Abstract
To date, inheritance of a mutant BRCA1 or BRCA2 gene is the best-established indicator of an increased risk of developing breast cancer. Sequence analysis of these genes is being used to identify BRCA1/2 mutation carriers, though these efforts are hampered by the high frequency of variants of unknown clinical significance (VUSs). Functional evaluation of such variants has been restricted due to lack of a physiologically relevant assay. In this study we developed a functional assay using mouse ES cells to study variants of BRCA1. We introduced BAC clones with human wild-type BRCA1 or variants into Brca1-null ES cells and confirmed that only wild-type and a known neutral variant rescued cell lethality. The same neutral variant was also able to rescue embryogenesis in Brca1-null mice. A test of several BRCT domain mutants revealed all to be deleterious, including a VUS. Furthermore, we used this assay to determine the effects of BRCA1 variants on cell cycle regulation, differentiation, and genomic stability. Importantly, we discovered that ES cells rescued by S1497A BRCA1 exhibited significant hypersensitivity after γ-irradiation. Our results demonstrate that this ES cell–based assay is a powerful and reliable method for analyzing the functional impact of BRCA1 variants, which we believe could be used to determine which patients may require preventative treatments.
Introduction
Breast cancer is one of the leading causes of mortality in women. It is estimated that 184,450 new cases of invasive breast cancer were diagnosed in the United States in 2008 (1). Of all the breast cancer cases, 5%–7% are familial, and mutations in BRCA1 and BRCA2 account for the majority of familial breast cancers (2, 3). BRCA1 or BRCA2 germline mutation carriers have a lifetime risk of up to 80% for developing the disease (4–6). In addition, BRCA1 mutation carriers also have a lifetime risk of up to 56% for developing ovarian cancer (4, 5, 7). Therefore, identification of BRCA1 and BRCA2 mutation carriers is important because it provides an opportunity to take preventive measures such as prophylactic chemotherapy or mastectomy to reduce the mortality and agony associated with the disease. To identify BRCA1 and BRCA2 mutation carriers, sequencing-based genetic tests are available (8). A major caveat of this approach lies in the interpretation of the actual risk(s) associated with mutations that do not clearly disrupt the gene and are considered as variants of unknown clinical significance (VUSs). The prevalence of VUSs in the population is high. A study conducted by Myriad Genetics Inc. reported that 13.0% of 10,000 individuals tested had a VUS (9). More than 800 BRCA1 and 1,100 BRCA2 VUSs, each with a single nucleotide substitution, are listed in the Breast Cancer Information Core (BIC) database (http://research.nhgri.nih.gov/bic/) database. Therefore, it is imperative to understand the functional significance of such variants.
At present, segregation of the mutation with the disease in families provides the most reliable information to assess cancer risk. However, such information is frequently not available. Recently, based on co-occurrence in trans with a known deleterious mutation, detailed analysis of personal and family history of cancer in probands and cosegregation of the variant with disease in pedigrees, 1,433 BRCA1 and BRCA2 variants were examined and their odds in favor of causality or neutrality reported (10). Such studies are valuable, but the predictions require validation.
Functional assays based on complementation by cDNAs in BRCA-deficient cell lines have been developed (11–13). Although these can distinguish between neutral and deleterious variants, they are of very limited use. An assay based on the transcriptional activation function of the C-terminal domain of BRCA1 is also frequently used (14). Because BRCA1 is known to be involved in multiple cellular processes, an assay that can be used to study various aspects of BRCA1 function is much needed. Recently, we reported the use of mouse ES cells and bacterial artificial chromosomes to develop a physiologically relevant method to study BRCA2 variants (15). Using this assay, we examined the functional significance of BRCA2 variants. Using a similar approach, we have now developed a functional assay to study BRCA1 variants. We have used the assay to classify clinically relevant variants that map to various functional domains. In addition, we have examined the physiological significance of multiple phosphorylation sites identified in BRCA1. Phosphorylation of BRCA1 in response to DNA damage is fundamental for its proper function. In vitro studies have revealed the presence of multiple phosphorylation sites (16–20). We have examined the consequence of disrupting these residues to validate our functional assay as well as to reveal the physiological relevance of these phosphorylation sites.
Results
Generating an ES cell–based functional assay for BRCA1.
BRCA1 is essential for the viability of mouse ES cells (21, 22). Based on this observation, we used mouse ES cells to generate an assay to study the functional significance of human BRCA1 variants (Figure 1A). We hypothesized that the neutral variants will rescue the lethality of Brca1-null (designated as Brca1KO/KO) ES cells and also complement known functions of BRCA1. Conversely, deleterious/hypomorphic variants may either fail to rescue the ES cell lethality or may not be fully functional when tested for known functions of BRCA1 in DNA repair, cell cycle regulation, transcriptional regulation, etc.
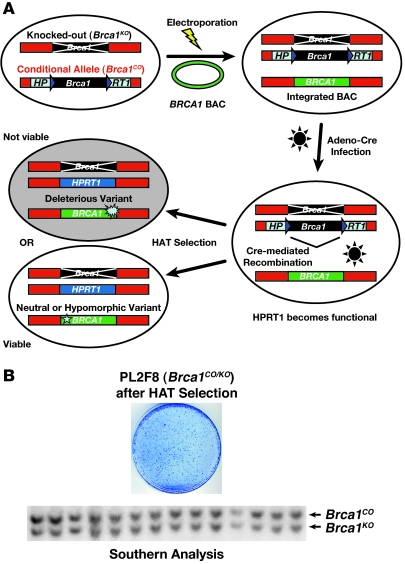
Figure 1. Outline of ES cell–based assay.
(A) Schematic representation of BRCA1 ES cell–based functional assay. Human BRCA1 BAC DNA with any mutation can be introduced into mouse PL2F8 ES cells containing a conditional allele of Brca1. After Cre-mediated recombination of the conditional allele, a functional copy of the human HPRT1 gene is generated and renders the cells resistant to HAT selection. Depending upon the impact of the mutation on the BRCA1 function, the cells will be viable or not after HAT selection. (B) ES cells lacking functional BRCA1 are not viable. Cre-mediated recombination of the conditional allele of Brca1 in PL2F8 ES cells results in few viable HAT-resistant colonies. Genotyping of these clones shows that they retain a conditional allele of Brca1 (lower panel, upper band).
To establish the functional assay, we engineered mouse ES cells by the gene targeting method. First, we modified one allele of Brca1 as a conditional allele (designated as Brca1CO) by flanking the locus with 2 loxP sites (Supplemental Figures 1 and 2; supplemental material available online with this article; doi: 10.1172/JCI39836DS1). We inserted two halves of the human HPRT1 minigene (5′HPRT1 and 3′HPRT1) along with the 2 loxP sites to allow selection of clones that undergo Cre-mediated recombination of the loxP sites in hypoxanthine aminopterin thymidine–containing (HAT-containing) medium (Figure 1A and Supplemental Figure 1). To allow HAT selection, we used Hprt-deficient AB2.2 ES cells. We also targeted a puromycin resistance gene (Puro) next to the 3′HPRT1 cassette as an additional selectable marker (Supplemental Figure 1). Next, we disrupted the second allele of Brca1 by deleting exons 3–7 and replacing the region with a blasticidin resistance gene (Blasti) (Supplemental Figure 3).
