Abstract
We describe an open source, portable, JavaScript-based genome browser, JBrowse, that can be used to navigate genome annotations over the web. JBrowse helps preserve the user's sense of location by avoiding discontinuous transitions, instead offering smoothly animated panning, zooming, navigation, and track selection. Unlike most existing genome browsers, where the genome is rendered into images on the webserver and the role of the client is restricted to displaying those images, JBrowse distributes work between the server and client and therefore uses significantly less server overhead than previous genome browsers. We report benchmark results empirically comparing server- and client-side rendering strategies, review the architecture and design considerations of JBrowse, and describe a simple wiki plug-in that allows users to upload and share annotation tracks.
In a genome, spatial relationships often indicate functional relationships. A genome browser (Stein et al. 2002; Kent et al. 2003; Stalker et al. 2004) visually conveys the spatial relationships between different pieces of genomic data, helping users form hypotheses about their functional relationships. Current mainstream web-based genome browsers help users understand the genomic data within a given region, but hinder the further development of that understanding by requiring users to navigate to other regions page-by-page. These discontinuous page transitions impair the user's intuitive understanding of which genomic locus they are viewing and how the displayed data points relate to one another.
A genome browser also allows a researcher to visually compare and correlate information from several different sources (Cline and Kent 2009); as such, it is a tool for evaluating multiple forms of evidence, looking at interesting biological cases, linking out to more detailed sources of information, such as genomic databases, communicating information to collaborators visually, preparing publication figures, and more. The availability of many genome browsers via the web allows scores of researchers to immediately dive into the data without the overhead of installing, configuring, or maintaining software, as well as provides the ability to link with a myriad of other web-based sources of information.
Most current web-based genome browsers are implemented using the Common Gateway Interface (CGI) protocol, which provides a mechanism for a web server to generate a web page to send to the user. For example, GBrowse (Stein et al. 2002), a genome browser commonly used for model organism databases, consists of a set of Perl scripts and libraries stationed on the server side. These scripts query a server-side database of genomic features, render the HTML and graphics files needed to display a region of the genome, and transmit them to the browser along with HTML form-based navigation controls. To scroll the displayed region, the user presses the “pan left” button or other navigation control; the browser transmits the changed coordinates to the server, and the process repeats itself. This use of CGI imposes a page-based model of viewing the data; that is, every action (such as moving to a different part of a chromosome or changing how the data are displayed) reloads the entire genome browser page, which incurs a delay and makes the user experience “choppy” (for example, the vertical scroll position and other state information is lost). This manner of progressing through a series of static pages results in disruption of user attention. Since navigating through large volumes of information requires these actions to be done frequently, the disruptions add up.
Another common implementation drawback (not directly having to do with CGI) is that the server generally does most of the work involved in showing genomic data to the user. Typically, a program running on the server has to query a database for genomic information in the region the user is viewing, and then render a static pictorial representation of that region, which the web browser passively displays. In this type of system, the server incurs the majority of the computational expense involved, which increases with the number of users and with the amount of genomic data. As that computational expense increases, so does the amount of time the user has to wait for each new page, unless the server computational resources (and therefore maintenance costs) are also increased.
Both of these issues are addressed by a class of web applications known as rich Internet applications. Rich Internet applications depart from page-based models by decoupling interactions with the user from interactions with the server. This decoupling enables the user to interact with the application without having to wait for the server; communication between the web browser and the server takes place asynchronously in the background. This functionality is generally implemented by techniques such as client-side scripting (using JavaScript and related “dynamic HTML” technologies) and structured data representation (using file formats like XML and JSON). These techniques, often collectively called Ajax (Asynchronous JavaScript and XML; http://www.adaptivepath.com/ideas/essays/archives/000385.php), work cooperatively with the CGI-based technologies on which page-based web applications rely. Such approaches generally have the effect of shifting overhead from the server to the client (the machine that the web browser is running on).
One of the earliest exemplars of Ajax applications was Google Maps (http://maps.google.com/), which broke from previous cartographic websites in responding to user drag requests with live panning of the map viewpoint. Google Maps achieves this by pre-rendering an image of the entire world map (or, at least, relevant parts of it) at multiple zoom levels, breaking this image into tiles of 256 × 256 pixels, and having a JavaScript client, which runs in the user's web browser, download only those tiles visible in the current view. The JavaScript client responds to click and drag events by dynamically changing the positional offset of these tiles, fetching new tile images when necessary (JavaScript provides for manipulation of the Document Object Model [http://www.w3.org/DOM/], the primary data structure by which a web browser represents an HTML page). This creates the effect of panning smoothly over a large pre-rendered image.
