Abstract
Utilizing the full power of next-generation sequencing often requires the ability to perform large-scale multiplex enrichment of many specific genomic loci in multiple samples. Several technologies have been recently developed but await substantial improvements. We report the 10,000-fold improvement of a previously developed padlock-based approach, and apply the assay to identifying genetic variations in hypermutable CpG regions across human chromosome 21. From ∼3 million reads derived from a single Illumina Genome Analyzer lane, ∼94% (∼50,500) target sites can be observed with at least one read. The uniformity of coverage was also greatly improved; up to 93% and 57% of all targets fell within a 100- and 10-fold coverage range, respectively. Alleles at >400,000 target base positions were determined across six subjects and examined for single nucleotide polymorphisms (SNPs), and the concordance with independently obtained genotypes was 98.4%–100%. We detected >500 SNPs not currently in dbSNP, 362 of which were in targeted CpG locations. Transitions in CpG sites were at least 13.7 times more abundant than non-CpG transitions. Fractions of polymorphic CpG sites are lower in CpG-rich regions and show higher correlation with human–chimpanzee divergence within CpG versus non-CpG sites. This is consistent with the hypothesis that methylation rate heterogeneity along chromosomes contributes to mutation rate variation in humans. Our success suggests that targeted CpG resequencing is an efficient way to identify common and rare genetic variations. In addition, the significantly improved padlock capture technology can be readily applied to other projects that require multiplex sample preparation.
For decades, DNA sequencing has been pivotal in understanding biology, yielding over 900 whole-genome sequences, and identifying genetic variations and somatic mutations that underlie human diseases (Frazer et al. 2007; Stenson et al. 2008). Recent sequencing-based studies suggest that a large panel of genes is mutated in various cancers (Sjoblom et al. 2006; Jones et al. 2008; Parsons et al. 2008). Individually rare but cumulatively frequent variations contribute to the inheritance of common multifactorial diseases (Cohen et al. 2004, 2006; Bodmer and Bonilla 2008; Ji et al. 2008). Recently, “deep sequencing” has been enabled by “next-generation” technologies that reduce sequencing costs by several orders of magnitude (Shendure and Ji 2008). However, it is still prohibitively expensive to sequence whole human genomes, particularly when sample sizes are large. Thus, multiplexed targeted amplification of many genomic regions of interest is crucial for rapid and cost-effective sequencing-based research projects.
Parallel targeted amplification of selected genome regions is a challenging task (Garber 2008). Two different categories of methods have been developed to enrich or capture desired genomic regions, such as exons. One category employs hybridization of sheared genomic DNA to probes complementary to targeted regions. The probes can be oligonucleotides on a microarray surface (Albert et al. 2007; Hodges et al. 2007; Okou et al. 2007) or in solution (Gnirke et al. 2009). Although most of the desired regions are captured, the specificity of enriched genomic DNA tends to be limited due to “off target” and “near target” capture. In addition, due to the low efficiency of hybridization on the surface of a microarray, large amounts of genomic DNA are needed. The other category of methods requires hybridization in regions flanking both sides of the target and subsequent circularization of the targets. One way is to use “selector” oligonucleotides to guide the directed circularization of the target sequences digested with restriction enzymes (Dahl et al. 2005, 2007). Another method, which is independent of the presence of flanking restriction enzyme sites and thus is more flexible, applies padlock (molecular inversion) probes that anchor targeted regions and are circularized after polymerization and ligation (Hardenbol et al. 2003; Porreca et al. 2007). In our initial study, we targeted 55,000 exons but observed only ∼10,000 unique sites in over 2 million end-sequencing reads, and most heterozygous loci were called incorrectly (Porreca et al. 2007), indicating the need for substantial improvement in capturing efficiency.
