Abstract
Heart formation is a complex morphogenetic process, and perturbations in cardiac morphogenesis lead to congenital heart disease. NKX2-5 is a key causative gene associated with cardiac birth defects, presumably because of its essential roles during the early steps of cardiogenesis. Previous studies in model organisms implicate NKX2-5 homologs in numerous processes, including cardiac progenitor specification, progenitor proliferation, and chamber morphogenesis. By inhibiting function of the zebrafish NKX2-5 homologs, nkx2.5 and nkx2.7, we show that nkx genes are essential to establish the original dimensions of the linear heart tube. The nkx-deficient heart tube fails to elongate normally: its ventricular portion is atypically short and wide, and its atrial portion is disorganized and sprawling. This atrial phenotype is associated with a surplus of atrial cardiomyocytes, whereas ventricular cell number is normal at this stage. However, ventricular cell number is decreased in nkx-deficient embryos later in development, when cardiac chambers are emerging. Thus, we conclude that nkx genes regulate heart tube extension and exert differential effects on ventricular and atrial cell number. Our data suggest that morphogenetic errors could originate during early stages of heart tube assembly in patients with NKX2-5 mutations.
Keywords: nkx2.5, nkx2.7, atrium, ventricle, zebrafish, heart development, heart tube assembly, chamber morphogenesis
INTRODUCTION
Heart formation is an intricate morphogenetic process that is necessary to engineer a dynamic and anatomically complex organ. Congenital heart disease (CHD) results when this process is disrupted during embryogenesis. Cardiac defects occur in more than 1–2% of live births and in 10% of spontaneously aborted fetuses (Hoffman, 1995; Hoffman and Kaplan, 2002). Increasing evidence for the genetic basis of CHD has been assembled through the study of recurrence risk in families, and mutations in several genes are now known to cause specific cardiac defects (Bruneau, 2008). The homeodomain transcription factor NKX2-5 is identified as one of the key causative genes associated with CHD, accounting for 1–4% of specific malformations (Benson et al., 1999; Elliott et al., 2003; McElhinney et al., 2003; Schott et al., 1998). Mutations in NKX2-5 have been recognized in patients with atrial and ventricular septal defects, outflow tract abnormalities (double outlet right ventricle and tetralogy of Fallot), hypoplastic left heart syndrome, and atrioventricular conduction block (Benson et al., 1999; Elliott et al., 2003; McElhinney et al., 2003; Schott et al., 1998). In order to understand how NKX2-5 mutations lead to this vast array of malformations, it is essential to explore the variety of roles played by this transcription factor during cardiogenesis.
Several studies in model organisms indicate that NKX2-5 homologs play important roles in cardiac progenitor specification. Cardiac precursor cells in fly, fish, frog, chick, and mouse embryos express NKX2-5 homologs (Azpiazu and Frasch, 1993; Bodmer, 1993; Evans et al., 1995; Komuro and Izumo, 1993; Lee et al., 1996; Lints et al., 1993; Newman et al., 2000; Schultheiss et al., 1995; Tanaka et al., 2000; Tonissen et al., 1994). Mutation of tinman in Drosophila leads to an absence of cardiac and visceral mesoderm (Azpiazu and Frasch, 1993; Bodmer, 1993). In Xenopus, expression of dominant negative XNkx2-3 and XNkx2-5 eliminates heart formation (Grow and Krieg, 1998). The potent effect of NKX2-5 and its homologs on cardiac specification is likely related to their roles in the transcriptional regulation of essential myocardial genes (Prall et al., 2002).
Work in mouse has highlighted additional roles for Nkx2-5 in promoting cardiac morphogenesis and maturation. Nkx2-5−/− embryos form a primitive heart tube; therefore, initial analysis of these mutants did not emphasize a role of Nkx2-5 during cardiac progenitor specification (Lyons et al., 1995; Tanaka et al., 1999). However, this tube fails to loop normally and exhibits impaired trabeculation and endocardial cushion formation (Lyons et al., 1995; Tanaka et al., 1999). A primitive heart tube also forms in Nkx2-5−/−;Nkx2-6−/− double mutant embryos, although these embryos display more severe morphological defects than those observed in Nkx2-5−/− mutants, including complete failure of atrial and ventricular septation (Tanaka et al., 2001). Nkx2-5+/− mice and mice carrying a hypomorphic Nkx2-5 allele exhibit less dramatic morphogenetic defects, resembling the variety of phenotypes associated with NKX2-5 mutations in patients (Biben et al., 2000; Prall et al., 2007; Stanley et al., 2002). Additional studies in Xenopus further illuminate morphogenetic functions for NKX2-5 homologs: reduction of XNkx2-10 leads to the formation of a small ventricle with impaired trabeculation (Allen et al., 2006), and expression of versions of XNkx2-5 carrying CHD-associated mutations results in problems with atrial septation, atrioventricular valve formation, and cardiac conduction (Bartlett et al., 2007). Together, these data identify a myriad of important roles for NKX2-5-related genes during cardiac morphogenesis.
In recent work, Prall and colleagues have begun to draw connections between the effects of Nkx2-5 on specification and morphogenesis (Prall et al., 2007). They revisited the phenotype of Nkx2-5−/− mutant mice in light of the recent identification of two discrete heart fields in amniotes: the first heart field (FHF), giving rise to the primary heart tube, and the second heart field (SHF), giving rise to the inflow and outflow poles of the heart (Buckingham et al., 2005). With awareness of the sequential differentiation of the FHF and SHF, it was possible to uncover temporally dynamic roles of Nkx2-5. At early stages, expression of cardiac progenitor markers is expanded in Nkx2-5−/− mutants, indicating excessive specification (Prall et al., 2007). In contrast, proliferation of SHF precursors is substantially impaired later in development (Prall et al., 2007). Interestingly, these differential effects of Nkx2-5 both appear to be mediated through the loss of feedback repression of Bmp2 signaling (Prall et al., 2007). The aberrations in progenitor pool specification and proliferation in Nkx2-5−/−mutants could underlie their morphogenetic defects; for example, it is likely that the inhibition of SHF proliferation is responsible for the narrowed, shortened, and malformed Nkx2-5−/− outflow tract. However, it remains unclear when the earliest morphogenetic defects arise in Nkx2-5-deficient embryos and whether these problems reflect an abnormal number of cardiomyocytes.
To address these open questions, we chose to inhibit the function of NKX2-5 homologs in zebrafish. While the zebrafish genome contains three NKX2-5-related genes -- nkx2.3, nkx2.5, and nkx2.7 -- only nkx2.5 and nkx2.7 are expressed in the precardiac mesoderm and in the developing myocardium (Chen and Fishman, 1996; Lee et al., 1996). We find that injection of anti-nkx2.5 and anti-nkx2.7 morpholinos causes striking defects in cardiac morphogenesis, beginning with abnormalities during heart tube assembly. Rather than elongating normally to create the linear heart tube, the nkx-deficient myocardium forms a dysmorphic structure, with a short and wide ventricular portion and a sprawling, enlarged atrial portion. nkx-deficient embryos exhibit an increased number of atrial cardiomyocytes at this stage, which may account for the aberrant shape of the atrium. In contrast, the number of ventricular cardiomyocytes is normal, indicating a cell number-independent impact of nkx genes on ventricular cell behavior during tube extension. At later stages, ventricular cell number is significantly reduced, contributing to the small and misshapen appearance of the emerging ventricle. Taken together, our data demonstrate that nkx2.5 and nkx2.7 play a previously unappreciated, crucial role in the earliest stages of heart tube assembly and exert differential effects on ventricular and atrial cell number.
MATERIALS AND METHODS
Zebrafish
We bred wild-type zebrafish or zebrafish carrying the transgene Tg(cmlc2:DsRed2-nuc) (Mably et al., 2003) to generate embryos for experiments. All care and use of zebrafish and embryos complied with relevant local animal welfare laws, guidelines, and policies as described in animal protocol #060911-01, approved by the New York University School of Medicine Institutional Animal Care and Use Committee.
