Abstract
Seborrheic keratoses (SKs) are common, benign epithelial tumors of the skin that do not, or very rarely, progress into malignancy, for reasons that are not understood. We investigated this by gene expression profiling of human SKs and cutaneous squamous cell carcinomas (SCCs) and found that several genes previously connected with keratinocyte tumor development were similarly modulated in SKs and SCCs, whereas the expression of others differed by only a few fold. In contrast, the tyrosine kinase receptor FGF receptor–3 (FGFR3) and the transcription factor forkhead box N1 (FOXN1) were highly expressed in SKs, and close to undetectable in SCCs. We also showed that increased FGFR3 activity was sufficient to induce FOXN1 expression, counteract the inhibitory effect of EGFR signaling on FOXN1 expression and differentiation, and induce differentiation in a FOXN1-dependent manner. Knockdown of FOXN1 expression in primary human keratinocytes cooperated with oncogenic RAS in the induction of SCC-like tumors, whereas increased FOXN1 expression triggered the SCC cells to shift to a benign SK-like tumor phenotype, which included increased FGFR3 expression. Thus, we have uncovered a positive regulatory loop between FGFR3 and FOXN1 that underlies a benign versus malignant skin tumor phenotype.
Introduction
In both clinical and experimental situations, the majority of benign tumors fail to progress into malignancy for reasons that are poorly understood. The skin provides an intensely studied model of self-renewing epithelial tissues, with distinct stem cell populations giving rise to tumors with different behavior (1). Among these, skin squamous cell carcinomas (SCCs) are among the most frequent human cancers (2, 3). Several genetic events have been implicated in the development and progression of this disease, including mutations and/or downmodulation of the tumor suppressor p53 (4, 5), upregulation of its homolog p63 (6), and amplification and/or overexpression of the gene for EGFR (7, 8). Downstream of EGFR and p53, the Notch1 gene, which plays an important function in promoting keratinocyte differentiation and suppressing tumorigenesis, is downmodulated (9–11).
Besides malignant tumors, keratinocyte subpopulations can give rise to benign skin tumors like seborrheic keratoses (SKs), which are not precursor lesions of malignant SCCs. SKs are very common lesions that develop with age in the vast majority of the human population. Common histological features are acanthosis, papillomatosis, and hyperkeratosis, along with varying degrees of pigmentation (12, 13). SKs have clinical similarities to the common wart. While human papillomaviruses have been implicated in the origin of these lesions (14), recent analyses have generally discounted a role for such viruses in the majority of cases (15). Patients can often have several independent SKs, and individuals developing a great number of these lesions (i.e., greater than 50) on a familial basis have been described previously (16). In spite of this multiplicity, SKs appear to be clonal in origin, which indicates that they do not result from a reactive epidermal hyperplasia, but from clonal expansion of somatically mutated cells (17). In fact, recent work has shown the presence of activating mutations in a specific transmembrane tyrosine kinase receptor, FGF receptor–3 (FGFR3; ref. 18–20), and/or in PI3K, a downstream effector of FGFR signaling (21), in a large fraction of sporadic SKs. A causative role of FGFR3 mutations is suggested by the fact that transgenic mice with keratinocyte-specific expression of an activated form of the receptor produce skin lesions histologically similar to SKs (20).
The FGFR family is composed of 4 tyrosine kinase receptors (FGFR1–FGFR4), each with several isoforms (22). A large number of FGF ligands have also been identified, some of which have restricted binding specificity for individual FGFR family members, while others have a more diverse spectrum of binding and activation (23). In the skin, FGFR1, FGFR2, and FGFR3 have all been implicated in the genesis of neoplastic lesions. The FGFR1 gene is amplified in SCCs, and its increased expression promotes keratinocyte proliferation and tumor progression (24). Levels of FGFR2 do not appear to change in SCCs. However, mice with keratinocyte-specific deletion of the FGFR2 gene show enhanced sensitivity to chemical carcinogenesis, pointing to a possible tumor suppression function (25). As mentioned, activating FGFR3 mutations have been linked to the development of SKs as well as, more recently, epidermal nevi, another type of benign lesions characterized mainly by epidermal thickening with hyperpigmentation (26). Importantly, identical FGFR3 mutations have also been associated, in other epithelial tumors including bladder carcinomas, with good prognosis and lack of malignant progression (27–29). Conversely, the same FGFR3 activations have been linked with disease progression of some hematopoietic malignancies, including multiple myeloma (30, 31). The basis for these cell type–specific consequences of FGFR3 activation on tumor development remains to be clarified.
FOXN1 is a forkhead transcription factor family member that plays an essential function in keratinocyte differentiation (32–35) and thymic epithelium development (36), with mutations of the FOXN1 gene resulting in the nude phenotype in mice, rats, and humans (37–39). FOXN1 functions as a transcriptional activator, as it contains a sequence-specific DNA-binding domain and a negatively charged transactivation motif (35, 40). FOXN1 promotes the early steps of the keratinocyte terminal differentiation program and suppresses the later steps, through an interplay with the PKC and/or Akt signaling pathways (32–35). Another novel function of FOXN1 in keratinocytes is its role as modulator of skin pigmentation (41). In contrast to the normal differentiation program of keratinocytes, nothing is known about the role of this transcription factor in tumor development. Using a combination of biochemical and functional approaches, we report the existence of a double-positive regulatory loop between FOXN1 expression and FGFR3 activation that lies at the basis of a benign versus malignant skin tumor phenotype.
Results
Deregulated gene expression in benign versus malignant skin tumors.
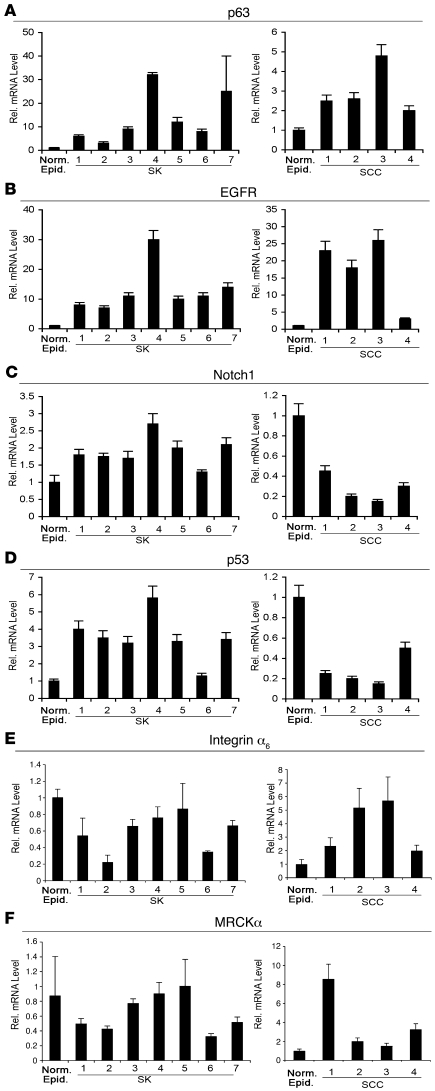
SCC of the skin is among the most frequent cancers in the Western world (1). Besides malignant tumors, keratinocyte subpopulations can give rise to benign skin tumors like SKs, which progress very rarely, if ever, into malignancy. By comparing patterns of gene expression, we found a number of genes involved in positive control of keratinocyte growth and carcinogenesis, like those for EGFR and p63 (6, 7), to be upregulated in both SKs and SCCs (Figure 1, A and B). Conversely, other important regulatory genes were differentially expressed in SKs versus SCCs. These include Notch1 (Figure 1C), which we have previously shown to be downmodulated in SCCs (9) but was instead upregulated in SKs, and p53 (Figure 1D), a key positive regulator of Notch1 expression in keratinocytes (9, 10, 42). Integrin α6 and CDC42 binding protein kinase α (MRCKα), 2 key negative Notch targets in these cells (9), were also upregulated in SCCs, yet exhibited normal or reduced levels in SKs (Figure 1, E and F).
