Abstract
Copy-number variants (CNVs) are substantial contributors to human disease. A central challenge in CNV-disease association studies is to characterize the pathogenicity of rare and possibly incompletely penetrant events, which requires the accurate detection of rare CNVs in large numbers of individuals. Cost and throughput issues limit our ability to perform these studies. We have adapted the Illumina BeadXpress SNP genotyping assay and developed an algorithm, SNP-Conditional OUTlier detection (SCOUT), to rapidly and accurately detect both rare and common CNVs in large cohorts. This approach is customizable, cost effective, highly parallelized, and largely automated. We applied this method to screen 69 loci in 1105 children with unexplained intellectual disability, identifying pathogenic variants in 3.1% of these individuals and potentially pathogenic variants in an additional 2.3%. We identified seven individuals (0.7%) with a deletion of 16p11.2, which has been previously associated with autism. Our results widen the phenotypic spectrum of these deletions to include intellectual disability without autism. We also detected 1.65–3.4 Mbp duplications at 16p13.11 in 1.1% of affected individuals and 350 kbp deletions at 15q11.2, near the Prader-Willi/Angelman syndrome critical region, in 0.8% of affected individuals. Compared to published CNVs in controls they are significantly (P = 4.7 × 10−5 and 0.003, respectively) enriched in these children, supporting previously published hypotheses that they are neurocognitive disease risk factors. More generally, this approach offers a previously unavailable balance between customization, cost, and throughput for analysis of CNVs and should prove valuable for targeted CNV detection in both research and diagnostic settings.
Recent technological developments have improved our ability to detect human genomic copy-number variants (CNVs), and it is clear that CNVs contribute substantially to disease pathogenesis. This is particularly true for neurocognitive disorders like intellectual disability (ID), autism, and schizophrenia (Vissers et al. 2003; de Vries et al. 2005; Friedman et al. 2006; Rosenberg et al. 2006; Sharp et al. 2006; Sebat et al. 2007; Szatmari et al. 2007; Christian et al. 2008; International Schizophrenia Consortium 2008; Kumar et al. 2008; Marshall et al. 2008; Stefansson et al. 2008; Walsh et al. 2008; Weiss et al. 2008; Kirov et al. 2009). Notably, these CNV-disease associations normally share two genetic features that have implications for future studies. First, pathogenic CNVs causing similar disease phenotypes are often independently generated, de novo mutations that arise as a consequence of nonallelic homologous recombination between flanking duplicated sequences. Direct detection of CNVs is therefore critical since many important variants are invisible to haplotype-based genotyping methods. Second, individual pathogenic CNVs are rare, being nearly absent in control populations and present in 1% or less of affected individuals. Therefore, studies of very large numbers (i.e., many thousands) of both affected and unaffected individuals are required to fully understand the phenotypic impact of known pathogenic variants and to identify new disease alleles.
Currently available platforms allow for the detection of rare CNVs in large sample collections, but cost and throughput obstacles limit our ability to effectively assess the contributions of CNVs to disease. An evaluation of the two most widely used technologies—array comparative genomic hybridization (array-CGH) and genome-wide single nucleotide polymorphism (SNP) microarrays—illustrates these limitations. Array-CGH platforms offer high resolution (millions of probes) and the ability to customize content. However, these experiments are expensive (hundreds of dollars per sample), labor intensive, and difficult to automate. SNP arrays offer similar levels of resolution as array-CGH along with better quality control and automation. Yet, these experiments are similar in cost to array-CGH platforms, are known to miss many known or potential CNVs (Cooper et al. 2008; Kidd et al. 2008), and offer limited or no customization options to target loci of interest. Other technologies are available for targeted CNV identification (e.g., quantitative PCR), but these assays generally cannot be multiplexed and impose significant locus-by-locus design challenges. Thus, there are limited effective options to detect CNVs at numerous targeted loci in the sample sizes required to identify rare, pathogenic CNVs.
Targeted SNP genotyping assays, in principle, offer the ability to merge customizable probe selection with speed and cost efficiency for CNV analysis. Several studies have used Illumina GoldenGate SNP genotyping data to detect CNVs (Carlson et al. 2006; McCarroll et al. 2006; Newman et al. 2006). However, these studies were unable to detect rare CNVs and required slow and labor-intensive analysis methods. We have adapted the Illumina BeadXpress SNP genotyping assay (which uses GoldenGate genotyping chemistry) to detect CNVs at many loci in large-scale studies. The BeadXpress assay allows customized selection of up to 384 probes, is substantially cheaper than other CNV platforms, offers considerable quality-control advantages, and is performed in 96-well plates. We developed an algorithm, SNP-Conditional OUTlier detection (SCOUT), that builds upon a previously developed method to genotype common CNVs (SCIMM) (Cooper et al. 2008), to provide reproducible, automatic data analysis. SCOUT performs an initial quality control pass to identify and remove low-quality data, and subsequently predicts copy-number changes at each targeted site by identifying samples with anomalous fluorescence intensity or allelic ratio measurements.
