Abstract
Interindividual variability in response to chemicals and drugs is a common regulatory concern. It is assumed that xenobiotic-induced adverse reactions have a strong genetic basis, but many mechanism-based investigations have not been successful in identifying susceptible individuals. While recent advances in pharmacogenetics of adverse drug reactions show promise, the small size of the populations susceptible to important adverse events limits the utility of whole-genome association studies conducted entirely in humans. We present a strategy to identify genetic polymorphisms that may underlie susceptibility to adverse drug reactions. First, in a cohort of healthy adults who received the maximum recommended dose of acetaminophen (4 g/d × 7 d), we confirm that about one third of subjects develop elevations in serum alanine aminotransferase, indicative of liver injury. To identify the genetic basis for this susceptibility, a panel of 36 inbred mouse strains was used to model genetic diversity. Mice were treated with 300 mg/kg or a range of additional acetaminophen doses, and the extent of liver injury was quantified. We then employed whole-genome association analysis and targeted sequencing to determine that polymorphisms in Ly86, Cd44, Cd59a, and Capn8 correlate strongly with liver injury and demonstrated that dose-curves vary with background. Finally, we demonstrated that variation in the orthologous human gene, CD44, is associated with susceptibility to acetaminophen in two independent cohorts. Our results indicate a role for CD44 in modulation of susceptibility to acetaminophen hepatotoxicity. These studies demonstrate that a diverse mouse population can be used to understand and predict adverse toxicity in heterogeneous human populations through guided resequencing.
Adverse reactions, such as liver injury, are prominent reasons for cessation of drug testing in clinical trials, restrictions on drug use, and the withdrawal of approved drugs (Shenton et al. 2004). Adverse reactions remain a significant safety concern since they occur at low rates, often undetectable in standard-sized clinical trials, and are not foreseen through traditional in vitro and animal safety testing paradigms (Larrey 2000). While it is widely recognized that better preclinical models are required to enable accurate prediction and identification of xenobiotic-induced toxicity (Collins et al. 2008), few experimental paradigms exist that provide preclinical population-wide testing.
The promise of personalized medicine and the accumulating knowledge of human genomic variation serve as potent catalysts for pharmacogenetics research (Ingelman-Sundberg 2008). Polymorphisms within genes encoding xenobiotic metabolizing enzymes and major histocompatibility complex proteins are promising genetic biomarkers that may predict the efficacy of drug treatment or identify individuals at risk of adverse reactions (Lanfear and McLeod 2007; Tomalik-Scharte et al. 2008). However, only a limited number of potentially useful biomarkers have been identified thus far. Furthermore, current research into pharmacogenetic biomarkers is largely focused on human studies where only a limited number of positive associations between a polymorphism and adverse drug reaction have been reproduced in independent cohorts.
For the past century, the mouse has been the most widely used model system for studying human disease and related phenotypes, often in ways that are not directly possible in humans (Paigen 2003). The major mouse genetic resource used for association studies of complex polygenic traits is the Laboratory Strain Diversity Panel (LSDP) (Paigen and Eppig 2000). Recent resequencing of 15 mouse inbred strains and the analysis of their polymorphism architecture (Roberts et al. 2007) have shown that LSDP contains as many or more single nucleotide polymorphisms (SNPs) than estimated to be present in humans, and minor allele frequency distribution in the LSDP is larger than that present in man. Thus, we postulated that a panel of inbred mouse strains can be used to model the phenotypic variation within the human population and to uncover susceptibility factors for drug-induced toxicities, thereby shortening the path to the discovery of pharmacogenetic biomarkers.
In this study, we tested this approach by investigating the genetic causes of variation in the hepatotoxicity of acetaminophen (N-acetyl-p-aminophenol). More than a third of all cases involving acute liver failure in the Unite States are due to overdose of this widely available medication (Lee 2003); about half of these cases are unintentional or involve chronic ingestion (Kaplowitz 2005). In addition, it has been estimated that 10% of patients experiencing liver failure due to acetaminophen were taking recommended doses of acetaminophen (Lee 2007). Liver injury due to acetaminophen is a complex phenotype, requiring accumulation of its reactive metabolite, N-acetyl-p-benzoquinone imine (NAPQI), covalent binding to cellular proteins, oxidative stress, and hepatocellular necrosis, as well as an imbalance between protective and injurious cytokines (James et al. 2005; Jaeschke and Bajt 2006). A recent placebo-controlled clinical study revealed that about a third of healthy adult volunteers who were administered the maximum therapeutic dose of acetaminophen (4 g/d for 14 d) exhibited transient, asymptomatic elevations in serum alanine aminotransferase (ALT) levels that were greater than three times the upper limit of normal (Watkins et al. 2006), indicating liver toxicity. Acetaminophen represents an intriguing model compound for pharmacogenetic studies, because, while subjects taking therapeutic doses of the drug exhibit transient serum ALT elevations, the drug has a good safety profile in long-term use (Kuffner et al. 2006). The same pharmacogenetic factors that predispose a person to transient low-dose ALT elevations may be responsible for decreasing that individual's hepatotoxic susceptibility threshold at higher doses. For these reasons, acetaminophen is an ideal compound for the validation of a human-to-mouse-to-human approach in pharmacogenetic research.
Results
Variability in acetaminophen-induced liver injury in humans
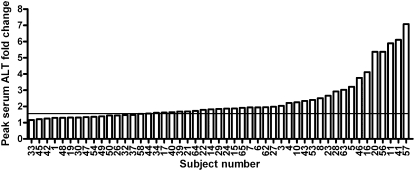
To confirm a prior report of differential sensitivity to acetaminophen hepatotoxicity among healthy human volunteers (Watkins et al. 2006), an independent cohort of 59 healthy subjects was enrolled in a double-blind, placebo-controlled study in which 49 received the maximum recommended therapeutic dose of acetaminophen (4 g/d for 7 d) and 10 subjects were randomly assigned to placebo. Elevations in ALT greater than 1.5-fold of individual baseline values were observed for 69% (34/49) of subjects receiving acetaminophen (Fig. 1; Supplemental Table 1) and values exceeding twofold baseline were observed in 37% (18/49), confirming that some healthy subjects experience mild liver injury in response to therapeutic doses of acetaminophen. In each subject, a 1.5-fold cut-off was confirmed to represent significant (P > 0.05) elevation in ALT from baseline by linear modeling. Interestingly, 31% (15/49) did not demonstrate ALT elevations greater than 1.5-fold of baseline and showed no meaningful differences from the placebo-control group (N = 10, P = 0.42). ALT levels were at baseline levels in all subjects 14 d after cessation of the treatment (Supplemental Table 1). Elevations in serum ALT correlate well with other markers of liver injury (e.g., alpha glutathione-S-transferase), in studies of acetaminophen-induced hepatotoxicity (Supplemental Fig. 1).