In the human genome, CpG dinucleotides are about fivefold less abundant than expected by chance (Sved and Bird 1990). This is due to the widespread methylation of cytosine in CpG and the deamination of 5-methylcytosine to thymidine (Wang et al. 1982); CpG is thus frequently mutated to TpG (or CpA on the complementary DNA strand). Overall, CpG elevates the mutation rate for transitions by 14- to 15-fold and for transversions by three- to fourfold (Kondrashov 2003; Hwang and Green 2004; Schmidt et al. 2008).
Mutations within the CpG context are a predominant cause of human diseases. At least one-third of mutations implicated in Mendelian diseases originated within CpG contexts (Cooper and Youssoufian 1988; Cooper and Krawczak 1993). Although CpG sites are depleted in the bulk of noncoding human DNA, they are selectively maintained in protein-coding genes and other functional genomic regions despite the elevated mutation rate (Subramanian and Kumar 2003; Kondrashov et al. 2006). Thus, the prevalence of CpG-induced mutations among disease mutations is much greater than among all mutations.
CpG context plays an important role in somatic mutations involved in human cancer. In the TP53 gene, which is mutated in >50% of all human tumors, ∼30% of all mutations occur at CpG dinucleotides, and all five major mutation hotspots are found at CpGs (Olivier et al. 2002). Recently, sequencing of nearly all protein-coding regions in cancer genomes revealed that 17%, 38%, 43%, and 48% of point mutations occur at CpGs in breast, pancreatic, brain, and colorectal cancers, respectively (Sjoblom et al. 2006; Jones et al. 2008; Parsons et al. 2008).
In this work, we describe improvements to our padlock capturing technology that yield an estimated 10,000-fold increase in capture efficiency over previous reports and significant improvements in sensitivity and uniformity. We designed 53,777 padlock probes to cover ∼24% of all CpGs on human chromosome 21, where each probe captured a 40-bp region containing at least one CpG, and applied them to discover genetic variants in the genomic DNA from one HapMap CEPH individual (NA10835) and five volunteers from the Personal Genome Project (PGP; http://www.personalgenomes.org). We report the improved performance and high reproducibility of our optimized methods, and demonstrate the utility of the data for identification of known and novel SNPs and unbiased analysis of CpG variation rates.
Results
Site selection, probe design, synthesis, and processing
We designed 53,777 padlock probes to capture CpGs on human chromosome 21. The probes were designed to maximize the number of CpGs in a nonoverlapping manner. Each probe flanks a 40-bp gapped target containing at least one CpG, with the entire probe set covering 90,158 CpGs (24%) of 383,729 total on chromosome 21. The gap size of 40 bp was chosen to simplify sequencing library construction, as this size is comparable to the read length of most next-generation technologies. The two anchoring sequences adjacent to the 40-bp targets are termed the extension and ligation arms. The extension arm is the anchoring sequence from which polymerization initiates, and the ligation arm is the sequence to which the polymerized DNA is ligated. In the gap, one CpG (the targeted CpG) was located right next to the ligation junction. Successful circularization of padlock probes depends on the fidelity of both polymerase and ligase. The melting temperature (Tm) of extension arms is generally 5°C lower than the Tm of ligation arms (Supplemental Fig. 1). This design may stabilize the ligation arm and minimizes polymerase displacement of the ligation arm at the ligation junction (Akhras et al. 2007). We also took measures to ensure the uniqueness of the arms by avoiding repetitive regions (see Methods for details).