It is natural to consider extending the Google Maps experience to the genome browser. Indeed, two recent papers have described the Ajax browsers XMap (Yates et al. 2008) and Genome Projector (Arakawa et al. 2009) that were built using the Google Maps API. XMap and Genome Projector follow the Google Maps approach of pre-rendering: each track (a collection of biological features of the same type, e.g., “genes” or “spliced ESTs”) is rendered and broken up into tiles, which are served up on demand depending on the region of the genome the user is viewing. However, this approach may be impractical for viewing eukaryotic genomes at a level of detail that shows individual DNA bases, due to the space required to store pre-rendered image tiles on the server. Genome Projector only demonstrates bacterial genomes, and neither browser demonstrates the ability to view individual bases; the highest zoom level in the XMap online demonstration is roughly 250-fold lower than the default per-base zoom level in GBrowse (Stein et al. 2002). The problems with pre-rendering would be compounded if users could upload custom tracks, a feature that exists in some genome browsers (Stein et al. 2002; Kent et al. 2003); supporting even a small number of users could require an orders-of-magnitude increase in the server storage space needed to hold graphics for their pre-rendered tracks.
Several recently developed genome browsers, the NCBI Sequence Viewer (http://www.ncbi.nlm.nih.gov/projects/sviewer), Anno-J (Lister et al. 2008), and version 2 of GBrowse, avoid the costs of pre-rendering in different ways. The NCBI Sequence Viewer renders an image on the server and sends it to the client, just like traditional CGI-based genome browsers. Unlike traditional genome browsers, it allows the user to drag the image from side-to-side as in Google Maps. Once the user does so, the server renders a new image representing the new region. Switching from the old image to the new image can be jarring for the user, though, especially if the feature layout has changed. This approach has server-side computational costs similar to those of traditional genome browsers. Anno-J renders genomic information on the client using the “canvas” HTML element supported by some web browsers. This relieves the server of the cost of rendering, but the server-side database query cost remains. This strategy also limits users to web browsers that support the canvas element, including Firefox and Safari, but excluding Internet Explorer.
GBrowse version 2.0 (L Stein, pers. comm.) improves on the original user interface by using Ajax to dynamically load, reorder, and update browser tracks without triggering a full page reload. It uses a “rubber band” interface to allow users to rapidly select and zoom into a region of interest. However, it does not support smooth zooming and panning and suffers the cost of server-side rendering, although the impact of the latter is somewhat lessened by the ability to spread out track rendering across multiple server-side machines.
Here, we consider a different approach, where the client does all the work of determining what genomic features are in the region of interest and then rendering those features using standard HTML and JavaScript functionality. Both the database-query and feature-rendering computational costs are borne by the client; the use of standard HTML and JavaScript features means that JBrowse will work for almost all modern web browsers. This approach is not without its drawbacks; the principal one being that the environments for server-side web applications are considerably more mature than those for client-side applications in their robustness, platform independence, existence of debuggers, and support for reusable code libraries. In addition, when we began, the server-side rendering code for existing genome browsers had already been written and debugged, while client-side rendering mechanisms were relatively untried. As a result, we found that the question of client-side vs. server-side rendering was difficult to answer definitively a priori. We implemented both client- and server-side rendering mechanisms and compared their performance both subjectively, and in terms of resource usage. Our empirical tests definitively favor client-side rendering. (It may be worth emphasizing that these results cannot be extrapolated to all client-server systems; decisions about how best to distribute work between client and server depend heavily on the nature of the task at hand.)
Based on these tests, we decided to implement a genome browser with client-side layout and rendering using Ajax technology. It implements essential genome browser features such as: searching for annotations by name or ID, track selection, quantitative (“wiggle”) tracks, exon–intron structure, and navigation bar. Panning from left to right, zooming in and out, and reordering tracks can all be done without communicating with the server; having this functionality on the client makes these operations faster and more fluid, which helps the user understand and compare data at different loci and from different sources. The client-side approach also achieves a significant reduction in server-side processing costs, making it easier to support large numbers of users and large amounts of data. In our implementation, no server-side CGI programs are required for browsing; aside from the work of preparing new data for use with JBrowse, the only work the server does is to send static files to the browser-side routines. These static files are organized in a way that enables the user's web browser to perform the work traditionally done on the server. In addition to the server-side computational savings enabled by this approach, it also takes advantage of the caching functionality built into the HTTP protocol. If a user repeatedly views the same genomic location, the data for that location will likely be cached on that user's computer, which almost entirely eliminates the time the user will spend waiting for the server to respond.
JBrowse is sufficiently compact and modular that it can be used as a drop-in component to a web content management system, or other web application, such as a wiki or blog. To illustrate this usage, we have developed an example “plug-in” for the TWiki wiki engine, allowing users to upload sequence annotation files as attachments to wiki pages; the resulting tracks then show up in a JBrowse browser embedded in the page.
In this paper, we describe the features and architecture of our client-rendered genome browser, as well as our earlier server-rendered model. We also report empirical results of comparing the architecture approaches (pre-rendering on server vs. live client-side rendering) on genome annotation data from Drosophila melanogaster. Online demos of our browser, client and server source code (freely distributed under an open source license), as well as support for users (such as tutorials and feature request) and developers, are available at our website: http://jbrowse.org/.
