transcriptional suppressor families, including the zinc finger proteins Snail 1 and Snail 2, the two-handed zinc finger δEF1 family factors (δEF1/Zeb1 and SIP1/Zeb2), and the basic helix–loop–helix factors Twist and E12/E47 (Eger et al. 2005; Aigner et al. 2007; Peinado et al. 2007). On the basis of these findings, investigators have begun to explore the upstream regulators of these transcriptional repressors.
Multiple groups have shown recently that Zeb1 and Zeb2 expression are regulated by the microRNA-200 family members (collectively referred to here as miR-200) (Burk et al. 2008; Gregory et al. 2008; Korpal et al. 2008; Park et al. 2008). miRs are small, noncoding RNAs that post-transcriptionally regulate gene expression (Bartel 2004). The miR-200 family consists of five members clustered in two genomic loci (200b–200a–429 and 200c–141). Induction of EMT by transforming growth factor-β (TGFβ) in multiple cell systems or by expression of the protein tyrosine phosphatase Pez in MDCK cells inhibited the miR-200 family, and forced expression of the miR-200 members reversed this process, inducing a mesenchymal-to-epithelial transition (MET) and abrogating TGFβ-induced EMT (Burk et al. 2008; Gregory et al. 2008; Korpal and Kang 2008; Korpal et al. 2008; Park et al. 2008). Given the growing body of evidence supporting a role for EMT in metastasis, studies are warranted to examine the contribution of the miR-200 family to metastasis.
Here, we postulated that the capacity of tumor cells to undergo EMT and metastasize requires changes in the expression of specific miRs, and we tested this hypothesis using a syngeneic tumor model in which tumor cell lines derived from mice that develop lung adenocarcinoma owing to expression of mutant Kras and p53 metastasize with defined (high or low) potential when injected subcutaneously into syngeneic mice. Of the miRs profiled, the miR-200 family had the most prominent differential expression in metastasis-prone tumors relative to metastasis-incompetent tumors, and forced expression of the miR-200b cluster in metastasis-prone tumor cells abrogated their capacity to undergo EMT and metastasize in syngeneic mice. This study provides the first in vivo demonstration that down-regulation of miR-200 has a causal role in metastasis.
Results
Creation of a syngeneic lung adenocarcinoma metastasis model
To have an in vivo system in which to identify specific genes that mediate metastases, we generated a panel of lung adenocarcinoma cell lines from KrasLA1/+; p53R172HΔG mice, named according to the mouse number and site of derivation (e.g., 393P denotes primary lung tumor, 393LN indicates lymph node metastasis, and 344SQ denotes subcutaneous metastasis). The cells expressed surfactant protein C, a marker of type II alveolar cells, which is a feature of lung adenocarcinoma cells in KrasLA1 mice (Johnson et al. 2001), and they were heterozygous for the p53R172HΔG and K-rasLA1 alleles (Supplemental Fig. S1). Furthermore, the cell lines exhibited genetic abnormalities, including centrosomal amplification, aberrant mitotic spindles, aneuploidy, chromosomal translocations, and small unidentified chromosomal fragments (Supplemental Fig. S2), which are similar in scope to those seen in human lung adenocarcinoma (Whang-Peng et al. 1991).
The cell lines were injected subcutaneously into syngeneic animals to determine their capacities to form tumors and to metastasize. We used immunocompetent, syngeneic mice as recipients in order to engage host-derived inflammatory and angiogenic cells in the metastatic process. The subcutaneous route of injection clearly separated the cell lines into two groups on the basis of their metastatic potential (high or low). Mice injected with 344SQ, 344P, 412P, or 531P developed metastatic disease, whereas mice injected with 393P or 393LN developed subcutaneous tumors, but by 6 wk exhibited no overt signs of morbidity or evidence of metastasis at necropsy (Supplemental Table S1; Supplemental Fig. S3).
Propensity of tumor cells to undergo EMT
To gain insight into the biologic processes driving tumor progression and metastasis in the syngeneic model, three cell lines that form subcutaneous tumors with differing metastatic potentials were transcriptionally profiled (four tumors per cell line), and gene expression differences between tumors were identified on the basis of their propensities to metastasize. Those classifications included incompetent (393P), prone (344SQ), and intermediate (393LN), given that 393LN produced lung metastases when injected by tail vein but not subcutaneously, whereas 393P cells did not form lung metastases with either injection route (data not shown). Using 393P as the reference tumor, 3002 transcripts (i.e., Affymetrix probe sets) were differentially expressed (fold change >1.5, P < 0.01) in 393LN cells, 344SQ cells, or both (Fig. 1A; Supplemental Table S2). It is noteworthy that, despite their distinct origins, 393LN and 344SQ tumors exhibited common differences from 393P tumors, whereas other differences from 393P were unique to either 344SQ or 393LN tumors (both increased and decreased).
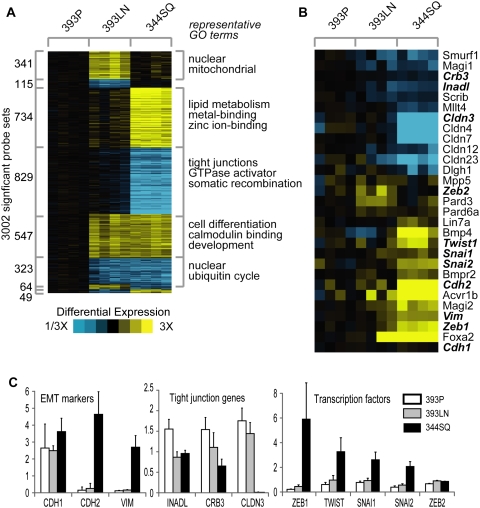
Figure 1.
mRNA profiling of tumors reveals changes consistent with EMT. (A) Differentially expressed genes as identified by mRNA expression profiling of syngeneic tumors, with yellow representing increased expression and blue indicating decreased expression versus the 393P control: (393P) metastasis-incompetent; (393LN) intermediate; (344SQ) metastasis-prone. (B) Differences in subsets of genes related to polarity and EMT. (C) Q-PCR validation of EMT markers, polarity genes, and EMT-inducing transcription factors. (White) 393P; (gray) 393LN; (black) 344SQ.
