Abstract
AIM: To understand the complex reaction of gastric inflammation induced by Helicobacter pylori (H pylori) in a systematic manner using a protein interaction network.
METHODS: The expression of genes significantly changed on microarray during H pylori infection was scanned from the web literary database and translated into proteins. A network of protein interactions was constructed by searching the primary interactions of selected proteins. The constructed network was mathematically analyzed and its biological function was examined. In addition, the nodes on the network were checked to determine if they had any further functional importance or relation to other proteins by extending them.
RESULTS: The scale-free network showing the relationship between inflammation and carcinogenesis was constructed. Mathematical analysis showed hub and bottleneck proteins, and these proteins were mostly related to immune response. The network contained pathways and proteins related to H pylori infection, such as the JAK-STAT pathway triggered by interleukins. Activation of nuclear factor (NF)-κB, TLR4, and other proteins known to function as core proteins of immune response were also found. These immune-related proteins interacted on the network with pathways and proteins related to the cell cycle, cell maintenance and proliferation, and transcription regulators such as BRCA1, FOS, REL, and zinc finger proteins. The extension of nodes showed interactions of the immune proteins with cancer-related proteins. One extended network, the core network, a summarized form of the extended network, and cell pathway model were constructed.
CONCLUSION: Immune-related proteins activated by H pylori infection interact with proto-oncogene proteins. The hub and bottleneck proteins are potential drug targets for gastric inflammation and cancer.
Keywords: Gastric cancer, Helicobacter pylori, Inflammation, Pathway, Protein interaction network
INTRODUCTION
Helicobacter pylori (H pylori) is a gram negative bacterium which infects about 50% of the world population[1-3]. It is known to cause various gastroduodenal diseases such as chronic active gastritis in experimental animals and in humans. In human volunteers, H pylori caused gastritis and hypochlorhydria[4]. Mongolian gerbils infected by H pylori also developed symptoms such as intestinal metaplasia and adenocarcinoma[5-9]. Many scholars have demonstrated a relationship between H pylori and gastric carcinoma[3], and the World Health Organization (WHO) and the International Agency for Research on Cancer consensus group have classified H pylori as a definite biological carcinogen[10].
H pylori colonization causes a strong systemic immune response[11]. It induces the production of interleukins (ILs) (Korean Society for Medical Microbiology, 2004), tumor necrosis factor (TNF)[12,13], and proinflammatory cytokines[14]. It also causes activation of nuclear factor κB (NF-κB)[15], activator protein-1 (AP-1), c-Jun, NH2-terminal kinase, mitogen-activated protein kinase/extracellular signal-regulated kinase, and other cell proliferation and survival factors[16]. Bacterial toxins, high levels of superoxides, radicals, and singlet oxygen are known to induce carcinogenesis in gastric cells. Bacterial virulence factors such as CagA and VacA[1,17,18] induce cell hyperproliferation and the expression of oncogenes. However, the exact mechanism between H pylori and gastric carcinoma is unclear[19].
Various tools have been employed to identify the relationship between H pylori and gastric cancer, including c-DNA microarrays[4,20]. However, most of these methods did not consider the systematic interaction of biological components. As an alternative, a network construction and analysis of protein-protein interactions[21] were applied to examine the inflammatory response to H pylori infection in a systematic manner.
MATERIALS AND METHODS
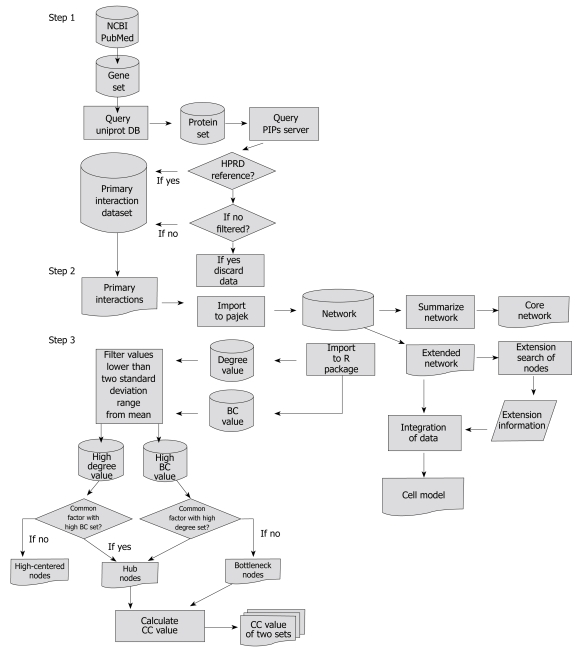
The research method used in this study mainly consisted of three steps. Step one: extraction of the genes which changed significantly during H pylori infection from the database and by querying web databases to gather protein-protein interactions. Step two: construction of a network and summarizing the constructed network. Step three: analysis and extension search of the network. A flow chart showing the data flow is described in Figure 1.
Figure 1.
Flow chart showing overall methods and data flow used in this study.
Searching genes related to H pylori infection (Step 1)
Genes related to H pylori infection were collected by searching PubMed. The expression of genes significantly changed (P < 0.05) by H pylori infection in the microarray[4,11,13,20] data was examined, and genes related to the immune response were identified and collected. A total of 39 filtered genes (Table 1) were obtained.
Table 1.
