Abstract
This paper explores the following question: what kind of statistical guarantees can be given when doing variable selection in high dimensional models? In particular, we look at the error rates and power of some multi-stage regression methods. In the first stage we fit a set of candidate models. In the second stage we select one model by cross-validation. In the third stage we use hypothesis testing to eliminate some variables. We refer to the first two stages as “screening” and the last stage as “cleaning.” We consider three screening methods: the lasso, marginal regression, and forward stepwise regression. Our method gives consistent variable selection under certain conditions.
Keywords: Lasso, Stepwise Regression, Sparsity
1. Introduction
Several methods have been developed lately for high dimensional linear regression such as the lasso (Tibshirani 1996), Lars (Efron et al. 2004) and boosting (Bühlmann 2006). There are at least two different goals when using these methods. The first is to find models with good prediction error. The second is to estimate the true “sparsity pattern,” that is, the set of covariates with nonzero regression coefficients. These goals are quite different and this paper will deal with the second goal. (Some discussion of prediction is in the appendix.) Other papers on this topic include Meinshausen and Bühlmann (2006), Candes and Tao (2007), Wainwright (2006), Zhao and Yu (2006), Zou (2006), Fan and Lv (2008), Meinshausen and Yu (2008), Tropp (2004, 2006), Donoho (2006) and Zhang and Huang (2006). In particular, the current paper builds on ideas in Meinshausen and Yu (2008) and Meinshausen (2007).
Let (X1, Y1),…,(Xn, Yn) be iid observations from the regression model
| (1) |
where ε ~ N(0, σ2), Xi = (Xi1,…, Xip)T ∈ ℝp and p = pn > n. Let X be the n × p design matrix with jth column X•j = (X1j,…, Xnj)T and let Y = (Y1,…, Yn)T. Let
be the set of covariates with nonzero regression coefficients. Without loss of generality, assume that D = {1,…, s}for some s. A variable selection procedure D̂ n maps the data into subsets of S = {1,…, p}.
The main goal of this paper is to derive a procedure D̂ n such that
| (2) |
that is, the asymptotic type I error is no more than α. Note that throughout the paper we use ⊂ to denote non-strict set-inclusion. Moreover, we want D̂ n to have nontrivial power. Meinshausen and Bühlmann (2006) control a different error measure. Their method guarantees lim supn→∞ ℙ(D̂ n ∩V ≠∅) ≤ α where V is the set of variables not connected to Y by any path in an undirected graph.
Our procedure involves three stages. In stage I we fit a suite of candidate models, each model depending on a tuning parameter λ,
In stage II we select one of those models Ŝ n using cross-validation to select λ̂ . In stage III we eliminate some variables by hypothesis testing. Schematically:
Genetic epidemiology provides a natural setting for applying screen and clean. Typically the number of subjects, n, is in the thousands, while p ranges from tens of thousands to hundereds of thousands of genetic features. The number of genes exhibiting a detectable association with a trait is extremely small. Indeed, for Type I diabetes only ten genes have exhibited a reproducible signal (Wellcome Trust 2007). Hence it is natural to assume that the true model is sparse. A common experimental design involves a 2-stage sampling of data, with stages 1 and 2 corresponding to the screening and cleaning processes, respectively.
In stage 1 of a genetic association study, n1 subjects are sampled and one or more traits such as bone mineral density are recorded. Each subject is also measured at p locations on the chromosomes. These genetic covariates usually have two forms in the population due to variability at a single nucleotide and hence are called single nucleotide polymorphisms (SNPs). The distinct forms are called alleles. Each covariate takes on a value (0, 1 or 2) indicating the number of copies of the less common allele observed. For a well designed genetic study, individual SNPs are nearly uncorrelated unless they are physically located in very close proximity. This feature makes it much easier to draw causal inferences about the relationship between SNPs and quantitative traits. It is standard in the field to infer that an association discovered between a SNP and a quantitative trait implies a causal genetic variant is physically located near the one exhibiting association. In stage 2, n2 subjects are sampled at a subset of the SNPs assessed in stage 1. SNPs measured in stage 2 are often those that achieved a test statistic that exceeded a predetermined threshold of significance in stage 1. In essence, the two stage design pairs naturally with a screen and clean procedure.
For the screen and clean procedure it is essential that Ŝ n has two properties as n → ∞
| (3) |
and
| (4) |
where |M| denotes the number of elements in a set M. Condition (3) ensures the validity of the test in stage III while condition (4) ensures that the power of the test is not too small. Without condition (3), the hypothesis test in stage III would be biased. We will see that the power goes to 1, so taking α= αn → 0 implies consistency: ℙ(D̂ n = D) → 1. For fixed α, the method also produces a confidence sandwich for D, namely,
To fit the suite of candidate models, we consider three methods. In Method 1,
where β̃j(λ) is the lasso estimator, the value of β that minimizes
In Method 2, take Ŝ n(λ) to be the set of variables chosen by forward stepwise regression after λ steps. In Method 3, marginal regression, we take
where μ̂ j is the marginal regression coefficient from regressing Y on Xj. (This is equivalent to ordering by the absolute t-statistics since we will assume that the covariates are standardized.) These three methods are very similar to basis pursuit, orthogonal matching pursuit and thresholding; see, for example, Tropp (2004, 2006) and Donoho (2006).
