Abstract
While altered expression of microRNAs (miRs) in tumors has been well documented, it remains unclear how the miR transcriptome intersects neoplastic progression. By profiling the miR transcriptome we identified miR expression signatures associated with steps in tumorigenesis and the acquisition of hallmark capabilities in a prototypical mouse model of cancer. Metastases and a rare subset of primary tumors shared a distinct miR signature, implicating a discrete lineage for metastatic tumors. The miR-200 family is strongly down-regulated in metastases and met-like primary tumors, thereby relieving repression of the mesenchymal transcription factor Zeb1, which in turn suppresses E-cadherin. Treatment with a clinically approved angiogenesis inhibitor normalized angiogenic signature miRs in primary tumors, while altering expression of metastatic signature miRs similarly to liver metastases, suggesting their involvement in adaptive resistance to anti-angiogenic therapy via enhanced metastasis. Many of the miR changes associated with specific stages and hallmark capabilities in the mouse model are similarly altered in human tumors, including cognate pancreatic neuroendocrine tumors, implying a generality.
Keywords: MicroRNA, pancreatic neuroendocrine tumor (PNET), cancer, transgenic mouse, mouse model of human cancer, miR-200
MicroRNAs (miRs) are a class of small RNAs with important regulatory functions in diverse biological processes (Ambros 2004; Bartel 2004; Kloosterman and Plasterk 2006). miRs bind partly complementary sequences in mRNAs, targeting them for degradation and/or inhibiting translation, thereby down-regulating protein expression. Recent studies have identified a number of miRs that are differentially regulated in tumor versus normal tissue (Michael et al. 2003; Calin et al. 2004, 2005; Ciafre et al. 2005; H He et al. 2005; Iorio et al. 2005; Lu et al. 2005; Calin and Croce 2006; Murakami et al. 2006; Pallante et al. 2006; Roldo et al. 2006; Volinia et al. 2006; Yanaihara et al. 2006; Bloomston et al. 2007; Lee et al. 2007; Szafranska et al. 2007). While a handful have been shown to act as bona fide oncogenes or tumor suppressors, the functions of most of these tumor-associated miRs remain unknown.
RIP-Tag2 (RT2) mice develop pancreatic neuroendocrine tumors (PNETs) as a consequence of SV40 T-antigen oncogene expression in pancreatic β cells. This model has proven useful for characterizing the process of multistep tumorigenesis, as multifocal, oncogene-expressing islets progress through several distinct stages as they acquire the hallmarks of cancer (Hanahan 1988; Folkman et al. 1989; Hanahan and Weinberg 2000). Tumorigenesis begins with the appearance of multiple hyperplastic/dysplastic islets, of which a subset undergo an “angiogenic switch” (Folkman et al. 1989; Hanahan et al. 1996) to a state of chronic angiogenesis, which persists in tumors. Tumors consist of encapsulated solid tumors (adenomas) that progress into invasive carcinomas, characterized by loss of E-cadherin (Perl et al. 1998); a separate pathway leading directly to highly invasive carcinomas has been implicated (Lopez and Hanahan 2002). Lymph node metastasis is infrequent, and distant metastasis is rare, likely due to the rapid time course of the disease, which culminates in an end stage at 15 wk, with two to 10 pancreatic tumors.
This study sought to address a hypothesis that the miR transcriptome was being altered in instructive ways in the distinctive stages of multistep tumorigenesis. We performed high-throughput miR profiling to audit the expression level of miRs in premalignant stages as well as in tumors and metastases. Each stage was found to be associated with a distinct miR expression signature. This study begins to segregate the functions of previously identified miRs that are altered in human cancers according to the stage of their initial alteration and the hallmark capability with which they are associated.
Results
Profiling miRs during multistage tumorigenesis
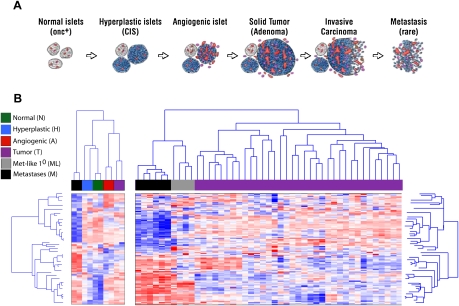
To assess the hypothesis that miRs were differentially regulated during multistage tumorigenesis, we profiled the expression level of all known mouse miRs in dissected tissues representing each of the histologically and temporally separable stages in the prototypical pathway to pancreatic neuroendocrine tumors in RT2 mice (Fig. 1A). The analysis involved two pools each of normal, hyperplastic/dysplastic, and angiogenic islets, as well as 39 individual tumors and six liver metastases. We also generated two pools of primary and metastatic tumors to facilitate comparisons with pretumor lesions. The miR transcriptome profile was similar in the independent representations of each stage. Thus, the two independent pools of normal, hyperplastic, and angiogenic islets each clustered together, as did tumors and metastases (Fig. 1B; Supplemental Table 1). The clustering analysis further revealed that each of the stages had marked differences in their miR expression profiles; each stage bore a distinct miR signature.
Figure 1.
Differential expression of the miR transcriptome in the distinctive stages of multistep tumorigenesis. (A) Schematic representation of the separable stages of multistep tumorigenesis in the RT2 transgenic mouse model of PNETs, which afforded the experimental approach to miR transcriptome profiling of discrete stages of carcinogenesis. (B) Clustering analysis of normal, hyperplastic, and angiogenic islet pools, along with pools of primary tumors and of metastases, are shown in the left panel. In the right panel, clustering analysis of 39 individual tumors and six liver metastases reveals that a subset of primary tumors is remarkably similar to metastases in its miR expression profile. Another subset of tumors up-regulates miRs in the Dlk1-Gtl2 imprinted cluster (miRs in red two-thirds of the way down in the left-most primary [purple] tumors as well as in metastases; see also Supplemental Fig. 2).
miR signatures of hyperproliferation and angiogenesis
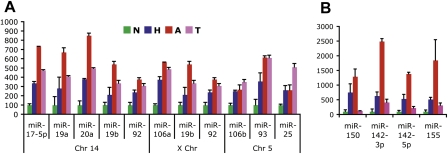
Hyperplastic/dysplastic islets are hyperproliferative but lack the latter-stage hallmarks of angiogenesis and invasiveness (Lopez and Hanahan 2002). Of the 430 miRs profiled, nine were up-regulated at least twofold at this stage, and one was down-regulated 2.5-fold (Table 1A). Six of the nine up-regulated miRs were from the well-characterized miR-17-92 clusters, previously demonstrated to have oncogenic properties and to be associated with increased proliferation and angiogenesis (L He et al. 2005; O'Donnell et al. 2005; Dews et al. 2006). Interestingly, in the present case, their up-regulation was first observed in hyperplastic islets and was modestly up-regulated further in angiogenic islets. All 11 of the 17-92 miRs, involving clusters on three chromosomes, exhibit a similar regulatory pattern, with each remaining elevated in subsequent stages of tumor progression, consistent with their association with the hyperproliferative capability (Fig. 2A). Two other hyperplastic signature miRs, miR-142-3p and miR-155, peak in angiogenic islets but then decrease in tumor stages (Fig. 2B). Although several miRs from the miR-17-92 clusters, as well as miR-150, did not reach statistical significance, these miRs are similarly regulated in the stages of tumorigenesis (Fig. 2), and arguably linked to the signature.