Morpholinos
Morpholinos (MOs; GeneTools, Philomath, Oregon, United States) were injected into embryos at the 1-cell stage. To block translation, embryos were injected with 4 ng of the anti-nkx2.5 ATG MO, with 2 ng of the anti-nkx2.7 ATG MO, or with a combination of 2 ng of the anti-nkx2.5 ATG MO and 1.5 ng of the anti-nkx2.7 ATG MO. To block splicing, embryos were injected with 2 ng of the anti-nkx2.5 splice MO, with 2 ng of the anti-nkx2.7 splice MO, or with a combination of 2 ng of the anti-nkx2.5 splice MO and 2 ng of the anti-nkx2.7 splice MO. MO sequences were:
anti-nkx2.5 ATG MO: 5′-TCATTTGGCTAGAGAACATTGCCAT-3′
anti-nkx2.7 ATG MO: 5′-TGGAGGTCACAGGACTCGGAAGCAT-3′
anti-nkx2.5 splice MO: 5′-TGTCAAGGCTCACCTTTTTTCTCTT-3′
anti-nkx2.7 splice MO: 5′-GACACATGCCTGAAAAAGAAAGCAA-3′
Intron inclusion caused by the anti-nkx2.5 splice MO would result in translation of the first 75 amino acids of Nkx2.5 and an additional 43 residues from intronic sequence, followed by a premature stop codon. Intron inclusion caused by the anti-nkx2.7 splice MO would result in translation of the first 98 amino acids of Nkx2.7 and an additional 7 residues from intronic sequence, followed by a premature stop codon. Neither protein is predicted to be functional, since they would lack both the homeodomain and the NK domain.
In situ hybridization and immunofluorescence
Whole-mount in situ hybridization for cmlc2, vmhc, amhc, and lefty2 was performed as previously described (Berdougo et al., 2003; Bisgrove et al., 1999; Yelon et al., 1999). Whole-mount immunofluorescence was also performed as previously described (Alexander et al., 1998), using primary monoclonal antibodies against sarcomeric myosin heavy chain (MF20) and atrial myosin heavy chain (S46). MF20 and S46 were obtained from the Developmental Studies Hybridoma Bank (Iowa City, Iowa, United States), maintained by the Department of Biological Sciences, University of Iowa, under contract NO1-HD-2-3144 from the NICHD. The secondary reagents goat anti-mouse IgG2b-TRITC and goat anti-mouse IgG1-FITC (Southern Biotechnology Associates, Inc., Birmingham, Alabama, United States) were used to recognize MF20 and S46, respectively.
RT-PCR
We performed RT-PCR to monitor splicing of nkx2.5 and nkx2.7 RNAs in 26 hours post fertilization (hpf) embryos injected with either anti-nkx2.5 or anti-nkx2.7 splice MOs. As illustrated in Figure 2, appropriate primers were used to amplify unspliced pre-mRNA and spliced mRNA for both nkx2.5 and nkx2.7. Primer sequences were:
Figure 2.
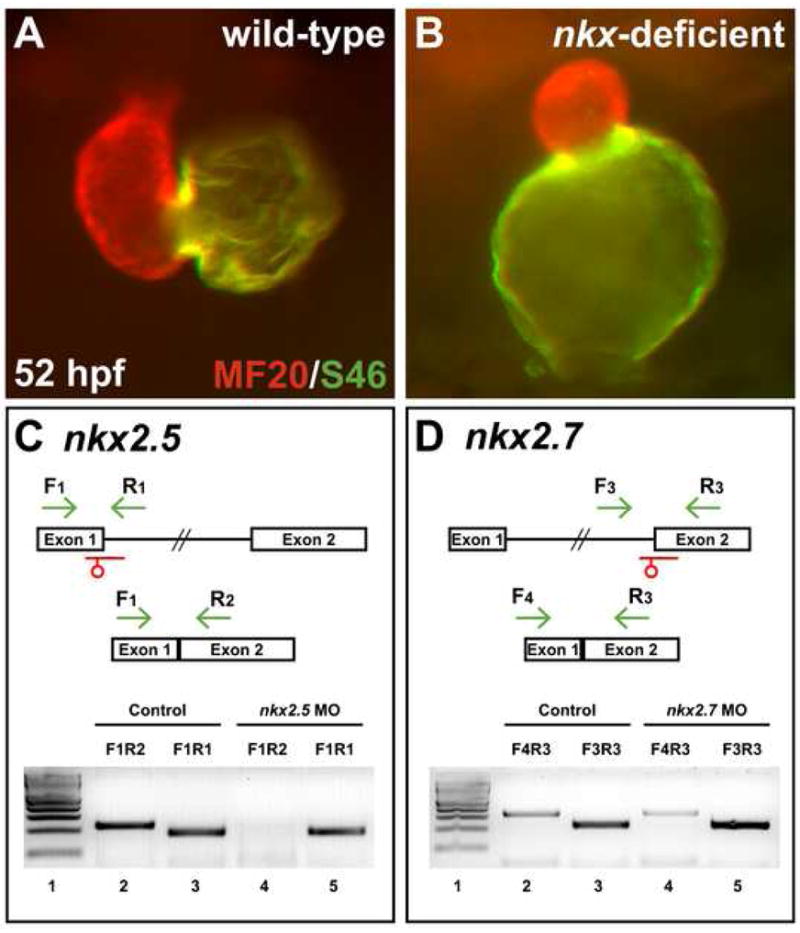
Splice MOs confirm specificity and efficacy of nkx2.5 and nkx2.7 knockdown. (A,B) Frontal views, dorsal to the top at 52 hpf. MF20/S46 immunofluorescence distinguishes ventricular myocardium (red) from atrial myocardium (yellow). Injection of anti-nkx2.5 and anti-nkx2.7 splice MOs has effects similar to that of injection of anti-nkx2.5 and anti-nkx2.7 ATG MOs (Fig. 1D). (C,D) MO injection impairs splicing of nkx2.5 and nkx2.7. Schematics depict the structures of the nkx2.5 (C) and nkx2.7 (D) pre-mRNA and mRNA. Locations of splice MOs are indicated in red. Primer pairs used for RT-PCR are indicated in green. Gels document RT-PCR amplification products, comparing detection of spliced (lanes 2 and 4) and unspliced (lanes 3 and 5) messages in cDNA from 26 hpf control embryos and embryos injected with anti-nkx2.5 splice MO or anti-nkx2.7 splice MO, respectively. Primer pair F1R1 amplifies 189 bp of unspliced nkx2.5 pre-mRNA, primer pair F1R2 amplifies 230 bp of spliced nkx2.5 mRNA, primer pair F3R3 amplifies 229 bp of unspliced nkx2.7 pre-mRNA, and primer pair F4R3 amplifies 323 bp of spliced nkx2.7 mRNA. In both gels, lane 1 contains a 100 bp molecular weight ladder (NEB, Ipswich, MA, USA). Embryos injected with anti-nkx2.5 splice MO exhibit a loss of spliced nkx2.5 mRNA, and embryos injected with anti-nkx2.7 splice MO demonstrate a partial reduction in spliced nkx2.7 mRNA with a concomitant increase in unspliced pre-mRNA. Note that the predicted PCR products with intronic inclusion for the unspliced nkx2.5 (1.4 kb) and nkx2.7 (2.2 kb) pre-mRNAs are absent, most likely because PCR conditions did not favor amplification of larger products.
F1: 5′-GCGAAGACCTTCAGGAGGACAAAGGCAAC-3′
R1: 5′-GTATTTTCAATTATATCGGTTTAAATGCCCG-3′
R2: 5′-CGAGGCTTCCTCCTCTTCCTCTGCTTGGG-3′
F3: 5′-GATGTCTTTCAACAACTTCCTGCGTG-3′
R3: 5′-CGGGGCCGAAAGGTATCTCTGC-3′
F4: 5′-GCTTCAGTGTATGCAGAACACCC-3′
β-actin F: 5′-CAGCTAGTGCGAATATCATCT-3′
β-actin R: 5′-TTTCTGTCCCATACCAACC-3′
Positive controls amplifying β-actin and negative controls omitting reverse transcriptase were examined for all cDNAs and primer pairs (Supplemental Fig. 1).
