Abstract
This study presents the detection of [2-13C]glucose metabolism in the carboxylic/amide region in the human brain, and demonstrates that the cerebral metabolism of [2-13C]glucose can be studied in human subjects in the presence of severe hardware constraints of widely available 3 T clinical scanners and with low power stochastic decoupling. In the carboxylic/amide region of human brain, the primary products of 13C label incorporation from [2-13C]glucose into glutamate, glutamine, aspartate, γ-aminobutyric acid, and N-acetylaspartate were detected. Unlike the commonly used alkanyl region where lipid signals spread over a broad frequency range, the carboxylic carbon signal of lipids was found to be confined to a narrow range centered at 172.5 ppm and present no spectral interference in the absence of lipid suppression. Comparison using phantoms shows that stochastic decoupling is far superior than the commonly used WALTZ sequence at very low decoupling power at 3 T. It was found that glutamine C1 and C5 can be decoupled using stochastic decoupling at 2.2 W although glutamine protons span a frequency range of ∼700 Hz. Detailed specific absorption rate analysis was also performed using finite difference time domain numerical simulation.
Keywords: decoupling, in vivo 13C MRS, human brain
Introduction
Proton decoupled 13C magnetic resonance spectroscopy (MRS) is a useful noninvasive method to study animal and human brain in vivo. With the enhanced 13C signal due to exogenous [1-13C]glucose infusion, the turnover of important metabolites in human brain can be quantitatively measured (1-14). Using 13C MRS as an investigative tool for human diseases has also been successful (15-17). However, in vivo 13C MRS in human brain has encountered more technical challenges than animal studies. One of the major difficulties has been associated with the need to decouple large 1H-13C scalar couplings (1JCH = 125−145 Hz) for alkanyl carbons of major brain metabolites. In order to achieve effective decoupling, the radio frequency (RF) field strength of decoupling pulses (γB2) has to be much greater than 1JCH. Because chemical shift dispersion is proportional to static magnetic field strength, the proton decoupling bandwidth and, therefore, the γB2 required for broadband decoupling, increase linearly with γB0. As a result, the RF power required for broadband proton decoupling increases as a function of (γB0)2 and has to be carefully controlled in order to keep the specific absorption rate (SAR) under the safety guidelines established by the U.S. Food and Drug Administration (FDA) (18) and the International Electrotechnical Commission (IEC) (19). To obtain necessary RF efficiency with acceptable RF power deposition, most human studies have used surface or half-volume transceiver coils for proton decoupling (20). As a result, nearly all of the human 13C studies have been performed in the occipital lobe of human brain, partially in order to avoid potential damage to the human eye as a result of poor perfusion due to RF heating from proton decoupling pulses (21-23).
Even at magnetic field strengths currently accessible to most clinical researchers (1.5−4 Tesla (T)), proton decoupling for in vivo 13C MRS still requires significant RF power deposition, often just under the safety limits for SAR. This issue has become one of the major obstacles to performing proton decoupled 13C spectroscopy using a high field scanner (e.g. 7 T). Using quadrature volume head coils for whole brain proton decoupling has previously been explored (24, 25). In one study that used 1.5 T, a forward power of 300 W was used to produce a γB2 of 500 Hz (>> 1JCH) (24). To avoid exceeding the SAR guidelines, a very short data sampling time of 85 ms was used, which significantly limited spectral resolution. In a 3 T study, the RF power transferred into the head was 70 W, which produces a γB2 of 350 Hz (25); 13C data sampling time was also relatively short (135 ms).
In addition to the obstacle of high decoupling power, strong background signal from subcutaneous lipid may also pose a challenge to 13C MRS of human brain. Baseline subtraction leads to penalty in Signal-to-Noise Ratio (SNR) and potential subtraction error due to subject movement and system instability. Spatial localization techniques for single-voxel spectroscopy are often used either using direct 13C localization and NOE enhancement (2, 6, 7, 11, 13) or using proton localization and heteronuclear polarization transfer (4, 5, 17). Localization combined with proton-to-carbon polarization transfer techniques offer many advantages including lipid suppression, small localization error and high SNR. Widely used clinical MRI scanners do not have a programmable second channel needed for heteronuclear polarization transfer. Implementation of heteronuclear polarization transfer techniques on these scanners requires significant hardware modification (26).
Our laboratory recently developed a new strategy for in vivo cerebral 13C MRS of monkey brain that uses a 4.7 T scanner aiming to overcome technical difficulties presented by the severe hardware constraints of clinical MRI scanners (27). Using this approach we infused [2-13C]glucose solution and detected the 13C signal of glutamate, glutamine, γ-aminobutyric acid (GABA), aspartate, and N-acetylaspartate (NAA) from the carboxylic/amide spectral region. The carboxylic/amide carbons are only coupled to protons via weak long-range 1H-13C couplings. We found that these carbons could be effectively decoupled using low RF power stochastic decoupling schemes. In contrast to the commonly used alkanyl spectral region, very little lipid signals were found in the carboxylic/amide region in the 4.7 T monkey study.
In this study, we demonstrate for the first time that this strategy for in vivo 13C spectroscopy can be extended to studying human brain. Expanding on a series of preliminary results (28, 29), we found that this method can be implemented on widely available broadband clinical 3 T scanners with a commercially available, standalone CW-type proton decoupler. Both WALTZ-4 and stochastic noise decoupling schemes were evaluated at 3 T. Incorporation of 13C labels from [2-13C]glucose into the carboxylic/amide carbons of glutamate, glutamine, aspartate, NAA, and GABA was detected in the human brain. To facilitate implementation of this method by others, a detailed description of all technical aspects of our work is provided. Additional issues regarding RF safety related to proton decoupling were investigated using a finite difference time domain (FDTD) numerical simulation method. The preliminary results of this work have been presented at The Fifteenth and Sixteenth Annual Meetings of International Society of Magnetic Resonance in Medicine (28, 29). Very recently, a 1.5 T study of the carboxylic/amide 13C spectral region using [1-13C]glucose infusion and the same low RF power stochastic decoupling sequence has also been reported (30).
Methods
Hardware
In vivo 13C MRS experiments were performed on a GE 3 Tesla Excite clinical scanner (GE Healthcare, Milwaukee, USA). A home-built RF coil system was used, which consisted of a single circular 13C coil and a proton quadrature surface coil (20). The proton coil comprised two overlapping octagon loops formed on a semi-cylindrical plastic tube with a diameter of 20.0 cm. Each proton resonance loop was made with 1.27 cm wide copper tape and its nominal inner length (or width) was 12.7 cm. The unloaded and loaded Q values were 125 and 30, respectively. These Q values indicate that 75% of the RF power forwarded into the proton coil was deposited into the human subject (31). All the experimental RF power values used in this report reflect the actual power deposited in the human subject, unless specified otherwise. The 13C coil was made with 1.27 cm wide copper tape (inner diameter = 7.5 cm) and formed on a separate semi-cylindrical plastic tube (diameter = 25.4 cm). It was placed 2.0 cm above the bottom of the semi-cylindrical tube of the proton coil. To prevent electromagnetic coupling between the 13C and proton coils, a parallel L-C tank circuit was inserted at the midpoint of the 13C loop, and its resonance frequency was tuned to the proton resonance frequency at 3 T. A foam pad was placed above the 13C coil to keep the subject's head at least 0.5 cm away from coil conductors.