Availability and installation
The JBrowse JavaScript genome browser can be test-driven at our website (http://jbrowse.org/) using Firefox (version 2 or later), Safari (version 3 or later), or Internet Explorer (version 6 or later). Browser demonstrations, source code (both client and server), bug- and request-tracking system, tutorials, and other support information are also available on our website.
All our code is available under a dual LGPL (http://www.gnu.org/licenses/lgpl.txt) /Artistic license (http://www.perlfoundation.org/artistic_license_2_0), free for academic and commercial use. Installation of JBrowse is straightforward, requiring an Internet-connected UNIX platform with a webserver (e.g., Apache) and Perl libraries BioPerl (Stajich et al. 2002) and JSON, available from CPAN (http://cpan.org/). (A copy of the JavaScript Dojo library is included with the JBrowse source code.) The JBrowse code repository can be browsed and code can be downloaded from http://jbrowse.org/. Preparation of a website from a data source is as straight forward as running a Perl script, as described in the tutorial distributed with the JBrowse code.
For convenience, we have prepared an Amazon Machine Image (a virtual machine image in which JBrowse and its prerequisites are already installed) for quick and easy installation of a JBrowse server on the Amazon EC2 cloud (http://aws.amazon.com/ec2/), a large computer cluster that can be rented by the machine hour.
Requests for new features and extensions can be entered into a tracking system using the Lighthouse web application, linked from http://jbrowse.org/.
Results
Browser capabilities
Basic genome browser functionality
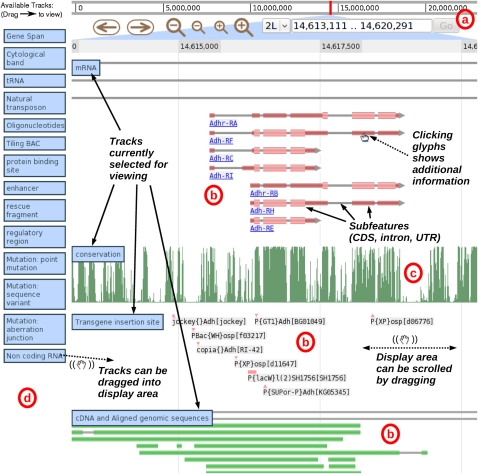
As seen in Figure 1, users can view genome annotations against a reference “ruler,” with an overhead bar giving a visual indication of chromosome position. The user can navigate by dragging the display left or right (which creates a smooth panning effect, with no page reload) or by clicking on navigation buttons; analogous buttons allow the display to be zoomed in or out (again, this is a smooth effect, with no page reload). Alternatively, users can navigate directly to a region (or feature) of interest by typing the region coordinates (or feature name) into a search box. Additional annotation tracks can be added to the current display by dragging them from a reservoir bar on the left of the screen, and can be removed by dragging them back off the main display. Tracks can, similarly, be reordered by dragging. All track manipulation, as with navigation, is live and requires no page reloads. Clicking on features triggers a configurable action (such as opening a feature-specific web page, or bringing up a pop-up window).
Figure 1.
Screenshot of JBrowse, illustrating the various parts of the screen. The navigation panel (a) includes an overview of the genome and the current location, together with navigation buttons for panning and zooming, a menu for selecting the current chromosome, and a text box for navigating directly to coordinates or named features. Below this are the currently active tracks, which can include feature tracks (b) or bar graph image tracks for displaying quantitative information (c). To the left of the active tracks is the reservoir of unused tracks (d), which can be dragged to the active track area, upon which they will be expanded.
Multiple types of track
The browser can display basic features (using a variety of simple glyphs), compound features (e.g., UTR/exon/intron structures), and quantitative tracks that have a value for every base. At lower zoom levels where too many features are displayed to be useful, feature tracks are displayed as histograms showing feature density instead, which presents the information contained in the data in a terser, more useful way.
Flexible data sources
Data can be drawn from GFF files (for simple feature tracks), or GFF3 files (for compound feature tracks), or from WIG files (“wiggle,” i.e., quantitative, tracks). Alternatively, data can be imported from a Bio∷DB database, building on the wide range of open-source applications that work with BioPerl libraries (Stajich et al. 2002) and related databases such as Chado (Mungall et al. 2007).
