Abstract
As the cost of DNA sequencing drops, we are moving beyond one genome per species to one genome per individual to improve prevention, diagnosis, and treatment of disease by using personal genotypes. Computational methods are frequently applied to predict impairment of gene function by nonsynonymous mutations in individual genomes and single nucleotide polymorphisms (nSNPs) in populations. These computational tools are, however, known to fail 15%–40% of the time. We find that accurate discrimination between benign and deleterious mutations is strongly influenced by the long-term (among species) history of positions that harbor those mutations. Successful prediction of known disease-associated mutations (DAMs) is much higher for evolutionarily conserved positions and for original–mutant amino acid pairs that are rarely seen among species. Prediction accuracies for nSNPs show opposite patterns, forecasting impediments to building diagnostic tools aiming to simultaneously reduce both false-positive and false-negative errors. The relative allele frequencies of mutations diagnosed as benign and damaging are predicted by positional evolutionary rates. These allele frequencies are modulated by the relative preponderance of the mutant allele in the set of amino acids found at homologous sites in other species (evolutionarily permissible alleles [EPAs]). The nSNPs found in EPAs are biochemically less severe than those missing from EPAs across all allele frequency categories. Therefore, it is important to consider position evolutionary rates and EPAs when interpreting the consequences and population frequencies of human mutations. The impending sequencing of thousands of human and many more vertebrate genomes will lead to more accurate classifiers needed in real-world applications.
Unshrouding the mysteries of human genome variation is the essential precursor to the development of personalized medicine where the aim is to relate the genotype with the phenotype in better understanding an individual's susceptibility to disease and response to treatment. Already, complete genomes from many individual humans have been sequenced, and projects are underway to expand that number to over a thousand genomes in the near future (Levy et al. 2007; Bentley et al. 2008; Wang et al. 2008; Wheeler et al. 2008). These projects have revealed that every individual carries thousands of amino acid–altering (nonsynonymous) nucleotide mutations and that a large number of these mutations are novel in terms of their location and the type of amino acid change induced. Experimental and other functional information are rarely available for the association of phenotypic effect with these mutations, so computational methods are used instead (e.g., Miller and Kumar 2001; Ramensky et al. 2002; Ng and Henikoff 2003; Shastry 2007; Tian et al. 2007; Lohmueller et al. 2008). These in silico predictions are of great interest in detecting variants for Mendelian and complex diseases, in prioritizing polymorphisms for experimental research in humans and other species, and in analyzing data from genome-wide association studies (e.g., Rudd et al. 2005; Bhatti et al. 2006; Kryukov et al. 2007; Doniger et al. 2008). Using various prediction tools, up to one-fourth of nonsynonymous mutations have been diagnosed to be not strictly neutral and are thus thought to harbor signatures of negative or positive selection (Yampolsky et al. 2005; Eyre-Walker et al. 2006; Levy et al. 2007; Shastry 2007; Bentley et al. 2008; Boyko et al. 2008; Wang et al. 2008; Wheeler et al. 2008).
The de novo prediction methods to predict functional effects of novel mutations often do not directly incorporate many biological attributes (e.g., interactions among multiple sites or genes, environmental influences on phenotypes, and allele state in the paired chromosome) because of the lack of information and the difficulty in modeling them mathematically. Still, these methods offer up to 80% accuracy for mutations in genes implicated in Mendelian diseases (for reviews, see Bhatti et al. 2006; Ng and Henikoff 2006; Bromberg and Rost 2007; Shastry 2007; Tian et al. 2007). PolyPhen is the most widely used method for estimating potential deleterious effects of amino acid mutations; it is available as a web-based service, and it relies on information from sequence conservation, physiochemical differences, proximity of mutations to predicted functional domains, and structural features (Sunyaev et al. 1999; Ramensky et al. 2002). PolyPhen and SIFT (Ng and Henikoff 2003) have been used in hundreds of studies, including the evaluation of nonsynonymous single nucleotide polymorphisms (nSNPs) found in complete genomes. Many other approaches have been proposed over the last decade, but these are not yet widely used (e.g., Bromberg and Rost 2007; Tian et al. 2007; Cheng et al. 2008).
In recent years, scientists have employed many strategies in efforts to build super-classifiers, using sophisticated computational approaches to improve the accuracy of computational prediction tools in diagnosing known disease-associated mutations (DAMs) to be function-altering (damaging; true-positives) and nSNPs to be neutral (benign; true-negatives). These strategies have resulted in some gains compared with classical methods such as PolyPhen and SIFT (e.g., Bromberg and Rost 2007; Tian et al. 2007). However, the anatomies of the misdiagnoses of different types of DAMs and nSNPs remain poorly understood, as the primary correlates of the observed failures are yet to be explored. With a focus on PolyPhen and the comparison of the observed patterns with those from SIFT, we have taken an evolutionary approach in examining the patterns of successes and failures of mutational diagnosis. Given that PolyPhen (and other methods) already considers a host of evolutionary and primary sequence attributes in making decisions, it is reasonable to work with the null hypothesis that the accuracy of correct prediction is similar for mutations occurring at positions evolving with different evolutionary rates and that the accuracy of the correct prediction is similar for different original and mutated amino acids. The choice of evolutionary conservation of positions, original amino acids, and mutant alleles reflects practical considerations, because these three attributes are readily available or calculable for all mutations. Other factors, such as secondary, tertiary, and higher structures undoubtedly play an important role, but that information is not yet available for an overwhelming proportion of known DAMs and nSNPs for human populations.