In this study, we targeted 69 regions of the genome predicted to be susceptible to rearrangement as a consequence of nearby flanking duplications, which we term rearrangement “hotspots” (Bailey et al. 2002). These include 25 hotspots known to be associated with genomic disorders (e.g., Prader-Willi, Williams, and Velo-Cardio-Facial (VCF) syndromes; Supplemental Table 1). After validation of the assay using patient samples with known pathogenic CNVs, we screened a cohort of 1105 individuals with unexplained intellectual disability (ID). We detected, and subsequently validated, pathogenic or possibly pathogenic CNVs in over 5% of these individuals. We show that this assay can be used to accurately detect both rare and common CNVs and has potential use in clinical diagnostics, population genetics studies, and genome-wide association studies.
Results
Detection of known CNVs
We selected 69 regions of the genome either known or predicted (based on genomic architecture) to undergo recurrent rearrangement resulting in deletion or duplication. For each region, we selected at least five SNP probes for genotyping. We also developed an automated computational method, SCOUT, to detect rare CNVs using SNP genotyping fluorescence data. SCOUT considers both the total intensity and the ratio of the two SNP alleles at each probe (Fig. 1). Scores from all probes within a given target region are summed to identify samples that are outliers across the entire interval (see Methods and Supplemental material).
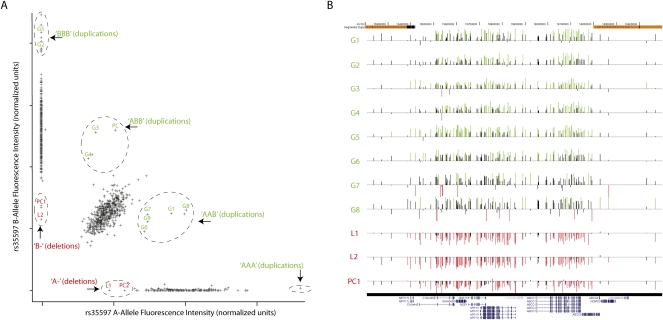
Figure 1.
CNVs at 16p13.11 in individuals with ID. (A) SCOUT uses fluorescence intensity data generated for SNP genotypes, in this case at rs35597, to identify samples that harbor deletions or duplications. Each sample is assigned a score based on its intensity relative to the center of the cluster of all samples with the same genotype. Samples that harbor duplications exhibit greater total intensity and potentially aberrant allelic ratio (inferred genotypes marked in green), while deletions exhibit lesser intensity and lack SNP heterozygosity (inferred genotypes labeled in red). Samples analyzed as positive controls are labeled PC, while validated predictions are labeled as L1–L2 (“loss”) or G1–G9 (“gain”). Dashed circles are drawn manually for visual aid. Unlabeled samples that appear to be outliers at this probe are either outliers only at this probe or samples with insufficient DNA for validation. (B) SCOUT predictions were validated with targeted array-CGH experiments. Shown are the resulting data for the labeled samples in panel A, with normalized log-transformed test/reference ratios at each tested probe presented as vertical bars. Those probes that are more than 1.5 standard deviations above (below) the average for the entire array are colored green (red). The samples shown include eight validated duplications, two validated deletions, and one positive control deletion. Note that G9 was analyzed with a different array-CGH platform and is not shown here. Genome coordinates and segmental duplications are annotated at the top. The bottom shows RefSeq genes/isoforms annotated in this region.
We first tested SCOUT's ability to predict deletions and duplications across the 69 hotspot regions using 39 DNA samples with ID and related phenotypes, each known to carry a deletion or duplication of one or more of the targeted regions in its entirety (Supplemental Table 2). Among these 39 samples were 35 deletions and 22 duplications involving 20 of the targeted regions, all of which had been previously confirmed by array-CGH. Four of the CNVs in the control set affect only part of a targeted hotspot (two deletions, two duplications), allowing us to test SCOUT's ability to detect variants when only a subset of the probes in a target is nondiploid. We found that CNVs were scored in the extreme positive (duplications) and negative (deletion) directions, with scores ranging in absolute value from 3.7 to 15.4 (median of 8.6), compared to a global distribution with a mean near zero and standard deviation 1.6 (Fig. 2). In fact, only three positive control CNVs score less than |5|, and each of these is at a locus containing only two successful probes (see Methods). Considering only hotspots with three or more probes (66 of 69 targeted loci; Supplemental Table 1), a threshold of |5| allows for 100% sensitivity with only two false-positives in these control samples. We detected an additional duplication in one control that was not previously known, but was subsequently validated using array-CGH. Finally, we were able to detect one of the two partial duplications (scores of 4.5 and 5.9) and one of the two partial deletions (scores of −3.3 and −5.6) at a threshold of |5|.
