Abstract
Expression of the smooth muscle α-actin gene in growth-activated vascular smooth muscle cells and stromal fibroblasts is negatively regulated by members of the Pur family of single-stranded DNA/RNA-binding proteins. In particular, Purα and Purβ are postulated to repress transcription by forming helix-destabilizing complexes with the sense strand of an asymmetric polypurine-polypyrimidine tract containing a canonical MCAT enhancer motif in the 5′ region of the gene. Herein, we establish the mechanism of Purβ binding to the purine-rich strand of the enhancer using quantitative methods and purified components. Initial evaluation of DNA-binding specificity and equilibrium stoichiometry via colorimetric-, autoradiographic-, and fluorescence-based assays suggested that Purβ interacts with two distinct G/A-rich sites within the nominal single-stranded enhancer element to form a high affinity 2:1 protein:DNA complex. Statistical mechanical analyses of band shift titrations of the nominal element in conjunction with DNase I footprint titrations of the extended smooth muscle α-actin 5′-flanking region demonstrated that assembly of the nucleoprotein complex likely occurs in a sequential, cooperative, and monomer-dependent fashion. Resolution of the microscopic energetics of the system indicated that monomer association with two non-identical sites flanking the core MCAT motif accounts for the majority of the intrinsic binding affinity of Purβ with intersite cooperativity contributing a ~12-fold increase to the stability of the nucleoprotein complex. These findings offer new insights into the mechanism, energetics, and sequence determinants of Purβ repressor binding to a biologically relevant, contractile phenotype-regulating cis-element while also revealing the thermodynamic confines of putative Purβ-mediated effects on DNA structure.
Textbook models of the transcription of eukaryotic protein-coding genes typically depict the recruitment of the RNA polymerase II machinery to a core promoter as coordinated by the pre-assembly of trans-acting factors at specific target sites in the 5′-flanking region of a gene in a manner dictated by nucleotide sequence and DNA accessibility. By-and-large, this classical view is predicated on the principle that recognition of specific DNA sequences by transcription factors involves chemical interactions between certain base-pairs oriented in the major or minor groove of duplex DNA and compatible amino acids arrayed on the surface of the protein. However, this conventional model of B-DNA-dependent activation or repression of transcription does not fully explain the regulatory intricacies of genes which harbor promoter elements that are prone to adopting non-B-DNA structures including single-stranded DNA (ssDNA)1 (1–6). Of particular interest from a thermodynamic perspective, is how sequence-specific ssDNA-binding proteins (ssBPs) participate in the formation and/or stabilization of these alternate structures to positively or negatively affect gene activity (7–12).
Among the relatively small group of ssDNA-binding transcription factors indentified to date, two members of the purine-rich element (PUR) binding protein family, Purα and Purβ, have been linked to the repression of genes encoding smooth muscle α-actin (SMαA) (13), α-myosin heavy chain (14), and β-myosin heavy chain (15). Consequently, these ssBPs likely serve as downstream effectors of muscle gene reprogramming induced by physiological stress or injury in cardiac, skeletal, or vascular cell types (14–18). In the case of the SMαA gene, Purα and Purβ appear to collaborate with another ssBP known as MSY1 (mouse YB-1) to suppress the activity of a highly-conserved muscle-CAT (MCAT)-containing enhancer in growth-activated fibroblasts and vascular smooth muscle cells (13). In keeping with the general composition and properties of other ssBP recognition sites, the SMαA enhancer possesses a high level of purine-pyrimidine asymmetry (19), and lies within a region of the promoter which is hypersensitive to ssDNA-specific chemical modification (20). An ensemble of biochemical studies conducted with fibroblast-derived, smooth muscle cell-derived, and recombinant proteins have established that Purα and Purβ specifically bind to the purine-rich strand, whereas MSY1 interacts with the opposing pyrimidine-rich strand of the nominal MCAT enhancer element (13, 21). On the basis of these and other nucleoprotein binding and promoter mutation analyses, a working model for enhancer encryption was proposed envisioning concurrent binding by all three ssBPs in such a way as to inhibit MCAT-dependent activation by transcription enhancer factor 1 (13, 19). Although this model assumed equal contribution by each ssBP, more recent gain-of-function, loss-of-function, and in vivo promoter occupancy studies have suggested that Purβ may be the dominant physical and functional effector of SMαA repression in certain cell types (22–24).
As a logical extension of prior work showing that 1) fibroblast-derived Purα and Purβ form deoxycholate-sensitive oligomeric complexes on the purine-rich strand of the MCAT-containing enhancer (21), 2) nucleotides near the 5′ and 3′ ends of the nominal enhancer element mediate high affinity binding by nuclear Purα and Purβ extracted from fibroblasts and vascular smooth muscle cells (13), and 3) purified recombinant Purβ reversibly self-associates in the absence of ssDNA (25), we hypothesized that dimerization may be a prerequisite to bidentate Pur protein interaction with two independent recognition sites flanking the core MCAT motif. In this study, we employed rigorous, quantitative methods to validate these putative recognition sites and to determine the mechanism of ssDNA-binding by Purβ. In contrast to obligate dimer-dependent association, our results indicate that protein binding to the purine-rich strand of the SMαA enhancer involves cooperative assembly of Purβ monomers at two slightly degenerate PUR sites to form a high affinity 2:1 protein:ssDNA complex. Moreover, the resolved energetics of strand- specific enhancer binding by Purβ suggests that formation and maintenance of an extended single-stranded state in the genomic SMαA promoter likely requires auxiliary factors, such as Purα and/or MSY1, to overcome the thermodynamic favorability of strand annealing.
EXPERIMENTAL PROCEDURES
Chemicals, recombinant Purβ protein, and oligonucleotide probes
All chemicals used in this study were of reagent grade or better. Recombinant Purβ was expressed as an amino-terminal hexahistidine-tagged fusion protein (referred to in this report as N-HisPurβ), purified from E. coli expression cultures, and quantified as described previously (25). All biotinylated, fluoresceinated, and non-labeled synthetic oligonucleotides were obtained from Sigma-Genosys.
Colorimetric ssDNA-Binding Immunoassay
The binding of recombinant Purβ to solid-phase biotinylated ssDNA probe (5′-GGGAGCAGAACAGAGGAATGCAGTGGAAGAGA-3′-biotin, dubbed PE32-bF) in the presence of selected oligonucleotide competitors was monitored by enzyme-linked immunosorbent assay (ELISA) as previously described (22) with modifications as detailed in Supporting Information. To facilitate the quantitative analysis of colorimetric datasets, wells containing the highest concentration of PE32-F self-competitor tested were used to determine minimum absorbance values (Amin), whereas wells with no competitor were used to obtain maximum absorbance values (Amax). These controls were included on each plate to permit normalization of absorbance values necessary for comparison of results from different plates. Normalized absorbance values (ANorm) were calculated using the expression ANorm,i = (Ai–Amin)/(Amax–Amin), where Ai is the absorbance at 405 nm of well i. Determinations of competitor concentrations necessary for 50% inhibition of complex formation, IC50, were performed by nonlinear least-squares fitting of data points to the following expression:
| (1) |
where αH is the Hill coefficient which permits variability of the slope of the transition. Data analysis was performed using Prism 5 software (GraphPad Software, Inc.).
Preparation of ssDNA for Quantitative DNase I footprinting
Methods for the synthesis and purification of ssDNA were adapted from established protocols (26, 27). A plasmid vector containing the full-length mouse SMαA promoter known as VSMP8 (28), was used as a template for polymerase chain reaction (PCR)-based generation of a 382 nucleotide probe (designated SMP382-F) corresponding to the forward or sense strand of the 5′ flanking region (−323 to +59 relative to the transcription start site). PCR primers were designed such that the reverse strand primer was biotinylated on the 5′ end (5′-biotin- GGCTACTTACCCTGACAGCGACT-3′ designated SMP8p1122s-R-5btn), whereas the forward strand primer was unmodified (5′-TTCTGAGGAATGTGCAAACCGTG-3′ designated SMP8p741s–F). PCR amplification of the 382 base-pair fragment from 1 ng/µL of VSMP8 template was carried out using Accuprime Supermix™ reagent (Invitrogen Corp.) according to the manufacturer’s instructions. Isolation of single-stranded SMP382-F was achieved through implementation of a biotin-streptavidin affinity based technique. Briefly, the double-stranded PCR product (typically 500 µL) was combined with 1 mg of Streptavidin MagneSphere® paramagnetic particles (Promega Corp.), pre-equilibrated in 20 mM Tris-HCl pH 8.8, and incubated at 4°C for 16 h. The PCR product-paramagnetic particle complex was washed twice with 20 mM Tris-HCl pH 8.8 and non-biotinylated SMP382-F was eluted by incubating the complex in 0.2 N NaOH for 5 min at 20±1°C. The eluate was then neutralized by the addition of 1/10 volume of 5 M ammonium acetate and SMP382-F was precipitated by the addition of 2 volumes of isopropanol at −20°C, washed with 70% v/v ethanol, and redissolved in nuclease-free water. The concentration of SMP382-F was approximated by absorbance measurement at 260 nm, assuming an extinction coefficient of 3,630,200 M−1cm−1 calculated using web-based software (http://biophysics.idtdna.com) (29, 30).
End-labeling of Oligonucleotides with [γ32-P]ATP
Oligonucleotides used in band shift assays (PE32-F), DNase I footprinting assays (SMP382-F), and dideoxy sequencing reactions (SMP8p741s-F) were labeled on their 5′ termini with 6000 Ci/mmole [γ32-P]ATP (Perkin-Elmer) using T4 polynucleotide kinase (New England Biolabs) as directed by the manufacturer. Reactions were performed at 37°C for 90 min. Upon completion, the enzyme was heat-inactivated by incubating reaction mixtures at 70°C for 10 min. Unincorporated [γ32- P]ATP was removed by buffer exchange over a G-25 Microspin column (Pharmacia). Specific activity was determined by scintillation counting of purified probes using a Perkin-Elmer Tri-Carb® scintillation counter. For purposes of clarity, oligonucleotides carrying a 5′-32PO4 radiolabel are marked with a “*” (e.g. PE32-F* denotes 5′-32PO4-labeled PE32-F).
Steady-State Fluorescence Anisotropy
A synthetic, 3′-fluorescein-labeled ssDNA probe (5′-GGGAGCAGAACAGAGGAATGCAGTGGAAGAGA-3′-fluorescein, dubbed PE32-F-3FLC) was combined with selected concentrations of N-HisPurβ in buffer consisting of 50 mM HEPES pH 7.8, 300 mM NaCl, 1.5 mM MgCl2, 2 mM β-mercaptoethanol, and allowed to equilibrate for approximately 16 h at 20±1 °C in the dark. Steady-state fluorescence anisotropy measurements were made on a Quantamaster-6 spectrofluorometer (Photon Technologies International) equipped with a 75 W xenon arc lamp excitation source, excitation and emission monochromators, and automatic excitation and emission polarizers in a T-format. Slit-widths were held at 5 nm. Parallel and perpendicular emission intensities were collected with horizontally polarized exciting light in order to first calculate the instrument correction factor, G, given by G = IHV/IHH where IHV and IHH are the intensities measured through the vertical and horizontal polarizers when excited with horizontally polarized light. Observed anisotropy values, robs, were calculated by the expression robs = (IVH – GIVH)/(IVV+ 2GIVH), where IVV and IVH are the intensities measured through the vertical and horizontal polarizers when excited with vertically polarized light. Changes in emission intensity as a function of protein concentration were not observed. Anistropy data were mathematically analyzed to estimate nucleoprotein complex stoichiometry as detailed in Supporting Information.