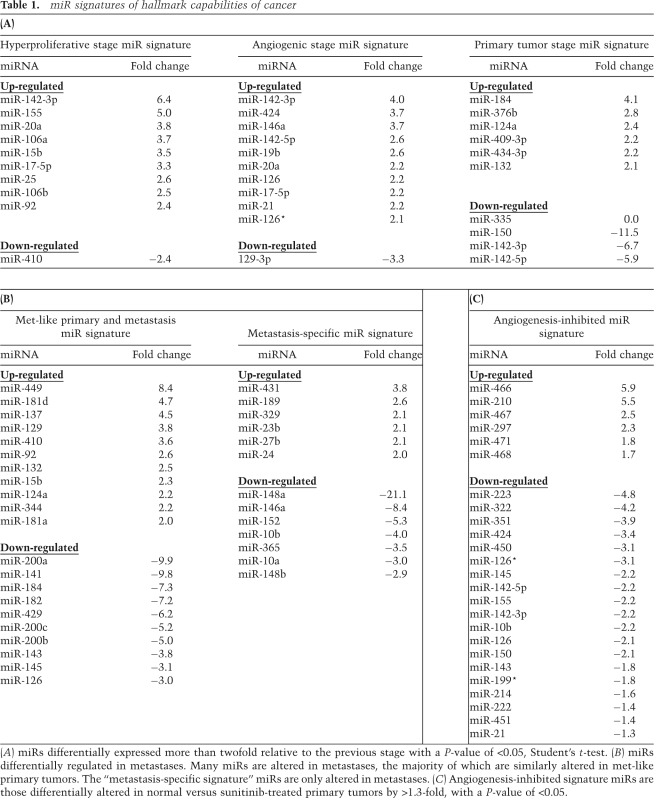
Table 1.
miR signatures of hallmark capabilities of cancer
(A) miRs differentially expressed more than twofold relative to the previous stage with a P-value of <0.05, Student's t-test. (B) miRs differentially regulated in metastases. Many miRs are altered in metastases, the majority of which are similarly altered in met-like primary tumors. The “metastasis-specific signature” miRs are only altered in metastases. (C) Angiogenesis-inhibited signature miRs are those differentially altered in normal versus sunitinib-treated primary tumors by >1.3-fold, with a P-value of <0.05.
Figure 2.
miR profiling reveals dynamic regulation and illuminates immune cell infiltration in pretumor stages in RT2 carcinogenesis. (A) Fold changes of the three miR-17-92 clusters by chromosomal location in normal (N), hyperplastic (H), and angiogenic (A) islets, and tumors (T). Average plus standard deviation are shown with expression in normal islets normalized to 100. (B) Expression of miR-150, miR-142-3p, miR-142-5p, and miR-155 in the neoplastic stages. Average plus standard deviation are shown with expression in normal islets normalized to 100%. Supplemental Figure 2 documents the immune cell specificity of these miRs in this pathway.
miR-483 was not included in the set of mouse miRs constituting the known miR transcriptome used in our initial profiling study. This miR came to our attention by virtue of its location within an intron of the IGF2 gene in both mice and humans, since IGF2 is up-regulated and functionally important for the hyperproliferative switch in the RT2 tumorigenesis pathway (Christofori et al. 1994). We therefore used quantitative PCR (Q-PCR) to assess miR-483 expression levels in pretumor stages, tumors, and metastases. We found that miR-483 is up-regulated at the hyperplastic stage and continues to be expressed in most tumors (Supplemental Fig 1A).
miR-142, miR-150, and miR-155 are highly expressed in hematopoietic cells (Shingara et al. 2005), and the islet tumorigenesis pathway is characterized by infiltration of macrophages and neutrophils in the angiogenic islet and tumor stages (Nozawa et al. 2006). We used flow cytometry (FACS) to fractionate hyperplastic islets, angiogenic islets, and tumors into populations of cells that were either leukocytes, as defined by expression of the pan-leukocyte marker CD45, or a mixture of mostly cancer cells, endothelial cells, and infrequent stromal fibroblasts, which were unlabeled by CD45. The CD45+ immune cell population was further fractionated using the activated macrophage marker F4/80, and the neutrophil cell surface antigens GR-1 and 7/4; the remaining single positive CD45 subpopulation likely contains dendritic cells and immature hematopoietic progenitors. We then performed Q-PCR for these miRs as well as for CSF1R, MMP25, and insulin to confirm the purity of macrophages, neutrophils, and cancer cells, respectively, in each sorted sample. miR-142-5p, miR-150, and miR-155 were not confined to any one immune cell population, but were expressed in all CD45+ cells (Supplemental Fig. 2A). They were, however, not expressed in the sorted cancer cell fraction, which did not contain innate immune cells, as evidenced by lack of CSF1R or MMP25 expression (data not shown).
miR-155 is notable in that it is up-regulated in many human cancers; moreover, miR-155 is induced in activated macrophages and is required for normal immune function (Calin et al. 2005; Eis et al. 2005; H He et al. 2005; Iorio et al. 2005; Taganov et al. 2006; Volinia et al. 2006; Yanaihara et al. 2006; Lee et al. 2007; O'Connell et al. 2007; Rodriguez et al. 2007; Szafranska et al. 2007; Thai et al. 2007). These results collectively suggest that up-regulation of miR-155 and similar miRs in tumor versus normal tissue lysates reflects the inflammatory response that changes the cellular composition, and hence, the levels of cell type-specific miR expression of the tumor tissue being profiled, therefore scoring as differential miR expression. Our results extend this concept beyond the late-stage tumors that are conventionally assessed, revealing stage-specific inflammatory cell population dynamics across the ontogeny of stages. The increase in hematopoietic cell-specific miR expression and in CD45+ immune cells in the hyperplastic stage (Fig. 2; Supplemental Fig. 2) may reflect early angiogenic lesions that have recruited proangiogenic neutrophils (Nozawa et al. 2006) but have yet to fully switch on angiogenesis with associated microhemorrhaging (“red islet phenotype”) that forms the basis for identifying and segregating angiogenic progenitor lesions.
Eleven miRs were differentially altered during the angiogenic switch from hyperproliferative islets with quiescent vasculature to angiogenic islets (Table 1A). Similar to the hyperproliferative signature, the majority of the signature miRs were up-regulated; only one, miR-129-3p, was down-regulated. miR-146, like miR-155, has high expression in activated macrophages (Taganov et al. 2006) and may be expressed in immune cell types implicated in the angiogenic switch. Another angiogenic signature miR, miR-21, is overexpressed in many tumor types, including human endocrine pancreatic tumors. A number of previous studies have implicated miR-21 by comparing tumors with cognate normal tissue (Chan et al. 2005; Si et al. 2007; Zhu et al. 2007). Our data now refine its association to a particular capability of tumors—angiogenesis—and to involvement in a premalignant stage where the angiogenic switch first occurs in many tumor types.
miR signatures of tumors and metastases
Only 10 miRs were altered in the majority cluster of end-stage primary tumors when compared with angiogenic islets (Table 1A). In contrast, if one compares normal islets with primary tumors, 34 miRs are differentially expressed. Thus, many of the miR expression changes that distinguish normal tissue from tumor—the standard experimental design in most previous studies—actually occurred during premalignant stages of tumorigenesis.