Figure 1.
Maximum serum ALT fold change measured in human volunteers taking daily oral doses of acetaminophen. The peak ALT fold change over baseline reached over the course of treatment by each subject in the UNC cohort is shown. Subjects were considered responders (N = 34) if peak serum ALT reached greater than 1.5-fold (line) higher than the subject's baseline value.
Differences in liver injury in mice following acetaminophen exposure
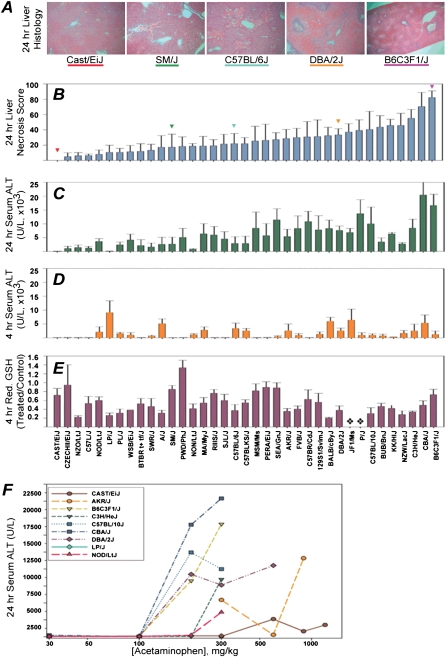
To determine whether genetic factors influence acetaminophen-associated liver toxicity, a panel of 36 inbred mouse strains was selected to represent the genetic variation present within humans (Beck et al. 2000). Liver toxicity was assessed at 4 and 24 h after administration of an acute dose (300 mg/kg) of acetaminophen. Hepatic necrosis was histologically quantified 24 h after treatment, and a dramatic interstrain variation in liver damage, exemplified by a characteristic centrilobular necrosis, was observed (Fig. 2A,B). CAST/EiJ mice were most resistant as they sustained no liver necrosis or alterations in serum ALT, while B6C3F1/J mice, which are commonly used to evaluate chemical toxicity, were the most sensitive strain.
Figure 2.
Response to the acute dose of acetaminophen in a panel of mouse strains. (A) Representative photomicrographs (100×) of the hematoxylin and eosin-stained sections of left liver lobe of mice 24 h after dosing with acetaminophen (300 mg/kg). (B) Liver necrosis score (mean ± SE, n = 3–4/strain) in mice treated with acetaminophen (300 mg/kg) for 24 h. (C) Serum ALT levels (mean ± SE) in acetaminophen-treated mice sacrificed 24 h after dosing. (D) Serum ALT levels (mean ± SE) in acetaminophen-treated mice sacrificed 4 h post-dosing. (E) Liver reduced glutathione (ratio between acetaminophen- and vehicle-treated animals in each strain, mean ± SE) 4 h post-dosing. ( ) Strains with no data. (F) Dose-response to acetaminophen-induced liver injury as measured by ALT release (n = 4/strain, mean ± SE) at 24 h after treatment.
) Strains with no data. (F) Dose-response to acetaminophen-induced liver injury as measured by ALT release (n = 4/strain, mean ± SE) at 24 h after treatment.
Serum ALT levels were measured at 4 and 24 h post-dosing (Fig. 2D,E). A Pearson correlation of 0.77 between serum ALT 24 h post-dosing with acetaminophen and extent of liver necrosis was noted, confirming that serum ALT is a good indirect marker for liver injury. However, comparison between ALT level at 4 and 24 h post-dosing shows that it may be difficult to predict injury outcome from ALT measured at early time points following acetaminophen doses. These data suggest that there are genetic factors that may independently affect the timing of acetaminophen-induced hepatocellular injury and ALT release.
It is well accepted that acetaminophen hepatotoxicity depends on metabolic activation, hepatic glutathione depletion, and protein binding of NAPQI as initiating events. However, it is not known whether variability in glutathione levels and drug metabolism enzymes contribute to differential toxicity outcomes between individuals. Therefore, we quantified the ratio of reduced to oxidized glutathione in livers from mice sacrificed 4 h post-dosing, a time when acetaminophen-induced glutathione depletion is still robust (Mitchell et al. 1973). There was no correlation between either reduced (Fig. 2C) or total (data not shown) glutathione pool at 4 h and liver necrosis at 24 h post-dosing, suggesting that liver glutathione is not a sensitive biomarker for predicting injury outcome across individuals. Similarly, protein levels of cytochrome p450 2E1 (CYP2E1), CYP1A2, catalase, and glutathione S-transferase pi (GSTP1) in liver microsomes from those mice sacrificed at 24 h did not correlate with acetaminophen-induced liver necrosis, or with serum ALT levels across individual strains (Supplemental Table 5).
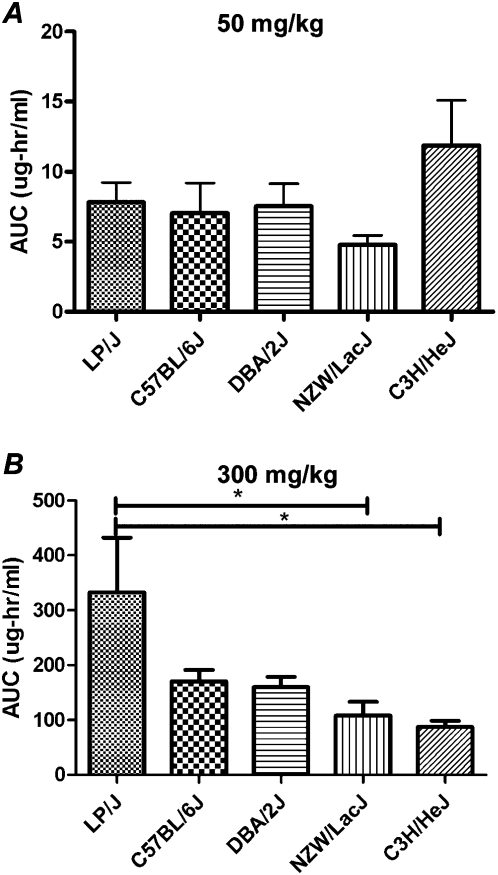
Interindividual differences in pharmacokinetics of acetaminophen were found to be not correlated with liver injury in the previous study of acetaminophen hepatotoxicity among healthy human volunteers (Watkins et al. 2006). To investigate the interstrain differences in metabolism of acetaminophen, we selected five strains (LP/J, C57BL/6J, DBA/2J, NZW/LacJ, and C3H/HeJ) from our panel based on the differences in sensitivity to acetaminophen-induced liver necrosis (Fig. 2A). The pharmacokinetics of the parent compound was assessed over a 6-h period following a bolus dose of 50 or 300 mg/kg by oral gavage using the area under the curve (AUC) (Fig. 3). After the 50 mg/kg dose, no differences between strains were observed (Fig. 3A). After the 300 mg/kg dose, LP/J mice showed a significantly different profile in exposure to acetaminophen (Fig. 3B). Despite the fact that metabolism of acetaminophen at high doses does vary between strains, this observation is insufficient to explain interindividual differences in liver injury in mice, similar to that in humans, since susceptible strains have a much lower plasma exposure to acetaminophen than the resistant strains.