Makefile-driven workflow
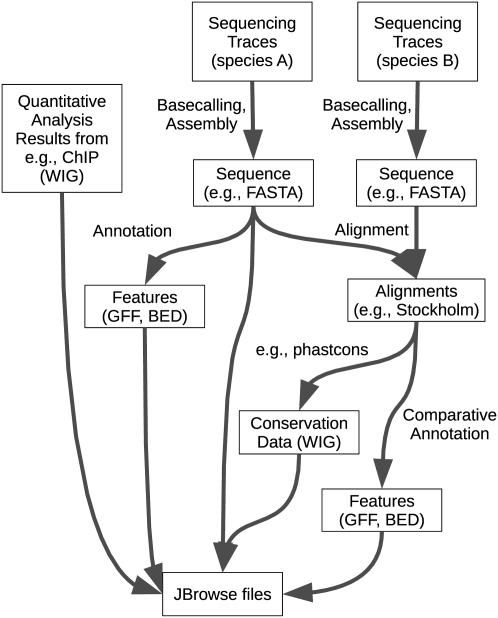
Assembly, alignment, annotation, and JBrowse viewing of genome sequences can be viewed as successive steps in a workflow, as shown in Figures 2 and 3. Here, we define a workflow as a set of logical rules for transforming datatypes automatically, such as a Makefile (Parker et al. 2003). We provide a Makefile that prepares sequence and annotation files on a server for viewing by a JBrowse client. Additionally, this Makefile defines simple file format conversions (e.g., conversion of annotation files from BED format [http://genome.ucsc.edu/FAQ/FAQformat] to GFF format [http://www.sequenceontology.org/gff3.shtml]). These Makefile rules can be applied by downloading the relevant FASTA, BED, GFF, and/or WIG files into the top-level directory and then executing “make jbrowse.”
Figure 2.
Schematic of an example workflow including JBrowse as the final step. This workflow includes base calling, assembly, annotation, alignment, and comparative annotation, with preparation of JBrowse files as the final step required before they can be viewed online. For more details on the preparation of the JBrowse files, see Figure 3.
Figure 3.
Schematic of JBrowse architecture showing the components of the server and the client. The “Server” area shows the data served by the web server (rectangles), the programs that generate that data (arrow labels), and the data sources used in turn by those programs (cylinders). The “Client” area shows the main pieces of code that run in the web browser, how they fit together, and what data they consume.
Preservation of state
HTTP cookies are used to preserve the navigation state and track selection/ordering preferences of individual users, so that a user can close a browser window and open a new one, and it will still show the same genome annotations.
Configuration files
In the manner of GBrowse, flexible configuration files allow the database administrator to customize the tracks and their behavior, including aspects such as glyphs and feature-click actions.
A “genome wiki”
JBrowse is compact, open source, and has a simple JavaScript API, so it is straightforward to embed JBrowse within other web applications, including rich Internet applications. To illustrate this capability, we have developed a JBrowse plug-in for the Perl wiki application, TWiki (http://twiki.org/). The core elements are Makefiles, situating JBrowse as part of an extensible workflow system whereby remote users of a wiki can trigger analyses by uploading sequence, annotation, or configuration files. This creates an open-source, portable, extensible wiki for uploading and sharing genome annotations. As discussed later, this plug-in is a step toward the kind of system imagined by Salzberg (2007) and others, providing rudimentary user account management, authentication, revision control, and bookmarking facilities. However, it lacks database robustness, fine-grained feature editing, or broad interoperability with other genome browsers and databases.
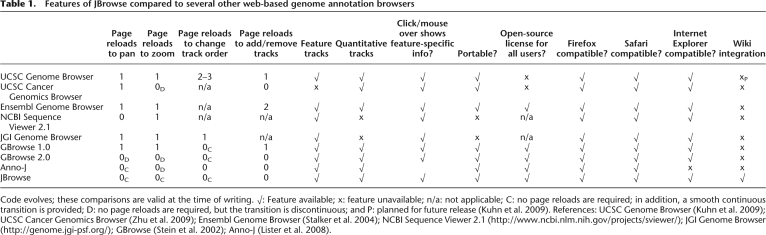
A screenshot of the browser is shown in Figure 1 and a comparison of the user experience to other browsers is presented in Table 1.
Table 1.
Features of JBrowse compared to several other web-based genome annotation browsers
Code evolves; these comparisons are valid at the time of writing. √: Feature available; x: feature unavailable; n/a: not applicable; C: no page reloads are required; in addition, a smooth continuous transition is provided; D: no page reloads are required, but the transition is discontinuous; and P: planned for future release (Kuhn et al. 2009). References: UCSC Genome Browser (Kuhn et al. 2009); UCSC Cancer Genomics Browser (Zhu et al. 2009); Ensembl Genome Browser (Stalker et al. 2004); NCBI Sequence Viewer 2.1 (http://www.ncbi.nlm.nih.gov/projects/sviewer/); JGI Genome Browser (http://genome.jgi-psf.org/); GBrowse (Stein et al. 2002); Anno-J (Lister et al. 2008).
Figure 3 shows the basic architecture of the JBrowse system. The client can display sequence tracks (only visible at the highest zoom level), simple feature tracks (essentially interval sets: each simple feature corresponds to a contiguous interval of sequence), compound feature tracks (where intervals are composed of subintervals; e.g., protein-coding gene intervals have exon, intron, and UTR subintervals) and quantitative tracks (currently represented as images in the style of bar graphs).