A stand-alone proton decoupler (GE Healthcare, Milwaukee, USA) was used to provide RF pulses for Nuclear Overhauser Enhancement (NOE) and proton decoupling. RF safety using this device had been previously evaluated (32). The decoupler produces a bi-level CW-type RF output: low level for NOE during relaxation delay and high level for proton decoupling during 13C data acquisition. Switching between these two levels was triggered via a Transistor-Transistor Logic (TTL) signal from the scanner. Proton RF pulse signal generated from a vector signal generator (E4438C, Agilent Technologies, Santa Clara, CA, USA) was used as input to a solid-state RF power amplifier (Model 3000, Herley Medical Products, Lancaster, PA, USA). Instant and average output power was measured using an RF power monitor (E4416A, Agilent Technologies, Santa Clara, CA, USA). VEE Pro (Agilent Technologies, Santa Clara, CA, USA) was used to generate WALTZ-4 and stochastic decoupling waveforms and graphic interface for controlling ancillary electrical devices. The stochastic waveform comprised a sequence of repeated short rectangular pulses. The amplitude of the pulse unit remained constant, with its phase randomly assigned to either 0° or 180° (27, 33). The decoupler was equipped with a 13C transmit-receive switch, a 13C preamplifier, and in-line RF filters. To further reduce extra noise injected into the 13C channel, a proton notch filter was placed immediately in front of the 13C digital receiver. Its insertion loss was less than 0.1 dB at 13C frequency, with rejection greater than 60 dB at proton frequency.
RF power of the 13C coil was calibrated using a 2-liter phantom bottle filled with distilled water and 6 g NaCl, which has approximately the same loading effect as a typical adult human head. A 2.0-cm diameter sphere filled with 1.1 M [1-13C]glucose solution was placed inside the bottle, ∼3 cm above the 13C coil. A spectral null of the undecoupled [1-13C]glucose peaks was used to determine the power of the nominal 180° flip angle for a 500-μs rectangular pulse. Power calibration for the proton coil (γB2) was performed using the same phantom and the WALTZ-4 waveform.
SAR simulation
A custom-designed FDTD program was developed in-house. Simulations were performed on a 2.0 GHz AMD Opteron processor (AMD, Sunnyvale, CA, USA). The human head model with 2 × 2 × 2 mm3 spatial resolution was based on the National Library of Medicine's Visual Man Project. Eighteen different bones and tissues and their electrical properties at 128 MHz were assigned (34). A sagittal view of the head/coil model is shown in Figure 1. RF coils were modeled using Yee cells according to their actual dimensions. In this simulation, the loop of the 13C coil was opened at the feeding point. Two proton loops were mutually decoupled. Each loop was tuned to the proton frequency at 3 T. A voltage source was applied to each loop, and the coil was driven in quadrature. The B1 field with a circularly-polarized component (B1+) and electric fields were calculated at 128 MHz, and local SAR was computed by averaging absorbed electric power within a volume of 1 g mass around the center of each cell. The results of B1+ and local SAR distributions were normalized to 1 W of absorbed RF power inside the head model with 100% duty cycle. Using the normalized results, B1+ intensity and local SAR at different power levels and duty cycles were computed. We summed the Yee cell mass from highest to lowest electric energy of the cells. The mass containing 90% of the total electric energy absorbed inside the head model (effective mass) was used to calculate the average SAR. The mass containing the remaining 10% of the total electric energy was omitted because the electric field in those cells was too weak to cause thermal heating inside living tissue in the presence of blood circulation. The effective mass in this model was 3.5 kg, and the mass of the whole head above the neck was 5.6 kg. Average SAR was calculated by taking 90% of the total energy absorbed inside the human model and dividing it by the effective mass of 3.5 kg. The SAR simulation procedures were validated using phantom samples by comparing simulated B1+ and RF power with experimentally determined values.
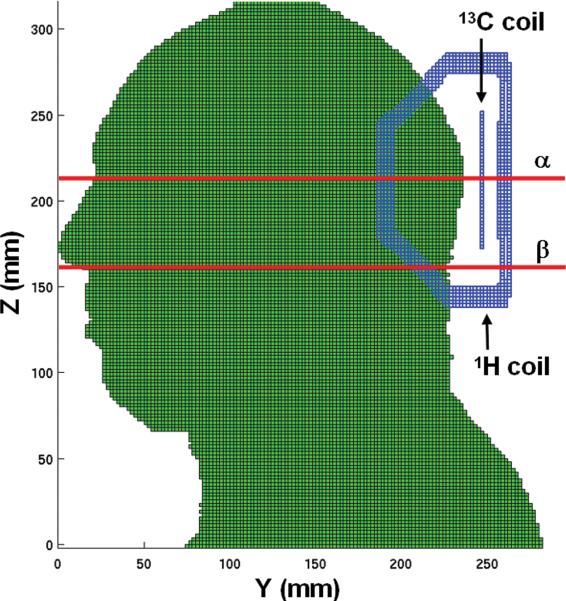
Figure 1.
A sagittal view of the computer model of the human head and RF coils for finite difference time domain (FDTD) simulation. The two red lines represent the α and β axial planes (β is 54 mm inferior to α) on which the calculated B1+ field and local SAR distributions are plotted, respectively, in Figure 2.
Plasma glucose analysis
Plasma proteins were precipitated using methanol. Proteins were pelleted by centrifugation; the solvent in supernatant was then removed under vacuum. The residue was dissolved using 2% hydroxylamine in dry pyridine and incubated for 1 hour followed by an additional 30 minutes after adding acetic anhydride. The resulting aldonitrile penta-acetate glucose derivative was analyzed using gas chromatography/electron impact-mass spectrometry (GC/EI-MS) operating in single ion monitoring (SIM) mode. The following signals related to the loss of 73 m/z or [CH3CO2CH2-]- from their molecular ions were recorded: 314 m/z, penta-O-acetyl-gluconitrile and 315 m/z, penta-O-acetyl-(2-13C)-gluconitrile. The fractional 13C enrichment of [2-13C]glucose was determined by measuring the peak area ratio of labeled glucose to the sum of labeled and unlabeled glucose from the extracted ion chromatogram.
Human subject preparation and in vivo 13C MRS
Healthy human subjects were examined in compliance with procedures approved by the Institutional Review Board (IRB) of the National Institute of Mental Health (NIMH). For 13C MRS infusion studies, subjects fasted at least 12 hours. Two antecubital veins were cannulated, one for administering [2-13C]glucose solution and the other for withdrawing blood to monitor glucose level using a SureStepFlexx Glucose Meter (LifeScan, Inc, Milpitas, CA, USA). Heart rate was monitored using MAGNITUDE (Invivo Corp., Orlando, FL, USA), a MR-compatible physiology monitor. A MR-compatible infusion pump, Continuum (MedRad, Indianola, PA), was used to infuse [2-13C]D-glucose solution (99% enrichment, 20% w/w, Cambridge Isotope Labs, Andover, MA). During glucose infusion, blood samples were withdrawn approximately every 10 minutes to measure blood glucose levels. Administration of the glucose solution began with a bolus infusion rate of 900 ml/hr followed by exponential decay to the rate of 100 ml/hr at the 15th minute of infusion. The subsequent infusion rate was adjusted accordingly to maintain glucose levels at 200 ± 20 mg/dL. The fractional enrichment of [2-13C]glucose reached 72 ± 6% at the end of the infusion period. A total of ∼40 g of [2-13C]glucose is typically used for a full time course study.