Figure 1. Expression profile of genes previously connected with keratinocyte tumor formation in SK versus SCC lesions.
RNAs extracted from 7 separate SK lesions, 4 separate surgically excised SCC lesions, and 3 normal, age-matched control epidermis were subjected to real-time RT-PCR for the indicated genes, with β-actin for internal normalization. SK and SCC values are expressed as folds of expression relative to the average of the 3 normal controls. Error bars denote SEM.
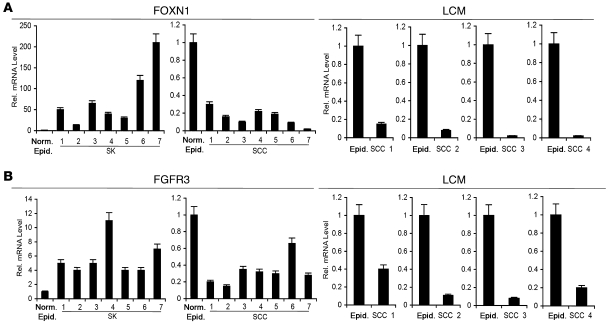
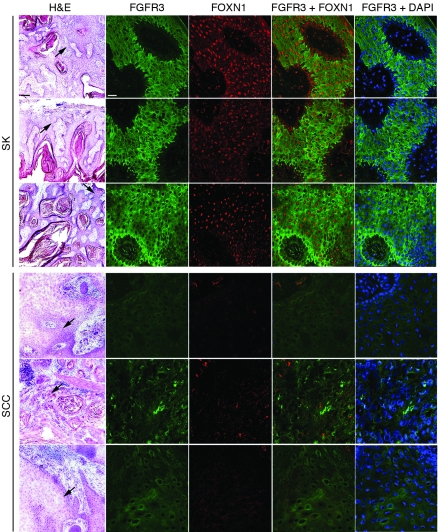
Although our previous work points to the functional importance of Notch1 and p53 in negative control of keratinocyte tumor development (9, 11), their expression in SKs was increased only a few fold. Upstream of these genes, we hypothesized that there may be others with more profound upregulation in these lesions. In fact, cDNA microarray analysis of multiple independent SK lesions versus normal epidermis controls pointed to a limited subset of genes with drastically increased expression in SKs (Supplemental Tables 1 and 2; supplemental material available online with this article; doi: 10.1172/JCI38543DS1). Besides genes of unknown or uncertain function in the skin, these included FGFR3, which has been previously implicated in the pathogenesis of SK (20), and FOXN1, a transcription factor with a key role in keratinocyte differentiation control (32, 33, 35). Real-time RT-PCR confirmed that these genes were strongly upregulated in SKs and showed that, in contrast, FOXN1 and FGFR3 were suppressed in SCCs (Figure 2, A and B). Paralleling the mRNA expression results, immunofluorescence analysis with an anti-FOXN1 antibody confirmed that SKs express high levels of this protein, which was instead detected in very few cells in SCCs (Figure 3 and Supplemental Figure 1). Immunofluorescence with FGFR3-specific antibodies produced a similarly high signal in SKs, while staining in SCCs was close to undetectable (Figure 3 and Supplemental Figure 1).
Figure 2. FOXN1 and FGFR3 mRNA levels are differentially modulated in SK versus SCC lesions.
(A) RNAs extracted from 7 separate SK lesions, 7 separate surgically excised SCC lesions, and 3 normal, age-matched control epidermis specimens were subjected to real-time RT-PCR with oligonucleotide primers for gene expression levels of FOXN1. The β-actin mRNA level was used for internal normalization. Similar results were obtained after normalization to 36B4 in this and other experiments. Gene expression values in SK and SCC specimens are expressed as fold expression relative to the average of the 3 normal controls. We similarly analyzed 4 other SCC samples obtained by laser capture microdissection (LCM) along with surrounding control epidermis. (B) Samples as in A were analyzed for FGFR3 mRNA expression by real-time RT-PCR, with corresponding specific primers and β-actin normalization. Error bars denote SEM.
Figure 3. Different expression levels of FOXN1 and FGFR3 in SK versus SCC tissue samples.
Frozen sections from 3 different surgically obtained SK and SCC tissue samples were stained with H&E or analyzed for expression of FGFR3 and FOXN1 by immunofluorescence analysis with corresponding primary antibodies and using FITC- (green) and rhodamine-conjugated (red) secondary antibodies, respectively, for detection. Concomitant staining patterns of FOXN1 and FGFR3 or of FGFR3 and DAPI (used for overall nuclear counterstaining) were visualized by image overlay. Fluorescent images correspond to the areas of the H&E sections indicated by arrows. No positive fluorescence signal was obtained when primary antibodies were preincubated with corresponding blocking peptides for 2 hours prior to incubation on sections (not shown). Scale bars: 200 μm (H&E); 20 μm (fluorescence).
Opposite control of FOXN1 expression by EGFR versus FGFR3 activation.