The JBrowse server requires some preprocessing of tracks before they can be served to the client (see the Methods section for details); Perl scripts to do this preprocessing are provided. Feature tracks are stored in Nested Containment Lists (NCLists) (Alekseyenko and Lee 2007), gene names and other text-navigable labels are stored in Patricia tries (Morrison 1968), and quantitative (wiggle) tracks are rendered as image files. The preprocessing outputs are stored as static files on the server; these files can then be served with minimal server load and they can also be cached by the user's web browser to reduce subsequent data traffic.
Comparison of server- and client-side rendering
As noted in the introduction, we first implemented a Google Maps version of GBrowse, the generic genome browser (Stein et al. 2002), that pre-rendered all tracks as statically served image files. The GBrowse Perl server renders graphics primitives using the GD graphics library (http://libgd.org/), for which we wrote a thin Perl proxy class that intercepted drawing primitives and stored them in an SQL database (or in-memory), allowing the primitives to be later “replayed” in order to generate individual tiles; thus, feature layout is performed once only, ensuring consistency between tiles. A JavaScript client then consults a tile index file to retrieve and organize tiles for display. This is a quite general approach that can be used to scale any pre-rendered graphics to arbitrarily large dimensions for interactive Google Maps-like exploration over the web; the GD proxy class (TiledImage.pm) and JavaScript client are available from our code repository.
In contrast, JBrowse represents genomic features graphically in the web browser using HTML elements. These elements are not typically used for graphics, because they can only take the form of horizontal or vertical rectangles. However, for displaying genomic features, rectangles are appropriate; they can represent a feature with a start and stop position on a chromosome, optionally, with additional rectangles for subfeatures (like exon/intron/UTR/CDS transcript structure). HTML rectangles can also use background images to indicate strand and phase information. Genomic data that does not fit into this rectangular mold (such as dense quantitative data) can be displayed in JBrowse through images (as with TiledImage) at the cost of extra storage space and processing time.
Both approaches (JBrowse and TiledImage) require the input genomic data to be preprocessed. This preprocessing is significant to the application's potential use, as slow preprocessing performance may limit interactivity when tracks can be uploaded by users; in addition, large index storage requirements on the server side may be prohibitive.
To compare resource utilization of server-side and client-side rendering of genome annotations, we conducted a benchmark of TiledImage (server-side) and JBrowse (client-side) running on the same hardware. We drew annotations from FlyBase version 5.1 (Tweedie et al. 2009). The platform used for benchmarks was a 2.2 GHz 64-bit AMD Opteron server with 2Gb of RAM running Centos 4.6 Linux with the ext3 file system. The genome annotation database was Bio∷DB∷SeqFeature∷Store from BioPerl version 1.6 (Stajich et al. 2002) with MySQL 4.1 on the back end.
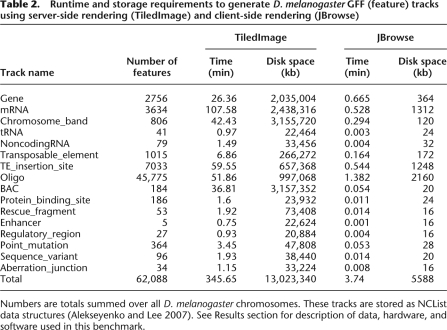
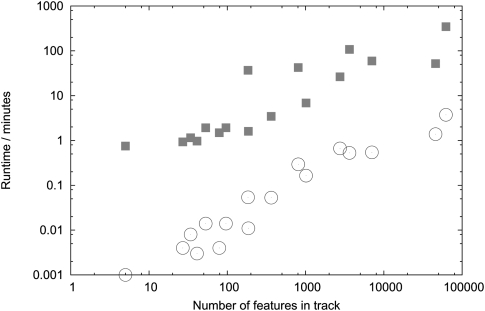
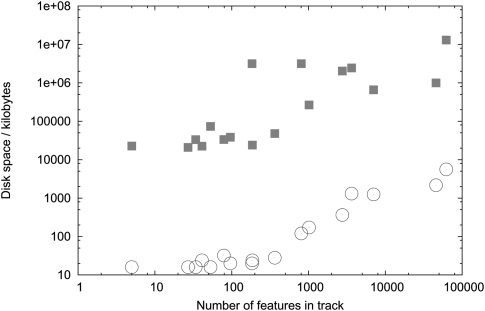
The raw data of our benchmark for feature tracks (e.g., GFF or BED files) are shown in Table 2. Resource utilization is plotted as a function of the number of features in the track in Figure 4 (runtime) and Figure 5 (storage). The figures show a monotonic increase in resource usage with respect to feature count; theoretically, putting N elements into a NCList takes at least as long as sorting those N elements, i.e., O(N log N); beyond this, no analytical results for the time complexity are known. Similarly, we expect at least O(N log N) time complexity for TiledImage to store, index, and retrieve graphics primitives for N features, though Table 2 and Figure 4 clearly show that, in practice, JBrowse is much faster at preprocessing than TiledImage. Generically, we would expect to need at least O(N) for storage in both cases, and potentially O(N log N) for TiledImage (due to indexing of the graphics primitives). Again, we find that JBrowse is far more efficient in practice, as shown by Table 2 and Figure 5. We speculate that the dramatic difference in performance between the TiledImage and the NCL preprocessing steps is due to overhead from GD graphics library's server-side rendering and layout operations, which were not designed to render chromosome-length track images. In contrast, the most time-consuming step in generating an NCL is the initial sort of the features.