Cardiomyocyte counting
We counted cardiomyocytes in wild-type and nkx-deficient embryos at 52 hpf using the transgene Tg(cmlc2:DsRed2-nuc) as previously described (Mably et al., 2003; Schoenebeck et al., 2007). For detection of Mef2 in cardiomyocyte nuclei at 26 hpf, we used anti-Mef2 polyclonal antisera (sc-313; 1:250; Santa Cruz Biotechnology, Santa Cruz, California, United States) and S46 (1:10; anti-Amhc), along with goat anti-rabbit IgG-Alexa 488 (1:250; Molecular Probes, Eugene, Oregon, United States) and goat anti-mouse IgG1-TRITC (1:100; Southern Biotechnology Associates, Inc.) as respective secondary reagents. Stained embryos were gently flattened under a cover slip and the fluorescent nuclei in each cardiac chamber were counted. S46 staining allowed us to distinguish atrial and ventricular cardiomyocytes.
Microscopy
Embryos were examined with Zeiss M2Bio and Axioplan microscopes and photographed with a Zeiss Axiocam digital camera (Carl Zeiss, Oberkochen, Germany). Images were processed with Zeiss AxioVision (v3.0.6) software and Adobe Photoshop 8 Creative Suite software (Adobe Systems, San Jose, California, United States).
RESULTS
nkx2.5 and nkx2.7 regulate cardiac chamber morphogenesis
In order to establish the roles of nkx genes during cardiac chamber formation in zebrafish, we set out to create a loss-of-function model using antisense morpholino oligonucleotides (MOs) (Draper et al., 2001; Nasevicius and Ekker, 2000). We designed MOs to block translation (ATG MOs) and splicing (splice MOs) of nkx2.5 and nkx2.7 in order to evaluate the individual and shared functions of nkx genes. Knockdown of nkx2.5 or nkx2.7 separately produces subtle and variable defects in cardiac morphogenesis. Injection of anti-nkx2.5 ATG MO or splice MO leads to the formation of a slightly small ventricular chamber and a slightly misshapen atrial chamber (Fig. 1A,B; phenotype observed in approximately 50% of injected embryos). Injection of anti-nkx2.7 ATG MO or splice MO causes similar effects on ventricular and atrial chamber morphologies (Fig. 1C; phenotype observed in approximately 50% of injected embryos).
Figure 1.

Knockdown of nkx2.5 and/or nkx2.7 causes prominent defects in cardiac chamber morphogenesis. (A–D) Frontal views, dorsal to the top at 52 hpf. MF20/S46 immunofluorescence distinguishes ventricular myocardium (red) from atrial myocardium (yellow). In comparison to wild-type embryos (A), injection of anti-nkx2.5 ATG MO (B) or anti-nkx2.7 ATG MO (C) causes subtle abnormalities in chamber size and shape. (D) Injection of anti-nkx2.5 and anti-nkx2.7 ATG MOs together reveals more striking defects in both ventricular and atrial morphology. (E,F) Lateral views of live embryos, anterior to the right, at 52 hpf. Other than their cardiac defects, pericardial edema, brain ventricle swelling, and jaw hypoplasia, nkx-deficient embryos appear morphologically normal.
In contrast, the effect of combining anti-nkx2.5 and anti-nkx2.7 ATG MOs or anti-nkx2.5 and anti-nkx2.7 splice MOs produces more dramatic errors in cardiac chamber formation than inhibition of nkx2.5 or nkx2.7 individually. In injected embryos, the ventricle is underdeveloped and can be either small and rounded (Fig. 1D; phenotype observed in >90% of embryos injected with anti-nkx2.5 and anti-nkx2.7 ATG MOs) or substantially diminished and rudimentary (Fig. 2B; phenotype observed in >50% of embryos injected with anti-nkx2.5 and anti-nkx2.7 splice MOs). In addition, we observed a decreased intensity of ventricular contraction in the injected embryos (KLT, TS, and DY, unpublished data). Furthermore, the atrium appears enlarged with a globular contour that is either slightly bulbous (Fig. 1D; phenotype observed in >90% of embryos injected with ATG MOs) or grossly dilated (Fig. 2B; phenotype observed in >50% of embryos injected with splice MOs). Taken together, our data suggest that nkx2.5 and nkx2.7 play overlapping roles in regulating cardiac chamber morphogenesis in zebrafish.
The similitude of phenotypes caused by the ATG MOs and the splice MOs corroborates the notion that our findings reflect the specific knockdown of nkx genes. Furthermore, aside from their cardiac phenotypes, all injected embryos appear morphologically intact, with no defects evident in body axis patterning or somitogenesis (Fig. 1E,F). Pericardial edema and mild swelling of brain ventricles develop secondary to cardiac dysfunction in injected embryos (Fig. 1E,F). In addition, embryos injected with anti-nkx2.7 MOs alone or in combination with anti-nkx2.5 MOs exhibit jaw abnormalities (TS and DY, unpublished data), consistent with the expression of nkx2.7 in the pharyngeal endoderm (Lee et al., 1996). While antibodies are not available to assess the effectiveness of ATG MOs, we were able to use RT-PCR as a measure of nkx gene knockdown achieved with splice MOs. Analysis of splicing efficiency in injected embryos indicates a complete loss of spliced nkx2.5 mRNA and a significant reduction of spliced nkx2.7 mRNA (Fig. 2C,D). The reduced penetrance of the phenotype caused by splice MOs, compared with the effectiveness of ATG MOs, may reflect the incomplete elimination of the spliced nkx2.7 transcript. Overall, our data validate that defects in cardiac chamber formation in our MO-injected embryos are a consequence of the specific and effective inhibition of nkx2.5 and nkx2.7.
Heart tube assembly begins normally in nkx-deficient embryos
In order to elucidate the mechanisms by which nkx genes regulate chamber morphology, we chose to investigate the origins of the morphogenetic problems in embryos injected with both anti-nkx2.5 and anti-nkx2.7 MOs (hereafter referred to as nkx-deficient embryos). First, we examined the bilateral populations of myocardial precursors that merge at the embryonic midline to assemble the heart tube. Chamber-specific molecular markers allow us to examine cardiac chamber precursor populations in the lateral plate mesoderm before the heart tube forms (Yelon et al., 1999). Expression of cardiac myosin heavy chain (cmlc2), the earliest known marker of myocardial differentiation (Yelon et al., 1999), and expression of ventricular myosin heavy chain (vmhc), the earliest known marker of ventricular cardiomyocytes (Yelon et al., 1999), are both normal in nkx-deficient embryos at the 18-somite stage (Fig. 3). Thus, nkx-deficient embryos do not seem to have significant defects in the initial specification of ventricular cardiomyocytes. However, we noted subtle defects in morphogenesis of the bilateral heart fields as they migrate medially: the nkx-deficient populations appear slightly delayed relative to wild-type (Fig. 3).
Figure 3.
