Abstract
Vaults are ubiquitous ribonucleoprotein complexes involved in a diversity of cellular processes, including multidrug resistance, transport mechanisms and signal transmission. The vault particle shows a barrel-shaped structure organized in two identical moieties, each consisting of 39 copies of the major vault protein MVP. Earlier data indicated that vault halves can dissociate at acidic pH. The crystal structure of the vault particle solved at 8 Å resolution, together with the 2.1-Å structure of the seven N-terminal domains (R1–R7) of MVP, reveal the interactions governing vault association and provide an explanation for a reversible dissociation induced by low pH. The structural comparison with the recently published 3.5 Å model shows major discrepancies, both in the main chain tracing and in the side chain assignment of the two terminal domains R1 and R2.
Keywords: major vault protein, ribonucleoprotein particle, signal transduction, vault, X-ray crystallography
Introduction
The vault complex, with a mass of 13 MDa and overall dimensions of 40 × 40 × 70 nm, is the largest ribonucleoprotein particle found in eukaryotes (Kedersha et al, 1990; Hamill and Suprenant, 1997; Herrmann et al, 1997). In mammals, vaults contain three proteins: the 100-kDa major vault protein MVP (Kedersha and Rome, 1986), the 193-kDa vault poly(ADP-ribosyl)ating polymerase VPARP (Kickhoefer et al, 1999a) and the 240-kDa telomerase-associated protein TEP1 (Kickhoefer et al, 1999b). Additionally, at least one small and untranslated RNA is found as a constitutive component (Kickhoefer et al, 1993). Approximately 75% of the vault particle mass is due to MVP (Kedersha et al, 1991). Vault-like particles (VLPs), similar to purified endogenous vaults, are observed when rat MVP is expressed in insect cells, indicating that MVP is sufficient to direct the formation of VLPs (Stephen et al, 2001). Earlier sequence analysis of the highly conserved MVP protein indicated the presence of seven 50-residue repeats at its N-terminus, whereas a two-stranded coiled coil was predicted to be present at the C-terminal half of the protein (van Zon et al, 2002).
Despite their diverse origin, vaults are uniform in size and morphology when imaged by electron microscopy presenting a barrel-like structure with an invaginated waist and two protruding caps (Kong et al, 1999). The N-terminal region of MVP forms the particle waist and accounts for the non-covalent interface at the vault midsection (Mikyas et al, 2004), whereas the C-terminus builds the cap as well as the cap/barrel junction.
The fact that the murine MVP was found to be orthologous to the earlier described human lung resistance-related protein, known to be overexpressed in multiple chemotherapy resistance models immediately associated vaults with intrinsic drug resistance (Scheffer et al, 1995). This particle has also been implicated in the regulation of several cellular processes including transport mechanisms, signal transmission and immune responses (Berger et al, 2009). An increasing number of proteins have been described as being bound and transported by MVP. Yu et al (2002) showed that MVP binds through N-terminal repeats R3 and R4 to the C2 domain of the tumour-suppressor phosphatase PTEN in a Ca2+-dependent manner. MVP has also been proposed to act as a scaffold for the epidermal growth factor-induced MAPK pathway (Kolli et al, 2004), and interactions have also been described with the estrogen receptor (Abbondanza et al, 1998). Another function for vaults was proposed by Herlevsen et al (2007) that found that MVP knockdown disrupted the lysosomal compartment. Interestingly, an independent set of experiments demonstrated vault dissociation at low pH (Goldsmith et al, 2007; Esfandiary et al, 2008). The former authors proposed that the acidic nature of the lysosomes may serve as an excellent microenviroment with which to trigger vault dissociation. Vault lability was also observed by Poderycki et al (2006) that managed to incorporate vault-associated proteins into preformed MVP-only recombinant vaults proving that they are not rigid, impenetrable boxes, but more a fluctuating dynamic structure presenting substantial flexibility. Their capsular structure and the occasional occurrence of a mass in the inner hollow cavity (Kong et al, 1999) led to the hypothesis that they might represent transport vehicles. However, both the cellular signals responsible for vault opening and the nature of this cargo have still to be determined.
Very recently, the structure of the rat vault at 3.5 Å resolution has been published (Tanaka et al, 2009). It shows that the vault shell is organized in two identical moieties, each consisting of 39 copies of MVP. The MVP monomers are folded into 12 distinct domains: nine repeat domains, a shoulder domain, a cap-helix and a cap ring. Here, we report the high resolution structures of a recombinant MVP fragment, containing the seven N-terminal domain repeats (R1–R7), in three different crystal forms and of the intact vaults from rat liver determined at 8 Å resolution. The comparison between the structures of R1–R7 and the equivalent region in the reported model (Tanaka et al, 2009) shows fundamental discrepancies in the tracing of domains R1 and R2. The discrepancies are most probably due to difficulties in model building, using a 3.5-Å map of the whole vault. The quality of the R1–R7 data allows unequivocal tracing of domains R1 and R2 that form the rims between the two vault halves. The positioning of the 2.1-Å structure of R1–R7 into the 8-Å map of the entire vault reveals the interactions stabilizing vault association and suggests a pH-dependent mechanism for the reversible disassembly of the particle.
