Abstract
Distinct catalytic mechanisms have been proposed for the Gcn5 and MYST histone acetyltransferases (HAT) families. Gcn5-like HATs utilize an ordered sequential mechanism involving direct nucleophilic attack of the N-ε-lysine on the enzyme-bound acetyl-CoA. Recently, MYST enzymes were reported to employ a ping-pong route of catalysis via an acetyl-cysteine intermediate. Here, using the prototypical MYST family member Esa1, and its physiological complex (piccolo NuA4), steady-state kinetic analyses revealed a kinetic mechanism that requires the formation of a ternary complex prior to catalysis, where acetyl-CoA binds first, and CoA is the last product released. In the absence of histone acceptor, slow rates of enzyme auto-acetylation (7 × 10−4 s−1, or ∼2500-fold slower than histone acetylation, kcat =1.6 s−1) and of CoA formation (0.0021 s−1) were inconsistent with a kinetically competent acetylated-enzyme intermediate. Previously, Cys-304 of Esa1 was the proposed nucleophile that forms an acetyl-cysteine intermediate. Here, mutation of this cysteine (C304A) in Esa1 or within the piccolo NuA4 complex yielded an enzyme that was catalytically indistinguishable from wild-type. Similarly, a pH rate (kcat) analysis of wild-type and C304A revealed an ionization (pKa = 7.6−7.8) that must be unprotonated. Mutation of a conserved active-site glutamate (E338Q) reduced kcat ∼200-fold at pH 7.5; however at higher pH, the E338Q displayed nearly wild-type activity. These data are consistent with Glu-338 (general base) activating the N-ε-lysine by deprotonation. Together the results suggest that MYST family HATs utilize a direct-attack mechanism within a ternary complex of Esa1•acetyl-CoA•histone.
Post-translational modification of histones is linked to a multitude of cellular processes including transcriptional regulation, DNA damage repair, and DNA replication (1-3). One prominent histone modification, N-ε-lysine acetylation, is dynamically controlled by the opposing actions of histone acetyltransferases (HATs1) and deacetylases (HDACs) (2). The two main families of HATs that comprise the Gcn5-related N-acetyltransferase (GNAT) superfamily are the Gcn5 family and the MYST (MOZ, Ybf2/Sas3, Sas2, Tip60) family (3-6). The Gcn5 family of HATs include Gcn5 and p/CAF which have well established sequential mechanisms of acetylation (7-10). Following the formation of a ternary complex between acetyl-CoA, histone and enzyme, an active-site base deprotonates lysine, allowing direct attack of the N-ε-lysine on the carbonyl carbon of acetyl-CoA. A direct acetyl-transfer mechanism contrasts with that recently proposed for the MYST family member Esa1 from S. cerevisiae (11).
The core structure of Esa1 displays significant similarity to that of the Gcn5 HAT family and accordingly, the first structural report proposed a catalytic mechanism analogous to that established for Gcn5-like HATs (7-10, 12-16). The most recent crystal structure of Esa1 truncated at residues 160−435 (Esa1160−435 ) revealed the presence of an acetylated cysteine residue (Cys-304) at the putative active site (11). This observation, and the fact that Cys-304 is invariant among MYST members led to the proposal that Esa1 and all MYST family HATs utilized a catalytic mechanism requiring the formation of a discrete acetyl-cysteine intermediate (4, 11). However, utilization of a poorly-active, truncated form of Esa1 and the structural similarity between Esa1 and Gcn5-like HATs raises significant questions about the catalytic mechanism.
Here, we investigate the catalytic mechanism using piccolo NuA4 (picNuA4), a trimeric complex composed of full-length Esa1, as well as two accessory proteins Epl1 and Yng2 that constitute the minimal core complex capable of efficient histone acetylation ((17, 18) and manuscript submitted). Previous biochemical analysis of a variety of Esa1 constructs indicated that catalytic activity is 100−1000-fold lower than picNuA4 (17). Therefore, we chose to examine this physiologically relevant enzyme complex, which displays similar catalytic activity to that observed for members of the Gcn5 HAT family (8-10). We demonstrate that Cys-304 is completely dispensable for substrate binding and catalysis, and provide evidence that Esa1 employs a direct-attack mechanism from a ternary complex. Moreover, we provide evidence that the conserved active-site Glu-338 deprotonates the N-ε-lysine of histone, facilitating the nucleophilic attack on the bound acetyl-CoA. This detailed functional analysis suggests that MYST family HATs utilize a direct-attack mechanism (sequential), similar to that established for the Gcn5 HAT family. These results have important implications for the rational design of mechanism-based inhibitors for the entire family of MYST HATs.
Experimental procedures
Chemicals and Reagents
DTT, Tris base, acetyl-CoA and other reagents were purchased from Sigma-Aldrich or Fisher, DTNB was purchased from Pierce, and Acrylamide/Bisacrylamide was purchased from Biorad. All reagents were of the highest quality and used without further purification.