Results
We begin with a report on the accuracy of correctly diagnosing known DAMs implicated in Mendelian diseases (>9000 DAMs from >500 genes) (Supplemental Fig. S1). These DAMs were subjected to the most recent version of the PolyPhen web service, which classifies them into three categories—benign, possibly-damaging, and probably-damaging—based on the logarithmic ratios of the likelihood of occurrence of a given DAM at the specific position and the likelihood of that amino acid occurring at any position (Ramensky et al. 2002). A probably-damaging designation indicates that the mutation's chance of affecting protein function is the highest, whereas a benign designation suggests little or no putative impact on the protein function.
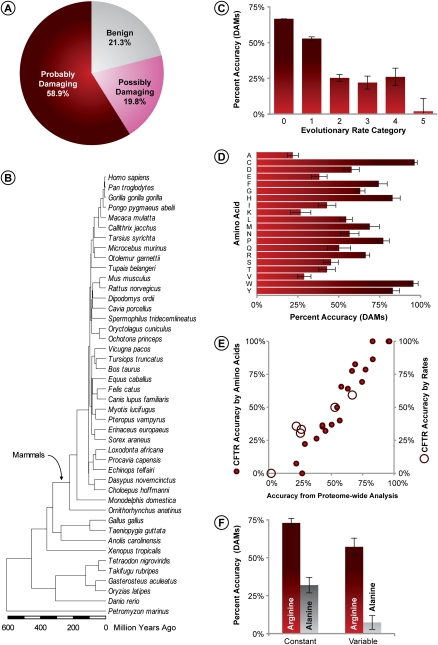
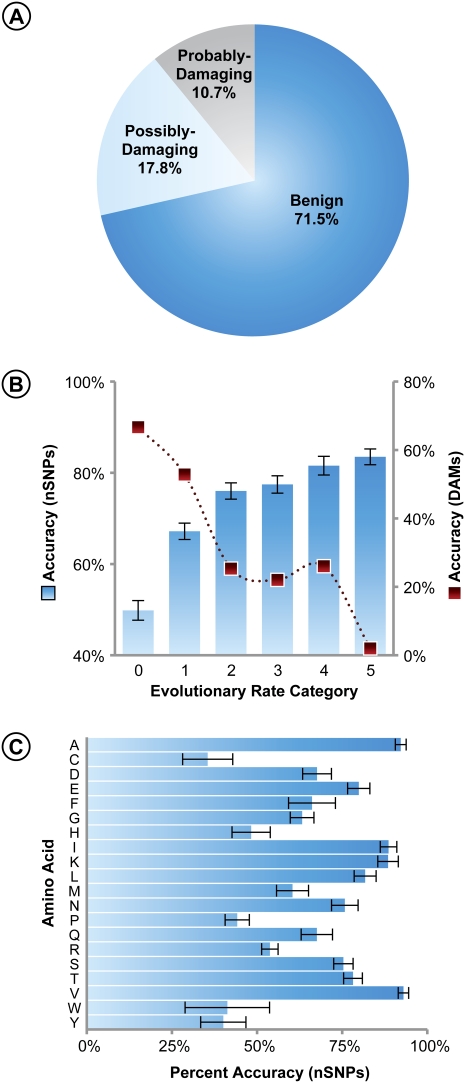
PolyPhen designated 60% of DAMs to be probably-damaging, which is the correct inference in this case (Fig. 1A). However, 21% of DAMs were identified to be benign, which provides a lower limit on the false-negative rate of inference. Similar accuracies are reported in other studies, as well (Ng and Henikoff 2006; Chan et al. 2007; Tian et al. 2007; Cheng et al. 2008; Lohmueller et al. 2008). Pooling of the benign and possibly-damaging (ambiguous) diagnoses increases the false-negative rate for DAMs to 41%, while the pooling of the possibly-damaging and probably-damaging categories increases the DAM-prediction accuracy of PolyPhen to 79%. However, it appears to be more prudent to use only the probably-damaging category to represent the correct inference for DAMs, because PolyPhen classified a very similar fraction of DAMs and nSNPs into the possibly-damaging category (20% and 18%, respectively) (cf. Figs. 1A and 2A). We compared PolyPhen results with those obtained from SIFT, which classifies mutations into only two categories: tolerant or not-tolerant (Ng and Henikoff 2006). SIFT designated 21% of DAMs to be tolerant, which is similar to the DAM misclassification rates in PolyPhen (Supplemental Fig. S2).
Figure 1.
Accuracy of PolyPhen diagnosis of 9460 DAMs. (A) Fraction of DAMs classified into benign, possibly-damaging, and probably-damaging categories. (B) Evolutionary timetree of 44 species used for estimating evolutionary rates. Relationship of evolutionary rates (C) and incident amino acids (D) with the correct diagnosis of DAMs (probably-damaging). Error bars, 95% confidence interval (two times the SE). (E) Correlation between the accuracy of DAM prediction from proteome-wide analysis, and one DAM-rich protein (cystic fibrosis transmembrane conductance regulator [CFTR]). Solid and open circles show data points for incident amino acids (r = 0.97; P << 0.01) and evolutionary rates (r = 0.95; P << 0.01), respectively. (F) Two examples showing the dependence of the accuracy of DAM diagnosis for constant and variables sites for arginine (an easy-to-diagnose amino acid; red bars) and alanine (a difficult-to-diagnose amino acid; gray bars). Error bars, 95% confidence interval based on the binomial variance of the fraction of sites in the plotted categories.
Figure 2.
PolyPhen classification of 12,421 nSNPs into benign, possibly-damaging, and probably-damaging categories. (A) Fraction of nSNPs classified into the three categories. The fraction of nSNPs designated to be benign at positions with different evolutionary rates (B) and original amino acids (C). Panel B also contains the accuracy of DAM inference from Figure 1B (filled squares). Error bars, 95% confidence interval based on the binomial variance of the fraction of sites.