Notation
Let ψ = minj∈D|βj|. Define the loss of any estimator β̂ by
| (5) |
where Σ̂ n = n−1XT X. For convenience, when β̂ ≡ β̂ (λ) depends on λ we write L(λ) instead of L(β̂ (λ)). If M ⊂ S, let XM be the design matrix with columns (X•j: j ∈ M) and let denote the least squares estimator, assuming it is well-defined. Note that our use of X•j differs from standard ANOVA notation. Write Xλ instead of XM when M = Ŝ n(λ). When convenient, we extend β̂ M to length p by setting β̂ M (j) = 0 for j ∉ M. We use the norms:
If C is any square matrix, let φ(C) and Φ(C) denote the smallest and largest eigenvalues of C. Also, if k is an integer define
We will write zu for the upper quantile of a standard Normal, so that ℙ(Z > zu) = u where Z ~ N (0, 1).
Our method will involve splitting the data randomly into three groups 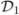 ,
, 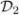 and
and 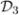 . For ease of notation, assume the total sample size is 3n and that the sample size of each group is n.
. For ease of notation, assume the total sample size is 3n and that the sample size of each group is n.
Summary of Assumptions
We will use the following assumptions throughout except in Section 8.
-
(A1) where εi ~ N (0, σ2), for i = 1, …, n.
(A2) The dimension pn of X satisfies pn → ∞ and pn ≤ c1enc2 for some c1 > 0 and 0 ≤ c2 < 1.
(A3) s ≡ |{j: βj ≠ 0}| = O(1) and ψ = min{|βj|: βj ≠ 0} > 0.
(A4) There exist positive constants C0, C1 and κ such that ℙ (lim supn→ ∞ Φn(n) ≤ C0) = 1 and ℙ(lim infn→ ∞ φn(C1 log n) ≥ κ) = 1. Also, ℙ(φn(n) > 0) = 1 for all n.
(A5) The covariates are standardized:
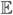 (Xij) = 0 and
. Also, there exists 0 < B < ∞ such that ℙ(|Xjk| ≤ B) = 1.
(Xij) = 0 and
. Also, there exists 0 < B < ∞ such that ℙ(|Xjk| ≤ B) = 1.
For simplicity, we include no intercepts in the regressions. The assumptions can be weakened at the expense of more complicated proofs. In particular, we can let s increase with n and ψ decrease with n. Similarly, the Normality and constant variance assumptions can be relaxed.
2. Error Control
Define the type I error rate q(D̂ n) = ℙ(D̂ n ∩ Dc ≠ ∅) and the asymptotic error rate lim supn→ ∞ q(D̂ n). We define the power π(D̂ n) = ℙ (D ⊂ D̂ n) and the average power
It is well known that controlling the error rate is difficult for at least three reasons: correlation of covariates, high dimensionality of the covariate and unfaithfulness (cancellations of correlations due to confounding). Let us briefly review these issues.
It is easy to construct examples where, q(D̂ n) ≤ α implies that π(D̂ n) ≈ α. Consider two models for random variables Z = (Y, X1, X2):
| Model 1 | Model 2 |
| X1 ~ N (0, 1) | X2 ~ N (0, 1) |
| Y = ψX1 + N (0, 1) | Y= ψX2 + N (0, 1) |
| X2= ρX1 + N (0, τ2) | X1= ρX2 + N (0, τ2). |
Under models 1 and 2, the marginal distribution of Z is P1 = N (0, Σ1) and P2 = N (0, Σ2) where
Given any ε > 0 we can choose ρ sufficiently close to 1 and τ sufficiently close to 0 such that Σ1 and Σ2 are as close as we like and hence where d is total variation distance. It follows that
Thus, if q ≤ α then the power is less than α + ε.
Dimensionality is less of an issue thanks to recent methods. Most methods, including those in this paper, allow pn to grow exponentially. But all the methods require some restrictions on the number s of nonzero βj’s. In other words, some sparsity assumption is required. In this paper we take s fixed and allow pn to grow.
False negatives can occur during screening due to cancellations of correlations. For example, the correlation between Y and X1 can be 0 even when β1 is huge. This problem is called unfaithfulness in the causality literature; see Spirtes, Glymour and Scheines (2001) and Robins, Spirtes, Scheines and Wasserman (2003). False negatives during screening can lead to false positives during the second stage.
Let μ̂ j denote the regression coefficient from regressing Y on Xj. Fix j ≤ s and note that
where ρkj = corr(Xk, Xj). If
then μj ≈ 0 no matter how large βj is. This problem can occur even when n is large and p is small.
For example, suppose that β = (10, −10, 0, 0) and that ρ(Xi, Xj) = 0 except that ρ(X1, X2) =ρ(X1, X3) = ρ(X2, X4) = 1 − ε where ε > 0 is small. Then
Marginal regression is extremely susceptible to unfaithfulness. The lasso and forward stepwise, less so. However, unobserved covariates can induce unfaithfulness in all the methods.
3. Loss and Cross-validation
Let Xλ = (X•j: j ∈ Ŝ n(λ)) denote the design matrix corresponding to the covariates in Ŝ n(λ) and let β̂ (λ) be the least squares estimator for the regression restricted to Ŝ n(λ), assuming the estimator is well defined. Hence, . More generally, β̂ M is the least squares estimator for any subset of variables M. When convenient, we extend β̂ (λ) to length p by setting β̂ j(λ) = 0 for j ∉ Ŝ n(λ).
3.1. Loss
Now we record some properties of the loss function. The first part of the following lemma is essentially Lemma 3 of Meinshausen and Yu (2008).