Electrophoretic Mobility Shift Assay (EMSA)
All binding reactions were carried in EMSA binding buffer consisting of 50 mM Tris-HCl pH 7.5, 100 mM KCl, 5% (v/v) glycerol, 0.5 mM dithiothreitol, 2 µg/mL dT32, 50 µg/mL BSA and allowed to equilibrate for 16 h at 20 ± 1 °C prior to non-denaturing electrophoresis. Complexes were resolved on a 1.5 mm thick 10% polyacrylamide gel (75:1 acrylamide:bisacrylamide) cast in running buffer consisting of 25 mM Tris, 25 mM boric acid, and 0.5 mM EDTA (TBE). Gels were pre-run at 4 W per gel for 1 h prior to loading and running for 45–75 min (depending upon application) at 4 W at room temperature. Upon completion, gels were disassembled and dried on Whatman filter paper in a Bio-Rad slab gel dryer for 45 min at 65°C. Dried gels were exposed to phosphor storage screens (Molecular Dynamics) for 24–48 h prior to digital image acquisition with a Bio-Rad PersonalFX™ phosphorimager. In certain cases, radioactive bands were visualized by exposure of dried gels to X-Omat film (Kodak) for 6–16 h at –80°C prior to development.
Quantitative Titration EMSA
A direct titration approach was used to visualize nucleoprotein complex assembly, to estimate macroscopic binding affinities, and to assess cooperativity of N-HisPurβ binding to PE32-F*. Briefly, 2× solutions of N-HisPurβ were prepared in EMSA binding buffer by 2/3-fold serial dilution of a 20 nM master stock solution. An equal volume of 2× PE32-F* in EMSA binding buffer was added to N-HisPurβ solutions so that final concentrations of each were at 1× with PE32-F* held constant across all binding reactions. In order to maintain validity of the assumption [Pfree] ≈ [Ptotal], PE32-F* concentration was kept at 25 pM for EMSAs used for rigorous thermodynamic interrogation. Free and bound probe was separated by non-denaturing electrophoresis as described above, with 5–10 µL of reaction mixture (usually 700 – 2000 cpm) loaded in each lane. Dried gels were exposed 72–96 h to phosphor storage screens. Quantification of binding results was carried out by measuring the optical density of each electrophoretic species using ImageQuant 5.2 software (Molecular Dynamics). Species density values were then used to determine the fraction of each species, Θi, where i is equal to the number of protein ligands bound to the ssDNA lattice (i = 0, 1, or 2 for a system with a finite stoichiometry of 2:1) by applying the following expression:
| (2) |
where Ii refers to the integrated optical density of the ith species, and the summation is over all of the bands in a particular lane of the gel. Data analysis was carried out by a statistical mechanical method assuming a general model involving two interacting DNA-binding sites and a single protein ligand (31) as detailed in Supporting Information. As an alternative approach to gauge the existence of cooperative interactivity in nucleoprotein complex formation, complete fractional saturation, Ȳ, was calculated by the relationship:
| (3) |
and then fit to the phenomenological Hill equation (32):
| (4) |
where αH is the Hill coefficient and Kd is the macroscopic dissociation constant (Kd = Ka−1). In both data analysis approaches, nonlinear least-squares fitting of datasets was performed using Prism 5 software (GraphPad Software, Inc.). Goodness of fit was assessed by visual inspection of residuals and by monitoring of fitting statistics.
Serial-dilution EMSA
The strategy used for the estimation of nucleoprotein stoichiometry by EMSA was based on the serial-dilution method (33). Briefly, solutions of N-HisPurβ (5.0 nM) and PE32-F* (1.0 nM) were serially-diluted 1.1:1 fold in EMSA binding buffer. The redistribution of nucleoprotein complex was then monitored by non-denaturing electrophoresis as described above with run times limited to 45 min to prevent streaking due to complex dissociation. Acquisition and analysis of densitometry data was done as described in (34) and as elaborated upon in Supporting Information.
Quantitative DNase I ssDNA Footprinting
To monitor the binding of N-HisPurβ to individual sites on the purine-rich strand of the SMαA enhancer element, quantitative DNase I footprinting was performed based on methods developed by Ackers and coworkers (35–38) with the following modifications. Each reaction contained 20,000 cpm of freshly labeled SMP382-F* template at a final concentration estimated to be well below N-HisPurβ binding affinity to maintain validity of the [Pfree] ≈ [Ptotal] assumption (< 25 pM). N-HisPurβ was added to each reaction at selected concentrations to cover a range from approximately 10−13 to10−8 M in a final volume of 200 µL. All binding reactions were carried out in buffer containing 50 mM Tris-HCl pH 7.5, 100 mM KCl, 1 mM dithiothreitol, 1 mM CaCl2, 2.5 mM MgCl2, 2 µg/mL dT32, 50 µg/mL BSA at 20 ± 1°C and allowed to equilibrate for 16 h. Template digestion was initiated by the addition of 5 µL of DNase I (Invitrogen Corp.) in assay buffer to a final concentration of 1.0 unit/mL. After a 2 min incubation at 20 ± 1°C, digestion was halted by the addition of 700 µL stop solution consisting of 97% v/v ethanol, 0.57 M ammonium acetate, and 50 µg/mL yeast tRNA (Sigma-Aldrich). Samples were incubated in a dry ice-ethanol bath for 30 min. Precipitated DNA was collected by centrifugation, washed twice with 70% v/v ethanol, and air dried. Pellets were dissolved in 5 µL of loading buffer containing 98% v/v formamide, 10 mM EDTA, 0.1% w/v bromophenol blue, and 0.1% w/v xylene cyanol. Samples were heated at 95°C for 5 min prior to electrophoresis for 130 min at 65 W on a 0.4 mm thick sequencing gel consisting of 8% polyacrylamide (29:1 acrylamide:bisacrylamide) and 8 M urea. Gels were cast and pre-run in TBE buffer at 65 W for at least 2 h or until a gel temperature of ≥50°C was attained prior to sample loading. End-labeled primer dideoxy-NTP sequencing reactions performed with double-stranded SMP382 PCR product as a template and SMP8p741s-F* as the extension primer were co-electrophoresed to identify sequences of interest on the resulting footprints. Ladders were generated using a Sequenase dideoxy-NTP kit (USB Corp.) as directed by the manufacturer. Typically, 10,000–15,000 cpm per sequencing reaction was loaded on the gels. After electrophoresis, gels were dried in a Bio-Rad slab gel dryer on Whatman filter paper at 75°C for 45 min. Dried gels were exposed to phosphor storage screens for 72–96 h and imaged as described for EMSA.
Densitometric analysis of phosphorimages was performed using ImageQuant 5.2 software (Molecular Dynamics) in order to determine values for fractional protection (Fp) of a given sequence using the following equation:
| (5) |
where I is the relative densitometric intensity, q refers to any lane of the gel with finite N-HisPurβ concentration, r refers to the reference lane containing no protein, site refers to the ssDNA binding site in question, and control refers to a region of the gel whose intensity is independent of protein ligand concentration (bases −208 to −201 of SMP382-F*). First line, crude analysis of fractional protection data was carried out by fitting data to the Langmuir isotherm:
| (6) |
where k refers to the microscopic association constant (assuming no interaction between sites), u and l refer to the upper and lower endpoints, respectively. This was done to determine the upper and lower endpoints of fractional protection since binding of protein ligand at specific sites, even at saturating conditions, does not provide complete protection. Fractional protection values were then converted to values of fractional saturation, Ȳ, using the following expression:
| (7) |
Resolution of Binding Mechanism and Microscopic Free Energies of Association
Individual binding site expressions were derived using a statistical thermodynamics approach that has been described previously (35). Briefly, the probability of the enhancer element existing in any one of several possible microscopic configurations, ƒs, can be expressed by the equation:
| (8) |
where AGs is the relative free energy change observed upon formation of the s configuration compared to the reference state, R is the gas constant, T is the absolute temperature, n is the number of N-HisPurβ monomers bound in the s configuration, and the summation is over all possible configurations. Since microscopic association constants are related to microscopic free energies through the relationship, ki= e−ΔGi/RT, equations describing the fractional saturation, Ȳ, of a given site can be derived for various two-site binding models in terms of microscopic association constants and N-HisPurβ monomer concentration as described in Supporting Information. Individual site isotherms were globally fit to equations describing each model using Prism 5 software (GraphPad Software, Inc.). Monte Carlo error simulations were also performed to evaluate model confidence and parameter constraints using Prism 5 software.
RESULTS
Nucleotide Determinants of Purβ Binding to the SMαA Enhancer Element
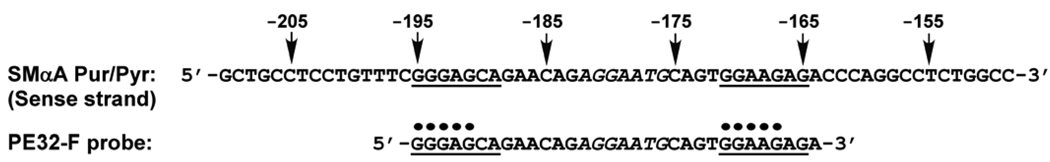
We first sought to confirm the specificity of nucleoprotein complex formation between recombinant Purβ and the purine-rich strand of the MCAT enhancer element (designated PE32-F) present in the mouse SMαA gene as previously documented with crude, mammalian cell extract-derived Purβ (13). A competitive ssDNA-binding ELISA was employed to gauge the ability of different fluid-phase oligonucleotides to compete for the binding of N-HisPurβ to immobilized, biotinylated PE32-F. Initial screening of a series of truncation mutants indicated that regions near the 5′ and 3′ ends of PE32-F contain critical determinants for N-HisPurβ binding. This conclusion was based on significant differences in the calculated IC50 values of 5′ and 3′ deletion mutants as compared to full-length PE32-F self-competition (Figs. S1 and S2 in Supporting Information). Closer inspection of the sequences near the termini of PE32-F led us to focus on two purine-rich stretches displaying remarkable similarity to a minimal Purα recognition motif, or PUR element, defined as GGGAGAG (39). These sequences, which exhibit only one or two nucleotide degeneracy with respect to the minimal PUR element, map to residues −195 to −189 (5′ site; GGGAGCA) and –171 to −165 (3′ site; GGAAGAG) relative to the transcription start site of the SMαA gene (Figure 1).