Enrichment analysis (Fisher's exact test using Gene Ontology [GO] terms) was performed to examine the predominant gene functions in the metastasis signature (Fig. 1A). The categories of up-regulated genes were, among others, “metal ion binding” (393LN and 344SQ, 79 genes, P = 5.0E-7) and “zinc ion binding” (393LN and 344SQ, 69 genes, P = 1.1E-6), whereas the categories of down-regulated genes included “tight junction” (344SQ, 13 genes, P = 1.9E-7), “cell differentiation” (393LN and 344SQ, 18 genes, P = 2.4E-6), and “multicellular organismal development” (393LN and 344SQ, 21 genes, P = 0.0002) (Fig. 1A; Supplemental Table S3). Within the expression profiles, 344SQ tumors exhibited loss of cell adhesion and polarity complex genes (e.g., CLDN3, CLDN4, CLDN7, CLDN12, CLDN23, INADL, DLGH1, SCRIB, CRB3, and PARD6), increases in the mesenchymal differentiation markers vimentin and N-cadherin (VIM and CDH2), and increases in transcription factors that promote these processes (ZEB1 and TWIST) (Fig. 1B). To confirm these findings, quantitative RT–PCR was performed on the tumor samples for ZEB1, TWIST, SNAI1, SNAI2, ZEB2, CDH1, CDH2, VIM, and tight junction-associated genes (INADL, CRB3, and CLDN3) (Fig. 1C). On the basis of these findings, 344SQ tumors exhibited features of EMT, whereas 393P and 393LN tumors did not. Although the syngeneic tumors expressed CDH1 at similar levels, the CDH1 to VIM ratio was 2:1 for 344SQ and 16:1 for 393P, consistent with a mesenchymal phenotype in the former and an epithelial phenotype in the latter. Of note, many of these EMT-related genes were differentially expressed in spontaneous metastases arising in KrasLA1/+; p53R172HΔG mice (Gibbons et al. 2009). Thus, the syngeneic tumors recapitulated the biologic features of spontaneous tumors in KrasLA1/+; p53R172HΔG mice and revealed gene expression changes involved in a variety of cellular functions, including EMT.
Sphere-forming capacity of metastasis-prone tumor cells in three-dimensional (3D) culture
Despite proliferating at similar rates when cultured in monolayer, the tumor cell lines exhibited sharp differences when grown on laminin-rich growth factor-reduced Matrigel. The metastasis-prone cells (e.g., 344P, 344SQ, and 531P) grew from single cells into small aggregate colonies, which formed spherical structures ∼80 μm in diameter over 7–10 d (Fig. 2A,B; Supplemental Fig. S5C). The formation of hollow cores was initiated by apoptosis on day 4 or 5, while the remaining cells on the rims continued to proliferate (Fig. 2C,D), forming a single cell layer by day 10–14 (Fig. 2A,G,H). The spheres expressed the epithelial markers β-catenin (Fig. 2E) and E-cadherin (Fig. 2F) on the cell membranes, formed tight junctions, and exhibited apical–basal polarity based on localization of Zonula occludens-1 (ZO-1) and partitioning defective-6B (ParD6B) to the apical surfaces at tight junctions (Fig. 2G,H), α6-integrin to the basal surfaces (Fig. 2G,H), and the luminal orientation of the golgi marker GM130 (Fig. 2I). Coincident with this morphogenesis was evidence that the cells underwent MET (Supplemental Fig. S4). In contrast, the metastasis-incompetent tumor cells (393P and 393LN) grew from single cells into disorganized aggregates in Matrigel cultures (Supplemental Fig. S5A,B,D–F), and did not display biochemical changes consistent with MET (data not shown).
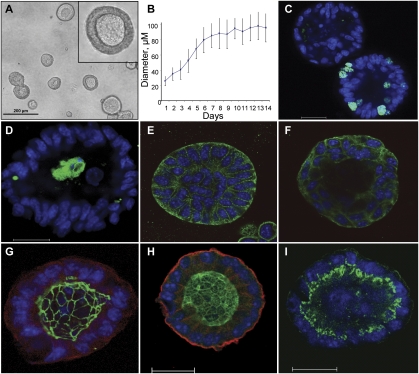
Figure 2.
Metastatic 344SQ cells form polarized epithelial spheres in 3D Matrigel culture. (A) Morphology of day 12 spheres by contrast microscopy. (B) Sphere growth versus time from plating onto Matrigel (MG); diameter measured in microns. (C) Ki-67 (green) and Topro-3 (blue) staining of developing sphere, visualized by confocal microscopy. (D) Cleaved caspase 3 staining (green) and Topro-3 (blue) of developing sphere. (E) β-catenin (green) and Topro-3 (blue) staining of developing sphere. (F) E-cadherin (green) and Topro-3 (blue) staining of developing sphere. (G) Sphere stained for ZO-1 (green), α6-integrin (red), and Topro-3 (blue). (H) Sphere stained for ParD6B (green), α6-integrin (red), and Topro-3 (blue). (I) Sphere stained for GM130 (green) and Topro-3 (blue). Bar in confocal images represents 20 μm.
To assess whether these polarized structures represented a terminal differentiation of the metastasis-prone cells, the structures were removed from the Matrigel beds (Fig. 3A) and either placed onto tissue culture plates or injected subcutaneously into syngeneic mice. Within 12 to 15 h of plating, the structures attached to the plastic and the adherent cells migrated onto the plate, assumed a monolayer growth pattern, and eventually grew to cover the entire surface (Fig. 3B,C). When these cells were trypsinized and replated as a single-cell suspension onto Matrigel, they reformed the growth-limited, polarized spherical structures (Fig. 3D). Spheres injected subcutaneously formed primary tumors and metastasized widely in all five syngeneic mice injected (Fig. 3E–H). Another metastatic cell line (344P) retained its sphere-forming potential after repeated cycling between tissue culture and syngeneic animals. Following four sequential cycles from adrenal metastasis to cell culture, tumor cells were still able to form TGFβ-responsive spheres on Matrigel (Supplemental Fig. S5G,H). We conclude that the metastasis-prone cells exhibit a reversible responsiveness to changes in cellular context both in vitro and in vivo.
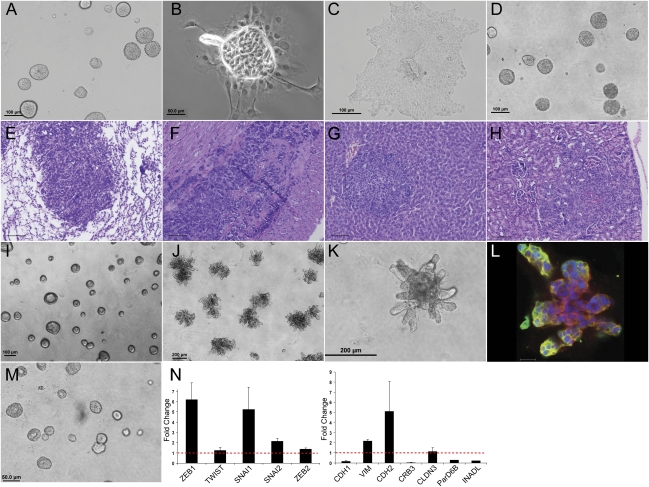
Figure 3.