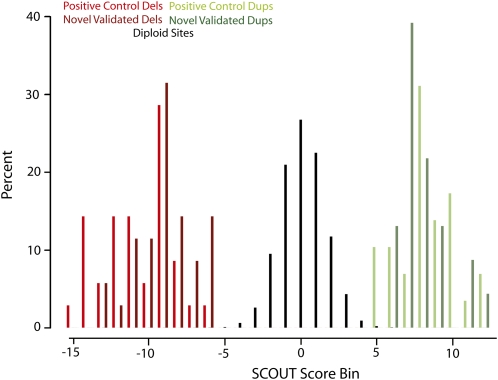
Figure 2.
SCOUT assigns highly negative scores to deletions (red), near-zero scores to diploid intervals (black), and highly positive scores to duplications (green). Normalized histograms (Y-axis indicates percent of each category) for known deletions (red, n = 35), predicted and validated deletions (dark red, n = 35), known duplications (green, n = 29), and predicted and validated duplications (dark green, n = 23) are shown. SCOUT scores are binned by 1-unit increments with the center of each bin labeled on the X-axis. The vast majority of scores correspond to diploid sites (n = 10,566 or 98.9%).
As additional proof-of-principle, we also placed probes within two common CNVs and used a previously described method, SCIMM (Cooper et al. 2008), to generate copy-number genotypes at these loci (see Methods). We also genotyped flanking SNPs with which the deletions are known to be tightly correlated (Supplemental Fig. 1). We found a near perfect correlation between each common deletion and its tag SNP (r2 of 1.0 and 0.994) across the entire sample collection (∼1100 samples for which both CNV and SNP genotypes were obtained). Since such a strong correlation between these independently generated genotypes for common variants is unlikely to occur in the absence of accurate genotypes, this shows that common CNVs can also be accurately detected with this assay.
Discovery and validation of CNVs in 1105 individuals with intellectual disability (ID)
We subsequently analyzed a series of 1105 DNA samples from individuals with unexplained ID (see Methods). Each 96-well plate included two controls (one individual with a known deletion and one individual with a known duplication). SCOUT automatically removed 95 samples (8.6%) due to excessive fluorescence intensity noise, leaving 1010 samples. Note that the quality-control measures we applied are more stringent than those applied for SNP genotyping (see Methods) and that failure to eliminate noisy samples results in an increased number of false-positive predictions (Supplemental Fig. 1).
We identified 69 potential duplications and 59 potential deletions in 98 samples that met a SCOUT threshold of |5|. We sought to validate as many of these predictions as possible and evaluated 59 samples that had at least one prediction at this threshold and for which sufficient DNA was available. These samples included 44 inferred deletions and 38 inferred duplications. We also evaluated 27 samples with predictions ranging in scores from |4| to |5| and 30 samples with no predictions above |4|. Each of these samples was analyzed using a custom oligonucleotide array-CGH platform with high-density probe coverage in each of the targeted hotspots (see Methods; Fig. 1). We found that 35 deletions (SCOUT scores from −6.0 to −13.4, median −8.9) and 23 duplications (SCOUT scores from 6.1 to 11.7, median 7.5) were validated by array-CGH (Table 1; Fig. 2). In addition, three deletions (SCOUT scores of −3.2, −5.8, and −5.9) and one duplication (score of 3.8) were found by array-CGH to be partial events. We identified one sample that had a trisomy of chromosome 15 (SCOUT scores from 3.9 to 7.5 at the nine tested chromosome 15 loci; this sample was eliminated from further consideration) and one sample that carried a large duplication spanning more than three hotspots on chromosome 16. Finally, we validated four duplication events at hotspots that contained only two successful probes (SCOUT scores from 3.7 to 6.6).
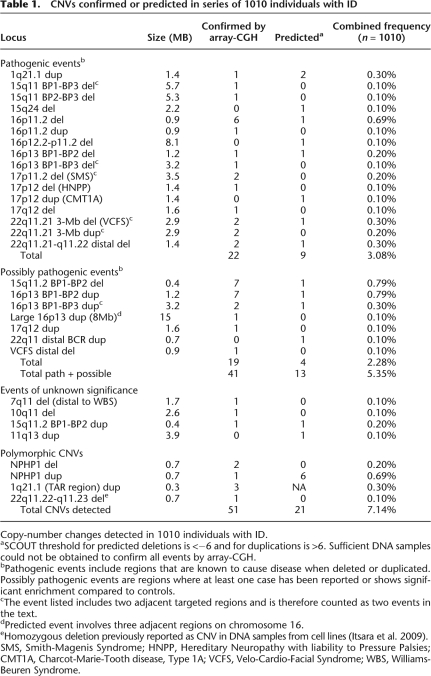
Table 1.