Figure 3.
Plasma AUC of acetaminophen (mean ± SE) measured across strains for 6 h post-dosing with 50 mg/kg (i.g.) (A) or 300 mg/kg (i.g.) (B) following an overnight fast. Asterisk indicates significant differences between strains by the Tukey post-hoc test.
Representative mouse strains were selected from across the hepatic injury gradient to examine whether genetic variation also affects the dose-response. We classified each strain into one of three groups by the degree of necrosis observed 24 h following administration of 300 mg/kg acetaminophen. Representative nonresponder (mean necrosis score less than 15%), mid-responder (mean necrosis score 15%–50%), and high-responder (mean necrosis score >50%) strains were tested at additional doses ranging from 30–1200 mg/kg (N = 4). Markedly different dose-response curves in response to acetaminophen were observed (Fig. 2F). High-responder strains CBA/J, DBA/2J, and B6C3F1/J and the mid-responder strain C57BL10/J have significant elevations in serum ALT at 24 h post-dosing with a 200 mg/kg dose. However, the high-responder strain C3H/HeJ and low-responder strain NOD/LtJ had no observable adverse response below 300 mg/kg. Of particular interest is strain CAST/EiJ, in which comparably small elevations in ALT were observed only at 600, 900, or 1200 mg/kg.
Identification of candidate genes for sensitivity to acetaminophen-induced liver injury
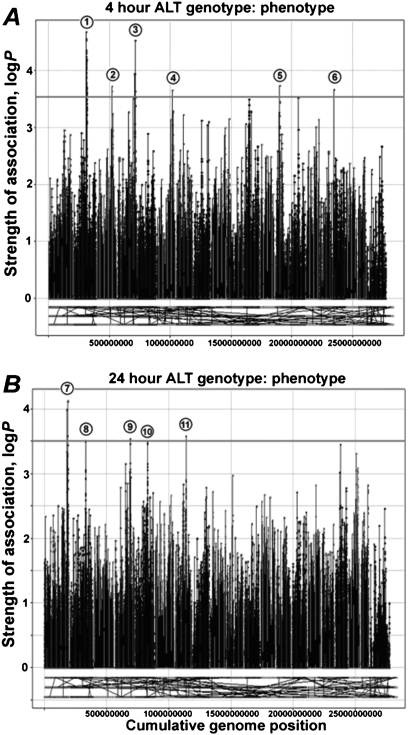
To uncover polymorphisms associated with sensitivity to acetaminophen toxicity, we performed haplotype-associated mapping utilizing a dense SNP map (McClurg et al. 2006). Association analyses were performed with mouse serum ALT levels for 4 h (Fig. 4A) and 24 h (Fig. 4B) post-dosing. Because the genomic intervals with the greatest computed association with toxicity contained several genes (Table 1), we selected candidate genes that could be reasonably linked to the propagation of oxidative- or immune-mediated stress responses following acetaminophen exposure. We chose Cd44, Cd59a, Ly86, Cat, and Capn8 as likely candidate genes responsible for strain-specific liver injury.
Figure 4.
Haplotype association mapping of acetaminophen-induced liver injury in the mouse. Serum ALT at 4 h (A) and 24 h (B) after acetaminophen (300 mg/kg) treatment was used to identify genomic intervals significantly associated with liver injury. Peaks (numbered, see Table 1) indicate a significant logP association score at each 3-SNP marker window. Marker colors indicate chromosome number across the mouse genome.
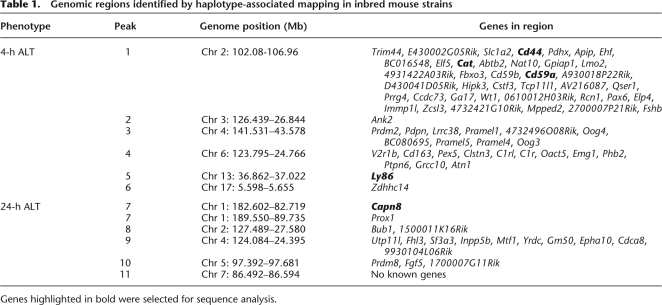
Table 1.
Genomic regions identified by haplotype-associated mapping in inbred mouse strains
Genes highlighted in bold were selected for sequence analysis.
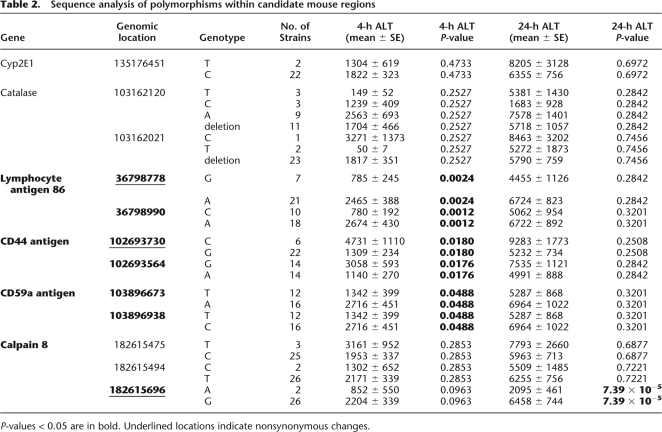
A 300- to 800-bp region from each gene that contained either known nonsynonymous coding SNPs or polymorphisms in intronic splice site regions was selected for resequencing. Also included in the analysis was Cyp2e1, which codes for a primary enzyme known to metabolize acetaminophen to its reactive metabolite, NAPQI (Gonzalez 2005). Ly86, Cd44, and Cd59a contain polymorphisms that, within the mouse diversity panel, correlate well with the degree of ALT release (P < 0.05) (Table 2). The Capn8 gene, which was implicated in the 24-h ALT phenotype genome scan, was found to have a nonsynonymous coding SNP that is highly correlative with 24-h ALT measurements (P = 7.39 × 10−5). The polymorphisms selected for genotyping in Cat or Cyp2e1 were not correlative with markers of liver injury.