Results and discussion
Structure of the MVP N-terminal fragment R1–R7
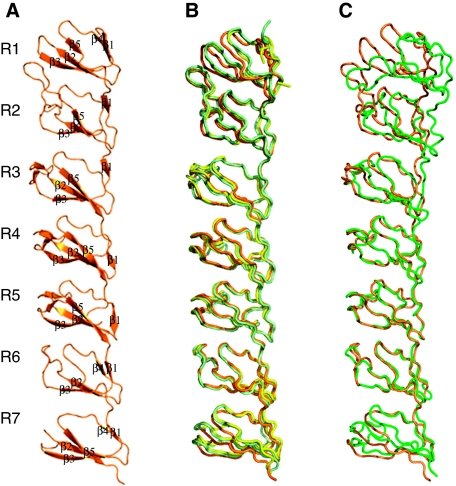
The N-terminal fragment of MVP (amino acid residues from Met1 to Asp383) containing the first seven domain repeats R1–R7 was crystallized in three different crystal forms: triclinic P1, diffracting to 2.1 Å resolution and two monoclinic crystal forms, P21A (3.0 Å) and P21B (2.5 Å) (Table I). The structure was determined by multiwavelength anomalous dispersion (MAD) of seleniomethionated protein from the monoclinic P21B crystals. The P1 and P21A structures were solved by molecular replacement, using the coordinates of the P21B crystals as a starting model. The seven MVP repeats share a similar fold, each consisting of five antiparallel β strands connected by loops (Figure 1). The main structural differences between domain repeats concentrate in the size and conformation of the loops, connecting strands. The loop between strands β2 and β3 shows the highest variability and appears partially disordered in repeat R7 in all crystal forms analysed. The central repeats R3, R4 and R5 possess an additional β-hairpin inserted in the protruding loop β2–β3 (Figure 1A).
Table 1.
R1–R7 data collection, phasing and refinement statistics
| Data collection | P1 | P21 Nat A | P21 Nat B | P21 Pk | P21 Rm |
|---|---|---|---|---|---|
| Wavelength (Å) | 0.979 | 0.979 | 0.979 | 0.979 | 0.976 |
| Resolution (Å) | 30–2.1 (2.2–2.1) | 50–3. (3.1–3.0) | 50–2.5 (2.6–2.5) | 42–2.4 (2.5–2.4) | 42–2.8 (2.9–2.8) |
| Space group | P1 | P21 | P21 | ||
| Unit cell (Å) | a=29.4 b=50.8 c=76.8 α=104.3 β=92.4 γ=99.8 | a=58.6 b=59.7 c=68.3 β=95.5 | a=36.5 b=98.7 c=140.8 β=97.2 | a=36.5 b=98.7 c=140.8 β=97.2 | a=36.5 b=98.7 c=140.8 β=97.2 |
| Total data | 26 128 | 38 501 | 96 598 | 84 955 | 59 212 |
| Unique data | 18 701 | 9260 | 34 051 | 28 695 | 24 204 |
| Mean (<I/σ>) | 14.3 (2.6) | 6.5 (1.6) | 10.1 (2.0) | 10.3 (1.1) | 5.2 (1.4) |
| Rmerge | 3.1 (12.9) | 11.8 (22.6) | 7.1 (37.2) | 4.9 (26.8) | 8.8 (38.1) |
| Completeness (%) | 99.9 (72.0) | 96.6 (95.5) | 96.9 (81.8) | 89.8 (73.6) | 99.2 (99.2) |
| Phasing power | 0.806 | 0.308/0.348 0.607/– | |||
| FOM acentric/centric | 0.249/0.155 | 0.249/0.155 | |||
| Rcullis iso/ano acentric/centric | −/− 0.879/− | 0.746/0.608 0.920/− | |||
| Refinement statistics | |||||
| Rwork (%) | 23.8 | 25.7 | 23.1 | ||
| Rfree (%) | 27.0 | 28.7 | 26.9 | ||
| Number of residues | |||||
| Protein | 363 | 371 | 751 | ||
| Solvent | 59 | 45 | |||
| RMSD | |||||
| Bond lengths (Å) | 0.006 | 0.005 | 0.005 | ||
| Bond Angles (deg) | 0.957 | 0.77 | 0.91 | ||
| Average temperature factors (Å) | 29.3 | 45.3 | 44.4 | ||
| Model quality (Ramachandran plot) | |||||
| Residues in most favoured regions (%) | 95 | 93.5 | 95 | ||
| Residues in additional allowed regions (%) | 5 | 6.5 | 5 | ||
| Values in parenthesis are for the highest resolution shell. | |||||
Figure 1.
The structure of the N-terminal fragment of MVP. (A) Ribbon representation of domain repeats R1–R7 showing the SSEs explicitly labelled. (B) Structural superimposition of the R1–R7 structures in the three different space groups P1 (orange), P21A (yellow) and P21B (light green). The Cα root mean square deviations are 0.42 and 0.45 Å for the superimposition of 220 residues, corresponding to the central R3–R6 domains, between the P1, P21A and P21B structures, respectively. (C) Structural superimposition of the R1–R7 fragments in the isolated R1–R7 structure (orange) and in the 3.5-Å structure of the complete vault (Tanaka et al, 2009) (green). Cα root mean square deviation is 0.9 Å for the superimposition of 150 Cα atoms, corresponding to the central R4–R6 domains.