Spheres are plastic in response to the in vitro and in vivo environment. (A) Spheres in Matrigel. (B) Sphere purified from Matrigel and plated on tissue culture plastic. Photo taken 20 h post-plating. (C) Sphere grown on tissue culture plastic for several days. (D) Second-generation spheres grown on Matrigel. Monolayer cells grown as in C were cultured in 3D Matrigel culture for 10 d. (E–H) Spheres purified from Matrigel were reimplanted by subcutaneous injection into syngeneic animals. Photos show H&E stain of tissue sections demonstrating metastases to the lung (E), heart (F), liver (G), and kidney (H). Bar, 100 μm. (I) Spheres in Matrigel prior to TGFβ treatment. (J) Spheres treated with TGFβ for 5 d. (K) Higher-power contrast image of spheres after 5 d of TGFβ treatment. (L) Confocal image of a plane through a TGFβ-treated structure, stained for ZO-1 (green), α6-integrin (red), and Topro-3 (blue). Bar, 20 μm. (M) Spheres were treated with the TGFβ inhibitor SB431542 (10 μM) for 3 h, followed by treatment with 5 ng/mL TGFβ in the presence of inhibitor for 5 d. (N) Quantitative RT–PCR of indicated markers, shown as fold change upon TGFβ treatment. Spheres were grown as in I and J, then harvested for analysis.
TGFβ induces EMT and produces invasive structures in 3D culture
Mining of expression array data demonstrated that primary lung tumors from KrasLA1/+; p53R172HΔG mice were enriched in TGFβ1, TGFβR1, and TGFβRII relative to those from KrasLA1/+ mice (data not shown). Furthermore, relative to syngeneic 393P tumors, 344SQ tumors had higher expression of TGFβ-related genes, including bone morphogenetic protein-4 (BMP4) (4.9-fold, P = 0.005), type II BMP receptor (BMPRII) (1.6-fold, P = 4.49 × 10−5), and type Ib activin A receptor (ACVRIb) (7.8-fold, P = 7.85 × 10−5) (Fig. 1A). Additionally, measurement of TGFβ1 by ELISA in 344SQ tumors versus 393P tumors showed average levels to be ∼2.5 times higher in the 344SQ tumors (Supplemental Fig. S6A). We therefore postulated that metastasis-prone tumor cells undergo EMT owing to TGFβ, a potent inducer of EMT (Gregory et al. 2008). TGFβ treatment of mature 344SQ spheres caused them to lose organization on the basis of apical–basal polarity markers, develop invasive outgrowths, and become larger (Fig. 3J–L). Coincident with these morphologic changes, the spheres expressed biochemical markers of EMT (decreased CDH1 and increased CDH2 and VIM) and higher levels of the transcriptional repressors that regulate them (ZEB1, SNAI1, and SNAI2) (Fig. 3N). Many of these changes were abrogated by cotreatment with SB431542, a TGFβ inhibitor (Fig. 3M). The nonmetastatic 393P cells were relatively resistant to TGFβ in both two-dimensional (2D) and 3D culture, requiring a greater time of exposure to undergo morphologic change (7 d vs. 2 d) (Supplemental Fig. S6, B–D vs. F–H; data not shown), and analysis of the EMT markers suggested only a partial EMT, with preservation of E-cadherin and the polarity gene levels (Supplemental Fig. S6E,I). The metastastasis-prone cells therefore displayed significantly higher levels of TGFβ1 in the tumors and a greater sensitivity to its EMT-inducing effects.
miR profiling of tumors reveals miR-200 family differences
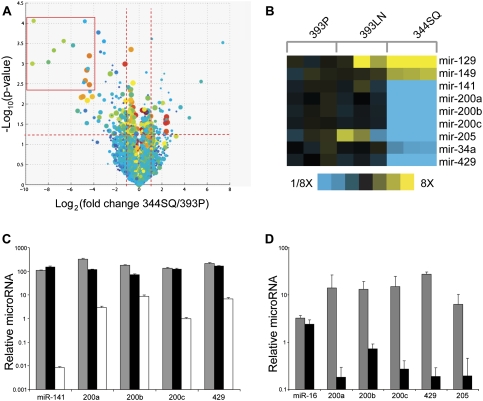
Given the reported role of the miR-200 family in regulating the expression of Zeb1 and Zeb2 (Burk et al. 2008; Gregory et al. 2008; Korpal et al. 2008; Park et al. 2008), we postulated that miR-200 expression regulates EMT and is responsive to TGFβ in this model. We analyzed RNA samples from the same syngeneic tumor samples analyzed in Figure 1 using the DiscovArray platform, and found that only a small number of miRs were differentially expressed, including miR-200, miR-205, miR-129, miR-149, and miR-34a (Fig. 4A,B; Supplemental Table S4). Relative to 393P and 393LN tumors, 344SQ tumors had lower expression of all of the mir-200 family members (miR-141, miR-200a, miR-200b, miR-200c, and miR-429) (Fig. 4B). Conspicuously absent from the list of differentially expressed miRs were let-7, miR-10b, miR-373, miR-21, miR-335, and miR-126, which have all been shown to function in tumor progression or metastasis (Ma and Weinberg 2008). Quantification of miR-200 family expression by TaqMan Q-PCR revealed a one- to four-log difference between 393P and 344SQ tumors (Fig. 4C). Furthermore, treatment of 344SQ spheres with TGFβ suppressed the levels of miR-200 family members by several logs (Fig. 4D). Collectively, these findings demonstrate a specific role for miR-200 in modulating EMT in this model.
Figure 4.
miR profiling of tumors reveals miR-200 family differences. (A) Volcano plot showing differences in the miRs detected by microarray for the 344SQ and 393P tumor samples profiled in Figure 1A. The −log10(P-value) is plotted against the fold change difference in expression between the samples for the indicated miR probes. The box indicates probes for the miR-200 family members. (B) Heat map showing differentially expressed miRs (P < 0.01, fold change >3, either 344SQ vs. 393LN or 393P vs. 393LN; average signal >100 U across tumors), from the three tumor types profiled in Figure 1A, with yellow representing increased expression and blue indicating decreased expression versus the 393P control. (C) TaqMan RT–PCR for the miR-200 family members as indicated, relative to a miR-16 control. (Gray) 393P; (black) 393LN.; (white) 344SQ. (D) RT–PCR assay of individual miR-200 members or miR-16 control, for 344SQ spheres in Matrigel (gray), or following TGFβ treatment (black). miR levels are normalized to snoRNA-135. Assay is of structures similar to those shown in Figure 3, I and J.