Purification of picNuA4, Esa1, and mutants
Purification of enzymes was performed as described by Selleck, et al. (18). Bradford and activity assays were used to determine concentration of enzyme (19).
Histone acetyltransferase assays
Assays were performed as described in Berndsen and Denu (20). All reactions contained 50 mM Tris, pH 7.5 and were performed at 25 °C using a peptide corresponding to the 20 N-terminal residues of histone H4, SGRGKGGKGLGKGGAKRHRK. Peptide concentration was determined by amino acid analysis (Molecular Structure Facility at the University of California-Davis) and by BCA assay.
Experiments with propionyl-CoA as the substrate were performed using the DTNB assay. Reactions were performed in 50 mM Tris, pH 7.5 at 25 °C with 1 mM EDTA with 0.1 μM picNuA4, 10 to 100 μM propionyl-CoA and 36 μM to 550 μM H41−20. Samples of reactions were quenched in 10% SDS and 4 mg/mL DTNB in 50 mM Tris, pH 7.5 and 1 mM EDTA. Absorbance was measured in a 384-well plate using a Multiskan Ascent plate reader (Thermo Scientific, Waltham, MA).
pH Profiles
Assays to determine the rate of catalysis over a range of pH were performed as described above with 75 μM acetyl-CoA and 1 mM H41−20 in each reaction. The assays were buffered with either 50 mM Tris, 50 mM bis-Tris, and 100 mM sodium acetate or 50 mM Tris, 50 mM ethanolamine, and 100 mM ACES. These buffers have been shown to have constant ionic strength over a wide range of pH (21). The picNuA4 complex showed similar levels of activity in both buffers.
Mutagenesis
The C304A, C304S and E338Q mutations were introduced separately into the Esa1 gene in the pST50Trc3-yEsa1 plasmid using the QuikChange mutagenesis procedure. The entire Esa1 coding region was verified by sequencing before the Esa1 translational cassette was subcloned to create pST44 bacterial expression vectors that co-express HIS-tagged Epl1(51−380), Yng2(1−218) and Esa1 containing the point mutation (18, 23).
Data fitting
Data from bi-substrate analysis and inhibition assays were fitted to the equations (eqn 1 and 2, respectively) of Cleland in Kinetasyst (Intellikinetics, State College, PA) as described previously (8, 9). The pH rate data were fitted to equation 3 (22), where C is the pH independent rate, H is the concentration of protons, and Ka is the acid dissociation constant. All data were displayed using Kaleidagraph (Synergy Software, Reading, PA).
| (Eqn. 1) |
| (Eqn. 2) |
| (Eqn. 3) |
Mass spectrometry of peptides
Esa1 C304A and C304S mutants were confirmed by mass spectrometry. The picNuA4 complex was resolved on SDS-PAGE and the band corresponding to Esa1 was excised. Gel pieces were reduced, alkylated, and trypsin digested using standard protocols and samples were further processed for MALDI by Omix tip (Varian, Inc.). MALDI TOF/TOF was performed at the University of Wisconsin-Madison Biotechnology center on a 4800 MALDI TOF/TOF (Applied Biosystems, Foster City, CA).
Results
Steady-state kinetic analysis of picNuA4 suggests an ordered sequential mechanism
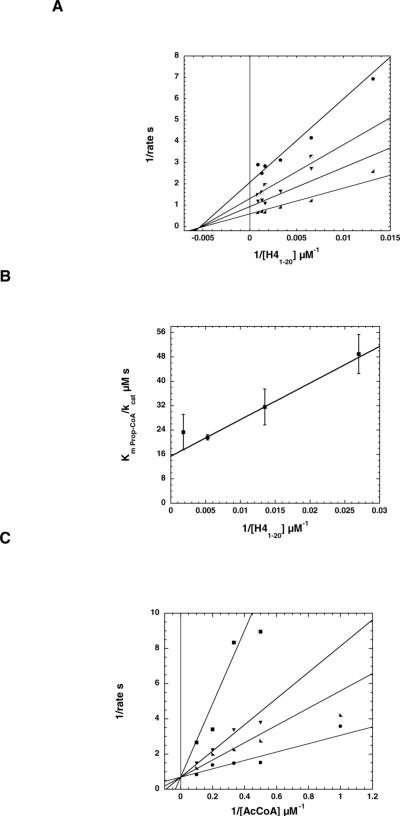
For the mechanistic investigation reported in this study, we utilized purified, recombinant piccolo trimeric complex picNuA4, containing the full-length catalytic subunit Esa1, and subunits Yng2 and Epl1, which is the minimal core enzyme capable of efficient nucleosomal acetylation (17, 18). Where appropriate, we also report results from full-length Esa1 that is not complexed with Yng2 and Epl1. To distinguish between direct-attack (sequential) and an obligate acetyl-cysteine enzyme intermediate (ping-pong) mechanisms, a steady-state kinetic analysis was performed. Initial rates of product formation were determined at varied concentrations of both substrates, acetyl-CoA and a histone H4 peptide (H41−20) corresponding to residues 1−20, and the rate data were plotted in double-reciprocal format (Lineweaver-Burk). Figure 1A shows a representative data set, demonstrating an intersecting line pattern, consistent with a sequential mechanism and the requirement for ternary complex formation prior to catalysis. The kcat value of 1.8 ± 0.6 s−1 was determined, with a Km for acetyl-CoA of 0.9 ± 0.3 μM (Kd= 1.2 ± 0.4 μM) and a Km for H4120 of 192 ± 63 μM (Kd = 251 ± 46 μM).