CNVs confirmed or predicted in series of 1010 individuals with ID
Copy-number changes detected in 1010 individuals with ID.
aSCOUT threshold for predicted deletions is <−6 and for duplications is >6. Sufficient DNA samples could not be obtained to confirm all events by array-CGH.
bPathogenic events include regions that are known to cause disease when deleted or duplicated. Possibly pathogenic events are regions where at least one case has been reported or shows significant enrichment compared to controls.
cThe event listed includes two adjacent targeted regions and is therefore counted as two events in the text.
dPredicted event involves three adjacent regions on chromosome 16.
eHomozygous deletion previously reported as CNV in DNA samples from cell lines (Itsara et al. 2009).
SMS, Smith-Magenis Syndrome; HNPP, Hereditary Neuropathy with liability to Pressure Palsies; CMT1A, Charcot-Marie-Tooth disease, Type 1A; VCFS, Velo-Cardio-Facial Syndrome; WBS, Williams-Beuren Syndrome.
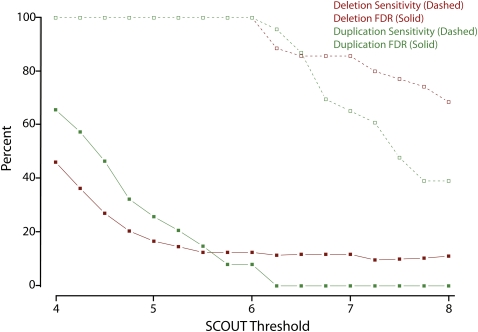
False discovery rate (FDR) and sensitivity estimation
Using the above data, we were able to generate estimates of our sensitivity and FDR using a range of thresholds (Fig. 3). We excluded loci with only two useful SNP probes and also eliminated CNVs that only affected part of a target locus. At a SCOUT threshold of |5|, we have a FDR of ∼21% (15 of 73). Importantly, we find that more stringent thresholds improved specificity with no drop in sensitivity. In fact, at a threshold of |6|, SCOUT detects all validated CNVs with a total estimated FDR of ∼11% (7 of 65; Fig. 3). We also note that three false-positive deletion predictions came from a single sample, with scores of −12.6, −8.7, and −7.1, each on a different chromosome. This is the only sample with three strongly scoring predictions from distinct chromosomes, suggesting the sample was likely to be of poor quality (see Methods). Elimination of this sample would reduce our FDR to ∼6% (4 of 62) at a threshold of |6|.
Figure 3.
Reliability of SCOUT predictions as a function of threshold. Sensitivity (dashed lines/open squares) and false discovery rates (FDR, solid lines/solid squares) to detect deletions (red) and duplications (green) are plotted as a function of the absolute value of the SCOUT threshold, incremented by units of 0.25. These estimates exclude regions covered by only two probes and also exclude CNVs in positive control samples.
Disease relevance of detected CNVs
We sought to assess the disease relevance of the CNVs detected in the 1010 individuals that we successfully screened, each of whom was affected by ID and related traits. In order to estimate the overall CNV discovery rate (independent of the availability of DNA for validation), we used a SCOUT threshold of |6| to predict the frequency of deletions and duplications, respectively, in this series (Table 1). Overall, we confirmed or predicted pathogenic or potentially pathogenic copy-number changes in 54 affected individuals. Many (31) of the discovered variants occurred at well-established disease loci, including both deletions (n = 3) and duplications (n = 2) of 22q11, and deletions of the Prader-Willi/Angelman (n = 2) and Smith-Magenis (n = 2) syndrome regions (Table 1). In addition, we found that seven (six confirmed) individuals carry deletions of 16p11.2, an event that was previously seen at similar frequencies (∼1%) in autism (Kumar et al. 2008; Marshall et al. 2008; Weiss et al. 2008).
We were also interested in those variants that are seen in multiple individuals of our cohort, but are of uncertain pathogenicity. We compared the frequency of each CNV detected here to a recent analysis of 2493 adult individuals that we conducted using genome-wide SNP genotyping data (Itsara et al. 2009). We note that each of the CNVs tested here are large and span many probes on the arrays used previously, suggesting that differential sensitivity is unlikely to be a major confounding factor in our analysis. Comparison against this panel identified two enriched CNVs in the group of affected individuals studied here. Deletions at 15q11.2 were detected here in eight of 1010 affected individuals contrasted with three events in 2493 controls (P = 0.003). In addition, duplications at 16p13 were detected in 11 of 1010 affected individuals contrasted with only two of 2493 controls (P = 4.7 × 10−5). We note that each of these enrichments remains significant even if we only include validated events (P = 0.0082 and 1.4 × 10−4 for 15q11.2 and 16p13, respectively).