A subset of tumors as well as the metastases were found to have up-regulated miRs located within the Dlk1-Gtl2 imprinted cluster located on mouse chromosome 12 (Fig. 1B; Supplemental Fig. 3; Lin et al. 2003). Such heterogeneity across panels of PNETs in this model has been noted previously in array CGH profiling that revealed recurrent chromosomal abnormalities in subsets of the tumors (Hodgson et al. 2001); notably, chromosome 12 was not among the affected chromosomes. Fifty-two miRs are located in the Dlk1-Gtl2 cluster, representing ∼10% of all known mouse miRs. Given the approximate twofold increase in expression of miRs from this region, it is tempting to speculate that loss of imprinting has occurred in these tumors.
The liver metastases were associated with a markedly distinct signature (Fig. 1B; Table 1B). In addition, of the 39 primary RT2 tumors profiled, four tumors clustered more closely with the metastases than with the other 35 primary tumors. Two of these tumors were from a mouse that developed metastases. A subset of primary neuroendocrine tumors, therefore, bore a metastasis-like miR signature and potentially harbored an increased proclivity to metastasize.
A notable alteration involves the miR-200 family, comprised of five members with clear sequence similarity: miR-200a, miR-200b, miR-200c, miR-141, and miR-429. The miR-200 genes are organized in two clusters on mouse chromosomes 4 and 6, with miR-200b and miR-200c being two of the top three most highly expressed miRs in normal islets (data not shown). This family is emerging as a critical regulator of differentiation and epithelial status in a plethora of cellular contexts including the brain, olfactory neurons, and adipocytes (Wienholds et al. 2005; Karres et al. 2007; Choi et al. 2008; Kennell et al. 2008). In tumor cells, the miR-200 family regulates an epithelial-to-mesenchymal transition (EMT) in vitro, and their expression was found to be lost in more invasive and metastatic cancer cell lines (Burk et al. 2008; Gregory et al. 2008; Park et al. 2008). We used quantitative RT–PCR (qRT–PCR) to verify several miR changes from the profiling experiment, including two representative miRs from this family: miR-200a and miR-200c. The majority cluster of primary tumors continues to express these miRs at high levels, whereas the four met-like primary tumors as well as the liver metastases had down-regulated these miRs fivefold to 10-fold (Fig. 3A), representing the largest absolute miR alteration in this study.
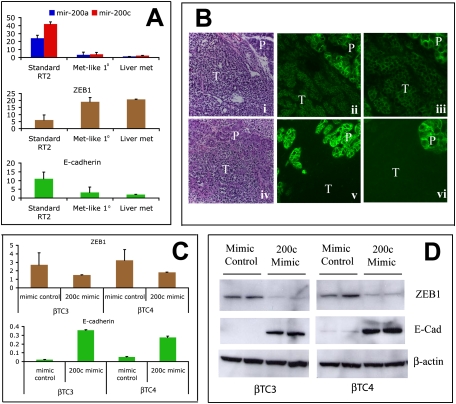
Figure 3.
The miR-200–Zeb1–E-cadherin axis is deregulated in metastases and a subset of primary tumors. (A) Expression levels for miR-200a and miR-200c by Q-PCR normalized to an unaffected miR, miR-16. The miR-200 (also known as “miR-8”) family is organized in two clusters in the human and mouse genome. miR-200a and miR-200c are located in separate clusters, and their expression is representative of all miRs from the two clusters. ZEB1 mRNA exhibits a reciprocal expression pattern compared with miR-200 in that it is low in standard RIP-Tag tumors but up-regulated in met-like primary tumors and liver metastases. E-cadherin expression mirrors miR-200 expression and is mutually exclusive with ZEB1. (B) H&E (panels i,iv) and E-cadherin immunofluorescence at 10× (panels ii,v) and 20× (panels iii,vi) magnification for a noninvasive RT2 tumor (panels i–iii) or a highly invasive IC2 tumor (panels iv–vi). (T) Tumor; (P) normal exocrine pancreas. Noninvasive RT2 tumors express E-cadherin, whereas expression is not detected in IC2s. Note the even higher levels of E-cadherin in normal exocrine pancreas as compared with noninvasive tumors, which are nevertheless positive. (C,D) RNA (C) and protein (D) levels of ZEB1 and E-cadherin following electroporation of a miR-200c mimic or mimic control oligos into βTC3 or βTC4 cells demonstrate their regulatory interconnection. Bar graphs show average plus standard deviation.
A fraction of islet tumors, termed invasive carcinoma grade 2 (IC2), are invasive on all fronts into the exocrine pancreas, and lack delineated/capsular margins (Lopez and Hanahan 2002). The question thus arose as to whether the IC2 grade of tumors identified histologically was one and the same as the met-like primary class distinguished by its miR signature. In a separate study (M Chun and D Hanahan, in prep.), the mRNA transcriptome has been comparatively analyzed using laser capture-microdissected tissue from IC2s versus noninvasive, adenoma grade tumors. We analyzed met-like primary and standard tumors for expression of several differentially expressed genes identified from the laser-captured microdissected study, and found that the met-like primary tumors from the present study exhibited near identical gene expression alterations compared with IC2s (Supplemental Table 3); hence, we infer the met-like primary tumors are IC2-grade PNET tumors. One gene up-regulated in met-like primary tumors and in microdissected IC2s is ZEB1 (Zfhx1a, TCF8) (Fig. 3A; data not shown). ZEB1 is one of the highest-ranked targets for the miR-200 family. ZEB1 mRNA is low in standard tumors, but high in met-like primary tumors, in the independently analyzed IC2 tumors, and in liver metastases (Fig. 3A; data not shown). ZEB1 is a master regulator of EMT, and acts as a transcriptional repressor by binding E-box elements in promoter sequences of target genes, including E-cadherin. Notably, E-cadherin is expressed in standard tumors but is extinguished in met-like primary tumors, in IC2-graded carcinomas, and in metastases (Fig. 3A,B; data not shown), mirroring miR-200 family expression and exhibiting a reciprocal expression pattern compared with ZEB1. While the miR-200 family has been conjectured to play a role in metastasis, this is, to our knowledge, the first in vivo demonstration of down-regulation of the family during de novo progression to a metastastic capability, and in addition reveals comparable down-regulation in a rare subset of primary tumors that may be selectively enabled to spawn metastases.
Seeking to substantiate the ascribed regulatory role of the miR-200 family, we asked whether the miR-200–ZEB1–E-cadherin axis was operative in cancer cells from this model. The βTC3 and βTC4 cancer cell lines, derived from PNET tumors in RT2 mice, express very low levels of miR-200 and E-cadherin, but appreciable levels of ZEB1. When a miR-200c mimic was introduced into these cancer cells, we observed down-regulation of ZEB1 by ∼50% at the RNA level and 80% at the protein level, compared with cells electroporated with a mimic control (Fig. 3C,D). Furthermore, E-cadherin expression was concomitantly increased fivefold to 10-fold at both the RNA and protein level. These data demonstrate that, indeed, the miR-200 family sits atop a regulatory pathway in which miR-200 expression normally blocks ZEB1 expression so as to maintain expression of E-cadherin, a known suppressor of invasiveness and metastasis in this model. As such, the observed down-regulation of the miR-200s represents a demonstrable means to switch off E-cadherin expression in the course of enabling invasion and metastasis. An accompanying study in this issue of Genes & Development (Gibbons et al. 2009) demonstrates a markedly reduced incidence of metastases in metastatic-prone lung cancer cell lines engineered to overexpress the miR-200a–200b–429 cluster. This result demonstrates the miR-200 family can prevent metastasis in vivo, congruent with our observations.