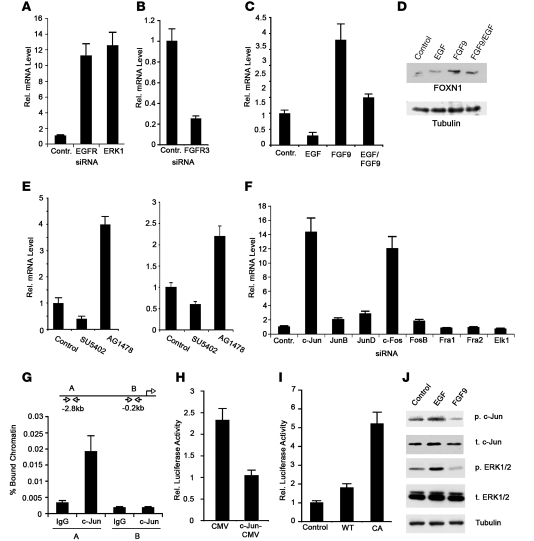
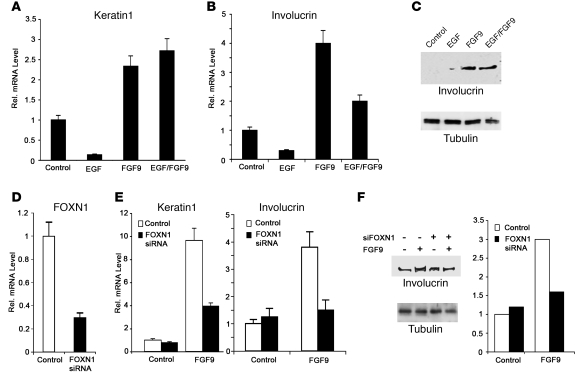
Little is known about the control of FOXN1 gene expression. Given that development of skin SCC is associated with an elevation of EGFR/ERK signaling (43), an attractive possibility was that suppressed FOXN1 expression in these lesions is linked, at least in part, to activation of the EGFR pathway. Conversely, a hallmark of SKs is activation and/or increased expression of FGFR3 (20), which suggested that elevated FOXN1 expression in SKs is caused, at least in part, by increased FGFR3 signaling. Consistent with these possibilities, FOXN1 expression in human keratinocytes was substantially induced by siRNA-mediated knockdown of the EGFR gene or its downstream effector ERK1, whereas it was suppressed by knockdown of FGFR3 (Figure 4, A and B). Mirroring these results, expression of FOXN1 was downmodulated by treatment with EGF, while it was markedly induced by FGF9, a FGFR ligand with highest affinity for FGFR3 (Figure 4, C and D, and refs. 44, 45). Importantly, when human keratinocytes were treated with EGF and FGF9 together, suppression of FOXN1 expression by EGFR was overcome by concomitant FGFR3 activation (Figure 4, C and D). These findings were further validated by treatment of cultured human primary keratinocytes or intact human skin explants with the FGFR- and EGFR-specific inhibitors SU5402 and AG1478, respectively: FOXN1 expression was reduced upon FGFR inhibition, while EGFR inhibition caused an induction (Figure 4E).
Figure 4. Opposite control of FOXN1 expression by EGFR and FGFR3.
(A and B) Human keratinocytes were transfected with siRNAs for EGFR, ERK1 (A), FGFR3 (B), or control siRNAs, and levels of FOXN1 mRNA were measured by real-time RT-PCR. For internal normalization, here and in all subsequent analyses, 36B4 was used. (C and D) Human keratinocytes were treated with 1 ng/ml EGF or 5 ng/ml FGF9 for 24 hours, followed by real-time RT-PCR analysis (C) or immunoblotting for FOXN1 expression (D). (E) Human keratinocytes (left) and fresh human skin explants (right) were treated with DMSO or with FGFR (SU5402) or EGFR (AG478) inhibitors and analyzed by real-time RT-PCR for FOXN1. (F) Human keratinocytes were transfected with siRNAs for the indicated genes or with control siRNA. Levels of FOXN1 mRNA were assessed by real-time RT-PCR. (G) Human keratinocytes were processed for ChIP with an antibody against c-Jun and control rabbit IgG. Real-time PCR was used to amplify regions of the human FOXN1 promoter using specific primers (Supplemental Table 3). (H) Human keratinocytes were cotransfected with a FOXN1 reporter together with a c-Jun plasmid (c-Jun–CMV; ref. 60) or empty vector control (CMV). Luciferase activity was determined using Renilla for normalization. (I) Human keratinocytes were cotransfected with a FOXN1 reporter with or without expression vectors for wild-type FGFR3, a constitutively active (CA) mutant (61), or control. Promoter activity was measured as described above. (J) Human keratinocytes were treated as in C and D, followed by immunoblot for phosphorylated (p) and total (t) c-Jun and ERK1/2. All error bars denote SEM.
A key AP-1 family member and mediator of EGFR/ERK effects on gene expression, c-Jun functions as either inducer or repressor of transcription, depending on target genes (46). We observed induction of endogenous FOXN1 expression similar to that caused by EGFR suppression after siRNA-mediated knockdown of c-Jun as well as c-Fos, which functions in association with c-Jun (47); however, little or no effect was observed after knockdown of other AP-1 family members, nor of Elk-1, a transcription factor activated by EGFR activation though an AP-1–independent mechanism (Figure 4F and ref. 48). The human FOXN1 promoter region contains a putative c-Jun binding site at positions –2.8 kb from the transcription initiation site to which, by chromatin immunoprecipitation (ChIP) experiments, endogenous c-Jun was found to bind (Figure 4G). Transcriptional activity of the FOXN1 promoter was substantially reduced by c-Jun overexpression, whereas it was increased in cells transfected with an expression vector for wild-type FGFR3 and, to a greater extent, vector for FGFR3 bearing an activating mutation G380R found in SKs (Figure 4, H and I, and ref. 20). Paralleling these results, levels of phospho–c-Jun as well as activated phospho-Erk1 were increased in human keratinocytes by EGF treatment and reduced by FGF9 (Figure 4J).
FGFR3-FOXN1 cross connection in control of keratinocyte differentiation.
FOXN1 was previously implicated in positive control of keratinocyte differentiation (32, 34, 49). In parallel with FOXN1 expression, real-time RT-PCR analysis showed that activation of FGFR3 by treatment of human keratinocytes with FGF9 caused induction of differentiation markers, like keratin1 and involucrin, that was suppressed by EGF treatment. Concomitant treatment with FGF9 and EGF showed that, as for FOXN1 expression, FGFR3 activation had a dominant role in induction of differentiation over suppression by EGFR (Figure 5, A–C). Importantly, induction of differentiation marker expression by FGF9 treatment was markedly suppressed by FOXN1 knockdown, which indicates that the prodifferentiating function of FGFR3 activation is linked to induction of FOXN1 (Figure 5, D–F).
Figure 5. FGFR3 activation induces keratinocyte differentiation through a FOXN1-dependent mechanism.