List of proteins extracted from the literary database showing significant change after H pylori infection
| Protein/gene name | Uniprot ID | HPRD reference |
| ITGB2 | P05107 | |
| LY96 | Q9Y6Y9 | X |
| TLR10 | Q9BXR5 | |
| TLR2 | O60903 | |
| TLR3 | O15455 | X |
| VCAM1 | P19320 | |
| HCK | P08631 | |
| MAPK8 | P45983 | |
| RAC2 | P15153 | |
| SOCS2 | O14508 | X |
| STAT6 | P42226 | |
| C2 | P06681 | X |
| C3 | P01024 | |
| C4A | P0C0L4 | X |
| CCL18 | P55774 | X |
| CCL19 | Q99731 | X |
| CCL3 | P10147 | X |
| CCL4 | P13236 | X |
| CRP | P02741 | |
| CXCL13 | O43927 | |
| CXCL2 | P19875 | X |
| CXCL9 | Q07325 | X |
| HLA-DMA | P28067 | X |
| HLA-DPB1 | Q30154 | X |
| HLA-DQB1 | P03992 | X |
| HLA-DRB5 | Q30154 | X |
| HSPH1 | Q92598 | |
| C11TA | P33076 | |
| PLAT | P00750 | |
| IFITM1 | P13164 | X |
| IRF4 | Q15306 | |
| MADCAM1 | Q13477 | |
| ALOX5 | P09917 | X |
| TLR5 | O60602 | X |
| CD53 | P19397 | X |
| TLR6 | Q9Y2C9 | X |
| SLAMF1 | Q13291 | |
| PTPRC | P08575 | |
| FAIM3 | O60667 | X |
| CD180 | Q99467 | |
| TLR4 | O00206 | |
| TLR1 | Q15399 | |
| CXCL3 | P19876 | X |
| CD47 | Q08722 | |
| IFNGR1 | P15260 | |
| IL10RA | Q13651 | X |
| IL18RAP | O95256 | X |
| ITGAX | P20702 | X |
| IL8 | P10145 |
Nodes with no HPRD reference were marked with x. HPRD: Human protein reference database.
Scanning protein interactions and construction of protein interaction networks (Step 2)
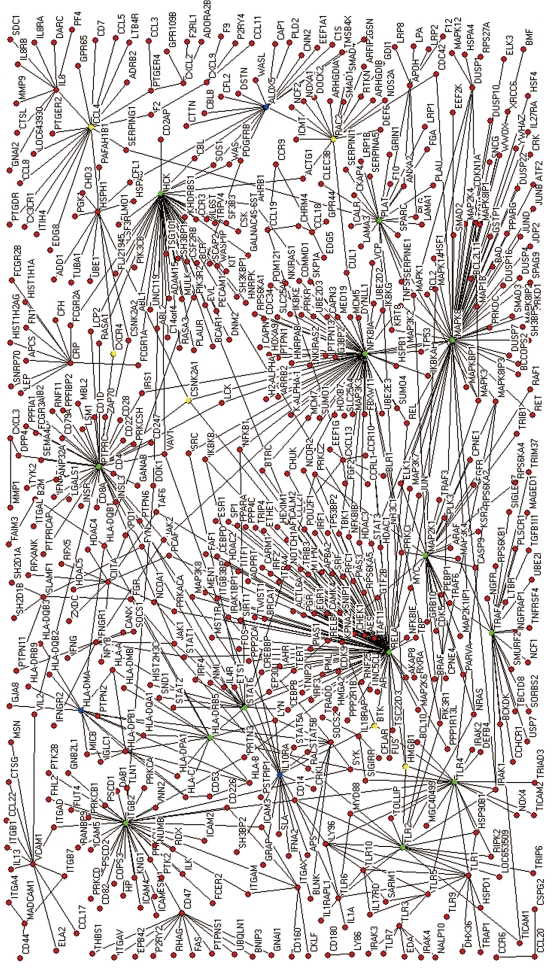
The protein interaction networks were constructed based on statistical prediction through the analysis of microarray data. Selected genes were queried to the Uniprot database to convert into proteins. The proteins were scanned by a human Protein-protein Interaction Prediction (PIPs) database (http://www.compbio.dundee.ac.uk/www-pips/). Protein links were then extracted from the Human Protein Reference Database reference (HPRD, http://www.hprd.org/index_html). Without HPRD references, any further search of the protein links was stopped. An extended network was constructed by integrating all results extracted from the PIPs server (Figure 2). Pajek (http://vlado.fmf.uni-lj.si/pub/networks/pajek/) was used for the construction of extended networks. Then, a core network showing simplified main pathways, major proteins, and subcellular location information was extracted from the extended network using Cytoscape (http://www.cytoscape.org/).
Figure 2.
The extended protein interaction network of a cell infected by H pylori. Green nodes (a large BC and degree), blue nodes (only a large degree), yellow nodes (only a large BC).
Analysis of protein interaction network (Step 3)
The protein interactions of an extended network were examined whether or not the network contained known pathways related to H pylori infection, inflammation, and carcinogenesis. The core network was not analyzed because it was just the simplified form of the extended network.
Four factors: Shortest paths, degree (connectivity), betweenness centrality (BC), and closeness centrality (CC), were adopted to analyze general mathematical properties of the extended network and to search topologically important proteins[21].
Degree, the most basic characteristic of a node, is defined as the number of links the node has with other nodes. Degree distribution is obtained by counting the number of nodes with a fixed degree value, which is variable from minimum to maximum degree, and dividing it by the total number of nodes of a network[22]. Highly concentrated nodes play a major role as a hub in a network. Degree was also used to check if an extended network was scale-free, which is frequently found in cellular networks[22,23]. The scale-free network follows a power-law degree distribution[22]. Power law is defined as:
P(x) = Cx-a
C = ec and P(x) is a probability that a selected node has exactly x links (degree value)[23]. α is the degree exponent which determines some properties of the network. Most of the networks found in nature are known to have degree exponent values between two and three[22]. In this study, cumulative distribution function, a superior method of plotting data[23], was used. The plot of log transformed probability distribution function P(x) in which x has a degree value greater than or equal to P(x), was drawn. is defined mathematically as[23]:
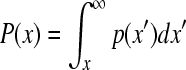 |
As the distribution follows power law,
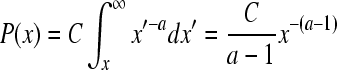 |
A cumulative plot also follows power law, but the degree exponent of the plot is one less than the original distribution[23]. The degree exponent was calculated by measuring the slope of the regression line and adding one to the exponent value. Other factors such as R square, standard error, and P-value were also computed.