Angiogenesis inhibition affects the angiogenic and metastatic signatures
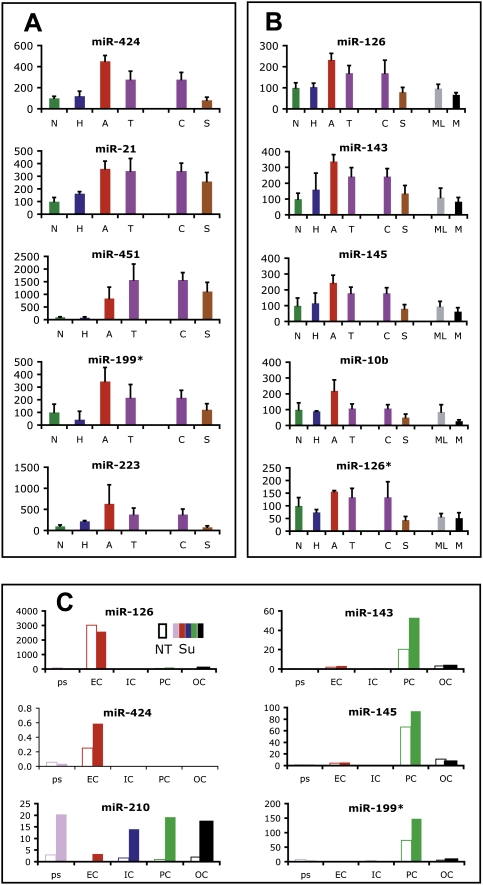
Reasoning that a miR signature ascribed to a particular hallmark capability, if bona fide, might be altered by pharmacological inhibition of that capability, we next asked whether targeting tumor angiogenesis would affect the associated angiogenic miR signature. We treated tumor-bearing RT2 mice with sunitinib, an angiogenesis inhibitor that targets VEGFR, PDGFR, and c-Kit. In previous studies, we had shown sunitinib was a potent inhibitor of angiogenesis and associated tumor growth in this mouse model (Pietras and Hanahan 2005; Paez-Ribes et al. 2009). Tumor-bearing RT2 mice were treated with sunitinib or vehicle daily for 7 d. Tumors were then profiled for miR expression. A subset of the miRs altered during the angiogenic switch was oppositely regulated, and indeed quasinormalized, in response to sunitinib treatment (Fig. 4A,B). miR-424, miR-126, and miR-21 are all up-regulated in angiogenic islets, and conversely restored in level by sunitinib treatment. Additionally, seven miRs that trend up in angiogenic islets—miR-451, miR-199*, miR-223, miR-143, miR-145, miR-10b, and miR-126*—were significantly down-regulated by sunitinib (Fig. 4A,B). Thus, the angiogenic miR signature is largely reversed by therapeutic treatment with a potent angiogenesis inhibitor, consistent with the signature reflecting salient aspects of this capability.
Figure 4.
Anti-angiogenic therapy quasinormalizes the angiogenic miR signature, and evokes elements of the metastatic miR signature. (A) miRs up-regulated in angiogenic islets and down-regulated by sunitinib treatment. (B) miRs up-regulated in angiogenic islets, down-regulated by sunitinib treatment, and similarly down-regulated in metastatic lesions. (N) Normal; (H) hyperplastic; (A) angiogenic; (T) tumor; (C) control-treated tumors; (S) sunitinib-treated tumors; (ML) met-like primary tumors; (M) metastases. (C) FACS of the constituent cell types of tumors followed by Q-PCR analysis reveals that affected miRs reflect either the differential abundance of the tumor vasculature and its component endothelial cells and pericytes, or altered regulation of miRs in the predominant population of cancer cells. (Open bars) (NT) Not treated. (Colored bar) (Su) Sunitinib treated; (ps) presort; (EC) endothelial cells; (IC) immune cells; (PC) pericytes; (OC) other cells.
Differential expression of a given miR in lesional stages could, in principle, reflect altered expression in similarly abundant cell populations (e.g., the cancer cells) or differential abundance of particular cell types characterized by high levels and cell type-restricted expression of the miR. To discriminate between these possibilities, we treated RT2 mice for 1 wk with sunitinib or vehicle, and FACS sorted CD31+ endothelial cells, PDGFRβ+ pericytes, CD45+ leukocytes, and nonlabeled cells, which are mostly cancer cells. Sorted populations were quite pure, as assessed by Q-PCR for cell type-specific genes (data not shown). We performed miR Q-PCR for angiogenic and sunitinib signature miRs and found miR-126 and miR-424 were endothelial cell-specific, whereas miR-143, miR-145, and miR-199* were expressed by pericytes (Fig. 4C). The expression levels for each of these miRs remained similar in the particular cell type found in normal or tumor tissue. What changed was the relative proportions of endothelial cells, pericytes, and inflammatory cells, which were all reduced in sunitinib-treated tumors (see also Paez-Ribes et al. 2009; data not shown); thus, we conclude the decreased levels observed following sunitinib treatment reflect decreased abundance of these cell types, consistent with histopathological analyses. One of the affected angiogenic signature miRs, miR-126, has been reported to be endothelial cell-specific and important for pathological angiogenesis (Fish et al. 2008; Wang et al. 2008). In another context, however, miR-126 has also been shown to function as a metastasis suppressor that is down-regulated in metastatic breast cancer cell lines (Tavazoie et al. 2008). In this neuroendocrine pancreatic tumor setting, miR-126 is not expressed in cancer cells, and is similarly expressed in treated versus untreated tumor endothelial cells (Fig. 4C). Therefore, the expression and roles of miR-126 may prove to be tumor type-specific. In contrast to the other angiogenic signature miRs that reflect vascular cell type-specific expression, miR-210 is up-regulated in all populations, including tumor cells, following sunitinib treatment. miR-210 is up-regulated by hypoxia (Kulshreshtha et al. 2007). This fits well with the phenotypic effects of sunitinib, in which angiogenesis inhibition results in tumor hypoxia (Paez-Ribes et al. 2009).
An increasing body of preclinical and clinical data indicates that potent angiogenesis inhibitors can, after a transitory period of response, elicit increased invasion and metastasis (Bergers and Hanahan 2008). Thus, treatment of human glioblastomas, and mouse models thereof, with angiogenic inhibitors can elicit more highly invasive tumors (Rubenstein et al. 2000; Blouw et al. 2003; Gomez-Manzano et al. 2008; Norden et al. 2008; Paez-Ribes et al. 2009). Other tumor types, including the RIP-Tag model, evidence a more invasive and metastatic phenotype in response to anti-angiogenic therapy (Casanovas et al. 2005b; Ebos et al. 2009; Paez-Ribes et al. 2009). Provocatively, many of the miRs that change following sunitinib treatment were similarly altered in metastases and met-like primaries (Fig. 4B; data not shown) suggesting the treated tumors have initiated a prometastatic evasive resistance program in response to angiogenic inhibition.