(A and B) Human keratinocytes were treated with 1 ng/ml EGF or 5 ng/ml FGF9 separately or in combination for 24 hours followed by real-time RT-PCR analysis of keratin1 (A) and involucrin (B). (C) Human keratinocytes treated as in A and B were analyzed for involucrin expression by immunoblotting with γ-tubulin as equal loading control. (D) Human keratinocytes were transfected with anti-FOXN1 siRNAs or scrambled siRNA control followed 48 hours later by real-time RT-PCR analysis to verify efficacy of gene knockdown. (E) Keratinocytes transfected with anti-FOXN1 siRNAs or scrambled siRNA control as in D were left untreated or treated with 5 ng/ml FGF9 for the last 24 hours of the experiment. Expression levels of keratin1 and involucrin were determined by real-time RT-PCR. (F) Primary human keratinocytes transfected with FOXN1 siRNA were left untreated or treated with FGF9 as in E, followed by immunoblot analysis of involucrin expression with γ-tubulin as equal loading control. Results were quantified by densitometric scanning of the immunoblots and normalization for γ-tubulin. All error bars denote SEM.
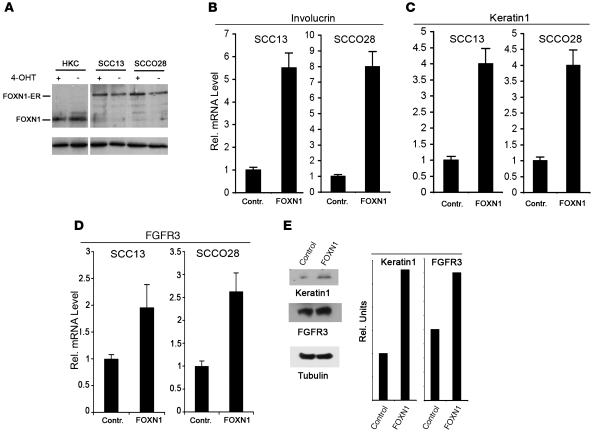
As in the case of cutaneous SCC from patients, FOXN1 expression was almost undetectable in keratinocyte-derived SCC cell lines (Supplemental Figure 2). Infection of these cells with a retroviral vector expressing the FOXN1 protein fused to an estrogen receptor (ER) domain (34) resulted in expression levels of the fusion protein comparable to those of endogenous FOXN1 in normal human keratinocytes (Figure 6A). Activation of FOXN1-ER by 4-hydroxytamoxifen (4-OHT) treatment was sufficient to induce differentiation markers as well as FGFR3 expression (Figure 6, B–E), confirming the existence of a positive cross-talk between FOXN1 and FGFR3 in control of keratinocyte differentiation. Similarly, transduction of SCCO28 cells with FOXN1 adenoviral construct resulted in positive regulation of FGFR3 and the differentiation marker keratin1 (Figure 6E).
Figure 6. Increased FOXN1 expression in SCC cells induces FGFR3 expression and promotes differentiation.
(A) The keratinocyte-derived SCC cell lines SCC13 and SCCO28 were infected with a retrovirus expressing FOXN1-ER fusion protein (34) or empty vector control. Cells with or without 200 nM 4-OHT treatment for 24 hours were analyzed, in parallel with control human keratinocytes, for levels of FOXN1 expression by immunoblotting, with γ-tubulin for equal loading control. Endogenous FOXN1 was detected at 69 kDa, and the higher–molecular weight band in the SCC cells corresponds to expression of the FOXN1-ER fusion protein. Lanes were run on the same gel but were noncontiguous (white line). (B–D) SCC13 and SCCO28 cells infected with the FOXN1-ER retrovirus or empty vector control were treated with 200 nM 4-OHT for 24 hours to induce activation of FOXN1-ER. Expression of involucrin (B), keratin1 (C), and FGFR3 (D) was analyzed by real-time RT-PCR using 36B4 for normalization. Error bars denote SEM. (E) SCCO28 cells were transduced with a FOXN1-expressing adenovirus (41) or GFP control and analyzed for expression of FGFR3 and keratin1 by immunoblotting, with γ-tubulin for equal loading control. Results were quantified by densitometric scanning of the immunoblots and normalization for γ-tubulin.
FOXN1 expression shifts the malignant behavior of SCC cells toward a benign SK-like phenotype.
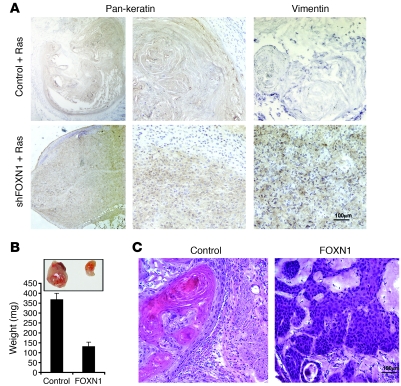
An inverse relationship exists between differentiation and tumor development. To assess whether suppression of FOXN1 expression can contribute to SCC development, expression of this gene was knocked down in human keratinocytes by shRNA retroviral vector infection. Real-time RT-PCR analysis confirmed efficient downmodulation of the gene by 2 different FOXN1 shRNA vectors versus empty vector control (Supplemental Figure 3). Cells were subsequently infected with an oncogenic RAS-expressing retrovirus followed by injection into mice at the dermal-epidermal junction, i.e., a location approximating that of malignant skin tumor formation. RAS-expressing control cells formed highly keratinized cystic lesions with limited cellularity (Figure 7A). Conversely, human keratinocytes with concomitant RAS expression and FOXN1 knockdown produced tumors with high cellularity and low grade of differentiation. There was also widespread positive staining for vimentin, a marker of aggressive epithelial tumors (50), which was absent in the lesions formed by the control cells (Figure 7A).
Figure 7. FOXN1 expression controls tumor formation in primary human keratinocytes and SCC cells.
(A) Human keratinocytes infected with 2 shRNA constructs targeting FOXN1 expression (shFOXN1) in parallel with empty vector control, were superinfected with a H-rasV12–expressing retrovirus (56). Cells (1 × 106) were injected at the dermal-epidermal junction of the back skin of nude mice. Tissue samples from 4 individual tumors and 2 controls were analyzed by immunohistochemistry with anti–pan-keratin and anti–human-specific vimentin antibodies. (B and C) SCC13 cells infected with a FOXN1-ER–expressing retrovirus or empty vector control were injected in parallel into the right and left suprascapular areas of NOD/SCID mice (5 × 105 cells per injection), After 1 week, animals were treated with 200 μg 4-OHT (see Methods) or vehicle control by i.p. injections for 5 weeks. Tumors (7 per condition) were separated from surrounding tissue for weight determination (B), and tissue samples were processed for H&E analysis (C). Similar results were obtained in a second similar experiment. Error bars denote SEM. Scale bars: 100 μm.
The positive connection between FGFR3 and FOXN1 expression and keratinocyte differentiation suggested that restored expression of these genes in SCC cells may be sufficient to counteract their malignant potential and induce a more benign phenotype. To assess this possibility, we generated a xenograft mouse model using the inducible FOXN1-ER SCC13 cell line. Control SCC cells subcutaneously injected into immunocompromised mice formed sizeable tumors within 6 weeks, whereas SCC cells expressing FOXN1 formed only small nodules (Figure 7B). Tumors formed by SCC13 cells, reflecting their origin from a skin SCC (51), showed a collection of large, epithelioid cells with striking nuclear atypia and areas of abrupt keratinization and keratin pearls, typical of moderately differentiated SCC. In contrast, tumors formed by these cells expressing FOXN1 showed pronounced histological features of SKs, including sheets of basaloid cells with large but monomorphic nuclei and easily appreciated intercellular bridges (Figure 7C).