For identifying the correlates of successes and failures in diagnosing DAMs, we first examined the accuracy of correct diagnoses for genes having different functions (as reflected in the gene ontology). We classified each gene into one or more of 13 major categories (plus a group of unannotated genes). The accuracy of predicting DAMs in these categories varied in a relatively narrow range, even though DAMs in some of the gene function categories were significantly easier to predict than others (e.g., translation) (see Supplemental Fig. S3).
In contrast to functional categories, the long-term evolutionary rates at DAM positions correlate strongly with the success in diagnosing DAMs in both PolyPhen (Fig. 1C) and SIFT (data not shown). DAMs in completely conserved positions (lowest evolutionary rate) were 1.5 times more likely to be correctly classified than those at positions harboring any interspecies variation (67% vs. 44%; P < 0.01), while more than 70% of DAMs in the fastest-evolving positions were misdiagnosed (benign). The accuracy of DAM prediction also varies tremendously and unexpectedly among the 20 amino acids, with accuracy ranging from 22%–96% in the proteome-wide analysis (Fig. 1D).
We examined the question of whether the observed relationship between the evolutionary rate and the accuracy of prediction is reproduced for DAMs of specific proteins, because PolyPhen scores the relative likelihood of a mutation to affect function in its protein context. Analysis of the cystic fibrosis transmembrane conductance regulator (CFTR) protein, which contributed the largest number of DAMs in our data set (444), produced patterns similar to those seen in the proteome-wide analysis for rates (Pearson's r = 0.95) as well as for original amino acids (r = 0.97) (Fig. 1E). Similar results were observed for other DAM-rich proteins (data not shown). Therefore, the dependence of correct inference of DAMs on evolutionary rate and amino acids is a fundamental attribute of positions, rather than an artifact of proteome-wide summarization of mutations in proteins evolving with vastly different conservation profiles and amino acid contents. Major differences among evolutionary rates and amino acids and were also observed in the SIFT analysis, and these differences were correlated with those observed for PolyPhen (r = 0.95 and 0.59, respectively; P << 0.01). Because different amino acids are known to evolve at intrinsically different rates, we also examined the relationship of evolutionary conservation on the DAM prediction accuracy for specific amino acids. In the easiest to diagnose amino acids (e.g., arginine), DAMs occurring at completely conserved positions were significantly harder to predict than those at positions with any site variability (P << 0.01) (Fig. 1F). A similar pattern is seen for amino acids whose mutations are difficult to diagnose (e.g., alanine) (P << 0.01) (Fig. 1F).
Next, we analyzed >12,000 nSNPs in order to examine the dependence of rate of evolution and the original amino acid for mutations not associated with any disease. The fraction of nSNPs identified as benign also depends strongly on evolutionary rates (Fig. 2B) and the original amino acids (Fig. 2C). However, DAMs and nSNPs show opposite patterns in terms of accuracy, assuming that a vast majority of nSNPs represent nondisease variations. For instance, alanine nSNPs are diagnosed as benign most often, and nSNPs at fast-evolving positions are also easily diagnosed to be benign.
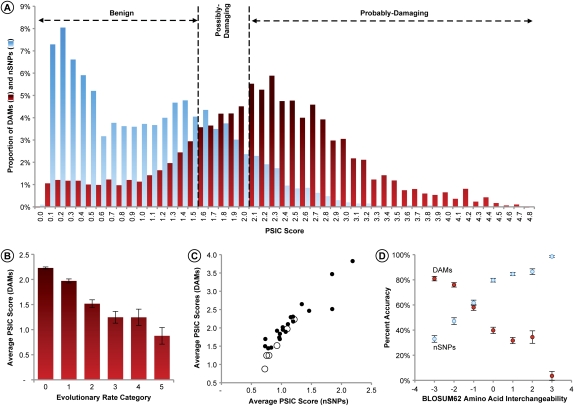
In order to investigate why DAMs and nSNPs show complementary patterns, we further analyzed results from PolyPhen, which uses a single score metric (position-specific independent counts [PSIC] score) in its decision making. Distributions of PSIC scores overlap extensively for DAMs and nSNPs proteome-wide (Fig. 3A) and for individual amino acids (Supplemental Fig. S4). Generally, DAMs exhibit a wider range of values and carry larger PSIC scores as compared with nSNPs. Underlying the wide PSIC distributions for DAMs and nSNPs are the relationships of PSIC scores with the evolutionary rates and amino acids involved. The lowest average PSIC scores are seen for the evolutionary rates and amino acids for which PolyPhen exhibited the worst performance for DAMs (Fig. 3B). In fact, nSNPs with these rates and incident amino acids show the lowest average PSIC scores, as well (P << 0.01) (Fig. 3C). This comparison of the PSIC scores for DAMs and nSNPs explains the inverse relationship between the accuracy in diagnosing nSNPs (to be benign) and DAMs (to be probably-damaging), because PolyPhen designates all mutants with PSIC ≤ 1.5 to be benign and with PSIC > 2.0 to be probably-damaging; PSIC scores between 1.5 and 2.0 yield the possibly-damaging diagnosis.
Figure 3.