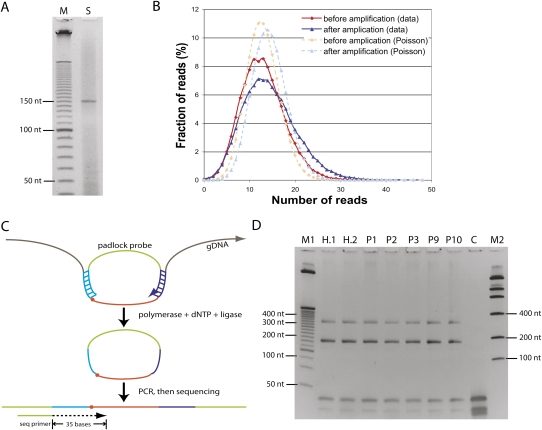
The padlock probes, flanked by amplifiable primers, were synthesized as 150-nucleotide (nt) oligonucleotides on and then released from a programmable microarray solid support. The padlock probe precursor oligonucleotides appeared as a single band of the desired size on a denaturing gel (Fig. 1A). To minimize PCR amplification bias while producing ample amounts of products, we performed two rounds of PCR with about 10 cycles each (see Methods). We sequenced the precursors before and after two rounds of amplification on an Illumina Genome Analyzer and found the number of reads per site was not very different from the Poisson distribution (Fig. 1B), suggesting that probe precursors were roughly evenly distributed both before and even after two rounds of PCR amplification. This result indicates that a small initial quantity of padlock precursors can be faithfully amplified to generate very large quantities of padlock probes, which greatly reduces the probe cost for analyzing large numbers of samples. In contrast to our previous method of using nicking enzymes whose recognition sites cannot be present on the capture arms (Porreca et al. 2007), we developed a novel approach that allows more flexibility in arm selection to generate single-stranded padlock probes (see Methods).
Figure 1.
Padlock probe capture of 53,777 CpG sites. (A) The raw probe precursor (150-mer) sample from Agilent (S) was loaded along with a 10-bp ladder (M) on a 6% denaturing PAGE gel. (B) The probe precursors before and after two rounds of PCR amplification were end-sequenced by Illumina Genome Analyzer. (C) The padlock probes were hybridized to the targeted genomic CpG sites with a uniform 40-nt size. To simplify library construction, a target CpG (dot) was located immediately next to the ligation arm of the probe. Enzymatic filling and ligation of the gap (brown) allowed a copy of the target site to form a circle with the padlock probe. The circles were then PCR amplified using the backbone sequences (green) as primers. The common backbone sequence immediately upstream of the ligation arm served as a sequencing primer. (D) Amplification of circles derived from padlock probes. PCR products were loaded on a 6% PAGE gel. The two upper DNA bands had the expected amplicon sizes: 184 bp (subject to gel purification and Illumina sequencing) and 334 bp (if polymerization extended around the circle twice); the lower bands below 50 nt were derived from PCR primers. (Lane M1) 25-bp DNA ladder (Invitrogen); (lanes H.1,H.2) technical replicates of HapMap sample NA10835; (lanes P1,P2,P3,P9,P10) Personal Genomes 1, 2, 3, 9, and 10, respectively; (lane C) no genomic DNA control; (lane M2) low mass DNA ladder (Invitrogen).
Improvement of padlock capturing
We previously reported an attempt to capture 55,000 exons in the human genome using padlock probes (Porreca et al. 2007). However, the capturing efficiency appeared to be very low—as a consequence, only ∼20% of the targeted sites could be detected, and most of the heterozygous SNPs were incorrectly called as homozygous. After experimentation aimed at increasing the efficiency, we identified three factors that, together with better probe design and synthesis, yielded substantial improvements: (1) The amount of time allowed for hybridization reactions between genomic DNA and padlock probes must be extended; (2) increased amounts of reactants are required to ensure adequate generation of circles—especially, increased amounts of padlock probes are required for reactions that use only low amounts of genomic DNA (0.5–1 μg); and (3) dNTP concentrations must be carefully adjusted to possibly minimize the strand displacement activity of the polymerase.