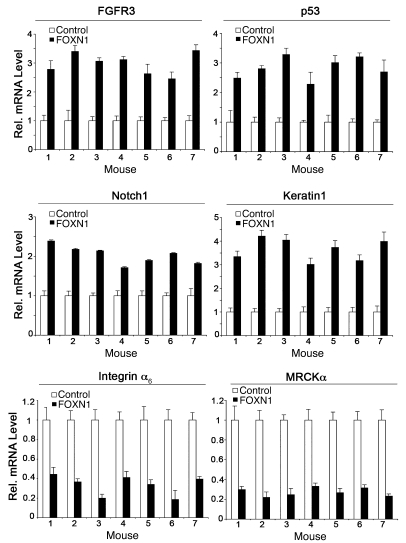
As described above, besides FOXN1, SK lesions were characterized by a concomitant increase in FGFR3, Notch1, and p53 expression, whereas the same genes were downmodulated in SCCs (Figures 1 and 2). Relative to the controls, in tumor nodules formed by SCC cells with induced FOXN1 expression, expression of all these genes was increased, with a parallel upregulation of keratin1 (Figure 8). Expression of integrin α6 and ΜRCKα, which are oppositely regulated in SCCs versus SKs (Figure 1), was negatively affected by FOXN1 expression (Figure 8).
Figure 8. Increased FOXN1 expression converts the gene signature of tumors formed by SCC cells.
The 7 individual pairs of tumors, obtained as described in Figure 7 by parallel injections of mice with FOXN1-ER–expressing SCC13 cells versus controls, were analyzed by real-time RT-PCR for expression of the indicated genes. In each case, gene expression levels in the tumor formed by FOXN1-ER–expressing cells are shown relative to those in the tumor formed by control cells in the same mouse. Error bars denote SEM.
Discussion
Many types of clinically occurring benign tumors fail to progress and invade, for yet-unknown reasons. SKs and SCCs are both derived from keratinocytes and provide an accessible system in which to study the dichotomy between benign and malignant behavior. By comparing the 2 lesions, we found that genes previously associated with keratinocyte tumor formation were similarly modulated or differed only few folds, making them unlikely independent determinants of malignant behavior. Instead, we identified a positive regulatory loop between FGFR3 and FOXN1 activity and expression that underlies the benign skin tumor phenotype.
The impact of FGFR3 signaling on tumor growth appears to be cell type specific. Activating FGFR3 mutations have been implicated in the pathogenesis and progression of hematopoietic malignancies like multiple myeloma (31), whereas similar activating mutations have been linked with development of benign tumors in the skin and with tumors with low likelihood of malignant progression in the bladder (18, 26, 28). In the skin, the fact that transgenic mice with keratinocyte-specific expression of activated FGFR3 develop SK-like lesions (20) indicates that this pathway elicits a positive growth-promoting function. However, we have shown here that FGFR activation also triggers a differentiation-promoting signal through induction of FOXN1 expression, a cell type–specific transcription factor with a positive role in the early stages of the keratinocyte differentiation process (32–35). Besides its direct impact on keratinocytes, FOXN1 plays an additional function in these cells, promoting melanocyte migration (41), which provides a possible explanation for another commonly observed feature of SKs: their increased pigmentation (12). This is also a characteristic of epidermal nevi, another type of benign lesion recently associated with FGFR3 mutations (26), suggesting that even in this case, FOXN1 expression is increased.
FGFR3 activation is specifically involved in positive control of FOXN1 expression, which is unaffected by other FGFR family members, such as FGFR1 and FGFR2 (our unpublished observations). Consistent with previous findings (49), we found that expression of the FOXN1 gene was under negative control of EGFR/ERK signaling. Importantly, suppression of FOXN1 expression by EGFR was overcome when FGFR3 was concomitantly activated. This provides an attractive explanation for the elevated FOXN1 expression in, as well as the differentiated features of, benign tumors in which both EGFR and FGFR3 activities are increased, as opposite to malignant tumors, in which only EGFR is active. ERK kinases are critical effectors of tyrosine kinase receptor function. In other cell types in which FGFR3 signaling induces oncogenic transformation, FGF ligand stimulation elicits ERK activation, whereas FGFR3 inhibition causes the opposite effects (52). The opposite regulation of ERK1 activity in keratinocytes is likely to be a crucial determinant of the cell type–specific consequences of FGFR3 in these cells, through a mechanism possibly involving inactivating phosphatases. Downstream of ERK1, we found that c-Jun activation was also negatively regulated by FGFR3 in keratinocytes. Depending on target genes and specific promoter context, c-Jun can function as either activator or suppressor of transcription (46, 53). Our observations establish FOXN1 as a direct negative target of c-Jun, with knockdown of this factor and its upstream regulators ERK1 and MEK1 leading to substantial FOXN1 upregulation. Similarly increased FOXN1 expression may occur in mice with keratinocyte-specific deletion of c-Jun, which share common phenotypic features with FOXN1-overexpressing mice, including curly whiskers, unfused eyelids, and increased keratin10 expression (54).
Little is known of transcriptional control of FGFR3 expression. Our findings establish FOXN1 as an inducer of this gene, which parallels the enhancing effects on differentiation. The resulting positive feedback loop — in which activating mutations of FGFR3 induce FOXN1 expression, which in turn leads to elevated FGFR3 expression — is likely to ensure that affected keratinocytes are locked in a prodifferentiation mode that prevents their malignant progression. This points to a possibly general approach to restraining malignant tumors by shifting their gene expression pattern toward that seen in corresponding benign lesions.
Methods
Microarray analysis, quantitative real-time RT PCR, promoter activity assays, ChIP, and immunodetection techniques.