To optimize static magnetic field homogeneity the FASTMAP automatic high-order shimming method (35) was implemented on the GE 3 T scanner using two-dimensional STEAM localization with 12 shots. TE/TR = 20/2000 ms, TM = 20 ms, echo delay = 5 ms, FOV = 24 cm, column thickness = 10 mm, bandwidth = 16 kHz, number of data point = 128. Raw field map data were automatically saved and processed using the host Linux computer of the scanner and an in-house program written in GNU C with a graphic user interface developed using the Motif toolkit. The field map data were Fourier-transformed followed by phase unwrapping. The resultant phase curves were subsequently fitted to a second-order polynomial, and the shim term coefficients were calculated according to the FASTMAP algorithm (35). In a final step, the shim currents were automatically downloaded into the shim coils using a system call incorporating the built-in shim access commands of the scanner.
Three-slice (horizontal, coronal, and sagittal) scout gradient-echo images (FOV = 24 × 24 cm2, slice thickness = 5 mm, TR/TE = 5.3/1.6 ms, 256 × 128 data matrix) were used to position the human head inside the magnet. Either the manufacturer provided high order shimming (HOS) tool, or the FASTMAP method was used to optimize B0 homogeneity. Proton spectra were acquired from a 5 × 5 × 5 cm3 cubical voxel using a STEAM sequence (TR/TE = 2000/50 ms, TM = 50 ms, SW = 5 kHz, number of data points = 2048, NEX = 2, and NS = 4). The quality of localized shimming was evaluated by measuring the linewidth of water peak reported in AutoPrescan of the STEAM protocol. With the application of the FASTMAP method, water linewidth acquired from the 125 cm3 voxel was typically 7−8 Hz. In vivo 13C MRS spectra during [2-13C]glucose infusion were acquired using the GE FID CSI pulse sequence without the phase encoding gradients. A 500-μs nominally 45° rectangular pulse was used for 13C excitation with a forward power of 65 W. The nominal flip angle of the 13C pulse was not adjusted on a per subject basis because of the small loading differences among the subjects. TR was 4 seconds with a 13C data sampling time of 205 ms (SW = 5 kHz, number of data points = 1024, and NS = 128). The 13C pulse angle of 45 degrees was empirically optimized to maximize SNR. Bi-level stochastic waveform was used for NOE and proton decoupling. The repetition unit of the stochastic noise was 1.2 ms. The RF power absorbed by the human head was 0.75 W for NOE and 7.5 or 15 W for proton decoupling, which corresponds to an average power of 1.09 (7.5 × 0.05 +0.75 × 0.95) or 1.46 W (15 × 0.05 + 0.75 × 0.95), respectively. The proton carrier frequency was centered at the water signal.
Results
SAR simulation analysis
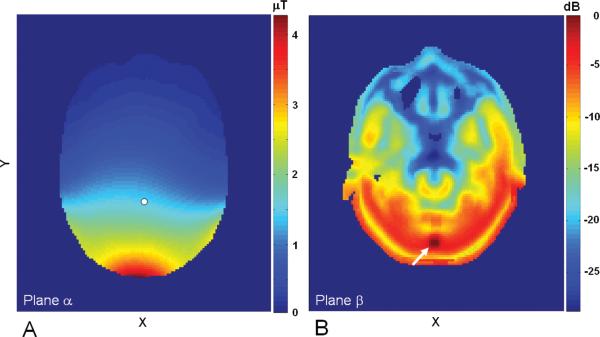
Figure 2A shows the normalized proton B1+ field distribution in the axial slice, plane αdepicted in Figure 1. B1+ field magnitude is given in μT units. Because of the geometric layout of the proton quadrature coils, the B1+ field was relatively uniform in the X direction and gradually decreased from 4.3 μT at the inferior surface of the head to 1.5 μT at 7.0 cm inside the brain (indicated by a white dot). The normalized local SAR distribution in the axial slice, plane β is shown in Figure 2B. This plot contains the maximum 1-g averaged local SAR of 1.66 W/kg in the head model under a normalized condition of a total of 1.0 W absorbed RF power inside the human head. Logarithm scale in dB for 1-g averaged local SAR was used. The location of the maximum local SAR (indicated by a white arrow) was inside the paraspinal muscle right below the occipital bone. We found that only about 30 cells had substantially high local SAR near the maximum value.
Figure 2.
(A) The B1+ field distribution in plane αdepicted in Figure 1. The field strength at the reference point (white dot) is 1.5 μT at the normalized condition of 1.0 W total RF absorption. (B) The normalized local SAR distribution in plane β that contains the maximum local SAR of 1.66 W/kg at the normalized condition. Local SAR is given in dB units, with the maximum local SAR at 1.66 W/kg. Location of maximum local SAR is marked by a white arrow.
In the in vivo experiments, the maximum RF power used was 7.5 or 15 W for decoupling and 0.75 W for NOE. With the 5% duty cycle for proton decoupling, the maximum 1-g averaged local SAR in the experiment was 2.43 W/kg [1.66 × (15 × 0.05 + 0.75 × 0.95)] for 15 W decoupling. For the average SAR calculation, 90% of the actual decoupling and NOE power, and an effective mass of 3.5 kg, were used as described in the Methods. Taking into account the actual duty cycles, the average SAR was 0.38 W/kg [(0.90 × (15 × 0.05 + 0.75 × 0.95))/3.5]. When the decoupling power of 7.5 W was used, the corresponding local and average SAR was further reduced to 2.0 W/kg and 0.31 W/kg, respectively. These SAR values are substantially below FDA and IEC safety thresholds.
Decoupling evaluations in vitro
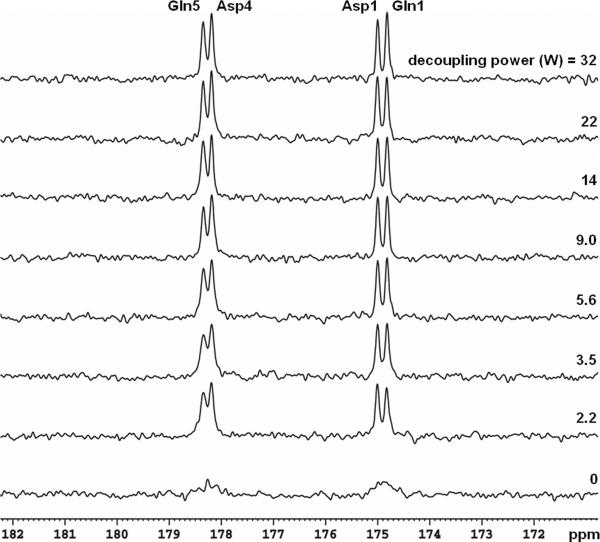
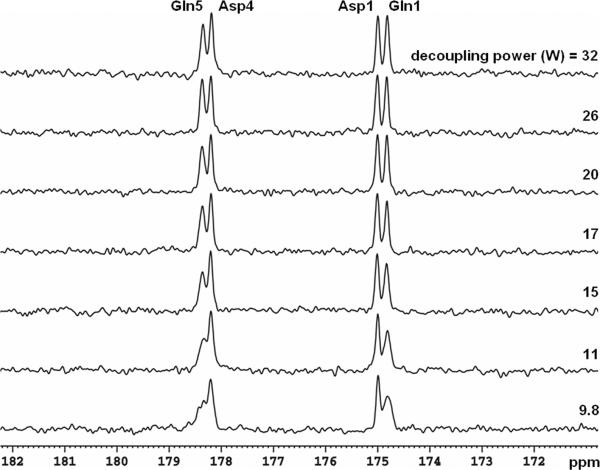
Among glutamate, glutamine, aspartate, NAA, and GABA, glutamine C5 and C1 peaks are most susceptible to imperfect decoupling and may overlap with aspartate C4 and C1 in the carboxylic/amide spectral region. To evaluate decoupling performance in vitro, a phantom of 2-liter bottle filled with distilled water and 6 g NaCl was used. Inside the bottle a spherical ball (diameter = 7 cm) was placed at the bottom of the cylinder, right above the 13C coil. The sphere contained 200 mM natural abundance glutamine and aspartate (pH = 7.0; SW = 5 kHz, number of data points = 2048, data acquisition time = 410 ms, and NS = 64). TR of 4 seconds and a 45° rectangular 13C pulse were used (500 μs, centered at 178 ppm, see Methods for nominal flip angle calibration). Spectra containing glutamine C5 (178.5 ppm) and C1 (175.4 ppm), as well as aspartate C4 (178.3 ppm) and C1 (175.0 ppm) acquired using stochastic decoupling at different decoupling power levels are shown in Figure 3. The free induction decay data were zero-filled to 16 K and apodized with exponential linebroadening (LB = 1.0 Hz). As the decoupling power was reduced, all resonances remained well separated, even at a very low decoupling power of 2.2 W (corresponding to a time-averaged power of (2.2 × 0.05 + 0.75 × 0.95) = 0.8 W). An undecoupled spectrum is shown at the bottom where all resonances become indiscernible. The results of WALTZ-4 decoupling are shown in Figure 4. As the decoupling power was reduced, resonance of glutamine C5 and, to a lesser extent, that of glutamine C1, were markedly broadened.
