Abstract
ClinSeq is a pilot project to investigate the use of whole-genome sequencing as a tool for clinical research. By piloting the acquisition of large amounts of DNA sequence data from individual human subjects, we are fostering the development of hypothesis-generating approaches for performing research in genomic medicine, including the exploration of issues related to the genetic architecture of disease, implementation of genomic technology, informed consent, disclosure of genetic information, and archiving, analyzing, and displaying sequence data. In the initial phase of ClinSeq, we are enrolling roughly 1000 participants; the evaluation of each includes obtaining a detailed family and medical history, as well as a clinical evaluation. The participants are being consented broadly for research on many traits and for whole-genome sequencing. Initially, Sanger-based sequencing of 300–400 genes thought to be relevant to atherosclerosis is being performed, with the resulting data analyzed for rare, high-penetrance variants associated with specific clinical traits. The participants are also being consented to allow the contact of family members for additional studies of sequence variants to explore their potential association with specific phenotypes. Here, we present the general considerations in designing ClinSeq, preliminary results based on the generation of an initial 826 Mb of sequence data, the findings for several genes that serve as positive controls for the project, and our views about the potential implications of ClinSeq. The early experiences with ClinSeq illustrate how large-scale medical sequencing can be a practical, productive, and critical component of research in genomic medicine.
Elucidating the sequence of the human genome (International Human Genome Sequencing Consortium 2001, 2004) and subsequent advances in DNA sequencing technologies (Mardis 2008) have the potential to dramatically improve the delivery of health care through the acquisition of genomic information about individual patients. However, much research will be needed to develop medical applications of genomics; for example, little is known about how to organize and implement large-scale medical sequencing (LSMS; i.e., systematic resequencing of human DNA) in a clinical context. Other approaches for applying high-throughput genomics to health care (e.g., assaying single-nucleotide polymorphisms and establishing gene-expression profiles) offer diagnostic promise; these are not further considered here, as our focus is on LSMS for studying the relationship of germline genomic variation to health and disease.
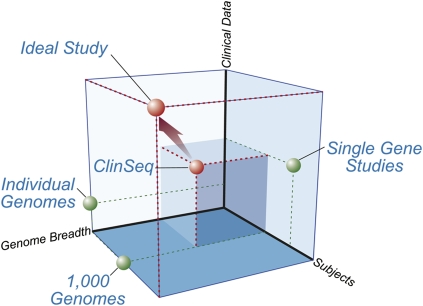
We recently launched ClinSeq (http://genome.gov/20519355), a project that aims to apply LSMS within a clinical research environment to answer questions about the genetic basis of health, disease, and drug response. The application of genomic approaches (in particular LSMS) in a clinical research context is associated with a number of considerations that define key “dimensions” of any study: the number of subjects, the associated clinical data, and the breadth of genome covered (Fig. 1). Numerous detailed studies of single genes have been carried out; while often performed on many participants with significant amounts of phenotypic information, they are focused on a very small portion of the genome. The flurry of papers that describe recently generated whole-genome sequences (Levy et al. 2007; Bentley et al. 2008; Wang et al. 2008; Wheeler et al. 2008) has provided the first true individual genome sequences, including a modest amount of associated clinical data; however, the number of examples is small to date. Greater numbers are promised by the 1000 Genomes Project (http://www.1000genomes.org/), although no phenotypic information will be available for the individuals being studied. ClinSeq aims to model a more ideal study with respect to these three dimensions (Fig. 1), with the potential to further move toward the ultimate ideal as technology advances.
Figure 1.
A spatial conceptualization of research studies in genomic medicine. There are three key “dimensions” to consider when applying genomics to clinical research: genome breadth (the fraction of the genome that is interrogated), number of subjects or participants, and the associated clinical data about those individuals (including its depth, breadth, and rigor). While the ideal study would acquire whole-genome sequences from large numbers of extensively phenotyped subjects, this is currently impractical. Single-gene studies can involve a few or numerous subjects and extensive clinical data, but by definition involve the examination of only a single gene and thus occupy one wall of this space. The individual genomes that have recently been sequenced (Levy et al. 2007; Bentley et al. 2008; Wang et al. 2008; Wheeler et al. 2008) provide nearly complete genome breadth, but with limited clinical data; further, their limited subject numbers place them on another wall of this space. The 1000 Genomes Project (http://www.1000genomes.org/) is providing large subject numbers and extensive genome breadth, but no clinical data—positioning it on the floor of this space. ClinSeq aims to reside in the center of this space, having attributes of substantial subject size (n = 1000 initially), moderate genome breadth (∼400 genes initially, with plans for expanding this breadth), and substantial clinical data.
The general aims of ClinSeq are to: (1) develop the infrastructure and approaches to acquire and analyze genome sequence from individual research participants; (2) pilot the use of LSMS to elucidate the genetic architecture underlying human traits; (3) provide an open, shared resource and environment for basic and clinical researchers to work collaboratively to perform research in genomic medicine; and (4) establish approaches for informed consent and the return of genetic information to subjects participating in LSMS studies. In pursuing these aims, our overriding goals include modeling whole-genome sequence acquisition in a manner that is practical for a clinical research setting, advancing our understanding of the genetic basis of important human diseases and traits, and establishing how to scale LSMS prior to the day when whole-genome sequencing becomes part of routine clinical practice. In this paper, we describe the ClinSeq study design, provide a snapshot of our very early data generation, and discuss the implications of this study for the nascent field of genomic medicine.