Conditions for microarray analysis, real-time RT-PCR analysis, ChIP, Western blot, immunofluorescence, and promoter activity assays were as previously described (9). Briefly, for microarrays, 15 μg total RNA from normal epidermis and SK and SCC lesions (all from different patients) were used for RNA probe preparation and hybridization to gene chips (Human Genome U133A 2.0 array; Affymetrix) according to the manufacturer’s recommendations. The list of gene-specific primers is provided in Supplemental Table 3. For promoter activity assays, a 4.5-kbp fragment containing the exon 1b (promoter) region of the mouse FOXN1 gene (55) was cloned into pT7blue vector, and the luciferase cDNA from pGL3.2 (Promega) was then inserted into the unique BstEII site located about 2,900 bp from the upstream end of the cloned genomic fragment and about 300 bp downstream from the transcription start site. FGFR3 constructs were a gift from D. Ornitz (Washington University School of Medicine, St. Louis, Missouri, USA; ref. 20). We used the following antibodies: FOXN1 (Santa Cruz Biotechnology Inc. G-20), keratin1 (Babco AF87), involucrin (Babco PRB140C), FGFR3 (Santa Cruz Biotechnology Inc. C-15), c-Jun (Transduction Labs), c-Jun for ChIP (Santa Cruz Biotechnology Inc.), total and phospho ERK (Cell Signaling), pan-Keratin (AbCam), Vimentin (DakoCytomation V9), and γ-tubulin (Sigma-Aldrich GTU87). Fluorescent images were acquired with a Zeiss LSM510 meta laser-scanning microscope (Carl Zeiss MicroImaging) using the same exposure and image capture conditions, with z stacks of 6–8 planes (1 mm each).
Cell culture and viruses.
Culturing of primary human keratinocytes, SCC13 cells, and SCCO28 cells — as well as infection with the pBabepuro FOXN1-ER (provided by D. Prowse, Cancer Research UK, London, United Kingdom; ref. 34), H-rasV12 (LZRS-rasV12; ref. 56), and control retroviruses; lentiviruses for FOXN1 shRNA; and adenoviruses for FOXN1 (41) — were as previously reported (9). Two individual shRNA constructs in pLKO.1 vector (TRCN0000013193 and TRCN0000013195; Open Biosystems) for FOXN1 were used to prepare lentiviruses. SCC cells were briefly selected for puromycin resistance prior to testing. For knockdown experiments, cells were transfected as described previously (57) with siRNAs for human EGFR and c-Jun (Invitrogen), FGFR3 and FOXN1 (Santa Cruz Biotechnology Inc.), ERK1, ERK2, JNK1, JNK2, c-Jun, JunB, JunD, c-Fos, FosB, Fra1, Fra2, and Elk1 (Gene Globe; Qiagen) in parallel with corresponding siRNA controls and analyzed 48–72 hours after transfection. The EGFR inhibitor AG1478 (LC Labs) and FGFR inhibitor SU5402 (Calbiochem) were dissolved in DMSO and used at concentrations of 2 and 10 μM, respectively; and recombinant EGF and FGF9 were purchased from R&D Systems and used at concentrations of 1.0 and 5.0 ng/ml, respectively.
SK, SCC, and normal skin samples.
Human SK and SCC samples were obtained, with Institutional Review Board approval from MGH, as material discarded during Mohs micrographic surgery. For normal epidermis controls, normal skin was shaved very superficially with a no. 15 scalpel blade with minimal dermal contamination. Alternatively, samples of SCC and surrounding normal epidermis were frozen in OCT and used for laser capture microdissection using an AutoPix Automated Laser Capture Microdissection System, followed by 1 round of linear RNA amplification as we previously described (58).
Organ cultures.
Discarded human skin samples from abdominoplasty procedures were obtained from W. Austen (MGH, Boston, Massachusetts, USA) under patients’ agreement and institutional approval. Skin samples, sterilized in 70% ethanol and cut — after removal of subcutaneous fat — into 1 × 1 cm pieces, were placed in keratinocyte serum-free medium (Gibco; Invitrogen) supplemented with EGF and bovine pituitary extract (BPE), in 0.25% agar (Sigma-Aldrich). The epidermis was maintained at the air-medium interface. For RNA collection, skin samples were placed in preheated PBS at 60°C for 45 seconds, then chilled (on ice) in 0.1M PBS for 1 minute, followed by mechanical separation of epidermis and dermis. The epidermis was homogenized in TRI Reagent (Sigma-Aldrich) for RNA preparation.
Tumorigenicity assays.
For in vivo tumorigenicity assays, control and FOXN1-ER–expressing SCC13 cells were brought into suspension, admixed with Matrigel (BD Biosciences), and injected (5 × 106 cells/injection) subcutaneously in 8-week-old female athymic nude mice. All studies performed in mice were approved by the Institutional Review Board of MGH. After 1 week, animals were treated 3 times (every other day) with 4-OHT (a dose of 200 μg in 100 μl sunflower oil) or vehicle control by i.p. injections for 5 weeks. Mice were sacrificed 2 days after the last treatment, and tumors were processed for RNA preparation and histochemical analysis.
Human keratinocytes were infected with FOXN1 shRNA or control lentiviruses, followed 48 hours later by a second infection with a H-rasV12 retrovirus (LZRS-rasV12; ref. 56). Then, 16 hours later, cells were brought into suspension, and 106 cells were injected at the dermal-epidermal junction of nude mice (2 side flank injections per mouse) using a technique previously used for hair follicle reconstitution assays (59).
Statistics.
Statistical significance of the gene expression studies analysis was assessed by unpaired 2-tailed Student’s t test, with P values less than 0.01 considered highly significant. All results of real-time RT-PCR, promoter activity assays, and tumor measurements are presented as mean ± SEM, as calculated by Excel software.
Supplementary Material
Acknowledgments
We thank W. Austen for human skin material, D. Prowse for the FOXN1-ER retrovirus, D. Ornitz for FGFR3 constructs, and V. Rajashekara for skillful technical help. This work was supported by NIH grants AR39190, AR054856, AR045284, CA16038, and CA73796; by the Swiss National Foundation; by a grant from the European Union (Epistem, Sixth Framework Program, LSHB-CT-2005-019067); and in part by the Cutaneous Biology Research Center through the MGH/Shiseido Co. Ltd. Agreement.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 119:3127–3137 (2009). doi:10.1172/JCI38543.