Figure 1. picNuA4 requires a ternary complex for histone acetylation.
(A) Double reciprocal plot from bi-substrate experiment. Acetyltransferase reaction conditions were 50 mM Tris, pH 7.5, 1 mM DTT, 0.1 μM picNuA4 with coupled assay conditions as described in Berndsen and Denu (20). Acetyl-CoA concentrations are 0.25 μM (circles), 0.5 μM (boxes), 1 μM (downward triangles), and 10 μM (right triangles) with H41−20 varied from 50 to 1800 μM. Data were fitted to equation 1 for a sequential mechanism in Kinetasyst and depicted in Kaleidagraph. Experiments were repeated in triplicate with representative experiment shown. (B)—Slope replot from bisubstrate experiment using propionyl-CoA and H41−20. DTNB assay conditions were 50 mM Tris, pH 7.5 with 1 mM EDTA with 0.1 μM picNuA4. Propionyl-CoA was varied from 10−104 μM with H41−20 varied from 36−550 μM. Curves were fitted to the Michaelis-Menten equation (v=Vm*[S]/(Km + [S])) in Kaleidagraph to determine kcat and kcat/Km. The 1/kcat/Km value with propionyl-CoA was then plotted versus 1/[H41−20]. Experiments were repeated in duplicate with average data ± standard deviation shown. (C)—Double reciprocal plot showing CoA inhibition when acetyl-CoA is varied at constant peptide concentration. Reaction conditions were 50 mM Tris, pH 7.5, 1 mM DTT, 0.03 μM picNuA4, and 500 μM H41−20. Acetyl-CoA was varied from 1 to 10 μM at 0 (circles), 2.5 (right triangles), 5 (downward triangles), and 18.2 (boxes) μM CoA. Data were fitted to equation 2 for competitive inhibition and depicted using Kaleidagraph. The Ki for CoA was determined to be 1.8 ± 0.8 μM. Experiments were repeated in duplicate with representative data shown.
To confirm the observed intersecting line pattern in the bi-substrate analysis, we utilized the approach of employing an alternate acyl-CoA substrate (higher Km), similar to that performed by Radika and Northrop, who determined a sequential mechanism for kanamycin acetyltransferase (24). Here proprionyl-CoA was used, revealing a higher Km (24 ± 11 μM) and Kd (130 ± 6 μM), compared to that of acetyl-CoA. The double reciprocal plot with varied H41−20 and proprionyl-CoA displayed a pronounced intersecting line pattern, demonstrating a strong effect of H41−20 concentration on the kcat/Km for propionyl-CoA. This dependence is represented in the re-plot of Km/kcat for propionyl-CoA vs. 1/[H41−20] in Figure 1B. The kcat for propionyl-CoA was 1.6 ± 0.3 s−1 with a kcat/Km of 6.5 ± 0.8 × 104 M−1 s−1, and for H41−20 the kcat/Km value was 5.5 ± 0.2 × 104 M−1 s−1.
A characteristic feature of the Gcn5-like HATs is the ability of the product CoA to act as a competitive inhibitor against the substrate acetyl-CoA (8, 9). Because acetyl-CoA is the first substrate to add, and CoA is the last product to leave in the kinetic mechanism, acetyl-CoA and CoA compete for the free enzyme form. We performed inhibition analysis using CoA as a product inhibitor of the picNuA4 reaction. A series of initial velocities were measured at saturating levels of H41−20, where acetyl-CoA concentrations were varied at different fixed levels of CoA. The resulting data shown in double reciprocal format (Fig 1C) produced an intersecting line pattern that converged on the y-axis, indicating competitive inhibition. The inhibition constant (Ki) for CoA was 1.8 ± 0.8 μM. Collectively, our steady state analyses are consistent with a sequential mechanism involving formation of a ternary complex (picNuA4•acetyl-CoA•H41−20) prior to any chemical step in the reaction mechanism.