The engineered ES cell clone used for the functional assay is referred to as PL2F8. Loss of the conditional allele is expected to yield no viable HAT-resistent colonies due to loss of BRCA1. To test this, we transiently expressed Cre recombinase in PL2F8 cells and selected the recombinant clones in the presence of HAT. We obtained 400–600 HAT-resistent colonies per 105 Cre-electroporated ES cells (Figure 1B). Genotyping of 200 clones by Southern blot analysis revealed all HAT-resistent clones to be Brca1 heterozygous (represented in Figure 1B; designated as Brca1CO/KO). We concluded that none of the HAT-resistent clones were Brca1-null (Brca1KO/KO) and all the viable HAT-resistent clones retained a copy of the conditional allele and were Brca1KO/+. The precise mechanism by which the HAT-resistent Brca1CO/KO clones are generated remains unknown. However, we hypothesize that some ES cells may carry trisomy of chromosome 11, giving rise to 2 copies of conditional allele, only one of which recombines to confer HAT resistance. This idea is supported by the karyotypic analysis of HAT-resistent clones, which showed the presence of 3 copies of chromosome 11 (Supplemental Figure 4). In addition, chromosomal anomalies are normally present in some ES cells, and trisomy of chromosome 11 has been reported in mouse ES cells (23).
If, indeed, Brca1KO/KO ES cells fail to survive due to the loss of Brca1, can the lethality of these cells be rescued by the presence of a BRCA1 transgene? To address this, we introduced a BAC clone (clone RPCI11-812-5O) containing a wild-type copy of human BRCA1 into PL2F8 cells. Expression of the BRCA1 transgene was confirmed by Western blot analysis (Figure 2A). This BAC was previously used to rescue the lethality of Brca1-deficient mice (24). When we expressed Cre in PL2F8 ES cells expressing the human BRCA1 transgene, we obtained about 1,000 HAT-resistent colonies (Figure 2B), and 44 of 96 (45.8%) were confirmed to be Brca1KO/KO (Figure 2C, WT). The remaining clones were Brca1CO/KO. These results demonstrated the ability of the human BRCA1 to rescue the lethality of Brca1KO/KO ES cells.
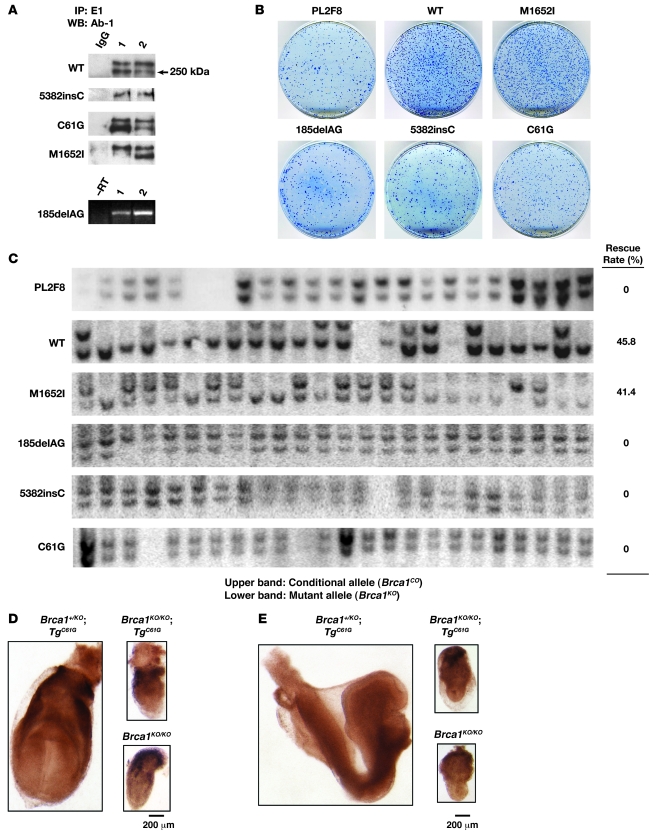
Figure 2. Validation of ES cell–based assay by using WT BRCA1 and variants of known clinical implication.
(A) Expression analysis of WT BRCA1, M1652I, 2 founder mutations (185delAG and 5382insC), and C61G variants using human BRCA1 BAC in mouse ES cell (2 clones for each). Protein expression was confirmed by immunoprecipitation using a human BRCA1-specific antibody (E1) followed by Western blot analysis using a commercial BRCA1-specific antibody (Ab-1), which gives 2 bands of about 250 kDa (48). Expression of 185delAG was detected by RT-PCR (lower panel). –RT, no RT, negative control. (B) Methylene blue staining of HAT-resistant ES cell colonies with no BAC (PL2F8), WT M1652I, 185delAG, 5382insC, and C61G BRCA1 BAC transgene. (C) Genotyping of HAT-resistant colonies by Southern blot analysis. The upper band representing the conditional allele was deleted in 45.8% of the cells with WT or 41.4% with M1652I BRCA1. In contrast, none of the colonies from PL2F8 cells or those expressing 185delAG, 5382insC, and C61G BRCA1 variants showed loss of the conditional allele. (D) Whole mount of a Brca1KO/KOBACTgC61G embryo at E7.5 showing retarded development, similar to the Brca1KO/KO embryo. Brca1KO/+BACTgC61G control embryo shows normal embryonic development at E7.5. (E) Whole mount of a Brca1KO/KOBACTgC61G embryo at E8.5 shows severe developmental defects compared with a Brca1KO/+BACTgC61G embryo (left).
Analysis of BRCA1 variants with known clinical impact.
Based on the ability of wild-type human BRCA1 to rescue the lethality of Brca1KO/KO ES cells, we proceeded to analyze various BRCA1 variants in PL2F8 ES cells (summarized in Table 1). First, we examined 3 variants known to be deleterious (185delAG, 5382insC, C61G) and a neutral variant (M1652I) to test the ability of the ES cell–based system to classify these variants. 185delAG and 5382insC are the 2 well-known founder mutations in BRCA1 that are associated with increased risk of cancer development (25, 26). 185delAG results in a frameshift at codon 23, causing a premature translation stop after codon 38. Similarly 5382insC is known to cause protein truncation at codon 1,829. The C61G amino acid change involves one of the conserved cysteine residues of the RING domain, and the presence of a glycine residue is predicted to disrupt the integrity of this domain (8). In contrast, the M1652I amino acid change is reported to be a neutral variant based on the observation that it is found at a frequency of 4.05% in the population of cancer-free controls of European ancestry (9). We verified the expression of each variant by Western blot analysis with human-specific BRCA1 antibody, with the exception of 185delAG, which is predicted to result in a truncated protein (Figure 2A). Expression of the 185delAG transcript was confirmed by RT-PCR (Figure 2A, lower panel).
Table 1 .