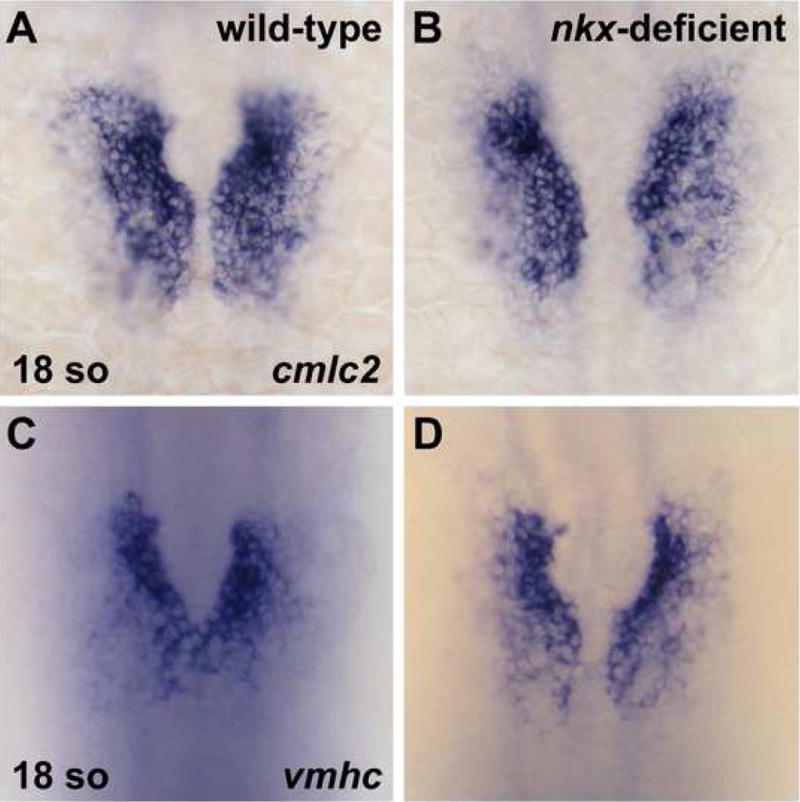
Bilateral populations of cardiomyocytes are normal in nkx-deficient embryos. In situ hybridization depicts expression of cmlc2 (A,B) and vmhc (C,D) in wild-type (A,C) and nkx-deficient (B,D) embryos. Embryos viewed dorsally, anterior to the top, at the 18-somite stage. Aside from a mild delay in cardiomyocyte migration toward the midline, nkx-deficient embryos appear to have the same patterns of cmlc2 and vmhc expression as are observed in wild-type embryos.
As cardiac fusion proceeds, the bilateral populations of cardiomyocytes merge to form a ring of cells, referred to as the cardiac cone (Glickman and Yelon, 2002). Once the cone forms, robust expression of the earliest known atrial cardiomyocyte marker, atrial myosin heavy chain (amhc), begins (Berdougo et al., 2003). We inspected the contours of the cardiac cone in nkx-deficient embryos but found no major morphological defects in either the atrial or ventricular precursor populations (Fig. 4). Cone formation occurs normally with appropriate anterior and posterior fusion of the bilateral heart fields. However, we often observed a slightly enlarged, elliptical lumen in the center of the cone in nkx-deficient embryos (Fig. 4B,D). This defect may be related to the mild impairment in myocardial migration observed at earlier stages (Fig. 3). Consistent with our findings at the 18-somite stage, we did not find any evidence for significant changes in cardiomyocyte number in nkx-deficient embryos at the 22-somite stage (Fig. 4); notably, amhc expression appears relatively normal (Fig. 4E,F). However, subtle increases in the number of amhc-expressing cells may be difficult to resolve precisely at this stage. Overall, our analysis of cone formation suggests that the initial phases of cardiac specification and morphogenesis proceed correctly in nkx-deficient embryos.
Figure 4.
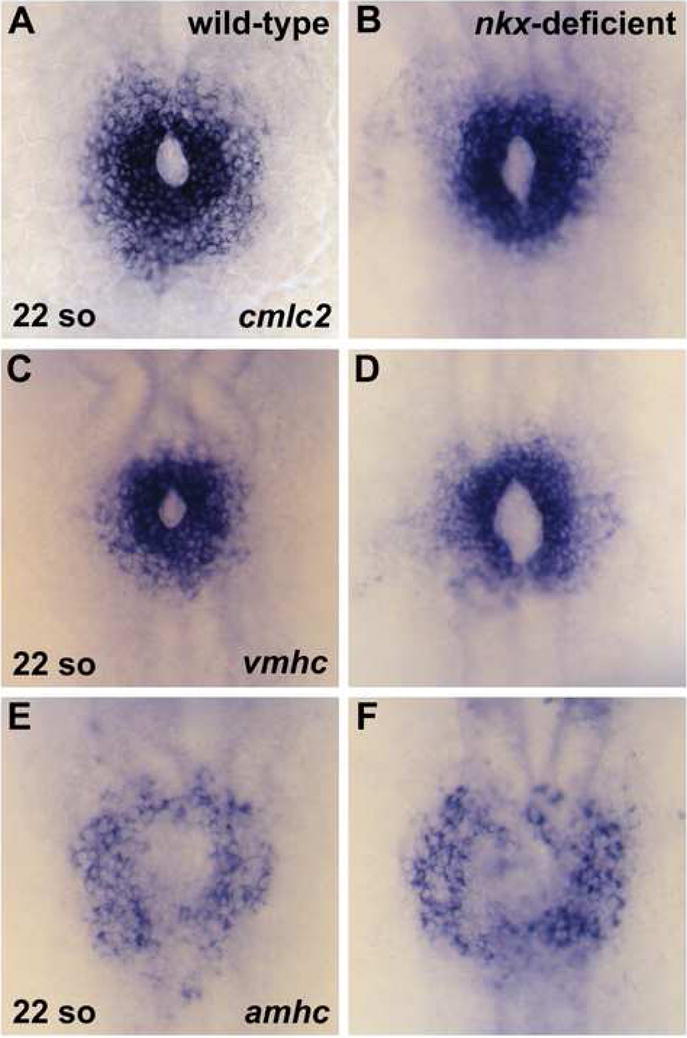
Cardiac cone formation occurs normally in nkx-deficient embryos. In situ hybridization depicts expression of cmlc2 (A,B), vmhc (C,D), and amhc (E,F) in wild-type and nkx-deficient embryos. Embryos viewed dorsally, anterior to the top, at the 22-somite stage. (A,C,E) Wild-type embryos exhibit normal fusion of the bilateral cardiac precursors, creating an intact cardiac cone. (B,D,F) nkx-deficient embryos mirror the expression patterns observed in wild-type, with the exception of a slightly enlarged and elongated lumen within the cardiac cone.
nkx genes play a critical role in early steps of heart tube extension
Since cone formation seems relatively unaffected by loss of nkx gene function, we proceeded to analyze the morphogenetic progression by which the cardiac cone becomes the heart tube. There is a critical transition that occurs during heart tube extension: the cardiac cone gradually elongates and narrows until its rings of vmhc-expressing and amhc-expressing cells become the ventricular and atrial portions of a linear tube (Berdougo et al., 2003; Glickman and Yelon, 2002). By 26 hours post fertilization (hpf), when the heart tube in wild-type embryos is fully elongated (Fig. 5A), there is a striking defect in heart tube extension in nkx-deficient embryos (Fig. 5B). The overall shape of the heart tube is dramatically dysmorphic, more resembling a pyramid than the linear, cylindrical structure of the wild-type heart. Additionally, the normal leftward displacement of the heart tube is disrupted in >75% of nkx-deficient embryos. This phenotype reflects a defect in heart tube morphogenesis rather than a defect in left-right axis patterning, since lefty2 expression is appropriately restricted to the left side of nkx-deficient embryos (KLT and DY, unpublished data). Furthermore, distinct morphogenetic defects appear in both the ventricular and atrial portions of the nkx-deficient heart tube. The vmhc-expressing portion of the nkx-deficient heart is substantially shorter and wider than the vmhc-expressing portion of the wild-type tube (Fig. 5C,D); in wild-type embryos, this portion of the heart tube is approximately twice as tall as it is wide, which is rarely achieved in nkx-deficient embryos. Additionally, atrial morphology is substantially disrupted: in wild-type embryos, the amhc-expressing portion of the heart tube becomes cohesive and tubular by 26 hpf, whereas nkx-deficient embryos display a spray of amhc-expressing cells that have not coalesced appropriately (Fig. 5E,F).
Figure 5.