Figure 3.
Phantom 13C spectra (LB = 1 Hz) of glutamine and aspartate decoupled using stochastic waveform with a unit repetition time of 1.2 ms. Decoupling power for each spectrum is shown, and the undecoupled spectrum is shown at the bottom. For a decoupling power of 2.2 W, the average total RF power is (2.2 × 0.05 + 0.75 × 0.95) = 0.8 W, corresponding to a nominal γB2 of 46 Hz. Gln: glutamine; Asp: aspartate.
Figure 4.
Phantom 13C spectra (LB = 1 Hz) of glutamine and aspartate decoupled using WALTZ-4 waveform. The 90° pulse length for the decoupling power of 9.8 W is 2.6 ms (γB2 = 96 Hz). Gln: glutamine; Asp: aspartate.
There was little change in the linewidth of aspartate C1 over the 2.2 − 32 W decoupling power range using either stochastic or WALTZ-4 decoupling waveforms. When the decoupling power for stochastic decoupling was reduced from 32 W to 2.2 W, the linewidth of aspartate C4 peak was increased by only 1.2 Hz. The relative insensitivity of aspartate C1 and C4 resonances to variations in decoupling power can be attributed to the narrow frequency range of aspartate H2 (3.90 ppm) and H3 (2.68 and 2.81 ppm). In contrast, resonances of glutamine C5 and C1 are coupled to protons over a much broader spectral range, from glutamine H2 at 2.14 ppm to HE at 7.60 ppm (36). Among glutamate, glutamine, aspartate, NAA, and GABA, the spectral span of glutamine protons is the largest. In the spectra decoupled using WALTZ-4 (Figure 4), the linewidth of glutamine C1 increased from 2.7 Hz at 32 W to 6.4 Hz at 9.8 W. At 178.5 ppm, the glutamine C5 peak became so broad at the decoupling power below 15 W that significant overlapping with the aspartate C4 peak at 178.3 ppm made it difficult to separate them at 3 T using WALTZ-4 decoupling. On the other hand, with stochastic decoupling, the linewidth of glutamine C5 and C1 were only increased by 1.2 and 0.6 Hz, respectively, when the decoupling power was reduced from 32 W to 2.2 W. The separation between glutamate C5 and aspartate C4 for stochastic decoupling power at 2.2 W was comparable to that for WALTZ-4 at 15 W.
In vivo 13C baseline MRS
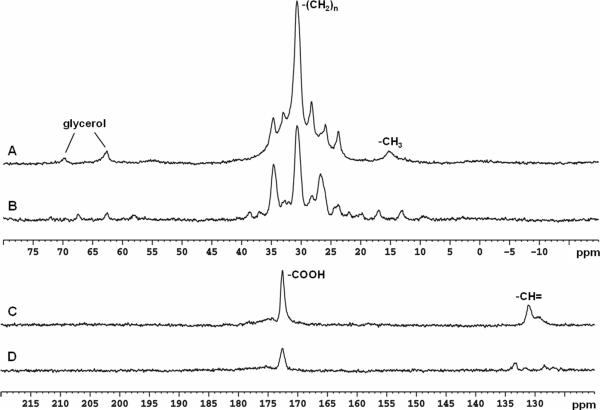
In vivo baseline spectra of the human head (NS = 128) obtained without infusion of [2-13C]glucose solution in the spectral region of alkanyl carbons (centered at 27.0 ppm) and carboxylic/amide region (centered at 178.0 ppm) are shown in Figure 5. FID data were zero-filled to 16 K and processed using exponential linebroadening (LB = 2.0 Hz). Zero and first order phase corrections were made. Baseline corrections were also made for the spectra acquired in the alkanyl spectral region. As observed previously, the alkanyl carbon region (0−75 ppm) is dominated by saturated carbons of the aliphatic chain and glycerol in lipids. For glutamate, glutamine, aspartate, and GABA methylene carbons resonating in the 24 − 40 ppm range, severe interference from unsuppressed lipid signals was present in both decoupled and undecoupled spectra in agreement with previous observations (Figures 5 A and B). In contrast to the wide spread of lipid signals in the alkanyl carbon region, the carboxyl signal of lipids was narrowly confined to a peak at 172.5 ppm (Figure 5 C and D). A low intensity broad hump downfield to that at 172.5 ppm was also observed. Neither interfered with detection of glutamate, glutamine, GABA, aspartate, or NAA carboxylic/amide signals.
Figure 5.
In vivo baseline 13C spectra (LB = 2 Hz, NS = 128) from human head without [2-13C]glucose infusion acquired in the spectral region of alkanyl carbons (A and B) and carboxylic/amide and alkenyl carbons (C and D). (A) WATLZ-4 decoupling at 32 W; (B) No decoupling; (C) Stochastic decoupling at 15 W; (D) No decoupling. In A and B, lipid methyl (15 ppm), methylene (20−40 ppm) and glycerol (62.5 and 69.5 ppm) carbon signals were detected. In C and D, lipid carboxylic (172.5 ppm) and alkenyl (131.9 ppm) carbon signals were detected.