Lemma 3.1
Let . Then,
| (6) |
Let . Then,
| (7) |
3.2. Cross-validation
Recall that the data have been split into groups 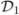 ,
, 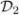 , and
, and 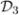 each of size n. Construct β̂ (λ) from
each of size n. Construct β̂ (λ) from 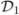 and let
and let
| (8) |
We would like L̂ (λ) to order the models the same way as the true loss L(λ) (defined after equation (5)). This requires that, asymptotically, L̂ (λ) − L(λ) ≈ δn where δn does not involve λ. The following bounds will be useful. Note that L(λ) and L̂ (λ) are both step functions that only change value when a variable enters or leaves the model.
Theorem 3.2
Suppose that maxλ∈Λn |Ŝ n(λ)| ≤ kn. Then there exists a sequence of random variables δn = OP (1) that do not depend on λ or X, such that, with probability tending to 1,
| (9) |
4. Multi-Stage Methods
The multi-stage methods use the following steps. As mentioned earlier, we randomly split the data into three parts 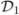 ,
, 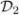 and
and 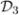 which we take to be of equal size.
which we take to be of equal size.
Stage I. Use
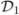 to find Ŝ n (λ) for each λ.
to find Ŝ n (λ) for each λ.Stage II. Use
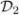 to find λ̂ by cross-validation and let Ŝ n = Ŝ n (λ̂ )
to find λ̂ by cross-validation and let Ŝ n = Ŝ n (λ̂ )Stage III. Use
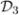 to find the least squares estimate β̂ for the model Ŝ n. Let
to find the least squares estimate β̂ for the model Ŝ n. Let
where Tj is the usual t-statistic, cn = zα/2m and m = |Ŝ n|
4.1. The Lasso
The lasso estimator (Tibshirani 1996) β̃(λ) minimizes
and let Ŝ n(λ) = {j: β̃j (λ) ≠ 0}. Recall that β̂ (λ) is the least squares estimator using the covariates in Ŝ n (λ).
Let kn = A log n where A > 0 is a positive constant.
Theorem 4.1
Assume that (A1)–(A5) hold. Let Λn = {λ: |Ŝ n(λ)| ≤ kn}. Then:
The true loss overfits: ℙ(D ⊂ Ŝ n(λ*)) → 1 where λ* = argminλ∈Λn L(λ).
Cross-validation also overfits: ℙ(D ⊂ Ŝ n(λ̂ )) →1 where λ̂ = argminλ∈ΛnL̂ (λ).
Type I error is controlled: lim supn→ ∞ ℙ(Dc ∩ D̂ n ≠ ∅) ≤ α
If we let α = αn → 0 then D̂ n is consistent for variable selection.
Theorem 4.2
Assume that (A1)–(A5) hold. Let αn → 0 and . Then, the multi-stage lasso is consistent,
| (10) |
The next result follows directly. The proof is thus omitted.
Theorem 4.3
Assume that (A1)–(A5) hold. Let α be fixed. Then (D̂ n; Ŝ n) forms a confidence sandwich:
| (11) |
Remark 4.4
This confidence sandwich is expected to be conservative in the sense that the coverage can be much larger than 1 − α.
4.2. Stepwise Regression
The version of stepwise regression we consider is as follows. Let kn = A log n for some A > 0.
Initialize: Res = Y, λ = 0, Ŷ = 0, and Ŝ n (λ) = ∅.
Let λ ← λ+ 1. Compute μ̂ j = n−1〈Xj, Res〉 for j = 1, …, p.
Let J= argmaxj |μ̂ j|. Set Ŝ n(λ) = {Ŝ n(λ −1), J}. Set Ŷ = Xλβ̂ (λ) where and let Res = Y − Ŷ .
If λ = kn stop. Otherwise, go to step 2.
For technical reasons, we assume that the final estimator xTβ̂ is truncated to be no larger than B. Note that λ is discrete and Λn = {0, 1, …, kn}.
Theorem 4.5
With Ŝ n(λ) defined as above, the statements of Theorems 4.1, 4.2 and 4.3 hold.
4.3. Marginal Regression
This is probably the oldest, simplest and most common method. It is quite popular in gene expression analysis. It is used to be regarded with some derision but has enjoyed a revival. A version appears in a recent paper by Fan and Lv (2008). Let Ŝ n(λ) = {j: |μ̂ j| ≥ λ} where μ̂ j =n−1 〈Y, X•j〉.
Let μj = 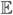 (μ̂ j) and let μ(j) denote the value of μ ordered by their absolute values:
(μ̂ j) and let μ(j) denote the value of μ ordered by their absolute values:
Theorem 4.6
Let kn → ∞ with . Let Λn = {λ: |Ŝ n(λ)| ≤ kn}. Assume that
| (12) |
Then, the statements of Theorems 4.1, 4.2 and 4.3 hold.
The assumption (12) limits the degree of unfaithfulness (small partial correlations induced by cancellation of parameters). Large values of kn weaken assumption (12) thus making the method more robust to unfaithfulness, but at the expense of lower power. Fan and Lv (2008) make similar assumptions. They assume that there is a C > 0 such that |μj| ≥ C|βj| for all j which rules out unfaithfulness. However, they do not explicitly related the values of μj for j ∈ D to the values outside D as we have done. On the other hand, they assume that Z = Σ−1/2 X has a spherically symmetric distribution. Under this assumption and their faithfulness assumption, they deduce that the μj’s outside D cannot strongly dominate the μj’s within D. We prefer to simply make this an explicit assumption without placing distributional assumptions on X. At any rate, any method that uses marginal regressions as a starting point must make some sort of faithfulness assumptions to succeed.