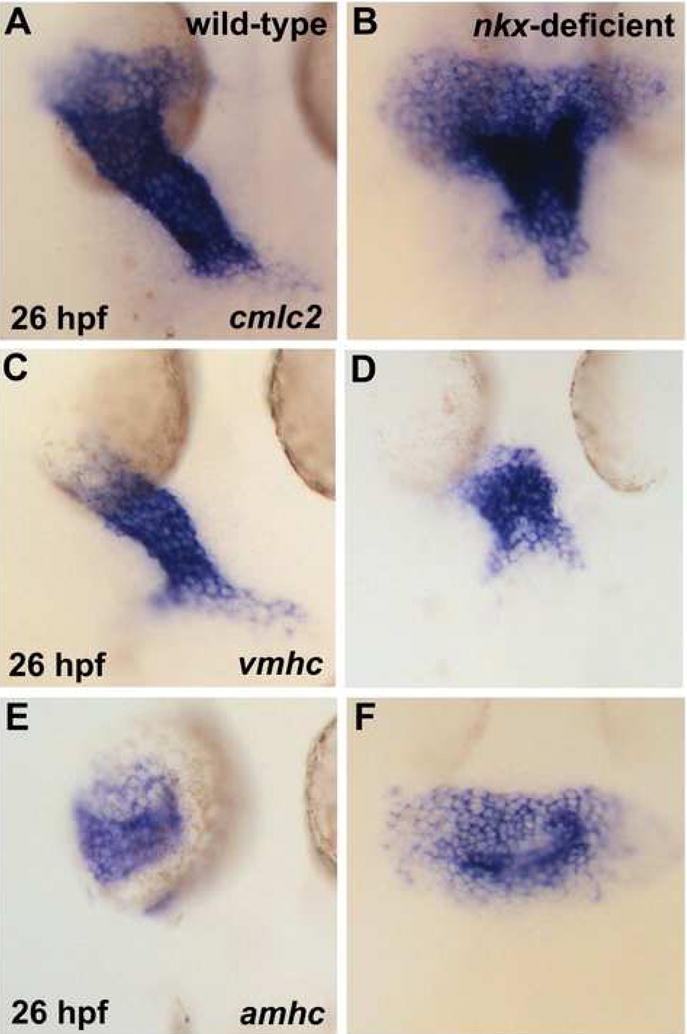
nkx genes play a critical role in early steps of heart tube extension. In situ hybridization depicts expression of cmlc2 (A,B), vmhc (C,D), and amhc (E,F) in wild-type (A,C,E) and nkx-deficient (B,D,F) embryos. Embryos viewed dorsally, anterior to the top, at 26 hpf. (A,B) nkx-deficient embryos exhibit prominent defects in heart tube extension, including a splayed and disorganized inflow region and an unusually compact outflow region. (C,D) In nkx-deficient embryos, the ventricular portion of the heart tube is abnormally short and wide. (E,F) In nkx-deficient embryos, the atrial portion of the heart tube is broad and sprawling.
Similar defects in heart tube extension were generated by injection of ATG MOs and by injection of splice MOs. For example, the stunted length and the broader width of the vmhc-expressing portion of the heart tube in embryos injected with splice MOs (Supplemental Fig. 2) recapitulate the characteristics observed in embryos injected with ATG MOs (Fig. 5C,D). Moreover, neither the ventricular nor the atrial defects can be explained simply by a developmental delay in nkx-deficient embryos, since the observed phenotypes are not equivalent to any stages during wild-type heart tube extension (Berdougo et al., 2003; Glickman and Yelon, 2002; Yelon et al., 1999). As was the case for the cardiac phenotype at 52 hpf (Fig. 1), individual anti-nkx2.5 and anti-nkx2.7 MOs are not as effective as the combination of both MOs at disrupting heart tube extension (KLT and DY, unpublished data). Thus, our data indicate that both nkx2.5 and nkx2.7 play critical roles in establishing the initial dimensions of the linear heart tube.
nkx genes limit atrial cell number during heart tube assembly
Next, we turned our attention to the possible cellular mechanisms underlying the defects in heart tube assembly in nkx-deficient embryos. Recent work has suggested that deficiency of Nkx2-5 in mice leads to an overspecification of cardiac progenitor cells and a failure of SHF cells to proliferate (Prall et al., 2007). In light of these data, we wondered whether aberrations in atrial and ventricular cell number could be responsible for the heart tube assembly defects observed in nkx-deficient embryos. Thus, we proceeded to quantify cardiomyocyte cell number in nkx-deficient hearts (Fig. 6).
Figure 6.
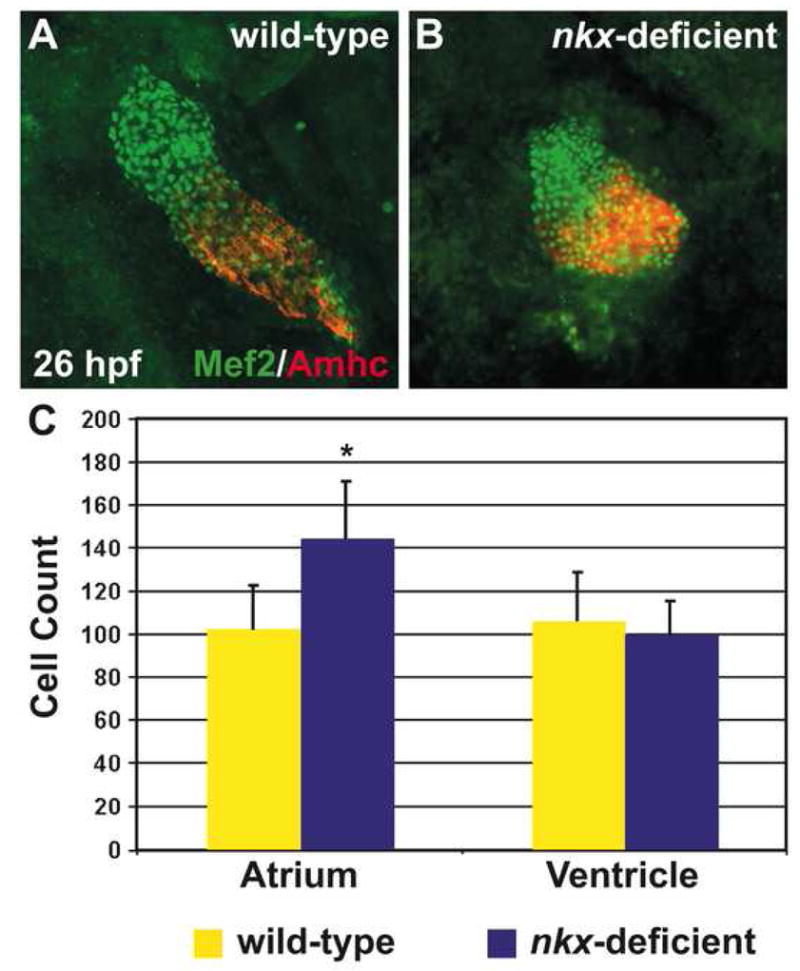
nkx genes limit atrial cell number during heart tube assembly. (A,B) Immunofluorescence indicates nuclear localization of Mef2 (green) throughout the heart tube, facilitating counting of differentiated cardiomyocytes at 26 hpf. Atrial cells are indicated by the anti-Amhc antibody, S46 (red). Hearts are flattened with a cover slip to improve visualization of cardiomyocyte nuclei. (C) Quantification of cardiomyocyte nuclei in wild-type (n=25) and nkx-deficient (n=11) embryos reveals a statistically significant increase in atrial cell number in nkx-deficient embryos (p<0.001, Student’s t-test) and no significant difference in ventricular cell number. Bar graph indicates mean and standard error of each data set; asterisk indicates statistically significant difference from wild-type.
Prior studies have employed the expression of a nuclear red fluorescent protein (DsRed2-nuc), driven by the transgene Tg(cmlc2:DsRed2-nuc), to count cardiomyocytes (Mably et al., 2003; Schoenebeck et al., 2007). However, the DsRed2-nuc protein takes a significant amount of time to fold and localize (Baird et al., 2000; Bevis and Glick, 2002), such that its nuclear fluorescence is not evident until approximately 24 hours after activation of the cmlc2 promoter (Lepilina et al., 2006). Therefore, the transgene Tg(cmlc2:DsRed2-nuc) is an excellent tool for detecting cardiomyocytes that began differentiation at least 24 hours earlier, but it is not appropriate for assessing cardiomyocyte cell number at 26 hpf. Instead, we visualized differentiated cardiomyocytes based on their nuclear localization of the transcription factor Mef2 (Fig. 6A,B). Two zebrafish mef2 genes, mef2a and mef2c, are expressed in precardiac mesoderm by the 18-somite stage, as cardiomyocytes are undergoing differentiation, and nuclear localization of Mef2 occurs as early as 24 hpf (Ticho et al., 1996).