Profiling and cross-filtering the miR signature of human PNETs
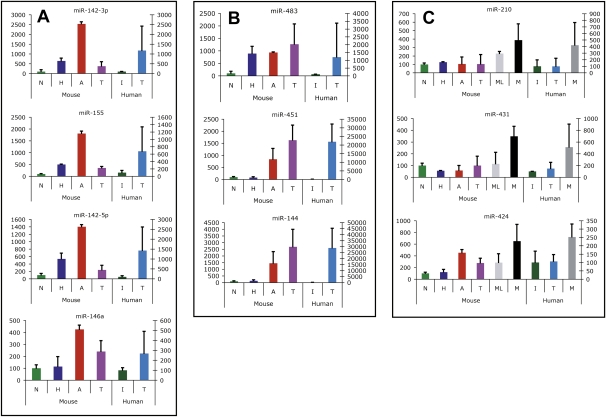
We surveyed published cancer miR profiling studies to determine how many miRs identified as differentially expressed in our study are altered in a variety of human tumors. Remarkably, the majority of altered miRs in these mouse neuroendocrine tumors are, in fact, similarly affected in a number of human tumor types (Supplemental Table 4), suggesting that the underlying mechanisms governing miR dynamics are consistent across disparate tumor types, oncogenic transformations, and cell types of origin. A pertinent question involves the possible correspondence with cognate human PNETs. Previous miR profiling studies for human PNETs have compared tumor to normal pancreas. This design precludes an accurate measurement of fold changes, given the fact that the normal tissue counterpart of a PNET, the endocrine pancreas, is embodied in the dispersed islets of Langerhans, comprising a few percent of the pancreatic mass. We therefore obtained and profiled normal human islet samples from three individuals, 19 pancreatic neuroendocrine tumors, and three liver metastases (Supplemental Table 2). Ten miRs showed concordant expression (Fig. 5). miR-142-3p, miR-142-5p, miR-155, and miR-146a were up-regulated in human PNETs as compared with normal human islets; these miRs were first up-regulated in pretumor stages (and then persisted in tumors) in the mouse model, and hence had been ascribed to either hyperplastic or angiogenic signatures (Fig. 5A; Table 1A). We infer that expression of hematopoietic-specific miRs reflects proangiogenic inflammation in the human PNETs, much as we documented in pretumor and tumor stages of the mouse (Fig. 2B; Supplemental Fig. 2). Additionally, three other miRs—miR-483, miR-451, and miR-144—were found to be up-regulated in mouse and human tumors compared with cognate normal islets (Fig. 5B). While these miRs did not meet our statistical criteria for inclusion into a signature, each is clearly up-regulated at a pretumor stage in the mouse. Hence, miR-483 is first up-regulated concomitant with the hyperproliferative switch in the mouse PNET pathway (Supplemental Fig. 1A), and is maintained at elevated levels in most tumors; interestingly, miR-483 is not up-regulated in metastases or met-like primary tumors. miR-483 is not expressed in normal human islets but is up-regulated in approximately half of the human tumors analyzed (Supplemental Fig. 1B). Both miR-451 and miR-144 are clearly up-regulated in mouse angiogenic islets as well as in tumors of both mice and humans. miR-451 and miR-144 are on the same cluster, consistent with their apparent coregulation. Finally, miR-210, miR-431, and miR-424 were up-regulated in metastases compared with tumors in both mice and humans (Fig. 5C). These cross-filtered miRs with concordantly altered expression in human and mouse tumors are thus suspected to play functional roles in pancreatic neuroendocrine tumorigenesis.
Figure 5.
miRs similarly regulated in mouse and human PNETs and metastases. (A) miRs differentially expressed between human PNETs and normal human islets, and identified as components of the hyperplastic or angiogenic miR signature in mice. (B) miRs differentially expressed between PNETs and normal human islets and similarly expressed differentially between normal islets and the panel of tumors in mice. (C) miRs differentially regulated between metastases and tumors in both mice and humans. (N) Normal islets; (H) hyperplastic islets; (A) angiogenic islets; (T) tumor; (I) human islets; (ML) met-like primary tumors; (M) liver metastases. Average plus standard deviation are shown, with expression in normal islets normalized to 100%. For C, P-value, average, and standard deviation for tumors, met-like primary tumors, and metastases were determined using values from individual tumors from each group.
Discussion
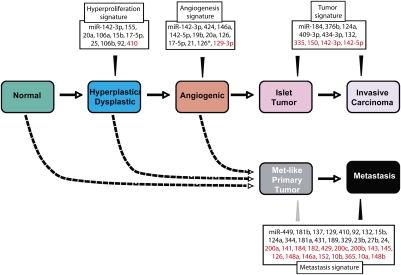
Following the initial implication that miRs might be regulating cancer phenotypes, inferred from the frequent deletion and down-regulation of miR-15/16 in chronic lymphocytic leukemia (Calin et al. 2002), a growing body of research has substantiated the realization that particular miRs are characteristically altered in their expression in different types of tumors. In turn, genetic manipulations are establishing the functional importance of the observed up-regulation and down-regulation of candidate miRs in tumors (L He et al. 2005; Costinean et al. 2006; Esquela-Kerscher et al. 2008; Kumar et al. 2008). The knowledge base has largely come from comparing miR expression in cancer cells versus normal cells, or tumor tissue versus normal tissue. In many cases, the number of characteristically altered miRs is substantial, posing a challenge to understanding the nature of their engagement. It is attractive to envision that the multiplicity of altered miRs reflects functional roles in different acquired capabilities—the hallmarks of cancer—necessary for manifestation of a tumor. The results of this study support that proposition (Fig. 6). We extended the approach by profiling the miR transcriptome in a prototypical mouse model of multistage tumorigenesis, involving the stepwise transformation of pancreatic islet β cells into pancreatic neuroendocrine carcinomas. We identified sets of miRs—stage-specific signatures—that characterize the onset of hyperproliferation in incipient neoplasias (hyperplastic/dysplastic pancreatic islets), or of the angiogenic switch in high-grade dysplasias, or of the formation of primary pancreatic tumors, or of metastases—the latter also reflected in a minor subset of primary tumors (Fig. 1B). An additional miR signature has been ascribed to tumors treated with the potent angiogenesis inhibitor sunitinib, incorporating miRs from the angiogenic and metastatic signatures, reflecting both the loss of vascularity and the adaptation to an “evasive resistance” state of heightened invasiveness and metastasis (Paez-Ribes et al. 2009). Notable among the metastatic miR signature is down-regulation of the miR-200 family. The miR-200 family is implicated, both by reciprocal expression and by their manipulation in cultured cancer cells from the PNETs, in the characteristic suppression of E-cadherin in the invasive/metastatic tumors, achieved via up-regulation of the mesenchymal transcriptional repressor ZEB1 in the absence of sufficient miR-200 family expression. The importance of this family was concurrently established in work described in the companion study (Gibbons et al. 2009), in which overexpression of one of the miR-200 family clusters in a metastatic lung cancer cell line dramatically reduced metastasis in vivo and obviated “stem-like” characteristics of the cancer cells in culture.