FIGURE 1.
Schematic of the polypurine-polypyrimidine tract containing a cryptic MCAT enhancer element in the 5′ flanking region of the mouse SMαA gene. Prior evidence for ssDNA-dependent structural rearrangements (20) in conjunction with ssBP-dependent repression of a core MCAT enhancer (italicized letters) (13) makes this region a focus of analysis in this study. The purine-rich probe, PE32-F, contains the minimum sequence supporting high affinity binding by Purβ and conferring MCAT enhancer repression. Nucleotides identified with a “•” were previously implicated in the ssDNA-binding specificity of Purβ extracted from cultured mammalian cell nuclei. Underlined sequences are homologous to a proposed minimal Purα recognition element (GGGAGAG) (39). Numbers denote nucleotide positions relative to the transcriptional start site.
Because nucleotides within these 5′ and 3′ sites had been previously identified as critical for physical and functional interaction by Purα and Purβ in cell-based assays (13, 19), a panel of oligonucleotides harboring heptadeoxythymidylate mutations within the 5′, 3′, and/or MCAT- sites were next evaluated as competitors for N-HisPurβ binding to wild type PE32-F (Table 1). Thymidylate mutations were used because preliminary experiments showed that polymeric deoxythymidylate (dT32) did not affect N-HisPurβ binding to biotinylated PE32-F in the competitive ELISA format (data not shown). The effectiveness of each mutant oligonucleotide as a fluid-phase competitor is shown in Figure 2. Consistent with deletion analysis, introduction of mutations within the putative 5′ GGGAGCA site (PE32-F-5T7) resulted in a modest loss of affinity compared to the PE32-F self-competitor, as judged by a nearly 4-fold increase in IC50. Interestingly, single mutation of the 3′ GGAAGAG site (PE32-F-3T7) or internal MCAT motif (PE32-F-IT7) produced no decrease in apparent binding affinity. However, double mutation of the 5′ and 3′ degenerate PUR elements (PE32-F-35T7) caused a drastic loss in affinity, evidenced by a greater than 50-fold increase in IC50. A similar but slightly less dramatic effect was seen by combined mutation of the MCAT and 5′ PUR element (PE32-F-5IT7) or the MCAT and 3′ PUR element (PE32-F-3IT7). The differential effects observed with the double mutants suggested that intact PUR elements are indeed critical for stable binding of N-HisPurβ, whereas the core MCAT motif may contribute as a non-specific site adding some limited stability to the nucleoprotein complex. In support of this contention, mutation of the 5′ and 3′ PUR elements along with the core MCAT motif (PE32-F-3I5T7) completely eliminated competition by this triple mutant over the concentration range tested. Although these findings alluded to a possible cooperative mode of nucleoprotein complex assembly, a more definitive account of the binding mechanism required a systematic evaluation of stoichiometry, affinity, and cooperativity.
Table 1.
Sequences of PUR element mutants tested in the Purβ ssDNA-binding ELISA.
| Competitora | Sequenceb | Site Diagram |
|---|---|---|
| PE32-Fc | GGGAGCAGAACAGAGGAATGCAGTGGAAGAGA | |
| PE32-F-IT7 | GGGAGCAGAACAGTTTTTTTCAGTGGAAGAGA | |
| PE32-F-5T7 | TTTTTTTGAACAGAGGAATGCAGTGGAAGAGA | |
| PE32-F-3T7 | GGGAGCAGAACAGAGGAATGCAGTTTTTTTTA | |
| PE32-F-35T7 | TTTTTTTGAACAGAGGAATGCAGTTTTTTTTA | |
| PE32-F-5IT7 | TTTTTTTGAACAGTTTTTTTCAGTGGAAGAGA | |
| PE32-F-3IT7 | GGGAGCAGAACAGTTTTTTTCAGTTTTTTTTA | |
| PE32-F-3I5T7 | TTTTTTTGAACAGTTTTTTTCAGTTTTTTTTA |
Designation of oligonucleotides used as fluid-phase competitors.
Underlined elements denote possible purine-rich binding sites. Italicized letters correspond to the core MCAT motif. Sequences are written in the 5′ to 3′ direction.
Wild-type oligonucleotide serving as the self-competition control.
FIGURE 2.
Analysis of the nucleotide determinants of PE32-F binding by N-HisPurβ. Results of fluid-phase competitor titrations in an ELISA-based ssDNA-binding assay are shown for a series of oligonucleotides harboring heptadeoxythymidylate (T7) mutations of putative purine-rich binding motifs in PE32-F. (A) Competition isotherms were fit to equation 1 to resolve IC50 values for each competitor. Sequences of oligonucleotides are presented in Table 1. Points represent individual measurements made in a representative assay. (B) IC50 values plotted as best fit value ± 67% confidence interval. The competitor marked with an asterisk indicates very low affinity as evidenced by an irresolvable IC50 value.
Characterization of Purβ-PE32-F Complex Assembly
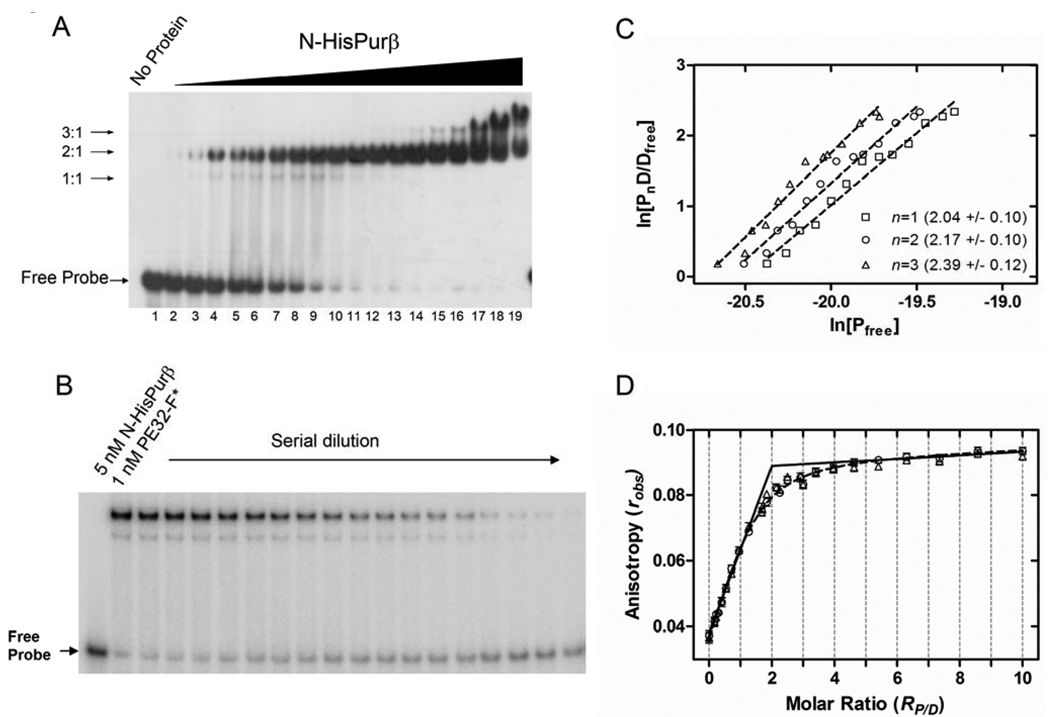
To estimate the number and distribution of nucleoprotein complexes formed between PE32-F and N-HisPurβ, we initially evaluated the dependence of recombinant protein concentration on complex assembly as visualized by qualitative EMSA. As shown in Figure 3A, the EMSA profile of nucleoprotein complexes formed after incubation of 0.41 to 400 nM N-HisPurβ with 5′ radiolabeled probe (PE32-F* at 2 nM) suggests that the protein binds to ssDNA in a sequential manner, as indicated by the detection of at least three discrete molecular species which progress from lower to higher apparent molecular weight as a function of N-HisPurβ concentration. The co-appearance of the two faster migrating species (speculated as adopting 1:1 and 2:1 protein:ssDNA stoichiometry) at lower protein concentrations coupled with the accumulation of the putative 2:1 complex at the expense of the weaker 1:1 complex at higher protein concentrations suggests a cooperative assembly mechanism leading to formation of a stable high affinity 2:1 complex. The emergence of an even slower migrating species (predicted to ≥3:1 in stoichiometry) at protein concentrations in excess of 100 nM implies that N-HisPurβ is capable of occupying other low affinity and/or non-specific binding sites on the ssDNA or associating indirectly with preformed nucleoprotein complexes via protein-protein interaction. Analysis of the electrophoretic mobility of nucleoprotein complexes generated with limiting concentrations of recombinant N-HisPurβ in comparison to complexes formed by fibroblast-derived Purβ suggests that the putative 2:1 species is the biologically relevant complex (Figure S3 in Supporting Information).
FIGURE 3.
Evaluation of the assembly and stoichiometry of N-HisPurβ:PE32-F complexes. (A) At least three electrophoretic species are detectable in band shift titrations of N-HisPurβ with 2.0 nM PE32-F* indicative of sequential formation of nucleoprotein complexes with differing putative stoichiometries (1:1, 2:1, and 3:1). Concentrations of N-HisPurβ in lanes 2 through 19 increase 1.5-fold from 0.41 nM (lane 2) to 400 nM (lane 19). (B) Limited serial dilution of a mixture of N-HisPurβ and PE32-F* (5.0 nM and 1.0 nM, respectively) was performed and samples were subjected to EMSA. The intensity of the Free Probe band was quantified by densitometry and standardized to known quantities of PE32-F* (lanes 1 and 20) to determine the concentration of free DNA, [Dfree], in each lane. The concentration of nucleoprotein complex [PnD] with stoichiometry n was determined from the equation [PnD] = [Dtotal] – [Dfree]. The concentration of free protein was estimated using the equation [Pfree] = [Ptotal] – n[PnD], in which n is an assumed integer value of 1, 2, or 3. (C) Isotherms of ln[PnD/Dfree] versus ln[Pfree] with assumed integer values of n were plotted. Each point represents the mean of duplicate experiments. Dashed lines represent the least-squares regression fits of each dataset to equation S11. Numbers in parentheses reflect the returned regression fit value of n ± s.d. (D) Fluorescence anisotropy analysis of the binding of N-HisPurβ to 50 nM PE32-F-3FLC. The dashed line represents a non-linear least squares fit of the data to equation S6. Fixing Kr at near-zero values (infinite affinity, solid line) verified the equivalency transition at an RP/D value of 2:1. Symbols show titrations using two different preparations of N-HisPurβ.