In vivo 13C MRS during [2-13C]glucose infusion
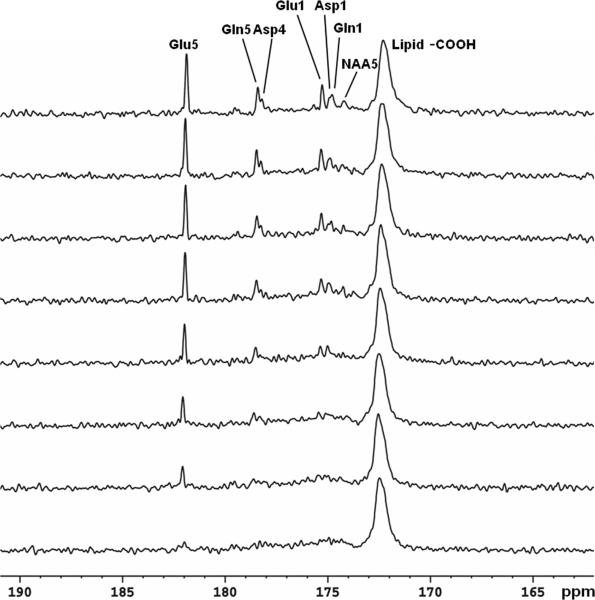
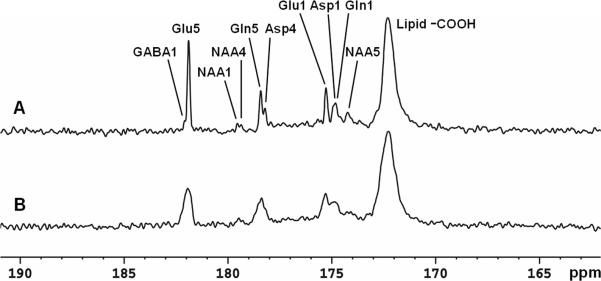
Figure 6 shows the time course spectra of glutamate, glutamine, aspartate, and NAA turnover from intravenously infused [2-13C]glucose in human brain. The FASTMAP shimming method was used to optimize B0 homogeneity in the occipital lobe. The time-averaged decoupling power was 1.46 W. Each spectrum corresponds to a 8.5 minute signal averaged with NS = 128. The natural abundance lipid carboxylic carbons (172.5 ppm), 13C-labeled glutamate C5 (182.0 ppm) and C1 (175.4 ppm), glutamine C5 (178.5 ppm) and C1 (174.9 ppm), aspartate C4 (178.3 ppm) and C1 (175.0 ppm), as well as NAA C5 (174.3 ppm) were shown. Glutamine C5 and aspartate C4 were spectrally resolved. When the last 17 minutes of data were summed (Figure 7A), GABA C1 (182.2 ppm), NAA C4 (179.4 ppm), and NAA C1 (179.6 ppm) were also detected. The [2-13C]glucose signals ([2-13C]-α-glucose at 72.5 ppm and [2-13C]-β-glucose at 75.2 ppm) were outside of the spectral window. The spectral assignments for the carboxylic/amide spectral region were based on literature reports (37-40). The assignments for GABA C1 and NAA C5 were further validated using in vivo animal 13C MRS studies performed at 11.7 Tesla (data not shown). The linewidth of glutamate C5 without apodization was 3.0 Hz in vivo. The spectral characteristics of the human 13C spectrum in the carboxylic/amide region were in excellent agreement with those observed from the monkey brain at 4.7 T except that the subcutaneous lipid signal at 172.5 ppm in the monkey spectra was much reduced (27). A 17-minute undecoupled spectrum acquired after completion of the decoupled spectra is shown in Figure 7B with all other parameters kept the same. The undecoupled spectrum shows that decoupling is necessary for proper spectral resolution in the carboxylic/amide region at 3 Tesla. The ratio of glutamate C5 amplitude in Figure 7A to that in Figure 7B is approximately 5:2. Additional NOE enhancement may be present in Figure 7A due to possible NOE carryover originated from proton decoupling.
Figure 6.
Time course spectra of glutamate, glutamine, and aspartate turnover detected in the occipital lobe during intravenous infusion of [2-13C]glucose. Lorentz-Gauss transformation (LB = −3 Hz, GB = 0.3) was applied. The time-averaged decoupling power was 1.46 W. Each spectrum corresponds to a 8.5-minute signal averaging with NS = 128. Glu C5 (182.0 ppm) and C1 (175.4 ppm), Gln C5 (178.5 ppm) and C1 (174.9 ppm), Asp C4 (178.3 ppm) and C1 (175.0 ppm), as well as NAA C5 (174.3 ppm) were detected. No baseline corrections were made. Glu: glutamate; Gln: glutamine; Asp: aspartate; NAA: N-acetylaspartate.
Figure 7.
(A) Spectrum summed from the last 17 minutes of data acquisition (LB = −3 Hz. GB = 0.3). Each spectrum corresponds to an average of 256 scans. GABA C1 (182.2 ppm), NAA C4 (179.4 ppm) and NAA C1 (179.6 ppm) were detected in addition to Glu C5 and C1, Gln C5 and C1, Asp C4 and C1, and NAA C5. (B) A 17-minute undecoupled spectrum from the same subject. Glu: glutamate; Gln: glutamine; Asp: aspartate; NAA: N-acetylaspartate; GABA: γ-aminobutyric acid.
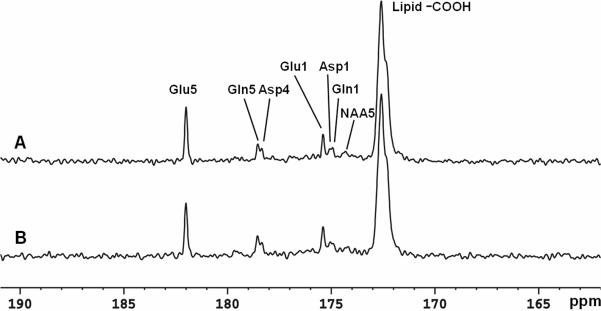
Spectra from a separate study on a different subject obtained at two different stochastic decoupling power levels are compared in Figure 8 during the 65th-102nd minute of [2-13C]glucose infusion. The manufacturer's HOS tool was used for this comparison. Each spectrum corresponds to an average of 256 scans in 17 minutes. The spectrum in Figure 8A was acquired using a decoupling power of 15 W (time-averaged power = 1.46 W). The spectrum in Figure 8B was acquired using a decoupling power of 7.5 W (time-averaged power = 1.09 W). Similar to the spectrum decoupled using 15 W, good spectral resolution between glutamate C5 and aspartate C4 was achieved with decoupling power of 7.5 W.
Figure 8.
Spectra obtained at two different stochastic decoupling power levels (LB = −3 Hz. GB = 0.3). Each spectrum corresponds to an average of 256 scans. (A) decoupling power of 15 W (time-averaged power decoupling = 1.46 W). (B) decoupling power of 7.5 W (time-averaged decoupling power = 1.09 W). Glu: glutamate; Gln: glutamine; Asp: aspartate; NAA: N-acetylaspartate.
Discussion
[2-13C]glucose infusion was previously used to measure anaplerotic flux rate in the human brain by detecting its label incorporation into the alkanyl spectral region in the 20−60 ppm range (14, 41). The primary metabolic intermediates of [1-13C]acetate were previously measured in the carboxylic/amide region of human brain (12, 42, 43). This report is the first to follow the primary metabolic fate of [2-13C]glucose in the human brain in vivo. Like in the case of switching 13C labels from [2-13C]acetate to [1-13C]acetate (12), the use of [2-13C]glucose greatly enhances the sensitivity of the carboxylic/amide carbons. The current study was performed using a typical clinical 3 T scanner and a commercially available, standalone CW-type proton decoupler. Our results demonstrate that it is feasible to perform in vivo human brain 13C MRS studies using low RF power for proton decoupling on widely available clinical 3 T scanners which do not have a programmable second channel.