BC for node k is defined as:
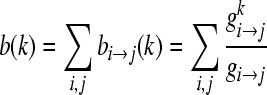 |
gi→j is the number of shortest paths from node i to j, while
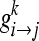 |
is the number of geodesics among gi→j that passes through node κ[21,24]. The BC value of all nodes in the network was examined to check for bottlenecks in the network.
CC is defined as the inverse of the average length of the shortest paths to/from all the other vertices in the graph[25]. It tells us the topological center of the network[25]. CC was calculated by adopting the core algorithm of the R igraph package (http://www.r-project.org/). CC values of the protein set with either large BC value or degree were measured and compared to total CC values to check topological centrality of hubs and bottlenecks in the network.
The shortest path (geodesics) is calculated by measuring the length of all the geodesics from or to the vertices in the network. The average shortest path was measured to see how many average steps were required to link two randomly selected nodes in the network.
After computing BC and the degree of all the nodes, nodes under two standard deviation ranges from the mean were filtered out and CC values of nodes larger than two standard deviation ranges from the mean were measured. As a result, nodes with a large BC value, a large degree, both a large BC and degree, and CC value were obtained. The R package was used to calculate and analyze these values.
The network was constructed by scanning primary interactions of significantly and differentially expressed genes compared to control. Thus, it may not include hidden interactions of protein nodes between the two major nodes. For example, only the primary interaction between node A and B is available by ordinary network analysis, although the two proteins are linked via node C in reality. However, by extending the network, a pathway passing through node C between A and B can be found.
RESULTS
Protein interaction networks
By integrating scanned primary interactions of previously selected nodes from the PIPs server, the extended network was constructed. A core network was then derived from the extended network.
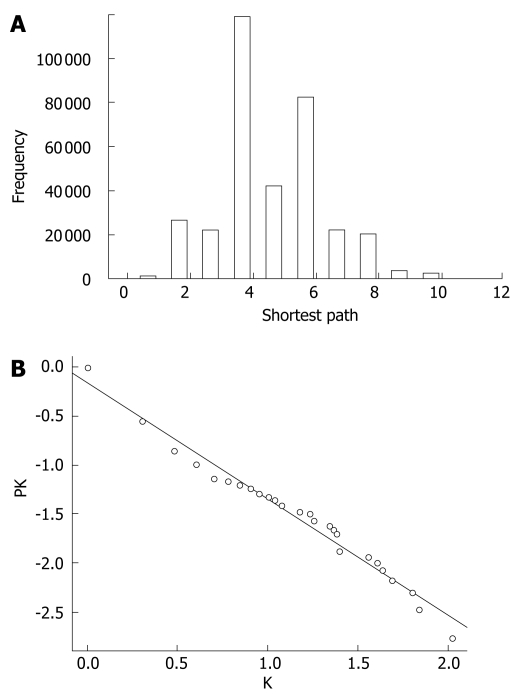
The extended network was composed of 604 nodes, connected via 808 edges (Figure 2). One giant network with 599 nodes and 805 edges, and two separate interactions were observed. Examining the shortest paths of the network showed that two randomly selected nodes on the network were connected via 4.89 links. This suggests that the nodes were very closely linked. In addition, a small world effect can be found[26]. The distribution of the shortest paths was plotted using histograms (Figure 3A). The average value (4.89) was similar to other values of human protein networks[21,26].
Figure 3.
Properties of the extended network. A: Histogram showing distribution of the shortest path. Two randomly selected nodes were connected via 4.9 links; B: Cumulative degree distribution plot of the extended network showing that degree distribution follows the power law. Line indicates the degree exponent of 1.2, which is one lower than the true degree exponent of 2.2.
The cumulative distribution plot showed clear evidence that the extended network follows scale-free distribution (Figure 3B). By measuring the slope of the regression line of the plot drawn on the basis of log transformed cumulative data, the α value of 1.1968 in the power law distribution was determined. As the degree exponent of the cumulative plot is one less than original distribution[23], the true degree exponent value should be 2.1968 (standard error = 0.04, coefficient of determination R square = 0.97, and P-value = nearly zero by the least square fit)[27]. It is known that networks with a degree exponent larger than three do not have features that scale-free networks have[22]. The degree exponent value of the extended network (2.1968) was lower than 3, which was similar to other networks following a scale-free distribution, rather than a random distribution.
Important nodes in the network
One of the properties of networks following scale-free distribution is the existence of a small number of highly connected nodes, called hubs which are more important than other less connected nodes[22,28]. The hub nodes are more critical to the survival of cells (Tables 2 and 3). The scale-free networks are prone to breakdown into fragments when nodes are attacked[29]. Other important nodes also have a large BC value. The node with a large BC functions as a bottleneck in the network, even when the node’s degree is low. Nodes with a degree or BC value larger than the mean plus two standard deviations were selected. Sixteen nodes were determined to have a large degree (Table 2) and 19 nodes had a large BC (Table 4). Twelve nodes had both a large degree and a large BC (Table 3). Six nodes: NF-κB3 (Nuclear factor κB p65 subunit), MAPK8 (Mitogen-activated protein kinase 8), NFKBIA (NF-κB inhibitor α), HCK (Hemopoietic cell kinase), PTPRC (Leukocyte common antigen CD45), and ITGB2 (Integrin β-2) were the top six nodes on both degree and BC values. STAT6 (Signal transducer and activator of transcription 6), TLRs (Toll-like receptor), TRAF4 (TNF receptor-associated factor 4), and PLAT (TPA, Tissue-type plasminogen activator) also had both a large BC and degree (Table 3). NF-κB3, NFKBIA, PTPRC, TLRs, and HLA-DRB5 are already well-known for having biological functions related to immune response[2,3,21]. STAT6 is related to the JAK-STAT pathway which sends signals from ILs directly to the nucleus[30]. MAPK8 of the MAPK signaling pathway can be found in the signaling of other inflammatory responses in asthma, and is related to cell proliferation[31]. TRAF4 is involved in tumor necrosis and TPA (PLAT) in plasminogen activation, respectively. Most of these nodes are related to immune response and signal transduction, suggesting that these nodes perform major functions against H pylori infection.