ClinSeq study design
Cohort enrollment and phenotypic characterization
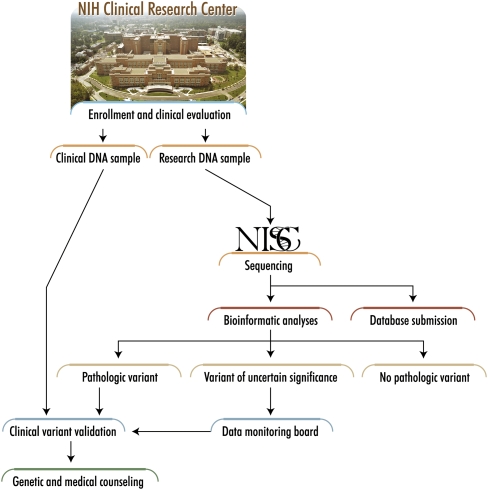
The initial ClinSeq cohort is planned to include 1000 individuals who will be enrolled and evaluated at the National Institutes of Health (NIH) Clinical Research Center in Bethesda, Maryland (Fig. 2). ClinSeq is designed to evolve into a study of all traits and the acquisition of whole-genome sequences. Because neither is presently practical, we sought to model key attributes of genomic medicine research by performing LSMS of candidate genes relevant to a common and broadly defined phenotype (initially, atherosclerotic heart disease), with consent to proceed to whole-genome sequencing and the study of other traits in the future. Cardiovascular disease will be a useful prototype for many other phenotypes that we plan to study eventually. Atherosclerotic heart disease is common, has quantitative subphenotypes, has an underlying genetic architecture that is known to be complex (Sing and Boerwinkle 1987), is already associated with a set of genes that can be readily interrogated with conventional sequencing technology, and offers various treatment options for mitigating identified risks. An initial cohort size of 1000 was selected to allow detection of gene variants with an effect comparable to that seen with PCSK9, a recently discovered, clinically important gene that plays a role in lipid regulation (Cohen et al. 2006). A cohort of 1000 subjects would have a power of 0.996 to detect an association of this nature at P = 0.05. In addition to rare variants, numerous common, apparently low-penetrance alleles have been associated with abnormal lipid levels (Kathiresan et al. 2007; Willer et al. 2008) and other phenotypes (Manolio et al. 2008); further, numerous rare, high-penetrance alleles have been shown to cause endo- or subphenotypes that comprise the spectrum of atherosclerosis (e.g., Jones et al. 2007; Mani et al. 2007).
Figure 2.
ClinSeq sample and data flow. DNA samples and clinical data emanate from the initial participant enrollment and clinical evaluation, and then flow through the indicated clinical and research processes (see text for details). Note the separate acquisition and handling of DNA samples for clinical and research purposes, respectively, with the former handled by a CLIA laboratory prior to any results being returned to participants. Further, variants identified in putative disease-causing genes must first be reviewed by a data-monitoring board before being reported back to participants.
ClinSeq participants are being selected to represent the spectrum of atherosclerotic heart disease using the Framingham score (Wilson et al. 1998) to balance accrual. The accrual target is three groups of 250 participants each who have a Framingham 10-yr coronary artery disease (CAD) risk of <5%, 5%–10%, and >10%, respectively, and an additional group of 250 participants who have a diagnosis of CAD based on a history of a myocardial infarction, coronary artery bypass graft surgery, stent placement, or other revascularization procedure. This accrual strategy was implemented so that the cohort would be enriched for detectable coronary atherosclerosis, as measured by computed tomography (see below).
Upon enrollment, each participant is evaluated (Fig. 2) for baseline information about his or her health status and family history. For acquiring a detailed family history, participants complete the Surgeon General's “My Family Health Portrait” (http://www.hhs.gov/familyhistory), after which a comprehensive family history is ascertained by a genetic counselor. An abbreviated health history, with a particular focus on cardiovascular disease, is also acquired by a nurse practitioner. The initial clinical assessment includes height, weight, head and waist circumferences, blood pressure, electrocardiogram, echocardiogram, and coronary artery calcification (assessed by multidetector computed tomography). We also perform a broad panel of blood and urine tests (see Supplemental Table S1), isolate genomic DNA and total RNA from peripheral blood, archive serum samples, and establish a lymphoblastoid cell line from each participant. Importantly, this is only an initial assessment, as the participants are consented for potential future clinical evaluation of many other traits (see Informed Consent section, below).
As of January 24, 2009, 586 participants have been enrolled in ClinSeq, including 259 males (44%) and 327 females (56%). Ninety-one percent of these participants self-identified as being of Caucasian descent and 97% self-identified as being non-Hispanic, reflecting the population of Bethesda, Maryland, where the NIH Clinical Research Center is located.
Informed consent
The ClinSeq study was reviewed and approved by the NIH National Human Genome Research Institute and Suburban Hospital (Bethesda, Maryland) Institutional Review Boards. In addition, a certificate of confidentiality was obtained for the study to provide additional protection from forced disclosure of research results to third parties. At the initial enrollment of each participant, both the consent form and the associated discussion make it clear that the goal of ClinSeq is to examine the entire genome and to study any and all phenotypes, including a wide spectrum of diseases. The participants are informed that they will be contacted to determine if they are interested in learning about clinically relevant results, if discovered. In addition, participants are consented at the initial visit for permission to contact them to initiate discussions of additional follow-up testing of themselves and/or their family members (the latter being limited to basic phenotype studies and genotyping for cosegregation analyses). To protect the interests of the participants and to provide the investigators with an independent source of advice and review, we plan to establish a sequence variant review panel (i.e., a data-monitoring board). This panel will be comprised of experts in medical and molecular genetics who are otherwise not involved in ClinSeq; their charge will be to periodically review data regarding genes that have not yet been proven to cause human disease. For selected genes, we will pursue hypothesis-testing clinical research to determine if a sufficient data set can be generated to support causation and the return of results to the participants. Such research may involve studies of the population frequency of sequence variants in cases and controls, in vitro or animal models, and participants with and without the variants in response to specific pathophysiologic perturbations. The resulting data will be presented to the sequence variant review panel to assess if the findings warrant the return of individual research information to the participants. This review by a designated data-monitoring board is integrated into the overall ClinSeq data analysis scheme (Fig. 2).
Sequence generation
The initial approach for data generation in ClinSeq involves sequencing a large set of candidate genes in search of high-penetrance variants that confer risk for coronary artery calcification and atherosclerosis (i.e., CAD). Genes are being selected by a number of methods based on a spectrum of evidence for their association with CAD; this includes genes already implicated in CAD, which serve as positive controls for the early phase of the study (see below). In addition, we are sequencing genes residing within genomic regions corresponding to genetic association peaks for CAD and related phenotypes, as well as those thought to be functionally related to such CAD genes. In this way, we hope to detect genetic variants that reflect a spectrum of relevance to CAD, from those that are obviously causative to those that are only weakly associated.