(A) Frequency distributions of DAM (red) and nSNPs (blue) PSIC scores. Vertical lines show the PolyPhen PSIC cut-offs for classification of variants in the absence of structural or other information; nSNPs and DAMs are from Subramanian and Kumar (2006a). (B) Mean PSIC values for DAMs in different evolutionary rate categories. The correlation (r) between mean PSIC values and mean evolutionary rate is −0.96 (P << 0.01). The 95% confidence intervals derived from the SEMs are shown. (C) Relation between mean PSIC scores for DAMs and mean PSIC scores for nSNPs, by amino acid types (solid circles) and evolutionary rates (open circles). (D) Inverse relationship of the accuracy of DAMs (probably-damaging) and nSNPs (benign) with the evolutionary interchangeability of amino acid pairs (original/variant pairs) as captured in the BLOSUM62 matrix. Each data point represents the average of all pairs for a given BLOSUM score, with the error bars displaying the 95% confidence intervals derived from binomial variance of the proportions. BLOSUM scores are log-odds substitution occurrences. Negative BLOSUM scores show amino acid pairs that are found to have a low probability of substitution, whereas a positive score indicates frequently observed amino acid pairs. Complete 20 × 20 matrices of DAM and nSNP accuracies (and their SEs) are given in the Supplemental material.
The estimation of PSIC scores also involves the use of an amino acid interchangeability matrix (evolutionary substitution matrix for each pair of amino acids), which is frequently inferred from multiple sequence alignments for a large number of proteins (e.g., BLOSUM log-odds substitution matrix). Amino acid interchangeability varies extensively, and we expect to see concordant differences in prediction accuracies. Indeed, the extensive heterogeneity in the accuracy of prediction for different original–mutant pairs is seen for DAMs and nSNPs, and it correlates with the BLOSUM62 amino acid interchangeability (Fig. 3D). The original–mutant pairs that occur with the highest frequency in nature are the hardest to diagnose when the mutant is disease-associated (Fig. 3D). In contrast, these pairs are the easiest to diagnose for nSNPs. Thus, nSNPs and DAMs show opposite relationships that are explained by the evolutionary properties of positions as well as the assumptions on amino acid interchangeability derived from long-term evolutionary patterns.
In addition to evolutionary rates, comparative genomics yields a set of amino acids observed among species in every position. Under the simplifying assumption that the function of a position has not changed significantly, these amino acids are evolutionarily permissible alleles (EPAs) at that position. Since EPAs are neutral alternatives at a position, they are not expected to be disease-associated. We inferred EPAs for each DAM and nSNP position by using multiple sequence alignments of 44 diverse vertebrate species (see Methods). A small fraction of DAMs are EPAs (∼9%), a finding that is similar to those reported elsewhere (e.g., Kondrashov 2003; Subramanian and Kumar 2006a). These DAMs occur preferentially at faster-evolving positions.
As expected, EPAs comprise a vast majority of nSNPs (59%). Still, many thousands of nSNPs are not EPAs. This result may not be attributed to a disproportionate number of alignment gaps and missing data at positions where nSNPs are not EPAs, because the fraction of species with alignment gaps and missing data were almost identical for EPA and non-EPA nSNP sites (32% and 33%, respectively). We expect the fraction of non-EPA nSNPs to increase in the future as more individual genomes are sequenced and rarer alleles are discovered. This increase will be counteracted by discovery of more nSNPs in EPA because the use of more species in the multiple sequence alignments would expand the list of EPAs at each position. Overall, the number of non-EPA nSNPs is likely to decline slowly, if at all. This conclusion is based on the observation that more than 80% of EPA nSNPs could be identified using only 33 nonhuman mammals, and a 30% increase in the number of species (nine additional species) led to the discovery of only a small fraction of nSNPs in expanded EPA lists for each site.
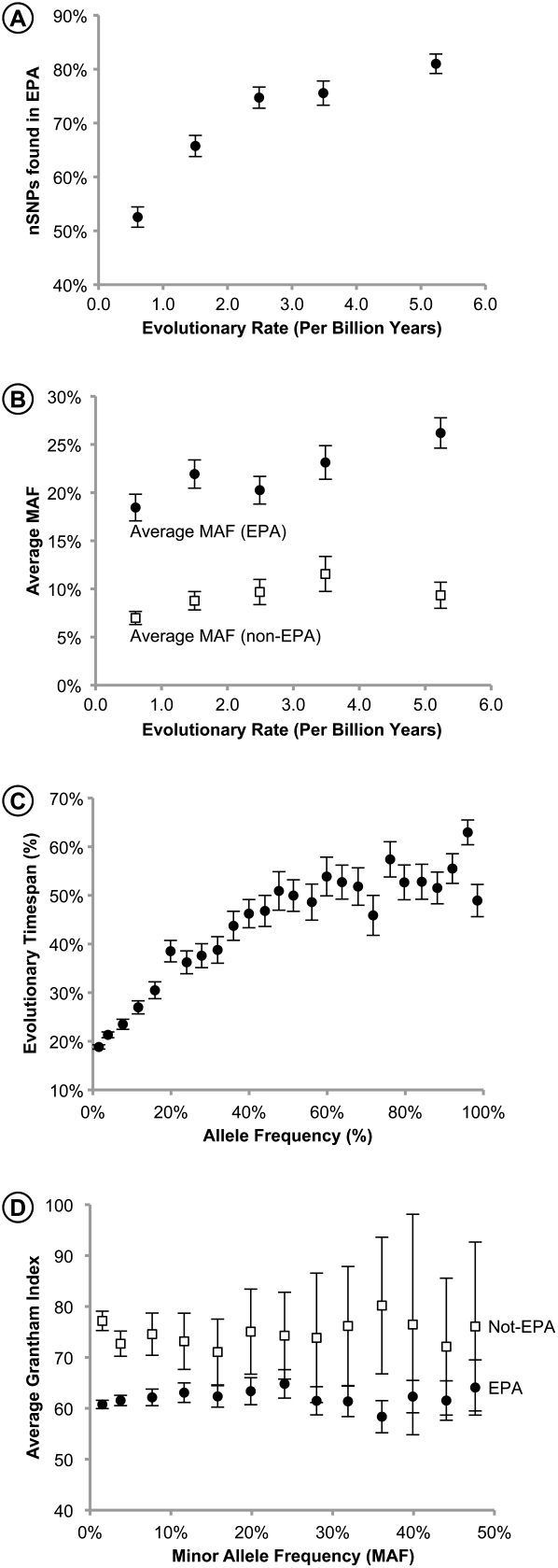
The frequency of nSNP occurrence in EPA shows a marked relationship with the evolutionary rate. The nonsynonymous polymorphisms in the fastest-evolving positions are EPAs significantly more frequently than those in the slowest-evolving positions (81% vs. 53%; P << 0.01) (Fig. 4A). Likely, this is because the strong purifying selection in the highly conserved sites would allow only recently emerged mutations to be found at those positions, and these nSNPs would occur with low frequencies. Furthermore, positions that evolve more slowly will have a smaller number of EPAs, which would result in a greater proportion of non-EPA nSNPs. This phenomenon is evident in the observation that non-EPA nSNPs occur with one-third the allele frequency of EPA nSNPs overall consistently across positions evolving with different rates (Fig. 4B).