We identified a prominent increase in the number of atrial cardiomyocytes in nkx-deficient embryos at 26 hpf: they exhibit 40% more atrial cells than wild-type embryos do at this stage (Fig. 6C). This dramatic difference in atrial cell number suggests that nkx genes could play a fundamental role in limiting the size of the atrial progenitor population. Furthermore, the excess atrial cardiomyocytes in nkx-deficient embryos may be responsible for the observed irregularities in atrial coalescence and elongation during heart tube extension (Fig. 5E,F). In contrast, we observed no significant difference in ventricular cell number when comparing wild-type and nkx-deficient embryos at 26 hpf (Fig. 6C). Therefore, cell number defects do not play a role in hindering ventricular elongation in nkx-deficient embryos, and this ventricular morphogenetic defect likely reflects other functions of nkx genes in regulating ventricular cell behavior.
nkx genes are necessary to establish sufficient ventricular cell number during cardiac chamber formation
Although the defects in ventricular elongation in nkx-deficient embryos appear to be independent of aberrations in ventricular cell number, we wondered whether cell number changes might underlie the dysmorphic nature of the nkx-deficient ventricle at 52 hpf (Figs. 1D, 2B). Since Nkx2-5 has distinct roles at different developmental stages in mouse -- an early role restricting cardiomyocyte specification and a later role promoting SHF proliferation (Prall et al., 2007) -- we hypothesized that zebrafish nkx genes might regulate the proper number of ventricular cardiomyocytes at stages following heart tube assembly. To count cardiomyocytes in wild-type and nkx-deficient embryos at 52 hpf, we chose to employ the transgene Tg(cmlc2:DsRed2-nuc) (Mably et al., 2003; Schoenebeck et al., 2007), since its expression is more robust and resolvable than Mef2 localization is at this stage.
Comparing wild-type and nkx-deficient hearts at 52 hpf, we found differences in cell number in both the atrium and the ventricle (Fig. 7). Analogous to our 26 hpf data, nkx-deficient embryos have nearly 30% more atrial cardiomyocytes than wild-type embryos (Figs. 6C, 7C). In contrast, nkx-deficient embryos have close to 30% fewer ventricular cells than wild-type embryos (Fig. 7C). This loss of cells is likely to contribute to the shrunken appearance of the nkx-deficient ventricle (Fig. 1D, 2B). Taken together, our analyses of cell number defects in nkx-deficient hearts indicate two distinct roles for nkx genes in regulating cardiomyocyte population size: an earlier role in restricting atrial cell number and a later role in establishing ventricular cell number.
Figure 7.
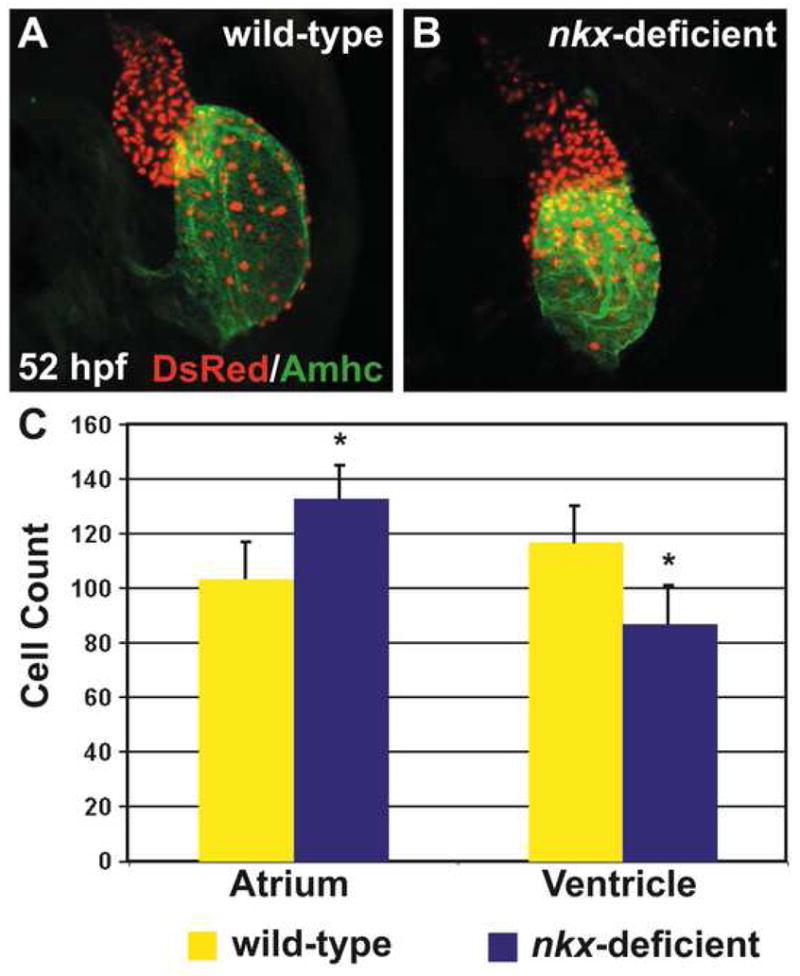
nkx genes are necessary to establish sufficient ventricular cell number during cardiac chamber formation. (A,B) Immunofluorescence indicates that both chambers express the transgene Tg(cmlc2:DsRed2-nuc) (red), facilitating cardiomyocyte counting at 52 hpf. Atria are labeled with the anti-Amhc antibody, S46 (green). Hearts are flattened with a cover slip to improve visualization of cardiomyocyte nuclei. (C) Quantification of cardiomyocyte nuclei in wild-type (n=10) and nkx-deficient (n=13) embryos reveals a statistically significant increase in atrial cell number in nkx-deficient embryos (p<0.001, Student’s t-test) and a statistically significant decrease in ventricular cell number in nkx-deficient embryos (p<0.001, Student’s t-test). Bar graph indicates mean and standard error of each data set; asterisks indicate statistically significant differences from wild-type.
DISCUSSION
Our data provide new insights into the roles played by nkx genes in determining the proper numbers of cardiomyocytes and in establishing the initial dimensions of the linear heart tube. nkx genes are required to limit atrial cell number during heart tube assembly and are necessary to establish sufficient ventricular cell number during chamber emergence. Additionally, nkx genes appear to control the extent of ventricular elongation during heart tube assembly without affecting cell number, suggesting a separate role of nkx genes in regulating cardiomyocyte cell behavior. Together, our results indicate that nkx genes have distinct roles in the ventricle and in the atrium, affecting cell number and morphogenesis in disparate manners and at different stages.
It is interesting to consider the interrelationship of the roles of nkx genes in controlling cardiomyocyte number and morphogenesis. For example, in the atrium, excess cells are likely to create challenges for efficient heart tube assembly. The dramatic increase in cell number prior to heart tube extension most likely contributes to the disorganized and sprawling atrial phenotype observed in nkx-deficient embryos. A similar correlation exists in cloche mutant embryos, which exhibit both an increased number of atrial cardiomyocytes and a sprawling, disorganized atrial portion of the heart tube (Holtzman et al., 2007; Schoenebeck et al., 2007). Furthermore, during atrial chamber formation, increased atrial cell number is a potential cellular mechanism underlying the enlarged, swollen atrium seen in both nkx-deficient and cloche mutant embryos (Schoenebeck et al., 2007). Therefore, we suggest that an excessive number of cardiomyocytes can disrupt heart morphogenesis from an early stage.