A critical aspect of ClinSeq is the expectation that during the course of the project, technical advances will allow us to broaden the scope of our gene set to the entire human exome and, eventually, to the entire human genome. Thus, the initial candidate gene set is only transiently important, and for this reason we are prospectively consenting all participants for whole-genome sequencing (see below).
Initially, we are generating data by the bidirectional sequencing of PCR products amplified from gene components, which we refer to as regions of interest (ROI), using Sanger-based chemistries and capillary electrophoresis-based detection methods (Wilson and Mardis 1997). Our current sequence-production scheme uses PrimerTile (Chines et al. 2005) to design primers for use in PCR assays that amplify ROIs corresponding to known or predicted exons (including 5 bp of the flanking introns), ∼1 kb of the 5′ untranslated region (UTR)/promoter region, and the 3′ UTR if it is evolutionarily conserved. Further, we PCR-amplify and sequence up to three of the most evolutionarily conserved regions of each gene that are not captured by the above features. Following amplification, PCR products are sequenced using 3730XL instruments (Applied Biosystems).
Quality control
We have implemented a number of steps for the quality control of ClinSeq-generated sequence data. For example, to ensure that a particular DNA sample being sequenced is, in fact, the same DNA sample analyzed at another point in time, we use a novel sample-tagging strategy that we developed. Specifically, upon arrival at the NIH Intramural Sequencing Center (NISC), each human DNA sample is “spiked” with an aliquot of a plasmid containing a unique segment of nonprimate DNA whose sequence is known not to be present in the human genome. Upon every use of that human DNA sample, the insert of the spiked plasmid is PCR-amplified and sequenced, so as to confirm that the correct spike DNA is present (and thus that the same ClinSeq participant DNA was used as previously). This approach does not tie the research DNA sample to the CLIA (Clinical Laboratory Improvement Amendments) DNA sample; in fact, ClinSeq has been designed with an information and data firewall that separates testing of the research DNA sample from the clinical data.
Our quality control measures for detecting sequence variants involve the routine analysis of HapMap (The International HapMap Consortium 2007) DNA samples, which are associated with extensive genotype data at known variant sites. Specifically, for every 30 ClinSeq participant DNA samples analyzed, we sequence the same genomic regions in one HapMap sample. The sequence generated with the HapMap samples is interrogated for the presence of known variants (from the available genotype data) to assess our overall false-negative rate. Such quality control steps facilitate the generation of reliable data, the estimation of false-negative rates, and accurate variant detection.
Data storage and sharing
All sequence chromatograms are being submitted to the public NCBI Trace Archive (http://www.ncbi.nlm.nih.gov/Traces/trace.cgi?,Query:CENTER_NAME=“NISC”andCENTER_PROJECT=“ClinSeq”) in batches of 10,000 traces. Each archived sequence is only correlated with its complementary sequence read (derived from the same PCR product) and not to any other sequences from that ClinSeq participant, thereby minimizing the possibility of breaching the confidentiality of the data (Homer et al. 2008). Further, the sequence chromatograms are being deposited at random from a larger pool to minimize inferences of identity based on the timing of deposition. The controlled access database dbGaP (http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gap) will provide qualified investigators access to both the sequence traces as well as links to other sequences and phenotype data from that individual.
Sequence analysis and variant detection
The primary sequence data are being analyzed using a customized suite of computational methods consisting of standard and novel components. Sequence trace quality is first assessed with the base-calling program phred (Ewing and Green 1998; Ewing et al. 1998). Each set of samples in a 384-well layout are resequenced if their corresponding reads in any quadrant fall below predetermined minimum quality thresholds. If the average number of bases with a quality score ≥20 is less than 450 in any quadrant of sequence reads, or if <70% of the reads in any quadrant have quality trim lengths ≥50, the entire plate is resequenced. All traces are included in subsequent analyses since insertion–deletion polymorphisms can mimic poor quality data detected by phred (but may contain valid sequence data). All sequences for a given PCR product are then assembled using consed (Gordon et al. 1998); groups of sequences derived from overlapping PCR products are assembled independently, which allows for independent cross-validation of the overlapping regions. Sequence variants, including single-nucleotide differences and short (<100 base) insertions and deletions, are identified using PolyPhred v6.11 (Bhangale et al. 2006; Stephens et al. 2006). Variants within genes are classified as residing within the untranslated regions (5′ and 3′ UTRs), within introns, or within protein-coding regions (with predicted gene structures based on UCSC Genome Browser annotations).
The above classifications, along with information about nonsense, missense, splice-site, and frame-shift variants, are being catalogued in a custom database. Missense variants are further classified and prioritized based on their predicted severity using a variety of amino acid substitution prediction methods (e.g., PolyPhen [Ramensky et al. 2002], SIFT [Ng and Henikoff 2003], and other customized tools). This first-pass ranking of missense variants, in conjunction with the identification of nonsense, splice-site, and frame-shift variants, is then used to create prioritized listings of sequence variants that undergo further analysis to establish their likely disease relevance and/or correlation with an observed phenotype. Once genotypes are validated by manual sequence trace review, variants of interest are examined for associated information in the public databases (e.g., dbSNP, OMIM, and HGMD) or in the published literature. Genotypes that do not pass manual quality control review are considered for re-sequencing by a customized finishing process (if a small fraction of the sequences are of poor quality and the putative variant is of high interest) or ROI redesign (if a large fraction of the sequences are of poor quality). A customized user interface has been developed for convenient review, sorting, and selection of specific variants in our growing data set.