Sequence analyses in earlier work (van Zon et al, 2002) predicted two possible EF hand domains in repeats R3 and R4 with the putative calcium-binding loop located at the β2–β3 hairpin mentioned above (positions 130–146 of R3 and 181–197 of R4; Supplementary Figure 1A and B). Further studies showed that MVP interactions with other proteins such as PTEN were mediated through the proposed EF hands and modulated by Ca2+ (Yu et al, 2002). The structures determined here, with all β-conformation, do not show EF hand domains, nor calcium-binding cavities. Moreover, no Ca2+ or other ions were found in any of the structures determined. These putative calcium-binding regions located within the β2–β3 hairpins contain several acidic residues (E130, E131, E135, D141, E146 in β2–β3 of R3 and E181, D184, D186, E189, E194 in β2–β3 of R4; Supplementary Figure 1C) totally exposed to the solvent. This large electronegative surface could be able to bind, either Ca2+ or the electropositive surface of the CBR3 loop of the PTEN C2 domain, or both in an alternative unknown mechanism.
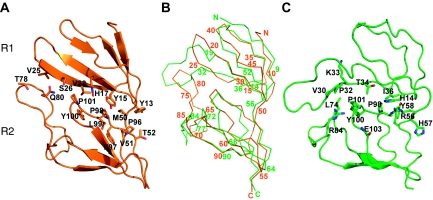
The overall structure of R1–R7 is maintained in the three crystal forms, showing only minor variations in the relative positioning of the repeats (Figure 1B). The seven modules are separated by a short linker and closely packed through a hydrophobic interface, formed by residues of strands β2 and β3 and loops β4–β5 of consecutive domains. The interdomain linker is also involved in interactions (Supplementary Table I). The contact surface is particularly extensive between domains R1 and R2 where, in addition to the interactions mentioned above, the long β2–β3 loop of R2 extends over R1 making both, hydrophobic and polar contacts with residues of the β3 strand (Figure 2; Supplementary Table I). The seven repeats are organized in an extended conformation, showing a characteristic bend between repeats R2 and R3 (Figure 1).
Figure 2.
R1–R2 interdomain interactions. (A) R1–R2 contacts found in the R1–R7 structure determined in this work. The contacting residues at the interface are shown as sticks and labelled. Strands β2 and β3 of domain R1 form polar and hydrophobic interactions with the R2 residues located in the β2–β3 loop, strand β4 and in the interdomain linker. (B) View of the α-carbon trace of the recombinant R1–R7 (orange) with every five residues numbered. The structure of the equivalent region in the 3.5-Å structure of the complete vault (Tanaka et al, 2009) is superimposed (green). The residue numbers are written when structural correspondence was observed. (C) R1–R2 contacts found in the 3.5-Å structure.
The structure of R1–R7 provides the building block for the central vault barrel
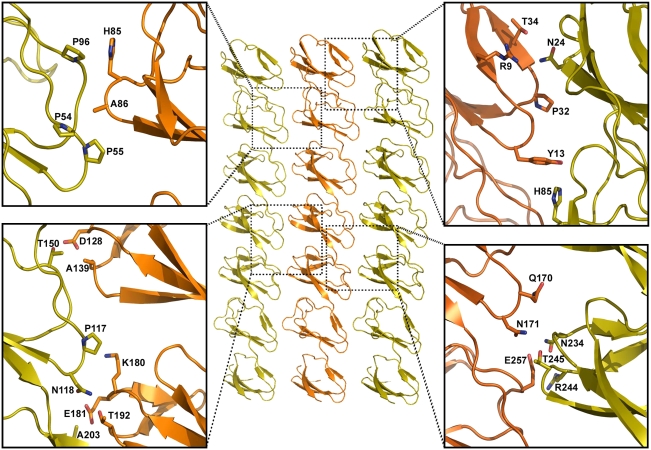
In the P1 crystals, the R1–R7 repeats are packed in a parallel arrangement, generating a flat wall of an ∼18 Å thickness (Figure 3) in good agreement with the dimensions estimated by cryo EM (Kong et al, 1999; Mikyas et al, 2004). With the exception of R7, all repeats participate in lateral packing contacts that are mediated by polar and hydrophobic interactions between 26 different residues that are located in loops β1–β2, β3–β4 and in the interdomain linker (Figure 3).
Figure 3.
R1–R7 packing contacts in the P1 crystal. Three R1–R7 fragments are represented in orange, the reference molecule and in yellow, the neighbour molecules in the crystal. The different boxes show the details of the interactions established between the different domains. Residues directly participating in hydrogen bonds or in hydrophobic contacts are shown as sticks and are explicitly labelled. The largest network of interactions involves R1 and R2 domains. R7 does not participate in lateral packing contacts.
Intact vaults, purified from rat liver were crystallized in different crystal forms, basically identical to the ones described by Querol-Audi et al (2005) and Kato et al (2008). Two of them, the monoclinic P21 and C2, were analysed by X-ray crystallography (Supplementary Table II). Data from the P21 crystals were used to solve the structure of the whole vault particle to 8 Å resolution by molecular replacement. Initial models were generated as rings of the R1–R7 structure with different rotational symmetries (see Materials and methods). The interactions observed in the triclinic crystal packing served as a restraint for building the central barrel of the vault. The bending between domains R2 and R3 could explain the invagination in the central part of the vault particle, with the R1 module located in this central part, as suggested earlier by cryo-EM and mutational studies (Mikyas et al, 2004). The positioning of the R1–R7 rings in both the C2 and P21 vault crystals indicate that only models with rotational symmetry of 39 allow intermolecular interactions between neighbour vault particles without introducing steric problems (Supplementary Figure 2), in complete agreement with the D39 symmetry suggested by Kato et al (2008).