Stoichiometry of the Purβ-PE32-F complex
Direct assessment of the stoichiometry of the predominant nucleoprotein complex generated from equilibration of 5 nM N-HisPurβ with 1 nM PE32-F* was performed using a serial dilution-coupled quantitative EMSA. Briefly, a reaction mixture containing N-HisPurβ and PE32-F* was serially diluted with buffer to create a series of reaction samples with the same molar ratio of components, but at differing concentrations and distribution of reversibly interacting species governed by the law of mass-action. Quantification of the free and bound molecular species by densitometric analysis subsequent to electrophoretic separation (Figure 3B) permitted the determination of the system stoichiometry by way of a value convergence approach outlined in Supporting Information. As shown in Figure 3C, implementation of this analytical method indicated that the major, persistent, and high affinity complex formed between N-HisPurβ and PE32-F* adopts a stoichiometry of 2:1, as convergence between estimated values of n and those obtained by linear regression of a plot of ln[PnD/Dfree] versus ln[Pfree] occurred at n = 2. However, as argued above, the presence of a faint faster-migrating species, suggests that formation of the terminal 2:1 complex likely proceeds through a transient 1:1 complex.
Interpretations of EMSAs conducted with limiting concentrations of 5′ end-labeled probe were corroborated by experiments in which the binding of N-HisPurβ to 3′ fluorescein-labeled PE32-F was monitored by measuring changes in fluorescence anisotropy under saturating binding conditions. The results of this analysis are shown in Figure 3D. Under binding conditions in which the probe was fixed at a concentration >10 times the apparent dissociation constant (23), N-HisPurβ appears to saturate PE32-F-3FLC to a specific terminal stoichiometry of 2:1, as indicated by the values of n returned from fitting two datasets to equation S6 (Supporting Information), and the estimated equivalency transition point where the apparent affinity constant, Kr, is fixed at values approaching zero (near-infinite affinity, solid line). The curvature in the best fit line at values of RP/D near the equivalence point when Kr is not fixed (dashed line) suggests that the total concentration of probe was not sufficiently high enough to ensure binding of every protein molecule at low protein concentrations. This condition is only satisfied when Dtotal/Kd≫10, in the case of a completely cooperative system (40). Another possible contributing factor to the absence of a sharp equivalency transition point is non-specific DNA-binding, which is clearly manifest at high Purβ concentrations (Figure 3A). Collectively, these results support the conclusion that N-HisPurβ has the capacity to interact with PE32-F beyond a 1:1 stoichiometry with a 2:1 complex being the predominant and most stable entity formed at limiting concentrations of ssDNA and protein.
Binding Affinity of Purβ for PE32-F
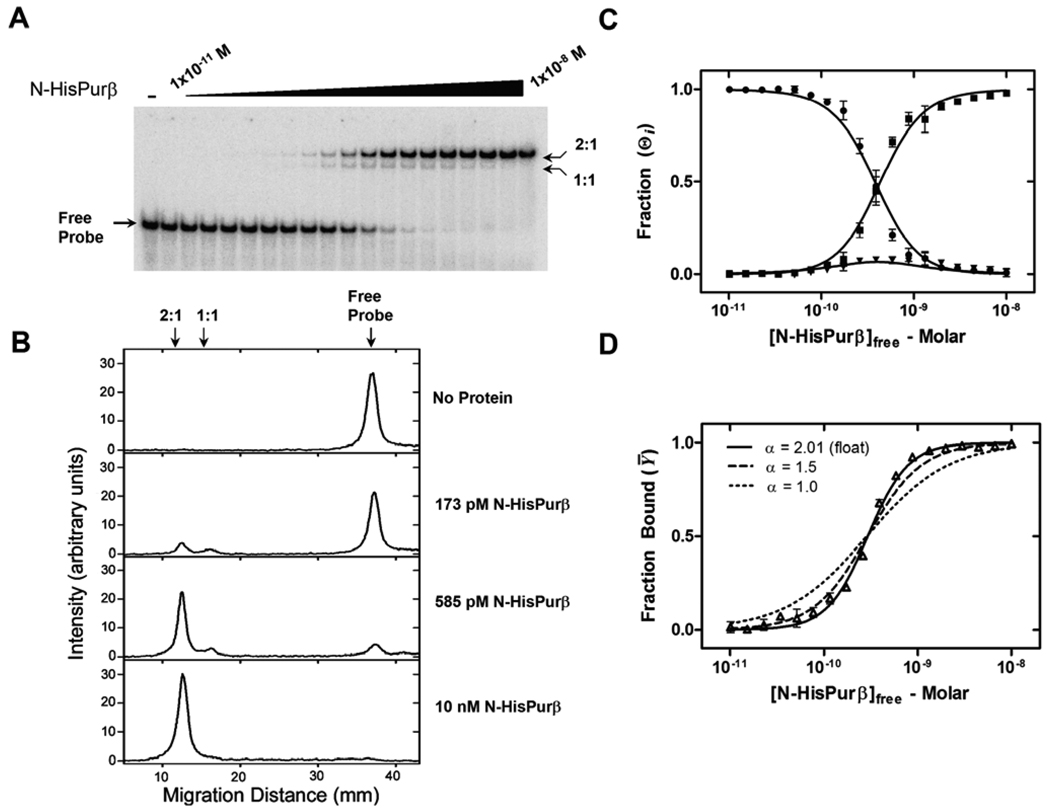
To determine the ssDNA-binding affinity of Purβ in a more thermodynamically rigorous fashion, we performed direct titrations of N-HisPurβ against 25 pM PE32-F*, a condition that maintains validity of the assumption [Pfree] ≈ [Ptotal], which is necessary for mathematical modeling of binding reactions (40). The distribution of molecular species at equilibrium was analyzed by non-denaturing electrophoresis and quantified as described in Experimental Procedures. Figure 4A shows a representative band shift profile of PE32-F* in the presence of increasing concentrations of N-HisPurβ. Visualization in concert with non-biased densitometric analysis indicates the presence of three separate bands or pixel intensity peaks (Figure 4B) corresponding to free probe, and two shifted complexes, namely (N-HisPurβ)1:PE32-F* (1:1 complex) and (N-HisPurβ)2:PE32-F* (2:1 complex). Of special interest is the transient nature of the 1:1 complex with respect to N-HisPurβ concentration. Comparing lane intensity profiles in Figure 4B reveals that the 1:1 complex is not a protein preparation contaminant as the peak intensity reaches a maximum at moderate N-HisPurβ concentrations and declines as PE32-F* becomes saturated. Moreover, this peak intensity pattern has been theoretically assigned to systems that adopt cooperative, two-site binding mechanisms (41).
FIGURE 4.
Titration analysis of N-HisPurβ binding to PE32-F by quantitative EMSA. (A) N-HisPurβ was diluted over a concentration range of 10−11 to 10−8 M and equilibrated with 25 pM PE32-F* prior to subjecting reaction mixtures to EMSA. (B) Densitometric analysis of each lane of the gel shown in (A) verifies the presence of three pixel intensity peaks indicative of separate electrophoretic species. Note that the putative 1:1 complex does not accumulate significantly compared to the free probe or the 2:1 complex, suggestive of a sequential and cooperative binding mechanism. (C) Individual band intensities are plotted as a function of [N-HisPurβ]free(●, Free Probe; ▼, 1:1 complex; ■, 2:1 complex). Each point represents the mean ± s.d. of quadruplicate experiments. Lines are global nonlinear least squares fits of individual species data points to equations S7–S9. (D) Band intensity data from (A) presented as Fraction Bound, Ȳ ([Dtotal] – [Dfree]) / [Dtotal]) versus [N-HisPurβ]free were fit to equation 4. The Hill coefficient, αH, was held constant at values of 1.5 (dashed line) and 1.0 (dotted line) to reflect the dependency of this variable on goodness of fit.
Peak integration values for each species were used to generate the isotherms presented in Figure 4C assuming a general two site cooperative DNA-binding model (31). Species specific isotherms for Θ0, Θ1, and Θ2 were globally fit to equations S7, S8, and S9 (Supporting Information), respectively, to resolve values for the macroscopic association constants, K1 and K2. This approach yielded K1 = 3.43 (± 0.368) × 108 M−1 and K2 = 6.06 (± 0.191) × 1018 M−1. It has been previously demonstrated that in instances where the nature of two-site binding of protein ligands to DNA is unknown (i.e. identical versus non-identical sites, positive or negative cooperativity) microscopic equilibrium constants cannot be definitively determined from the resolved macroscopic terms, regardless of their precision (31). Despite this theoretical barrier, inferences on the nature of binding can be made. For instance, Senear and Brenowitz showed that whenever K2 > K12/4, as is observed here, it can be inferred that ligand binding is cooperative (31).
As a further assessment of positive cooperativity, EMSA titrations were analyzed by the Hill binding equation. In order to circumvent quantification issues arising from the presence of multiple shifted complexes and the streaking of bands due to system reversibility, the extent of binding was determined based on the amount of free probe in each lane, which likely represents the extent of binding at equilibrium prior to electrophoresis (41). Non-linear least-squares fitting of fractional saturation data to the Hill equation generated the isotherm presented in Figure 4D and returned a macroscopic dissociation constant of ~0.3 nM, which is in close agreement to previously reported values for Purβ interaction with purine-rich ssDNA (23, 39). The returned Hill coefficient, αH (2.01 ± 0.07), implies that binding of N-HisPurβ to PE32-F* is cooperative, since αH converges at a value close to the experimentally determined value of n (Figure 3). It has been shown that values of αH approach the system stoichiometry only in cases where positive cooperativity is present (32). Fixing the value of αH at 1.5 or 1.0 exposes the strict dependence of the goodness of fit on this variable (Figure 4D). Collectively, these results suggest that N-HisPurβ binds in a sequential and cooperative manner to the purine-rich strand of the SMαA enhancer element with an apparent affinity in the sub-nanomolar range. However, microscopic binding constants could not be resolved by EMSA due to the non-identity of the two putative binding sites in PE32-F*.