Among the carboxylic/amide carbons, glutamine is most demanding for effectiveness of decoupling schemes. This is because both alkanyl (H2 at 3.78 ppm, H3 at 2.14 ppm and H4 at 2.46 ppm) and amide (HZ at 6.83 ppm and HE at 7.60 ppm) protons are coupled to glutamine C5 and C1. Overall, the glutamate, glutamine, aspartate, NAA, and GABA protons span a spectral range of 1.91−7.60 ppm. Although stochastic decoupling schemes have been considered obsolete since the arrival of coherent decoupling methods such as WALTZ-4, a previous 4.7 T study conducted by our laboratory showed that at 4.7 T, effective decoupling of the carboxylic/amide carbon region can be achieved with a nominal γB2 of 100 Hz using stochastic decoupling methods; WALTZ-4 had few decoupling effects at the same decoupling power level. At the lower field strength of 3 T used in the current study, the proton chemical shift range was reduced from 1138 Hz at 4.7 T to 726 Hz. As a result, even lower decoupling power (and lower γB2) was required for stochastic decoupling, as evidenced by the phantom (Figure 3) and in vivo (Figure 8) results. Compared with the 4.7 T results, the effectiveness of WALTZ-4 was also improved because of the lower field strength (Figure 4). However, the performance of the stochastic decoupling scheme still overwhelms that of WALTZ-4 for the carboxylic/amide carbons at 3 T. Therefore, we chose the stochastic decoupling scheme for our in vivo applications at 3 T.
In previous 13C studies of human brain, the typical proton decoupling power was 15 W at 1.5 T (8) or 2.1 T (6) and 30 W at 4.0 T (4) when the quadrature surface coil was used to decouple the large 1H-1C couplings for alkanyl carbons. Taking advantage of weak long-range 1H-13C coupling for carboxylic/amide carbons, in this study we reduced the decoupling power to 7.5 or 15 W at 3 T. The phantom spectra shown in Figure 4 indicate that the noise decoupling power may be further reduced below 7.5 W. This advantage may allow us to perform proton decoupled in vivo 13C spectroscopy in the frontal lobe of human brain in future studies.
One significant advantage of detecting the carboxylic/amide carbons, as shown in Figure 5, is the lack of interference from the intense subcutaneous lipid signals despite the reduced sensitivity, due to longer T1 of carboxylic/amide carbons as noted previously (27 and references therein). In our previous study performed using a 4.7 Tesla Bruker scanner which has a programmable second channel for heteronuclear polarization transfer, a comparison of SNR was made between the surface-coil localized carboxylic/amide 13C spectra and proton localized alkanyl 13C spectra from the same monkey, the latter were acquired using a single-shot, mostly adiabatic polarization transfer technique from a 30.6 mL voxel in the monkey brain with a large white matter content (44). The overall sensitivity of the surface-coil localized glutamate C5 signal, which originated from a larger volume including cortical gray matter close to the 13C receive coil, was found to be ∼35% lower than that of glutamate C4 using proton-carbon polarization transfer. From a separate study of rat brain at 11.7 Tesla, the in vivo T1 of glutamate C5 was found to be ∼16 seconds. A similar T1 is anticipated at 3 Tesla in human brain. Under the conditions of the present study (TR = 4s and 45 deg excitation pulse angle, which is close to the optimal Ernst angle), about 65% signal loss is expected theoretically due to T1 saturation. The sensitivity of the carboxylic/amide carbons signals is therefore lower than their alkanyl counter parts, but this SNR penalty is partially compensated by less susceptibility to B1 inhomogeneity and more access to high sensitivity cortical gray matter close to the 13C receive coil.
The efficiency of NOE enhancement was estimated in vivo at 3 T by interleaving proton-decoupled scans with and without NOE RF output. At a TR of 16.5 seconds and a nominal flip angle of 65°, the ratio of glutamate C5 signal intensity with and without NOE RF output is ∼1.9:1. Single voxel-type localization for carboxylic/amide carbons from the 13C channel is possible using T1-independent outer volume saturation schemes. Since the scalp lipid signal at 172.5 ppm is of no concern, only moderate suppression of outer volume signals would suffice to define a large voxel for 13C MRS. Either windowed noise pulses (45) or slice-selection with crusher gradients (46) could be used for this purpose to overcome the current limitation in spatial localization. The lack of lipid interference also makes it possible to use techniques capable of localizing arbitrarily shaped regions to extract signals from cortical gray matter close to the 13C receive coil with significantly enhanced sensitivity. A better defined volume and a full time course are very desirable for metabolic modeling studies although many important discoveries have been made using localization provided by the 13C surface transceiver coil only and/or with very short scan time (e.g., 8, 9, 12, 15, 16 and 47). Because of the close proximity between glutamine and aspartate signals excellent shimming is needed to resolve glutamine C5 and aspartate C5 as well as glutamine C1 and aspartate C1. For the full time course studies, excellent control of subject movement is also necessary to maintain good shimming over a prolonged period of time.
Because of RF heating associated with decoupling of the alkanyl carbons, proton-localized heteronuclear polarization transfer 13C MRS of the alkanyl carbons without proton decoupling has been proposed and successfully demonstrated in a study of rat brain at 9.4 Tesla (48). The SNR reduction due to the splitting of the undecoupled 13C signals can be partially compensated by the use of LCModel analysis (40). This ingenious no decoupling strategy may be readily extended to studying human brain at lower field strength (e.g., 3 Tesla) on scanners equipped with a programmable second channel or after hardware modification to execute both proton and carbon pulses from a single broadband channel (26). The Deelchand et al's no decoupling method could also be used with other localized 13C detection method such as direct 13C MRS with ISIS localization. The SNR penalty from line-broadening of 13C signals by undecoupled long-range protons at 3 Tesla, however, is expected to be larger than at 9.4 Tesla because of the narrower intrinsic linewidth at lower field strength.
Experimentally, we found that the intensity of the main lipid carboxylic signal at 172.5 ppm varied from subject to subject, presumably depending on the amount of fat in the scalp. However, the spectral pattern of this lipid signal remained unchanged. Therefore, unlike the commonly used alkanyl region where lipid interference may present a challenge unless excellent spatial localization is achieved, the carboxylic/amide carbons are essentially free from spectral contamination associated with extracranial signals. This key combination of low decoupling power requirement and lack of lipid contamination presents an exciting opportunity to integrate brain 13C MRS into the framework of modern parallel imaging techniques that rely on multiple small surface coils and gradients for spatial localization. In contrast, in single-voxel spectroscopy localization techniques, the dominant lipid signal in the 10−40 ppm region is suppressed using slice selection and outer volume suppression methods. The use of multiple small surface coils would also significantly increase sensitivity for brain regions close to the coils and overcome the current limitation on spatial localization. Furthermore, the RF safety of using a short birdcage coil for decoupling has been evaluated via FDTD analysis (49). The B1 field efficiency of a birdcage head coil appears to be approximately 50% of that of the quadrature surface coil used in this study. Therefore, the volume coil would need four times the decoupling power of the surface coil, assuming the same B1 distribution. Even under these conditions, the simulated local and average SAR values are still well within safety guidelines (49). Notably, the homogenous B1 distribution rendered by a volume coil undoubtedly improves the decoupling efficiency of stochastic decoupling schemes. The actual increase in stochastic decoupling power using volume coils is therefore expected to be much less. This postulation has been proven in a series of preliminary in vivo 13C spectroscopy experiments using a short birdcage volume coil (Li and Shen, unpublished results).
The overall information content in the carboxylic/amide spectral region is less than the alkanyl region if the bicarbonate signal at 161.0 ppm is excluded. It should be pointed out, however, that the bicarbonate signal is directly related to the TCA cycle rate regardless of the rate of exchange between mitochondrial and cytosolic pools. This is because, unlike keto- and amino acids, the nonpolar CO2 is freely diffusible across mitochondrial membranes. Therefore, it could be used as an independent constraint in metabolic modeling when quantification of absolute metabolic flux rates is necessary.
In summary, the use of [2-13C]glucose infusion and detection of 13C label incorporation into glutamate, glutamine, aspartate, NAA, and GABA offer a strategy to significantly reduce decoupling power and an exciting potential to integrate 13C MRS into the framework of modern parallel imaging. This study demonstrates for the first time that this novel strategy can be extended to performing human brain 13C MRS using widely available clinical 3 T scanners.