Figure 4.
Analysis of EPAs. (A) The relation of the evolutionary rate with the proportion of nSNPs present in the set of EPAs in the variable sites (r = 0.91, P < 0.02). (B) The average allele frequencies of nSNPs present in EPAs (closed circles) and absent from the set of EPAs (open squares) in variable positions evolving with different rates. Mean allele frequencies are significantly different between two EPA categories for each rate class (P << 0.01). (C) Relationship between nSNPs frequency and the percentage of evolutionary time span (%ETS) of the corresponding EPA (r = 0.90, P << 0.01). All non-EPA nSNPs have an ETS of 0. (D) The biochemical severities of nSNPs present in EPAs (closed circles) and absent from EPAs (open squares). Error bars, 95% confidence intervals derived from the SEMs.
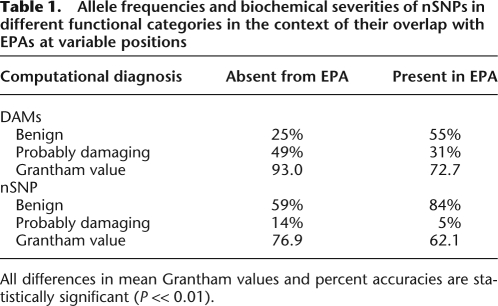
A stratification of PolyPhen results based on the EPA status of the analyzed mutations shows the importance of EPAs (Table 1). At variable positions, DAMs are diagnosed to be probably-damaging twice as often as benign when they do not overlap EPAs. This accuracy declines to 31% when DAMs are EPAs, and DAMs are diagnosed to be probably-damaging much less frequently than benign. Therefore, DAM accuracy prediction depends strongly on their overlap with EPAs. There is also a great influence of EPAs on the prediction of functional classification of nSNPs. nSNPs are much easier to categorize as benign if they appear as EPAs. In fact, nSNPs are designated to be probably-damaging only 5% of the time if they are EPAs; from this rarity and from the above-mentioned results, we can infer that the observed EPAs at a position are important indicators of the accuracy with which functional impact of novel mutations can be predicted.
Table 1.
Allele frequencies and biochemical severities of nSNPs in different functional categories in the context of their overlap with EPAs at variable positions
All differences in mean Grantham values and percent accuracies are statistically significant (P << 0.01).
Discussion
In proteome-wide analyses, we have shown that evolutionary rate and positional amino acid composition correlates extensively with the computational assessment of a mutation's functional effects. A large number of DAMs are found in positions that vary among species, and a majority of DAMs in these positions are misdiagnosed. Similarly, a large number of nSNPs occur in positions that are highly conserved, which, in many cases, are predicted by computational tools to carry functional consequences. Correlation between classification accuracies and PSIC scores suggests that we could improve prediction by tailoring PSIC classification thresholds to individual classes of variants (e.g., by amino acid type and by rate class). However, such efforts would likely suffer the handicap of the classical trade-off between the false-negative and false-positive prediction rates. That is, while changes in PSIC diagnostic thresholds for individual amino acids and/or rate classes might reduce false-negatives for DAMs, they might simultaneously increase false-positives for nSNPs. In such cases, it is prudent to associate a reliability indicator with inferences produced using computational methods (e.g., Bromberg and Rost 2007).
We suggest that a reliability of inference (RoI) measure be included with functional predictions to reflect their uncertainty. The RoI measure is the average of probability of true-positives (PTP) and the probability of true-negatives (PTN). The former is calculated by applying the given computational method on all available DAMs, while the latter is calculated by using all available strictly “neutral” nSNP data. By design, the RoI does not depend on the inference made. Rather, it captures how difficult it will be to make a correct prediction for a given type of change in its evolutionary context. The RoI may only be improved by improving true-positive and true-negative rates (such efforts are already underway for PolyPhen) (S Sunyaev, pers. comm.). Of course, PTP and PTN may be weighted unequally in calculating the RoI when analyzing nonsynonymous mutations from the genomes of “healthy” individuals, because they are expected to carry a large number of neutral mutations. In this case, RoI = (PTN + ωPTP)/(1 + ω), where ω is the expected ratio of DAMs to nSNPs and will generally be less than one. Furthermore, single and multidimensional RoI matrices may be constructed, with amino acid pair and rate classes as additional dimensions, because the accuracy of diagnosis differs among classes for the same amino acid. We anticipate that sufficient data will become available in the future from the profiling of an expanded number of diseases, individuals, and populations to build such matrices.