References
- 1.Owens D.M., Watt F.M. Contribution of stem cells and differentiated cells to epidermal tumours. Nat. Rev. Cancer. 2003;3:444–451. doi: 10.1038/nrc1096. [DOI] [PubMed] [Google Scholar]
- 2.Alam M., Ratner D. Cutaneous squamous-cell carcinoma. N. Engl. J. Med. 2001;344:975–983. doi: 10.1056/NEJM200103293441306. [DOI] [PubMed] [Google Scholar]
- 3.Albert M.R., Weinstock M.A. Keratinocyte carcinoma. CA Cancer J. Clin. 2003;53:292–302. doi: 10.3322/canjclin.53.5.292. [DOI] [PubMed] [Google Scholar]
- 4.Brash D.E., et al. Sunlight and sunburn in human skin cancer: p53, apoptosis, and tumor promotion. J. Investig. Dermatol. Symp. Proc. 1996;1:136–142. [PubMed] [Google Scholar]
- 5.Boyle J.O., et al. The incidence of p53 mutations increases with progression of head and neck cancer. Cancer Res. 1993;53:4477–4480. [PubMed] [Google Scholar]
- 6.Di Como C.J., et al. p63 expression profiles in human normal and tumor tissues. Clin. Cancer Res. 2002;8:494–501. [PubMed] [Google Scholar]
- 7.Brinkman B.M., Wong D.T. Disease mechanism and biomarkers of oral squamous cell carcinoma. Curr. Opin. Oncol. 2006;18:228–233. doi: 10.1097/01.cco.0000219250.15041.f8. [DOI] [PubMed] [Google Scholar]
- 8.Rho O., Beltran L.M., Gimenez-Conti I.B., DiGiovanni J. Altered expression of the epidermal growth factor receptor and transforming growth factor-alpha during multistage skin carcinogenesis in SENCAR mice. Mol. Carcinog. 1994;11:19–28. doi: 10.1002/mc.2940110105. [DOI] [PubMed] [Google Scholar]
- 9.Lefort K., et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes Dev. 2007;21:562–577. doi: 10.1101/gad.1484707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mandinova A., et al. The FoxO3a gene is a key negative target of canonical Notch signalling in the keratinocyte UVB response. EMBO J. 2008;27:1243–1254. doi: 10.1038/emboj.2008.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolev V., et al. EGFR signalling as a negative regulator of Notch1 gene transcription and function in proliferating keratinocytes and cancer. Nat. Cell Biol. 2008;10:902–911. doi: 10.1038/ncb1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stulberg D.L., Clark N., Tovey D. Common hyperpigmentation disorders in adults: Part II. Melanoma, seborrheic keratoses, acanthosis nigricans, melasma, diabetic dermopathy, tinea versicolor, and postinflammatory hyperpigmentation. Am. Fam. Physician. 2003;68:1963–1968. [PubMed] [Google Scholar]
- 13.Toussaint S., Salcedo E., Kamino H. Benign epidermal proliferations. Adv. Dermatol. 1999;14:307–357. [PubMed] [Google Scholar]
- 14.zur Hausen H. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J. Natl. Cancer Inst. 2000;92:690–698. doi: 10.1093/jnci/92.9.690. [DOI] [PubMed] [Google Scholar]
- 15.Lee E.S., Whang M.R., Kang W.H. Absence of human papillomavirus DNA in nongenital seborrheic keratosis. J. Korean Med. Sci. 2001;16:619–622. doi: 10.3346/jkms.2001.16.5.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pariser R.J. Benign neoplasms of the skin. Med. Clin. North Am. 1998;82:1285–1307, v–vi. doi: 10.1016/S0025-7125(05)70416-8. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura H., et al. Clonal nature of seborrheic keratosis demonstrated by using the polymorphism of the human androgen receptor locus as a marker. J. Invest. Dermatol. 2001;116:506–510. doi: 10.1046/j.1523-1747.2001.01289.x. [DOI] [PubMed] [Google Scholar]
- 18.Hafner C., et al. Spectrum of FGFR3 mutations in multiple intraindividual seborrheic keratoses. J. Invest. Dermatol. 2007;127:1883–1885. doi: 10.1038/sj.jid.5700804. [DOI] [PubMed] [Google Scholar]
- 19.Hafner C., et al. High frequency of FGFR3 mutations in adenoid seborrheic keratoses. J. Invest. Dermatol. 2006;126:2404–2407. doi: 10.1038/sj.jid.5700422. [DOI] [PubMed] [Google Scholar]
- 20.Logie A., et al. Activating mutations of the tyrosine kinase receptor FGFR3 are associated with benign skin tumors in mice and humans. Hum. Mol. Genet. 2005;14:1153–1160. doi: 10.1093/hmg/ddi127. [DOI] [PubMed] [Google Scholar]
- 21.Hafner C., et al. Oncogenic PIK3CA mutations occur in epidermal nevi and seborrheic keratoses with a characteristic mutation pattern. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13450–13454. doi: 10.1073/pnas.0705218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohammadi M., Olsen S.K., Ibrahimi O.A. Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev. 2005;16:107–137. doi: 10.1016/j.cytogfr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Burke D., Wilkes D., Blundell T.L., Malcolm S. Fibroblast growth factor receptors: lessons from the genes. Trends Biochem. Sci. 1998;23:59–62. doi: 10.1016/S0968-0004(97)01170-5. [DOI] [PubMed] [Google Scholar]
- 24.Freier K., et al. Recurrent FGFR1 amplification and high FGFR1 protein expression in oral squamous cell carcinoma (OSCC). Oral Oncol. 2007;43:60–66. doi: 10.1016/j.oraloncology.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 25.Grose R., et al. The role of fibroblast growth factor receptor 2b in skin homeostasis and cancer development. EMBO J. 2007;26:1268–1278. doi: 10.1038/sj.emboj.7601583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hafner C., et al. Mosaicism of activating FGFR3 mutations in human skin causes epidermal nevi. J. Clin. Invest. 2006;116:2201–2207. doi: 10.1172/JCI28163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knowles M.A. Role of FGFR3 in urothelial cell carcinoma: biomarker and potential therapeutic target. World J. Urol. 2007;25:581–593. doi: 10.1007/s00345-007-0213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Junker K., et al. Fibroblast growth factor receptor 3 mutations in bladder tumors correlate with low frequency of chromosome alterations. Neoplasia. 2008;10:1–7. doi: 10.1593/neo.07178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Black P.C., Dinney C.P. Growth factors and receptors as prognostic markers in urothelial carcinoma. Curr. Urol. Rep. 2008;9:55–61. doi: 10.1007/s11934-008-0011-6. [DOI] [PubMed] [Google Scholar]
- 30.Onwuazor O.N., et al. Mutation, SNP, and isoform analysis of fibroblast growth factor receptor 3 (FGFR3) in 150 newly diagnosed multiple myeloma patients. Blood. 2003;102:772–773. doi: 10.1182/blood-2003-04-1204. [DOI] [PubMed] [Google Scholar]
- 31.Chesi M., Bergsagel P.L., Kuehl W.M. The enigma of ectopic expression of FGFR3 in multiple myeloma: a critical initiating event or just a target for mutational activation during tumor progression. Curr. Opin. Hematol. 2002;9:288–293. doi: 10.1097/00062752-200207000-00005. [DOI] [PubMed] [Google Scholar]