Table 2.
Sequence analysis of polymorphisms within candidate mouse regions
P-values < 0.05 are in bold. Underlined locations indicate nonsynonymous changes.
Mouse genes associated with acetaminophen-induced liver injury translate to humans
To evaluate the human relevance of the susceptibility genes identified in mice, we tested if polymorphisms in orthologous genes correlate with interindividual variability in acetaminophen toxicity in humans. We sequenced 300- to 650-bp regions of CD44, CD59, CAPN10 (human ortholog of mouse Capn8), and LY86 that included SNPs reported by the HapMap Project (http://www.hapmap.org) as having a minimal r2 of 0.8 and a minor allele frequency greater than 0.05. Genomic DNA from two independent human cohorts were available for these experiments: a UNC cohort reported here and the Purdue Pharma cohort (Watkins et al. 2006).
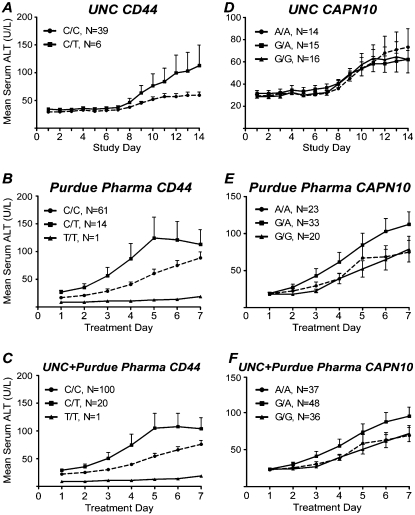
Within the UNC cohort, we observed an association between an individual's genotype at a CD44 SNP (rs1467558) and the elevation in serum ALT reached during the 14-d study (P = 0.02) (Fig. 5A). The polymorphism is nonsynonymous, encoding an amino acid change from an isoleucine (C allele) to a threonine (T allele) residue in the CD44 protein. In order to test whether this association is replicable, we evaluated DNA from 76 subjects enrolled in the Purdue Pharma cohort. Because the duration of acetaminophen administration was 14 d in the Purdue Pharma cohort (vs. 7 d in the UNC cohort), data analysis was limited to the first 7 d of treatment. Within the Purdue Pharma cohort, a C/T genotype at the same CD44 SNP (rs1467558) was also found to be associated with ALT elevations during treatment (P = 0.01) (Fig. 5B). When the two cohorts were combined, the association was more significant (P = 0.002) (Fig. 5C). The Cohen's d effect size (0.44) indicates that the SNP has a moderate effect on the toxicity outcome following acetaminophen exposure. Interestingly, the allele effect size in almost identical within the mouse strain panel (0.42).
Figure 5.
Polymorphisms in CD44 (A–C) and CAPN10 (D–F) associated with susceptibility to acetaminophen-induced liver injury in humans. Data from UNC (A,D), Purdue Pharma (B,C) and a combined cohort (C,F) are shown. Average mean (±SE) serum ALT per genotype is plotted for each matching study day.
To further assess the functional relevance of this finding to acetaminophen-induced liver injury in mice and humans, we performed experiments in Cd44-null mice and performed in silico prediction of the effect of the amino acid substitution resulting from the polymorphism at CD44 SNP rs1467558. Indeed, Cd44-null mice exhibit significantly greater liver injury 24 h following administration of acetaminophen (300 mg/kg) compared with their wild-type (C57BL/6J) counterparts (i.e., the mean liver necrosis ± SE was 40% ± 4% for the wild type and 61% ± 7% in the Cd44-null). Furthermore, the structural ramification of the change from isoleucine (C allele) to threonine (T allele) in the human CD44 protein due to SNP rs1467558 was predicted in silico to be possibly damaging to the function of the protein due to the potential creation of a cavity within a buried site (with a PolyPhen PSIC score difference between the variant proteins of 1.711).
A polymorphism within CAPN10 (rs3749166) displayed a trend across both cohorts in which individuals having the G/A allele tended to be more sensitive to acetaminophen-induced ALT elevations in the first 7 d of treatment (Fig. 5D–F). While the trend remained consistent across sample populations, the data were only marginally significant when analyzed in the combined cohorts (P = 0.045). It is interesting to note that while rs3749166 is a synonymous coding SNP, it is a tag SNP for rs2975766, a nonsynonymous polymorphism that alters coding from isoleucine to valine.
There was no correlation between increased serum ALT and genotyped polymorphisms within the CD59 (rs10538602) or LY86 (rs5874047) genes in the data collected. There was also no statistical difference between sensitivity to acetaminophen and genotype when all pairs of gene–gene interactions were examined (data not shown).
Discussion
One of the major reasons that efficacious drugs fail to advance through late stages of development, or are removed or restricted after entering the marketplace, is because of rare adverse health events that were not predicted using current preclinical testing paradigms (Ingelman-Sundberg 2008). Consequently, being able to identify which drugs cause, and more importantly which individuals are likely to develop, adverse reactions is a major challenge preventing informed deployment of new medicines. The experimental approach we describe here, using acetaminophen as a model compound, bypasses the limitations of humans-only pharmacogenetics studies by showing that a population of mouse strains can be used to predict genetic biomarkers of toxicity sensitivity.
A traditional genome-wide pharmacogenetic investigation (Nelson et al. 2008) into the genetic factors linked to the liver toxicity of acetaminophen would require a much larger number of individuals due to greatly reduced power associated with P-value correction in whole-genome SNP analyses. Conversely, a so-called “candidate gene” analysis (Kindmark et al. 2007) may be equally challenging due to the complexity of the mechanism of action of acetaminophen (Kaplowitz 2005). We instead narrowed the set of potential gene targets by first utilizing haplotype-associated mapping to determine significant genetic loci. Interestingly, well-characterized genes known to be essential for acetaminophen toxicity did not correlate with liver injury in the panel of mouse strains. While a priori knowledge of the toxicant's mode of action can be useful in the selection of genes for follow-up analysis, validation of susceptibility-modulating genes in the laboratory is essential. By using this approach, the candidate susceptibility genes identified through genetic studies in the mouse translated to two independent human cohorts despite small numbers of individuals available.