Summary of Brca1 mutants analyzed in this study
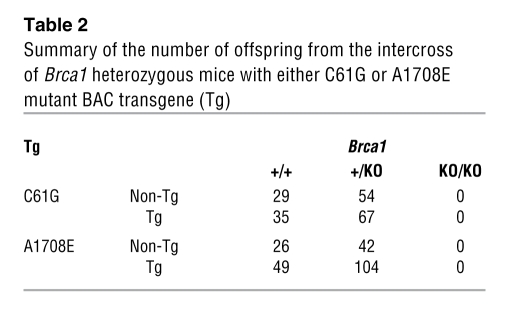
After Cre expression, 185delAG, 5382insC, and C61G resulted in a background level of HAT-resistent colonies (Figure 2B, compare with PL2F8), whereas M1652I yielded colonies similar to those expressing wild-type BRCA1 (Figure 2B). Genotyping of these clones confirmed that 185delAG, 5382insC, and C61G are unable to rescue the lethality of Brca1KO/KO cells and are therefore deleterious (Figure 2C). In contrast, approximately 41% of the HAT-resistent colonies obtained by introducing the M1652I variant were Brca1KO/KO (Figure 2C). Furthermore, when we tested the sensitivity of Brca1KO/KO ES cells rescued by M1652I to various DNA-damaging agents, we observed no differences compared with the control cells (Supplemental Table 1, bottom line), confirming that it is a neutral variant. To further validate our findings and demonstrate the in vivo relevance of our ES cell–based approach to studying BRCA1, we examined the effect of the M1652I and C61G variants in mice. We used the BACs expressing the 2 variants to generate transgenic mice. The expression of mutant BRCA1 in mouse was confirmed by RT-PCR and Western blot analysis (Supplemental Figure 5 and data not shown). The transgenic mice were crossed to Brca1 heterozygous mice. The resulting Brca1KO/+BACTgM1652I and Brca1KO/+BACTgC61G mice were next crossed to Brca1KO/+ mice to test the ability of the two variants to rescue the lethality of Brca1KO/KO, as shown previously for wild-type BRCA1 (24). While we obtained viable Brca1KO/KOBACTgM1652I mice, we failed to obtain Brca1KO/KOBACTgC61G mice. Brca1KO/KO mice rescued by the M1652I variants are phenotypically indistinguishable from control littermates expressing a wild-type copy of Brca1 (data not shown), suggesting that this BRCA1 variant is functionally similar to the wild-type allele. In contrast, the C61G variant failed to rescue the lethality of Brca1KO/KO mice (Table 2). Next, we examined the phenotype of mutant embryos to determine whether the C61G variant is partially functional in mice. At E7.5 and E8.5, the Brca1KO/KOBACTgC61G embryos were phenotypically similar to the Brca1KO/KO embryos (Figure 2, D and E), with a marked defect in embryogenesis compared with the control embryos. The mutant embryos were much smaller in size than control embryos and exhibited no signs of gastrulation or primitive streak formation. In contrast, there was no difference between Brca1KO/KOBACTgM1652I and Brca1+/KOBACTgM1652I embryos at E7.5 (data not shown), providing additional evidence that this variant is neutral. These results suggested that the ES cell–based system reflects the functional impact of BRCA1 variants and is able to recapitulate the in vivo effects of the mutation.
Table 2 .
Summary of the number of offspring from the intercross of Brca1 heterozygous mice with either C61G or A1708E mutant BAC transgene (Tg)
Functional analysis of BRCT domain variants.
We next generated 3 variants that mapped to the BRCT domain of BRCA1 and are either predicted to be deleterious or are of unknown clinical significance based on BIC database, structural analysis, transcriptional activity assay, and in silico prediction (11, 27–30). The first variant is A1708E, which maps to the first BRCT domain and has been shown to be critical for transcriptional activity. It is considered to be a deleterious, cancer-causing variant. Another variant, V1804D, present in the second BRCT domain, is predicted to be a neutral variant based on the yeast small colony assay and transcriptional activity assay (27, 29). However, the transcriptional activity was estimated to about 50% of the wild-type BRCA1, and structural analysis of the BRCT domain predicted this variant to be deleterious (31). Thus, the predicted role of this variant is controversial. The third variant (R1737X) we tested results in a truncated protein that retains the first BRCT but lacks the second BRCT domain. A mouse model (Brca11700T) expressing such a truncated protein has been generated and predicted to retain some BRCA1 functions based on the observation that the mutant embryos progress a little further in development than the Brca1-null embryos (32).
To determine whether these BRCA1 variants were defective in their transcriptional activation function, we examined their ability to bind to the promoters regulated by BRCA1. Failure to bind to the promoter will abrogate the transcriptional activation of its target genes, such as Gadd45a and p21 (33, 34). ChIP results showed that V1804D, A1708E, and R1737X BRCA1 exhibited marked reduction in their binding to the Gadd45a and p21 promoters compared with the wild-type BRCA1, suggesting that the access to these promoters is blocked or reduced (Figure 3A). Therefore, these variants are likely to be defective in their transcriptional activation function. The precise mechanism of the binding defect is unclear. One possibility is that the interaction between these variants and RNA polII transcriptional machinery is disrupted, as previously shown for M1775E BRCA1 (35).
Figure 3. Functional evaluation of BRCT domain variants of BRCA1.
(A) A1708E, R1737X, and V1804D BRCA1 variants were analyzed by ChIP to test their binding to Gadd45a and p21 promoters. Wild-type human BRCA1 was used as positive control. Binding of p53 to the p21 promoter was used as an internal control. Quantitation of ChIP-PCR products is shown in the graph. (B) Methylene blue staining of HAT-resistant ES cell colonies with no BAC (PL2F8), WT BRCA1, and A1708E, R1737X, and V1804D variants of BRCA1. (C) Southern blot analysis of HAT-resistant ES cell clones reveals that none of the clones expressing deleterious BRCA1 variants (A1708E, R1737X, V1804D) lost the conditional allele (upper band). Half of clones rescued with WT BRCA1 retained only the mutant band (lower band). (D) Whole mount of Brca1KO/KOBACTgA1708E embryo at E7.5 shows retarded development, similar to the phenotype of Brca1KO/KO embryos at this stage. Brca1+/+ control littermates with or without the A1708E transgene show normal development at E7.5.
We next tested their ability to rescue the lethality of Brca1KO/KO ES cells. In all cases, we obtained only background level of colonies, and none were Brca1KO/KO (Figure 3, B and C), suggesting that these variants are all deleterious. These results also demonstrate the essential role of transcriptional control and, possibly, phosphopeptide binding activity of BRCA1. The lethal effect of V1804D mutation in the ES cell–based assay suggests that this variant is deleterious, probably due to the structural disruption of BRCT domain, as previously predicted (29).
To test the in vivo relevance of the results obtained in ES cells, we generated BAC transgenic mice expressing the A1708E variant. As shown in Table 2, the BAC transgenic mice failed to survive on a Brca1-null background. Furthermore, the Brca1KO/KOBACTgA1708E embryos exhibited a phenotype similar to that of the Brca1KO/KO embryos at E7.5 (Figure 3D). Thus, the results obtained from ES cell are physiologically relevant and were corroborated by our studies in mice.
Analysis of BRCA1 phosphorylation site variants reveals a role for S1497.
To further demonstrate the usefulness of the ES cell-based approach to study BRCA1 function, we examined the physiological significance of various phosphorylation sites identified in BRCA1 by in vitro studies (16–20). We analyzed BRCA1 variants with the changes in serine residues (S308A, S988A, S1497A, S1423/S1524A double mutant, and S1423/S1387/S1524 triple mutant) that are phosphorylated by several kinases including Aurora, ATM, ATR, Chk2, and Cdk2. With the exception of the S1423/S1387/S1524 triple mutant (Supplemental Figure 6A), all other phosphorylation site mutants could rescue the lethality of Brca1KO/KO ES cells. The rescue rate ranged from 10.5% to 50.0% (Supplemental Figure 6B). Although the mRNA of the triple mutant BRCA1 was expressed at levels similar to other variants (data not shown), the protein was barely detectable (Supplemental Figure 6C). We reasoned that the mutant protein might be unstable because of the change of 3 serine residues, explaining its inability to rescue the ES cell lethality. We next performed various assays to determine whether the phosphorylation mutants had any effect on the known functions of BRCA1.