Data generation to date
We report here a preliminary summary of a small portion of the initial ClinSeq data to illustrate the scope and nature of the findings that will eventually emanate from the project. The current list of candidate genes totals 219, from which we selected ROIs for PCR primer design and sequencing. These genes were selected for a variety of reasons as described above. To date, we have completed PCR primer design for 140 genes (i.e., genes for which there were 100% coverage of ROIs after a single round of PCR primer design or that have undergone two rounds of PCR primer design). Coverage statistics for these 140 genes show that the 403,646 targeted ROI bases have thus far yielded sequence data from at least one participant for 357,912 of these bases, corresponding to 88.7% “design coverage” (see Supplemental Table S2A,B). For 48 of these 140 genes, we have achieved 100% design coverage. Of the initial 219 genes, 79 have undergone only one round of PCR primer design. An additional 101 genes have already been selected for future sequencing.
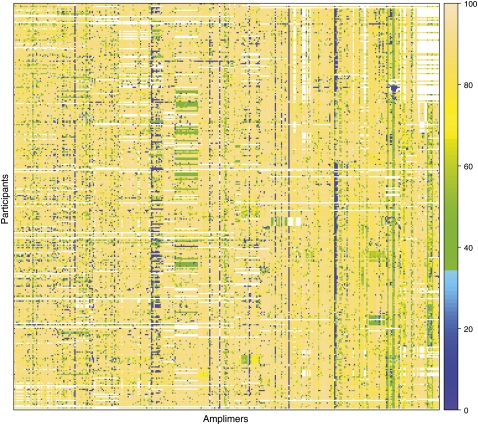
For the 219 genes for which we have at least some sequence data, we have already generated over 1.7 million sequence reads from 354 individuals (including 28 HapMap control samples) using 2444 PCR primer pairs. These reads together comprise 826 Mb of sequence that aligns to the expected genomic regions, and of this, 305 Mb falls within the ROIs (since the PCR products often include regions outside the ROIs themselves). A “heat map” overview of the yield of the interrogated sequences among ClinSeq participants is shown in Figure 3 and Supplemental Figure S1.
Figure 3.
Snapshot of ClinSeq sequence coverage. This “heat map” provides an overview of the targeted sequence coverage for 27 genes selected at random from the set of 140 genes with completed PCR primer design. The figure illustrates the range and variability in the yield of sequence data for a subset of the analyzed genes. These 27 genes are being sequenced using 343 amplimers (of 2444 total) represented by columns; the data are shown for 326 enrolled participants (of 586 total) represented by rows. The colors represent the percent sequence coverage (see scale on right) of the corresponding PCR products at or above a threshold of phred Q20, with white indicating the absence of data at this time. Such heat-map results are used to monitor overall quality of the ClinSeq sequencing pipeline, whereas more direct quality measures (see text) are used to assess the suitability of individual sequence data for inclusion in subsequent analyses and/or return to participants. The complete heat map showing all amplimers and all participants for the data set discussed here is provided in Supplemental Figure S1.
Within the generated sequences for the ROIs, we have detected 3353 variants with at least one genotype score of 99 (the maximum genotype score given by PolyPhred). Since this analysis is based on a snapshot of an active project that is continuously collecting data, sequence coverage varies depending on the region being analyzed, as shown in Figure 3. To perform certain analyses with a consistent number of observed alleles, we examined only those variants residing in regions for which sequence data were available from at least 250 participants, and then randomly removed data to yield a data set corresponding to exactly 250 participants (or 500 alleles, as we limited this analysis to autosomal genes). This reflects 25% of the initial ClinSeq project goal of sequencing DNA from 1000 individuals, and reduced the number of variants to analyze from 3353 (3288 on the autosomes) to 2161 (2107 of which were autosomal). The number of ROI bases meeting the coverage threshold of 500 chromosomes totaled 390,577 (380,714 on the autosomes). From this, we can calculate heterozygosity per nucleotide site, giving a theta of 0.000814 for this sampling of the genome, in agreement with a prior, smaller sample set (Halushka et al. 1999). The following initial set of analyses was performed on this subset of autosomal variants.
We examined several different categories of variants: (1) variants within ROIs and observed in only one participant (ROI-Singletons); (2) variants within ROIs and observed in two or more participants (ROI-Multiples); (3) variants within ROIs that were also within coding sequences (ROI-CDS); (4) variants within ROIs that were also in conserved noncoding regions (ROI-CNC); and (5) variants outside of ROIs (nonROI-Control, which were used as controls for comparison to other regions). To assess the false-positive rate of our variant-detection approach, we manually reviewed the sequence traces for randomly selected variants from each of these five categories. The ROI-Singletons were associated with the highest false-positive rate (10/100), considerably higher than that for the ROI-Multiples (3/100). Since 51% of the detected variants in this initial data set are singletons, the overall within-ROI false-positive rate appears to be roughly 6.5% (13/200). Of note, the other categories were associated with the following false-positive rates: 5/145 for ROI-CDS, 3/100 for ROI-CNC, and 6/100 for nonROI-Control.
Implications of ClinSeq
The ClinSeq project occupies an important niche in genome research and aims to catalyze the development of a clinical research facet of genomics, which has been primarily a basic science activity. The project has been deliberately engineered to capitalize on the unique infrastructure of the NIH Intramural Research Program, which is designed to function as a prospectively funded incubator for high-risk research and studies that might not align with traditional funding mechanisms. In addition, the NIH Clinical Research Center, the largest clinical research hospital in the world, provides the ability to pursue cutting-edge and exploratory clinical research; it is our hope that ClinSeq will stimulate other genomic-oriented clinical research studies through more traditional funding mechanisms. Although ClinSeq has been designed to allow the study of any human phenotype, the initial focus on atherosclerosis reflects both the key clinical and genetic attributes of this disease and the strong interest of the National Heart, Lung, and Blood Institute to advance the utility of genomics in diagnosing and treating CAD. Finally, the direct interface of a state-of-the-art sequencing facility (NISC) with a large clinical research project should provide important insights about applying LSMS to contemporary problems in genomic medicine.
ClinSeq was designed to model LSMS, initially using PCR- and capillary-based sequencing methods. While the technical challenges we have encountered to date do not fully reflect all aspects of LSMS using “next-generation” DNA sequencing technologies, many of the critical components are being examined. For example, we are already dealing with the significant challenges associated with informed consent, patient recruitment, clinical informatics, pathogenicity assessments, and interactions with study participants; such issues are independent of the sequencing platform. Some other important implications of ClinSeq are detailed below.