Table 2.
List of proteins with a large degree value and their CC values
| Protein | Degree | CC value |
| RELA/NF-κB3 | 105 | 0.047675522 |
| MAPK8 | 68 | 0.046693511 |
| NFKBIA | 63 | 0.047164646 |
| HCK | 49 | 0.046599691 |
| PTPRC | 43 | 0.046388184 |
| ITGB2 | 40 | 0.045643782 |
| MAP2K1 | 36 | 0.045681818 |
| PLAT | 25 | 0.045451119 |
| STAT6 | 24 | 0.046513422 |
| HLA-DMA | 24 | 0.04396646 |
| TRAF4 | 24 | 0.044062843 |
| TLR4 | 24 | 0.046527778 |
| HLA-DRB5 | 23 | 0.04573032 |
| TLR2 | 22 | 0.046083301 |
| IL10RA | 18 | 0.046206897 |
| ALOX5 | 18 | 0.044056404 |
CC: Closeness centrality.
Table 3.
List of proteins with both a large BC and degree, and their functions
| Protein name | Function |
| RELA/NF-κB3 | NF-κB is a pleiotropic transcription factor which is present in almost all cell types and is involved in many biological processes such as inflammation, immunity, differentiation, cell growth, tumorigenesis and apoptosis |
| MAPK8 | Responds to activation by environmental stress and pro-inflammatory cytokines by phosphorylating a number of transcription factors, primarily components of AP-1 such as JUN, JDP2 and ATF2 and thus regulates AP-1 transcriptional activity |
| NFKBIA | Inhibits the activity of dimeric NF-κB/REL complexes by trapping REL dimers in the cytoplasm, masking their nuclear localization signals |
| HCK | May serve as part of a signaling pathway coupling the Fc receptor to activation of the respiratory burst. May also contribute to neutrophil migration and regulate the degranulation process of neutrophils |
| PTPRC | Required for T-cell activation through the antigen receptor |
| ITGB2 | Receptor for ICAM1, ICAM2, ICAM3 and ICAM4 |
| TLR4 | Cooperates with LY96 and CD14 to mediate the innate immune response to bacterial lipopolysaccharide (LPS). Acts via MYD88, TIRAP and TRAF6, leading to NF-κB activation, cytokine secretion and the inflammatory response |
| PLAT | Converts the abundant, but inactive, zymogen plasminogen to plasmin by hydrolyzing a single Arg-Val bond in plasminogen. By controlling plasmin-mediated proteolysis, it plays an important role in tissue remodeling and degradation, in cell migration and many other physiopathological events |
| TRAF4 | Adapter protein and signal transducer that links members of the tumor necrosis factor receptor family to different signaling pathways by association with the receptor cytoplasmic domain and kinases |
| MAP2K1 | Catalyzes the concomitant phosphorylation of a threonine and a tyrosine residue in a Thr-Glu-Tyr sequence located in MAP kinases. Activates ERK1 and ERK2 MAP kinases |
| TLR2 | Cooperates with LY96 to mediate the innate immune response to bacterial lipoproteins and other microbial cell wall components |
| STAT6 | Carries out a dual function: signal transduction and activation of transcription. Involved in interleukin-4 signaling |
Table 4.
List of proteins with a large BC value and their CC values
| Protein | BC | CC |
| RELA/NF-κB3 | 62240.75685 | 0.047675522 |
| MAPK8 | 37123.82691 | 0.046693511 |
| PTPRC | 34614.23746 | 0.046388184 |
| HCK | 33484.99898 | 0.046599691 |
| NFKBIA | 28771.33132 | 0.047164646 |
| ITGB2 | 27901.91607 | 0.045643782 |
| TLR4 | 19587.75501 | 0.046527778 |
| PLAT | 19339.65216 | 0.045451119 |
| CCL4 | 18441.86414 | 0.043654528 |
| HMGB1 | 15263.85652 | 0.046466826 |
| MAP2K1 | 14047.72141 | 0.045681818 |
| STAT6 | 13376.61128 | 0.046513422 |
| CXCR4 | 11471.71166 | 0.044769471 |
| CSNK2A1 | 11279.76894 | 0.046574496 |
| HLA-DRB5 | 11088.56681 | 0.045730320 |
| BTK | 11016.20767 | 0.046621308 |
| TLR2 | 10824.15886 | 0.046083301 |
| RAC2 | 10692.38019 | 0.044872749 |
| TRAF4 | 10472.80579 | 0.044062843 |
BC: Betweenness centrality.
Not only nodes with both a large degree and BC, but also nodes with a large BC and a small degree were considered important in previous research[21], since these nodes function as bottlenecks in the network, even without the role of hubs. Six nodes: HMGB1 (High mobility group protein B), BTK (Bruton tyrosine kinase), CSNK2A1 (CSK2A1, Casein kinase II subunit alpha), RAC2 (Ras-related C3 botulinum toxin substrate 2), CCL4 (C-C motif chemokine 4), and CXCR4 (C-X-C chemokine receptor type 4) had a large BC but a low degree. Large BC nodes such as CXCR4, CCL4, BTK, CSNK2A1, and RAC2 with the exception of HMGB1 are related to immune response and signal transduction (Table 5). HMGB1 unwinds double-stranded DNA and binds preferentially to single-stranded DNA, which may be related to the gene regulation of immune response. As expected these large BC nodes were linked to important nodes, such as hubs. HMGB1 was linked to NF-κB3, TLR4, TLR2, and PLAT, which have a large BC degree (Figure 2). BTK interacted with NFKBIA, TLR4, HCK, and IL10RA. NFKBIA, TLR4, and HCK had a large BC and degree, while IL10RA had a large degree only. CSNK2A1 was linked to NF-κB3, NFKBIA, PTPRC, and HSPH1. RAC2 interacted with NFKBIA, HCK, and ALOX5. Lastly, CCL4 and CXCR4 were linked to PTPRC and PLAT. Thus, it was demonstrated that the nodes with large BC play important roles in the connection and communication of nodes including hubs.