Cysteine 304 does not mediate acetyl-transfer
To directly investigate the role of Cys-304 in picNuA4- and Esa1-catalyzed acetyl-transfer, we replaced Cys-304 with either alanine or serine in full-length Esa1 and within the trimeric picNuA4 complex. These amino-acid substitutions were confirmed by DNA sequencing of the expression plasmids and by mass spectrometry of the expressed, purified protein (Table 1). Mass spectral analysis of tryptic peptides revealed the correct mass of 2198 Da (2141 + alkylation) for wild type (ESADGYNVACILTLPQYQR), of 2109 Da for C304A (ESADGYNVAAILTLPQYQR), and of 2125 Da for C304S (ESADGYNVASILTLPQYQR). The picNuA4 mutants (C304A and C304S) were subjected to steady-state kinetic analysis to determine their effect on all kinetic parameters (Table 1). Strikingly, the C304A mutant of picNuA4 displayed kcat and kcat/Km values within 3-fold of those determined for wild-type picNuA4 (Table 1). The C304S mutation yielded a 10-fold lower kcat compared to that of wild-type picNuA4, although the Km values for both substrates were not appreciably affected. Together, these data indicate that Cys-304 does not play an essential catalytic role in the efficient histone acetylation mediated by picNuA4.
Table 1.
Kinetic Constants for piccolo NuA4 and catalytic mutants
| Enzyme | Peptide sequence by MS (Mass, Da) | kcat, s−1a | Km AcCoA, μMa | Km peptide, μMa | kcat/Km AcCoA, M−1 s−1 | kcat/Km peptide, M−1 s−1 |
|---|---|---|---|---|---|---|
| WT | ESADGYNVACiltlpqyqr (2141 + alkylation = 2198) | 1.6 ± 0.1 | 2.5 ± 0.3 | 216 ± 28 | 6.4 ± 0.9 × 105 | 7.4 ± 1.1 × 103 |
| C304A | ESADGYNVAAiltlpqyqr (2109) | 0.76 ± 0.1 | 1.5 ± 0.2 | 372 ± 27 | 5.1 ± 1.0 × 105 | 2.0 ± 0.3 × 103 |
| C304S | ESADGYNVASiltlpqyqr (2125) | 0.14 ± 0.02 | 2.0 ± 0.3 | 182 ± 34 | 7.0 ± 1.5 × 104 | 7.7 ± 1.8 × 102 |
| E338Q | 9.1 ± 1 × 10−3 | 1.2 ± 0.2 | 135 ± 10 | 7.6 ± 1.5 × 103 | 6.7 ± 0.9 × 101 |
Table Legend
All values are averages determined from duplicate or triplicate independent experiments shown ± standard deviation
To determine if the picNuA4 complex had a compensatory mechanism when Cys-304 was substituted, we determined the effect of the C304A substitution on Esa1 alone. The rate (apparent kcat) of peptide acetylation for Esa1 and the C304A mutant was 0.0019 ± 0.0001 s−1 and 0.0015 ± 0.0001 s−1, respectively (data not shown). These results are consistent with the lack of involvement of the Cys-304 sulfhydryl in catalysis by either Esa1 or picNuA4.
Acetylated piccolo NuA4 is not kinetically competent
To characterize the auto-acetylation of picNuA4, a variety of biochemical and kinetic experiments were performed. In the presence of [14C]-acetyl-CoA, all three proteins, Esa1, Yng2 and Epl1 were labeled with [14C]-acetate. Yng2 displayed the highest level of acetyl incorporation, followed by Esa1, then Epl1 (Supplemental Fig 1A). The incorporated acetate into picNuA4 could not be removed with heat, DTT, β-mercaptoethanol, or CoA, suggesting the formation of a stable linkage that is not consistent with an acetyl-cysteine (Supplemental Figure 1B and data not shown). Additional attempts to observe picNuA4-catalyzed acetyl-exchange between acetyl-CoA and a CoA analog, 3’-dephospho-CoA, yielded no significant exchange above background levels (data not shown). We next measured the rates of CoA formation in the absence of histone peptide, under either catalytic (nM) or non-catalytic (μM) amounts of picNuA4 (Supplemental Figure 1C and D). With catalytic levels of picNuA4, the rate of CoA formation is not significantly higher than that observed for the spontaneous hydrolysis of acetyl-CoA. With μM levels of picNuA4, the rate of CoA formed was 0.0021 ± 0.001 s−1. Similarly, the rate of picNuA4 auto-acetylation was 7 ± 0.2 × 10−4 s−1, or ∼2500-fold slower than histone acetylation (Supplemental Fig 1E).
Glutamate 338 deprotonates the substrate lysine
Having provided strong evidence that Esa1 and picNuA4 employ a direct-attack mechanism from a ternary complex, we next sought to identify the general base responsible for activating the attacking lysine nucleophile. In Gcn5, a conserved glutamate residue (Glu-173) was shown to function as a general base, deprotonating the lysine residue for direct attack on the carbonyl of bound acetyl-CoA (7, 12, 25, 26). Previously proposed mechanisms for acetyl-transfer by Esa1 implicated Glu-338 as critical for deprotonation of the substrate lysine (11, 12). We constructed an Esa1 E338Q mutant of picNuA4 and investigated its potential role as the general base.