Discussion
Numerous recent studies show that CNVs contribute substantially to the etiology of human disease, with rare variants and recurrent de novo mutations of large effect being particularly important (McCarroll 2008; Mefford and Eichler 2009). Identification and characterization of such disease associations require reliable ascertainment of rare variants in large cohorts, which cannot be accomplished efficiently by current methods for discovery of CNVs in individual samples. We have used Illumina BeadXpress assays in conjunction with a newly developed algorithm (SCOUT) to interrogate known and putative mutational hotspots in a collection of over 1000 individuals affected by ID and related traits. The resulting data demonstrate the capacity of this technology to rapidly and affordably analyze CNVs in large collections of phenotyped samples. Our results also provide insights specific to the genetic basis for ID. We address both the technical and biological results in turn.
The approach that we describe combines the flexibility, speed, and throughput of a SNP-based genotyping assay with the ability to accurately identify rare CNVs (singletons in arbitrarily large sample collections) and genotype common CNVs in a single experiment. To our knowledge, this is the first automated application of this technology applied to the analysis of rare and common CNVs. In this study, we analyzed 69 custom-selected loci in more than 1200 samples in less than 1 mo of processing time. We estimate that a single technician could genotype a customized set of more than 100 CNV loci in 600–700 samples/wk with per-sample reagent costs five- to 20-fold less than that of array-CGH or genome-wide SNP experiments. Since these samples were collected in a clinical setting and, in most cases, stored for many years, this cohort is likely to provide a reliable indication as to the performance of this method when applied to other clinical sample collections. Additionally, the fact that BeadXpress assays have the potential for CLIA certification may simplify development of clinical diagnostic assays to genotype CNVs of known effect, including all known recurrent microdeletion/duplication syndromes. With refinement to provide improved sensitivity and quality control, such assays could be applied as a cost-effective screen before using more expensive genome-wide chips, similar to the way that we efficiently diagnosed causal variation for dozens of individuals in this study (Table 1). In general, targeted SNP-based CNV detection provides a powerful and previously unavailable combination of customizability, cost efficiency, and throughput for the discovery of CNVs that contribute to disease.
With respect to the genetic basis for ID, we identified pathogenic or potentially pathogenic variants in 5.4% of the 1010 individuals studied here. Many of these individuals carried variants of known effect and could be given a clinically relevant diagnosis. Of note, at least six of the individuals with a deletion of 16p11.2 in our cohort do not have a diagnosis of autism, which was the disease originally associated with this locus (Weiss et al. 2008). Our results therefore widen the phenotypic spectrum to include ID without autism, consistent with a recent study that reported the same deletion in 0.3% of 4284 individuals with ID but not autism (Bijlsma et al. 2009).
We also detected a high frequency of 15q11.2 deletions between BP1 and BP2 (eight of 1010 individuals, 0.79%), near the Prader-Willi and Angelman syndromes critical region (BP2–BP3). While more definitive proof will require even larger sample sizes and ideally an analysis of cases and controls on the same platform, we observed that 15q11.2 deletions are significantly enriched (P = 0.003) in our cohort in comparison to a recently published genome-wide analysis of control individuals (Itsara et al. 2009). Additional contextual data also suggest that this variant is pathogenic. This region is deleted in some individuals with Prader-Willi and Angelman syndromes, and there is evidence that individuals with Prader-Willi or Angelman syndrome that carry the larger deletion (BP1–BP3) are more severely affected than those with only deletions of the critical interval (Butler et al. 2004; Hartley et al. 2005; Sahoo et al. 2006). Furthermore, deletions of this region were found more frequently in individuals with schizophrenia compared to controls in two independent studies (Stefansson et al. 2008; Kirov et al. 2009). Combination of our data and these other studies suggests that deletions of this region are a risk factor for neurocognitive disease.
We also identified 11 individuals (∼1.1%) with duplications of 16p13.11 (eight BP1–BP2 and three BP1–BP3) and found it to be significantly (P = 4.7 × 10−5) enriched in comparison to the same published control panel used above. One additional individual was confirmed to harbor a larger duplication event spanning this entire region (Table 1). Deletions at this locus are known to be pathogenic (Ullmann et al. 2007; Hannes et al. 2008), and duplications were associated with autism and ID in one study (Ullmann et al. 2007). In a separate study of ID, the duplications were found to be enriched in affected individuals, but this enrichment was statistically nonsignificant, suggesting the variant was either benign or incompletely penetrant (Hannes et al. 2008). Interestingly, these duplications were also reported to be nonsignificantly enriched in individuals with schizophrenia in two independent studies (International Schizophrenia Consortium 2008; Kirov et al. 2009). Thus, similar to deletions at 15q11.2, our data combined with previous literature suggest that duplications at 16p13 are, in fact, pathogenic but incompletely penetrant. In any case, future studies of neurocognitive illness, which may benefit from both the technical and biological results presented here, are warranted to confirm the influence of CNVs at these loci and more clearly delineate their phenotypic consequences.