Table 2.
Runtime and storage requirements to generate D. melanogaster GFF (feature) tracks using server-side rendering (TiledImage) and client-side rendering (JBrowse)
Numbers are totals summed over all D. melanogaster chromosomes. These tracks are stored as NCList data structures (Alekseyenko and Lee 2007). See Results section for description of data, hardware, and software used in this benchmark.
Figure 4.
Time to generate tracks: TiledImage vs. JBrowse. Time required to generate each D. melanogaster feature annotation track, plotted as a function of the number of features in the track, for both server-side rendering (TiledImage, squares) and client-side rendering (JBrowse, circles). See Table 2 for raw data. See Results section for description of data, hardware, and software used in this benchmark.
Figure 5.
Disk space required to store track: TiledImage vs. JBrowse. Disk space required to store each D. melanogaster feature annotation track, plotted as a function of the number of features in the track, for both server-side rendering (TiledImage, squares) and client-side rendering (JBrowse, circles). See Table 2 for raw data. See Results section for description of data, hardware, and software used in this benchmark.
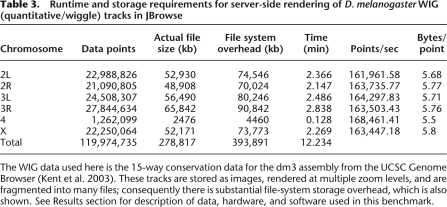
We also benchmarked the resource utilization of JBrowse for pre-rendering quantitative track images from WIG files. Results are shown in Table 3, indicating a rate of data-point generation (∼1.6 × 106 points/sec) and file-system usage (∼6 bytes/point) that is roughly independent of the size of the track. No comparison to TiledImage was necessary here, since JBrowse also uses a server-side pre-rendering approach for such tracks. The rate-limiting step in this image rendering procedure appears to be image compression of the resulting monochromatic bar graph images, which are stored in PNG format (data not shown). Alternate compact visualization formats, e.g., single-pixel grayscales, rather than multipixel bar graphs, might therefore save some time, since the compression step would have fewer pixels to deal with. Conceivably, rendering these tracks on demand would also save time during track preparation (although preparation would still involve reading the entire WIG file).
Table 3.
Runtime and storage requirements for server-side rendering of D. melanogaster WIG (quantitative/wiggle) tracks in JBrowse
The WIG data used here is the 15-way conservation data for the dm3 assembly from the UCSC Genome Browser (Kent et al. 2003). These tracks are stored as images, rendered at multiple zoom levels, and are fragmented into many files; consequently there is substantial file-system storage overhead, which is also shown. See Results section for description of data, hardware, and software used in this benchmark.
In addition to the server-side costs, the client-side costs must also be considered. Since JBrowse moves some costs from the server to the client, the capabilities of the client may impose scaling limitations on JBrowse. It is difficult to quantify those limitations, however, because they are heavily dependent on the user's web browser and hardware, and the limits are subjective, manifesting mainly as slower animations, longer download waits, and garbage collection pause times. To qualitatively investigate those limits, we created a JBrowse instance using the hg19 human data from the University of California at Santa Cruz (Kuhn et al. 2009).
JBrowse can accommodate large amounts of sequence data by breaking it up into easily handled segments, but the JBrowse NCList implementation used to store features is currently incapable of breaking down feature sets into pieces smaller than one per track per chromosome. On our circa-2005 hardware, JBrowse easily handles feature tracks with tens of thousands of features in a track/chromosome, and can go up to the low hundreds of thousands with some performance loss. This is enough to accommodate all the tracks we have encountered except for the human EST and SNP tracks. We believe that accommodating those tracks are straightforward engineering, and that work is high on our list of priorities for the future. In the near term, large feature sets might be loaded into JBrowse as subsets. For example, rather than having all SNPs in the same track, they could be broken down into coding/intronic/intergenic, etc., or filtered to only include confirmed SNPs, or SNPs that correspond to mutations recorded in OMIM.
Discussion
We have implemented an open source genome browser, JBrowse; the server can be readily downloaded and installed on UNIX systems, while the client is actively tested on Firefox, Safari, and Internet Explorer. We report benchmark comparisons to a server-side rendering implementation derived from GBrowse, and outline a broad summary of the complexity issues and other challenges we encountered in developing this prototype. We demonstrate that client-side rendering of genome annotation data, with minimal server-side preprocessing (to facilitate fast queries by the client), requires significantly less up-front work than bulk server-side pre-rendering.