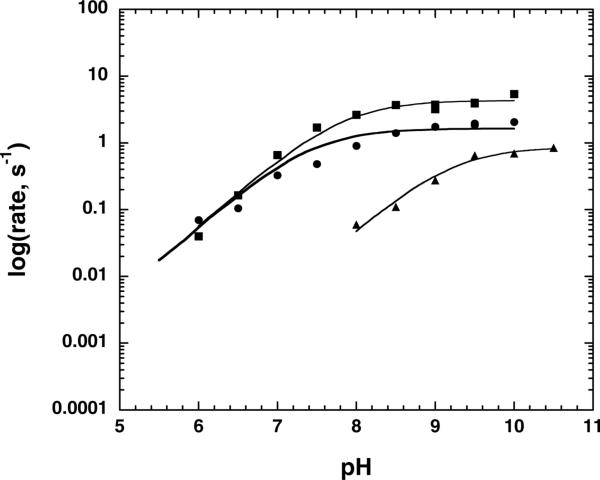
Steady-state kinetic analysis of the E338Q mutant of picNuA4 indicated that while substrate Km values for both substrates were similar between the E338Q mutant and wild-type complexes, the kcat of the E338Q mutant was reduced 175-fold at pH 7.5 (Table 1). We next determined the effect of pH on the kcat for wild-type, C304A, and E338Q picNuA4 complexes (Fig 2). The pH profiles for wild-type and C304A complexes were similar, with a pKa of 7.8 ± 0.1 and 7.6 ± 0.2, respectively, indicating the occurrence of a single ionization that must be unprotonated for catalysis. In contrast, the E338Q mutant displayed a much higher pKa value of 9.2 ± 0.1 for a single ionization that must be unprotonated for activity. As is evident in Figure 2, the kcat value of the E338Q mutant begins to approach that of the wild-type enzyme at very high pH values. The E338Q mutant data revealed a pH-independent rate of 0.8 ± 0.1 s−1, while those from the C304A and wild-type complexes yielded pH-independent values of 1.8 ± 0.2 s−1 and 3.9 ± 0.5 s−1, respectively. These results are consistent with a role for Glu-338 as general base. At physiological pH values, Glu-338 deprotonates the N-ε-lysine, however the requirement for Glu-338–catalyzed activation of lysine becomes unnecessary for efficient catalysis at higher pH because the lysine side chain is unprotonated (apparent pKa of 9.2) (27).
Figure 2. pH profile of picNuA4 and mutants.
Assays of picNuA4 activity were performed under saturating conditions for both substrates (75 μM acetyl-CoA, 1 mM H41−20). Reactions were performed in either 50 mM Tris, 50 mM bis-Tris, 100 mM sodium acetate or 50 mM Tris, 50 mM ethanolamine, 100 mM ACES buffer from pH 5.5 to 10.5. Data for the Wild-type enzyme are denoted with boxes, C304A with circles, and E338Q with triangles. Data were fitted to equation 3 and depicted using Kaleidagraph. The pKa value determined for the wild-type complex was determined to be 7.8 ± 0.1, for C304A 7.6 ± 0.2, and for E338Q 9.2 ± 0.1. Experiments were repeated in duplicate or triplicate with representative curves for each enzyme shown.
Discussion
The catalytic mechanism for the MYST family of HATs has been unresolved (11, 12). Esa1 is used as the prototypical enzyme for understanding the structure and mechanism of the MYST family. Based primarily on structural similarity to the Gcn5 family of HATs, initial structure determination of the truncated Esa1160−435 enzyme suggested a sequential mechanism of acetyl-transfer (12). However, a subsequent X-ray structure of the same truncated form showed an acetylated cysteine near the active site, serving as primary evidence for the proposal that an acetyl-Cys-304 intermediate is required for catalysis (11). Here, we provide detailed functional data demonstrating that the mechanism of Esa1 and picNuA4 does not involve the formation of an acetyl-cysteine-304 intermediate. Instead, the results fully support a ternary complex mechanism involving a direct attack of a deprotonated N-ε-lysine on the bound acetyl-CoA. Like the Gcn5 HATs, the conserved Glu-338 of Esa1 functions to abstract a proton from lysine to promote the nucleophilic attack on the acetyl carbonyl-carbon of acetyl-CoA.