It is noteworthy that the top candidate genes suggested by the analysis of the inbred mouse strains are related to the immune response, and not to metabolism and detoxification of acetaminophen. The traditional view on the mechanisms of toxicity, the approach widely utilized to predict individual responses to xenobiotics, would imply that metabolism of acetaminophen to the reactive electrophile NAPQI and/or detoxification of the latter by glutathione conjugation should explain, at least to a considerable degree, the variability in responses. However, no apparent correlation between levels of major metabolizing enzymes, glutathione, or acetaminophen plasma exposure in select strains and liver injury was observed in the mouse population. Similarly, in several cytokine knockout mouse models of acetaminophen toxicity, the sensitivity to liver necrosis due to acetaminophen was largely independent of covalent binding of NAPQI to proteins or glutathione depletion (Kaplowitz 2005). Furthermore, we found no correlation with sensitivity for polymorphisms in the genes encoding catalyses or cytochrome P450 2E1, implying that variation at these key mediators of acetaminophen toxicity cannot explain differential susceptibility to acetaminophen. This conclusion does not refute the molecular mechanism of APAP toxicity via bioactivation by CYP2E1. In contrast, our data form a basis by which we show that the end outcome of the toxicity response is not directly correlative with interindividual differences in the basic metabolism of acetaminophen. This indicates that other cellular processes leading to tissue injury, in addition to metabolism and pathways-involved cell damage, are involved in final determination of liver necrosis following treatment with acetaminophen. This raises a critical distinction between genes (enzymes/proteins) that are essential mediators of toxicity but that do not functionally vary (i.e., CYP2E1) and those whose activity or function may vary considerably among individuals and determine susceptibility to toxicity (i.e., CD44, see below).
While events downstream of the consumption of hepatic intracellular glutathione are not as well described as those upstream of acetaminophen metabolism, these downstream events have been shown to be a major mediator of the toxicity response. Indeed, the presence of inflammatory mediators released from nonparenchymal cells in the liver, including interleukin (IL)6 (James et al. 2003), IL10 (Bourdi et al. 2007), interferon-gamma (Ishida et al. 2002), and tumor necrosis factor-alpha (Gardner et al. 2002), have been shown to affect liver sensitivity to acetaminophen. Furthermore, neutrophil-mediated necrosis (Liu et al. 2006) and Kupffer cell recruitment (Ju et al. 2002) have also been implicated as important factors in progression of liver injury; however, their precise role and timing of involvement are debated (Knight and Jaeschke 2004; Jaeschke 2006).
Our data support the notion that variation in immune response may be the most critical of the complex events that determine susceptibility to acetaminophen toxicity since a number of candidates from this pathway were significantly associated with strain-specific injury in response to acetaminophen. Within the mouse diversity panel, ALT release at 4 h was shown to be affected by polymorphisms in lymphocyte antigen 86 (Ly86, also known as MD-1), CD44 antigen (Cd44), and CD59a antigen (Cd59a), which are involved in B-cell responsiveness to lipopolysaccharide, lymphocyte adhesion and activation, and regulation of complement deposition, respectively. Subtle, transient alterations in immunogenic signaling during acetaminophen toxicity may also play a role in the development of idiosyncratic toxicities in an individual; however, more data are needed to fully characterize this relationship.
Capn8, a gene identified by association mapping of the 24-h ALT phenotype, was the only nonimmune gene found to be associated with sensitivity to acetaminophen (an exonic A-to-G base change). This observation is intriguing given that calpain released from necrotic hepatocytes has been associated with the progression of acetaminophen-induced liver injury (Limaye et al. 2003). In addition, calpastatin, a specific inhibitor of calpain, was recently shown to play a role in attenuating liver injury and increasing survival of mice following an acute dose (Limaye et al. 2006).
The ability of the panel of mouse strains to predict sensitivity to acetaminophen-induced liver injury in humans was supported by sequencing of the orthologous genes positively associated with liver injury in mice. Consistent with the data in the mouse population, we found CD44 to be a marker of sensitivity in two independent human cohorts. The genotypes at CD44 allowed partitioning of subjects based upon susceptibility to acetaminophen-induced hepatic toxicity and implicate variation in immunogenic cell surface antigens as potential mediators of acetaminophen sensitivity. It is noteworthy that heterozygous (C/T) individuals are more susceptible, since (1) in silico prediction of the effect of this nonsynonymous coding SNP suggests a disruption in the protein function and (2) Cd44-null mice are more susceptible to liver necrosis due to acetaminophen. These data are intriguing given that Cd44-deficient mice exhibit greater liver injury at 24 h following administration of another classic hepatotoxicant, carbon tetrachloride (Kimura et al. 2008). Interestingly, inflammatory response to carbon tetrachloride was considerably shifted in the Cd44-deficient mice compared with wild type (C57BL/6 mice), an effect that may be mediated by temporal differences in liver NFKB activity. Therefore, it is possible that variations in CD44 may significantly affect liver necrosis through effects on leukocyte signaling via cytokine modulation. However, owing to the many physiologic and pathologic roles of CD44 isoforms in vivo (Rouschop et al. 2006), including cell–cell matrix interaction, lymphocyte extravasation, wound healing, scar formation, cell migration, and the binding and presentation of growth factors, the precise mechanistic role of this gene in conferring sensitivity to acetaminophen-induced ALT elevations remains to be determined.
Collectively, our results indicate that the use of an inbred mouse strain panel is a valuable tool for evaluating drug safety and for the development of biomarkers to prescreen individuals prior to therapeutic drug treatment with potential toxicities. The identification of the genes associated with differential susceptibility to toxicity in a preclinical phase, exemplified by the finding that CD44 may be involved in modulation of susceptibility to acetaminophen hepatotoxicity, has potential to focus pharmacogenetics research, overcome the challenges of small human cohorts, and shorten the validation period. The data acquired with this model could therefore be influential in the analysis of individual risk to pharmaceutical agents and may facilitate both drug development and human safety endeavors. One of the limitations of this approach, however, lies in the uncertainties of whether the associations between SNPs and modest increases in ALT observed with “therapeutic” doses would also predict individuals susceptible to more severe toxicity seen in overdose situations. Additional research into the mechanisms of predisposition to minor forms of liver injury and those that lead to more severe organ damage is needed.