Figure 6.
miR signatures of the stages in multistep tumorigenesis, ascribed to hallmark capabilities. The orchestrated stepwise progression from normality to metastasis during tumorigenesis of the pancreatic islets is marked by distinctive miR signatures of the temporally and histologically separable neoplastic stages, correlating with the acquisition of hallmark capabilities of cancer. In addition, miR profiling uncovered strong correlative evidence that a distinct class of primary tumors spawns the metastases, in contradistinction to expectations of the alternative hypothesis that enabling mutations occur in a few cells of primary tumors, endowing those cells with a metastatic capability. Gray arrowhead from Metastasis signature to Met-like tumor indicates the majority of the metastasis signature is present in met-like primary tumors.
The first step in the PNET tumorigenesis pathway that unfolds in RT2 oncomice involves a switch whereby quiescent oncogene-expressing islets become hyperproliferative, with consequent development of hyperplasias and preangiogenic dysplasias. The induction of the growth/survival factor IGF-2 is the most well-validated component of the “hyperproliferative switch” (Christofori et al. 1994). The concomitant up-regulation of miR-483, located within an IGF2 intron, adds an intriguing layer of complexity to this locus. miRs located within introns are presumably functionally linked, but how? It will be of interest then to parse out which functions ascribed to this locus are mediated through miR-483 versus IGF2. The strong up-regulation of miR-483 in half of human PNETs further argues this locus performs a conserved and functional role in neuroendocrine tumor formation.
As for other factors that mediate the hyperproliferative switch, we can now provisionally add the miR-17-92 family, clearly established in other systems to be proproliferative and regulated by E2F transcription factors and, indirectly, by c-Myc (L He et al. 2005; O'Donnell et al. 2005; Woods et al. 2007; Petrocca et al. 2008; Ventura et al. 2008). The SV40 Tag oncoprotein functionally abrogates the Rb tumor suppressor, albeit incompletely in the RT2 model (Casanovas et al. 2005a); Rb normally acts to repress the promitotic E2F3 transcription factor (Hiebert et al. 1992). Notably, however, Tag oncoprotein expression is not sufficient to switch on hyperproliferation in every islet that expresses it (Teitelman et al. 1988; Casanovas et al. 2005a), implicating stochastic cellular alterations in the hyperproliferative switch. It may well prove that up-regulation of 17-92 miRs, along with the Igf2 locus, are functionally involved in inducing and sustaining this first step in tumorigenesis.
As for the subsequent angiogenic switch, our miR profiling of preangiogenic versus angiogenic lesions (and of the constituent cell types therein) in comparison with tumors treated with a potent angiogenesis inhibitor has revealed two distinct variables: vascular-specific miRs whose levels of expression in whole lesions reflect the relative abundance of endothelial cells and pericytes of the tumor blood vessels, and miRs, in particular miR-210, that are differentially expressed in the cancer cells populating nonangiogenic versus angiogenic lesional states. The results indicate, therefore, that experimental designs involving miR profiling of multicellular tissue biopsies (both the normal and the distinctive stages of neoplastic progression) must factor in changes in the abundance of the constituent cell types. Concordant with recent studies implicating increased invasion and metastasis as an adaptive response to angiogenesis inhibition (Paez-Ribes et al. 2009), we found that the angiogenesis inhibitor sunitinib altered expression of several miRs from the metastatic signature similarly to what was observed in bona fide metastases. Thus, metastatic signature miRs may be involved in regulating the invasive/metastatic program following angiogenesis inhibition, a possibility worthy of future investigation.
The identification of a metastatic miR signature also present in a subset of primary tumors is provocative in another context. A current debate has centered on the origin of metastatic tumor cells. One possibility is that rare cancer cells evolve during malignant progression, producing metastatic clones that are genetically distinct from the majority of cancer cells in the primary tumor. The alternate possibility is that pathways of tumorigenesis toward a particular tumor type can diverge, with one route producing primary tumors having enhanced metastatic capability, and another less so (Bernards and Weinberg 2002). Our results suggest that mouse PNETs arising via the branched pathway to highly invasive IC2 tumors are the subset most prone to spawning metastases, reflected in the relatively infrequent met-like primary tumors identified in the present study. While these observations do not exclude late-stage clonal evolution to a metastatic capability in some tumor types, they indicate the metastatic phenotype can be endowed early in tumorigenesis, such that miR profiling of primary tumors may be predictive of metastasis. Not only might such knowledge have prognostic value, it could foster studies into metastatic mechanisms, and possibly guide the use of therapeutic strategies targeting prospective metastasis.
While it is not unreasonable to question the generality of results forthcoming from this prototypical mouse model of multistage tumorigenesis, there are reasons to suggest applicability to human cancer. Thus, a majority of the miRs ascribed to stage-specific signatures in this mouse model show a correspondence in a variety of human tumors (Supplemental Table 4). We infer that the miR transcriptome of solid tumors represents a compendium of the distinctive stages in tumorigenesis and acquired capabilities each step reflects, which we subdivided in the mouse by separately analyzing incipient proliferative lesions and angiogenic neoplasias as well as primary tumors. Additionally, we detected similarities in affected miRs in cognate human PNETs and metastases. There is only modest overlap in the miR signatures of mouse and human PNETs, perhaps reflecting increased histologic and genetic complexity in this heterogeneous human tumor class. Such partial overlaps between human cancer and mouse models have been observed in comparisons of mRNA transcriptomes and of genome-wide chromosomal aberrations, and have been exploited in an approach referred to as “cross-filtering,” in which it is inferred that commonalities between mice and humans reflect alterations with conserved, functional importance (Sweet-Cordero et al. 2005; Maser et al. 2007; Zender and Lowe 2008). As such, the commonly affected miRs between mouse and human PNETs (Fig. 5) may prove relevant to the biology of this disease. Irrespective, the larger commonality of the signature miRs affected in the stages of tumorigenesis in this prototypical model seem likely to have general relevance to the multistep ontogeny of other human malignances.
Materials and methods
RT2 mouse care and handling
The generation of RT2 mice has been described previously (Hanahan 1985). All mice in this study were on a C57Bl6/CR background. All animal research was performed under the auspices of animal protocols approved by the University of California at San Francisco Institutional Animal Care and Use Committee.
Tumor and pretumor lesion isolation
Pancreatic tumors and liver metastases were dissected out of 14- to 16-wk-old RT2 mice and snap-frozen in liquid nitrogen. Normal, hyperplastic, and angiogenic islets were isolated as described previously (Parangi et al. 1995) and snap-frozen in liquid nitrogen.
RNA extraction reverse transcription and Q-PCR analysis
RNA was TRIzol-purified (Invitrogen) using a homogenizer according to the manufacturer's protocol. RNA quality was assessed using the Lab-on-a-Chip 2100 Bioanalyzer (Agilent) platform. miR and standard gene expression TaqMan assays were purchased from Applied Biosystems and performed according to the manufacturer's protocol. For gene expression analysis, 0.1–1 μg of RNA was DNase I-treated (Promega) and reverse-transcribed using 250 ng of random primers and SuperScript III (Invitrogen) according to the manufacturer's protocol. For mRNA analysis, data were normalized using L19 or cyclophilin, each of which yielded similar data. For miR TaqMan, data were normalized using either mir-16 or let-7a, both of which yielded similar data. TaqMan assays for L19 and cyclophilin (control genes), insulin, and ApoB were used to determine the percentage of contamination of liver tissue in the liver metastases, which was ∼2%. TaqMan assays for CSF1R (macrophage-specific), MMP25 (neutrophil-specific), CD31 (endothelial cell-specific), Collagen 1A and PDGFRβ (pericyte-specific), Ptprt (pan-leukocyte-specific), and insulin (β-cell-specific) were used to assess the purity of sorted cell populations.
miR profiling
miRs were profiled as described in Lu et al. (2008). One microgram to 2 μg of total RNA were used in each analysis. Briefly, miRs were first bound to antisense capture probes immobilized in 96-well plates, and were ligated to a 3′ adaptor and 5′ adaptor consecutively, before reverse transcription and adaptor-mediated amplification using PCR to incorporate biotin labels. Biotinylated PCR products were hybridized with color-coded beads coupled with detection probes and were detected on a Luminex 100S machine. Median fluorescence intensity was used in data analysis.