4.4. Modifications
Let us now discuss a few modifications of the basic method. First, consider splitting the data only into two groups 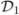 and
and 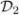 . Then do these steps:
. Then do these steps:
Stage I. Find Ŝ n(λ) for λ ∈ Λn where |Ŝ n(λ)| ≤ kn for each λ ∈ Λn using
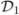 .
.Stage II. Find λ̂ by cross-validation and let Ŝ n = Ŝ n(λ̂ ) using
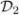 .
.Stage III. Find the least squares estimate β̂ Ŝ n using
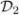 . Let D̂ n = {j ∈ Ŝ n: |Tj| > cn} where Tj is the usual t-statistic.
. Let D̂ n = {j ∈ Ŝ n: |Tj| > cn} where Tj is the usual t-statistic.
Theorem 4.7
Choosing
| (13) |
controls asymptotic type I error.
The critical value in (13) is hopelessly large and it does not appear it can be substantially reduced. We present this mainly to show the value of the extra data-splitting step. It is tempting to use the same critical value as in the tri-split case, namely, cn = zα/2m where m = |Ŝ n| but we suspect this will not work in general. However, it may work under extra conditions.
5. Application
As an example we illustrate an analysis based on part of the Osteoporotic Fractures in Men Study (MrOS, Orwoll et al. 2005). A sample of 860 men were measured at a large number of genes and outcome measures. We consider only 296 SNPs which span 30 candidate genes for bone mineral density. An aim of the study was to identify genes associated with bone mineral density that could help in understanding the genetic basis of osteoporosis in men. Initial analyses of this subset of the data revealed no SNPs with a clear pattern of association with the phenotype; however, three SNPs, numbered (67, 277, 289) exhibited some association in the screening of the data. To further explore the effacacy of the lasso screen and clean procedure we modified the phenotype to enhance this weak signal and then reanalyzed the data to see if we could detect this planted signal.
We were interested in testing for main effects and pairwise interactions in these data; however, including all interactions results in a model with 43,660 additional terms, which is not practical for this sample size. As a compromise we selected 2 SNPs per gene to model potential interaction effects. This resulted in a model with a total of 2066 potential coefficients, including 296 main effects and 1770 interaction terms. With this model our initial screen detected 10 terms, including the three enhanced signals, 2 other main effects and 5 interactions. After cleaning, the final model detected the 3 enhanced signals, and no other terms.
6. Simulations
To further explore the screen and clean procedures, we conducted simulation experiments with four models. For each model where the measurement errors, εi and , are iid Normal(0, 1) and the covariates Xij’s are Normal(0, 1) (except for model D). Models differ in how Yi is linked to Xi and the dependence structure of the Xi’s. Models A, B and C explore scenarios with moderate and large p, while Model D focuses on confounding and unfaithfullness.
Null model: β = (0,…,0) and the Xij’s are iid.
Triangle model: βj = δ(10 − j), j = 1,…, 10, βj = 0, j > 10 and Xij’s are iid.
Correlated Triangle model: as B, but with for j > 1, and ρ = 0.5.
Unfaithful model: Yi = β1Xi1 + β2Xi2 + εi, for β1 = − β2 = 10, where the Xij’s are iid for j = {1, 5, 6, 7, 8, 9, 10}, but , and , for τ = 0.01 and ρ = 0.95.
We used a maximum model size of kn = n1/2 which technically goes beyond the theory but works well in practice. Prior to analysis the covariates are scaled so that each has mean 0 and variance 1. The tests were initially performed using a third of the data for each of the three stages of the procedure (Table 1, top half, 3 splits). For models A, B and C each approach has Type I error less than ρ, except the stepwise procedure which has trouble with model C when n = p = 100. We also calculated the false positive rate and found it to be very low (about 10−4 when p = 100 and 10−5 when p = 1000) indicating that even when a Type I error occurs, only a very small number of terms are included erroneously. The lasso screening procedure exhibited a slight power advantage over the stepwise procedure. Both methods dominated the marginal approach. The Markov dependence structure in model C clearly challenged the marginal approach. For Model D none of the approaches controlled the Type I error.
Table 1.
Size and Power of Screen and Clean Procedures using Lasso, Stepwise and Marginal regression for the screening step. For all procedures α = 0.05. For p = 100, δ = 0.5 and for p = 1000, δ = 1.5. Reported power is πav. The top 8 rows of simulations were conducted using three stages as described in section 4, with a third of the data used for each stage. The bottom 8 rows of simulations were conducted splitting the data in half, using the first portion with leave-one-out cross validation for stages 1 and 2 and the second portion for cleaning.