Analysis of Purβ:PE32-F Complex Formation by DNase I Footprinting
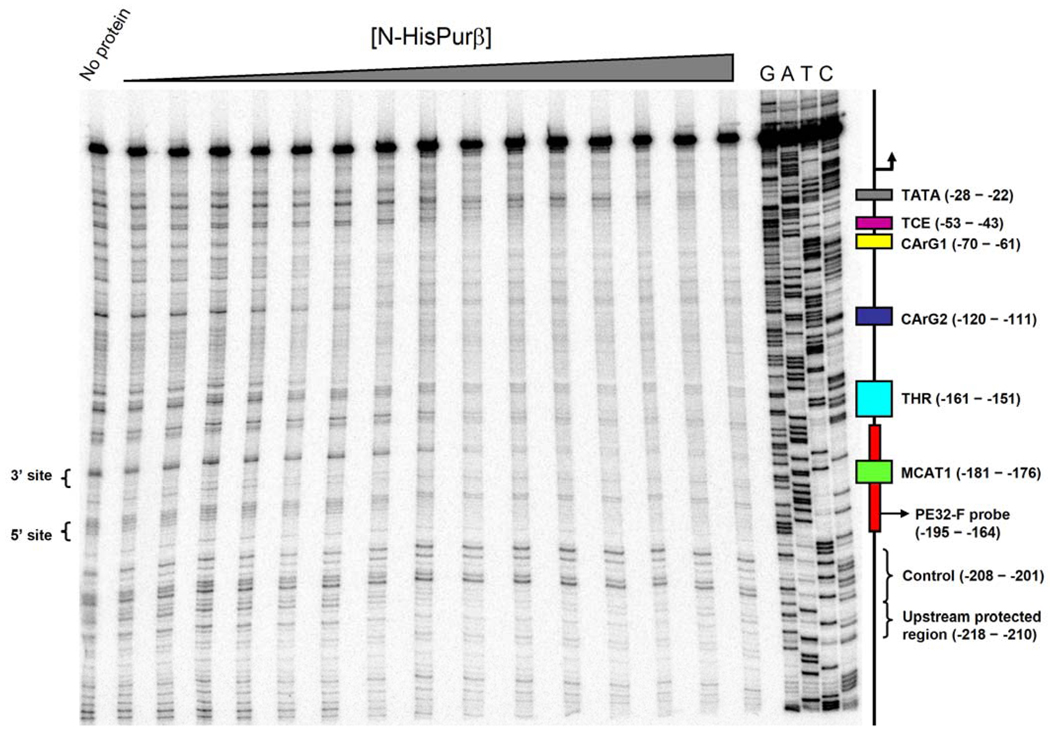
As an independent test of ssDNA-binding specificity and cooperativity, a quantitative DNase I footprinting method was used to measure site-specific fractional saturation of the coding strand of the SMαA 5′ flanking region between nucleotides −323 to +59 (SMP382-F) as a function of N-HisPurβ concentration. The power afforded by this technique is in the ability to quantify the interaction of a protein with sites of interest on a DNA template, thus providing a means to determine microscopic binding constants and to discriminate between possible binding mechanisms. The net effect of increasing N-HisPurβ concentration on the DNase I protection profile of SMP382-F* is shown in Figure 5. Several sites of protection, and by inference Purβ binding, can be seen either within or nearby cis-elements that have been reported to be directly or indirectly regulated by Purα or Purβ. These include the TGFβ1 control element, TCE (17), the TGFβ1 hypersensitive region, THR (42), CArG elements 1 and 2 (43) and, of course, the proximal MCAT enhancer element (13), as represented by PE32-F. Despite the relatively high apparent affinity of these interactions, we cannot exclude the possibility that the protection of some these regions could be due to opportunistic, non-specific binding by N-HisPurβ owing to the complete single-strandedness of the template. However, it is unlikely that the observed protection pattern is an artifact generated by protein-induced formation of secondary structures, as it has been previously reported that DNase I exhibits substrate preference and enhanced cleavage rates for dsDNA over ssDNA (44, 45).
FIGURE 5.
Titration analysis of N-HisPurβ binding to SMP382-F by quantitative DNase I footprinting. Representative footprint titration analysis of N-HisPurβ binding to SMP382-F* shows two regions of protection adjacent to the core MCAT motif and within the nominal PE32-F sequence (marked as 3′ site and 5′ site). The protected sites within the PE32-F sequence are separated by a band with protein-independent pixel intensity, when normalized to pixel intensity of the control region (–208 to –201) suggesting this intervening region is not protected by N-HisPurβ. Other sites of protection are evident within or near the THR, TCE, and CArG boxes 1 and 2 as well as a previously uncharacterized upstream region (−218 to −210), and a region adjacent to the TATA box. A positional map of the above noted cis-elements is shown on the right next to the lanes containing dideoxy NTP sequencing reactions (G, A, T, and C).
Irrespective of these technical considerations, our primary goal was to quantitatively interrogate the binding of N-HisPurβ to the region encompassing the PE32-F sequence containing the core MCAT motif (−195 to −164). Careful examination of this region by visual inspection and densitometry revealed two sites of protection by N-HisPurβ. These sites have been termed the 3′ and 5′ sites, and are labeled as such in Figure 5. It should be noted that these sites are located within or near to the two degenerate PUR elements identified by competitive ELISA (Figure 2) and are separated by a span that is only weakly protected when compared to the control region (−208 to −201). Based on the experimentally determined stoichiometry of the nucleoprotein complex formed between N-HisPurβ and this sequence (i.e. 2:1), the nature of protection of two sites flanking the core MCAT motif appears appropriate.
Resolution of the Microscopic Interaction Free Energies of Purβ:PE32-F
Mathematical expressions describing various models of interaction between N-HisPurβ and the promoter-embedded PE32-F element were generated using a statistical mechanical approach (35). For complexes with a saturating stoichiometry of 2:1, five possible models can be proposed. 1) The first possibility is that of a preformed (obligate) dimer assembling on a single binding site. Other possible models involve sequential assembly of monomers on the ssDNA lattice in which binding sites are: 2) identical and independent, 3) identical and interacting, 4) non-identical and independent, or 5) non-identical and interacting. Several simplifying assumptions were made in order to constrain and mathematically define the models described above. First, the binding of N-HisPurβ to the MCAT enhancer region (−195 to −164) is independent of the binding of N-HisPurβ to other regions of the promoter, outside of this vicinity. Second, DNA-independent self association of N-HisPurβ, as defined by the previously determined equilibrium constant of kdi = 8.85 × 105 M−1 (25), is negligible in cases of sequential monomer assembly, where half saturation of sites is in the sub-nanomolar range. Third, identical sites exhibit equal intrinsic binding free energy changes upon ligand binding, and as such, are denoted by ΔG1 for both sites. Fourth, non- identical sites exhibit non-equivalent intrinsic binding free energy changes upon ligand binding, and consequently, are designated by ΔG1 and ΔG2 to reflect this prediction. Fifth, in binding pathways possessing intersite interaction, the difference in the change in free energy for a monomer binding to each site and the total free energy change due to facilitated binding is represented by ΔGc. The various macromolecular configurations for each ligation state allowed by the restrictions of each binding model along with the corresponding microscopic free energy terms and equilibrium constants used for constructing expressions of N-HisPurβ binding are summarized in Tables S2 and S3 in Supporting Information.
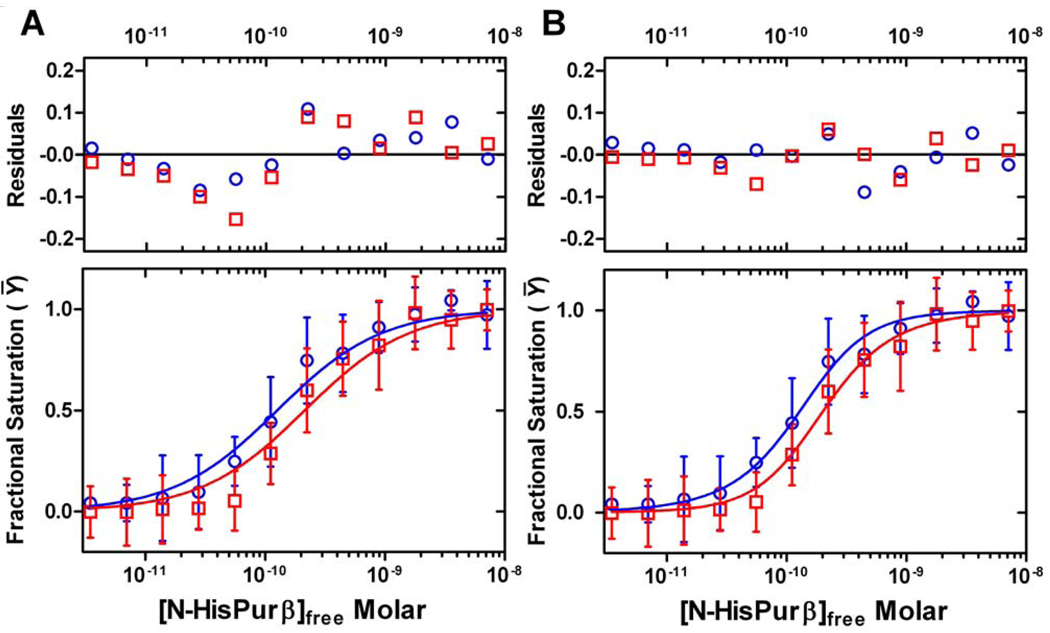
Quantification of N-HisPurβ binding to the identified 3′ and 5′ sites flanking the core MCAT enhancer motif was performed by densitometry to generate distinct binding site isotherms. Global fitting of data points corresponding to each putative site to the model-specific expressions shown in Table S3, provided a means to further discriminate between models based on goodness of fit. Isotherms generated by the two seemingly most relevant models, which assume non-identical binding sites, are shown Figure 6. A comprehensive comparison of the isotherms and fitting statistics for all five models, including the rejected obligate dimer and identical binding site scenarios, is depicted in Figure S4. From this approach, it is apparent that the binding of N-HisPurβ to PE32-F embedded within the extended promoter likely proceeds in a monomer-dependent fashion to 3′ and 5′ sites which are non-identical and interacting, as judged by random distribution of residuals and fit statistics (Figure 6, compare panel A to B and Figure S4). This model is in line with results of the qualitative and quantitative EMSAs with the nominal PE32-F probe, which also pointed to a cooperative mechanism of nucleoprotein complex assembly (Fig. 3 and Fig. 4). The resolved free energy parameters obtained from fitting of individual site isotherms to a non-identical interacting model are as follows with 67% confidence intervals noted in parentheses: ΔG1 (3′ site) = −12.8 (−12.9 to −12.7) kcal/mole, ΔG2 (5′ site) = −12.0 (−12.2 to −11.5) kcal/mole, and ΔGc, = −1.46 (−1.77 to −0.75) kcal/mole. These values indicate that even though Purβ binds to each site with high affinity, intersite cooperativity produces a ~12-fold enhancement in the stability of the nucleoprotein complex.
FIGURE 6.
Delineation of a cooperative binding model describing the interaction of N-HisPurβ with the MCAT-containing enhancer based on DNase I footprint data. Individual 3′ and 5′ site data points implying differential N-HisPurβ affinity were systematically and globally fit to equations (Table S3) describing a two-site model for either non-identical, independent binding sites (A) or non-identical, interacting binding sites (B). Blue symbols represent N-HisPurβ binding to the 3′ site. Red symbols represent binding to the 5′ site. Each point represents the mean ± s.d. of five independent experiments and the lines are best fit isotherms. Residual analysis and fit statistics support a cooperative binding model involving two non-identical binding sites.