Methods
Selection of loci
The coordinates of all regions targeted in this assay are listed in Supplemental Table 1. We selected regions of unique sequence (>50 kb) that are flanked by large (>10 kb), highly similar (>95% identity) blocks of duplicated sequence, which we term rearrangement “hotspots” (Bailey et al. 2002). Regions with this architecture have an increased propensity to generate de novo deletion or duplication events involving the entire region due to nonallelic homologous recombination. We selected 69 hotspots out of an estimated total of 110 nonredundant loci in the human genome reference assembly. We chose 25 hotspots that are known to associate with disease; the remaining 44 were chosen based upon size and gene content. We also selected two control intervals, which are common, resequenced deletion events that we have previously genotyped using multiple assays (Newman et al. 2006; Cooper et al. 2008; Kidd et al. 2008). Each of these deletions is tightly correlated with nearby “tag” SNPs, which were also included in the assay design.
Selection of SNP probes
Within each targeted region, we selected HapMap (International HapMap Consortium 2005) SNPs to genotype in the assay based on a number of criteria. First, we prioritized probes with a high Illumina design score (optimally “1.1”). Second, since allelic ratios at heterozygous SNPs provide valuable information beyond absolute intensities (Cooper et al. 2008), we favored SNPs with a minor allele frequency above 10% in both the CEU and YRI HapMap populations (our disease panel consists of both European and African-American individuals). Third, we eliminated probes within segmental duplications to minimize cross-hybridization artifacts. Finally, we optimized spacing between probes to achieve maximal coverage of the hotspots. Based upon previous experience (Newman et al. 2006; Cooper et al. 2008) and simulations using gender tests at X-linked SNPs genotyped previously (Carlson et al. 2006; data not shown), we estimated that five probes per target would provide an acceptable compromise between accuracy and probe density. In addition, for troubleshooting and control purposes, we placed ten probes within each of several known disease loci (1q21.1 [Brunetti-Pierri et al. 2008; Mefford et al. 2008], 15q13.3 [Miller et al. 2008; Sharp et al. 2008; Pagnamenta et al. 2009; van Bon et al. 2009], and 17q12 [Mefford et al. 2007]).
After the samples were genotyped, scatterplots of intensity data were generated for each probe and manually inspected (note that this step only needs to be performed once for any given set of probes). Probes with excessive noise or anomalous clustering patterns, such as the presence of multiple heterozygote clusters (Cooper et al. 2008), were excluded from further analysis. After this filtering, 25 of the original 69 targets had five or more useful probes, 52 had four or more useful probes, and 66 had at least three useful probes. Hotspots with only two probes (three loci) were analyzed but are ignored in the accuracy statistics (Fig. 3). The SNPs assigned to each target, including failed and successful probes, are provided in Supplemental Table 1.
BeadXpress genotyping
The BeadXpress assay was performed in accordance with manufacturer's protocols (http://www.illumina.com). DNA concentration was determined using a NanoDrop ND-1000 Spectrophotometer (Thermo Scientific). Ninety-six well plates were prepared with DNA at uniform concentration (either 50 or 100 ng/μL, depending on DNA availability). BeadXpress raw data were processed using Illumina's BeadStudio software suite (genotyping module 3.3.7), producing report files containing normalized intensity data and SNP genotypes. All SNP genotypes were inferred using a genotyping cluster file automatically generated by BeadStudio from an initial 96-sample control plate containing 49 HapMap DNA samples, 11 ID patient samples (see Disease Cohort below), and 36 positive control samples (Supplemental Table 2).
Algorithm: SNP-Conditional OUTlier detection (SCOUT)
SCOUT is an outlier detection algorithm that analyzes fluorescence data and SNP genotype calls to produce a per-site score for every sample at each SNP. Both the total intensity and allelic ratio (position relative to the center of the heterozygote cluster) are incorporated into the scoring function. Under the null hypothesis that all samples have the same copy number (generally assumed to be a diploid copy number, although this is not, in principle, necessary), these scores are normally distributed with mean zero. Individuals that have increased (duplication event) or decreased (deletion event) copy numbers are likely to have positive or negative intensity scores, respectively, relative to the population average (Fig. 2). The statistical model used by SCOUT is an extension of the model used by SNP-Conditional Mixture Modeling (SCIMM) for common CNV genotyping (Cooper et al. 2008), with the most important distinctions being SCOUT's assumption that most samples have the same copy number at a given SNP, and SCOUT's use of fluorescence intensity and allelic ratio information at SNP-heterozygous sites to predict duplications. After scores are generated for each probe, all probes within a targeted region are summed to determine whether a given sample carries a CNV in the targeted region. Thus, a sample must generally be an outlier in the same direction across multiple probes in a target to be flagged as a potential CNV carrier. To provide robustness against artifactual variation caused by poor-quality samples, SCOUT first performs an automated filtering step to eliminate samples with an assay-wide excess of extreme outlier scores (see Supplemental Methods and Supplemental Fig. 1). SCOUT sample filtering and scores and SCIMM genotypes were calculated independently for each 96-sample plate.