| Splits | n | p | model | Lasso | Size Step | Marg | Lasso | Power Step | Marg |
|---|---|---|---|---|---|---|---|---|---|
| 2 | 100 | 100 | A | 0.005 | 0.001 | 0.004 | 0.00 | 0.00 | 0.00 |
| 2 | 100 | 100 | B | 0.01 | 0.02 | 0.03 | 0.62 | 0.62 | 0.31 |
| 2 | 100 | 100 | C | 0.001 | 0.01 | 0.01 | 0.77 | 0.57 | 0.21 |
| 2 | 100 | 10 | D | 0.291 | 0.283 | 0.143 | 0.08 | 0.08 | 0.04 |
|
| |||||||||
| 2 | 100 | 1000 | A | 0.001 | 0.002 | 0.010 | 0.00 | 0.00 | 0.00 |
| 2 | 100 | 1000 | B | 0.002 | 0.020 | 0.010 | 0.17 | 0.09 | 0.11 |
| 2 | 100 | 1000 | C | 0.02 | 0.14 | 0.01 | 0.27 | 0.15 | 0.11 |
| 2 | 1000 | 10 | D | 0.291 | 0.283 | 0.143 | 0.08 | 0.08 | 0.04 |
|
| |||||||||
| 3 | 100 | 100 | A | 0.040 | 0.050 | 0.030 | 0.00 | 0.00 | 0.00 |
| 3 | 100 | 100 | B | 0.02 | 0.01 | 0.02 | 0.91 | 0.90 | 0.56 |
| 3 | 100 | 100 | C | 0.03 | 0.04 | 0.03 | 0.91 | 0.88 | 0.41 |
| 3 | 100 | 10 | D | 0.382 | 0.343 | 0.183 | 0.16 | 0.18 | 0.09 |
|
| |||||||||
| 3 | 100 | 1000 | A | 0.035 | 0.045 | 0.040 | 0.00 | 0.00 | 0.00 |
| 3 | 100 | 1000 | B | 0.045 | 0.020 | 0.035 | 0.57 | 0.66 | 0.29 |
| 3 | 100 | 1000 | C | 0.06 | 0.070 | 0.020 | 0.74 | 0.65 | 0.19 |
| 3 | 1000 | 10 | D | 0.481 | 0.486 | 0.187 | 0.17 | 0.17 | 0.13 |
To determine the sensitivity of the approach to using distinct data for each stage of the analysis, simulations were conducted screening on the first half of the data and cleaning on the second half (2 splits). The tuning parameter was selected using leave-one-out cross validation (Table 1, bottom half). As expected this approach lead to a dramatic increase in the power of all the procedures. More surprising is the fact that the Type I error was near α or below for models A, B and C. Clearly this approach has advantages over data splitting and merits further investigation.
A natural competitor to screen and clean procedure is a two-stage adaptive lasso (Zou, 2006). In our implementation we split the data and used one half for each stage of the analysis. At stage one, leave-one-out cross validation lasso screens the data. In stage two, the adaptive lasso, with weights wj = |β̂ j|−1, cleans the data. The tuning parameter for the lasso was again chosen using leave-one-out cross validation. Table 2 provides the size, power and false positive rate (FPR) for this procedure. Naturally, the adaptive lasso does not control the size of the test, but the FPR is small. The power of the test is greater than we found for our lasso screen and clean procedure, but this extra power comes at the cost of a much higher Type I error rate.
Table 2.
Size, Power and False Positive Rate (FPR) of Two-stage Adaptive Lasso Procedure
| n | p | model | Size | Power | FPR |
|---|---|---|---|---|---|
| 100 | 100 | A | 0.93 | 0 | 0.032 |
| 100 | 100 | B | 0.84 | 0.97 | 0.034 |
| 100 | 100 | C | 0.81 | 0.96 | 0.031 |
| 100 | 10 | D | 0.67 | 0.21 | 0.114 |
| 100 | 1000 | A | 0.96 | 0 | 0.004 |
| 100 | 1000 | B | 0.89 | 0.65 | 0.004 |
| 100 | 1000 | C | 0.76 | 0.77 | 0.002 |
| 1000 | 10 | D | 0.73 | 0.24 | 0.013 |
7. Proofs
Recall that if A is a square matrix then φ(A) and Φ(A) denote the smallest and largest eigenvalues of A. Throughout the proofs we make use of the following fact. If v is a vector and A is a square matrix then
| (14) |
We use the following standard tail bound: if Z ~ N(0, 1) then ℙ(|Z| > t) ≤ t−1e−t2/2. We will also use the following results about the lasso from Meinshausen and Yu (2008). Their results are stated and proved for fixed X but, under the conditions (A1)–(A5), it is easy to see that their conditions hold with probability tending to one and so their results hold for random X as well.
Theorem 7.1 (Meinshausen and Yu, 2008)
Let β̃(λ) be the lasso estimator.
-
The squared error satisfies:
(15) where m = |Ŝ n(λ)| and c > 0 is a constant.
-
The size of Ŝ n(λ) satisfies
(16) where .
Proof of Lemma 3.1
Let D ⊂ M and . Then
where . Conditional on X, where . Let . By Hoeffding’s inequality, (A2) and (A5), ℙ(En) → 1 where . So
But and (6) follows.
Now we lower bound L(β̂ M). Let M be such that D ⊄ M. Let A = {j: β̂ (j) ≠ 0} ∪ D. Then |A| ≤ m + s. Therefore, with probability tending to 1,
Proof of Theorem 3.2
Let Ỹ denote the responses, and X̃ the design matrix, for the second half of the data. Then Ỹ = X̃β + ε̃. Now
and
where δn = ||ε̃||2/n, and and Σ̃n = n−1 X̃T X̃. By Hoeffding’s inequality
for some c > 0 and so
Choose εn = 4/(cn1−c2). It follows that
Note that
Hence, with probability tending to 1,
for all λ ∈ Λn, where
and
. Now
since ||β̂ (λ)||2 = OP (kn/φ(kn)). Thus, ||β̂ (λ) − β||1 ≤ C(kn + s) with probability tending to 1, for some C > 0. Also, |μi(λ)| ≤ B||β̂ (λ) − β||1 ≤ BC(kn + s) with probability tending to 1. Let W ~ N (0, 1). Conditional on 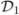 ,
,
so .