Acknowledgements
The authors thank Dr. Jerry Sanacora for help with glucose infusion technique, Mrs. Renee Hill for acquiring clinical images and Ms. Ioline Henter for editing the manuscript. This work is supported by the Intramural Research Program of the NIMH-NIH.
References
- 1.Beckmann N, Turkalj I, Seelig J, Keller U. 13C NMR for the assessment of human brain glucose metabolism in vivo. Biochem. 1991;30:6362–6366. doi: 10.1021/bi00240a002. [DOI] [PubMed] [Google Scholar]
- 2.Gruetter R, Novotny EJ, Boulware SD, Mason GF, Rothman DL, Shulman GI, Prichard JW, Shulman RG. Localized 13C NMR spectroscopy in the human brain of amino acid labeling from D-[1-13C]glucose. J Neurochem. 1994;63:1377–1385. doi: 10.1046/j.1471-4159.1994.63041377.x. [DOI] [PubMed] [Google Scholar]
- 3.Mason GF, Gruetter R, Rothmann DL, Behar KL, Shulman RG, Novotny EJ. Simultaneous determination of the rates of the TCA cycle, glucose utilization, α-ketoglutarate/glutamate exchange, and glutamine synthesis in human brain by NMR. J Cereb Blood Flow Metab. 1995;15:12–25. doi: 10.1038/jcbfm.1995.2. [DOI] [PubMed] [Google Scholar]
- 4.Gruetter R, Adriany G, Merkle H, Andersen PM. Broadband decoupled, 1H-localized 13C MRS of the human brain at 4 Tesla. Magn Reson Med. 1996;36:659–664. doi: 10.1002/mrm.1910360503. [DOI] [PubMed] [Google Scholar]
- 5.Gruetter R, Seaquist ER, Kim S, Ugurbil K. Localized in vivo 13C-NMR of glutamate metabolism in the human brain: initial results at 4 Tesla. Dev Neurosci. 1998;20:380–388. doi: 10.1159/000017334. [DOI] [PubMed] [Google Scholar]
- 6.Shen J, Petersen KF, Behar KL, Brown P, Nixon TW, Mason GF, Petroff OAC, Shulman GI, Shulman RG, Rothman DL. Determination of the rate of the glutamate/glutamine cycle in the human brain by in vivo 13C NMR. Proc Natl Acad Sci USA. 1999;96:8235–8240. doi: 10.1073/pnas.96.14.8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan JW, Stein DT, Telang F, Lee JH, Shen J, Brown P, Cline G, Mason GF, Shulman GI, Rothman DL, Hetherington HP. Spectroscopic imaging of glutamate C4 turnover in human brain. Magn Reson Med. 2000;44:673–679. doi: 10.1002/1522-2594(200011)44:5<673::aid-mrm3>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 8.Bluml S, Moreno A, Hwang JH, Ross BD. 1-13C glucose magnetic resonance spectroscopy of pediatric and adult brain disorders. NMR Biomed. 2001;14:19–32. doi: 10.1002/nbm.679. [DOI] [PubMed] [Google Scholar]
- 9.Moreno A, Bluml S, Hwang JH, Ross BD. Alternative 1-13C glucose infusion protocols for clinical 13C MRS examinations of the brain. Magn Reson Med. 2001;46:39–48. doi: 10.1002/mrm.1158. [DOI] [PubMed] [Google Scholar]
- 10.Shulman RG, Rothman DL. Brain Energetics and Neuronal Activity. John Wiley & Sons; England: 2004. [Google Scholar]
- 11.Lebon V, Petersen K, Cline GW, Shen J, Mason GF, Dufour S, Behar KL, Shulman GI, Rothman DL. Astroglial contribution to brain energy metabolism in humans revealed by 13C nuclear magnetic resonance spectroscopy: elucidation of the dominant pathway for neurotransmitter glutamate repletion and measurement of astrocytic oxidative metabolism. J Neurosci. 2002;22:1523–1531. doi: 10.1523/JNEUROSCI.22-05-01523.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bluml S, Moreno-Torres A, Shic F, Nguy CH, Ross BD. Tricarboxylic acid cycle of glia in the in vivo human brain. NMR Biomed. 2002;15:1–5. doi: 10.1002/nbm.725. [DOI] [PubMed] [Google Scholar]
- 13.Mason GF, Petersen KF, de Graaf RA, Kanamatsu T, Otsuki T, Rothman DL. A comparison of 13C NMR measurements of the rates of glutamine synthesis and the tricarboxylic acid cycle during oral and intravenous administration of [1- 13C]glucose. Brain Research Protocols. 2003;10:181–190. doi: 10.1016/s1385-299x(02)00217-9. [DOI] [PubMed] [Google Scholar]
- 14.Mason GF, Petersen KF, de Graaf RA, Shulman GI, Rothman DL. Measurements of the anaplerotic rate in the human cerebral cortex using 13C magnetic resonance spectroscopy and [1-13C] and [2-13C]glucose. J Neurochem. 2007;100:73–86. doi: 10.1111/j.1471-4159.2006.04200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang JH, Blum S, Leaf A, Ross BD. In vivo characterization of fatty acids in human adipose tissue using natural abundance 1H decoupled 13C MRS at 1.5T: clinical applications to dietary therapy. NMR Biomed. 2003;16:160–167. doi: 10.1002/nbm.824. [DOI] [PubMed] [Google Scholar]
- 16.Lin AP, Shic F, Enriquez C, Ross BD. Reduced glutamate neurotransmission in patients with Alzheimer's disease – an in vivo 13C magnetic resonance spectroscopy study. MAGMA. 2003;16:29–42. doi: 10.1007/s10334-003-0004-x. [DOI] [PubMed] [Google Scholar]
- 17.Otsuki T, Nakama H, Kanamatsu T, Tsukada Y. Glutamate metabolism in epilepsy: 13C-magnetic resonance spectroscopy observation in the human brain. NeuroReport. 2005;16:2057–2060. doi: 10.1097/00001756-200512190-00018. [DOI] [PubMed] [Google Scholar]
- 18.Criteria for significant risk investigations of magnetic resonance diagnostic devices. Center for Devices and Radiological Health, Food and Drug Administration; USA: 2003. http://www.fda.gov/cdrh/ode/guidance/793.html. [Google Scholar]
- 19.International Electrotechnical Commission International standard, medical equipment – part 2: particular requirements for the safety of magnetic resonance equipment for medical diagnosis, 2nd revision. Geneva: International Electrotechnical Commission. 2002;601:2–33. [Google Scholar]
- 20.Adriany G, Gruetter R. A half-volume coil for efficient proton decoupling in human at 4 Tesla. J Magn Reson. 1997;125:178–184. doi: 10.1006/jmre.1997.1113. [DOI] [PubMed] [Google Scholar]
- 21.Shellock FG, Crues JV. Corneal temperature changes induced by high-field-strength MR imaging with a head coil. Radiology. 1988;167:809–811. doi: 10.1148/radiology.167.3.3363146. [DOI] [PubMed] [Google Scholar]
- 22.Shellock FG, Schatz CJ. Increased corneal temperature caused by MR imaging of the eye with a dedicated local coil. Radiology. 1992;185:697–699. doi: 10.1148/radiology.185.3.1438747. [DOI] [PubMed] [Google Scholar]