In a previous kinetic bi-substrate analysis, Yan, et al.reported a parallel line pattern when Esa1160−435 was employed and acetyl-CoA was the acyl donor (11). The exact nature of this discrepancy is unclear, however, the low rates of acetylation by the truncated Esa1160−435 (∼60-fold lower in kcat/Km for peptide) and the narrow range in substrate concentrations used in the previous study may have contributed to this apparent difference and to an assignment of a ping-pong mechanism (11). In the present study, we observe intersecting line patterns using both acetyl-CoA and proprionyl-CoA as a varied substrate. It is likely that the low Km and Kd for acetyl-CoA and the technical limitations this presents for the HAT assays may contribute to more subtle intersection line patterns with acetyl-CoA. This issue is alleviated with a poorer (higher Km) substrate such as propionyl-CoA (Fig 1B), which displayed a pronounced effect on the apparent kcat/Km for propionyl-CoA at varied peptide concentrations (Fig 1B). Interestingly, the kcat using propionyl-CoA (1.6 s−1) was nearly the same as that for acetyl-CoA (1.8 s−1), suggesting that the rate-limiting step is similar at saturating substrate concentrations. The ability of CoA to competitively inhibit acetyl-CoA provides additional evidence for a sequential mechanism in which acetyl-CoA is the first substrate to bind and CoA is the last product released. In a classical ping-pong mechanism, acetyl-CoA binds first, followed by acetyl-transfer to an enzyme nucleophile (28). CoA must leave before the peptide lysine binds and reacts with acetyl-enzyme intermediate, thereby releasing acetylated peptide in the last step. In such a scenario, CoA should display noncompetitive inhibition, which was not observed in our study.
Moreover, if the MYST family HATs utilized an obligate acetyl-enzyme intermediate, there are several predictable outcomes upon reaction of enzyme with acetyl-CoA. These include the kinetic competence of the acetylated enzyme and the corresponding formation of CoA in the absence of peptide acceptor (27, 29). While Esa1 is capable of auto-acetylation, this acetylation rate in the absence of peptide was over 2500-fold slower than the steady- state rate of acetylation (7 × 10−4 s−1 vs. 1.6 s−1), indicating that the observed acetylation cannot serve as a catalytically competent species (Supplemental Figure 1E). Furthermore, the auto-acetylation of picNuA4 was not reversible by addition of CoA, suggesting that this inter- or intra-molecular acetylation is occurring on lysine residues (Supplemental Fig 1B). Lastly, the rate of CoA formation (0.0021 s−1) upon reaction of high levels of picNuA4 and acetyl-CoA does not account for a kinetically competent rate (Supplemental Fig 1C). Together these results argue against an acetylated-enzyme intermediate involved in the catalytic mechanism.
Contrary to a previously proposed catalytic mechanism, Cys-304 of Esa1 does not function as the nucleophile in a covalent catalysis-type mechanism (11). A complete steady-state analysis of the C304A mutant of picNuA4 revealed that an alanine substitution at this site leads to an almost insignificant change in all kinetic parameters (Table 1). Similarly, the C304A mutant in Esa1 alone displayed no significant change in maximum rate of acetylation, compared to full-length wild-type Esa1. Lack of an effect of the C304A substitution suggests that both Esa1 alone and picNuA4 utilize the same catalytic mechanism, one that does not require an acetyl-Cys-304 intermediate. Yan, et al. (2002) reported that the C304A mutation in the truncated Esa1160−435 resulted in a protein with no detectable activity (11). Given our recent finding that Epl1 and Yng2 function to stabilize full-length Esa1 in the picNuA4 complex (manuscript submitted), it appears likely that the C304A mutation may further destabilize Esa1160−435, generating an enzyme with background levels of activity (11). The slight reduction in turnover of the C304S complex may be due to this enzyme destabilization. The crystal structure of Esa1160−435 with a C304S substitution shows significant structural changes to the loop between α2 and β7, residues 257−271 (PDB ID 1MJ9)(11). Mutations in this loop reduce HAT activity, indicating that structural alterations to this loop impact the efficiency of catalysis (30). The observation of an acetyl-Cys-304 in the most recent Esa1160−435 crystal structure may have resulted from non-catalyzed reaction of this sulfhydryl with the acetyl-CoA added during crystallization (11). Cysteine reactivity is well established and non-enzymatic acylation of cysteine residues within peptides by acyl-CoAs has been observed previously (31, 32).
Probing the catalytic role of conserved glu-338 provides further insight into the mechanism utilized by Esa1. Previously, this residue was proposed to deprotonate Cys-304 and the substrate lysine (11). Mutation of this residue to glutamine reduced turnover by nearly 200-fold at pH 7.5, while the Km values for both substrates were essentially unchanged (Table 1). However, the E338Q mutant approached near wild-type kcat values (Fig 2) at higher pH, similar to the behavior observed previously for the general base mutant E173Q of the Gcn5 HAT (7). At most physiological pH values, Glu-338 functions as the critical general base by deprotonating the substrate lysine and enhancing catalysis ∼200-fold. In the E338Q mutant, restoration of activity occurs as the N-ε-lysine (pKa ∼10) becomes unprotonated at higher pH, relinquishing the need for general-base catalysis. The pH profiles of both wild-type and the C304A mutant reveal a critical ionization that must be unprotonated for catalysis. Consistent with the general base role of Glu-338, this ionization (pKa = 7.6−7.8) likely represents the hydrogen-bonded ion pair between Glu-338 and the N-ε-lysine within the ternary enzyme complex. When this carboxylate is removed in the E338Q mutant, the turnover rate is now dictated by the ionization of the substrate lysine, leading to the higher apparent pKa of 9.2 (Fig 2).