Analyzing human genetic variation
The ClinSeq data set has great promise to supplement and complement other studies of human genetic variation, such as the 1000 Genomes Project. The initial sequencing of 300–400 candidate genes in 1000 phenotyped participants should provide insight into the relationship between genetic variants and disease, as well as enhance our understanding of normal variation. In this preliminary analysis of a subset of genes in about 25% (250 subjects or 500 autosomes) of our initial cohort size, we have begun to see the outline of these data. The following variant analyses examined sequence data from precisely 500 autosomes; this was accomplished by removing data in those cases where sequence across an ROI was generated from more than 500 chromosomes and by not including data from an ROI if sequence from less than 500 chromosomes was available.
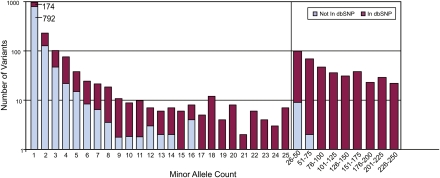
The total number of variants in the data set of 250 samples described above was 2107. We reduced this total by extrapolation to 1984 variants by accounting for the predicted false-positive results (based on the manual data review described above). Figure 4 shows the number of times each variant was detected. Also shown are the variants with a dbSNP Build 129 Reference SNP identifier and those without one. It is striking that roughly half of the variants (966 of 1984) discovered to date were only detected once among the 500 chromosomes examined, and of these 966 variants, only 174 were present in dbSNP.
Figure 4.
Distribution of variant counts. An estimated total of 1984 variants (corrected for false-positives from the observed 2107 autosomal ROI variants) were identified in ROIs among 250 ClinSeq participants (see text for details). The number of times each variant was detected is depicted, in each case broken down relative to its presence or absence in dbSNP. Note that the x-axis is discontinuous beyond a count of 25, and the allele counts greater than 25 are presented in bins of 25; also note that the y-axis uses a logarithmic scale. The data show that 966 variants are unique (i.e., a minor allele count of 1), and, in fact, comprise about half of the variants detected in this data set (792 not in dbSNP and 174 in dbSNP).
The routine analysis of HapMap samples as controls in the ClinSeq sequencing pipeline provides the ability to measure sensitivity and concordance between the HapMap phase II genotypes (http://ftp.hapmap.org/genotypes/2008-03/forward/non-redundant/) and ClinSeq sequence-derived genotypes. For the four HapMap samples analyzed to date (NA07345, NA07357, NA11839, and NA12812), we have found 985 genotypes that are concordant between HapMap and ClinSeq, 81 that are discordant, and 84 that could not be classified due to low sequence quality. This corresponds to a sensitivity of 92% and a discordance rate of 7.6%. Therefore, the overall sensitivity (including PCR primer design failures) is 92% × 88.7%, or 82%.
Validating variant-detection methodologies
Because ClinSeq is an ongoing project with nonuniform sequencing progress with respect to genes and DNA samples at the present time, we developed several metrics to illustrate our progress thus far and our approaches to quality control. Our efforts to date have involved sequencing 219 genes, and then screening 305 Mb of generated sequence originating within ROIs for variants (205 Mb from coding regions and 100 Mb from adjacent noncoding regions [untranslated regions, conserved noncoding regions, and introns]). We categorized all the single-nucleotide variants (SNVs) that fell within ROIs, identifying 3353 SNVs; these consist of 2198 coding SNVs and 1155 noncoding SNVs. We defined a variant as a nucleotide difference relative to the reference sequence detected in at least one read in the forward and the reverse orientation (from the same DNA sample) with a genotype PolyPhred score of ≥99. These 2198 coding SNVs could be grouped into 1179 nonsynonymous, 999 synonymous, and 20 nonsense SNVs. As noted above, the project is at an early stage, with each DNA sample having undergone different degrees of sequencing and each gene having been variably interrogated. We have thus captured information about a subset of the results that illustrate our progress and quality control. For this, we restricted the analysis to coding variants with exactly 500 genotypes. If a variant was interrogated in fewer than 250 DNA samples, it was excluded. If a variant was interrogated in more than 250 DNA samples, pairs of sequence reads (forward and reverse derived from a single sample) were randomly removed to yield a remaining set of data from precisely 250 samples. This resulted in the total number of coding variants being reduced from 2198 to 1389. Minor allele counts shown in Supplemental Table S3 were tabulated using this high-depth coverage of autosomal variants. Of note, we also identified 15 variants that affected one of the four nucleotides at the exon–intron splice junction.
We looked in greater detail at the data for a small subset of genes (LDLR, APOB, ANGPTL4, SLC12A1, SLC12A3, and KCNJ1) previously shown to harbor rare variants that cause clinically important phenotypes (Boren et al. 2001; Sukonina et al. 2006; Soutar and Naoumova 2007; Ji et al. 2008). These genes exemplify the approach of using LSMS to detect individually rare variants that can potentially explain clinically significant phenotypic variation, which may provide opportunities to practice personalized medicine in the future. Unlike the prior analyses, which were limited to 250 samples (500 autosomal alleles), all sequences were included in this analysis. All the detected coding variants in this subset of genes were reviewed manually to validate the genotype calls and to calculate the false-positive rate for these genes (5/145).
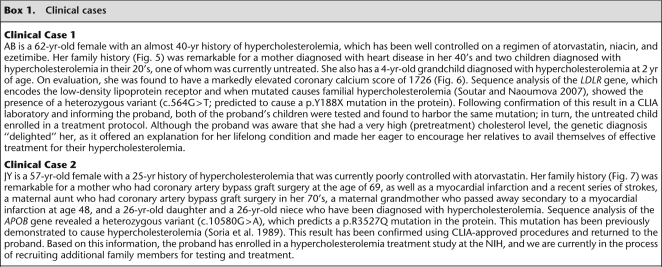
LDLR encodes a cell surface protein involved in receptor-mediated endocytosis of low-density lipoprotein. Mutations in LDLR cause familial hypercholesterolemia (for review, see Soutar and Naoumova 2007). In our data set, we detected 22 variants in LDLR: 21 coding (six nonsynonymous, 12 synonymous, one nonsense, and two frame-shift) and one splice-site (Supplemental Table S4). Five of these LDLR variants were determined to be causative of familial hypercholesterolemia (e.g., see Box 1, Clinical Case 1; Supplemental Clinical Cases S1–S4).