EMT and the miR-200 family in human lung cancer cells
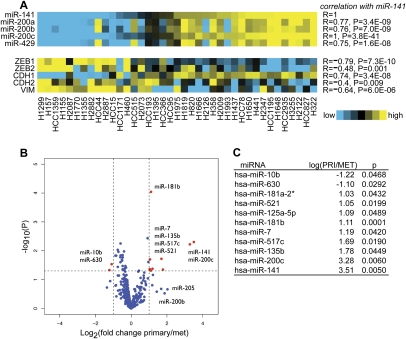
To determine whether the above findings in the syngeneic tumors are relevant to human lung cancer, we profiled basal expression of miRs and mRNAs in a panel of 39 lung cancer cell lines. The cell lines grouped on the basis of miR-200 family expression, demonstrating strong correlations between miR-141 and ZEB1, ZEB2, CDH1, CDH2, and VIM (Fig. 5A). Further analysis revealed that miR-141 and miR-200c best distinguished the cell lines on the basis of their site of origin, with ∼11-fold higher levels of these miRs in cells from a primary lung tumor than in cell lines derived from metastatic sites (Fig. 5B,C). When the expression fold changes of the genes with the motif content of their 3′ untranslated regions (UTRs) was correlated using the program miReduce (Bussemaker et al. 2001; Sood et al. 2006), we found significant overrepresentation of the target site 7-mer for miR-200b, miR-200c, and miR-429 in the 3′UTRs of genes with reduced expression in cell lines derived from primary tumors relative to those derived from metastases (Supplemental Fig. S7).
Figure 5.
miR-200 family and EMT marker levels in a panel of non-small-cell lung cancer cell lines. (A) Heat map of expression patterns for miRNAs in the miR-200 family and markers of EMT in a panel of 40 human lung cancer cell lines. Correlation of each gene or miR expression with that of miR-141 is indicated by Pearson's R- and P-values to the right. (B) Volcano plot showing the log-transformed P-values plotted against the log-transformed fold change in expression between cell lines derived from primary tumors relative to those derived from metastatic sites. miRs in red are differentially expressed more than twofold, with P < 0.05. (C) Table with x- and P-values corresponding to the highlighted miRs in B.
Forced miR-200 expression blocks EMT and metastasis
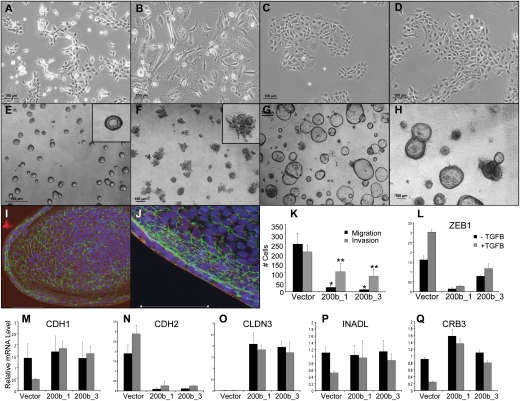
We postulated that syngeneic tumors metastasize as a consequence of decreased miR-200 expression, and tested this hypothesis by constitutively expressing the miR-200b/a/429 (200b) cluster in 344SQ cells. Quantification by Q-PCR of miR-200 levels in the 200b stable transfectants revealed elevated levels of the miR-200b cluster, the miR-200c cluster, and miR-205, which is functionally related to the miR-200 family (Supplemental Table S5; Gregory et al. 2008; Park et al. 2008), suggesting that forced expression of 200b activated a feed-forward loop that regulated the expression of the other endogenous miRs. Compared with parental cells and control transfectants (Fig. 6A,B), the 200b transfectants grew in monolayer more tightly clustered and with increased cell–cell contacts (Fig. 6C) and did not respond to TGFβ treatment even after exposure for 10 d (Fig. 6D). In Matrigel, they produced polarized spheres that were generally larger than those made by control transfectants and were refractory to TGFβ treatment (Fig. 6E–J). In Transwell plates, they exhibited a decreased capacity to migrate and invade (Fig. 6K). Quantitative RT–PCR measurements of ZEB1 (Fig. 6L), CDH1, CDH2 (Fig. 6M,N), and the polarity genes CLDN3, INADL, and CRB3 (Fig. 6O–Q) revealed that the 200b transfectants had a more epithelial baseline phenotype in monolayer than the control transfectants had, and this phenotype was refractory to TGFβ treatment. Interestingly, the other EMT-inducing transcription factors ZEB2, SNAI1, SNAI2, and TWIST did not differ between the transfectants (data not shown), suggesting that miR-200 targeted specific transcriptional repressors in 344SQ cells.
Figure 6.
Constitutive miR-200 expression converts 2D morphology, blocks TGFβ reponsiveness, and abrogates in vitro migration and invasion. (A) 344SQ cells grown on tissue culture plastic. (B) 344SQ cells treated for 10 d with TGFβ. (C) 344SQ_200b_1 transfectants on tissue culture plastic. (D) 344SQ_200b_1 transfectants treated for 10 d with TGFβ. (E) 344SQ_vector cells grown on Matrigel. (F) 344SQ_vector cells grown on Matrigel and stimulated with TGFβ for 5 d. (G) 344SQ_200b_1 cells grown on Matrigel. (H) 344SQ_200b_1 cells grown on Matrigel and stimulated with TGFβ for 5 d. (I,J) 344SQ_200b_1 cells grown on Matrigel were imaged by confocal microscopy after staining for ZO-1 (green), α6-integrin (red), and Topro-3 (blue). Bar, 50 μm. (K) In vitro Transwell migration (black) and invasion (gray) assays for the three cell lines. (L–Q) Q-PCR analysis of indicated EMT-related genes at baseline (black) or after TGFβ treatment (gray) for the 344SQ_vector cells or two different 344SQ_200b transfectants grown in monolayer culture as in A–D.
To assess the biologic effects of constitutive miR-200 expression on tumor growth and metastasis in vivo, the 200b transfectants were injected subcutaneously into syngeneic mice. Control transfectants formed tumors in six of eight mice, all of which developed metastases, whereas the 200b transfectants formed primary tumors in six of 12 mice, none of which developed metastases as assessed by necropsy and microscopic evaluation of hematoxylin and eosin (H&E)-stained tissue sections from the lung, heart, liver, and kidney. The absence of metastatic disease was noteworthy given that the primary tumors had comparable growth rates to the parental cells (Supplemental Fig. S3) and grew to sizes that necessitated sacrifice of the animals.