The source of the atrial cardiomyocyte surplus in nkx-deficient embryos is not immediately apparent. Possible explanations for the observed accumulation of cells include excess proliferation or augmented progenitor specification. While it is possible that atrial cells undergo excessive proliferation in nkx-deficient embryos, phospho-histone-3 immunohistochemistry rarely reveals labeled cardiomyocytes in either the wild-type or nkx-deficient heart tubes (KLT and DY, unpublished data), consistent with previous studies of cardiomyocyte proliferation at these stages (Rohr et al., 2006). It is therefore appealing to consider that the atrial surplus in nkx-deficient embryos reflects an overspecification of cardiac progenitors, as observed in Nkx2-5−/− mice (Prall et al., 2007). As it is technically challenging to resolve subtle differences in amhc expression at early stages, enhanced atrial specification may begin earlier than the 22-somite stage in nkx-deficient embryos, with recruitment of atrial cells continuing through 26 hpf. Although the origins of additional atrial progenitor cells are ambiguous, it is unlikely that the increase in atrial cells arises from a fate transformation of ventricular cells. The timing of alterations in cell number is incompatible with this idea: the boost in atrial cardiomyocytes during heart tube assembly significantly precedes the decline in ventricular cardiomyocytes during cardiac chamber formation.
Our analysis of the ventricular phenotype in nkx-deficient embryos also indicates a connection between cell number and morphogenesis. Whereas the number of ventricular cardiomyocytes in wild-type embryos increases between the first and second days of cardiac development, the number of ventricular cardiomyocytes in nkx-deficient embryos does not exhibit this trend. This depletion of ventricular cell number is likely to limit the expansion of the maturing nkx-deficient chamber. Yet, the etiology of the ventricular cell shortage remains unclear and could reflect roles of nkx genes in ventricular cell proliferation, survival, or recruitment. Little is known about the contribution of these mechanisms to the maintenance or augmentation of the expanding ventricle in wild-type embryos. Proliferation does not seem to be a critical mechanism for regulation of ventricular cell number during the emergence of chambers from the heart tube (Rohr et al., 2006; S. Marques and DY, unpublished data). Furthermore, TUNEL analysis of nkx-deficient embryos did not reveal evidence of cardiomyocyte apoptosis during these stages (KLT and DY, unpublished data). Perhaps the decrease in ventricular cardiomyocytes instead reflects a problem with recruitment of cells from an additional heart field. Although the identification of a zebrafish equivalent to the amniote second heart field is still under inquiry (Meilhac et al., 2004), it is interesting to consider that nkx genes could influence the specification, growth, or recruitment of such a population.
While cell number abnormalities are linked to the defects in ventricular chamber architecture, another cellular mechanism must be invoked to explain the ventricular morphogenetic problems during heart tube assembly in nkx-deficient embryos. The ventricular component of the nkx-deficient heart tube is unusually short and wide, yet ventricular cardiomyocyte number is normal at this stage. Studies of the zebrafish mutants nagie oko (nok/mpp5) and heart and soul (has/prkci) have highlighted the importance of apicobasal polarity and cell shape changes during heart tube extension (Rohr et al., 2006), suggesting that downstream targets of nkx genes could be essential for ventricular cardiomyocyte polarity or shape. Alternatively, targets of nkx genes could be important for cardiomyocyte motility, as recent work demonstrates that a characteristic speed and direction of cell movements is necessary to drive heart tube extension (Rohr et al., 2008; Smith et al., 2008). In this regard, the ventricular defects in nkx-deficient embryos could be secondary to the atrial surplus: perhaps the movement of ventricular cells is encumbered by their connection to an excess of atrial cells. Further studies will be necessary to elucidate the cellular mechanisms by which nkx genes regulate extension of the ventricular portion of the heart tube.
Altogether, our data reveal functions of nkx genes in zebrafish that complement and extend previous studies of Nkx2-5 in mouse. Our observation of an early increase in atrial cell number in nkx-deficient embryos parallels the excess cardiac progenitor specification seen at early stages in Nkx2-5−/− mice (Prall et al., 2007). Later, the decrease in ventricular cell number in our nkx-deficient embryos mimics the collapse in SHF proliferation in Nkx2-5−/− mice (Prall et al., 2007). The evident analogies between nkx-deficient phenotypes in zebrafish and mouse highlight the different functions of nkx genes during temporally discrete aspects of cardiogenesis. It is intriguing that, in zebrafish, the distinct functions of nkx genes also appear to be chamber-specific, underscoring the independent regulation of the atrial and ventricular lineages, which separate from each other in the midblastula (Stainier et al., 1993). Additionally, our work extends prior understanding of the roles of nkx genes by illuminating their contribution to heart tube extension and their particular influence on ventricular elongation through cell number-independent mechanisms. Given the similarities observed in zebrafish and mouse loss-of-function models, we propose that these newly recognized functions of nkx genes could be conserved across species.
In conclusion, our data have illuminated the unique roles that nkx genes play in distinct cardiomyocyte populations at critical transitions in cardiac morphogenesis. Our results suggest two possible mechanisms through which loss of nkx gene function could lead to congenital heart disease. An excessive atrial cell number from an early stage in development could disrupt later atrial morphogenesis, potentially interfering with proper septation. Furthermore, defects in ventricular extension during heart tube assembly or loss of ventricular cells during chamber formation could produce a dysmorphic ventricle, as seen in hypoplastic left heart syndrome. Ultimately, these data provide a basis for further investigation of the essential cellular functions of NKX2-5 and of its role as a key causative gene in congenital heart disease. Through the future identification of the molecular targets and downstream effectors of nkx genes, we will greatly enhance our ability to provide essential risk stratification, improve prognosis, and therapeutically intervene for patients with NKX2-5 mutations.
Supplementary Material
Supplemental Figure 1. Positive and negative controls for the RT-PCR experiment shown in Figure 2C,D. (A) Positive controls using primers for β-actin demonstrate similar efficacy of cDNA synthesis in all samples. (B) Negative controls omitting reverse transcriptase from the cDNA synthesis reaction demonstrate absence of contaminating genomic DNA.
Supplemental Figure 2. Splice MOs recapitulate the phenotype resulting from injection of ATG MOs. In situ hybridization depicts expression of vmhc in (A) wild-type and (B) anti-nkx2.5 and anti-nkx2.7 splice MO-injected embryos. Embryos viewed dorsally, anterior to the top, at the 26 hpf stage. The vmhc-expressing portion of the nkx-deficient heart tube is substantially shorter and wider than the vmhc-expressing portion of the wild-type heart tube.