Proof of Theorem 4.1
(1) Let , M = Ŝ n(λn) and m = |M |. Then, ℙ(m ≤ kn) → 1 due to (16). Hence, ℙ(λn ∈ Λn) → 1. From (15),
Hence, . So, for each j ∈ D,
and hence ℙ(minj∈D|β̃j(λn)| > 0) → 1. Therefore, Γn = {λ ∈ Λn: D ⊂ Ŝ n(λ)} is nonempty. By Lemma 3.1,
| (17) |
On the other hand, from Lemma 3.1,
| (18) |
Now, nφn(kn)/(kn log pn) → ∞ and so, (17) and (18) imply that
Thus, if λ* denotes the minimizer of L(λ) over Λn, we conclude that ℙ(λ* ∈ Γn) → 1 and hence, ℙ(D ⊂Ŝ n(λ*)) → 1.
(2) This follows from part (1) and Theorem 3.2.
(3) Let A = Ŝ n ∩ Dc. We want to show that
Now,
Conditional on (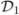 ,
, 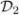 ), β̂ A is Normally distributed with mean 0 and variance matrix
when D ⊂ Ŝ n. Recall that
), β̂ A is Normally distributed with mean 0 and variance matrix
when D ⊂ Ŝ n. Recall that
where M = Ŝ n, and ej= (0, …, 0, 1, 0, …, 0)T where the 1 is in the jth coordinate. When D ⊂ Ŝ n, each Tj, for j ∈ A, has a t-distribution with n − m degrees of freedom where m = |Ŝ n|. Also, cn/tα/2m → 1 where tu denotes the upper tail critical value for the t-distribution. Hence,
where an = o(1), since |A| ≤ m. It follows that
Proof of Theorem 4.2
From Theorem 4.1, ℙ(D̂ n ∩ Dc ≠ ∅) ≤ αn and so ℙ(D̂ n ∩ Dc ≠ ∅) → 0. Hence, ℙ(D̂ n ⊂ D) → 1. It remains to be shown that
| (19) |
The test statistic for testing βj = 0 when Ŝ n = M is
For simplicity in the proof, let us take σ̂ = σ, the extension to unknown σ being straightforward. Let j ∈ D, ℳ = {M: |M| ≤ kn, D ⊂ M}. Then,
Conditional on 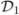 ∪
∪ 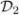 , for each M ∈ ℳ, Tj(M) = (βj/sj) + Z where Z ~ N (0, 1). Without loss of generality assume that βj > 0. Hence,
, for each M ∈ ℳ, Tj(M) = (βj/sj) + Z where Z ~ N (0, 1). Without loss of generality assume that βj > 0. Hence,
Fix a small ε > 0. Note that . It follows that, for all large n, . So,
The number of models in ℳ is
where we used the inequality
So,
by (A2). We have thus shown that ℙ(j ∉ D̂ n) → 0 for each j ∈ D. Since |D| is finite, it follows that ℙ(j ∉ D̂ n for some j ∈ D) → 0 and hence (19).
Proof of Theorem 4.5
A simple modification of Theorem 3.1 of Barron, Cohen, Dahmen and DeVore (2008) shows that
(The modification is needed because Barron, Cohen, Dahmen and DeVore (2008) require Y to be bounded while we have assumed that Y is Normal. By a truncation argument, we can still derive the bound on L(kn).) So
Hence, for any ε > 0, with probability tending to 1, ||β̂ (kn) − β||2 < ε so that |β̂ j| > ψ/2 > 0 for all j ∈ D. Thus, ℙ(D ⊂ Ŝ n(kn)) → 1. The remainder of the proof of part 1 is the same as in Theorem 4.1. Part 2 follows from the previous result together with Theorem 3.2. The proof of Part 3 is the same as for Theorem 4.1.
Proof of Theorem 4.6
Note that . Hence, . So, for any δ > 0,
By (12), conclude that D ⊂ Ŝ n(λ) when λ = μ̂ (kn). The remainder of the proof is the same as the proof of Theorem 4.5.
Proof of Theorem 4.7
Let A = Ŝ n ∩ Dc. We want to show that
For fixed A, β̂ A is Normal with mean 0 but this is not true for random A. Instead we need to bound Tj. Recall that
where M = Ŝn,
and ej = (0, …, 0, 1, 0, …, 0)T where the 1 is in the jth coordinate. The probabilities that follow are conditional on 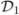 but this is supressed for notational convenience. First, write
but this is supressed for notational convenience. First, write
When D ⊂ Ŝ n,
where , and βŜ n (j) = 0 for j ∈A. Now, so that
for j ∈ Ŝ n. Therefore,
Let γ = n−1XTε. Then,
It follows that
since κ > 0. So,
Note that γj ~ N (0, σ2/n) and hence
There exists εn → 0 such that ℙ(Bn) → 1 where Bn = {(1 − εn) ≤ σ̂ /σ ≤ (1 + ε)}. So,
8. Discussion
The multi-stage method presented in this paper successfully controls type I error while giving reasonable power. The lasso and stepwise have similar performance. Although theoretical results assume independent data for each of the three stages, simulations suggest that leave-one-out cross-validation leads to valid Type I error rates and greater power. Screening the data in one phase of the experiment and cleaning in a followup phase leads to an efficient experimental design. Certainly this approach deserves further theoretical investigation. In particular, the question of optimality is an open question.