Table 5.
List of proteins with only a large BC and their functions
| Protein name | Function |
| HMGB1 | Binds to preferentially single-stranded DNA and unwinds double-stranded DNA |
| BTK | Plays a crucial role in B-cell ontogeny |
| CSNK2A1 | Casein kinases are operationally defined by their preferential utilization of acidic proteins such as caseins as substrates |
| CXCR4 | Transduces a signal by increasing the intracellular calcium ion level |
| CCL4 | Monokine with inflammatory and chemokinetic properties |
| RAC2 | Plasma membrane-associated small GTPase which cycles between an active GTP-bound and inactive GDP-bound state. In active state binds to a variety of effector proteins to regulate cellular responses, such as secretory processes, phagocytosis of apoptotic cells and epithelial cell polarization. Seems to be involved in the regulation of NADPH oxidase |
The CC values of nodes with a large degree or BC were checked to see if these proteins were near to the topological center of the network. The larger the CC value is, the closer the node is to the center of the network[21]. NF-κB3 was closest to the topological center, and NFKBIA was the second closest in the network (Tables 2 and 4).
Biological functions of pathways and nodes in the network
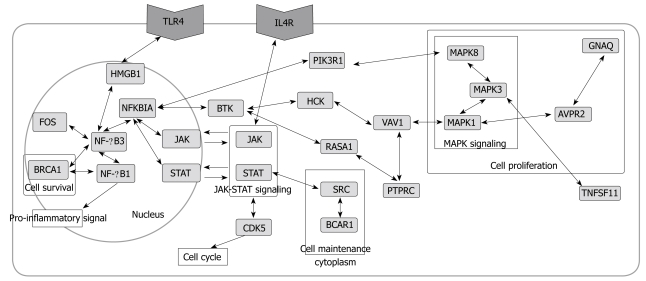
Pathways related to immune response and other biological phenomena were observed in the network (Figures 4 and 5). The network contained previously known pathways which were involved in H pylori infection and inflammation.
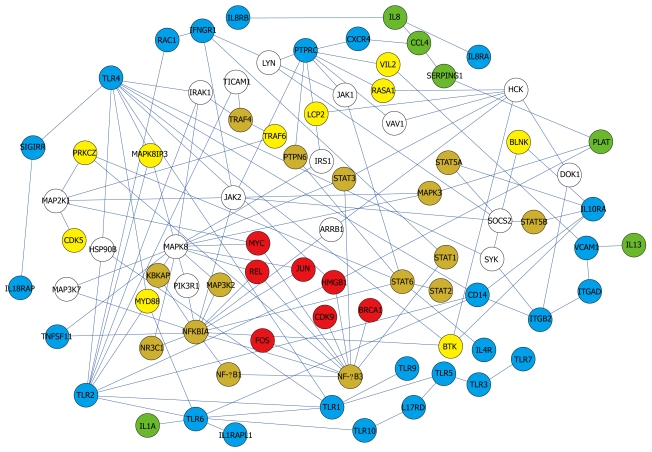
Figure 4.
Core network showing simplified interactions and major pathways. Color of the nodes indicates subcellular location. Red (nucleus), orange (nucleus and cytoplasm), yellow (cytoplasm), blue (membrane), green (extracellular), and white (unknown).
Figure 5.
Cell model showing major interactions after H pylori infection.
The network (Figures 2 and 4) showed interactions of IL 1, 4, 8, 10, 13, 17, and 18 receptors with JAKs and STATs that send signals from cell-surface receptors to the nucleus[30]. IL 8 increases significantly during H pylori infection, thus it was used as a standard to determine the pathogenicity of different H pylori strains[19]. IL 1, 10, and 18 changed significantly, which was demonstrated by microarray analysis or Western blotting data[11,13,32]. IL 4 and 13 are proinflammatory cytokines. While IL 4 induces eosinophilic inflammation and differentiation of Th2 cells, IL 13 produces immunoglobulin E (IgE)[33].
Interactions of Toll-like receptors (TLRs), also known to be immune-related, were observed. The TLR4 signaling pathway is associated with an immune response by interacting with MYD88 and IRAK1[34,35] in the network (Figure 4). They were linked to proteins in the nucleus through MAPKs.
Another pathway in the network was found among the MAPKs. Interactions among MAPK 1, 3, and 8 in the network were observed. In immune-related diseases such as asthma, the activation of MAPK due to infection has also been reported[21,36,37].
Besides full pathways, the presence of single or a few interactions having biological functions were informative. NF-κB and AP-1 are two key regulatory factors of inflammation[38-40]. NF-κB1-NF-κB3 linkage and JAK-NFKBIA-STAT linkage were found (Figure 5). The regulation of NF-κB by AP-1(JUN) and NFKBIA was also observed (Figure 4).
Although activation of TNFα[13] was not found in the network, TNFSF11 (Tumor necrosis factor ligand superfamily member 11) and TRAFs (TNF receptor associated factor), related to TNF, were found. Tumor necrosis factors induce cell proliferation by activating anti-apoptosis[16]. Cell proliferation and carcinogenesis are one of the well-known characteristics of cells infected by H pylori[19]. In addition, BRCA1 (Breast cancer type 1 susceptibility protein), FOS (c-fos, Proto-oncogene protein)[41], REL (C-Rel proto-oncogene protein), and VAV1 (Proto-oncogene vav), which are oncogenes, were found. The presence of TNF and the oncogenes in the network suggests that H pylori infection may be related to carcinogenesis.