Global gene expression changes in the 200b transfectants
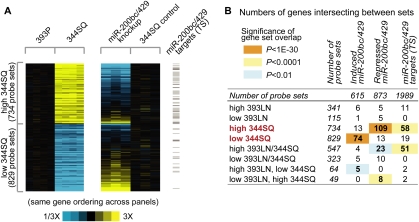
To examine the transcriptional basis for these biologic changes, the 200b and control transfectants (triplicate RNA samples of each) were subjected to mRNA and miR profiling. The most highly changed miRs were the miR-200 family members (miR-429, >1000-fold change, P < 0.0001; miR-200b, >30-fold, P < 0.005; miR-200a, >20-fold, P < 0.005). Among the mRNAs profiled, 615 probe sets were increased and 873 probe sets were decreased (P < 0.01, >1.5-fold) in the miR-200b transfectant cells compared with the vector-transfected cells (Fig. 7A; Supplemental Table S6A,B). Strikingly, these differentially expressed genes overlapped significantly with those differentially expressed in 393P and 344SQ tumors (Supplemental Table S7A,B), representing a broad range of biologic functions by GO categorization. For instance, 109 of the 734 genes that were highly expressed in 344SQ tumors were decreased in the 200b transfectants (enrichment P-value = 1.7E-62), and 74 of the 829 genes that were decreased in 344SQ tumors were increased in the 200b transfectants (P value = 1.25E-37) (Fig. 7B). Concordant with the RT–PCR data for the 200b transfectants (Fig. 6L–Q), N-cadherin levels were markedly lower, while expression of the tight junction genes INADL and multiple CLDN isoforms was restored. Among the genes with decreased expression in 200b transfectants, a significant number were predicted miR-200 targets on the basis of TargetScan analysis (Fig. 7B, last column; Supplemental Table S6; Lewis et al. 2005). Collectively, these findings provide compelling evidence that miR-200 regulated many of the biologic differences between metastasis-prone and metastasis-incompetent tumors in this syngeneic tumor model.
Figure 7.
Forced miR-200 expression reverts the cells to a nonmetastatic mRNA expression pattern and prevents metastasis. (A) Expression patterns for the genes significant between 344SQ and 393P (from Fig. 1A) or between the mir-200bc/429-overexpressing 344SQ cells (344SQ_200b_1) and the 344SQ_vector control. For 344SQ/393P data set, genes are centered on 393P group; for the 200bc/429 knockup data set, genes are centered on 344SQ_vector control group. Genes represented in both heat maps are the same and have the same ordering. Gray bars to the right highlight genes predicted to be miR-200 family targets, using Targetscan (version 5.0). (B) Table of gene probe sets intersecting between the 393P/393LN/344SQ gene signatures (rows, from Fig. 1A) and the genes induced or repressed by mir-200bc/429 expression. The last column indicates the number of genes predicted to be miR-200bc/429 targets using Targetscan (version 5.0). P-values by one-sided Fisher's exact test (using the population of ∼45,000 probe sets).
Discussion
Comparison of miR expression from normal versus human tumor tissues has established a strong link between tumorigenesis and aberrant miR regulation, with the subsequent classification of many miRs as either oncogenes or tumor suppressors (Lu et al. 2005; Hammond 2007). A growing body of evidence from work on a variety of mouse models of cancer driven by common genetic lesions supports a role for miRs in malignant progression (Ma and Weinberg 2008). The accompanying study by Olson et al. (2009) in this issue of Genes & Development reveals the dynamic nature of these changes in the RIP-Tag2 model of pancreatic neuroendocrine carcinoma, which exhibited unique expression patterns associated with early and late stages of malignant progression, including a prominent suppression of the miR-200 family in late-stage or metastatic lesions. In this study, we performed an in-depth analysis of a single aspect of tumorigenesis (namely, metastasis) in a syngeneic tumor model using a panel of cell lines derived from mice that develop metastatic lung adenocarcinoma owing to expression of mutant K-ras and p53. Surprisingly, we found that metastasis-prone tumor cells had the capacity to transit reversibly between epithelial and mesenchymal states, a property that was regulated by a single miR cluster. Forced expression of the miR-200b cluster in 344SQ cells was sufficient to lock these cells into an epithelial state, abrogate their metastatic capacity in syngeneic mice, and confer transcriptional features that resembled those of metastasis-incompetent tumors. Biological reprogramming of this magnitude supports a central role for the miR-200 family in metastasis.
A growing body of evidence indicates that K-ras mutations are important in the development of lung adenocarcinoma. Somatic mutations that activate K-ras have been detected in ∼30% of lung adenocarcinomas, the most common subtype of lung cancer, and in atypical alveolar hyperplasia lesions, a histologic change thought to precede the development of lung adenocarcinoma (Mitsudomi et al. 1991; Mori et al. 1996). Mice expressing mutant K-ras develop lung adenocarcinoma rapidly and with high penetrance (Fisher et al. 2001; Jackson et al. 2001; Johnson et al. 2001; Guerra et al. 2003). However, the long latency periods and low metastatic potential of these tumors suggest that other somatic events are necessary to complete the development of invasive disease. One candidate is mutant p53, which confers metastatic potential to K-ras-initiated lung adenocarcinomas (Liu et al. 2000; Olive et al. 2004; Jackson et al. 2005; Zheng et al. 2007). To query the mechanistic basis of metastasis, we established lung adenocarcinoma cell lines from these mice and characterized their metastatic potential following injection into immunocompentent, syngeneic mice. Because all the cell lines had mutant K-ras and p53 alleles, but varied in their metastatic capacity, these findings suggest that K-ras and p53 mutations were not sufficient to confer metastatic capacity. Although multiple genetic events might have contributed to the metastatic capacity of these cells, findings presented here support the argument that silencing of miR-200 expression is one of those events. The relevance of these findings to human lung cancer was supported by evidence from a large panel of human lung cancer cell lines that miR-200 expression correlated with changes in the EMT markers and distinguished these cell lines on the basis of their site of origin (lung primary tumor vs. metastasis).
Previous comparative expression profiling of spontaneous metastases versus primary lung tumors from the KrasLA1/+;p53R172HΔG/+ animals suggested a role for EMT (Gibbons et al. 2009), a finding that was confirmed here by comparison of metastasis-prone and metastasis-incompetent syngeneic tumors. Although frequently invoked as a necessary step for tumor invasion and metastasis, the factors controlling the EMT switch are poorly understood. One hypothesis is that metastatic cells accumulate widespread chromosomal changes, producing somatic mutations that predispose the cells to undergo EMT and metastasize. Although the highly aberrant genomes of the tumor cells studied here provide general support for this hypothesis, the metastasis-prone tumor cells also exhibited a marked responsiveness to their cellular context both in vitro and in vivo, raising the possibility that somatic mutations are not the sole drivers of malignant progression in this model. Furthermore, aspects of this plasticity may be unique to metastatic cells, as we found that the metastasis-prone tumor cells formed spheres in Matrigel and in nonadherent tissue culture plates, whereas the nonmetastatic cells did not (data not shown). We therefore propose that our data support an alternative model in which a subset of tumor cells either originates from, or acquires features of, progenitor cells and metastasizes as a consequence of their unique responsiveness to diverse microenvironmental cues. In support of this model was evidence that the differentially expressed genes in 344SQ tumors were enriched for genes identified in a population of breast cancer stem cells (Shipitsin et al. 2007; data not shown). Furthermore, lung tumorigenesis in several K-ras-driven mouse models is preceded by expansion of a progenitor cell population called bronchoalveolar stem cells (Kim et al. 2005; Yang et al. 2008). Evidence for this hypothesis was reported recently in human mammary epithelial cells, showing that EMT resulted in up-regulation of multiple stem cell markers and produced cells with biologic features of progenitor cells (Mani et al. 2008; Morel et al. 2008).