- 32.Li J., et al. Foxn1 promotes keratinocyte differentiation by regulating the activity of protein kinase C. Differentiation. 2007;75:694–701. doi: 10.1111/j.1432-0436.2007.00176.x. [DOI] [PubMed] [Google Scholar]
- 33.Lee D., Prowse D.M., Brissette J.L. Association between mouse nude gene expression and the initiation of epithelial terminal differentiation. Dev. Biol. 1999;208:362–374. doi: 10.1006/dbio.1999.9221. [DOI] [PubMed] [Google Scholar]
- 34.Janes S.M., Ofstad T.A., Campbell D.H., Watt F.M., Prowse D.M. Transient activation of FOXN1 in keratinocytes induces a transcriptional programme that promotes terminal differentiation: contrasting roles of FOXN1 and Akt. J. Cell Sci. 2004;117:4157–4168. doi: 10.1242/jcs.01302. [DOI] [PubMed] [Google Scholar]
- 35.Brissette J., Li J., Kamimura J., Lee D., Dotto G.P. The product of the mouse nude locus, Whn, regulates the balance between epithelial cell growth and differentiation. . Genes Dev. 1996;10:2212–2221. doi: 10.1101/gad.10.17.2212. [DOI] [PubMed] [Google Scholar]
- 36.Su D.M., Navarre S., Oh W.J., Condie B.G., Manley N.R. A domain of Foxn1 required for crosstalk-dependent thymic epithelial cell differentiation. Nat. Immunol. 2003;4:1128–1135. doi: 10.1038/ni983. [DOI] [PubMed] [Google Scholar]
- 37.Segre J.A., Nemhauser J.L., Taylor B.A., Nadeau J.H., Lander E.S. Positional cloning of the nude locus: genetic, physical, and transcription maps of the region and mutations in the mouse and rat. Genomics. 1995;28:549–559. doi: 10.1006/geno.1995.1187. [DOI] [PubMed] [Google Scholar]
- 38.Nehls M., Pfeifer D., Schorpp M., Hedrich H., Boehm T. New member of the winged-helix protein family disrupted in mouse and rat nude mutations. Nature. 1994;372:103–107. doi: 10.1038/372103a0. [DOI] [PubMed] [Google Scholar]
- 39.Frank J., et al. Exposing the human nude phenotype. Nature. 1999;398:473–474. doi: 10.1038/18997. [DOI] [PubMed] [Google Scholar]
- 40.Schlake T., Schorpp M., Nehls M., Boehm T. The nude gene encodes a sequence-specific DNA binding protein with homologs in organisms that lack an anticipatory immune system. Proc. Natl. Acad. Sci. U. S. A. 1997;94:3842–3847. doi: 10.1073/pnas.94.8.3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiner L., et al. Dedicated epithelial recipient cells determine pigmentation patterns. Cell. 2007;130:932–942. doi: 10.1016/j.cell.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 42.Yugawa T., et al. Regulation of Notch1 gene expression by p53 in epithelial cells. Mol. Cell. Biol. 2007;27:3732–3742. doi: 10.1128/MCB.02119-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scholl F.A., Dumesic P.A., Khavari P.A. Mek1 alters epidermal growth and differentiation. Cancer Res. 2004;64:6035–6040. doi: 10.1158/0008-5472.CAN-04-0017. [DOI] [PubMed] [Google Scholar]
- 44.Ornitz D.M., et al. Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 45.Jin C., et al. Directionally specific paracrine communication mediated by epithelial FGF9 to stromal FGFR3 in two-compartment premalignant prostate tumors. Cancer Res. 2004;64:4555–4562. doi: 10.1158/0008-5472.CAN-03-3752. [DOI] [PubMed] [Google Scholar]
- 46.Schreiber M., et al. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 1999;13:607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Dam H., Castellazzi M. Distinct roles of Jun: Fos and Jun: ATF dimers in oncogenesis. Oncogene. 2001;20:2453–2464. doi: 10.1038/sj.onc.1204239. [DOI] [PubMed] [Google Scholar]
- 48.Friday B.B., Adjei A.A. Advances in targeting the Ras/Raf/MEK/Erk mitogen-activated protein kinase cascade with MEK inhibitors for cancer therapy. Clin. Cancer Res. 2008;14:342–346. doi: 10.1158/1078-0432.CCR-07-4790. [DOI] [PubMed] [Google Scholar]
- 49.Baxter R.M., Brissette J.L. Role of the nude gene in epithelial terminal differentiation. J. Invest. Dermatol. 2002;118:303–309. doi: 10.1046/j.0022-202x.2001.01662.x. [DOI] [PubMed] [Google Scholar]
- 50.Banks-Schlegel S.P., Gazdar A.F., Harris C.C. Intermediate filament and cross-linked envelope expression in human lung tumor cell lines. Cancer Res. 1985;45:1187–1197. [PubMed] [Google Scholar]
- 51.Rheinwald J.G., Beckett M.A. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultures from human squamous cell carcinomas. Cancer Res. 1981;41:1657–1663. [PubMed] [Google Scholar]
- 52.Masih-Khan E., et al. MIP-1alpha (CCL3) is a downstream target of FGFR3 and RAS-MAPK signaling in multiple myeloma. Blood. 2006;108:3465–3471. doi: 10.1182/blood-2006-04-017087. [DOI] [PubMed] [Google Scholar]
- 53.Dunn C., Wiltshire C., MacLaren A., Gillespie D.A. Molecular mechanism and biological functions of c-Jun N-terminal kinase signalling via the c-Jun transcription factor. Cell Signal. 2002;14:585–593. doi: 10.1016/S0898-6568(01)00275-3. [DOI] [PubMed] [Google Scholar]
- 54.Li G., et al. c-Jun is essential for organization of the epidermal leading edge. Dev. Cell. 2003;4:865–877. doi: 10.1016/S1534-5807(03)00159-X. [DOI] [PubMed] [Google Scholar]
- 55.Schorpp M., Hofmann M., Dear T.N., Boehm T. Characterization of mouse and human nude genes. Immunogenetics. 1997;46:509–515. doi: 10.1007/s002510050312. [DOI] [PubMed] [Google Scholar]
- 56.Lazarov M., et al. CDK4 coexpression with Ras generates malignant human epidermal tumorigenesis. Nat. Med. 2002;8:1105–1114. doi: 10.1038/nm779. [DOI] [PubMed] [Google Scholar]
- 57.Nguyen B.C., et al. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 2006;20:1028–1042. doi: 10.1101/gad.1406006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mammucari C., et al. Integration of Notch 1 and calcineurin/NFAT signaling pathways in keratinocyte growth and differentiation control. Dev. Cell. 2005;8:665–676. doi: 10.1016/j.devcel.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 59.Zheng Y., et al. Organogenesis from dissociated cells: generation of mature cycling hair follicles from skin-derived cells. J. Invest. Dermatol. 2005;124:867–876. doi: 10.1111/j.0022-202X.2005.23716.x. [DOI] [PubMed] [Google Scholar]
- 60.Musti A.M., Treier M., Bohmann D. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science. 1997;275:400–402. doi: 10.1126/science.275.5298.400. [DOI] [PubMed] [Google Scholar]
- 61.Henderson J.E., et al. Expression of FGFR3 with the G380R achondroplasia mutation inhibits proliferation and maturation of CFK2 chondrocytic cells. J. Bone Miner. Res. 2000;15:155–165. doi: 10.1359/jbmr.2000.15.1.155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.