Disease cohort
After obtaining informed consent, we evaluated DNA from 1105 individuals who received services through the South Carolina Department of Disabilities and Special Needs. Common causes of ID had been excluded by fragile X testing, chromosome analysis, amino acid and organic acid analyses, and urinary metabolic screening (n = 1105, 49.8% female, 50.2% male, 53% African American, 46% European American, 1% unstated or other ethnicity). Eight hundred forty-seven of these samples were also previously screened using TaqMan quantitative PCR assays for deletions or duplications of 15q13 (Sharp et al. 2008), 1q21.1 (Mefford et al. 2008), 17q21.3 (data not shown), and 15q24 (data not shown).
Array-CGH validation of predicted CNVs
We performed oligonucleotide array comparative genomic hybridization (array-CGH) using a custom 12×135K array (Roche NimbleGen) with 135,000 oligonucleotide probes. Each of the hotspots targeted by the BeadXpress assay described here were represented on the array with an average spacing of one probe per 2650 bp, allowing for validation of putative deletions and duplications involving all or part of each hotspot. Hybridizations were performed as previously described (Sharp et al. 2008) using a single common reference sample (NA15724). In a few cases, samples were validated using a whole-genome tiling array (NimbleGen HG18 12×135K WG Tiling v2.0) that also offers sufficient coverage to validate potential CNVs at all targeted hotspots.
Acknowledgments
This work was supported, in part, by an NIH grant HD043569 to E.E.E. H.C.M. received support from an NIH K12 grant HD043376. T.Z. and D.A.N. acknowledge support from NIH grants HL66682 and HL66642. G.M.C. is supported by a Merck, Jane Coffin Childs Memorial Fund Fellowship. E.E.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.094987.109.
References
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Bijlsma EK, Gijsbers AC, Schuurs-Hoeijmakers JH, van Haeringen A, Fransen van de Putte DE, Anderlid BM, Lundin J, Lapunzina P, Perez Jurado LA, Delle Chiaie B, et al. Extending the phenotype of recurrent rearrangements of 16p11.2: Deletions in mentally retarded patients without autism and in normal individuals. Eur J Med Genet. 2009;52:77–87. doi: 10.1016/j.ejmg.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, Sahoo T, Lalani SR, Graham B, Lee B, Shinawi M, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG, Bittel DC, Kibiryeva N, Talebizadeh Z, Thompson T. Behavioral differences among subjects with Prader-Willi syndrome and type I or type II deletion and maternal disomy. Pediatrics. 2004;113:565–573. doi: 10.1542/peds.113.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CS, Smith JD, Stanaway IB, Rieder MJ, Nickerson DA. Direct detection of null alleles in SNP genotyping data. Hum Mol Genet. 2006;15:1931–1937. doi: 10.1093/hmg/ddl115. [DOI] [PubMed] [Google Scholar]
- Christian SL, Brune CW, Sudi J, Kumar RA, Liu S, Karamohamed S, Badner JA, Matsui S, Conroy J, McQuaid D, et al. Novel submicroscopic chromosomal abnormalities detected in autism spectrum disorder. Biol Psychiatry. 2008;63:1111–1117. doi: 10.1016/j.biopsych.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Zerr T, Kidd JM, Eichler EE, Nickerson DA. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nat Genet. 2008;40:1199–1203. doi: 10.1038/ng.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries BB, Pfundt R, Leisink M, Koolen DA, Vissers LE, Janssen IM, Reijmersdal S, Nillesen WM, Huys EH, Leeuw N, et al. Diagnostic genome profiling in mental retardation. Am J Hum Genet. 2005;77:606–616. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JM, Baross A, Delaney AD, Ally A, Arbour L, Armstrong L, Asano J, Bailey DK, Barber S, Birch P, et al. Oligonucleotide microarray analysis of genomic imbalance in children with mental retardation. Am J Hum Genet. 2006;79:500–513. doi: 10.1086/507471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannes FD, Sharp AJ, Mefford HC, de Ravel T, Ruivenkamp CA, Breuning MH, Fryns JP, Devriendt K, Van Buggenhout G, Vogels A, et al. Recurrent reciprocal deletions and duplications of 16p13.11: The deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J Med Genet. 