- 23.Elder JA. Ocular effects of radiofrequency energy. Bioelectomagnetics. 2003;6:S148–S161. doi: 10.1002/bem.10117. [DOI] [PubMed] [Google Scholar]
- 24.Barker PB, Golay X, Artemov D, Ouwerkerk R, Smith MA, Shaka AJ. Broadband proton decoupling for in vivo brain spectroscopy in humans. Magn Reson Med. 2001;45:226–232. doi: 10.1002/1522-2594(200102)45:2<226::aid-mrm1031>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 25.Klomp DWJ, Renema WKJ, van der Graaf M, de Galan BE, Kentgens APM, Heerschap A. Sensitivity-enhanced 13C MR spectroscopy of the human brain at 3 Tesla. Magn Reson Med. 2006;55:271–278. doi: 10.1002/mrm.20745. [DOI] [PubMed] [Google Scholar]
- 26.Klomp DWJ, Kentgens APM, Heerschap A. Polarization transfer for sensitivity-enhanced MRS using a single radio frequency transmit channel. NMR Biomed. 2008;21:444–452. doi: 10.1002/nbm.1208. [DOI] [PubMed] [Google Scholar]
- 27.Li S, Yang J, Shen J. Novel strategy for cerebral 13C MRS using very low RF power for proton decoupling. Magn Reson Med. 2007;57:265–271. doi: 10.1002/mrm.21148. [DOI] [PubMed] [Google Scholar]
- 28.Li S, Zhang Y, Shen J. Implementing cerebral 13C MRS using low RF power for proton decoupling on a 3 Tesla clinical scanner.. Proceedings of the 15th Annual Meeting of ISMRM; Berlin, Germany. 2007.p. 535. [Google Scholar]
- 29.Li S, Zhang Y, Yang J, Maria Ferraris Araneta, Innis R, Shen J. In vivo 13C spectroscopy of human brain on a clinical 3T scanner using [2-13C]glucose infusion.. Proceedings of the 16th Annual Meeting of ISMRM; Toronto, Ontario, Canada. 2008.p. 1560. [Google Scholar]
- 30.Sailasuta N, Robertson LW, Harris KC, Gropman AL, Allen PS, Ross BD. Clinical NOE 13C MRS for neuropschiatric disorders of the frontal lobe. J Magn Reson. 2008;195:219–225. doi: 10.1016/j.jmr.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bottomley PA, Hardy CJ, Roemer PB, Mueller OM. Proton-decoupled, Overhauser-enhanced, spatially localized carbon-13 spectroscopy in humans. Magn Reson Med. 1989;12:348–363. doi: 10.1002/mrm.1910120307. [DOI] [PubMed] [Google Scholar]
- 32.Saito M, Matsuda T, Tropp J, Inubushi T, Nakai T. Assessment of the specific absorption rate and calibration of decoupling parameters for proton decoupled carbon-13 MR spectroscopy at 3.0 T. Euro J Radiology. 2005;55:289–293. doi: 10.1016/j.ejrad.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 33.Ernst RR. Nuclear magnetic double resonance with an incoherent radio-frequency field. J Chem Phys. 1966;45:3845–3861. [Google Scholar]
- 34.Gabriel C. Compilation of the dielectric properties of body tissues at RF and microwave frequencies, Technical Report, Brooks Air Force AL/OE-TR-1996−0037.
- 35.Gruetter R. Automatic, localized in vivo adjustment of all first- and second-order shim coils. Magn Reson Med. 1993;29:804–811. doi: 10.1002/mrm.1910290613. [DOI] [PubMed] [Google Scholar]
- 36.Kanamori K, Ross BD. Glial alkalinization detected in vivo by 1H-15N heteronuclear multiple-quantum coherence-transfer NMR in severely hyperammonemic rat. J Neurochem. 1997;18:1209–2119. doi: 10.1046/j.1471-4159.1997.68031209.x. [DOI] [PubMed] [Google Scholar]
- 37.Barany M, Arus C, Chang YC. Natual-abundance 13C NMR of brain. Magn Reson Med. 1985;2:289–295. doi: 10.1002/mrm.1910020312. [DOI] [PubMed] [Google Scholar]
- 38.Kunnecke B, Cerdan S, Seelig J. Cerebral metabolism of [1,2-13C2]glucose and [U-13C4]3-hydroxybutyrate in rat brain as detected by 13C NMR spectroscopy. NMR Biomed. 1993;6:264–277. doi: 10.1002/nbm.1940060406. [DOI] [PubMed] [Google Scholar]
- 39.Fan TWM. Metabolite profiling by one- and two-dimensional NMR analysis of complex mixtures. Progress in Nucl Magn Reson Spectroscopy. 1996;28:161–219. [Google Scholar]
- 40.Henry PG, Oz Gulin, Provencher S, Gruetter R. Toward dynamic isotopomer analysis in the rat brain in vivo: automatic quatitation of 13C NMR spectra using LCModel. NMR Biomed. 2003;16:400–412. doi: 10.1002/nbm.840. [DOI] [PubMed] [Google Scholar]
- 41.Mason GF, Petersen K, Shen J, Behar KL, Petroff OAC, Shulman GI, Rothman DL. Measurement of the rate of pyruvate carboxylase in human brain by 13C NMR.. Proceedings of the 8th Annual Meeting of ISMRM; 2000.p. 422. [Google Scholar]
- 42.Ross B, Lin A, Harris K, Bhattacharya P, Schweinsburg B. Clinical experience with 13C MRS in vivo. NMR Biomed. 2003;16:358–369. doi: 10.1002/nbm.852. [DOI] [PubMed] [Google Scholar]
- 43.Shic F, Ross B. Automated data processing of {1H-decoupled} 13C MR spectra acquired from human brain in vivo. J Magn Reson. 2003;162:259–268. doi: 10.1016/s1090-7807(03)00117-4. [DOI] [PubMed] [Google Scholar]
- 44.Li S, Chen Z, Zhang Y, Lizak M, Bacher J, Innis RB, Shen J. In vivo single-shot, proton-localized 13C MRS of rhesus monkey brain. NMR Biomed. 2005;18:560–569. doi: 10.1002/nbm.993. [DOI] [PubMed] [Google Scholar]
- 45.Ordidge RJ. Random noise selective excitation pulses. Magn Reson Med. 1987;5:93–98. doi: 10.1002/mrm.1910050113. [DOI] [PubMed] [Google Scholar]
- 46.Choi IY, Tkac I, Gruetter R. Single-shot, three-dimensional “non-echo” localization method for in vivo NMR spectroscopy. Magn Reson Med. 2000;44:387–394. doi: 10.1002/1522-2594(200009)44:3<387::aid-mrm8>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 47.Chhina N, Kuestermann E, Halliday j, Simpson LJ, Macdonald IA, Bachelard HS, Morris PG. Measurement of human tricarboxylic acid cycle rates during visual activation by 13C magnetic resonance spectroscopy. J Neurosci Res. 2001;66:737–746. doi: 10.1002/jnr.10053. [DOI] [PubMed] [Google Scholar]
- 48.Deelchand DK, Ugurbil K, Henry PG. Investigating brain metabolism at high fields using localized 13C NMR spectroscopy without 1H decoupling. Magn Reson Med. 2006;55:279–286. doi: 10.1002/mrm.20756. [DOI] [PubMed] [Google Scholar]
- 49.Li S, Wang S, Shen J. Safety evaluation for 1H decoupled 13C spectroscopy at 3 T in human frontal lobe: SAR analysis using numerical simulation.. Proceedings of the 16th Annual Meeting of ISMRM; Toronto, Ontario, Canada. 2008.p. 783. [Google Scholar]