Box 1.
Clinical cases
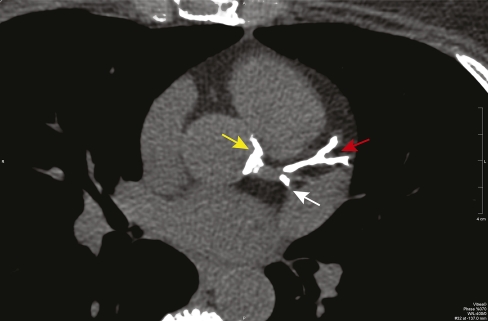
APOB encodes apolipoprotein B, the primary lipoprotein of LDL cholesterol. In our data set, we detected 142 variants in APOB, which included 74 coding variants (53 nonsynonymous and 21 synonymous) (Supplemental Table S5). One of these variants was determined to be causative of familial deficiency of apolipoprotein B100, a milder form of familial hypercholesterolemia (see Box 1, Clinical Case 2). Although APOB is a larger gene and our data set includes more variants in this gene than in LDLR, the number of participants with disease-causing variants in LDLR was greater than for APOB because only a small fraction of APOB variants cause a hypercholesterolemia phenotype (Boren et al. 2001).
ANGPTL4 encodes ANGPTL4, which inhibits the enzymatic activity of lipoprotein lipase, a central enzyme in lipoprotein metabolism (Sukonina et al. 2006). Loss-of-function mutations in ANGPTL4 have been reported to lower plasma triglyceride levels (Romeo et al. 2009). In our data set, we detected 30 variants in ANGPTL4, including 11 coding variants (seven nonsynonymous and four synonymous) (Supplemental Table S6). We found one novel variant (p.P27R) in a single participant that had not been previously reported; this individual's fasting plasma triglyceride levels were in the lowest quartile of our participant population, similar to the results of Romeo et al. (Romeo et al. 2009). The previously reported p.E40K variant has also been associated with lower plasma triglyceride levels; we found this variant in a heterozygous state in three participants.
Rare heterozygous coding variants in SLC12A3, SLC12A1, and KCNJ1 have been associated with a reduction in blood pressure and protection from hypertension (Ji et al. 2008). SLC12A1 and SLC12A3 encode Na–K–Cl cotransporters; KCNJ1 encodes an inward-rectifying apical potassium channel that regulates resting membrane potential and cell excitability. By analyzing the sequence data for these three genes in our data set, we detected 15 unique nonsynonymous coding variants among 14 participants: five in SLC12A1 (Supplemental Table S7), seven in SLC12A3 (Supplemental Table S8), and three in KCNJ1 (Supplemental Table S9). Interestingly, the majority of these 14 participants were being treated for hypertension, and the untreated participants in this group did not have low blood pressures for their age.
Studying issues relating to the return of individual sequence results
For several reasons, ClinSeq has been designed to allow the return of selected individual sequence results to participants, but not to return all sequence data to participants. First, the primary sequence data in ClinSeq are not CLIA validated, and our interpretation of the CLIA and IRB regulations indicates that our individual LSMS research data cannot be returned to participants. Second, only a tiny fraction of the generated sequence data can be interpreted because of our limited knowledge about most of the genes being studied (and the influence of variants therein). Third, we predict that the future use of LSMS might well involve individuals prospectively acquiring (but banking or storing) their genome sequence, with subsequent bioinformatic interrogations of that sequence used to address specific health care questions.
ClinSeq is designed to model this future use of whole-genome sequence data where health care researchers will work with individuals to determine what kinds of questions should be asked and the nature of the results that are returned to the individual. ClinSeq aims to return information about disease-causing variants to the participants, but not to return all sequence data. In fact, for known disease-causing genes, the return of mutation results is not subject to review by the sequence variant review panel as the IRB concluded that it was standard practice to return such results. There is ample reason to be concerned that even the restricted return of data to ClinSeq participants may be overwhelming, counterproductive, or even useless. For this reason, an ancillary research goal of the project is to learn from the participants what results they find to be useful, with the hope that this information might guide future clinical research practice. We thus felt it was most reasonable and practical to initially focus on high-penetrance, Mendelian variants and then later evolve toward progressively less-penetrant variants, using the participants as a barometer to establish when we had crossed a threshold from utility to nonutility and the attributes of the subjects and the sequence data that correlated with the subject's assessments of utility.
In our initial screen, six of the variants described in the prior section were deemed to be high-penetrance, disease-causing mutations. For five of these six variants, the results were confirmed in a CLIA-certified (http://www.cms.hhs.gov/clia/) testing laboratory, and the corresponding information returned to the participants and their personal physicians. In addition, genetic and medical counseling was provided, and family studies were initiated both to diagnose potentially affected relatives as well as to test for the segregation of the variants with the relevant phenotypes (see Box 1, Clinical Cases 1 and 2; Supplemental Clinical Cases S1–S4). Interestingly, the one result that we did not confirm reflects a case where the participant indicated that he/she was unsure about receiving an individual result (as noted below, we do not CLIA-validate results unless a participant indicates an interest in receiving that information).
The clinical cases we describe in Box 1 and the supplemental clinical cases illustrate that it is feasible and medically useful to return research results from a LSMS project to individual participants. This is particularly the case for rare, high-penetrance variants, whereas the situation for common variants that confer a small increased relative risk to a disease is less clear. The challenge is deciding which results should be returned to research participants and defining the appropriate boundary within the spectrum of variants between these two extremes. We propose that a sensible way to approach this situation is to generate a data set that includes the full spectrum of such variants, and then work with the study participants to define the attributes of results that should and should not be returned to interested individuals. In this regard, we hope that ClinSeq extends the clinical practice of medical geneticists from what they already do well (dealing with rare, high-penetrance variants) into novel territory (dealing with common, lower-penetrance variants).