For now, we used the estimates of PTP and PTN based on the DAM and nSNP data analyzed (see 20 × 20 matrices in the Supplemental Figs. S5, S6), respectively, to estimate the RoI for 682 mutations found in the disease-associated genes of one individual (Levy et al. 2007). The average RoI for these mutations is 57.5% when PTP and PTN are equally weighted. It rises to 71% when PTN is given a weight 10 times that to PTP (i.e., ω = 0.1). This ad hoc ratio may be justifiable, because ∼10% of nonsynonymous mutations are found to be fixed among species in comparative genomic analysis involving humans and chimpanzees (e.g., Subramanian and Kumar 2006b). While a 71% success rate may appear reasonably good for some academic research, it is presently too low to be useful in real-world applications (especially in making health decisions).
In addition to helping us understand the factors that modulate the accuracy of computational methods, evolutionary rates and frequencies of EPAs at positions involved in DAMs and nSNPs supply null expectations for interpreting the observed population frequencies of alleles. For example, computational methods have been used to predict the functional effects of nSNPs (benign, possibly-damaging, and probably-damaging) found in genome-scale population surveys and the distributions of frequencies of alleles in the three functional categories compared (Lohmueller et al. 2008). Lohmueller et al. (2008) noted that the mean derived allele frequency (MAF) for the benign alleles is significantly higher than that for the damaging alleles. The direction and magnitude of this difference is predictable based on the average evolutionary rates of positions in the three functional categories, because the long-term evolutionary rates at any given position will modulate allele frequencies within populations under the principles of the neutral theory (Kimura 1983; Subramanian and Kumar 2006a). Indeed, rates of evolution and the MAF are highly correlated over all nSNPs and when considering EPA and non-EPA nSNPs separately (r = 0.88; P < 0.05). The evolutionary rate ratio for probably-damaging and benign positions is quite similar to that reported for the MAF (0.49 and 0.40, respectively), but a second-degree polynomial fits the relationship between rate and the MAF better than a linear regression (r2 = 0.86 and 0.77, respectively). Furthermore, the neutral theory predicts that EPAs found in a larger number of species would occur with higher frequencies in the human population. Significant correlation is found between the nSNP frequency in the human population and the evolutionary time span (%ETS) when all nSNPs are divided into 25 allele frequency categories (r = 0.90; P << 0.01) (Fig. 4C) and for raw data (r = 0.41, P << 0.01). With the upcoming sequencing of a large number of individuals, it will be possible to estimate allele frequencies in different populations more reliably and to examine the predictive power of ETS in generating expected allele frequencies of nSNPs for use in functional genomics.
In addition to frequencies, the biochemical severity of mutations is an important factor to consider, particularly as it relates to the DAMs found in EPAs and nSNPs absent from EPAs. At variable sites, DAMs that overlap EPAs are biochemically much less severe than other DAMs (average Grantham values of 73 vs. 93; P << 0.01) (Table 1) but are more severe than nSNPs and inter-specific differences (average Grantham value of 68). Compensatory evolution is thought to be one of the mechanisms to explain this observation for DAMs and is discussed elsewhere (e.g., Kondrashov et al. 2002; Gao and Zhang 2003; Subramanian and Kumar 2006a). On the other hand, non-EPA nSNPs show a higher biochemical severity than other SNPs (77 vs. 62; P << 0.01). Furthermore, many non-EPA nSNPs are observed with relatively high frequencies in the Lohmueller et al. (2008) data, and these higher frequency alleles show a greater biochemical severity than EPA nSNPs with similar frequencies (Fig. 4D). These observations suggest that some non-EPA nSNPs may be involved in adaptive evolution or persist due to compensatory changes. We are currently investigating biological properties of a large number of nSNPs that occur with high frequency in human populations but have significantly smaller than expected ETS. These mutations are excellent candidates for potential (ancient or modern) lineage-specific adaptations (and compensations), and they will be discussed elsewhere (S Kumar and A Filipski, in prep.). In the meantime, it is clear that the interpretations of rare and common alleles with different functional predictions need to account for evolutionary rates, the amino acids involved, and the EPA status of mutations and their resident positions when determining their genome-wide associations with the phenotypes.
In conclusion, with decreasing costs for sequencing personal genomes and variants, it is quickly becoming feasible to use individual genetic novelties for learning about predisposition to diseases and to better carry out optimally informed treatments based on personal genomic profiles. In such efforts, computational methods that predict the propensity of novel mutations to cause disease will play a critical role, because it is not possible to investigate the effects of individual rare (or even common) mutations in the laboratory and because each individual carries many unique mutations. Our findings show that some amino acid mutations will be easier to diagnose with high accuracy because of the amino acids involved and because of the evolutionary properties of the positions they afflict when we apply genome-wide observations to individual positions. The availability of thousands of human genomes will reveal nonsynonymous mutations even at positions where DAMs are known to occur, which will make it possible to develop position-specific estimators that diagnose novel mutations with a high RoI. With the knowledge of information on the genotypes of nonsynonymous mutations and SNPs, the copy number variation of the protein (including paralogs), and the availability of more protein structures, it will become possible to build more accurate mutation classifiers to diagnose disease propensities of novel mutations, select and prioritize variants for experimental research, and develop baseline patterns of novel allele frequencies within populations.