To examine the effect of phosphorylation site mutants on cell cycle regulation, we analyzed the viable Brca1KO/KO ES cells expressing S308A, S988A, S1497A, and S1423/S1524A BRCA1 variants by checking their cell cycle profiles with BrdU/PI double staining. We did not observe any significant differences in phosphorylation site mutants, compared with the Brca1KO/KO ES cells rescued by the wild-type BRCA1 (Supplemental Table 2). These rescued clones also did not reveal any significant change in sensitivity to DNA-damaging agents (Supplemental Table 1). However, when the ES cells rescued by S1497A and S1423/S1524A BRCA1 were γ-irradiated, they exhibited significant hypersensitivity (Figure 4A). The hypersensitivity of S1423/1524 BRCA1 was consistent with previous results indicating that S1423 and S1524 are phosphorylated by the DNA damage–dependent kinases ATM and ATR (16, 18). In contrast, the role of S1497 phosphorylation in DNA repair was previously unknown. The serine residue at position 1,497 is known to be phosphorylated by the cell cycle–dependent Cdk2 kinase (17); therefore, it was predicted to be involved in cell cycle regulation. Surprisingly, this variant had no significant effect on cell cycle progression in the absence of S1497 phosphorylation (Figure 4B and Supplemental Table 2); instead, it resulted in hypersensitivity to irradiation. This hypersensitivity was further confirmed by the clonogenic survival assay (Figure 4, C and D), as well as by the presence of more TUNEL-positive cells after γ-irradiation compared with control cells expressing wild-type BRCA1 (Figure 4E). Interestingly, Cdk2-null MEFs were recently reported to be sensitive to γ-irradiation (36). Although the mechanism remains unknown, a defect in S1497 phosphorylation may account for this phenotype.
Figure 4. Role of S1497 residue of BRCA1 in irradiation-induced DNA damage repair.
(A) Relative survival after γ-irradiation (IR) of 2 independent clones expressing S1497A compared with ES cells expressing no transgene (PL2F8), WT BRCA1, and S1423/1524A double mutant. *P < 0.006 for WT and S1497A-1. Error bars represent ± SEM. *P < 0.006 (B) Western blot analysis of 3 clones expressing S1497A and 1 with WT BRCA1 using phospho-S1497 antibody (upper panel). The lower panel shows expression of BRCA1. Rabbit IgG was used as negative control (NC); HeLa cells were used as positive control (PC). (C) Clonogenic survival assay to examine hypersensitivity of S1497A ES cells in response to 0.2 and 0.5 Gy of IR. (D) Quantitation of the number of colonies in C. (E) Quantitation of TUNEL-positive ES cells expressing S1497A BRCA1 after IR (2 Gy). ES cells expressing WT BRCA1 were used as a control. Con, control, before IR. Error bars represent ± SEM. (F) Phosphorylation of the S1497 residue affects the hyperphosphorylation of BRCA1 after IR. Wild-type or S1497A cells were left untreated or pretreated with an inhibitor of Cdk2 kinases. After IR, BRCA1 phosphorylation was analyzed by IP–Western blotting with a BRCA1 antibody and a phospho-S1497–specific antibody. Densitometric quantitation of the S1497 residue is indicated below the left panel. HEK293 cells were used as a control to show the difference in mobility of phosphorylated and unphosphorylated BRCA1. (G) Quantitation of chromosomal abnormalities in response to IR in ES cells (n = 100) expressing WT or S1497A BRCA1. (H) Representative picture of metaphase spreads of WT and S1497A cells after IR. Inset shows magnified view of 3 chromosomal aberrations. The scale bar on the left applies to the insets; the scale bar on the right applies to the larger images.
To determine the role of S1497 in radiation-induced DNA repair, we examined the phosphorylation of the 1,497 serine residue in response of γ-irradiation using a phospho-specific antibody. As shown in Figure 4F, after irradiation, we observed a 9.1-fold increase in phosphorylation of wild-type BRCA1, with a specific 3.5-fold increase in S1497 phosphorylation. However, the S1497A variant displayed virtually no (1.3-fold) hyperphosphorylation after irradiation, even though the amount of BRCA1 was increased (Figure 4F). Therefore, the hypersensitivity of S1497A mutant ES cells upon IR treatment is due not to the protein stability or other defects in BRCA1 function, but rather to the abrogated phosphorylation of BRCA1, which is required for the proper recruitment of the protein to the site of damaged DNA. More interestingly, the hyperphosphorylation of BRCA1 was inhibited by pretreatment with 2(bis-(hydroxyethyl)amino)-6-(4-methoxybenzylamino)-9-isopropyl-purine, a Cdk2 inhibitor, suggesting a role of Cdk2 in radiation-induced phosphorylation of BRCA1 (Figure 4F).
To determine the functional consequence of impaired phosphorylation of S1497 on overall genomic integrity in response to DNA damage, we examined the karyotype of ES cells expressing wild-type and S1497A BRCA1 in response to 1 Gy of γ-irradiation. We examined 100 nuclei of each genotype for the presence of various chromosomal abnormalities and found a marked increase in the number of gaps, radials, and breaks in the S1497A ES cells (Figure 4, G and H). In untreated ES cells of both genotypes, 0%–2% of cells showed these abnormalities. However, after irradiation, while only 5.5% of ES cells expressing wild-type BRCA1 showed gaps, radials, and breaks, we found that 15.5% of S1497A ES cells exhibited such abnormalities. These results clearly demonstrate that the S1497 residue plays an important role in maintaining the genomic stability in response to IR-induced DNA damage. We next tested whether the S1497A variant had any effect on homologous recombination, a process known to involve BRCA1. We measured the gene targeting efficiency at the Rosa26 locus in ES cells as described previously (15). We observed no difference in the homologous recombination activity of S1497A variant compared with wild-type BRCA1 (Supplemental Table 3). These results suggest that although S1497A mutant ES cells exhibited an increase in genomic instability in response to IR, the homologous recombination function of the ES cells was not compromised.
The role of S308 residue in differentiation of ES cells.
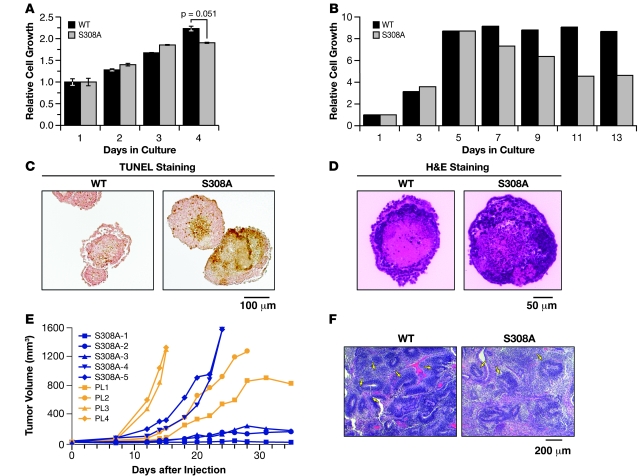
The S308 residue is known to be phosphorylated by Aurora-A kinase, and its phosphorylation is suggested to play a role in G2/M phase progression and centrosome microtubule nucleation (20, 37). Although the rescue of Brca1KO/KO ES cells by S308A BRCA1 was reduced (10% compared with 40%–50% by M1652I or wild-type; Figure 2C and Supplemental Figure 6B), cell cycle analysis revealed no significant defects (Supplemental Table 2). Surprisingly, when we cultured the S308A mutant ES cells without feeder cells, we observed a marked reduction in cell growth by day 4 (Figure 5A). We reasoned that this reduction in proliferation rate might be associated with cell differentiation because we observed clear changes of the S308A mutant ES cell morphology (Supplemental Figure 7) in the absence of feeder. Based on these results, we further analyzed the differentiation potential of S308A mutant cells by generating embryoid bodies. As shown in Figure 5B, the number of cells in embryoid bodies gradually increased and then became constant when control ES cells were used. In contrast, S308A ES cells exhibited a significant reduction in cell number in the embryoid bodies after day 7. The decrease in cell number was due to a relative increase in apoptotic cells, as marked by an increase in TUNEL-positive cells in the center of the embryoid body (Figure 5C). In addition, the embryoid bodies generated from control and S308A ES cells displayed a clear difference in their morphology based on histological analysis (Figure 5D). Wild-type embryoid bodies at day 7 showed the presence of well-defined layers of primitive ectoderm and mesoderm, whereas the embryoid bodies derived from S308A ES cells exhibited irregular structures, with many apoptotic cells in the center. From these observations we concluded that the S308A variant results in a differentiation defect of the ES cells.