Density averaging and solvent flattening with DM (CCP4, 1994) were applied for the P21 crystals, using the independent positioning of the two R1–R7 rings as starting phases and rotational symmetry 39 (see Materials and methods). The final averaged maps at 8 Å resolution (Supplementary Figure 3) showed unambiguous density, fitting the complete MVP protein. In particular, two additional β domains (R8 and R9) and the long helical domain, for which no information was included in the initial model were clearly defined (Supplementary Figure 3). The small changes required in the orientation of repeats R1, R2 and R7, departing from the R1–R7 model used, were also visible in the averaged maps. Furthermore, to automatically evaluate the quality of the 8-Å averaged density of the vault particle, the secondary structural elements (SSEs) were quantitatively detected and annotated with the program SSEHunter (Baker et al, 2007), rendering the same overall structure to that reported by Tanaka et al (2009) (Supplementary Figure 4).
Structural comparisons of the seven N-terminal domains in the R1–R7 structure and in the 3.5-Å particle
Overall comparisons between the R1–R7 structures determined here with the same region in the intact vault particle (Tanaka et al, 2009) showed a good superimposition of the central domains, from R3 to R6, but different changes in the relative positioning of domains R1, R2 and to a lesser extent R7, affecting the relative curvature of the fragment (Figure 1C). Structural superimpositions of the individualized domains revealed close similarities in repeats R3 to R7, as indicated by the root mean square deviations ranging from 0.6 to 1.3 Å, for the superimposition of all the residues within the repeats. However, critical differences were observed when repeats R1 and R2 from the R1–R7 structures were compared with those reported by Tanaka et al (Figure 2B). Repeat R1 shows well-defined SSEs (strands R1β1–R1β5; Figure 1A) in all three crystal forms determined. In the 3.5-Å vault structure, most of the SSEs of R1 are lost and the main chain is out of register from residue Ile10 to Gln88. The largest discrepancies concentrate in the β2–β3 loops of both, R1 and R2 domains (Figure 2B) and affect most of the R1–R2 and R1–R1 interdomain interactions, including those responsible for the contacts between the two vault halves (Figures 2 and 4). Real space map correlation, calculated with SFcheck (CCP4, 1994) for R1R2 domains in the P1 structure, show an overall value of 93.1% with minor deviations (Supplementary Figure 5). The overall density correlation for the same region in the 3.5-Å vault structure is 88.5% with the largest deviations concentrated in the β2–β3 loop of R2 (Supplementary Figure 5). The loops β2–β3 of both, R1 and R2 domains show well-defined electron density in all R1–R7 structures determined (Supplementary Figure 6).
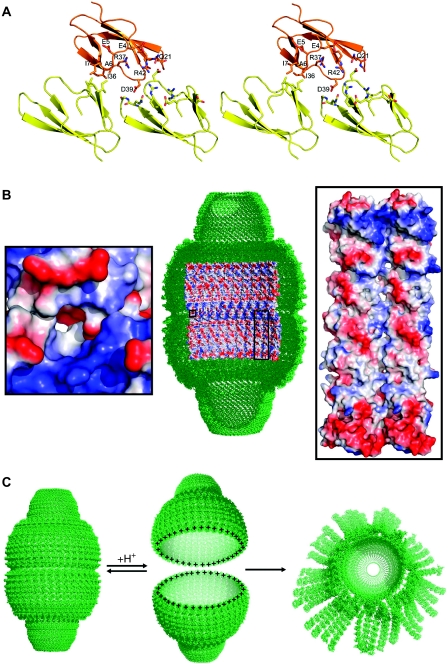
Figure 4.
Interdomain interactions at the interface between vault halves. (A) Ribbon diagram of the R1–R1 contacts at the half vault interface. The reference R1 domain (orange) contacts two consecutive R1 molecules (yellow) through the molecular two-fold axis. Interacting residues are shown in sticks and labelled. (B) Thirty-nine-fold averaged density of the vault particle at 8 Å resolution and inner view of the molecular surface of the central vault barrel. The electrostatic potential is represented in blue and red for the positive and negative charges, respectively. Nine R1–R7 protomers per half vault subunit are shown. The left side inset shows a close-up of the contacting residues at the interface between the two vault halves. The right side inset shows a close-up of the contacting interfaces between MVP protomers. Two consecutive R1–R7 molecules are shown. The amino acids involved in these lateral contacts are the same as that are shown in Figure 3. The electrostatic potential was calculated and rendered with PyMOL (DeLano, 2002), with colouring levels ranging from −66.7 to 66.7. (C) Schematic drawing, showing the mechanism of vault opening. At low pH, the acidic residues at the vault interface would become neutral, leaving a highly electropositive charge and inducing the disassembly of the vault particle by charge repulsion. At higher pH, the aspartate and glutamate residues, would present their acidic state establishing attractive electrostatic interactions between the two vault halves. The half vault moiety at the right side of the figure represents the flower-like structures described in Kedersha et al (1991).