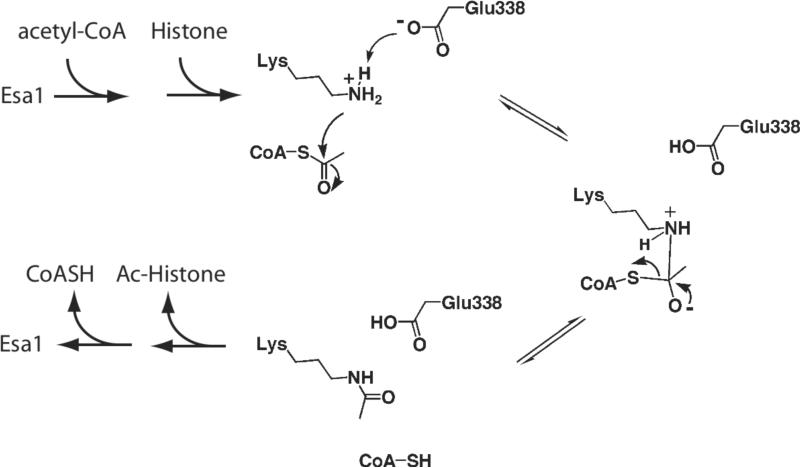
From the detailed biochemical analysis presented here, we propose the following mechanism for Esa1 and picNuA4-catalyzed acetyl-transfer to histones. After ternary complex formation, Glu-338 abstracts a proton from the substrate lysine, allowing for direct attack on the acetyl-moiety of acetyl-CoA (Fig 3). A putative tetrahedral intermediate is formed, which collapses to release CoA and acetylated substrate (Fig 3). These findings reconcile the previously-held conclusion that MYST HATs might employ a different catalytic mechanism than other members of the GNAT superfamily (11). Instead, the present study supports the notion that, despite the disparities in primary structure and substrate specificity between the diverse members of transferases, GNAT superfamily enzymes use an evolutionarily conserved mechanism of N-acyl-transfer (4).
Figure 3. Direct attack mechanism for acetyl-transfer by Esa1.
After binding acetyl-CoA and peptide substrate to form a ternary complex, Glutamate 338 of Esa1 deprotonates the ε-amine of lysine in the substrate. Lysine attacks the carbonyl carbon of the acetyl moiety of acetyl-CoA forming a tetrahedral intermediate, which then collapses to form CoA-SH and acetylated product.
Supplementary Material
Acknowledgements
We wish to thank Dr. Alvan Hengge and members of the Denu lab for helpful comments and assistance especially Dr. Suzi Lee and Brian Smith. Additionally, we wish to thank Jim Brown for assistance with analysis of MALDI spectra.
Footnotes
This work was supported in part NIH Grant GM059785 to JMD and NIH Grant GM064089 to ST
Abbreviations used in this work: HAT, Histone Acetyltransferase; DTT, dithiothreitol; GNAT, Gcn5 related N-acetyltransferase; MYST, MOZ, Ybf2/SAS3, SAS2, Tip60; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; picNuA4, piccolo NuA4 histone acetyltransferase complex; MALDI TOF/TOF, matrix assisted laser desorption ionization time-of-flight/time-of-flight mass spectrometry; AcCoA, acetyl-CoA; HDAC, Histone Deacetylase
Supplemental Information
Supplemental information containing data on the characterization of picNuA4 auto-acetylation available online at http://pubs.acs.org.