Figure 5. Role of S308 residue in ES cell differentiation.
(A) Growth analysis of S308A ES cells compared with WT BRCA1 cells cultured without feeders. Error bars represent ± SEM. (B) Growth of embryoid bodies generated from ES cells expressing WT human BRCA1 and S308A, measured by trypsinizing the embryoid bodies and counting the total number of cells at each indicated time points. (C) TUNEL staining of embryoid bodies generated from S308A and WT ES cells after 7 days in culture. (D) H&E staining of embryoid bodies generated from S308A and PL2F8 ES cells after 14 days in culture. (E) Growth of teratomas generated from PL2F8 (n = 4) and S308A (n = 5) ES cells. A single ES cell clone of PL2F8 and S308A was analyzed for each genotype. (F) Histological analysis of teratomas generated from WT and S308A ES cells. The WT teratomas show numerous neural rosettes (arrows) that were greatly reduced in S308A teratomas.
To test the differentiation potential of S308A mutant ES cells in vivo, we injected one S308A ES cell clone subcutaneously into 5 nude mice and monitored the formation of teratomas that represent a differentiated mass of cells. While all the teratomas from control cells (PL2F8, n = 4) grew rapidly within a few weeks, only 2 teratomas from S308A cells (n = 5) grew at a rate comparable to that of the control teratomas. Interestingly, the other 3 teratomas from S308A cells grew extremely slowly, suggesting a proliferation or differentiation defect (Figure 5E). Furthermore, histological analysis of the two S308A teratomas that grew rapidly revealed the presence of more differentiated neural cells, compared with the immature, fast-growing cells present as a neural rosette structure in the control teratomas (Figure 5F). These results suggested a role for the S308 residue in stem cell differentiation, in addition to its known role in the G2/M phase transition by Aurora-A kinase–mediated phosphorylation.
Analysis of the RXRXH mutant revealed a marginal effect on cell cycle progression.
The 358LXCXE motif was initially predicted to be one of the RB-binding domains in BRCA1 (38). Subsequently, it was reported that the disruption of the 358LXCXE motif by changing it to 358RXRXH resulted in functional alteration of BRCA1, including misregulation of RB family proteins (39). Based on this finding, we tested the effect of 358RXRXH BRCA1 in our functional assay. The Brca1KO/KO ES cells expressing this variant were viable (Supplemental Figure 6). To find out the functional defect related to this mutation, we first examined the cell cycle profiles of these cells. As shown in Supplemental Figure 8, we found an elevated percentage of S phase cells and a correspondingly decreased percentage of G2/M phase cells in 2 mutant ES clones. The differences are not dramatic but are statistically significant and reproducible. It is notable that the changes in cell cycle profile observed in the mutant ES cells are similar to the changes in the prostate cancer cell line DU-145 (39), suggesting that the mouse ES cells can phenocopy the defects observed in human cancer cell lines. We also observed increased RB level and slower growth rate of the mutant ES cells, but the differences were marginal or not statistically significant (data not shown).
Discussion
ES cells have been previously used to study the role of BRCA1 in homology-directed DNA repair (40). Recently a knock-in approach was used in mouse ES cells to examine the effect of a mutation, I26A, that disrupts the interaction between BRCA1 and E2 conjugating enzyme (41). It was shown to have no effect on the viability and genomic stability of the ES cells. We report here the generation and validation of a powerful mouse ES cell–based functional assay for performing comprehensive functional analysis of human BRCA1 variants. Combined with the recombineering-based method, which enables easy introduction of any subtle mutations into the BAC DNA, we have used mouse ES cells to test 13 variants that map to various functional domains of BRCA1. Our approach of using BACs and ES cells has several advantages over the previous methods that have relied on complementation by cDNAs in BRCA-deficient cell lines (11–13). Use of BACs allows the expression of the transgene at physiological levels. We found the expression of BRCA1 variants in mouse ES cells to be comparable to the level of BRCA1 in human cancer cell lines (Supplemental Figure 9). Moreover, it also allows examination of variants that affect expression (regulatory mutations), splicing, as well as transcript stability. We have gone on to demonstrate the reliability and physiological relevance of our assay by examining 3 variants in humanized mouse models. We have shown that variants that result in ES cell lethality (C61G and A1708E) are also deleterious for mouse embryonic development. In contrast, a variant that is known to be neutral (M1652I) does not show any apparent defect in BRCA1 function in ES cells and is also able to fully rescue the lethality of Brca1-null mice. Even though the embryos with either one of the two variants (C61G and A1708E) showed early embryonic lethality similar to that of BRCA1-deficient embryos, it is highly possible that other variants can partially or fully rescue the lethal effects in embryo. Also, it will be interesting to see whether the embryonic lethality is delayed or rescued by introduction of the p53 mutation, as previously reported (42). Based on our findings, we believe that the ES cell–based assay is a reliable tool for understanding the functional significance of BRCA1 VUSs. We have streamlined the procedure, which allows us to test the ability of a BRCA1 variant to rescue Brca1KO/KO ES cell lethality in 7 weeks (see Figure 6 for the various steps involved). We also demonstrate that the mutant ES cells we generated are very useful to gain mechanistic insight into the role of BRCA1 in different biological processes.
Figure 6. Schematic representation of the ES cell–based assay to study BRCA1 variants.
(A) Flow diagram showing the experimental steps involved in the BRCA1 variant analysis and time required for each phase. (B) Functional analysis of BRCA1 mutant ES cells. For variants that rescue Brca1-null ES cells, a series of assays to test various functions of BRCA1 is performed. HATR, HAT-resistant.
Functional analysis of variants that result in viable ES cells.