In the 3.5-Å structure of the vault particle, the N-terminal residues (from Met1 to Glu4) were organized in an intermolecular antiparallel β sheet around the two-fold axis. An ionic interaction was also observed at the interface (Tanaka et al, 2009). The R1–R7 structures reported here show that the most N-terminal residues of MVP are highly flexible. In P21A and P21B crystals, the seven first residues point out of the domain core and participate in crystal packing interactions. In the P1 crystal form, the first four amino acids are disordered and from residue Glu5 onward the region runs parallel to the R1 core, following similar trajectory to that found in the equivalent domain of the entire particle (Figure 1).
Hypothetical mechanism for vault opening
The correct positioning of the 2.1-Å R1–R7 structure into the 8-Å map of the complete particle (this work) as well as in the 3.5-Å averaged map, calculated from the deposited structure factors (PDB id; 2ZV4) of the Tanaka structure (Tanaka et al, 2009), reveals important charge complementarity at the interface between the two vault halves (Figure 4). R1–R1 interactions involve two R1 subunits in each half vault moiety (Figure 4A and B). Side chain of residue Asp39 forms a salt bridge with Arg42 and contacts the main chain N atoms of Ala21 and Gln22. The charged amino acids Glu4, Glu5 and Arg37 and a cluster of hydrophobic residues (Ala6, Ile7 and Ile36), interacting through the two-fold axis of the particle, also determine the contact surface (Figure 4A).
Recent data obtained by combining fluorescence spectroscopy, multiangle laser light scattering and electron microscopy, provided evidences that vaults dissociate into halves at low pH (Goldsmith et al, 2007). The interactions determined in this work let us propose a reversible mechanism of dissociation of the vault particle induced by a pH change (Figure 4C). At low pH, the 234 acidic residues at the vault interface would become neutral (Grimsley et al, 2009), leaving a highly electropositive charge and inducing the disassembly of the vault particle by charge repulsion (Figure 4). At higher pH, the aspartate and glutamate residues would recover their acidic state and re-establish the electrostatic interactions, allowing the re-association between the two vault halves. Subsequently, the hydrophobic interactions would contribute to stabilize the locked conformation of the particle.
Among all the acidic residues located at the interface between the two vault halves, Asp39 seems to be directly involved in the opening (Figure 4). Crystallization of the R1–R7 fragment occurred at relatively low pH (pH=5.6) probably leading to protonation of some acidic side chains in particular environments.
Examination of the packing interactions shows that, for all the R1–R7 structures determined, Asp39 is in fact protonated, in close contact with the peptide oxygen of Gly354 from the R7 repeat of a neighbour R1–R7 subunit (Supplementary Figure 7).
Early experiments, using freeze-etch electron microscopy, in polylysine-coated mica supports, showed that vaults can change to an opened state appearing as flower-like structures, which were usually seen in pairs (Kedersha et al, 1991). On close examination, each flower appeared to be composed of eight rectangular petals surrounding a central ring and with the corner of each petal connected to the ring by a short hook (Kedersha et al, 1991). After the separation of the two vault halves, the external acidic surfaces observed in the R1–R7 region (Supplementary Figure 1C), would try to maximize their electrostatic interactions with the polylysine-coated support. About eight petals, with four or five MVP protomers each, seem to provide suited flattened interacting surfaces, while still retaining a high percentage of the lateral interactions among MVP protomers (Figure 4C). The vault cap regions, stabilized by extensive hydrophobic interactions (Tanaka et al, 2009), are expected to retain native-like conformations, which might correspond to the central ring of the flowers.
Understanding the mechanisms governing vault opening constitutes an important step towards unravelling vault functions. Moreover, exploiting recombinant vaults as nanocapsules for the delivery of biomolecules (Esfandiary et al, 2008) is a promising therapeutic application that would benefit from such understanding.
Materials and methods
Production and purification of the N-terminal fragment of MVP
The coding sequence for the N-terminus region of MVP, spanning residues 1–383, was amplified by PCR using the commercially available Mus Musculus MVP cDNA (RIKEN full-length enriched library, clone: E430002N03) as a template. This fragment was cloned into the pGemT easy vector system (Promega). pGex_R1–R7 plasmid was generated by subcloning this insert into the pGex-6P1 vector (Amersham Biosciences), an expression vector that incorporates an amino-terminal GST-tag to the expressed protein. The resulting fusion protein contains a Prescission protease (Promega) specific cleavage site. BL21 DE3 Escherichia coli cells (Stratagene) were transformed with pGex_R1–R7 and grown at 310K in LB medium supplemented with 100 μM ampycillin until an OD600 of 0.4. The expression of R1–R7 fusion protein was induced by adding 0.5 mM IPTG to the medium. After 24 h at 293K cells were harvested by centrifugation and resuspended in PBS supplemented with complete EDTA-free protease inhibitor cocktail (Roche) and lysed by sonication. The soluble fraction was applied to a GST affinity column following manufacturer's recommendations. Immobilized R1–R7 fusion protein was subjected to an overnight cleavage with Prescission protease (GE Healthcare). After elution, R1–R7 protein was purified to homogeneity by two additional chromatographic steps through MonoQ and Superdex200 columns (GE Healthcare). Selenometionyl R1–R7 was expressed in E. coli BL21 DE3 in minimal media supplemented with Selenomethionine and purified in the same way.