FACS sorting of hyperplastic and angiogenic islets and tumor samples
Tumors from 14.5-wk-old RT2 mice were incubated for 13 min at 37°C in 10 mL of FACS buffer (1% BSA in Ca, Mg-free PBS) plus 0.025 g of Collagenase Type II (Worthington), 0.025 g of Collagenase Type IV (Worthington), and 0.0025 g of DNase I (Worthington) after mincing with scissors in ice-cold PBS for 5 min. Cells were strained through a 70-μm filter into a 50-mL conical tube and , after cold FACS buffer was added, were spun at 1550 rpm for 5 min at 4°C. Hyperplastic and angiogenic islets were obtained as described (Parangi et al. 1995). Collagenase-treated tumor cell preps along with hyperplastic and angiogenic islets were resuspended in 1 mL of cell dissociation buffer (Sigma) + 1.5 mL of 0.25% trypsin (GIBCO-BRL) for 3 min at 37°C with pipetting. Trypsinization was stopped by addition of 0.5 mL of FBS (JRS Scientific), cells were spun down, the supernatant was removed, and cells were resuspended in Pharmlyse buffer for 10 min at room temperature. Cells were spun down and resuspended in an Fc block for 10 min on ice. Aliquots were removed for presort samples, and samples were incubated with the following directly conjugated antibodies: CD45-PE, 7/4-FITC, GR-1-FITC, and F4/80-APC. After sorting CD45+; CD45+, 7/4, GR-1+; and CD45+, F4/80+ cells along with triple-negative cells, cells were spun down and resuspended in 1 mL of TRIzol and RNA was purified according to the manufacturer's protocol. RNase-free Glycogen (Invitrogen) was added before the RNA precipitation step.
Short-term anti-angiogenic intervention trial
RT2 mice (13.5 wk old) were given either 40 mg/kg sunitinib or vehicle alone for 7 d, three mice per treatment. Mice were injected via oral gavage. Three hours to 4 h after the last injection, mice were euthanized and three to four tumors per mouse were dissected and snap-frozen, and RNA was purified as described above.
FACS sorting of vehicle or sunitinib-treated tumors
RT2 mice (13.5 wk old) were given either 40 mg/kg sunitinib or vehicle alone for 7 d, four to eight mice per treatment. Mice were injected via oral gavage. One hour to 2 h after the last injection, mice were euthanized and tumors were dissected. Tumors were collagenase-treated and prepared as described above and incubated with the following directly conjugated antibodies: CD31-FITC, CD45-APC, and PDGFRβ-PE. Single-channel or triple-negative cells were collected and RNA was purified using TRIzol.
Profiling data analysis
Profiling data were normalized using the sum of all probes normalization method. miRs were included in a signature if there was at least a twofold difference relative to the previous stage, and if they had a P-value of <0.05 and at least one stage in which the miR had a normalized expression value of >50. For comparison, the most highly expressed miR from the study was just under 10,000 normalized fluorescence. The met-like primary and metastasis signature consists of miRs that were differentially expressed more than twofold between primary tumor and liver metastases pools (P-value, <0.05) and, in addition, were similarly differentially expressed (P-value, <0.05) between standard RT2 tumors that did not cluster with the metastases and met-like primary tumors. The metastasis-specific signature consists of miRs that were differentially expressed between metastases and tumor pools but were not differentially expressed between standard RT2 tumors and met-like primary tumors. For sunitinib signature miRs, miRs were included if the average normalized expression value was altered >1.3-fold compared with control-treated tumors and had a P-value of <0.05. The sunitinib-treated tumors were profiled in a separate profiling run. To compare miR changes between experiments, tumors from the first profiling experiment were subdivided into standard RT2 tumors if they did not cluster with the metastases and met-like primary RT2 tumors. Standard RT2 tumors were then normalized to vehicle-treated tumors from the sunitinib trial, and the expression level in normal islets was set to 100%.
Immunofluorescence of E-cadherin in RT2 tumors
Ten-micron tumor sections were incubated with an antibody against E-cadherin (Zymed, Invitrogen) followed by an FITC-conjugated secondary antibody. Slides were mounted with Vectashield with DAPI to stain nuclei and were analyzed on a fluorescent scope.
Electroporation of RT2-derived tumor cell lines
β-Tumor cells were electroporated using the Amaxa Nucleofector System (Lonza). Three-million cells were electroporated in 100 μL with 5 μL of a 20 μM solution of either a mimic control or miR-200c mimic (Dharmacon, Thermo Fisher) using kit V and protocol G-20. All treatments were performed in duplicate. Following electroporation, cells were plated in 12-well plates and were harvested 48 h later in TRIzol for RNA and RIPA buffer plus complete protease inhibitor tablets (Roche) for protein analysis.
Acknowledgments
We thank Bob Horvitz for encouraging this project and our collaboration, and for comments on the manuscript. We thank Gary Ruvkun and Michael McManus for critical reading of the manuscript and James Christensen (Pfizer, Inc.) for providing sunitinib. We thank Marina Vayner, Susan Cacacho, and Ehud Drori for excellent technical assistance. We acknowledge access to the University of California at San Francisco Diabetes Center's Microscopy and Islet Isolation Cores. Q-PCR was performed at the Genome Analysis Core, Helen Diller Family Comprehensive Cancer Center, UCSF. The research at UCSF was supported by a grant from the National Cancer Institute. D.H. is an American Cancer Society Research Professor. P.O. and A.S. acknowledge salary support from NCI Training Grant T32 CA09043 (Director J. Michael Bishop), Molecular Analysis of Tumor Viruses. M.G.C. was supported by a fellowship from the NSF.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1820109.
Supplemental material is available at http://www.genesdev.org.