While variants that fail to rescue the lethality of Brca1KO/KO ES cells can be easily classified as deleterious, those that result in viable ES cells have to be evaluated further by using various functional assays in order to understand their functional significance as outlined in Figure 6. In an effort to evaluate various functions of BRCA1, we have examined the effect of such variants on DNA repair and genomic stability, cell cycle regulation, ES cell proliferation, differentiation, and transcriptional regulation. Hypomorphic variants that result in viable ES cells are also useful for functional dissection of BRCA1. We have uncovered a role for S1497 and S308 phosphorylation sites of BRCA1. The hypersensitivity of S1497A mutant ES cells to γ-irradiation suggests that the S1497 residue plays a direct role in DNA damage repair, similar to other phosphorylation residues of DNA damage–dependent kinases. This is supported by our observation that the phosphorylation of S1497 is induced in response to γ-irradiation (Figure 4F). It is also possible that the phosphorylation of the S1497 residue can facilitate the phosphorylation of neighboring residues, such as S1423 or S1524. This is supported by the reduced hyperphosphorylation of BRCA1 in S1497A mutant (Figure 4F), and a similar effect was observed with pretreatment with a Cdk2 inhibitor. These two possible roles of the S1497 are not mutually exclusive. Considering that the S1497 mutant neither showed sensitivity to any of the DNA damaging drugs (Supplemental Table 1) nor exhibited any effect on homologous recombination efficiency (Supplemental Table 3), we believe that the phosphorylation of S1497 has a specific role in radiation-induced DNA damage repair. We have also uncovered a novel role for the S308 residue in ES cell differentiation. We show that the abrogation of the phosphorylation at the S308 residue results in increased cell death by apoptosis (Figure 5C) during differentiation.
While our results clearly demonstrate the ability of ES cell–based assay to distinguish between deleterious and neutral variants, as with any functional assay, we cannot rule out the possibility that we may get false (negative) results. This is most likely due to the fact that we do not fully understand all the biochemical functions of BRCA1. We also cannot rule out the possibility that there are phenotypes that are not apparent in ES cells. We have attempted to address this by differentiating the ES cells in vitro into embryoid bodies. For a protein with many interacting partners and involvement in multiple biological processes, no single approach can be completely relied upon to determine the biological significance of VUSs. We believe that in combination with the family linkage and other epidemiological data, as well as from structural and evolutionary conservation analyses, our assay can provide meaningful results that can be valuable in assessing the biological significance of any BRCA1 variant.
In conclusion, we have described here what we believe to be a novel, but simple and tractable approach to analyzing BRCA1 variants. As illustrated in Figure 6, it takes less than 2 months to test whether a variant is likely to be neutral or deleterious. By using standard molecular biology and tissue culture techniques, it is possible to simultaneously examine 4–5 variants. Results obtained from such functional assays are likely to help genetic counselors and physicians in making clinical decisions in the future. It is also believed that this approach will be useful for study of variants identified in other human disease genes.
Methods
Reagents.
Human BRCA1-specific antibodies (E1) were generated by using a peptide sequence of 768–793 (SISLVPGTDYGTQESISLLEVSTLGK) in human BRCA1. A cysteine residue was added at the N terminus for affinity column coupling, and the antibody was purified from the rabbit antisera immunized with the peptide. The other antibodies used were anti-BRCA1 (Ab-1; Calbiochem, OP92), p1497-specific BRCA1 (Millipore, 07-007), Rb (Santa Cruz Biotechnology Inc., sc50) and phospho-Rb (S608, Cell Signaling Technology, 2181), p53 (for ChIP, mix of Santa Cruz Biotechnology Inc. sc100 and sc1315), p21 (for ChIP, Santa Cruz Biotechnology Inc., sc471), actin (Santa Cruz Biotechnology Inc., sc8431).
Generation of mutations in BRCA1 in a BAC clone.
A BAC clone (RPCI11-812-5O) containing full-length human BRCA1 was used to generate mutations as described previously (43). The sequence of oligonucleotides used to generate the mutations in the BAC is described in Supplemental Table 4.
ES cell culture and BAC transfection.
The PL2F8 ES cell line was generated as described in Supplemental Figures 1–3 using selection marker cassettes provided by P. Liu (Wellcome Trust Sanger Institute, Hinxton, United Kingdom). The ES cell clones were expanded, genotyped, and analyzed as described previously (15).
Adeno-Cre infection and selection of HAT-resistant ES cell clones.
Purified adeno-Cre virus (1 μl; 1010 CFU/ml; NCI-Frederick Viral Technology Laboratory) was added to 105 ES cells and incubated for 30 minutes in a CO2 incubator. The 1,000 infected cells were then plated onto a 60-mm feeder plate, followed by HAT selection for 5 days and HT selection for 2 days. After 2 days in M15 media, colonies were picked up for the analysis and the remaining plate was stained with methylene blue solution for visualization.
Expression analysis of BRCA1 transgene.
To identify ES cell clones expressing the human BRCA1 transgene, RNA was extracted from ES cells in 96-well plates using RNeasy-96 kit (QIAGEN) and subsequently analyzed by Titan one-step RT-PCR kit (Roche), as according to the manufacturer’s protocol.
For real-time analysis of BRCA1 in ES cells and human cancer cell lines, cDNA was synthesized from total RNA of the cell lines first. To quantitate BRCA1 expression levels, we used a Brilliant II SYBR Green QPCR kit. Ribosomal protein13a (RPL13a) was used as an internal control for both ES cells and human cancer cell lines. Sequences of primers used are described in Supplemental Table 5.
For detection of BRCA1 protein, ES cells in a 6-well plate were lysed in IP buffer (20 mM HEPES [pH 7.5], 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM NaF, 1 mM DTT, 0.1% Triton X-100, protease inhibitor cocktail [Roche]), and human BRCA1 was immunoprecipitated with anti-BRCA1 rabbit polyclonal antibody (human BRCA1 specific). The immunoprecipitated proteins were separated in 6% SDS-PAGE gels, and BRCA1 was detected by mouse monoclonal antibody for BRCA1 (Ab-1). Phospho-specific antibody for S1497 (Upstate Biotechnology) was used for the detection of phosphorylated S1497 of BRCA1.
Drug and radiation sensitivity assay.
For the drug sensitivity assay, 104 cells were plated in 96-well plates without feeder cells. On the next day, cells were treated with M15 medium containing appropriate concentrations of different drugs (cisplatin [1–10 μM], hydroxyurea [25–200 μM], mitomycin C [20–200 ng/ml], methyl methanesulfonate [5–50 μg/ml], MNNG [10–100 μM]). After 48 hours of drug treatment, the metabolic activity of the cells was measured by Alamar Blue assay (Invitrogen), according to the manufacturer’s instructions. All samples were analyzed in triplicate. For sensitivity to γ-irradiation, the cells in 96-well plates were irradiated at varying doses (1–10 Gy). UV light treatment of the cells was performed in a UV-crosslinker (Stratagene), with a dose of 10–100 J/m2.
Generation and expression analysis of the BAC transgenic mice.
BAC DNA with a given BRCA1 mutation was prepared by QIAGEN Maxiprep kit. The purified DNA was diluted in TE buffer to 10 ng/ml concentration and used for the microinjection. Mice were genotyped by Southern analysis as described previously (44). The BAC transgenic founder mice were mated with mice carrying a null allele of Brca1 (Brca1KO/+) to obtain BAC transgenic mice on a Brca1 heterozygous background (Brca1KO/+;Tg). The expression of mutant BRCA1 was examined in several tissues by RT-PCR with the same primers used for transgene expressions in ES cells. The protein expression was analyzed as described for transgene expression in ES cells. All animals were housed and handled under guidelines approved by the NCI-Frederick Animal Care and Use Committee.
Embryoid body culture and histological analysis.