Exploring issues surrounding informed consent in genomics research
The ClinSeq study was designed to explore some of the challenges of informed consent associated with LSMS and to blaze a new path for consenting and working with research participants.6 As noted above, one goal of the project is to ascertain a cohort of participants whose DNA samples can be used for whole-genome sequencing. The regulations for human subjects research in the United States specify that participants must be informed of “reasonably foreseeable risks” (US Department of Health and Human Services 1991). We have implemented a conceptual consent form and process for ClinSeq participants, with opportunities during the study to decline receipt of individual results. With this “opt-in, opt-out” model, we are exercising respect for study participants over the course of the study (Fernandez et al. 2003). If a ClinSeq participant is interested in learning about his/her results, those results are then validated by a CLIA-certified clinical laboratory. Only after this validation is the participant provided the results, which is then followed by genetic and medical counseling; appropriate follow-up studies, referrals, and testing of family members are also discussed and arranged. Most of the initial evaluation and care is started at the NIH Clinical Research Center, with subsequent care transitioned to the participant's primary care provider.
We give the participants control over their genetic information. By first eliciting their interests, participants can maintain control of the information that they wish to receive and have entered into their medical records. Without their consent, results are not validated by the CLIA-certified clinical laboratory, and without that confirmatory testing, it is against federal regulations to associate the results with their medical records. By documenting the different choices made by participants, we should gain insights into the trends and preferences relating to LSMS.
Clinical molecular testing versus hypothesis-generating clinical research
ClinSeq represents a radical shift from the typical use of molecular genetic testing. Clinical investigators and clinicians have traditionally tested a given patient for genetic variants in one or a few gene(s) based upon a hypothesis (or in the clinical realm, a differential diagnosis). This can be considered hypothesis-testing clinical genomic interrogation. Such an approach to medical practice is deeply ingrained in clinical training; indeed, it is generally held that no test should be performed on a patient unless the ordering physician understands the test, knows how to interpret the result, and will change diagnosis or management based on the alternative results. Whole-genome studies violate essentially all of these dicta. ClinSeq and other clinical genomic projects aim to interrogate the genome and generate thousands of “test results” per patient, with only a few of the detected variants being readily interpreted or clinically useful for some time. Yet, one can readily envision how such data sets could be useful for performing clinical research.
Full-genome interrogation for clinical genomic studies is now a reality, with the resulting data sets analyzed to produce lists of identified variants and grouping of participants based on shared genomic attributes. For example, a cohort could be subjected to genome sequencing, with participants then binned into three groups for each gene (or pathway): those lacking sequence variants, those with variants thought to be pathogenic, and those with variants of uncertain significance. A subsequent study could then be pursued that compares the physiology of the participants in each bin, with the study design based on knowledge and predictions about the function of the interrogated gene (or pathway). Such an approach bypasses some of the biases that limit clinical research studies, such as rigid eligibility criteria. This may provide an opportunity to identify phenotypes based on genetic variants in a fashion that might otherwise be missed using hypothesis-testing approaches to phenotypic analysis. These and other new approaches to clinical genomics research need to be piloted, developed, and refined. ClinSeq thus provides an opportunity to explore the use of genomics for hypothesis-generating clinical research studies.
Summary
The ClinSeq project is pioneering approaches for applying high-throughput genomics to clinical research and confronting many of the challenges that will be faced as we approach ideal studies involving whole-genome analyses of large numbers of well-phenotyped individuals. By bringing contemporary genomic approaches into the clinical research arena, we also seek to develop improved models for informed consent, human research subject engagement, and interacting with participants as they receive genomic information about themselves. The cardiovascular aspects of this study are only an entrée into the clinical research utility of the ClinSeq cohort. We fully expect that DNA samples from this cohort will be among those used for whole-genome sequencing, which in turn will catalyze explorations of other important human phenotypes, including numerous diseases. Toward that end, we are already transitioning the analysis of ClinSeq DNA samples to “next-generation” sequencing instruments, initially in conjunction with methods for selecting targeted regions of the genome (Turner et al. 2009), but with the full expectation that something approaching true whole-genome sequencing is inevitable and desirable. The fact that ClinSeq participants consent to such whole-genome sequencing and are available for future interactions and study should make this cohort a productive part of the genomic medicine research landscape for years to come.
Figure 5.
Pedigree for Clinical Case 1 (Box 1). Standard pedigree nomenclature is used. Abbreviations used: d. 70: died in his/her 70s, in addition, the cause of death may be specified—MI, myocardial infarction, cancer, etc.; CABG 60, coronary artery bypass graft in his/her 60s; HL 20, hyperlipidemia of unknown type, diagnosed in his/her 20s; and HL 2, hyperlipidemia of unknown type diagnosed at age 2. For the patients who have undergone mutation testing, the LDLR mutation status is indicated; c.564G>T/+ indicates heterozygosity for the mutation.
Figure 6.
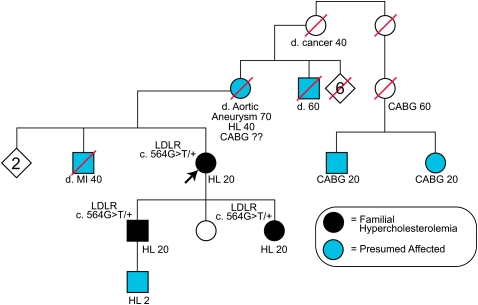
Single axial slice of the coronary calcium scan from the patient described in Clinical Case 1 (Box 1) that shows severe calcification of the left anterior descending coronary artery (red arrow), the portion of the circumflex coronary artery within the imaging plane (white arrow), and the aortic root around the origin of the left main coronary artery (yellow arrow).
Figure 7.
Pedigree for Clinical Case 2 (Box 1). Abbreviations used are similar to those in Figure 5.