Methods
Acetaminophen administration to human subjects
Study volunteers were healthy men and women between 18–45 yr of age and were not receiving concomitant medications. Prescreening was performed 14 d prior to admission to confirm health as previously described (Watkins et al. 2006). Written informed consent was obtained and approved by the UNC Institutional Review Board. Participants remained in the General Clinical Research Center at UNC for the duration of the 14-d study, during which they received a controlled diet of standardized whole-food meals. From days 4–11, subjects received either Extra Strength Tylenol (two 500-mg tablets of acetaminophen, commercial product; n = 49) or placebo (n = 10) orally every 6 h. Blood samples were taken at 08:00 h daily prior to dosing and analyzed for aspartate aminotransferase (AST), ALT (Supplemental Table 1), total bilirubin, alkaline phosphatase, blood urea nitrogen, alpha-glutathione-S-transferase, and creatinine. Dosing was discontinued for subjects in whom serum ALT or AST reached more than three times the upper limit of normal. Baseline serum ALT was determined as the mean of the values obtained prior to the start of dosing. Blood was collected from study participants for DNA isolation. Leukocytes were isolated from whole blood and DNA was extracted using the Qiagen MidiPrep kit (Qiagen) and the manufacturer's protocol. The protocol for the Purdue Pharma L.P. cohort study has been as previously described (Watkins et al. 2006).
Acetaminophen administration to mice
Toxicity studies
Male mice aged 7–9 wk were obtained from the Jackson Laboratory (Bar Harbor, ME) and housed in polycarbonate cages on Sani-Chips irradiated hardwood bedding (P.J. Murphy Forest Products Corp.). Animals were fed NTP-2000 diet (Zeigler Brothers, Inc.) and water ad libitum, and maintained on a 12-h light-dark cycle. Mice utilized in this study comprised 36 inbred strains that are priority strains for the Mouse Phenome Project (Bogue and Grubb 2004); B6C3F1/J hybrid mice were also used. Care of mice followed institutional guidelines under a protocol approved by the Institutional Animal Care and Use Committee. Mice were singly housed and fasted 18 h prior to intragastric dosing with acetaminophen (30, 100, 300, 600, 900, or 1200 mg/kg; 99% pure, Sigma-Aldrich) or vehicle (0.5% methyl 2-hydroxyethyl cellulose, Sigma-Aldrich) with a dosing volume of 10 mL/kg for all doses. Dosing was performed at the same time of day throughout the study to avoid diurnal variability (Boorman et al. 2005). Feed was returned 3 h after dosing, and animals were sacrificed at 4 or 24 h. Blood was collected from the vena cava from animals anesthetized with Nembutal (100 mg/kg intraperitoneally [i.p.], Abbott Laboratories). Samples were assayed for serum markers by standard enzymatic procedures (Bergmeyer et al. 1986). Livers were quickly excised, and sections of the left lobes were placed in 10% phosphate buffered formalin for histological analyses. Remaining liver was snap-frozen in liquid nitrogen and stored at -80°C.
Metabolism studies
Adult (aged 7 wk) male mice of strains C3H/HeJ, C57BL/6J, DBA/2J, LP/J, and NZW/LacJ were selected for metabolism studies based on their wide variation of liver toxicity observed at 24 h following a 300 mg/kg dose. Mice were fed overnight prior to dosing with 50 mg/kg or fasted for 18 h overnight prior to dosing with 300 mg/kg APAP (N = 5 per strain per dose). Blood (45 μL) was collected sequentially from the tail artery at 0, 0.5, 1, 2, and 3 h post-dosing. At 6 h, blood was collected by exsanguination at 6 h for metabolite measurements and ALT quantification and livers collected as described above.
CD44 knockout studies
To test the ability of CD44 protein to modulate APAP toxicity, Cd44 knockout mice, B6.Cg-Cd44tm1Hbg/J (N = 6), and wild-type mice, C57BL/6J (N = 6), were dosed with 300 mg/kg APAP (i.g.) and sacrificed at 24 h as described in toxicity studies. An interim blood sample was collected from the tail artery at 4 h post-dosing for ALT analysis.
Glutathione quantification
Liver samples were homogenized in borax/EDTA (pH 9.3) solution, precipitated with chloroform, and centrifuged. Reduced glutathione was derived in liver samples, calibration standards, and QC samples with 7-fluorobenzofurazan-4-sulfonic acid ammonium salt (SBD-F) and analyzed by HPLC with fluorescence detection. Concentrations were calculated using the glutathione response, sample weights, and a regression line constructed from the concentrations and peak responses of the appropriate calibration standards (Sigma).
Enzyme-linked immunosorbent assay
Quantitative determinations of protein levels of CYP2E1, CYP1A2, catalase, and GST was performed using microsomes isolated from the left liver lobe using the Protein Detector ELISA kit protocol (KPL, Inc.) as detailed by the manufacturer.
Liver histopathology
Formalin-fixed liver specimens were embedded in paraffin, and 5-μm sections cut in duplicate were applied to each slide. Sections were stained with hematoxylin and eosin (H&E), and liver injury was blindly scored. Necrosis was quantified by a point counting technique (Mouton 2002). Scores were independently verified by a veterinary pathologist.
Serum metabolite quantification
The procedure used for the quantification of APAP is similar to that previously described (Shenton et al. 2004). Briefly, a reversed-phase HPLC assay was used, in which the mobile phase was 5% acetonitrile and 95% 5 mM sodium sulfate/20 mM potassium phosphate buffer (pH 3.2) with a flow rate of 1.2 mL/min. Retention times for APAP and the internal standard (3-acetamidophenol) detected at 254 nm were 7 and 11 min, respectively. APAP standard (Sigma) and the internal standard were spiked into naive mouse plasma to generate standard curves. The AUC was calculated by using noncompartmental analysis in WinNonLin (Pharsight). A one-way ANOVA with a Tukey post-hoc test was used to assess significantly different AUC across mouse strains (P < 0.05).
Haplotype association mapping
Haplotype association mapping was performed as described elsewhere (Schadt et al. 2003). Briefly, haplotype associations were calculated using a modified F-statistic based upon genotype–phenotype pairings at each 3-SNP window across a 160,000 SNP data set. Strains excluded from association analysis due to lack of polymorphism data were C57BL/10J, NZO/H1LtJ, and P/J. LogP values were plotted across the mouse genome using SpotFire (SpotFire, Inc.). Genomic intervals with association scores greater than 3.5 were considered significant. Genes within significant intervals were identified with the BioMart feature of Ensembl using NCBI build 36 (http://www.ensembl.org).