Crystallization, data collection and processing
R1–R7 recombinant proteins, both the native form and the seleniomethionine (SeMet) derivative, were crystallized using the hanging drop vapour diffusion method. Typically, 1 μl of protein solution (4 mg/ml) in 40 mM Tris–HCl, pH 8.5, 0.2 M NaCl was mixed with an equal volume of reservoir solution containing 18–20% PEG 5K, 0.1 M sodium citrate, pH 5.6. Triclinic crystals, with cell parameters of a=29.39 Å, b=50.79 Å, c=76.85 Å, α=104.32°, β=92.44° and γ=99.75°, and containing one R1–R7 molecule per asymmetric unit (a.u) were obtained for both native and SeMet derivative when plates were incubated at 20°C. However, two different monoclinic crystals belonging to space group P21 were obtained using the same crystallization conditions when plates were incubated at 4°C: crystal form A (P21A; a=58.6 Å, b=59.7 Å, c=68.3 Å and β=95.5°; one molecule in the a.u) and crystal form B (P21B; a=36.5 Å, b=98.8 Å, c=140.6 Å and β=97.2°; two molecules in the a.u.). X-ray data of native and seleniomethionine-R17 crystals were collected using synchrotron radiation (beamlines ID23-EH1, ESRF, France and X06SA, SLS, Switzerland).
Seven datasets were recorded at 100K from the R1–R7 crystals, in the different space groups (Table I). Diffraction images were processed using MOSFLM and SCALA (CCP4, 1994). Anisotropic scaling was then applied to the merged datasets using Xprep (Sheldrick, 2008) to compensate both the anisotropy and scaling problems derived from radiation damage and the low symmetry space group. The anisotropically scaled data were used throughout (Supplementary Figure 8).
R1–R7 structure solution and refinement
The structure was solved using the MAD technique, combining one MAD and two SAD data from three isomorphous SeMet derivatives, using the crystal form P21B (Table I). This data yielded phasing information to 2.7 Å using program SHARP (Bricogne et al, 2003). Phase improvement with both RESOLVE (Terwilliger, 2001) and ARP/WARP (CCP4, 1994) allowed an automatic tracing of ∼400 Ala residues out of the 766 total amino acids. Initial maps were improved and extended to 2.5 Å with program DM (CCP4, 1994), using solvent flattening and non-crystallographic symmetry restraints for the two molecules found in the a.u. The appropriate masks were calculated by combining the initial 400 Ala residues model with a partial molecular replacement solution, obtained with the program MOLREP (CCP4, 1994), using the earlier reported NMR structure of repeat 4 (Kozlov et al, 2006). This procedure resulted in an electron density map of sufficient quality to position accurately mostly the backbone of the two independent R1–R7 molecules in the a.u. Some particular features in the map, especially the presence of bulky densities that would correspond to highly conserved Trp residues allowed the sequence assignment of about 20% of the residues. Several cycles of manual rebuilding using COOT (Emsley and Cowtan, 2004) and refinement using both CNS (Brunger et al, 1998) and REFMAC5 (Murshudov et al, 1997) were performed until model reached an Rfactor and Rfree below 30%. This model was then used to solve the structure of the R1–R7 protein within the two other crystal forms by molecular replacement using MOLREP. Additional rounds of manual rebuilding and refinement produced the present model for the three crystal forms. A summary of the refinement statistics is given in Table I. Coordinates and structure factors have been deposited at PDB with accession codes 3GNF, 3GF5 and 3GNG for the P1, P21A and P21B, respectively.
Crystallization and X-ray analysis of the vault particle
3D crystals of native vault particles purified from rat liver were obtained as described earlier (Querol-Audi et al, 2005). Purified vault particles were subjected to extensive crystallization trials and a diversity of crystals were obtained, though only for two of them X-ray diffraction data have been collected (Supplementary Table II). The first crystals were monoclinic, space group C2, with unit cell parameters a=726.2 Å, b=391.4 Å, c=607.6 Å and β=124.1° and diffracted to a maximum of about 10 Å resolution. The second type of crystals, that in a few cases diffracted well beyond 7 Å resolution, were also monoclinic, space group P21, with unit cell parameters a=601.1 Å, b=386.6 Å, c=627.1 Å and β=108.6°. Data collection was performed at 100K, using synchrotron radiation (beamlines ID23-EH1, ESRF, France and X06SA, SLS, Switzerland). Crystals were very sensitive to radiation, being stable to the X-ray for only 10–15 exposures (0.3° per exposure). Diffraction images were processed and scaled using the programs MOSFLM and SCALA (CCP4, 1994). Large number of crystals were analysed but most of them resulted non-isomorphous. The P21 data, 50.1% complete, were obtained from five crystals and the C2 data, with 48% completeness, were obtained form four crystals (Supplementary Table II). A detailed comparison of the crystal packing modes of vaults in the two space groups is provided as Supplementary data.