SRC (Proto-oncogene tyrosine-protein kinase) in the network is involved in cell maintenance and communication[21]. CDK5 (Cyclin-dependent kinase 5), RASA1 (Ras GTPase-activating protein 1) and RASA3 are related to cell growth effect[30].
Not only protein nodes related to inflammation and carcinogenesis, but also proteins related to stress resistance were found. Infection of H pylori increases levels of superoxide and singlet oxygen. The stress-resistance protein, HSPH1 (Heat shock protein 105 kDa), HSPA8 (Heat shock cognate 71 kDa protein), and HSPB1 (Heat shock protein β-1) were found.
Generally, stimulation and regulation of the immune system through their receptors were found in the network. Activation of cell signaling, cell proliferation, cell survival, proto-oncogenes, and stress resistance were observed. These functions are reminiscent of the observed response of cells infected by H pylori. The virtual network analysis in this study reflects the real protein-interaction-network in the cell.
Extension search of the network
A biologically important protein can be missed, as the network is constructed by searching only the primary interactions of selected genes. To overcome this problem, further interactions of nodes regardless of their degree or BC were examined. The extension of SRC led to BCAR1 (Figure 5). Thus, the role of cell maintenance[42] was connected with that of carcinogenesis in BCAR1. ADRB2 (β-2 adrenergic receptor) extension was linked to PRKCE, PRKACA, MAPK1, and MAPK3 (Figure 2). This pathway has not previously been reported in H pylori infection, but has been found in immune-related diseases such as asthma[21]. BRCA1 was further linked to CDK2, 4, 7, and CDC2 (Cell division control protein 2 homolog) (Figure 5). CDKs are activated proceeding to the cell cycle. The extension of BRCA1 was linked to JUND (transcription factor jun D), which binds to an AP-1 site and stimulates its promoter activity. BRCA1 extension led to ZNF467 (Zinc finger protein 467), a transcription regulator which has a possible relationship with cancer (Figure 2). The extension of MAPK1 led to GNAQ (Guanine nucleotide-binding protein G(q) subunit α) via GNAS (Guanine nucleotide-binding protein G(s) subunit α isoforms short) and AVPR2 (Vasopressin V2 receptor)[21] (Figure 5). The proteins in this pathway contribute to cell proliferation, a well-known characteristic of cells infected by H pylori[19]. STAT1-CREBBP (CREB-binding protein) linkage was related to G1 arrest of a cell[21] (Figure 5).
DISCUSSION
The correlation between inflammation caused by H pylori infection and gastric cancer has been studied and supported by many researchers. It is important to understand the relationship between inflammation and the carcinogenesis mechanism. Microarray data were used to determine the global gene expression of infected cells. Microarray data showed up/down regulation of gene expression related to immune response, cell cycle, cell growth, and signal transduction, which may support the hypothesis that H pylori infection causes cancer development[4,11,13,20]. However, the data did not present a clear mechanism of carcinogenesis in a systematic manner. In this study, network analysis methods were applied to integrate previous data and construct the network model which shows the relationship between inflammation and cancer development.
The extended network showing primary interactions of significantly expressed genes (proteins in the network) was constructed. The network contained many protein nodes related to immune response and signal transduction induced by extracellular signals such as cytokines. The important nodes selected based on large BC and degree values were mostly involved in immune response and signal transduction. For example, the p65 subunit of NF-κB (NF-κB3), one of the most important regulatory factors of inflammation, was the node with the largest degree and BC value. Large BC nodes, the bottlenecks in the network, were linked to important nodes with a large degree, a large BC, or both. Like large BC and degree nodes, a large BC node was mostly related to immune response and signal transduction, with the exception of HMGB1. The constructed network also contained many pathways related to immune response and signal transduction. TLR4, JAK-STAT, and MAPK8 pathways are major pathways found in the network. Not only the pathways, but important nodes such as NF-κB and AP-1 (JUN) were also found in the network.
The network also showed many nodes related to carcinogenesis. Tumor related proteins such as BRCA1, FOS, REL, VAV1, TNFSF11, and TRAFs were found. The extension search of nodes was also linked to pathways related to cell proliferation, cell survival, and the cell cycle. The extracellular signal from ILs and TLRs goes to NF-κB, NFKBIA, and AP-1 in the nucleus via the JAK-STAT and MAPK signaling pathways. The signal then goes to proteins in the cytoplasm via the JAK-STAT pathway and BTK, promoting cell proliferation and proceeding to the cell cycle. These activated processes are one of the characteristics of cells infected by H pylori. In addition, H pylori infection is known to increase levels of radicals and oxides. Radicals and oxides are widely thought to be possible mutagens. Oxidative stress may be an additional mechanism of carcinogenesis.
Another important factor of hub and bottleneck protein nodes is that they are potential drug targets. By inhibiting the functions of hubs and bottlenecks by small molecules, the function of the network can be shut down, meaning that the inflammatory and carcinogenesis processes can be stopped, theoretically. Traditionally, antibiotics have been used to treat gastric inflammation caused by H pylori infection[14]. However, this treatment has the potential problem of antibiotic resistance in the bacteria. As a potential alternative, this study presented the hub and bottleneck nodes as a drug target of gastric inflammation, cancer, and other diseases caused by H pylori infection.
The analysis of protein network interactions showed immune response and carcinogenesis-related cell responses in a bigger picture. The extension search of nodes also demonstrated key signal transductions linking inflammatory response and carcinogenesis. This study showed how a systematic approach such as the network construction produces meaningful information. It also offered a relatively easy and simple framework to understand the complexity of cellular interactions having functional importance. Therefore, the application of this tool may be an alternative to find important genes and drug targets in other diseases and in complex biological systems.
COMMENTS
Background
The correlation between inflammation caused by Helicobacter pylori (H pylori) infection and gastric cancer has been studied and supported by many researchers.