Methods
We analyzed two large-scale data sets of DAMs and nSNPs (Subramanian and Kumar 2006a; Lohmueller et al. 2008). The Subramanian and Kumar (2006a) data set consisted of 10,685 DAMs and 5308 human nSNPs. This data set was constructed by downloading the human proteome from GenBank (build 34.1) with associated RefSeq identifiers for each gene. Of all available DAMs in 1307 human genes from HGMD (http://archive.uwcm.ac.uk/ uwcm/mg/hgmd0.html) and all putatively-benign nSNP sites in 11,753 human genes from various genome projects (see Subramanian and Kumar 2006a), genes containing no DAMs or nSNPs were discarded. Complete proteomes of four diverse species (Homo sapiens, Mus musculus, Gallus gallus, and Takifugu rubripes) were obtained for the remaining genes from the Ensembl web server (http://www.ensembl.org/), along with orthologs identified via a reciprocal BLASTP search with each RefSeq gene (Altschul et al. 1990; Waterston et al. 2002). Additionally, the BLOSUM substitution matrix was employed using appropriate threshold scores (Subramanian and Kumar 2004). If any of the three vertebrate orthologs could not be determined for any human gene, then that gene and all DAMs and nSNPs contained within it were excluded from the data set. Each ortholog was aligned to the homologous human sequence with CLUSTALW using default settings (Thompson et al. 1994), and all sites (and thus associated DAMs and nSNPs) containing indels or missing data at homologous sites in any of the three vertebrate species were excluded in order to represent at least four species.
From the Lohmueller et al. (2008) data, we extracted all nSNPs by removing all synonymous, noncoding and redundant SNPs. Then, we used dbSNP rsIDs for each nSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) to generate a RefSeq identifier (Pruitt et al. 2007). This information was used to map each nSNP onto the 44-species protein alignments available in the UCSC Genome Browser (Kuhn et al. 2009). During this process, a substantial number of nSNPs was eliminated because either dbSNP records did not contain a map from rsIDs to RefSeq identifiers, not all human RefSeq identifiers were present in the UCSC data set, or the wild-type amino acid in the Lohmueller data set was not the human representative in the UCSC data set. The outcome was a set of 12,712 nSNPs with allele frequencies as reported by Lohmueller et al. (2008), and the 44-species alignment for each nSNP position. The 44-species alignments were also generated by using the RefSeq identifiers in UCSC for all the DAMs. We discarded all positions where the amino acid state of any of the species in the original four sequence alignment disagreed between the Subramanian and Kumar (2006a) data set and the UCSC alignment. This produced a total of 8696 DAMs with 44-species alignments.
We estimated the evolutionary rate for each amino acid site separately using the amino acids found in the 44-species alignment. The number of substitutions at each site was obtained by using the known phylogeny of the species (Fig. 1B) and applying the Fitch (1971) algorithm. The total of the substitutions was divided by the total time elapsed on the tree to obtain the evolutionary rate in the units of the number of substitutions per site per billion years. Species divergence times were obtained from an advanced version of the TimeTree resource (www.timetree.org, version 2.0 prerelease) (Hedges et al. 2006). For each position, all species containing alignment gaps or missing data were pruned from the tree before calculating the number of substitutions and the total evolutionary time. We repeated this procedure to calculate the evolutionary rate using only 33 mammalian species. Vertebrate and mammal rates were highly correlated for all sites used (r = 0.92; P << 0.01), and we employed the latter rates, as mammalian genomes are more appropriate models for the human genome as compared to more distantly related species. Furthermore, we have previously shown that maximum likelihood estimates of relative evolutionary rates are very highly correlated with rates obtained using the Fitch algorithm (Miller and Kumar 2001), as each site contains data from many closely and distantly related species. This was confirmed in our analysis of DAM positions for which rates from four species ML analysis from Subramanian and Kumar (2006a) and the 44-species analysis in this study showed significant correlation (r = 0.70; P << 0.01). Because the calculation of rates by our current method only requires the amino acids in all other species at a given site, it is more suitable for application in personalized diagnostics. We quantized evolutionary rates into six discrete categories such that sites showing no variation across all species comprise the slowest-evolving group (category 0), and the cut-off rates for the other five categories (1–5) were such that they each contained a similar number of sites when applied to the Lohmueller et al. (2008) nSNPs. The five categories of evolutionary rate of variable positions had average evolutionary rates of 0.6, 1.5, 2.5, 3.5, and 5.3 with standard deviations of 0.2, 0.3, 0.3, 0.3, and 1.5, respectively.
These UCSC Genome Browser alignments were also used to generate EPAs at each position, because they cover 44 diverse vertebrate species, including agnathans, fishes, amphibians, birds, and mammals (http://genome.ucsc.edu/). Under the principles of the neutral theory of molecular evolution, a vast majority of EPAs are expected to represent neutral variants at a site. For each DAM/nSNP, we estimated the percentage of evolutionary time span (%ETS) in the 44-species tree, which is the total branch length (times) in the tree obtained after pruning all nonhuman species lacking the variant allele divided by the total branch length of the tree after pruning all species containing an alignment gap or missing data. For each variant at a site, the ETS varies from 0%–100%, with constant sites containing a single EPA with an ETS of 100% and non-EPA mutations producing an ETS of 0%. A smaller ETS is frequently associated with variation that has occurred recently in species closely related to humans.
The PolyPhen web resource was used to classify mutations into benign, possibly-damaging, and probably-damaging categories for DAMs and nSNPs (Ramensky et al. 2002). After removing duplicate entries and sites for which PolyPhen returned “unknown” or was unable to return any result, the final data set contained 9460 DAMs and 4020 nSNPs for the Subramanian and Kumar (2006a) data sets. We noticed that while PolyPhen attempts to incorporate information from the protein structure (when available from databases such as Protein Data Bank) and available functional data from site annotations, the final diagnosis for this data set was rooted solely in primary sequences for >97% of the mutations we tested. Inclusion and exclusion of these mutations produce the same results, so we did not consider nonsequence attributes in any of our analyses. Because of the slowness of the web resource (http://sift.cchmc.org/), SIFT analyses are based on a subset of these DAMs and nSNPs (approximately one-third each, 2375 and 1439, respectively). Supplementary information available from Lohmueller et al. (2008) provided the PolyPhen diagnosis for all the nSNPs.