We have also implemented a simple wiki plug-in that allows users to embed fully interactive JBrowse instances into wiki pages and configure them via wiki file uploads. In 2007, Steven Salzberg called for a genome wiki, commenting that “A wiki would allow the community of experts to work out the best name for each gene, to indicate uncertainty where appropriate and to discuss alternative annotations” (Salzberg 2007). Our technical interpretation of such a wiki, discussed at greater length on our website (http://biowiki.org/GenomeWiki), includes the following five desiderata: core wiki functions (i.e., an intuitive user interface offering the ability to upload, browse, share, and revise annotation tracks), bioinformatic granularity (i.e., common wiki functions, such as editing or browsing revision histories, should work down to the level of individual features and subfeatures), a robust database back end, portability of open-source client and server code, and compatibility with standard web applications (e.g., search engines), protocols (e.g., Google PageRank), bioinformatics file formats, established biological databases and browsers, and other bioinformatics terminologies and standards (e.g., the Gene Ontologies [Ashburner et al. 2000]). The example JBrowse wiki plug-in has core wiki functions, portability, and rudimentary standards-compliance (it speaks common bioinformatic file formats). Basic user authentication and revision control mechanisms are provided by the underlying wiki engine (TWiki; http://twiki.org/). However, TWiki's underlying database is not particularly robust (it uses the server file system), and the JBrowse plug-in offers edit operations only at the atomic level of entire tracks, not finer-grained annotation elements such as genes or exons. For comparison, the Genboree system (http://www.genboree.org/) satisfies most of our criteria for a genome wiki, but lacks portability (it is designed to be a hosted service, run only on one site) and has a page-based CGI user interface.
To demonstrate how JBrowse can be embedded in other applications, we have provided a plug-in implementing a genome wiki, whereby tracks can be uploaded and shared by users (in the manner of Google Calendar or Google Documents). Our future plans for this include APIs for advanced search (e.g., on track metadata, such as authorship or experimental information) and client notification of newly uploaded tracks.
Methods
In this section, we first outline the general issues and design principles that motivated the choice of data structures for the system, then discuss in detail our specific implementation.
JBrowse is a client-server application operating over the Internet, where client and server are implemented in different languages and operate on data asynchronously. The client side is implemented in JavaScript using the Dojo library (http://dojotoolkit.org/); the server, in Perl with the BioPerl library (Stajich et al. 2002).
Issues that motivated the choice of novel data structures for the system
Ease of implementation
Typically, due to the difference in operating environments and programming languages, each data structure must be implemented twice: once on the server and once on the client. A serialized representation for network transmission must also be implemented. This strongly favors simple data structures that are intuitively easy to understand and debug.
Efficiency of data structure preparation and readout
Efficient access to data structures is important for a responsive client; efficient construction and serialization of those same structures is important for an interactive server application, particularly in the context of a wiki-like application (such as our TWiki plug-in) where genome annotation tracks can be uploaded and shared.
Storage and CPU usage on the server
As noted in the Results section, inefficient server implementations could require terabytes of storage and hours or days to preprocess data.
Resident memory size and CPU usage on the client
Since the client is running within a web browser, a somewhat limited environment compared to a full operating system, it is essential that it not overburden the web browser with excessive demands on memory or CPU usage.
Size on the wire
As well as the “traditional” analyses of time, memory, and disk usage noted above, every effort must be taken to minimize the byte size of the serialized form of the data. Otherwise, network latency will become prohibitive for interactive browsing.
Tracks can be added by running Perl scripts on the server to preprocess raw annotation files (GFF/GFF3 for simple/compound feature tracks, WIG for quantitative tracks, and FASTA for sequence tracks) and generate the relevant files that are downloaded by the client (JSON files for simple/compound feature tracks, PNG images for quantitative tracks, and chunked strings for sequence tracks). Alternatively, the annotations can be fetched from a BioPerl genome database (Bio∷DB, Bio∷DasI, etc.), in which case a configuration file (somewhat styled after the GBrowse con figuration file [Stein et al. 2002], but using the JSON format) identifies the database and determines the track listing and types.
In the case of feature tracks, the generated files are JSON representations of Nested Containment Lists, or NCLists. An NCList is an efficiently queryable data structure for an interval set (Alekseyenko and Lee 2007). The interval set is decomposed into disjoint subsets, arranged in a hierarchical tree. Within each subset, there are no two intervals such that one interval is fully contained by the other interval (condition 1). Each subset is sorted by interval startpoint; since there are no containment relationships within the subset, the subset is also sorted by an interval endpoint. Furthermore, all the intervals in a subset are fully contained within one interval of the parent subset (condition 2). These two conditions allow efficient querying for intervals overlapping a given range (Alekseyenko and Lee 2007). An example NCList is illustrated in Figure 3. The asymptotic time complexity to construct an NCList in place from an unsorted interval set empirically appears to be O(N log N); the time complexity of querying it for intervals overlapping a given range is known to be O(n + log N), where N is the cardinality of the interval set and n is the number of intervals returned by the query. While this performance is similar to competing interval set containers, such as R-trees (Guttman 1984) or quad-trees (Finkel and Bentley 1974), NCLists in practice are considerably simpler to implement, with lower storage overhead. In benchmarks, NCLists compared very favorably to competitors, running 10–50 times faster than R-trees (Alekseyenko and Lee 2007). One drawback of NCLists is that they currently lack published algorithms for in-place modification or partial loading; these would clearly be useful (e.g., for incremental loading of big tracks), motivating future development of such algorithms.