Research frontiers
To explain the relationship between H pylori infection and cancer development, microarray analysis was used. Microarray data showed the regulatory patterns of gene expression related to immune response, cell cycle, cell growth, and signal transduction. However, the data obtained did not show the mechanism of carcinogenesis in a systematic manner.
Innovations and breakthroughs
In this study, protein network analysis, one of the bioinformatic tools, was applied to integrate previous microarray data, and a network model was constructed showing the relationship between inflammation and cancer development. The network contained many proteins related to immune response and signal transduction induced by extracellular cytokines. Some tumor-related proteins (BRCA1, FOS, REL, VAV1, TNFSF11, TRAF) were found.
Applications
This article offered a relatively easy and simple framework to understand the complexity of cellular interactions having functional importance. This tool may be used as an alternative to find important genes and drug targets in gastric inflammation and cancer and in complex biological systems.
Peer review
This study about protein interaction network in H pylori infection is potentially interesting and informative.
Acknowledgments
The authors thank Sohyun Hwang, Seung-woo Son, and Doheon Lee of Korea Advanced Institute of Science and Technology (KAIST) for their kind advice and aid.
Footnotes
Peer reviewer: Hidekazu Suzuki, Assistant Professor, Department of Internal Medicine, Keio University School of Medicine, 35 Shinanomachi, Shinjuku-ku, Tokyo 160-8582, Japan
S- Editor Cheng JX L- Editor Webster JR E- Editor Yin DH
References
- 1.Suzuki H, Hibi T, Marshall BJ. Helicobacter pylori: present status and future prospects in Japan. J Gastroenterol. 2007;42:1–15. doi: 10.1007/s00535-006-1990-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brooks GF, Janet SB, Ornston LN. Medical microbiology. 20th ed. Vol. 42. East Norwalk: Appleton & Lange; 1995. [Google Scholar]
- 3.Martinon F, Holler N, Richard C, Tschopp J. Activation of a pro-apoptotic amplification loop through inhibition of NF-kappaB-dependent survival signals by caspase-mediated inactivation of RIP. FEBS Lett. 2000;468:134–136. doi: 10.1016/s0014-5793(00)01212-6. [DOI] [PubMed] [Google Scholar]
- 4.Kitadai Y, Sasaki A, Ito M, Tanaka S, Oue N, Yasui W, Aihara M, Imagawa K, Haruma K, Chayama K. Helicobacter pylori infection influences expression of genes related to angiogenesis and invasion in human gastric carcinoma cells. Biochem Biophys Res Commun. 2003;311:809–814. doi: 10.1016/j.bbrc.2003.10.077. [DOI] [PubMed] [Google Scholar]
- 5.Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, Sibley RK. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 6.Peek RM Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2:28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 7.Sugiyama A, Maruta F, Ikeno T, Ishida K, Kawasaki S, Katsuyama T, Shimizu N, Tatematsu M. Helicobacter pylori infection enhances N-methyl-N-nitrosourea-induced stomach carcinogenesis in the Mongolian gerbil. Cancer Res. 1998;58:2067–2069. [PubMed] [Google Scholar]
- 8.Talley NJ, Zinsmeister AR, Weaver A, DiMagno EP, Carpenter HA, Perez-Perez GI, Blaser MJ. Gastric adenocarcinoma and Helicobacter pylori infection. J Natl Cancer Inst. 1991;83:1734–1739. doi: 10.1093/jnci/83.23.1734. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe T, Tada M, Nagai H, Sasaki S, Nakao M. Helicobacter pylori infection induces gastric cancer in mongolian gerbils. Gastroenterology. 1998;115:642–648. doi: 10.1016/s0016-5085(98)70143-x. [DOI] [PubMed] [Google Scholar]
- 10.Schistosomes , liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7-14 June 1994. IARC Monogr Eval Carcinog Risks Hum. 1994;61:1–241. [Google Scholar]
- 11.Wen S, Felley CP, Bouzourene H, Reimers M, Michetti P, Pan-Hammarström Q. Inflammatory gene profiles in gastric mucosa during Helicobacter pylori infection in humans. J Immunol. 2004;172:2595–2606. doi: 10.4049/jimmunol.172.4.2595. [DOI] [PubMed] [Google Scholar]
- 12.Noach LA, Bosma NB, Jansen J, Hoek FJ, van Deventer SJ, Tytgat GN. Mucosal tumor necrosis factor-alpha, interleukin-1 beta, and interleukin-8 production in patients with Helicobacter pylori infection. Scand J Gastroenterol. 1994;29:425–429. doi: 10.3109/00365529409096833. [DOI] [PubMed] [Google Scholar]
- 13.Shibata W, Hirata Y, Yoshida H, Otsuka M, Hoshida Y, Ogura K, Maeda S, Ohmae T, Yanai A, Mitsuno Y, et al. NF-kappaB and ERK-signaling pathways contribute to the gene expression induced by cag PAI-positive-Helicobacter pylori infection. World J Gastroenterol. 2005;11:6134–6143. doi: 10.3748/wjg.v11.i39.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korea Society for Medical Microbioloy. Medical Microbiology. 3rd ed. Vol. 11. Seoul: Hyunmoon; 2004. [Google Scholar]
- 15.Keates S, Hitti YS, Upton M, Kelly CP. Helicobacter pylori infection activates NF-kappa B in gastric epithelial cells. Gastroenterology. 1997;113:1099–1109. doi: 10.1053/gast.1997.v113.pm9322504. [DOI] [PubMed] [Google Scholar]