Pioneering work from Bissell and colleagues (Streuli et al. 1991; Weaver et al. 1997; Wang et al. 1998; Muthuswamy et al. 2001) has powerfully demonstrated the critical role for cell–extracellular matrix interactions in normal breast epithelial differentiation and tumor cell morphogenesis. The findings reported here extend on that concept by showing that tumor cell morphogenesis (EMT vs. MET) is controlled by expression of the miR-200 family, which is influenced by the sum total of interactions with the extracellular matrix (e.g., laminin-rich Matrigel) and soluble factors (e.g., TGFβ). This suggests the possibility that the phenotypic differences between tumor cells might arise as much from their differing abilities to direct the stromal components generated around them as from somatic genetic changes. This implies the potential ability to convert metastasis-prone tumor cells to a metastasis-incompetent state by manipulating their interactions with the extracellular environment, an idea with significant therapeutic implications that requires additional investigation.
Identifying the miR-200 targets that regulate epithelial tumor cells may lead to a deeper understanding of the transcriptional events that control their metastatic potential. Based on prior studies that used either transient expression of miR-200 family members (Park et al. 2008) or transient knockdown of ZEB1 (Aigner et al. 2007) in human cancer cells to study their effects on EMT, we expected changes in a relatively limited number of genes upon constitutive expression of the miR-200b cluster. Surprisingly, stable constitutive miR-200b expression caused global changes in mRNA expression and produced a cell that was strikingly similar on a transcriptional level to metastasis-incompetent 393P cells. Given that Zeb1 regulates the levels of classical EMT markers, tight junctional elements, and various polarity genes, certainly some of the changes observed with stable miR-200b expression are attributable to the double negative feedback loop with ZEB1 (Bracken et al. 2008; Burk et al. 2008). However, the number of genes up-regulated in the 200b transfectants that are presumably secondary effects, rather than direct, indicates broader changes in expression regulation likely due to interactions with transcription factors that have not yet been defined. These unidentified miR-200 targets are likely to be critical mediators of EMT and metastasis in the context of signals originating from soluble and structural factors in the extracellular matrix.
Materials and methods
Cell lines and culture
The human lung cancer cell lines were established at the National Cancer Institute (NCI-H series) or at the Hamon Center for Therapeutic Oncology Research, University of Texas Southwestern Medical Center (HCC series). These cell lines have been deposited for distribution in the American Type Culture Collection (ATCC) (http://www.atcc.org). Cells were grown in a humidified atmosphere with 5% CO2 at 37°C in RPMI 1640 with 5% fetal bovine serum (FBS).
Cell lines from p53R172HΔg/+ K-rasLA1/+ mice were derived from tumor tissues at necropsy from different mice, as indicated by the cell line number (e.g., #344 and #393). Tissues were minced, placed in culture, and passed serially in RPMI 1640 with 10% FBS, which yielded mass populations of tumor cells derived from primary lung tumors (designated as “P”; e.g., 393P), mediastinal lymph nodes (“LN”; e.g., 393LN), and a subcutaneous site (“SQ”; e.g., 344SQ) (see Supplemental Table S1 for complete list). Recombinant human TGFβ1 (Calbiochem) was used at 5 ng/mL. The TGFβ inhibitor SB431542 was purchased from Sigma-Aldrich and used at a final concentration of 10 μM. TGFβ1 levels were measured with the Quantikine immunoassay, according to the manufacturer's instructions (R&D Systems).
Stable 344SQ cell lines expressing the miR-200b–200a–429 cluster or control vector were generated by transduction with the described lentivirus vectors (Gregory et al. 2008), generated in the Trans-Lentiviral pLEX packaging system with TLA-HEK293T cells (Open Biosystems). GFP-positive transfectant pools were selected by growth in RPMI 1640 with 10% FBS and puromycin.
Syngeneic tumor cell injections
All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee at The University of Texas M.D. Anderson Cancer Center. Wild-type 129Sv mice from our colony (males and females) of at least 8 wk of age were used for the syngeneic tumor experiments. Subcutaneous injections of 1 million cells in single-cell suspension were placed in the posterior flank in a volume of 100 μL of PBS or complete media. Animals were monitored regularly and euthanized when they exhibited signs of morbidity or when the size of the subcutaneous tumor required sacrifice (typically at 5–6 wk time).
Total RNA isolation and quality control
Total RNA for miR expression profiling of the human cell lines was prepared using the mirVana miRNA Isolation Kit (Ambion). RNA purity was assessed by spectrophotometric measurement of the OD260/OD280 ratio with acceptable values falling between 1.8 and 2.1, and the OD260/OD230 ratio with acceptable values falling between 1.9 and 2.1. RNA integrity was assessed by Bioanalyzer (Agilent Technologies), with an RNA integrity number (RIN) of >7.0 considered acceptable for expression profiling by microarray.
mRNA expression profiling
For the syngeneic murine tumors, snap-frozen samples were shipped on dry ice to Asuragen for processing and analysis on Affymetrix Mouse Expression Array 430A 2.0 chips. Total RNA was isolated by the Ambion kit. RNA quality and quantity were evaluated on a NanoDrop spectrophotometer and the Agilent 2100 Bioanalyzer. Gene expression data sets have been made available through the Gene Expression Omnibus (GEO) (accession numbers GSE14458 and GSE15741).
After scanning and low-level quantification using Microarray Suite (Affymetrix), DNA Chip (dChip) analyzer (Li and Wong 2001) was used to estimate expression values, using the PM/MM difference model and invariant set normalization. Two-sided t-tests using log-transformed data determined significant differences in mean gene mRNA levels between groups of samples. Fold changes between groups were estimated as described in Creighton et al. (2008a). Expression patterns were visualized as color maps using the Java TreeView software (Saldanha 2004). GO annotation terms were searched within gene sets using SigTerms (Creighton et al. 2008b). Genes were clustered (Fig. 1A) using the technique described in Creighton et al. (2008a).
For the human non-small-cell lung cancer cell lines, RNA fluorescent labeling reaction and hybridization were performed using the Affymetrix HG-U133A and HG-U133B Gene Chips according to the manufacturer's instructions (http://www.affymetrix.com), as described (Zhou et al. 2006). The arrays consist of probes for 23,583 unique genes based on Unigene build 173. Microarray analysis was performed using Affymetrix Microarray Suite 5.0 and MATRIX 1.26. Array data were median-normalized, with replicate measurements averaged together. Samples (or averages of samples) were then compared by calculating log ratios for each gene, and statistical significance was calculated by Student's t-test. The microarray data can be downloaded from the GEO, accession number GSE-4824.
miR expression profiling
The murine tumors and cell lines were profiled using the DiscovArray platform by Asuragen, after preparation and evaluation of total RNA samples as described for the mRNA analysis. From the 1086 probes in the Sanger miRBase version 9.2 covering humans, rats, and mice, all but 91 are represented, and an additional 12,894 exploratory probes covering multiple other species are included.