Statistical Caveats of the Cooperative Binding Model
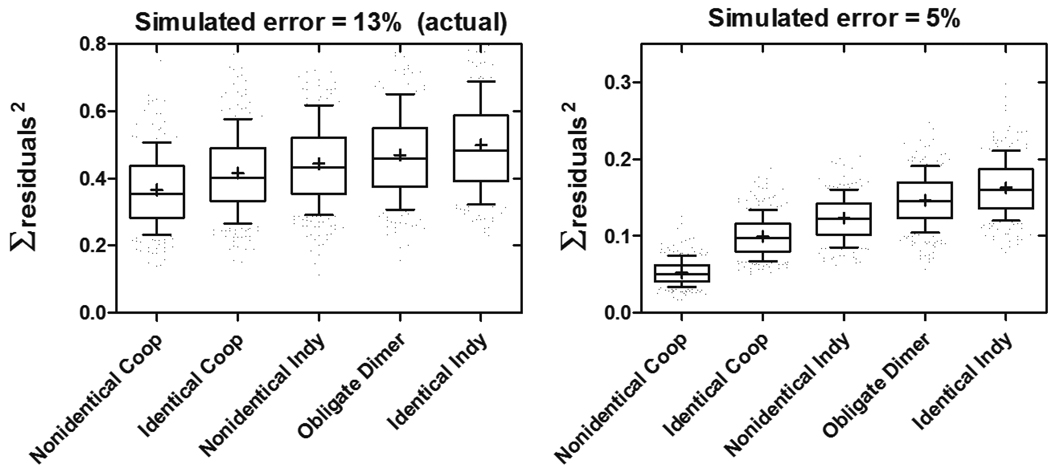
Despite the experimental power afforded by quantitative footprinting techniques, a shortcoming is the low level of precision that is attainable. Typical footprint titrations yield precision in the ± 10% range (37, 38, 46, 47). This trend appears to be amplified in experiments utilizing ssDNA templates, as performed here, with error settling at ± 13%. The exact reasons for this imprecision are unknown, but likely reflect differences in preference of the nuclease for single-stranded and double-stranded substrates. Due to the error level obtained, we opted to further assess confidence in our ability to discriminate between the possible binding models. To do this, we used a Monte Carlo simulation approach to test the effects of randomly introduced (Gaussian distributed) error at a level of ± 13% to datasets describing a non-identical interacting model, with binding parameters identical to those obtained with our actual data, on the goodness of global fits of the resulting isotherms. This process was performed for 1000 iterations, and the goodness of fit to each model was judged by the sum of residuals squared of the fit for each iteration. A box-and-whisker representation of this analysis is depicted in Figure 7 indicating that accurate model estimation for a non-identical interacting system is possible and likely when the possessed binding energetics are similar to those observed here, based on significant differences in the mean and median values of the sum of residuals squared for each model. However, estimation of the incorrect model is also theoretically possible, a conclusion based on the considerable overlap in the 25–75th percentile boxes for all of the models. It can be seen that complete resolution of these binding models would require a simulated error as low as ± 5%, a level that is not achievable experimentally using ssDNA templates.
FIGURE 7.
Monte Carlo error simulations to assess model confidence. Reiterative error simulations (1000) were performed on individual site isotherms shown in Figure 6 to yield error-incorporated isotherms that were then globally fit to various two site models. Box-and-whisker plots representing the distributions of fitting statistics (∑residuals2 ) for each model are shown when error is introduced at the level observed in experiments described herein (±13%, left panel). Reducing error to ±5% leads to higher model confidence as indicated by resolution of box-and-whisker plots (right panel). Boxes represent 25th-75th percentiles, whiskers represent 10th-90th percentiles. Median is marked by a line across the box, and mean is denoted as (+). Coop, cooperatively-interacting binding sites; Indy, independent non-interacting binding sites.
DISCUSSION
In this report, we endeavored to determine the mechanism of ssDNA-binding by Purβ, as this protein has been implicated in non-B-DNA-dependent repression of genes encoding muscle-restricted isoforms of actin and myosin in growth-activated vascular, cardiac, and skeletal muscle cell types (13–15). In the case of the SMαA gene, an ensemble of physical and functional studies has pointed to the association of Purβ with the purine-rich strand of a MCAT-containing enhancer element as a critical factor in promoter repression (22–24). Recent hydrodynamic studies conducted with purified recombinant Purβ indicated that the protein has the intrinsic ability to reversibly dimerize (25). The identification of this unique biophysical feature in conjunction with earlier work showing that the binding of cell extract-derived Purβ to ssDNA requires protein-protein interaction (21), led us to speculate that dimerization may be essential to site-specific DNA recognition. To test this hypothesis, we initially sought to validate the determinants of recombinant Purβ (N-HisPurβ) binding to a MCAT-containing cis-element (PE32-F) previously established using Purβ extracted from mammalian cells (13, 19, 23). As predicted, results of oligonucleotide competition assays pointed to two degenerate PUR elements near the 5′ and 3′ ends of the probe interacting in a seemingly cooperative fashion with the core MCAT motif playing an accessory role in protein binding. However, the measured 2:1 stoichiometry of the major N-HisPurβ:PE32-F nucleoprotein complex detected by band shift and fluorescence anisotropy assays was consistent with binding by either a bidentate dimer or two monomers.
Ultimately, the quaternary state of the ssDNA-binding competent Purβ species was clarified using two independent approaches both of which support a nucleoprotein assembly mechanism involving cooperatively interacting monomers as opposed to a preformed obligate dimer. Quantitative EMSA experiments revealed that half-saturation of PE32-F by N-HisPurβ occurs at a concentration (~0.3 nM) where the protein would be almost entirely monomeric. Accordingly, self-association at 0.3 nM total protein concentration assuming a Kd of ~1 µM (25) would give rise to N-HisPurβ dimer levels of ~75 fM. While a limiting concentration of preformed dimer does not in and of itself preclude the potential for obligate dimer-ssDNA interactions, a dissociation constant in the femtomolar range would seem to be unrealistically low for any reversibly associating system. Moreover, close inspection of the EMSA titration data showed not only the existence of an intermediate complex at concentrations near the half-saturation point, but that the species distribution patterns followed those predicted for a cooperative system (41). Importantly, data analysis using a statistical mechanics approach, assuming a two-site system, also indicated positive intersite cooperativity, a conclusion also supported by fitting the same EMSA data to the more phenomenological Hill equation. Despite the congruence of these data analysis methods, we could not strictly exclude the possibility of a coupled protein dimerization reaction owing to an inability to unequivocally detect a stable intermediate ligation state because of anomalous electrophoretic behavior of the T7 mutants relative to PE32-F (data not shown). This technical roadblock necessitated the use of an alternative method to independently assess protein binding to multiple sites.
To resolve the microscopic energetics of N-HisPurβ binding to the MCAT-containing PE32-F element, we turned to quantitative DNase I footprinting of a 382 nucleotide fragment of the coding strand of the SMαA promoter to detect and measure fractional occupation of each putative binding site. Results achieved using the footprinting approach provided corroborative evidence for a model whereby N-HisPurβ binds to the purine-rich strand of the PE32-F sequence via a cooperative mechanism involving two non-identical binding sites. The validity of this conclusion was substantiated by Monte Carlo error simulations which showed that we could distinguish between binding mechanisms with a degree of statistical confidence consistent with the reduced level of precision inherent in footprint analysis of ssDNA. However, it is also apparent that some of the resolved thermodynamic parameters featured relatively broad value constraints. This outcome was dictated by a combination of experimental imprecision, as well as parameter cross-correlation owing to the mathematical expressions from which the free energy values were obtained (Figure S5 in Supporting Information). Despite this statistical limitation, the interaction free energies we determined (ΔG1 = −12.8 kcal/mole, ΔG2 = −12.0 kcal/mole, ΔGc = −1.46 kcal/mole) are comparable to values reported for canonical transcription factors that adopt similar binding mechanisms (35–37, 47–50). Furthermore, the relative affinity of Purβ for individual sites is on par with the sub-nanomolar Kd values reported for the relaxase domain of TraI, an exceptionally sequence-specific ssDNA-binding protein, which plays an important role in the transfer of plasmid F factor during bacterial conjugation (51, 52).
The issue of parameter cross-correlation is not unique to the situation described herein but is endemic to all multisite cooperative systems, and is typically dealt with by analysis of DNA templates with mutated sites, so as to remove uncertainty caused by cooperativity (31, 37, 38). It has been shown by Brenowitz and colleagues (37) that without microscopic values determined by way of reduced valency templates, resolved cooperative free energy terms represent a lower limit to the actual cooperative free energy of the system, and the greater the cooperative free energy that exists in a system, the more difficult it is to resolve individual site interaction free energies. These considerations underlie the need to better define the composition of individual binding sites so that more accurate thermodynamic values of nucleoprotein assembly can be obtained. Although our results clearly point to two degenerate PUR elements in mediating the binding of Purβ to the SMαA enhancer element, we cannot at present exclude the possibility that other nucleotides outside these elements also contribute to protein recognition. Interestingly, a recent study assessing the binding of GST-Purα and GST-Purβ to a c-myc gene derived sequence containing a consensus PUR element and 92% purine residues reported that both proteins interact with this 24mer probe with sub-nanomolar affinity and 1:1 stoichiometry (39). While differences in the ssDNA template studied, recombinant protein preparations used, and analytical methods implemented may explain discrepancies, it remains to be determined to what extent lattice length, polynucleotide shape, and PUR element spacing affects cooperativity in nucleoprotein complex assembly.
Cooperative binding mechanisms are common for non-specific ssBPs, particularly those involved in DNA replication and recombination such as T4 bacteriophage gp32 (53) and E. coli SSB protein (54). In the case of sequence-specific ssBPs, there are relatively few examples in the literature where cooperative binding has been identified as a signature biochemical feature. Curiously, similar to what we have determined for N-HisPurβ, the binding of the yeast telomere protection protein Pot1 has been ascribed a cooperative binding mechanism in which sequential monomer binding to specific telomeric sites proceeds in a 3′ to 5′ direction (48). Cooperative binding of Pot1 to yeast telomeres has been functionally linked to nucleoprotein filament assembly and, in turn, protection of chromosome ends from damage by cellular nucleases. Hence, it is possible that cooperative binding of Pur proteins to transiently-formed ssDNA sequences in the SMαA promoter may serve an analogous function in terms of reducing the likelihood of trans-activator recognition of dsDNA target sites. A cooperative ssDNA-binding mechanism of this type would seemingly provide a regulatory advantage to the cell in that a relatively small change in cellular Purβ concentration could have a significant impact on gene transcription.