Embryoid bodies were generated and cultured as described previously (45). For the growth test, the embryoid bodies were harvested, trypsinized, and dissociated into single cells by pipetting. Cells were counted using a Coulter counter. For histological analysis, 7- to 14-day-old embryoid bodies were harvested and fixed in 10% neutralized formalin. Paraffin-embedded sections were stained with H&E. For the TUNEL staining, the DeadEnd colorimetric TUNEL system (Promega) was used. The stained embryoid bodies were observed and photographed under an Axioplan2 upright microscope (Zeiss).
Analysis of mouse embryos.
Transgenic mice (C61G, M1652I, A1708E) on a Brca1 heterozygous background (Brca1KO/+;Tg) were intercrossed. Brca1-null mice used in the study were generated by T. Ludwig (Columbia University, New York, New York, USA) (46). Embryos at days 7.5 and 8.5 of gestation were dissected under a microscope (Leica MZ8) and photographed. Embryos were genotyped by PCR using primers as listed in Supplemental Table 4.
ChIP assay.
ChIP was performed as described previously (47). Briefly, ES cells in 60-mm plates were fixed in 1% formaldehyde for 10 minutes. The cross-linking was stopped by addition of 1 M glycine, to final concentration of 125 mM. After wash in PBS, the nuclear fraction was isolated using hypotonic cell lysis buffer (5 mM HEPES, 85 mM KCl, 0.5% NP40). The isolated nuclei were lysed and sonicated to yield DNA fragments in the size range of approximately 600 bp. After preclearing with ssDNA-coated agarose beads for 1 hour, 1 μg of the indicated antibodies was added and incubated overnight at 4°C. After washing, the purified complex was eluted in elution buffer (1% SDS, 0.1 M NaHCO3) and then reverse cross-linked in a 65°C water bath for 5 hours followed by proteinase K treatment for 2 hours. The final DNA was purified by phenol/chloroform extraction and ethanol precipitation with 1 μg of yeast tRNA. The sequences of primers for ChIP are listed in Supplemental Table 5.
Homologous recombination assay.
Homologous recombination efficiency was measured by gene targeting at the Rosa 26 locus as described previously (15). The Rosa26 targeting vector was provided by S. Kuznetsov (Institute for Molecular Medicine Finland, University of Helsinki, Helsinki, Finland).
Teratoma assay.
ES cells were harvested and washed with PBS and resuspended in PBS at a concentration of 5 × 107 cells/ml. A total of 100 μl of cell suspension was injected subcutaneously into 6-week-old athymic nude mice (C3H/HeNCr-nu). Five mice were used for each BRCA1 variant. Mice were monitored for teratoma formation a week after the injection. The tumor size was measured every other day until any dimension of the tumor reached 1.5 cm. The tumors were dissected out and analyzed further by generating paraffin-embedded sections stained with H&E. Tumor volume (in mm3) was calculated as the product of 2 × length × width.
Statistics.
All data are expressed as mean ± SEM. Differences between 2 groups were compared using a 2-tailed unpaired Student’s t test (Microsoft Excel for Mac). P values less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank J. Acharya, I. Daar, K. Reilly, and L. Tessarollo for helpful discussions and critical review of the manuscript. We also thank Kyung-Ae Kim and Susan Lynn North for technical assistance; Deborah Swing, Yongping Yang, Prajakta Varadkar, and Lionel Feigenbaum for help with BAC transgenic mice; Sandra Burkett for cytogenetic analysis; Allen Kane, Jiro Wada, and Richard Frederickson (NCI-Frederick Publications Department) for illustrations; Serguei Kozlov for help with in vitro ES cell differentiation; and Diana Haines for histolopathological analysis. The research was sponsored by the Center for Cancer Research, NCI, NIH.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 119:3160–3171 (2009). doi:10.1172/JCI39836.
See the related Commentary beginning on page 2895.
References
- 1.Mahoney M.C., Bevers T., Linos E., Willett W.C. Opportunities and strategies for breast cancer prevention through risk reduction. CA Cancer J. Clin. 2008;58:347–371. doi: 10.3322/CA.2008.0016. [DOI] [PubMed] [Google Scholar]
- 2.Yang X., Lippman M.E. BRCA1 and BRCA2 in breast cancer. Breast Cancer Res. Treat. 1999;54:1–10. doi: 10.1023/A:1006189906896. [DOI] [PubMed] [Google Scholar]
- 3.Futreal P.A., et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266:120–122. doi: 10.1126/science.7939630. [DOI] [PubMed] [Google Scholar]
- 4.Merajver S.D., et al. Germline BRCA1 mutations and loss of the wild-type allele in tumors from families with early onset breast and ovarian cancer. Clin. Cancer Res. 1995;1:539–544. [PubMed] [Google Scholar]
- 5.Gayther S.A., et al. Germline mutations of the BRCA1 gene in breast and ovarian cancer families provide evidence for a genotype-phenotype correlation. Nat. Genet. 1995;11:428–433. doi: 10.1038/ng1295-428. [DOI] [PubMed] [Google Scholar]
- 6.Rahman N., Stratton M.R. The genetics of breast cancer susceptibility. Annu. Rev. Genet. 1998;32:95–121. doi: 10.1146/annurev.genet.32.1.95. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi H., et al. Mutation analysis of the BRCA1 gene in ovarian cancers. Cancer Res. 1995;55:2998–3002. [PubMed] [Google Scholar]
- 8.Friedman L.S., et al. Confirmation of BRCA1 by analysis of germline mutations linked to breast and ovarian cancer in ten families. Nat. Genet. 1994;8:399–404. doi: 10.1038/ng1294-399. [DOI] [PubMed] [Google Scholar]
- 9.Deffenbaugh A.M., Frank T.S., Hoffman M., Cannon-Albright L., Neuhausen S.L. Characterization of common BRCA1 and BRCA2 variants. Genet. Test. 2002;6:119–121. doi: 10.1089/10906570260199375. [DOI] [PubMed] [Google Scholar]
- 10.Easton D.F., et al. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am. J. Hum. Genet. 2007;81:873–883. doi: 10.1086/521032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayes F., Cayanan C., Barilla D., Monteiro A.N. Functional assay for BRCA1: mutagenesis of the COOH-terminal region reveals critical residues for transcription activation. Cancer Res. 2000;60:2411–2418. [PMC free article] [PubMed] [Google Scholar]
- 12.Vallon-Christersson J., et al. Functional analysis of BRCA1 C-terminal missense mutations identified in breast and ovarian cancer families. Hum. Mol. Genet. 2001;10:353–360. doi: 10.1093/hmg/10.4.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carvalho M.A., Couch F.J., Monteiro A.N. Functional assays for BRCA1 and BRCA2. Int. J. Biochem. Cell Biol. 2007;39:298–310. doi: 10.1016/j.biocel.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glover J.N. Insights into the molecular basis of human hereditary breast cancer from studies of the BRCA1 BRCT domain. Fam. Cancer. 2006;5:89–93. doi: 10.1007/s10689-005-2579-z. [DOI] [PubMed] [Google Scholar]