Molecular replacement and density modification
Initial models corresponding to the R1–R7 region of the vaults were generated as rings of the R1–R7 structure with different rotational symmetries. Interactions between neighbour R1–R7 chains are essentially identical in the ring and in the P1 crystal, except for the small departures introduced by the curvature of the ring (Supplementary Figure 1). In the modelled rings, the average radius of the modules differs only for the R1 module, which allows the definition of the internal face of the ring as the one with the R1 module having the smallest radius. This would explain the invagination in the central part of the vault, implying also that the R1 module is located in the central part, in full agreement with what had been reported by EM and mutational studies. When the R1–7 rings were placed in both the C2 and P21 vault crystals, the models with a rotational symmetry of 38 or smaller result in unfeasible, loose packings in which no direct interactions can be established between neighbour vault particles. In turn, models with rotational symmetries of 40 or higher result in unacceptable steric clashes between neighbour vaults in the crystal (Supplementary Figure 2). A rotational symmetry of 39 allows interaction between neighbour vault particles without introducing steric problems (Supplementary Figure 2).
Density averaging and solvent flattening were applied, for the P21 crystals, using the program DM. The averaging (and corresponding solvent flattening) masks were initially defined as cylindrical shells following the external profile of the vaults reported from EM (Kong et al, 1999). Shrinkage factors for the profile (with a final value of 0.95) were used to minimize the overlaps between masks from neighbour vaults. The masks were also limited by the middle planes of the straight line joining (and perpendicular) to the longest axis of the close neighbour vaults, similarly to what is often done in virus studies. The thickness of the shells were initially set very large (even values of >100 Å in the central part of the vault were tested) to avoid any limiting assumption. The solvent averaging masks were taken, in general, as the complementary of the averaging masks, though buffer regions were also tested.
Initial phases were either taken from models of continuous density within the mask, by placing the R1–R7 rings in different orientations or by combinations of both. Cycles of averaging and phase extension, performed using for averaging only the rotational symmetry, consistently resulted in a thin shell of continuous density with details differing depending on the initial phasing and in the phase extension protocol used. Therefore, a new averaging mask was then defined adjusted to the thin shell of density. The volume inside the averaging mask represented a density of 20%, close to the one estimated using the Matthew coefficient for 2 × 39 MVP subunits.
With these new averaging and solvent flattening masks, cycles of phase extensions were preformed with the initial phases obtained using only one R1–R7 ring whose orientation around and positioning along the vault axis were systematically screened. To reduce model bias, the no-combine mode in DM was used and starting figures of merit were set to 0.3. The best result gave a final electron density map in which the second R1–R7 ring was clearly visible in the second half of the vault, even despite no model had been introduced in that region. This density required only minor changes in the available R1–R7 ring model with a rotational symmetry of 39. Then, the process was repeated using the positioning of only the second R1–R7 for initial phasing. The corresponding electron density map showed again the density corresponding to the R1–R7 ring in the first half of the vault, where now no model had been introduced. Therefore, the two R1–R7 rings have been well fitted in the vaults density derived without any model bias. The process was then repeated using as starting phases the two R1–R7 rings and either the 39 rotational symmetry or the complete point group symmetry of the vault, 39 2, where the position of the vault two-fold axis was determined from the relationships between the chains in the two R1–R7 rings.
Supplementary Material
Supplementary Data
Review Process File
Acknowledgments
This work was supported by grants BFU2005-08686-C02-01 to IF and BFU2005-02376/BMC and BIO2008-2556 to NV. JQA and AC were supported by I3P contracts from CSIC. X-ray data were collected at the protein crystallography beam lines ID23.1 at ESRF (France) and X06SA, SLS, Switzerland. Financial support was provided by the ESRF and SLS.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abbondanza C, Rossi V, Roscigno A, Gallo L, Belsito A, Piluso G, Medici N, Nigro V, Molinari AM, Moncharmont B, Puca GA (1998) Interaction of vault particles with estrogen receptor in the MCF-7 breast cancer cell. J Cell Biol 141: 1301–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Ju T, Chiu W (2007) Identification of secondary structure elements in intermediate-resolution density maps. Structure 15: 7–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger W, Steiner E, Grusch M, Elbling L, Micksche M (2009) Vaults and the major vault protein: novel roles in signal pathway regulation and immunity. Cell Mol Life Sci 66: 43–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W (2003) Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D Biol Crystallogr 59: 2023–2030 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System. California, USA: DeLano Scientific, San Carlos [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Esfandiary R, Kickhoefer VA, Rome LH, Joshi SB, Middaugh CR (2008) Structural stability of vault particles. J Pharm Sci 98: 1376–1386 [DOI] [PubMed] [Google Scholar]