Genetic sequence analysis
For sequence-based genotyping, primers were designed using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). For each reaction, genomic DNA from pedigreed mice or from human subjects was diluted to 100 ng/μL and 1 μL of DNA mixed with 12.5 μL of 2× PCR Master Mix (Promega), 2.5 μL of 10 μM upstream primer, 2.5 μL of 10 μM downstream primer, and 6.5 μL of nuclease-free water. Primers and conditions used for PCR amplification are listed in Supplemental Table 2. Sequencing reactions were performed using the ABI PRISM BigDye Terminator version 1.1 Cycle Sequencing Ready Reaction Kit with the AmpliTaq DNA polymerase (Applied Biosystems). DNA was sequenced on a 3730 DNA analyzer (Applied Biosystems) (Supplemental Table 3). Sequence alignment was performed using Vector NTI version 10 (Invitrogen).
Statistical methods
Phenotypic values were expressed as the mean ± SEM. Differences were considered significant when the P-value < 0.05. The Pearson correlation coefficient was used to determine correlation between phenotypic toxicity measurements. Genotype-to-phenotype associations for the mouse data were performed using the two-tailed Student's t-test (for two variants) or ANOVA (for more than two variants). P-values were adjusted for multiple comparisons using a false discovery rate of 5% for the total number of SNPs (12) genotyped in mouse strains across the six genes. P-value corrections were performed using the p.adjust module in R (version 2.4.0). P-value corrections were not determined for the four SNPs tested in humans due to the small number of gene comparisons. Correlation between human genotype data for CD44 and phenotypic responses across time was performed in Partek Genomics Suite (Partek) using repeated measures ANOVA across the first 7 d of acetaminophen treatment in which the study centers were coded as random effects (Supplemental Table 4). In determining the effect of genotype to influence serum ALT increases in acetaminophen-treated human subjects during treatment at UNC, we excluded subjects whose average baseline was 55 U/L or greater (four subjects). Elevations in ALT level within each subject were analyzed using linear modeling in which the daily ALT of each individual over time was assigned a P-value using lm{stats} module in R (version 2.4.0). To determine the effect size of a SNP, we calculated the Cohen's d effect size; we used either the serum ALT measured at 24 h for mouse Cd44 or the maximum serum ALT measured within the first 7 d of Tylenol dosing for human CD44.
Acknowledgments
We thank Blair Bradford, Oksana Kosyk, Lorraine Balletta, Cindy Lodestro, Daekee Lee, Keili Meyer, and David Malarkey for technical assistance. This work was supported, in part, by grants and contracts from the National Institutes of Health (T32-ES07126, U19-ES11391, P30-ES10126, R37-GM38149, RR00046, N01-ES35513, N01-ES25497, and N01-ES65406) and the EPA(STAR-RD832720).
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.090241.108.
References
- Beck JA, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig JT, Festing MF, Fisher EM. Genealogies of mouse inbred strains. Nat Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- Bergmeyer HU, Horder M, Rej R. International Federation of Clinical Chemistry (IFCC) Scientific Committee, Analytical Section: Approved recommendation (1985) on IFCC methods for the measurement of catalytic concentration of enzymes. Part 3. IFCC method for alanine aminotransferase (L-alanine: 2-oxoglutarate aminotransferase, EC 2.6.1.2) J Clin Chem Clin Biochem. 1986;24:481–495. [PubMed] [Google Scholar]
- Bogue MA, Grubb SC. The Mouse Phenome Project. Genetica. 2004;122:71–74. doi: 10.1007/s10709-004-1438-4. [DOI] [PubMed] [Google Scholar]
- Boorman GA, Blackshear PE, Parker JS, Lobenhofer EK, Malarkey DE, Vallant MK, Gerken DK, Irwin RD. Hepatic gene expression changes throughout the day in the Fischer rat: Implications for toxicogenomic experiments. Toxicol Sci. 2005;86:185–193. doi: 10.1093/toxsci/kfi166. [DOI] [PubMed] [Google Scholar]
- Bourdi M, Eiras DP, Holt MP, Webster MR, Reilly TP, Welch KD, Pohl LR. Role of IL-6 in an IL-10 and IL-4 double knockout mouse model uniquely susceptible to acetaminophen-induced liver injury. Chem Res Toxicol. 2007;20:208–216. doi: 10.1021/tx060228l. [DOI] [PubMed] [Google Scholar]
- Collins FS, Gray GM, Bucher JR. Toxicology: Transforming environmental health protection. Science. 2008;319:906–907. doi: 10.1126/science.1154619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner CR, Laskin JD, Dambach DM, Sacco M, Durham SK, Bruno MK, Cohen SD, Gordon MK, Gerecke DR, Zhou P, et al. Reduced hepatotoxicity of acetaminophen in mice lacking inducible nitric oxide synthase: Potential role of tumor necrosis factor-alpha and interleukin-10. Toxicol Appl Pharmacol. 2002;184:27–36. [PubMed] [Google Scholar]
- Gonzalez FJ. Role of cytochromes P450 in chemical toxicity and oxidative stress: Studies with CYP2E1. Mutat Res. 2005;569:101–110. doi: 10.1016/j.mrfmmm.2004.04.021. [DOI] [PubMed] [Google Scholar]
- Ingelman-Sundberg M. Pharmacogenomic biomarkers for prediction of severe adverse drug reactions. N Engl J Med. 2008;358:637–639. doi: 10.1056/NEJMe0708842. [DOI] [PubMed] [Google Scholar]
- Ishida Y, Kondo T, Ohshima T, Fujiwara H, Iwakura Y, Mukaida N. A pivotal involvement of IFN-gamma in the pathogenesis of acetaminophen-induced acute liver injury. FASEB J. 2002;16:1227–1236. doi: 10.1096/fj.02-0046com. [DOI] [PubMed] [Google Scholar]
- Jaeschke H. How relevant are neutrophils for acetaminophen hepatotoxicity? Hepatology. 2006;43:1191–1194. doi: 10.1002/hep.21246. [DOI] [PubMed] [Google Scholar]
- Jaeschke H, Bajt ML. Intracellular signaling mechanisms of acetaminophen-induced liver cell death. Toxicol Sci. 2006;89:31–41. doi: 10.1093/toxsci/kfi336. [DOI] [PubMed] [Google Scholar]