- 15.Kuznetsov S.G., Liu P., Sharan S.K. Mouse embryonic stem cell-based functional assay to evaluate mutations in BRCA2. Nat. Med. 2008;14:875–881. doi: 10.1038/nm.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cortez D., Wang Y., Qin J., Elledge S.J. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- 17.Ruffner H., Jiang W., Craig A.G., Hunter T., Verma I.M. BRCA1 is phosphorylated at serine 1497 in vivo at a cyclin-dependent kinase 2 phosphorylation site. Mol. Cell. Biol. 1999;19:4843–4854. doi: 10.1128/mcb.19.7.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J. Ataxia telangiectasia-related protein is involved in the phosphorylation of BRCA1 following deoxyribonucleic acid damage. Cancer Res. 2000;60:5037–5039. [PubMed] [Google Scholar]
- 19.Zhang J., et al. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol. Cell. Biol. 2004;24:708–718. doi: 10.1128/MCB.24.2.708-718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ouchi M., et al. BRCA1 phosphorylation by Aurora-A in the regulation of G2 to M transition. J. Biol. Chem. 2004;279:19643–19648. doi: 10.1074/jbc.M311780200. [DOI] [PubMed] [Google Scholar]
- 21.Gowen L.C., Johnson B.L., Latour A.M., Sulik K.K., Koller B.H. Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat. Genet. 1996;12:191–194. doi: 10.1038/ng0296-191. [DOI] [PubMed] [Google Scholar]
- 22.Hakem R., et al. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell. 1996;85:1009–1023. doi: 10.1016/S0092-8674(00)81302-1. [DOI] [PubMed] [Google Scholar]
- 23.Wiener F., Coleman A., Mock B.A., Potter M. Nonrandom chromosomal change (trisomy 11) in murine plasmacytomas induced by an ABL-MYC retrovirus. Cancer Res. 1995;55:1181–1188. [PubMed] [Google Scholar]
- 24.Chandler J., Hohenstein P., Swing D.A., Tessarollo L., Sharan S.K. Human BRCA1 gene rescues the embryonic lethality of Brca1 mutant mice. Genesis. 2001;29:72–77. doi: 10.1002/1526-968X(200102)29:2<72::AID-GENE1007>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 25.Castilla L.H., et al. Mutations in the BRCA1 gene in families with early-onset breast and ovarian cancer. Nat. Genet. 1994;8:387–391. doi: 10.1038/ng1294-387. [DOI] [PubMed] [Google Scholar]
- 26.Miki Y., et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 27.Coyne R.S., McDonald H.B., Edgemon K., Brody L.C. Functional characterization of BRCA1 sequence variants using a yeast small colony phenotype assay. Cancer Biol. Ther. 2004;3:453–457. doi: 10.4161/cbt.3.5.809. [DOI] [PubMed] [Google Scholar]
- 28.Williams R.S., et al. Detection of protein folding defects caused by BRCA1-BRCT truncation and missense mutations. J. Biol. Chem. 2003;278:53007–53016. doi: 10.1074/jbc.M310182200. [DOI] [PubMed] [Google Scholar]
- 29.Carvalho M.A., et al. Determination of cancer risk associated with germ line BRCA1 missense variants by functional analysis. Cancer Res. 2007;67:1494–1501. doi: 10.1158/0008-5472.CAN-06-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karchin R., Monteiro A.N., Tavtigian S.V., Carvalho M.A., Sali A. Functional impact of missense variants in BRCA1 predicted by supervised learning. PLoS Comput. Biol. 2007;3:e26. doi: 10.1371/journal.pcbi.0030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams R.S., Green R., Glover J.N. Crystal structure of the BRCT repeat region from the breast cancer-associated protein BRCA1. Nat. Struct. Biol. 2001;8:838–842. doi: 10.1038/nsb1001-838. [DOI] [PubMed] [Google Scholar]
- 32.Hohenstein P., et al. A targeted mouse Brca1 mutation removing the last BRCT repeat results in apoptosis and embryonic lethality at the headfold stage. Oncogene. 2001;20:2544–2550. doi: 10.1038/sj.onc.1204363. [DOI] [PubMed] [Google Scholar]
- 33.Harkin D.P., et al. Induction of GADD45 and JNK/SAPK-dependent apoptosis following inducible expression of BRCA1. Cell. 1999;97:575–586. doi: 10.1016/S0092-8674(00)80769-2. [DOI] [PubMed] [Google Scholar]
- 34.Somasundaram K., et al. Arrest of the cell cycle by the tumour-suppressor BRCA1 requires the CDK-inhibitor p21WAF1/CiP1. Nature. 1997;389:187–190. doi: 10.1038/38291. [DOI] [PubMed] [Google Scholar]
- 35.Anderson S.F., Schlegel B.P., Nakajima T., Wolpin E.S., Parvin J.D. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat. Genet. 1998;19:254–256. doi: 10.1038/930. [DOI] [PubMed] [Google Scholar]
- 36.Satyanarayana A., Hilton M.B., Kaldis P. p21 inhibits Cdk1 in the absence of Cdk2 to maintain the G1/S phase DNA damage checkpoint. Mol. Biol. Cell. 2008;19:65–77. doi: 10.1091/mbc.E07-06-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sankaran S., Crone D.E., Palazzo R.E., Parvin J.D. Aurora-A kinase regulates breast cancer associated gene 1 inhibition of centrosome-dependent microtubule nucleation. Cancer Res. 2007;67:11186–11194. doi: 10.1158/0008-5472.CAN-07-2578. [DOI] [PubMed] [Google Scholar]
- 38.Aprelikova O.N., et al. BRCA1-associated growth arrest is RB-dependent. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11866–11871. doi: 10.1073/pnas.96.21.11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan S., et al. Disruption of BRCA1 LXCXE motif alters BRCA1 functional activity and regulation of RB family but not RB protein binding. Oncogene. 2001;20:4827–4841. doi: 10.1038/sj.onc.1204666. [DOI] [PubMed] [Google Scholar]
- 40.Moynahan M.E., Chiu J.W., Koller B.H., Jasin M. Brca1 controls homology-directed DNA repair. Mol. Cell. 1999;4:511–518. doi: 10.1016/S1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 41.Reid L.J., et al. E3 ligase activity of BRCA1 is not essential for mammalian cell viability or homology-directed repair of double-strand DNA breaks. Proc. Natl. Acad. Sci. U. S. A. 2008;105:20876–20881. doi: 10.1073/pnas.0811203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu X., et al. Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nat. Genet. 2001;28:266–271. doi: 10.1038/90108. [DOI] [PubMed] [Google Scholar]
- 43.Yang Y., Sharan S.K. A simple two-step, “hit and fix” method to generate subtle mutations in BACs using short denatured PCR fragments. Nucleic Acids Res. 2003;31:e80. doi: 10.1093/nar/gng080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Y., Swaminathan S., Martin B.K., Sharan S.K. Aberrant splicing induced by missense mutations in BRCA1: clues from a humanized mouse model. Hum. Mol. Genet. 2003;12:2121–2131. doi: 10.1093/hmg/ddg222. [DOI] [PubMed] [Google Scholar]
- 45.Hopfl G., Gassmann M., Desbaillets I. Differentiating embryonic stem cells into embryoid bodies. Methods Mol. Biol. 2004;254:79–98. doi: 10.1385/1-59259-741-6:079. [DOI] [PubMed] [Google Scholar]
- 46.Ludwig T., Chapman D.L., Papaioannou V.E., Efstratiadis A. Targeted mutations of breast cancer susceptibility gene homologs in mice: lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev. 1997;11:1226–1241. doi: 10.1101/gad.11.10.1226. [DOI] [PubMed] [Google Scholar]
- 47.Orlando V., Strutt H., Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 48.Coene E.D., et al. Phosphorylated BRCA1 is predominantly located in the nucleus and mitochondria. Mol. Biol. Cell. 2005;16:997–1010. doi: 10.1091/mbc.E04-10-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.