2008;46:223–232. doi: 10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley SL, Maclean WE, Jr, Butler MG, Zarcone J, Thompson T. Maladaptive behaviors and risk factors among the genetic subtypes of Prader-Willi syndrome. Am J Med Genet A. 2005;136:140–145. doi: 10.1002/ajmg.a.30771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsara A, Cooper GM, Baker C, Girirajan S, Li J, Absher D, Krauss RM, Myers RM, Ridker PM, Chasman DI, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009;84:148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P, Craddock N, Owen MJ, O'Donovan MC. Support for the involvement of large CNVs in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, Gilliam TC, Nowak NJ, Cook EH, Jr, Dobyns WB, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA. Extending genome-wide association studies to copy-number variation. Hum Mol Genet. 2008;17:R135–R142. doi: 10.1093/hmg/ddn282. [DOI] [PubMed] [Google Scholar]
- McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, Barrett JC, Dallaire S, Gabriel SB, Lee C, Daly MJ, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- Mefford HC, Eichler EE. Duplication hotspots, rare genomic disorders, and common disease. Curr Opin Genet Dev. 2009;19:196–204. doi: 10.1016/j.gde.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Clauin S, Sharp AJ, Moller RS, Ullmann R, Kapur R, Pinkel D, Cooper GM, Ventura M, Ropers HH, et al. Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am J Hum Genet. 2007;81:1057–1069. doi: 10.1086/522591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, Huang S, Maloney VK, Crolla JA, Baralle D, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DT, Shen Y, Weiss LA, Korn J, Anselm I, Bridgemohan C, Cox GF, Dickinson H, Gentile J, Harris DJ, et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatirc disorders. J Med Genet. 2008;46:242–248. doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman TL, Rieder MJ, Morrison VA, Sharp AJ, Smith JD, Sprague LJ, Kaul R, Carlson CS, Olson MV, Nickerson DA, et al. High-throughput genotyping of intermediate-size structural variation. Hum Mol Genet. 2006;15:1159–1167. doi: 10.1093/hmg/ddl031. [DOI] [PubMed] [Google Scholar]
- Pagnamenta AT, Wing K, Akha ES, Knight SJ, Bolte S, Schmotzer G, Duketis E, Poustka F, Klauck SM, Poustka A, et al. A 15q13.3 microdeletion segregating with autism. Eur J Hum Genet. 2009;17:687–692. doi: 10.1038/ejhg.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg C, Knijnenburg J, Bakker E, Vianna-Morgante AM, Sloos W, Otto PA, Kriek M, Hansson K, Krepischi-Santos AC, Fiegler H, et al. Array-CGH detection of micro rearrangements in mentally retarded individuals: Clinical significance of imbalances present both in affected children and normal parents. J Med Genet. 2006;43:180–186. doi: 10.1136/jmg.2005.032268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo T, Peters SU, Madduri NS, Glaze DG, German JR, Bird LM, Barbieri-Welge R, Bichell TJ, Beaudet AL, Bacino CA. Microarray based comparative genomic hybridization testing in deletion bearing patients with Angelman syndrome: Genotype-phenotype correlations. J Med Genet. 2006;43:512–516. doi: 10.1136/jmg.2005.036913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ, Hansen S, Selzer RR, Cheng Z, Regan R, Hurst JA, Stewart H, Price SM, Blair E, Hennekam RC, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- Sharp AJ, Mefford HC, Li K, Baker C, Skinner C, Stevenson RE, Schroer RJ, Novara F, De Gregori M, Ciccone R, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmann R, Turner G, Kirchhoff M, Chen W, Tonge B, Rosenberg C, Field M, Vianna-Morgante AM, Christie L, Krepischi-Santos AC, et al. Array-CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat. 2007;28:674–682. doi: 10.1002/humu.20546. [DOI] [PubMed] [Google Scholar]
- van Bon BWM, Mefford HC, Menten B, Koolen DA, Sharp AJ, Nillesen WM, Innis JW, de Ravel TJL, Mercer CL, Fichera M, et al. Further delineation of the 15q13 microdeletion and duplication syndromes: A clinical spectrum varying from non-pathogenic to a severe outcome. J Med Genet. 2009 doi: 10.1136/jmg.2008.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LE, de Vries BB, Osoegawa K, Janssen IM, Feuth T, Choy CO, Straatman H, van der Vliet W, Huys EH, van Rijk A, et al. Array-based comparative genomic hybridization for the genomewide detection of submicroscopic chromosomal abnormalities. Am J Hum Genet. 2003;73:1261–1270. doi: 10.1086/379977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]