For miR expression profiling of the human non-small-cell lung cancer cell lines, total RNA meeting quality control criteria were sent to MOgene, where the samples were purified further using Micro Bio-Spin six columns (Bio-Rad) and dried in a Speed-Vac. RNA was quantified using the Nanodrop-1000 spectrophotometer (Nanodrop Technologies) and the quality of total RNA was determined on an Agilent Bioanalyzer (Agilent Technologies). Aliquots (100 ng) of each RNA sample were treated with calf intestine phosphatase (GE Healthcare) for 30 min at 37°C, denatured using 100% DMSO (Sigma) for 5 min at 100°C, and then transferred to an ice-water bath to prevent RNA reannealing. RNA samples were then labeled with pCp-Cy3 using T4 RNA ligase (GE Healthcare) by incubation for 2 h at 16°C. Labeled samples were hybridized to Agilent Human Genome V2 miRNA microarrays, which contain probes for 723 human and 76 human viral miRs cataloged in the Sanger database version 10.1 (http://microrna.sanger.ac.uk). Hybridizations were performed in SureHyb chambers (Agilent) for 24 h at 55°C. Arrays were then washed using Agilent-prepared buffers and scanned using an Agilent scanner (G2505B). Feature extraction was performed using Agilent FE software. Raw data were thresholded to 1, and arrays were normalized using a median shift to the 75th percentile.
Analysis of non-small-cell lung cancer cell line miR and mRNA expression
Correlations between expression profiles of individual miRs, and between expression of miR-141 and that of EMT markers ZEB1, ZEB2, CDH1, CDH2, and VIM were calculated by Pearson product–moment correlation coefficient. The significance of the correlation was estimated as the fractional probability of the Student's t-distribution, giving the probability that a correlation at least as good will be obtained at random. Cell lines were divided on the basis of origin (primary tumor or metastatic site), with mean expression in each group and the significance of the differential expression between groups determined by t-test, using log-transformed data. Finally, to identify miR target sites enriched in the set of genes expressed at lower levels in cell lines derived from primary tumors relative to those derived from metastatic sites, expression fold changes of the genes were correlated with the motif content of their 3′UTRs using miReduce (Sood et al. 2006).
Quantitative RT–PCR analysis
RNA from samples grown in culture was purified by either the RNeasy Kit (Qiagen) or the RiboPure Kit (Applied Biosystems) using the manufacturers' instructions. Analysis of mRNA levels was performed on a 7500 Fast Real-Time PCR System (Applied Biosystems) with SYBR Green-based real-time PCR, using primers designed with Primer Express (Applied Biosystems). Serially diluted cDNA samples were amplified to establish a standard curve for calculation of relative target concentrations. The housekeeping gene L32 ribosomal gene was used as an internal control. Data were analyzed with the SDS 2.1 software (Applied Biosystems).
Analysis of miR levels was similarly performed using the TaqMan miR assays according to the manufacturer's instructions (Applied Biosystems). All miR data are expressed relative to a miR-16 or snoRNA-135 control reaction (as indicated) performed simultaneously on the same sample.
3D culture and indirect immunofluorescent staining
Cells were grown in 3D culture on Matrigel in eight-well glass chamber slides, essentially as described (Debnath et al. 2003). In brief, a single-cell suspension containing 500–5000 cells (depending on the assay) was plated onto a layer of growth factor-reduced Matrigel (BD Biosciences). The cells were grown in RPMI 1640 with 10% FBS and 2% Matrigel, which was changed every 2–3 d. Cell growth and morphology were followed by phase contrast microscopy on an Olympus IX 71 microscope.
Indirect immunofluorescent staining and analysis were performed as outlined previously (Debnath et al. 2003). Briefly, cells grown on Matrigel were washed with phosphate-buffered saline (PBS) and fixed with 2% formalin. The cells were permeablized with PBS/0.5% TX-100 for 10 min at 4°C and washed three times with 100 mM glycine in PBS for 10–15 min at room temperature. Cells were blocked with immunofluorescence buffer (PBS containing 7.7 mM sodium azide, 0.1% bovine serum albumin, 0.2% TX-100, 0.05% Tween-20) containing 10% goat serum for 1–2 h at room temperature. The cells were incubated with the indicated primary antibodies in the same blocking buffer overnight at 4°C, and washed three times for 20 min each in the immunofluorescence buffer. The fluorescent-conjugated secondary antibodies (1:200 dilution) in the same blocking buffer were incubated with the cells for 45–60 min at room temperature, followed by three washes of 20 min each in immunofluorescence buffer. The nuclei were stained with a combination of TOPRO-3 (5 μM) and DAPI (0.5 ng/mL) for 15–30 min. The slides were mounted with Prolong Antifade Gold prior to imaging on a Zeiss LSM 510 confocal microscope. We used primary antibodies against ZO-1 and Ki-67 (Zymed); β-catenin, E-cadherin, and GM-130 (BD Transduction Laboratories); ParD6B (Santa Cruz Biotechnologies); α6-integrin (Chemicon) and cleaved caspase 3 (Cell Signaling); and Alexa-fluor-conjugated secondary antibodies (Molecular Probes).
Migration and invasion assays
Cells in serum-free RPMI 1640 (5 × 104) were seeded onto Transwell plates coated with 0.1% gelatin or GFR-Matrigel (BD Biosciences), with RPMI 1640/5% FBS in the lower well as the chemoattractant. Each condition was performed in triplicate or quadruplicate. After 16–18 h of incubation, the medium was removed and the cells were fixed with 90% ethanol. The migrated cells were stained with 0.1% crystal violet and washed with ddH2O, and five microscopic fields (10×) were counted per filter.
Cytogenetic analysis
Primary cells isolated from K-rasLA/+ p53R172HΔg/+ mice were cultured for five or fewer passages before cytogenetic analysis. Metaphase spreads were prepared and evaluated in the M.D. Anderson Cancer Center Molecular Cytogenetics Core Facility for quantitative cytogenetics and for spectral karyotyping (SKY), by methods described previously (Hingorani et al. 2005).