- James LP, Lamps LW, McCullough S, Hinson JA. Interleukin 6 and hepatocyte regeneration in acetaminophen toxicity in the mouse. Biochem Biophys Res Commun. 2003;309:857–863. doi: 10.1016/j.bbrc.2003.08.085. [DOI] [PubMed] [Google Scholar]
- James LP, Simpson PM, Farrar HC, Kearns GL, Wasserman GS, Blumer JL, Reed MD, Sullivan JE, Hinson JA. Cytokines and toxicity in acetaminophen overdose. J Clin Pharmacol. 2005;45:1165–1171. doi: 10.1177/0091270005280296. [DOI] [PubMed] [Google Scholar]
- Ju C, Reilly TP, Bourdi M, Radonovich MF, Brady JN, George JW, Pohl LR. Protective role of Kupffer cells in acetaminophen-induced hepatic injury in mice. Chem Res Toxicol. 2002;15:1504–1513. doi: 10.1021/tx0255976. [DOI] [PubMed] [Google Scholar]
- Kaplowitz N. Idiosyncratic drug hepatotoxicity. Nat Rev Drug Discov. 2005;4:489–499. doi: 10.1038/nrd1750. [DOI] [PubMed] [Google Scholar]
- Kimura K, Nagaki M, Kakimi K, Saio M, Saeki T, Okuda Y, Kuwata K, Moriwaki H. Critical role of CD44 in hepatotoxin-mediated liver injury. J Hepatol. 2008;48:952–961. doi: 10.1016/j.jhep.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Kindmark A, Jawaid A, Harbron CG, Barratt BJ, Bengtsson OF, Andersson TB, Carlsson S, Cederbrant KE, Gibson NJ, Armstrong M, et al. Genome-wide pharmacogenetic investigation of a hepatic adverse event without clinical signs of immunopathology suggests an underlying immune pathogenesis. Pharmacogenomics J. 2007;8:186–195. doi: 10.1038/sj.tpj.6500458. [DOI] [PubMed] [Google Scholar]
- Knight TR, Jaeschke H. Peroxynitrite formation and sinusoidal endothelial cell injury during acetaminophen-induced hepatotoxicity in mice. Comp Hepatol. 2004;3:S46. doi: 10.1186/1476-5926-2-S1-S46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuffner EK, Temple AR, Cooper KM, Baggish JS, Parenti DL. Retrospective analysis of transient elevations in alanine aminotransferase during long-term treatment with acetaminophen in osteoarthritis clinical trials. Curr Med Res Opin. 2006;22:2137–2148. doi: 10.1185/030079906x148346. [DOI] [PubMed] [Google Scholar]
- Lanfear DE, McLeod HL. Pharmacogenetics: Using DNA to optimize drug therapy. Am Fam Physician. 2007;76:1179–1182. [PubMed] [Google Scholar]
- Larrey D. Drug-induced liver diseases. J Hepatol. 2000;32:77–88. doi: 10.1016/s0168-8278(00)80417-1. [DOI] [PubMed] [Google Scholar]
- Lee WM. Acute liver failure in the United States. Semin Liver Dis. 2003;23:217–226. doi: 10.1055/s-2003-42641. [DOI] [PubMed] [Google Scholar]
- Lee WM. Acetaminophen toxicity: Changing perceptions on a social/medical issue. Hepatology. 2007;46:966–970. doi: 10.1002/hep.21926. [DOI] [PubMed] [Google Scholar]
- Limaye PB, Apte UM, Shankar K, Bucci TJ, Warbritton A, Mehendale HM. Calpain released from dying hepatocytes mediates progression of acute liver injury induced by model hepatotoxicants. Toxicol Appl Pharmacol. 2003;191:211–226. doi: 10.1016/s0041-008x(03)00250-3. [DOI] [PubMed] [Google Scholar]
- Limaye PB, Bhave VS, Palkar PS, Apte UM, Sawant SP, Yu S, Latendresse JR, Reddy JK, Mehendale HM. Upregulation of calpastatin in regenerating and developing rat liver: Role in resistance against hepatotoxicity. Hepatology. 2006;44:379–388. doi: 10.1002/hep.21250. [DOI] [PubMed] [Google Scholar]
- Liu ZX, Han D, Gunawan B, Kaplowitz N. Neutrophil depletion protects against murine acetaminophen hepatotoxicity. Hepatology. 2006;43:1220–1230. doi: 10.1002/hep.21175. [DOI] [PubMed] [Google Scholar]
- McClurg P, Pletcher MT, Wiltshire T, Su AI. Comparative analysis of haplotype association mapping algorithms. BMC Bioinformatics. 2006;7:61. doi: 10.1186/1471-2105-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Jollow DJ, Potter WZ, Gillette JR, Brodie BB. Acetaminophen-induced hepatic necrosis. IV. Protective role of glutathione. J Pharmacol Exp Ther. 1973;187:211–217. [PubMed] [Google Scholar]
- Mouton PR. Principles and practices of unbiased stereology: An introduction for bioscientists. The Johns Hopkins University Press; Baltimore, MD: 2002. [Google Scholar]
- Nelson MR, Bacanu SA, Mosteller M, Li L, Bowman CE, Roses AD, Lai EH, Ehm MG. Genome-wide approaches to identify pharmacogenetic contributions to adverse drug reactions. Pharmacogenomics J. 2008;9:23–33. doi: 10.1038/tpj.2008.4. [DOI] [PubMed] [Google Scholar]
- Paigen K. One hundred years of mouse genetics: an intellectual history. II. The molecular revolution (1981–2002) Genetics. 2003;163:1227–1235. doi: 10.1093/genetics/163.4.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K, Eppig JT. A mouse phenome project. Mamm Genome. 2000;11:715–717. doi: 10.1007/s003350010152. [DOI] [PubMed] [Google Scholar]
- Roberts A, Pardo-Manuel de Villena F, Wang W, McMillan L, Threadgill DW. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: Implications for QTL discovery and systems genetics. Mamm Genome. 2007;18:473–481. doi: 10.1007/s00335-007-9045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouschop KM, Claessen N, Pals ST, Weening JJ, Florquin S. CD44 disruption prevents degeneration of the capillary network in obstructive nephropathy via reduction of TGF-beta1-induced apoptosis. J Am Soc Nephrol. 2006;17:746–753. doi: 10.1681/ASN.2005080808. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- Shenton JM, Chen J, Uetrecht JP. Animal models of idiosyncratic drug reactions. Chem Biol Interact. 2004;150:53–70. doi: 10.1016/j.cbi.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Tomalik-Scharte D, Lazar A, Fuhr U, Kirchheiner J. The clinical role of genetic polymorphisms in drug-metabolizing enzymes. Pharmacogenomics J. 2008;8:4–15. doi: 10.1038/sj.tpj.6500462. [DOI] [PubMed] [Google Scholar]
- Watkins PB, Kaplowitz N, Slattery JT, Colonese CR, Colucci SV, Stewart PW, Harris SC. Aminotransferase elevations in healthy adults receiving 4 grams of acetaminophen daily: A randomized controlled trial. JAMA. 2006;296:87–93. doi: 10.1001/jama.296.1.87. [DOI] [PubMed] [Google Scholar]