- 16.Yanai A, Hirata Y, Mitsuno Y, Maeda S, Shibata W, Akanuma M, Yoshida H, Kawabe T, Omata M. Helicobacter pylori induces antiapoptosis through buclear factor-kappaB activation. J Infect Dis. 2003;188:1741–1751. doi: 10.1086/379629. [DOI] [PubMed] [Google Scholar]
- 17.Hirata Y, Maeda S, Mitsuno Y, Tateishi K, Yanai A, Akanuma M, Yoshida H, Kawabe T, Shiratori Y, Omata M. Helicobacter pylori CagA protein activates serum response element-driven transcription independently of tyrosine phosphorylation. Gastroenterology. 2002;123:1962–1971. doi: 10.1053/gast.2002.37044. [DOI] [PubMed] [Google Scholar]
- 18.Prinz C, Schöniger M, Rad R, Becker I, Keiditsch E, Wagenpfeil S, Classen M, Rösch T, Schepp W, Gerhard M. Key importance of the Helicobacter pylori adherence factor blood group antigen binding adhesin during chronic gastric inflammation. Cancer Res. 2001;61:1903–1909. [PubMed] [Google Scholar]
- 19.Yoon YJ PhD thesis. Study on the correlation of oncogenic expression with Helicobactor pylori virulence factors based on RpoB polymorphism. Seoul National University, 2007 . [Google Scholar]
- 20.Hofman VJ, Moreilhon C, Brest PD, Lassalle S, Le Brigand K, Sicard D, Raymond J, Lamarque D, Hébuterne XA, Mari B, et al. Gene expression profiling in human gastric mucosa infected with Helicobacter pylori. Mod Pathol. 2007;20:974–989. doi: 10.1038/modpathol.3800930. [DOI] [PubMed] [Google Scholar]
- 21.Hwang S, Son SW, Kim SC, Kim YJ, Jeong H, Lee D. A protein interaction network associated with asthma. J Theor Biol. 2008;252:722–731. doi: 10.1016/j.jtbi.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 22.Barabási AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 23.Newman MEJ. Power laws, Pareto distributions and Zipf’s law. Contemporary Physics. 2005;46:323–351. [Google Scholar]
- 24.Brandes U. A faster algorithm for betweenness centrality. J Math Sociol. 2001;25:163–177. [Google Scholar]
- 25.Freeman LC. A set of measures of centrality based on betweenness. Sociometry. 1997;40:35–41. [Google Scholar]
- 26.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 27.Son SW, Jeong H. Reconstruction of a genetic network from gene perturbation data. J Kor Phys Soc. 2006;48:S208. [Google Scholar]
- 28.Jeong H, Mason SP, Barabási AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 29.Albert R, Jeong H, Barabasi AL. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- 30.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P, editors . Molecular Biology of the Cell. 4th ed. Vol. 406. New York: Garland Science; 2002. [Google Scholar]
- 31.Han J, Lee JD, Bibbs L, Ulevitch RJ. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science. 1994;265:808–811. doi: 10.1126/science.7914033. [DOI] [PubMed] [Google Scholar]
- 32.Tomita T, Jackson AM, Hida N, Hayat M, Dixon MF, Shimoyama T, Axon AT, Robinson PA, Crabtree JE. Expression of Interleukin-18, a Th1 cytokine, in human gastric mucosa is increased in Helicobacter pylori infection. J Infect Dis. 2001;183:620–627. doi: 10.1086/318541. [DOI] [PubMed] [Google Scholar]
- 33.Wills-Karp M, Luyimbazi J, Xu X, Schofield B, Neben TY, Karp CL, Donaldson DD. Interleukin-13: central mediator of allergic asthma. Science. 1998;282:2258–2261. doi: 10.1126/science.282.5397.2258. [DOI] [PubMed] [Google Scholar]
- 34.Jiang H, Harris MB, Rothman P. IL-4/IL-13 signaling beyond JAK/STAT. J Allergy Clin Immunol. 2000;105:1063–1070. doi: 10.1067/mai.2000.107604. [DOI] [PubMed] [Google Scholar]
- 35.Nakashima K, Hirota T, Obara K, Shimizu M, Jodo A, Kameda M, Doi S, Fujita K, Shirakawa T, Enomoto T, et al. An association study of asthma and related phenotypes with polymorphisms in negative regulator molecules of the TLR signaling pathway. J Hum Genet. 2006;51:284–291. doi: 10.1007/s10038-005-0358-1. [DOI] [PubMed] [Google Scholar]
- 36.Lee JC, Laydon JT, McDonnell PC, Gallagher TF, Kumar S, Green D, McNulty D, Blumenthal MJ, Heys JR, Landvatter SW. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature. 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 37.Keates S, Keates AC, Warny M, Peek RM Jr, Murray PG, Kelly CP. Differential activation of mitogen-activated protein kinases in AGS gastric epithelial cells by cag+ and cag- Helicobacter pylori. J Immunol. 1999;163:5552–5559. [PubMed] [Google Scholar]
- 38.Baeuerle PA. IkappaB-NF-kappaB structures: at the interface of inflammation control. Cell. 1998;95:729–731. doi: 10.1016/s0092-8674(00)81694-3. [DOI] [PubMed] [Google Scholar]
- 39.Barnes PJ, Karin M. Nuclear factor-kappaB: a pivotal transcription factor in chronic inflammatory diseases. N Engl J Med. 1997;336:1066–1071. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- 40.Kim T, Yoon J, Cho H, Lee WB, Kim J, Song YH, Kim SN, Yoon JH, Kim-Ha J, Kim YJ. Downregulation of lipopolysaccharide response in Drosophila by negative crosstalk between the AP1 and NF-kappaB signaling modules. Nat Immunol. 2005;6:211–218. doi: 10.1038/ni1159. [DOI] [PubMed] [Google Scholar]
- 41.Mitsuno Y, Maeda S, Yoshida H, Hirata Y, Ogura K, Akanuma M, Kawabe T, Shiratori Y, Omata M. Helicobacter pylori activates the proto-oncogene c-fos through SRE transactivation. Biochem Biophys Res Commun. 2002;291:868–874. doi: 10.1006/bbrc.2002.6530. [DOI] [PubMed] [Google Scholar]
- 42.Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]