The text box on the JBrowse page may be used to navigate directly to named features of interest. To locate features quickly, the client uses a Patricia Trie or radix tree (Morrison 1968), which is prepared on the server concurrently with the NCLists, and lazily loaded by the client (only required nodes are fetched, so that the entire feature dictionary does not have to be downloaded whenever the user tries to search for a feature). Extraction of feature names from input files is controlled by the configuration file.
WIG files are rendered into PNG images at multiple zoom levels. These images are then broken into tiles and delivered on request to the client. The client displays these images agnostically with respect to their content, so in fact any fixed-height image track (not just bar graphs or other representations of wiggle tracks) can be displayed along the genome, as long as sufficient disk space exists to store them on the server. Thus, JBrowse effectively subsumes server-side-rendering genome browsers such as Genome Projector (Arakawa et al. 2009) and TiledImage. (Note, however, that pre-rendering WIG tracks is considerably less computationally expensive than pre-rendering GFF tracks; cf. Tables 2 and 3.)
Acknowledgments
This work was funded by NIH grant HG004483. We thank the attendees of the 2006 Biology of Genomes and 2007 RECOMB meetings for helpful feedback about our posters, which presented the first (TiledImage) prototypes of our browser.
Footnotes
[The JBrowse source code (freely licensed), live demonstrations, mailing list, documentation, bug-tracking, and virtual machine images are available at http://jbrowse.org/.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.094607.109.
References
- Alekseyenko A, Lee C. Nested containment list (NCList): A new algorithm for accelerating interval query of genome alignment and interval databases. Bioinformatics. 2007;23:1386–1393. doi: 10.1093/bioinformatics/btl647. [DOI] [PubMed] [Google Scholar]
- Arakawa K, Tamaki S, Kono N, Kido N, Ikegami K, Ogawa R, Tomita M. Genome Projector: Zoomable genome map with multiple views. BMC Bioinformatics. 2009;10:31. doi: 10.1186/1471-2105-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Kent WJ. Understanding genome browsing. Nat Biotechnol. 2009;27:153–155. doi: 10.1038/nbt0209-153. [DOI] [PubMed] [Google Scholar]
- Finkel R, Bentley JL. Quad trees: A data structure for retrieval on composite keys. Acta Informatica. 1974;4:1–9. [Google Scholar]
- Guttman A. R-trees: A dynamic index structure for spatial searching. Proceedings of the 1984 ACM SIGMOD International Conference on Management of Data; Boston, MA: ACM Press; 1984. pp. 47–57. [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2003;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al. The UCSC Genome Browser Database: Update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D. Patricia-practical algorithm to retrieve information coded in alphanumeric. J ACM. 1968;15:514–534. [Google Scholar]
- Mungall CJ, Emmert DB The FlyBase Consortium. A Chado case study: An ontology-based modular schema for representing genome-associated biological information. Bioinformatics. 2007;23:i337–i346. doi: 10.1093/bioinformatics/btm189. [DOI] [PubMed] [Google Scholar]
- Parker DS, Gorlick MM, Lee CJ. Evolving from bioinformatics in-the-small to bioinformatics in-the-large. OMICS. 2003;7:37–48. doi: 10.1089/153623103322006580. [DOI] [PubMed] [Google Scholar]
- Salzberg SL. Genome re-annotation: A wiki solution? Genome Biol. 2007;8:102. doi: 10.1186/gb-2007-8-1-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich J, Block D, Boulez K, Brenner S, Chervitz S, Dagdigian C, Fuellen G, Gilbert J, Korf I, Lapp H. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalker J, Gibbins B, Meid P, Smith J, Spooner W, Hotz H-R, Cox AV. The Ensembl web site: Mechanics of a genome browser. Genome Res. 2004;14:951–955. doi: 10.1101/gr.1863004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein L, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich J, Harris T, Arva A, et al. The generic genome browser: A building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. Flybase: Enhancing Drosophila gene ontology annotations. Nucleic Acids Res. 2009;36:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates T, Okoniewski M, Miller C. X:Map: Annotation and visualization of genome structure for Affymetrix exon array analysis. Nucleic Acids Res. 2008;36:D780–D786. doi: 10.1093/nar/gkm779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Sanborn JZ, Benz S, Szeto C, Hsu F, Kuhn RM, Karolchik D, Archie J, Lenburg ME, Esserman LJ, et al. The UCSC cancer genomics browser. Nat Methods. 2009;6:239–240. doi: 10.1038/nmeth0409-239. [DOI] [PMC free article] [PubMed] [Google Scholar]