References
- 1.Doyon Y, Cote J. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev. 2004;14:147–54. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 3.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–59. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vetting MW, LP S. d. C., Yu M, Hegde SS, Magnet S, Roderick SL, Blanchard JS. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys. 2005;433:212–26. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Marmorstein R. Structure of histone acetyltransferases. J Mol Biol. 2001;311:433–44. doi: 10.1006/jmbi.2001.4859. [DOI] [PubMed] [Google Scholar]
- 6.Marmorstein R. Structure and function of histone acetyltransferases. Cell Mol Life Sci. 2001;58:693–703. doi: 10.1007/PL00000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanner KG, Trievel RC, Kuo MH, Howard RM, Berger SL, Allis CD, Marmorstein R, Denu JM. Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J Biol Chem. 1999;274:18157–60. doi: 10.1074/jbc.274.26.18157. [DOI] [PubMed] [Google Scholar]
- 8.Tanner KG, Langer MR, Kim Y, Denu JM. Kinetic mechanism of the histone acetyltransferase GCN5 from yeast. J Biol Chem. 2000;275:22048–55. doi: 10.1074/jbc.M002893200. [DOI] [PubMed] [Google Scholar]
- 9.Tanner KG, Langer MR, Denu JM. Kinetic mechanism of human histone acetyltransferase P/CAF. Biochemistry. 2000;39:15652. doi: 10.1021/bi005121q. [DOI] [PubMed] [Google Scholar]
- 10.Lau OD, Courtney AD, Vassilev A, Marzilli LA, Cotter RJ, Nakatani Y, Cole PA. p300/CBP-associated factor histone acetyltransferase processing of a peptide substrate. Kinetic analysis of the catalytic mechanism. J Biol Chem. 2000;275:21953–9. doi: 10.1074/jbc.M003219200. [DOI] [PubMed] [Google Scholar]
- 11.Yan Y, Harper S, Speicher DW, Marmorstein R. The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate. Nat Struct Biol. 2002;9:862–9. doi: 10.1038/nsb849. [DOI] [PubMed] [Google Scholar]
- 12.Yan Y, Barlev NA, Haley RH, Berger SL, Marmorstein R. Crystal structure of yeast Esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases. Mol Cell. 2000;6:1195–205. doi: 10.1016/s1097-2765(00)00116-7. [DOI] [PubMed] [Google Scholar]
- 13.Smith ER, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook RG, Lucchesi JC, Allis CD. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci U S A. 1998;95:3561–5. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarke AS, Lowell JE, Jacobson SJ, Pillus L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol. 1999;19:2515–26. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–5. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- 16.Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–96. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boudreault AA, Cronier D, Selleck W, Lacoste N, Utley RT, Allard S, Savard J, Lane WS, Tan S, Cote J. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 2003;17:1415–28. doi: 10.1101/gad.1056603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selleck W, Fortin I, Sermwittayawong D, Cote J, Tan S. The Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the Enhancer of Polycomb A domain and chromodomain to acetylate nucleosomes. Mol Cell Biol. 2005;25:5535–42. doi: 10.1128/MCB.25.13.5535-5542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 20.Berndsen CE, Denu JM. Assays for mechanistic investigations of protein/histone acetyltransferases. Methods. 2005;36:321–31. doi: 10.1016/j.ymeth.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 21.Ellis KJ, Morrison JF. Buffers of constant ionic strength for studying pH-dependent processes. Methods Enzymol. 1982;87:405–26. doi: 10.1016/s0076-6879(82)87025-0. [DOI] [PubMed] [Google Scholar]
- 22.Cleland WW. The use of pH studies to determine chemical mechanisms of enzyme-catalyzed reactions. Methods Enzymol. 1982;87:390–405. doi: 10.1016/s0076-6879(82)87024-9. [DOI] [PubMed] [Google Scholar]
- 23.Tan S, Kern RC, Selleck W. The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expr Purif. 2005;40:385–95. doi: 10.1016/j.pep.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Radika K, Northrop DB. The kinetic mechanism of kanamycin acetyltransferase derived from the use of alternative antibiotics and coenzymes. J Biol Chem. 1984;259:12543–6. [PubMed] [Google Scholar]
- 25.Rojas JR, Trievel RC, Zhou J, Mo Y, Li X, Berger SL, Allis CD, Marmorstein R. Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature. 1999;401:93–8. doi: 10.1038/43487. [DOI] [PubMed] [Google Scholar]
- 26.Clements A, Rojas JR, Trievel RC, Wang L, Berger SL, Marmorstein R. Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A. Embo J. 1999;18:3521–32. doi: 10.1093/emboj/18.13.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fersht A. Structure and mechanism in protein science: a guide to enzyme catalysis and protein folding. W.H. Freeman; New York: 1999. pp. 170, 216–242. [Google Scholar]
- 28.Segel IH. Enzyme kinetics: behavior and analysis of rapid equilibrium and steady-state enzyme systems. Wiley; New York: 1975. pp. 653–656. [Google Scholar]
- 29.Purich DL. Covalent enzyme-substrate compounds: detection and catalytic competence. Methods Enzymol. 2002;354:1–27. doi: 10.1016/s0076-6879(02)54003-9. [DOI] [PubMed] [Google Scholar]
- 30.Adachi N, Kimura A, Horikoshi M. A conserved motif common to the histone acetyltransferase Esa1 and the histone deacetylase Rpd3. J Biol Chem. 2002;277:35688–95. doi: 10.1074/jbc.M204640200. [DOI] [PubMed] [Google Scholar]
- 31.Quesnel S, Silvius JR. Cysteine-containing peptide sequences exhibit facile uncatalyzed transacylation and acyl-CoA-dependent acylation at the lipid bilayer interface. Biochemistry. 1994;33:13340–8. doi: 10.1021/bi00249a021. [DOI] [PubMed] [Google Scholar]
- 32.Sang SL, Silvius JR. Novel thioester reagents afford efficient and specific S-acylation of unprotected peptides under mild conditions in aqueous solution. J Pept Res. 2005;66:169–80. doi: 10.1111/j.1399-3011.2005.00285.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.