Acknowledgments
We thank all of the members of the Kurie and Pertsemlidis laboratories for assistance and comments on the work; Dror Berel for assistance with the mRNA and miR expression data analysis; Hank Adams of the Department of Molecular Genetics Microscopy Core for advice and assistance with the confocal microscopy; Dr. Gigi Lozano for the KrasLA1/+; p53R172HΔG mice and cell lines; Dr. Asha Multani of the M.D. Anderson Cytogenetics Core for assistance with the cytogenetic analyses; Dr. Waun Ki Hong for financial support; and Jeff Schageman and Cheng Hui Lee for assistance in analyzing the miR expression profiles of the non-small-cell lung cancer cell lines. We also thank Dr. Peter Olson and Dr. Doug Hanahan for sharing data prior to publication and for helpful comments on our manuscript. Grant support: NIH grants 5 T32 CA009666 (to D.L.G and W.L.); P50 CA70907 (Lung Cancer SPORE), R01 grants CA132608 and CA117965, David M. Sather Memorial Fund, and The Armour Family Lung Cancer Research Fund (to J.M.K.); P30 CA125123 and the Dan L. Duncan Cancer Center at Baylor College of Medicine (to C.J.C.); and P50 CA70907 and R01 CA129632 (to A.P.). D.L.G. was also supported by a Young Investigator Award from The ASCO Cancer Foundation and an International Association for the Study of Lung Cancer (IASLC) Fellow Grant. J.M.K. is the Elza A. and Ina Shackelford Freeman Endowed Professor in Lung Cancer.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1820209.
Supplemental material is available at http://www.genesdev.org.
References
- Aigner K, Dampier B, Descovich L, Mikula M, Sultan A, Schreiber M, Mikulits W, Brabletz T, Strand D, Obrist P, et al. The transcription factor ZEB1 (δEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26:6979–6988. doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussemaker HJ, Li H, Siggia ED. Regulatory element detection using correlation with expression. Nat Genet. 2001;27:167–171. doi: 10.1038/84792. [DOI] [PubMed] [Google Scholar]
- Creighton CJ, Casa A, Lazard Z, Huang S, Tsimelzon A, Hilsenbeck SG, Osborne CK, Lee AV. Insulin-like growth factor-I activates gene transcription programs strongly associated with poor breast cancer prognosis. J Clin Oncol. 2008a;26:4078–4085. doi: 10.1200/JCO.2007.13.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Nagaraja AK, Hanash SM, Matzuk MM, Gunaratne PH. A bioinformatics tool for linking gene expression profiling results with public databases of microRNA target predictions. RNA. 2008b;14:2290–2296. doi: 10.1261/rna.1188208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, Berx G, Cano A, Beug H, Foisner R. δEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24:2375–2385. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- Fisher GH, Wellen SL, Klimstra D, Lenczowski JM, Tichelaar JW, Lizak MJ, Whitsett JA, Koretsky A, Varmus HE. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes & Dev. 2001;15:3249–3262. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons DL, Lin W, Creighton CJ, Zheng S, Berel D, Yang Y, Raso MG, Liu DD, Wistuba II, Lozano G, et al. Expression signatures of metastatic capacity in a genetic mouse model of lung adenocarcinoma. PLoS One. 2009;4:e5401. doi: 10.1371/journal.pone.0005401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Guerra C, Mijimolle N, Dhawahir A, Dubus P, Barradas M, Serrano M, Campuzano V, Barbacid M. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell. 2003;4:111–120. doi: 10.1016/s1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- Hammond SM. MicroRNAs as tumor suppressors. Nat Genet. 2007;39:582–583. doi: 10.1038/ng0507-582. [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Huber MA, Kraut N, Beug H. Molecular requirements for epithelial–mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes & Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, Jacks T. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- Ji H, Houghton AM, Mariani TJ, Perera S, Kim CB, Padera R, Tonon G, McNamara K, Marconcini LA, Hezel A, et al. K-ras activation generates an inflammatory response in lung tumors. Oncogene. 2006;25:2105–2112. doi: 10.1038/sj.onc.1209237. [DOI] [PubMed] [Google Scholar]
- Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT, Jacks T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial–mesenchymal transition and cancer metastasis. RNA Biol. 2008;5:115–119. doi: 10.4161/rna.5.3.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial–mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc Natl Acad Sci. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, McDonnell TJ, Montes de Oca Luna R, Kapoor M, Mims B, El-Naggar AK, Lozano G. High metastatic potential in mice inheriting a targeted p53 missense mutation. Proc Natl Acad Sci. 2000;97:4174–4179. doi: 10.1073/pnas.97.8.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Ma L, Weinberg RA. MicroRNAs in malignant progression. Cell Cycle. 2008;7:570–572. doi: 10.4161/cc.7.5.5547. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsudomi T, Viallet J, Mulshine JL, Linnoila RI, Minna JD, Gazdar AF. Mutations of ras genes distinguish a subset of non-small-cell lung cancer cell lines from small-cell lung cancer cell lines. Oncogene. 1991;6:1353–1362. [PubMed] [Google Scholar]
- Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial–mesenchymal transition. PLoS One. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Tezuka F, Chiba R, Funae Y, Watanabe M, Nukiwa T, Takahashi T. Atypical adenomatous hyperplasia and adenocarcinoma of the human lung: Their heterology in form and analogy in immunohistochemical characteristics. Cancer. 1996;77:665–674. [PubMed] [Google Scholar]
- Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol. 2001;3:785–792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Olson P, Lu J, Zhang H, Shai A, Chun MG, Wang Y, Libutti SK, Nakakura EK, Golub TR, Hanahan D. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes & Dev. 2009 doi: 10.1101/gad.1820109. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes & Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview—Extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streuli CH, Bailey N, Bissell MJ. Control of mammary epithelial differentiation: Basement membrane induces tissue-specific gene expression in the absence of cell–cell interaction and morphological polarity. J Cell Biol. 1991;115:1383–1395. doi: 10.1083/jcb.115.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Weaver VM, Petersen OW, Larabell CA, Dedhar S, Briand P, Lupu R, Bissell MJ. Reciprocal interactions between β1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: A different perspective in epithelial biology. Proc Natl Acad Sci. 1998;95:14821–14826. doi: 10.1073/pnas.95.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whang-Peng J, Knutsen T, Gazdar A, Steinberg SM, Oie H, Linnoila I, Mulshine J, Nau M, Minna JD. Nonrandom structural and numerical chromosome changes in non-small-cell lung cancer. Genes Chromosomes Cancer. 1991;3:168–188. doi: 10.1002/gcc.2870030303. [DOI] [PubMed] [Google Scholar]
- Yang Y, Iwanaga K, Raso MG, Wislez M, Hanna AE, Wieder ED, Molldrem JJ, Wistuba II, Powis G, Demayo FJ, et al. Phosphatidylinositol 3-kinase mediates bronchioalveolar stem cell expansion in mouse models of oncogenic K-ras-induced lung cancer. PLoS One. 2008;3:e2220. doi: 10.1371/journal.pone.0002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, El-Naggar AK, Kim ES, Kurie JM, Lozano G. A genetic mouse model for metastatic lung cancer with gender differences in survival. Oncogene. 2007;26:6896–6904. doi: 10.1038/sj.onc.1210493. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Peyton M, He B, Liu C, Girard L, Caudler E, Lo Y, Baribaud F, Mikami I, Reguart N, et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell. 2006;10:39–50. doi: 10.1016/j.ccr.2006.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]