Acknowledgments
We thank Revak Raj Tyagi for his help with UCSC Genome browser data extraction, Antoine Ah-Foune and Veronica Shi for some early analyses, and Kristi Garboushian for providing editorial support. We thank David Cooper (HGMD) for permitting us to use the disease-associated mutation data of Subramanian and Kumar (2006a). This research was supported by a research grant from NIH HG2096 (S.K.).
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.091991.109.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatti P, Church DM, Rutter JL, Struewing JP, Sigurdson AJ. Candidate single nucleotide polymorphism selection using publicly available tools: A guide for epidemiologists. Am J Epidemiol. 2006;164:794–804. doi: 10.1093/aje/kwj269. [DOI] [PubMed] [Google Scholar]
- Boyko AR, Williamson SH, Indap AR, Degenhardt JD, Hernandez RD, Lohmueller KE, Adams MD, Schmidt S, Sninsky JJ, Sunyaev SR, et al. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 2008;4:e1000083. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg Y, Rost B. SNAP: Predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35:3823–3835. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PA, Duraisamy S, Miller PJ, Newell JA, McBride C, Bond JP, Raevaara T, Ollila S, Nystrom M, Grimm AJ, et al. Interpreting missense variants: Comparing computational methods in human disease genes CDKN2A, MLH1, MSH2, MECP2, and tyrosinase (TYR) Hum Mutat. 2007;28:683–693. doi: 10.1002/humu.20492. [DOI] [PubMed] [Google Scholar]
- Cheng TM, Lu YE, Vendruscolo M, Lio P, Blundell TL. Prediction by graph theoretic measures of structural effects in proteins arising from non-synonymous single nucleotide polymorphisms. PLoS Comput Biol. 2008;4:e1000135. doi: 10.1371/journal.pcbi.1000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniger SW, Kim HS, Swain D, Corcuera D, Williams M, Yang SP, Fay JC. A catalog of neutral and deleterious polymorphism in yeast. PLoS Genet. 2008;4:e1000183. doi: 10.1371/journal.pgen.1000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Woolfit M, Phelps T. The distribution of fitness effects of new deleterious amino acid mutations in humans. Genetics. 2006;173:891–900. doi: 10.1534/genetics.106.057570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WM. Toward defining the course of evolution: Minimum change for a specific tree topology. Syst Zool. 1971;20:406–416. [Google Scholar]
- Gao L, Zhang J. Why are some human disease-associated mutations fixed in mice? Trends Genet. 2003;19:678–681. doi: 10.1016/j.tig.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S. TimeTree: A public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge University Press; Cambridge, UK: 1983. [Google Scholar]
- Kondrashov AS. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Hum Mutat. 2003;21:12–27. doi: 10.1002/humu.10147. [DOI] [PubMed] [Google Scholar]
- Kondrashov AS, Sunyaev S, Kondrashov FA. Dobzhansky-Muller incompatibilities in protein evolution. Proc Natl Acad Sci. 2002;99:14878–14883. doi: 10.1073/pnas.232565499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: Implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al. The UCSC Genome Browser Database: Update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller KE, Indap AR, Schmidt S, Boyko AR, Hernandez RD, Hubisz MJ, Sninsky JJ, White TJ, Sunyaev SR, Nielsen R, et al. Proportionally more deleterious genetic variation in European than in African populations. Nature. 2008;451:994–997. doi: 10.1038/nature06611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MP, Kumar S. Understanding human disease mutations through the use of interspecific genetic variation. Hum Mol Genet. 2001;10:2319–2328. doi: 10.1093/hmg/10.21.2319. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: Server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd MF, Williams RD, Webb EL, Schmidt S, Sellick GS, Houlston RS. The predicted impact of coding single nucleotide polymorphisms database. Cancer Epidemiol Biomarkers Prev. 2005;14:2598–2604. doi: 10.1158/1055-9965.EPI-05-0469. [DOI] [PubMed] [Google Scholar]
- Shastry BS. SNPs in disease gene mapping, medicinal drug development and evolution. J Hum Genet. 2007;52:871–880. doi: 10.1007/s10038-007-0200-z. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Kumar S. Gene expression intensity shapes evolutionary rates of the proteins encoded by the vertebrate genome. Genetics. 2004;168:373–381. doi: 10.1534/genetics.104.028944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Kumar S. Evolutionary anatomies of positions and types of disease-associated and neutral amino acid mutations in the human genome. BMC Genomics. 2006a;7:306. doi: 10.1186/1471-2164-7-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Kumar S. Higher intensity of purifying selection on >90% of the human genes revealed by the intrinsic replacement mutation rates. Mol Biol Evol. 2006b;23:2283–2287. doi: 10.1093/molbev/msl123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyaev SR, Eisenhaber F, Rodchenkov IV, Eisenhaber B, Tumanyan VG, Kuznetsov EN. PSIC: Profile extraction from sequence alignments with position-specific counts of independent observations. Protein Eng. 1999;12:387–394. doi: 10.1093/protein/12.5.387. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Wu N, Guo X, Guo J, Zhang J, Fan Y. Predicting the phenotypic effects of non-synonymous single nucleotide polymorphisms based on support vector machines. BMC Bioinformatics. 2007;8:450. doi: 10.1186/1471-2105-8-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Zhang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- Yampolsky LY, Kondrashov FA, Kondrashov AS. Distribution of the strength of selection against amino acid replacements in human proteins. Hum Mol Genet. 2005;14:3191–3201. doi: 10.1093/hmg/ddi350. [DOI] [PubMed] [Google Scholar]