The literature on high dimensional variable selection is growing quickly. The most important deficiency in much of this work, including this paper, is the assumption that the model Y = XTβ + ε is correct. In reality, the model is at best an approximation. It is possible to study linear procedures when the linear model is not assumed to hold as in Greenshtein and Ritov (2004). We discuss this point in the appendix. Nevertheless, it seems useful to study the problem under the assumption of linearity to gain insight into these methods. Future work should be directed at exploring the robustness of the results when the model is wrong.
Other possible extensions include: dropping the Normality of the errors, permitting non-constant variance, investigating the optimal sample sizes for each stage, and considering other screening methods besides cross-validation.
Finally let us note that the example involving unfaithfulness, that is, cancellations of parameters to make the marginal correlation much different than the regression coefficient, pose a challenge for all the methods and deserve more attention even in cases of small p.
Acknowledgments
The authors are grateful for the use of a portion of the sample from the Osteo-porotic Fractures in Men (MrOS) Study to illustrate their methodology. MrOs is supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), the National Institute on Aging (NIA), and the National Cancer Institute (NCI) through grants U01 AR45580, U01 AR45614, U01 AR45632, U01 AR45647, U01 AR45654, U01 AR45583, U01 AG18197, and M01 RR000334. Genetic analyses in MrOS were supported by R01-AR051124. This work was supported by NIH grant MH057881. We also thank two referees and an AE for helpful suggestions.
Appendix
Prediction
Realistically, there is little reason to believe that the linear model is correct. Even if we drop the assumption that the linear model is correct, sparse methods like the lasso can still have good properties as shown in Greenshtein and Ritov (2004). In particular, they showed that the lasso satisfies a risk consistency property. In this appendix we show that this property continues to hold if λ is chosen by cross-validation.
The lasso estimator is the minimizer of . This is equivalent to minimizing subject to ||β||1 ≤ Ω, for some Ω. (More precisely, the set of estimators as λ varies is the same as the set of estimators as Ω varies.) We use this second version throughout this section.
The predictive risk of a linear predictor ℓ(x) = xTβ is R(β) = 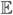 (Y − ℓ(x))2 where (X, Y) denotes a new observation. Let γ = γ(β) = (−1, β1, …, βp)T and let Γ =
(Y − ℓ(x))2 where (X, Y) denotes a new observation. Let γ = γ(β) = (−1, β1, …, βp)T and let Γ = 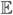 (ZZT) where Z = (Y, X1, …, Xp). Then we can write R(β) = γTΓγ. The lasso estimator can now be written as β̂ (Ωn) = argminβ∈B(Ωn) R̂ (β) where R̂ (β) = γTΓ̂ γ and
.
(ZZT) where Z = (Y, X1, …, Xp). Then we can write R(β) = γTΓγ. The lasso estimator can now be written as β̂ (Ωn) = argminβ∈B(Ωn) R̂ (β) where R̂ (β) = γTΓ̂ γ and
.
Define
where
Thus, ℓ*(x) = xT β* is the best linear predictor in the set B(Ωn). The best linear predictor is well defined even though 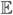 (Y | X) is no longer assumed to be linear. Greenshtein and Ritov (2004) call an estimator β̂ n persistent, or predictive risk consistent, if
(Y | X) is no longer assumed to be linear. Greenshtein and Ritov (2004) call an estimator β̂ n persistent, or predictive risk consistent, if
as n → ∞.
The assumptions we make in this section are:
(B1) pn ≤ enξ for some 0 ≤ ξ < 1 and
-
(B2) The elements of Γ̂ satisfy an exponential inequality:
for some c3, c4 > 0 and
(B3) There exists B0 < ∞ such that, for all n, maxj;k
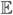 (|ZjZk|) ≤ B0.
(|ZjZk|) ≤ B0.
Condition (A2) can easily be deduced from more primitive assumptions as in Greenshtein and Ritov (2004) but for simplicity we take (A2) as an assumption. Let us review one of the results in Greenshtein and Ritov (2004). For the moment, replace (A1) with the assumption that pn ≤ nb for some b. Under these conditions, it follows that
Hence,
The latter term is oP (1) as long as Ωn = o((n/log n)1/4). Thus we have:
Theorem 8.1 (Greenshtein and Ritov 2004)
If Ωn = o((n/log n)1/4) then the lasso estimator is persistent.
For future reference, let us state a slightly different version of their result that we will need. We omit the proof.
Theorem 8.2
Let γ > 0 be such that ξ + γ < 1. Let Ωn = O(n(1−ξ−γ)/4). Then, under (B1) and (B2),
| (20) |
for some c > 0.
The estimator β̂ (Ωn) lies on the boundary of the ball B(Ωn) and is very sensitive to the exact choice of Ωn. A potential improvement—and something that reflects actual practice—is to compute the set of lasso estimators β̂ (ℓ) for 0 ≤ ℓ ≤ Ωn and then select from that set based on cross validation. We now confirm that the resulting estimator preserves persistence. As before we split the data into 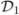 and
and 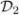 . Construct the lasso estimators {β̂ (ℓ): 0 ≤ ℓ ≤ Ωn}. Choose ℓ̂ by cross validation using
. Construct the lasso estimators {β̂ (ℓ): 0 ≤ ℓ ≤ Ωn}. Choose ℓ̂ by cross validation using 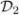 . Let β̂ = β̂ (ℓ̂ ).
. Let β̂ = β̂ (ℓ̂ ).