A challenging biochemical problem that remains to be solved is how members of the Pur family recognize and bind to ssDNA sequences in chromatin. Some have proposed that binding of sequence-specific ssBPs to larger, more complex DNA molecules is facilitated by transient formation of alternate structures in sequences prone to adopting non-B-DNA configurations as a consequence of topological stress. This theory has been substantiated previously for the far upstream element binding protein, a trans-activator, which requires transcription-induced negative supercoiling and unwinding for exposure of its single-stranded recognition element (55, 56). However, a RNA polymerase-coupled mechanism of this type may not be applicable to the cryptic MCAT enhancer element since the transcriptional activity of the SMαA gene is repressed rather than activated when the promoter is occupied by Purα, Purβ, and MSY1 in vivo (23, 24). This presents somewhat of a thermodynamic paradox. Based on results described herein, if the binding of Purβ to localized ssDNA regions in the context of a dsDNA lattice proceeds via direct competition (bubble formation), target sequences would have to possess less than −26 kcal/mole of annealing free energy. The average free energy of annealing for a single base-pair in dsDNA of infinite length is approximately −1.8 kcal/mole at 20°C (57). Thus, in the absence of other destabilizing factors, sequences of ~14–15 base-pairs might be prone to strand displacement by direct competition with Purβ. This theoretical range is remarkably similar to the size of sequence elements that have been empirically shown to be sensitive to Purα-mediated strand separation in vitro (39, 58). Nonetheless, Purβ binding alone falls short of the free energy needed to overcome the preferred annealing of the entire PE32 duplex (ΔG20˚C = −55.8 kcal/mol). Taking into account the fact that the genomic SMαA 5′-flanking region from −210 to −150 region appears to susceptible to structural alteration in living cells (20), it stands to reason that validation of the putative sequence-specific helix-destabilizing properties of Purα and Purβ will require analysis of larger and more topologically-strained DNA molecules with nuclease and chemical footprinting techniques that are sensitive to ssDNA formation.
As mentioned previously, repression of MCAT enhancer-dependent transcription of the SMαA gene appears to rely on the combined ssDNA-binding activities of Purα, Purβ, and MSY1, as well as a network of protein-protein interactions between the three factors (13, 21, 23). While limited, ATP-independent strand separation activity has been ascribed to Purα and Purβ using small chimeric probes (39, 58), YB-1/MSY1 is reportedly capable of promoting strand displacement from a more diverse set of oligonucleotides including short blunt-ended and Y-box-containing duplexes, cisplatin-modified duplexes, as well as engineered fork and bubble structures (59, 60). Unlike Purα, the strand separation activity for YB-1 is apparently elevated in the presence of ATP in manner attributable to an allosteric effect on the quaternary structure of the protein (60). Interestingly, the ssDNA-binding specificity of Purβ has been shown to be modulated by MSY1 in vitro (22). Hence, the progression from DNA site recognition to strand separation and repression of the SMαA MCAT enhancer in vivo may require physical and functional collaboration of all three ssBPs. Resolution of the biochemical role of each protein in this process is a credible undertaking as functional interplay between Pur and Y-box proteins has been reported in other systems (61, 62).
In conclusion, we have found that purified recombinant Purβ interacts with the purine-rich strand of the promoter-proximal SMαA enhancer in a cooperative manner mediated by two slightly degenerate PUR elements that flank the core consensus MCAT motif. Rigorous thermodynamic interrogation suggests that Purβ binding alone is insufficient to stably disrupt enhancer base-pairing. Defining the specific nucleotide binding determinants of Purβ interaction and the effect of co-repressors Purα and MSY1 on enhancer structure are goals of future studies.
Supplementary Material
Tables S1 to S3, Figures S1 to S5, and associated methods, results, and discussion. This material is available free of charge via the Internet at http://pubs.acs.org.
ACKNOWLEDGMENT
The authors would like to thank Keith Connaghan-Jones and David Bain for technical assistance with quantitative DNase I footprinting experiments and helpful discussions regarding data analysis.
Footnotes
This work was supported by grants R01 HL054281 and T32 HL007594 from the National Institutes of Health, National Heart, Lung and Blood Institute.
Abbreviations: ssDNA, single-stranded DNA; ssBP, single-stranded DNA binding protein; dsDNA, double-stranded DNA; SMαA, smooth muscle α-actin; PUR, purine-rich element; MCAT, muscle-CAT; PCR, polymerase chain reaction; EMSA, electrophoretic mobility shift assay; ELISA, enzyme-linked immunosorbent assay.
REFERENCES
- 1.Bergemann AD, Johnson EM. The HeLa pur factor binds single-stranded DNA at a specific element conserved in gene flanking regions and orgins of DNA replication. Mol. Cell. Biol. 1992;12:1257–1265. doi: 10.1128/mcb.12.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang ZY, Lin XH, Nobyuoshi M, Qui QQ, Deuel TF. Binding of single-stranded oligonucleotides to a non-B-form DNA structure results in loss of promoter activity of the platelet-derived growth factor A-chain gene. J. Biol. Chem. 1992;267:13669–13674. [PubMed] [Google Scholar]
- 3.Michelotti GA, Michelotti EF, Pullner A, Duncan RC, Eick D, Levens D. Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Mol. Cell. Biol. 1996;16:2656–2669. doi: 10.1128/mcb.16.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen S, Supakar PC, Vellanoweth RL, Song CS, Chatterjee B, Roy AK. Functional role of a conformationally flexible homopurine/homopyrimidine domain of the androgen receptor gene promoter interacting with Sp1 and a pyrimidine single strand DNA-binding protein. Mol. Endocrinol. 1997;11:3–15. doi: 10.1210/mend.11.1.9868. [DOI] [PubMed] [Google Scholar]
- 5.Rustighi A, Tessari MA, Vascotto F, Sgarra R, Giancotti V, Manfioletti G. A polypyrimidine/polypurine tract within the Hmga2 minimal promoter: a common feature of many growth-related genes. Biochemistry. 2002;41:1229–1240. doi: 10.1021/bi011666o. [DOI] [PubMed] [Google Scholar]
- 6.Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takimoto M, Tomonaga T, Matunis M, Avigan M, Krutzsch H, Dreyfuss G, Levens D. Specific binding of heterogeneous ribonucleoprotein particle protein K to the human c-myc promoter in vitro. J. Biol. Chem. 1993;268:18249–18258. [PubMed] [Google Scholar]
- 8.Negishi Y, Nishita Y, Saegusa Y, Kakizaki I, Galli I, Kihara F, Tamai K, Miyajima N, Iguchi-Ariga SM, Ariga H. Identification and cDNA cloning of single-stranded DNA binding proteins that interact with the region upstream of the human c-myc gene. Oncogene. 1994;9:1133–1143. [PubMed] [Google Scholar]
- 9.Michelotti EF, Tomonaga T, Krutzsch H, Levens D. Cellular nucleic acid binding protein regulates the CT element of the human c-myc protooncogene. J. Biol. Chem. 1995;270:9494–9499. doi: 10.1074/jbc.270.16.9494. [DOI] [PubMed] [Google Scholar]
- 10.MacDonald GH, Itoh-Lindstrom Y, Ting JP. The transcriptional regulatory protein, YB-1, promotes single-stranded regions in the DRA promoter. J. Biol. Chem. 1995;270:3527–3533. doi: 10.1074/jbc.270.8.3527. [DOI] [PubMed] [Google Scholar]
- 11.Swamynathan SK, Nambiar A, Guntaka RV. Role of single-stranded DNA regions and Y-box proteins in transcriptional regulation of viral and cellular genes. Faseb J. 1998;12:515–522. doi: 10.1096/fasebj.12.7.515. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Q, Pedigo N, Shenoy S, Khalili K, Kaetzel DM. Purα activates PDGF-A gene transcription via interactions with a G-rich, single-stranded region of the promoter. Gene. 2005;348:25–32. doi: 10.1016/j.gene.2004.12.050. [DOI] [PubMed] [Google Scholar]
- 13.Carlini LE, Getz MJ, Strauch AR, Kelm RJ., Jr Cryptic MCAT enhancer regulation in fibroblasts and smooth muscle cells. Suppression of TEF-1 mediated activation by the single-stranded DNA-binding proteins, Purα, Purβ, and MSY1. J. Biol. Chem. 2002;277:8682–8692. doi: 10.1074/jbc.M109754200. [DOI] [PubMed] [Google Scholar]
- 14.Gupta M, Sueblinvong V, Raman J, Jeevanandam J, Gupta MP. Single-stranded DNA-binding proteins Purα and Purβ bind to a purine-rich negative regulatory element of the α-myosin heavy chain gene and control transcriptional and translational regulation of gene expression. J. Biol. Chem. 2003;278:44935–44948. doi: 10.1074/jbc.M307696200. [DOI] [PubMed] [Google Scholar]
- 15.Ji J, Tsika GL, Rindt H, Schreiber KL, McCarthy JJ, Kelm RJ, Jr, Tsika R. Purα and Purβ collaborate with Sp3 to negatively regulate β-myosin heavy chain gene expression during skeletal muscle inactivity. Mol. Cell. Biol. 2007;27:1531–1543. doi: 10.1128/MCB.00629-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian SV, Kelm RJ, Jr, Polikandriotis JA, Orosz CG, Strauch AR. Reprogramming of vascular smooth muscle α-actin gene expression as an early indicator of dysfunctional remodeling following heart transplant. Cardiovasc. Res. 2002;54:539–548. doi: 10.1016/s0008-6363(02)00270-5. [DOI] [PubMed] [Google Scholar]
- 17.Subramanian SV, Polikandriotis JA, Kelm RJ, Jr, David JJ, Orosz CG, Strauch AR. Induction of vascular smooth muscle α-actin gene transcription in transforming growth factor β1-activated myofibroblasts mediated by dynamic interplay between the Pur repressor proteins and Sp1/Smad coactivators. Mol. Biol. Cell. 2004;15:4532–4543. doi: 10.1091/mbc.E04-04-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang A, David JJ, Subramanian SV, Liu X, Fuerst MD, Zhao X, Leier CV, Orosz CG, Kelm RJ, Jr, Strauch AR. Serum response factor neutralizes Purα- and Purβ-mediated repression of the fetal vascular smooth muscle α-actin gene in stressed adult cardiomyocytes. Am. J. Physiol. Cell. Physiol. 2008;294:C702–C714. doi: 10.1152/ajpcell.00173.2007. [DOI] [PubMed] [Google Scholar]
- 19.Sun S, Stoflet ES, Cogan JG, Strauch AR, Getz MJ. Negative regulation of the vascular smooth muscle α-actin gene in fibroblasts and myoblasts: disruption of enhancer function by sequence-specific single-stranded DNA-binding proteins. Mol. Cell. Biol. 1995;15:2429–2436. doi: 10.1128/mcb.15.5.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becker NA, Kelm RJ, Jr, Vrana JA, Getz MJ, Maher LJ., III Altered sensitivity to single-strand-specific reagents associated with the genomic vascular smooth muscle α-actin promoter during myofibroblast differentiation. J. Biol. Chem. 2000;275:15384–15391. doi: 10.1074/jbc.M909687199. [DOI] [PubMed] [Google Scholar]