- Goldsmith LE, Yu M, Rome LH, Monbouquette HG (2007) Vault nanocapsule dissociation into halves triggered at low pH. Biochemistry 46: 2865–2875 [DOI] [PubMed] [Google Scholar]
- Grimsley GR, Scholtz JM, Pace CN (2009) A summary of the measured pK values of the ionizable groups in folded proteins. Protein Sci 18: 247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill DR, Suprenant KA (1997) Characterization of the sea urchin major vault protein: a possible role for vault ribonucleoprotein particles in nucleocytoplasmic transport. Dev Biol 190: 117–128 [DOI] [PubMed] [Google Scholar]
- Herlevsen M, Oxford G, Owens CR, Conaway M, Theodorescu D (2007) Depletion of major vault protein increases doxorubicin sensitivity and nuclear accumulation and disrupts its sequestration in lysosomes. Mol Cancer Ther 6: 1804–1813 [DOI] [PubMed] [Google Scholar]
- Herrmann C, Zimmermann H, Volknandt W (1997) Analysis of a cDNA encoding the major vault protein from the electric ray Discopyge ommata. Gene 188: 85–90 [DOI] [PubMed] [Google Scholar]
- Kato K, Tanaka H, Sumizawa T, Yoshimura M, Yamashita E, Iwasaki K, Tsukihara T (2008) A vault ribonucleoprotein particle exhibiting 39-fold dihedral symmetry. Acta Crystallogr D Biol Crystallogr 64: 525–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Heuser JE, Chugani DC, Rome LH (1991) Vaults. III. Vault ribonucleoprotein particles open into flower-like structures with octagonal symmetry. J Cell Biol 112: 225–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Miquel MC, Bittner D, Rome LH (1990) Vaults. II. Ribonucleoprotein structures are highly conserved among higher and lower eukaryotes. J Cell Biol 110: 895–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Rome LH (1986) Isolation and characterization of a novel ribonucleoprotein particle: large structures contain a single species of small RNA. J Cell Biol 103: 699–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kickhoefer VA, Searles RP, Kedersha NL, Garber ME, Johnson DL, Rome LH (1993) Vault ribonucleoprotein particles from rat and bullfrog contain a related small RNA that is transcribed by RNA polymerase III. J Biol Chem 268: 7868–7873 [PubMed] [Google Scholar]
- Kickhoefer VA, Siva AC, Kedersha NL, Inman EM, Ruland C, Streuli M, Rome LH (1999a) The 193-kD vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J Cell Biol 146: 917–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kickhoefer VA, Stephen AG, Harrington L, Robinson MO, Rome LH (1999b) Vaults and telomerase share a common subunit, TEP1. J Biol Chem 274: 32712–32717 [DOI] [PubMed] [Google Scholar]
- Kolli S, Zito CI, Mossink MH, Wiemer EA, Bennett AM (2004) The major vault protein is a novel substrate for the tyrosine phosphatase SHP-2 and scaffold protein in epidermal growth factor signaling. J Biol Chem 279: 29374–29385 [DOI] [PubMed] [Google Scholar]
- Kong LB, Siva AC, Rome LH, Stewart PL (1999) Structure of the vault, a ubiquitous cellular component. Structure 7: 371–379 [DOI] [PubMed] [Google Scholar]
- Kozlov G, Vavelyuk O, Minailiuc O, Banville D, Gehring K, Ekiel I (2006) Solution structure of a two-repeat fragment of major vault protein. J Mol Biol 356: 444–452 [DOI] [PubMed] [Google Scholar]
- Mikyas Y, Makabi M, Raval-Fernandes S, Harrington L, Kickhoefer VA, Rome LH, Stewart PL (2004) Cryoelectron microscopy imaging of recombinant and tissue derived vaults: localization of the MVP N termini and VPARP. J Mol Biol 344: 91–105 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Poderycki MJ, Kickhoefer VA, Kaddis CS, Raval-Fernandes S, Johansson E, Zink JI, Loo JA, Rome LH (2006) The vault exterior shell is a dynamic structure that allows incorporation of vault-associated proteins into its interior. Biochemistry 45: 12184–12193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querol-Audi J, Perez-Luque R, Fita I, Lopez-Iglesias C, Caston JR, Carrascosa JL, Verdaguer N (2005) Preliminary analysis of two and three dimensional crystals of vault ribonucleoprotein particles. J Struct Biol 151: 111–115 [DOI] [PubMed] [Google Scholar]
- Scheffer GL, Wijngaard PL, Flens MJ, Izquierdo MA, Slovak ML, Pinedo HM, Meijer CJ, Clevers HC, Scheper RJ (1995) The drug resistance-related protein LRP is the human major vault protein. Nat Med 1: 578–582 [DOI] [PubMed] [Google Scholar]
- Sheldrick GM (2008) A short history of SHELX. Acta Crystallogr A 64: 112–122 [DOI] [PubMed] [Google Scholar]
- Stephen AG, Raval-Fernandes S, Huynh T, Torres M, Kickhoefer VA, Rome LH (2001) Assembly of vault-like particles in insect cells expressing only the major vault protein. J Biol Chem 276: 23217–23220 [DOI] [PubMed] [Google Scholar]
- Tanaka H, Kato K, Yamashita E, Sumizawa T, Zhou Y, Yao M, Iwasaki K, Yoshimura M, Tsukihara T (2009) The structure of rat liver vault at 3.5 angstrom resolution. Science 323: 384–388 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC (2001) Maximum-likelihood density modification using pattern recognition of structural motifs. Acta Crystallogr D Biol Crystallogr 57: 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zon A, Mossink MH, Schoester M, Scheffer GL, Scheper RJ, Sonneveld P, Wiemer EA (2002) Structural domains of vault proteins: a role for the coiled coil domain in vault assembly. Biochem Biophys Res Commun 291: 535–541 [DOI] [PubMed] [Google Scholar]
- Yu Z, Fotouhi-Ardakani N, Wu L, Maoui M, Wang S, Banville D, Shen SH (2002) PTEN associates with the vault particles in HeLa cells. J Biol Chem 277: 40247–40252 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Review Process File