Acknowledgments
The authors thank Francis Collins, Robert Nussbaum, Lawrence Brody, Barbara Biesecker, Benjamin Wilfond, Colleen McBride, Elaine Ostrander, Paul Meltzer, and various other members of the genomics community for providing advice and support during the early conception and development of this project. We thank Stephanie Brooks, Jennifer Johnston, Amy Linn, Paul Gobourne, and numerous staff of the NIH Clinical Research Center for ongoing participant recruitment and evaluation; Betty Benjamin, Shelise Brooks, and Jyoti Gupta for quality review of sequence traces; and Eugene Passamani and the staff of Suburban Hospital for clinical support. Most importantly, we thank the ClinSeq study participants—past, present, and future. This study is supported by funds from the Intramural Research Program of the National Institutes of Health. The opinions expressed here are those of the authors and do not necessarily reflect the opinions of their affiliated institutions.
Footnotes
The ClinSeq consent form is available by request from the corresponding author.
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.092841.109.
References
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhangale TR, Stephens M, Nickerson DA. Automating resequencing-based detection of insertion-deletion polymorphisms. Nat Genet. 2006;38:1457–1462. doi: 10.1038/ng1925. [DOI] [PubMed] [Google Scholar]
- Boren J, Ekstrom U, Agren B, Nilsson-Ehle P, Innerarity TL. The molecular mechanism for the genetic disorder familial defective apolipoprotein B100. J Biol Chem. 2001;276:9214–9218. doi: 10.1074/jbc.M008890200. [DOI] [PubMed] [Google Scholar]
- Chines PS, Swift AJ, Bonnycastle LL, Erdos MR, Mullikin JC, NIH Intramural Sequencing Center, Collins FS. PrimerTile: Designing overlapping PCR primers for resequencing. Am J Hum Genet. 2005;77:A1257. [Google Scholar]
- Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automater sequencer traces using Phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Fernandez CV, Kodish E, Taweel S, Shurin S, Weijer C. Disclosure of the right of research participants to receive research results: An analysis of consent forms in the Children's Oncology Group. Cancer. 2003;97:2904–2909. doi: 10.1002/cncr.11391. [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Halushka MK, Fan JB, Bentley K, Hsie L, Shen N, Weder A, Cooper R, Lipshutz R, Chakravarti A. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat Genet. 1999;22:239–247. doi: 10.1038/10297. [DOI] [PubMed] [Google Scholar]
- Homer N, Szelinger S, Redman M, Duggan D, Tembe W, Muehling J, Pearson JV, Stephan DA, Nelson SF, Craig DW. Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet. 2008;4:e1000167. doi: 10.1371/journal.pgen.1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Ji W, Foo JN, O'Roak BJ, Zhao H, Larson MG, Simon DB, Newton-Cheh C, State MW, Levy D, Lifton RP. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40:592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C, Garuti R, Michaely P, Li WP, Maeda N, Cohen JC, Herz J, Hobbs HH. Disruption of LDL but not VLDL clearance in autosomal recessive hypercholesterolemia. J Clin Invest. 2007;117:165–174. doi: 10.1172/JCI29415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, Manning AK, Demissie S, D'Agostino RB, Surti A, Guiducci C, Gianniny L, Burtt NP, Melander O, Orho-Melander M, et al. A genome-wide association study for blood lipid phenotypes in the Framingham Heart Study. BMC Med Genet. 2007;8:S17. doi: 10.1186/1471-2350-8-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani A, Radhakrishnan J, Wang H, Mani MA, Nelson-Williams C, Carew KS, Mane S, Najmabadi H, Wu D, Lifton RP. LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science. 2007;315:1278–1282. doi: 10.1126/science.1136370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: Server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo S, Yin W, Kozlitina J, Pennacchio LA, Boerwinkle E, Hobbs HH, Cohen JC. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J Clin Invest. 2009;119:70–79. doi: 10.1172/JCI37118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing CF, Boerwinkle EA. Genetic architecture of inter-individual variability in apolipoprotein, lipoprotein and lipid phenotypes. Ciba Found Symp. 1987;130:99–127. doi: 10.1002/9780470513507.ch7. [DOI] [PubMed] [Google Scholar]
- Soria LF, Ludwig EH, Clarke HR, Vega GL, Grundy SM, McCarthy BJ. Association between a specific apolipoprotein B mutation and familial defective apolipoprotein B-100. Proc Natl Acad Sci. 1989;86:587–591. doi: 10.1073/pnas.86.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutar AK, Naoumova RP. Mechanisms of disease: Genetic causes of familial hypercholesterolemia. Nat Clin Pract Cardiovasc Med. 2007;4:214–225. doi: 10.1038/ncpcardio0836. [DOI] [PubMed] [Google Scholar]
- Stephens M, Sloan JS, Robertson PD, Scheet P, Nickerson DA. Automating sequence-based detection and genotyping of SNPs from diploid samples. Nat Genet. 2006;38:375–381. doi: 10.1038/ng1746. [DOI] [PubMed] [Google Scholar]
- Sukonina V, Lookene A, Olivecrona T, Olivecrona G. Angiopoietin-like protein 4 converts lipoprotein lipase to inactive monomers and modulates lipase activity in adipose tissue. Proc Natl Acad Sci. 2006;103:17450–17455. doi: 10.1073/pnas.0604026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner EH, Lee C, Ng SB, Nickerson DA, Shendure J. Massively parallel exon capture and library-free resequencing across 16 genomes. Nat Methods. 2009;6:315–316. doi: 10.1038/nmeth.f.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US Department of Health and Human Services. Protection of human subjects, section 45CFR46. Federal Register; Washington, D.C: 1991. http://hhs.gov/ohrp/humansubjects/guidance/45crf46.htm. [Google Scholar]
- Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Guo Y, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RK, Mardis ER. Fluorescence-based DNA sequencing. In: Birren B, et al., editors. Analyzing DNA. genome analysis: A laboratory manual series 1. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. [Google Scholar]
- Wilson PW, D'Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97:1837–1847. doi: 10.1161/01.cir.97.18.1837. [DOI] [PubMed] [Google Scholar]