Theorem 8.3
Let γ > 0 be such that ξ + γ < 1. Under (A1), (A2) and (A3), if Ωn = O(n(1−ξ−γ)/4). then the cross validated lasso estimator β̂ is persistent. Moreover,
| (21) |
Proof
Let β*(ℓ) = argminβ∈B(ℓ)R(β). Define h(ℓ) = R(β*(ℓ)), g(ℓ) = R(β̂ (ℓ)) and c(ℓ) = L̂ (β̂ (ℓ)). Note that, for any vector b, we can write R(b) = τ2 + bTΣb −2bT ρ where ρ = (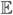 (Y X1), …,
(Y X1), …, 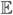 (Y Xp))T.
(Y Xp))T.
Clearly, h is monotone nonincreasing on [0, Ωn]. We claim that |h(ℓ + δ) − h(ℓ)| ≤ cΩnδ where c depends only on Γ. To see this, let u = β*(ℓ), v = β*(ℓ + δ) and a = ℓ β*(ℓ + δ)/(ℓ + δ) so that a ∈ B(ℓ). Then,
where C = maxj,k |Γj,k| = O(1).
Next we claim that g(ℓ) is Lipschitz on [0, Ωn] with probability tending to 1. Let β̂ (ℓ) = argminβ∈B̂ (ℓ)R̂ (β) denote the lasso estimator and set û = β̂ (ℓ) and v̂ = β̂ (ℓ + δ). Let εn = n−γ/4. From (20), the following chain of equations hold except on a set of exponentially small probability:
A similar argument can be applied in the other direction. Conclude that
except on a set of small probability.
Now let A = {0, δ, 2δ, …, mδ} where m is the smallest integer such that mδ ≥ Ωn. Thus, m ~ Ωn/δn. Choose δ = δn = n−3(1−ξ−γ)/8. Then Ωnδn → 0 and Ωn/δn ≤ n3(1−ξ−γ)/4. Using the same argument as in the proof of Theorem 3.2,
where σn = oP (1). Then,
and persistence follows. To show the second result, let β̃ = argmin0≤ℓ≤Ωn g(ℓ) and β̄ = argminℓ∈A g(ℓ). Then,
and the claim follows.
References
- Barron A, Cohen A, Dahmen W, DeVore R. Approximation and learning by greedy algorithms. The Annals of Statistics. 2008;36:64–94. [Google Scholar]
- Bühlmann P. Boosting for high-dimensional linear models. The Annals of Statistics. 2006;34:559–583. [Google Scholar]
- Candes E, Tao T. The Dantzig selector: statistical estimation when p is much larger than n. The Annals of Statistics. 2007;35:2313–2351. [Google Scholar]
- Donoho D. For Most Large Underdetermined Systems of Linear Equations, the minimal l1-norm near-solution approximates the sparsest near-solution. Communications on Pure and Applied Mathematics. 2006;59:797–829. [Google Scholar]
- Efron B, Hastie T, Johnstone I, Tibshirani R. Least angle regression. The Annals of Statistics. 2004;32:407–499. [Google Scholar]
- Fan J, Lv J. Sure independence screening for ultra-high dimensional feature space. To appear: Journal of the Royal Statistical Association, Series B. 2008 doi: 10.1111/j.1467-9868.2008.00674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenshtein E, Ritov Y. Persistence in high-dimensional linear predictor selection and the virtue of overparametrization. Bernoulli. 2004;10:971–988. [Google Scholar]
- Hoeffding W. Probability inequalities for sums of bounded random variables. Journal of the American Statistical Association. 1963;58:13–30. [Google Scholar]
- Meinshausen N. Relaxed Lasso. Computational Statistics and Data Analysis. 2007;52:374–393. [Google Scholar]
- Meinshausen N, Bühlmann P. High dimensional graphs and variable selection with the lasso. The Annals of Statistics. 2006;34:1436–1462. [Google Scholar]
- Meinshausen N, Yu B. Lasso-type recovery of sparse representations of high-dimensional data. To appear: The Annals of Statistics 2008 [Google Scholar]
- Orwoll E, Blank JB, Barrett-Connor E, Cauley J, Cummings S, Ensrud K, Lewis C, Cawthon PM, Marcus R, Marshall LM, McGowan J, Phipps K, Sherman S, Stefanick ML, Stone K. Design and baseline characteristics of the osteoporotic fractures in men (MrOS) study–a large observational study of the determinants of fracture in older men. Contemp Clin Trials. 2005;26:569–585. doi: 10.1016/j.cct.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Robins J, Scheines R, Spirtes P, Wasserman L. Uniform consistency in causal inference. Biometrika. 2003;90:491–515. [Google Scholar]
- Spirtes P, Glymour C, Scheines R. Causation, Prediction, and Search. MIT Press; 2001. [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society Series B. 1996;58:267–288. [Google Scholar]
- Tropp JA. Greed is good: algorithmic results for sparse approximation. IEEE Transactions on Information Theory. 2004;50:2231–2242. [Google Scholar]
- Tropp JA. Just relax: convex programming methods for identifying sparse signals in noise. IEEE Transactions on Information Theory. 2006;52:1030–1051. [Google Scholar]
- Wainwright M. Sharp thresholds for high-dimensional and noisy recovery of sparsity.arxiv.org/math.ST/0605740 2006 [Google Scholar]
- Wellcome Trust. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Yu B. On model selection consistency of lasso. Journal of Machine learning research. 2006;7:2541–2563. [Google Scholar]
- Zhang CH, Huang J. Model selection consistency of the lasso in high-dimensional linear regression. To appear: The Annals of Statisstics 2006 [Google Scholar]
- Zou H. The adaptive lasso and its oracle properties. Journal of the American Statistical Association. 2006;101:1418–1429. [Google Scholar]