Acknowledgments
We are grateful for input from Richard Harvey, Owen Prall, the Yelon laboratory, the Torres-Vázquez laboratory, and the Skirball Developmental Genetics Program. This work was supported by grants to DY from the National Institutes of Health, the American Heart Association, and the March of Dimes. KLT received support from the National Institutes of Health (K12 HD043389 and K08 HL088002).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander J, et al. Screening mosaic F1 females for mutations affecting zebrafish heart induction and patterning. Dev Genet. 1998;22:288–299. doi: 10.1002/(SICI)1520-6408(1998)22:3<288::AID-DVG10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Allen BG, et al. Reduction of XNkx2-10 expression leads to anterior defects and malformation of the embryonic heart. Mech Dev. 2006;123:719–729. doi: 10.1016/j.mod.2006.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiazu N, Frasch M. tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 1993;7:1325–1340. doi: 10.1101/gad.7.7b.1325. [DOI] [PubMed] [Google Scholar]
- Baird GS, et al. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci U S A. 2000;97:11984–11989. doi: 10.1073/pnas.97.22.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett HL, et al. Transient early embryonic expression of Nkx2-5 mutations linked to congenital heart defects in human causes heart defects in Xenopus laevis. Dev Dyn. 2007;236:2475–2484. doi: 10.1002/dvdy.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DW, et al. Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. J Clin Invest. 1999;104:1567–1573. doi: 10.1172/JCI8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdougo E, et al. Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development. 2003;130:6121–6129. doi: 10.1242/dev.00838. [DOI] [PubMed] [Google Scholar]
- Bevis BJ, Glick BS. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed) Nat Biotechnol. 2002;20:83–87. doi: 10.1038/nbt0102-83. [DOI] [PubMed] [Google Scholar]
- Biben C, et al. Cardiac septal and valvular dysmorphogenesis in mice heterozygous for mutations in the homeobox gene Nkx2-5. Circ Res. 2000;87:888–895. doi: 10.1161/01.res.87.10.888. [DOI] [PubMed] [Google Scholar]
- Bisgrove BW, et al. Regulation of midline development by antagonism of lefty and nodal signaling. Development. 1999;126:3253–3262. doi: 10.1242/dev.126.14.3253. [DOI] [PubMed] [Google Scholar]
- Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 1993;118:719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943–948. doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- Buckingham M, et al. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- Chen JN, Fishman MC. Zebrafish tinman homolog demarcates the heart field and initiates myocardial differentiation. Development. 1996;122:3809–3816. doi: 10.1242/dev.122.12.3809. [DOI] [PubMed] [Google Scholar]
- Draper BW, et al. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis. 2001;30:154–156. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- Elliott DA, et al. Cardiac homeobox gene NKX2-5 mutations and congenital heart disease: associations with atrial septal defect and hypoplastic left heart syndrome. J Am Coll Cardiol. 2003;41:2072–2076. doi: 10.1016/s0735-1097(03)00420-0. [DOI] [PubMed] [Google Scholar]
- Evans SM, et al. tinman, a Drosophila homeobox gene required for heart and visceral mesoderm specification, may be represented by a family of genes in vertebrates: XNkx-2.3, a second vertebrate homologue of tinman. Development. 1995;121:3889–3899. doi: 10.1242/dev.121.11.3889. [DOI] [PubMed] [Google Scholar]
- Glickman NS, Yelon D. Cardiac development in zebrafish: coordination of form and function. Semin Cell Dev Biol. 2002;13:507–513. doi: 10.1016/s1084952102001040. [DOI] [PubMed] [Google Scholar]
- Grow MW, Krieg PA. Tinman function is essential for vertebrate heart development: elimination of cardiac differentiation by dominant inhibitory mutants of the tinman-related genes, XNkx2-3 and XNkx2-5. Dev Biol. 1998;204:187–196. doi: 10.1006/dbio.1998.9080. [DOI] [PubMed] [Google Scholar]
- Hoffman JI. Incidence of congenital heart disease: II. Prenatal incidence. Pediatr Cardiol. 1995;16:155–165. doi: 10.1007/BF00794186. [DOI] [PubMed] [Google Scholar]
- Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39:1890–1900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- Holtzman NG, et al. Endocardium is necessary for cardiomyocyte movement during heart tube assembly. Development. 2007;134:2379–2386. doi: 10.1242/dev.02857. [DOI] [PubMed] [Google Scholar]
- Komuro I, Izumo S. Csx: a murine homeobox-containing gene specifically expressed in the developing heart. Proc Natl Acad Sci U S A. 1993;90:8145–8149. doi: 10.1073/pnas.90.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, et al. A new tinman-related gene, nkx2.7, anticipates the expression of nkx2.5 and nkx2.3 in zebrafish heart and pharyngeal endoderm. Dev Biol. 1996;180:722–731. doi: 10.1006/dbio.1996.0341. [DOI] [PubMed] [Google Scholar]
- Lepilina A, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Lints TJ, et al. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development. 1993;119:419–431. doi: 10.1242/dev.119.2.419. [DOI] [PubMed] [Google Scholar]
- Lyons I, et al. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- Mably JD, et al. heart of glass regulates the concentric growth of the heart in zebrafish. Curr Biol. 2003;13:2138–2147. doi: 10.1016/j.cub.2003.11.055. [DOI] [PubMed] [Google Scholar]
- McElhinney DB, et al. NKX2.5 mutations in patients with congenital heart disease. J Am Coll Cardiol. 2003;42:1650–1655. doi: 10.1016/j.jacc.2003.05.004. [DOI] [PubMed] [Google Scholar]
- Meilhac SM, et al. The clonal origin of myocardial cells in different regions of the embryonic mouse heart. Dev Cell. 2004;6:685–698. doi: 10.1016/s1534-5807(04)00133-9. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Newman CS, et al. Transient cardiac expression of the tinman-family homeobox gene, XNkx2-10. Mech Dev. 2000;91:369–373. doi: 10.1016/s0925-4773(99)00291-9. [DOI] [PubMed] [Google Scholar]
- Prall OW, et al. Developmental paradigms in heart disease: insights from tinman. Ann Med. 2002;34:148–156. [PubMed] [Google Scholar]
- Prall OW, et al. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell. 2007;128:947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr S, et al. Heart and soul/PRKCi and nagie oko/Mpp5 regulate myocardial coherence and remodeling during cardiac morphogenesis. Development. 2006;133:107–115. doi: 10.1242/dev.02182. [DOI] [PubMed] [Google Scholar]
- Rohr S, et al. Asymmetric involution of the myocardial field drives heart tube formation in zebrafish. Circ Res. 2008;102:e12–19. doi: 10.1161/CIRCRESAHA.107.165241. [DOI] [PubMed] [Google Scholar]
- Schoenebeck JJ, et al. Vessel and blood specification override cardiac potential in anterior mesoderm. Dev Cell. 2007;13:254–267. doi: 10.1016/j.devcel.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott JJ, et al. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- Schultheiss TM, et al. Induction of avian cardiac myogenesis by anterior endoderm. Development. 1995;121:4203–4214. doi: 10.1242/dev.121.12.4203. [DOI] [PubMed] [Google Scholar]
- Smith KA, et al. Rotation and asymmetric development of the zebrafish heart requires directed migration of cardiac progenitor cells. Dev Cell. 2008;14:287–297. doi: 10.1016/j.devcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Stanley EG, et al. Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3′UTR-ires-Cre allele of the homeobox gene Nkx2-5. Int J Dev Biol. 2002;46:431–439. [PubMed] [Google Scholar]
- Tanaka M, et al. Nkx2.5 and Nkx2.6, homologs of Drosophila tinman, are required for development of the pharynx. Mol Cell Biol. 2001;21:4391–4398. doi: 10.1128/MCB.21.13.4391-4398.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, et al. Complex modular cis-acting elements regulate expression of the cardiac specifying homeobox gene Csx/Nkx2.5. Development. 1999;126:1439–1450. doi: 10.1242/dev.126.7.1439. [DOI] [PubMed] [Google Scholar]
- Tanaka M, et al. Phenotypic characterization of the murine Nkx2.6 homeobox gene by gene targeting. Mol Cell Biol. 2000;20:2874–2879. doi: 10.1128/mcb.20.8.2874-2879.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticho BS, et al. Three zebrafish MEF2 genes delineate somitic and cardiac muscle development in wild-type and mutant embryos. Mech Dev. 1996;59:205–218. doi: 10.1016/0925-4773(96)00601-6. [DOI] [PubMed] [Google Scholar]
- Tonissen KF, et al. XNkx-2.5, a Xenopus gene related to Nkx-2.5 and tinman: evidence for a conserved role in cardiac development. Dev Biol. 1994;162:325–328. doi: 10.1006/dbio.1994.1089. [DOI] [PubMed] [Google Scholar]
- Yelon D, et al. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev Biol. 1999;214:23–37. doi: 10.1006/dbio.1999.9406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Positive and negative controls for the RT-PCR experiment shown in Figure 2C,D. (A) Positive controls using primers for β-actin demonstrate similar efficacy of cDNA synthesis in all samples. (B) Negative controls omitting reverse transcriptase from the cDNA synthesis reaction demonstrate absence of contaminating genomic DNA.
Supplemental Figure 2. Splice MOs recapitulate the phenotype resulting from injection of ATG MOs. In situ hybridization depicts expression of vmhc in (A) wild-type and (B) anti-nkx2.5 and anti-nkx2.7 splice MO-injected embryos. Embryos viewed dorsally, anterior to the top, at the 26 hpf stage. The vmhc-expressing portion of the nkx-deficient heart tube is substantially shorter and wider than the vmhc-expressing portion of the wild-type heart tube.