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bergers G, Hanahan D. Modes of resistance to anti-angiogenic therapy. Natl Rev. 2008;8:592–603. doi: 10.1038/nrc2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards R, Weinberg RA. A progression puzzle. Nature. 2002;418:823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- Blouw B, Song H, Tihan T, Bosze J, Ferrara N, Gerber HP, Johnson RS, Bergers G. The hypoxic response of tumors is dependent on their microenvironment. Cancer Cell. 2003;4:133–146. doi: 10.1016/s1535-6108(03)00194-6. [DOI] [PubMed] [Google Scholar]
- Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Natl Rev. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Casanovas O, Hager JH, Chun MG, Hanahan D. Incomplete inhibition of the Rb tumor suppressor pathway in the context of inactivated p53 is sufficient for pancreatic islet tumorigenesis. Oncogene. 2005a;24:6597–6604. doi: 10.1038/sj.onc.1208823. [DOI] [PubMed] [Google Scholar]
- Casanovas O, Hicklin DJ, Bergers G, Hanahan D. Drug resistance by evasion of antiangiogenic targeting of VEGF signaling in late-stage pancreatic islet tumors. Cancer Cell. 2005b;8:299–309. doi: 10.1016/j.ccr.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Choi PS, Zakhary L, Choi WY, Caron S, Alvarez-Saavedra E, Miska EA, McManus M, Harfe B, Giraldez AJ, Horvitz HR, et al. Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron. 2008;57:41–55. doi: 10.1016/j.neuron.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofori G, Naik P, Hanahan D. A second signal supplied by insulin-like growth factor II in oncogene-induced tumorigenesis. Nature. 1994;369:414–418. doi: 10.1038/369414a0. [DOI] [PubMed] [Google Scholar]
- Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebos JM, Lee CR, Cruz-Munoz W, Bjarnason GA, Christensen JG, Kerbel RS. Accelerated metastasis after short-term treatment with a potent inhibitor of tumor angiogenesis. Cancer Cell. 2009;15:232–239. doi: 10.1016/j.ccr.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkman J, Watson K, Ingber D, Hanahan D. Induction of angiogenesis during the transition from hyperplasia to neoplasia. Nature. 1989;339:58–61. doi: 10.1038/339058a0. [DOI] [PubMed] [Google Scholar]
- Gibbons DL, Lin W, Creighton CJ, Rizvi ZH, Gregory PA, Goodall GJ, Thilaganathan N, Du L, Zhang Y, Pertsemlidis A, et al. Contextual cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes & Dev. 2009 doi: 10.1101/gad.1820209. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Manzano C, Holash J, Fueyo J, Xu J, Conrad CA, Aldape KD, de Groot JF, Bekele BN, Yung WK. VEGF Trap induces antiglioma effect at different stages of disease. Neuro-oncol. 2008;10:940–945. doi: 10.1215/15228517-2008-061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Heritable formation of pancreatic β-cell tumours in transgenic mice expressing recombinant insulin/simian virus 40 oncogenes. Nature. 1985;315:115–122. doi: 10.1038/315115a0. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Dissecting multistep tumorigenesis in transgenic mice. Annu Rev Genet. 1988;22:479–519. doi: 10.1146/annurev.ge.22.120188.002403. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Christofori G, Naik P, Arbeit J. Transgenic mouse models of tumour angiogenesis: The angiogenic switch, its molecular controls, and prospects for preclinical therapeutic models. Eur J Cancer. 1996;32A:2386–2393. doi: 10.1016/s0959-8049(96)00401-7. [DOI] [PubMed] [Google Scholar]
- He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiebert SW, Chellappan SP, Horowitz JM, Nevins JR. The interaction of RB with E2F coincides with an inhibition of the transcriptional activity of E2F. Genes & Dev. 1992;6:177–185. doi: 10.1101/gad.6.2.177. [DOI] [PubMed] [Google Scholar]
- Hodgson G, Hager JH, Volik S, Hariono S, Wernick M, Moore D, Nowak N, Albertson DG, Pinkel D, Collins C, et al. Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nat Genet. 2001;29:459–464. doi: 10.1038/ng771. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007;131:136–145. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- Kennell JA, Gerin I, MacDougald OA, Cadigan KM. The microRNA miR-8 is a conserved negative regulator of Wnt signaling. Proc Natl Acad Sci. 2008;105:15417–15422. doi: 10.1073/pnas.0807763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SP, Youngson N, Takada S, Seitz H, Reik W, Paulsen M, Cavaille J, Ferguson-Smith AC. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet. 2003;35:97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- Lopez T, Hanahan D. Elevated levels of IGF-1 receptor convey invasive and metastatic capability in a mouse model of pancreatic islet tumorigenesis. Cancer Cell. 2002;1:339–353. doi: 10.1016/s1535-6108(02)00055-7. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Lu J, Guo S, Ebert BL, Zhang H, Peng X, Bosco J, Pretz J, Schlanger R, Wang JY, Mak RH, et al. MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev Cell. 2008;14:843–853. doi: 10.1016/j.devcel.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser RS, Choudhury B, Campbell PJ, Feng B, Wong KK, Protopopov A, O'Neil J, Gutierrez A, Ivanova E, Perna I, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael MZ, O'Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- Norden AD, Young GS, Setayesh K, Muzikansky A, Klufas R, Ross GL, Ciampa AS, Ebbeling LG, Levy B, Drappatz J, et al. Bevacizumab for recurrent malignant gliomas: Efficacy, toxicity, and patterns of recurrence. Neurology. 2008;70:779–787. doi: 10.1212/01.wnl.0000304121.57857.38. [DOI] [PubMed] [Google Scholar]
- Nozawa H, Chiu C, Hanahan D. Infiltrating neutrophils mediate the initial angiogenic switch in a mouse model of multistage carcinogenesis. Proc Natl Acad Sci. 2006;103:12493–12498. doi: 10.1073/pnas.0601807103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Paez-Ribes M, Allen E, Hudock J, Takeda T, Okuyama H, Vinals F, Inoue M, Bergers G, Hanahan D, Casanovas O. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell. 2009;15:220–231. doi: 10.1016/j.ccr.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallante P, Visone R, Ferracin M, Ferraro A, Berlingieri MT, Troncone G, Chiappetta G, Liu CG, Santoro M, Negrini M, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13:497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- Parangi S, Dietrich W, Christofori G, Lander ES, Hanahan D. Tumor suppressor loci on mouse chromosomes 9 and 16 are lost at distinct stages of tumorigenesis in a transgenic model of islet cell carcinoma. Cancer Res. 1995;55:6071–6076. [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes & Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl AK, Wilgenbus P, Dahl U, Semb H, Christofori G. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature. 1998;392:190–193. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]
- Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, et al. E2F1-regulated microRNAs impair TGFβ-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Pietras K, Hanahan D. A multitargeted, metronomic, and maximum-tolerated dose “chemo-switch” regimen is antiangiogenic, producing objective responses and survival benefit in a mouse model of cancer. J Clin Oncol. 2005;23:939–952. doi: 10.1200/JCO.2005.07.093. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- Rubenstein JL, Kim J, Ozawa T, Zhang M, Westphal M, Deen DF, Shuman MA. Anti-VEGF antibody treatment of glioblastoma prolongs survival but results in increased vascular cooption. Neoplasia. 2000;2:306–314. doi: 10.1038/sj.neo.7900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingara J, Keiger K, Shelton J, Laosinchai-Wolf W, Powers P, Conrad R, Brown D, Labourier E. An optimized isolation and labeling platform for accurate microRNA expression profiling. RNA. 2005;11:1461–1470. doi: 10.1261/rna.2610405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- Sweet-Cordero A, Mukherjee S, Subramanian A, You H, Roix JJ, Ladd-Acosta C, Mesirov J, Golub TR, Jacks T. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, Labourier E, Hahn SA. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitelman G, Alpert S, Hanahan D. Proliferation, senescence, and neoplastic progression of β cells in hyperplasic pancreatic islets. Cell. 1988;52:97–105. doi: 10.1016/0092-8674(88)90534-x. [DOI] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Zender L, Lowe SW. Integrative oncogenomic approaches for accelerated cancer-gene discovery. Curr Opin Oncol. 2008;20:72–76. doi: 10.1097/CCO.0b013e3282f31d5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 Targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]