- 21.Kelm RJ, Jr, Cogan JJ, Elder PK, Strauch AR, Getz MJ. Molecular interactions between single-stranded DNA-binding proteins associated with an essential MCAT element in the mouse smooth muscle α-actin promoter. J. Biol. Chem. 1999;274:14238–14245. doi: 10.1074/jbc.274.20.14238. [DOI] [PubMed] [Google Scholar]
- 22.Kelm RJ, Jr, Wang SX, Polikandriotis JA, Strauch AR. Structure/function analysis of mouse Purβ, a single-stranded DNA-binding repressor of vascular smooth muscle α-actin gene transcription. J. Biol. Chem. 2003;278:38749–38757. doi: 10.1074/jbc.M306163200. [DOI] [PubMed] [Google Scholar]
- 23.Knapp AM, Ramsey JE, Wang SX, Godburn KE, Strauch AR, Kelm RJ., Jr Nucleoprotein interactions governing cell type-dependent repression of the mouse smooth muscle α-actin promoter by single-stranded DNA-binding proteins Purα and Purβ. J. Biol. Chem. 2006;281:7907–7918. doi: 10.1074/jbc.M509682200. [DOI] [PubMed] [Google Scholar]
- 24.Knapp AM, Ramsey JE, Wang SX, Strauch AR, Kelm RJ., Jr Structure-function analysis of mouse Purβ II. Conformation altering mutations disrupt single-stranded DNA and protein interactions crucial to smooth muscle α-actin gene repression. J. Biol. Chem. 2007;282:35899–35909. doi: 10.1074/jbc.M706617200. [DOI] [PubMed] [Google Scholar]
- 25.Ramsey JE, Daugherty MA, Kelm RJ., Jr Hydrodynamic studies on the quaternary structure of recombinant mouse Purβ. J. Biol. Chem. 2007;282:1552–1560. doi: 10.1074/jbc.M609356200. [DOI] [PubMed] [Google Scholar]
- 26.Hultman T, Stahl S, Hornes E, Uhlen M. Direct solid phase sequencing of genomic and plasmid DNA using magnetic beads as solid support. Nucleic Acids Res. 1989;17:4937–4946. doi: 10.1093/nar/17.13.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kasuga T, Cheng J, Mitchelson KR. Metastable single-strand DNA conformational polymorphism analysis results in enhanced polymorphism detection. PCR Methods Appl. 1995;4:227–233. doi: 10.1101/gr.4.4.227. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Niu W, Nikiforov Y, Naito S, Chernausek S, Witte D, LeRoith D, Strauch A, Fagin JA. Targeted overexpression of IGF-I evokes distinct patterns of organ remodeling in smooth muscle cell tissue beds of transgenic mice. J. Clin. Invest. 1997;100:1425–1439. doi: 10.1172/JCI119663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tataurov AV, You Y, Owczarzy R. Predicting ultraviolet spectrum of single stranded and double stranded deoxyribonucleic acids. Biophys. Chem. 2008;133:66–70. doi: 10.1016/j.bpc.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Owczarzy R, Tataurov AV, Wu Y, Manthey JA, McQuisten KA, Almabrazi HG, Pedersen KF, Lin Y, Garretson J, McEntaggart NO, Sailor CA, Dawson RB, Peek AS. IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. 2008;36:W163–W169. doi: 10.1093/nar/gkn198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Senear DF, Brenowitz M. Determination of binding constants for cooperative site-specific protein-DNA interactions using the gel mobility-shift assay. J. Biol. Chem. 1991;266:13661–13671. [PubMed] [Google Scholar]
- 32.Weiss JN. The Hill equation revisited: uses and misuses. Faseb J. 1997;11:835–841. [PubMed] [Google Scholar]
- 33.Fried M, Crothers DM. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasimas JJ, Pegg AE, Fried MG. DNA-binding mechanism of O6-alkylguanine-DNA alkyltransferase. Effects of protein and DNA alkylation on complex stability. J. Biol. Chem. 2003;278:7973–7980. doi: 10.1074/jbc.M211854200. [DOI] [PubMed] [Google Scholar]
- 35.Ackers GK, Johnson AD, Shea MA. Quantitative model for gene regulation by lambda phage repressor. Proc. Natl. Acad. Sci. U S A. 1982;79:1129–1133. doi: 10.1073/pnas.79.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Senear DF, Brenowitz M, Shea MA, Ackers GK. Energetics of cooperative protein-DNA interactions: comparison between quantitative deoxyribonuclease footprint titration and filter binding. Biochemistry. 1986;25:7344–7354. doi: 10.1021/bi00371a016. [DOI] [PubMed] [Google Scholar]
- 37.Brenowitz M, Senear DF, Shea MA, Ackers GK. “Footprint” titrations yield valid thermodynamic isotherms. Proc. Natl. Acad. Sci. U S A. 1986;83:8462–8466. doi: 10.1073/pnas.83.22.8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brenowitz M, Senear DF, Shea MA, Ackers GK. Quantitative DNase footprint titration: a method for studying protein-DNA interactions. Methods Enzymol. 1986;130:132–181. doi: 10.1016/0076-6879(86)30011-9. [DOI] [PubMed] [Google Scholar]
- 39.Wortman MJ, Johnson EM, Bergemann AD. Mechanism of DNA binding and localized strand separation by Purα and comparison with Pur family member, Purβ. Biochem. Biophys. Acta. 2005;1743:64–78. doi: 10.1016/j.bbamcr.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 40.Rippe K. Analysis of protein-DNA binding at equilibrium, B.I.F. Futura. 1997;12:20–26. [Google Scholar]
- 41.Cann JR. Phenomenological theory of gel electrophoresis of protein-nucleic acid complexes. J. Biol. Chem. 1989;264:17032–17040. [PubMed] [Google Scholar]
- 42.Cogan JG, Subramanian SV, Polikandriotis JA, Kelm RJ, Jr, Strauch AR. Vascular smooth muscle α-actin gene transcription during myofibroblast differentiation requires Sp1/3 protein binding proximal to the MCAT enhancer. J. Biol. Chem. 2002;277:36433–36442. doi: 10.1074/jbc.M203232200. [DOI] [PubMed] [Google Scholar]
- 43.Gupta M, Sueblinvong V, Gupta MP. The single-strand DNA/RNA-binding protein, Purβ, regulates serum response factor (SRF)-mediated cardiac muscle gene expression. Can. J. Physiol. Pharmacol. 2007;85:349–359. doi: 10.1139/y07-009. [DOI] [PubMed] [Google Scholar]
- 44.Suck D, Oefner C. Structure of DNase I at 2.0 A resolution suggests a mechanism for binding to and cutting DNA. Nature. 1986;321:620–625. doi: 10.1038/321620a0. [DOI] [PubMed] [Google Scholar]
- 45.Sutton DH, Conn GL, Brown T, Lane AN. The dependence of DNase I activity on the conformation of oligodeoxynucleotides. Biochem. J. 1997;321:481–486. doi: 10.1042/bj3210481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brenowitz M, Senear DF, Kingston RE. DNase I footprint analysis of protein-DNA binding. Curr. Protoc. Mol. Biol. 2001:14. doi: 10.1002/0471142727.mb1204s07. Chapter 12, Unit 12. [DOI] [PubMed] [Google Scholar]
- 47.Heneghan AF, Connaghan-Jones KD, Miura MT, Bain DL. Cooperative DNA binding by the B-isoform of human progesterone receptor: thermodynamic analysis reveals strongly favorable and unfavorable contributions to assembly. Biochemistry. 2006;45:3285–3296. doi: 10.1021/bi052046g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lei M, Baumann P, Cech TR. Cooperative binding of single-stranded telomeric DNA by the Pot1 protein of Schizosaccharomyces pombe. Biochemistry. 2002;41:14560–14568. doi: 10.1021/bi026674z. [DOI] [PubMed] [Google Scholar]
- 49.Heneghan AF, Connaghan-Jones KD, Miura MT, Bain DL. Coactivator assembly at the promoter: efficient recruitment of SRC2 is coupled to cooperative DNA binding by the progesterone receptor. Biochemistry. 2007;46:11023–11032. doi: 10.1021/bi700850v. [DOI] [PubMed] [Google Scholar]
- 50.Connaghan-Jones KD, Heneghan AF, Miura MT, Bain DL. Thermodynamic analysis of progesterone receptor-promoter interactions reveals a molecular model for isoform-specific function. Proc. Natl. Acad. Sci. U S A. 2007;104:2187–2192. doi: 10.1073/pnas.0608848104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stern JC, Schildbach JF. DNA recognition by F factor TraI36: highly sequence-specific binding of single-stranded DNA. Biochemistry. 2001;40:11586–11595. doi: 10.1021/bi010877q. [DOI] [PubMed] [Google Scholar]
- 52.Harley MJ, Toptygin D, Troxler T, Schildbach JF. R150A mutant of F TraI relaxase domain: reduced affinity and specificity for single-stranded DNA and altered fluorescence anisotropy of a bound labeled oligonucleotide. Biochemistry. 2002;41:6460–6468. doi: 10.1021/bi011969i. [DOI] [PubMed] [Google Scholar]
- 53.Alberts BM, Frey L. T4 bacteriophage gene 32: a structural protein in the replication and recombination of DNA. Nature. 1970;227:1313–1318. doi: 10.1038/2271313a0. [DOI] [PubMed] [Google Scholar]
- 54.Ruyechan WT, Wetmur JG. Studies on the cooperative binding of the Escherichia coli DNA unwinding protein to single-stranded DNA. Biochemistry. 1975;14:5529–5534. doi: 10.1021/bi00696a023. [DOI] [PubMed] [Google Scholar]
- 55.Kouzine F, Liu J, Sanford S, Chung HJ, Levens D. The dynamic response of upstream DNA to transcription-generated torsional stress. Nat. Struct. Mol; Biol. 2004;11:1092–1100. doi: 10.1038/nsmb848. [DOI] [PubMed] [Google Scholar]
- 56.Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat. Struct. Mol. Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 57.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. U S A. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Darbinian N, Gallia GL, Khalili K. Helix-destabilizing properties of the human single-stranded DNA- and RNA-binding protein Purα. J. Cell. Biochem. 2001;80:589–595. [PubMed] [Google Scholar]
- 59.Ise T, Nagatani G, Imamura T, Kato K, Takano H, Nomoto M, Izumi H, Ohmori H, Okamoto T, Ohga T, Uchiumi T, Kuwano M, Kohno K. Transcription factor Y-box binding protein 1 binds preferentially to cisplatin-modified DNA and interacts with proliferating cell nuclear antigen. Cancer Res. 1999;59:342–346. [PubMed] [Google Scholar]
- 60.Gaudreault I, Guay D, Lebel M. YB-1 promotes strand separation in vitro of duplex DNA containing either mispaired bases or cisplatin modifications, exhibits endonucleolytic activities and binds several DNA repair proteins. Nucleic Acids Res. 2004;32:316–327. doi: 10.1093/nar/gkh170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Safak M, Gallia GL, Khalili K. Reciprocal interaction between two cellular proteins, Purα and YB-1, modulates transcriptional activity of JCVCY in glial cells. Mol. Cell. Biol. 1999;19:2712–2723. doi: 10.1128/mcb.19.4.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lasham A, Lindridge E, Rudert F, Onrust R, Watson J. Regulation of the human fas promoter by YB-1, Purα and AP-1 transcription factors. Gene. 2000;252:1–13. doi: 10.1016/s0378-1119(00)00220-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3, Figures S1 to S5, and associated methods, results, and discussion. This material is available free of charge via the Internet at http://pubs.acs.org.