We used a systematic approach to optimize padlock capture. To quantitate the capturing efficiency, we developed a simple SYBR Green-based real-time PCR assay to measure the number of circularized padlock probes. We define 100% capturing efficiency as the condition in which one padlock probe is circularized at each genomic copy of a target locus. When we hybridized 1 μg of human genomic DNA (0.5 amol or ∼300,000 copies of haploid genomes) and 10× excess of probes at each target site at 60°C for up to 1 h, the capturing efficiency is ∼0.0025%. This translates to an average of 7.5 circles formed per site (0.0025% × 300,000 = 7.5). When we used the previous 55k exon set (Porreca et al. 2007), the efficiency was only 0.0001% (an average of 0.3 circles per site). The 25-fold improvement in this work, even under the same reaction conditions, was clearly due to better probe design and synthesis that led to higher and more uniform hybridization efficiency. An additional ∼10-fold increase was obtained by extending the reaction time from 1 h to at least 24 h (Fig. 2, left panel). We then explored further conditions with larger amounts of padlock probes. When we increased the amount of probes to 50×, 100×, and 250× excess, the capturing efficiency climbed to as high as 0.6% (an average of 1800 circles per site) and did not yet seem to saturate (Fig. 2, middle panel).
Figure 2.
Improvement of padlock capturing efficiency with longer hybridization time (left), more probes (middle), and appropriate dNTP concentration (right). The ratios (10:1, 50:1, 100:1, and 250:1) are molar ratios between each of the padlock probes and genomes. dNTP (1×) is defined as the minimum amount of dNTP needed to capture all genomic copies at each target region. The fold improvement (vertical axis at right) is relative to the reaction with 10:1 probe ratio, hybridized for 15 min at 60°C, and with 100× dNTP. Similar results were observed in independent experiments.
In addition to the complex hybridization reaction of the human genome and thousands of probes in a single tube, another challenge is to assure efficient circularization through polymerization and ligation. After the polymerase finishes filling the gap, it has to dissociate from the DNA to enable the ligase to close the gap. We used the Stoffel fragment of the AmpliTaq DNA polymerase (Applied Biosystems) that lacks 5′→3′ exonuclease activity. Fewer circles were formed when 10× more or 10× less amount of Stoffel was used (data not shown). We further tested the effect of dNTP concentration on the polymerase strand displacement ability, and found that there seemed to be a narrow range of dNTP concentration that gave rise to the highest circularization efficiency (Fig. 2, right panel). With the optimal dNTP concentration along with at least 24 h hybridization and a probe to genome molar ratio of 100:1, we were able to form padlock circles from >1% of genomic DNA copies (i.e., >3000 circles) at each target site on average, which should virtually guarantee detection of each allele of a diploid DNA locus. This is a combined 435-fold improvement compared with the least optimal reaction condition using the optimized probe set (Fig. 2). When the additional 25-fold increase due to refined probe design and synthesis is factored in, we achieved a total of >10,000-fold improvement after the optimization. This suggests that even smaller amounts of genomic DNA can be analyzed successfully in cases where input sample DNA is limiting, e.g., when DNA is from tumor samples or microdissected tissue: 10 ng of genomic DNA would generate an average of more than 30 circles per site, which is sufficient for seeing at least three copies of each allele with a false-negative rate of 10−5.
Performance of improved padlock capturing
Single-end sequencing of the ligation arm (25 bp) and 10 bases of the adjacent polymerized extension region (35 bp total) were obtained with an Illumina Genome Analyzer 1 (Fig. 1C,D) (“run 1”); subsequently, for reasons described below (and in the Supplemental Text), we resequenced these libraries on an updated Illumina Genome Analyzer 2 (“run 2”). The 10 bases of the polymerized extension region (which we called the “Target10” region) are the only parts of each sequence read that are copied from the subject genomes versus synthesized as parts of the padlock probes. The Target10 region thus comprises read positions 26–35, with each probe's target CpG occupying positions 26–27. Although there is an option to trim the ligation arm with the Type IIS restriction enzyme EcoP15I built into the probe design and then ligate with sequencing adapters, we chose the simple end-sequencing approach (Fig. 1C), which focuses on the CpGs at or near the ligation junction by design and that fall within the sequencing read length. Filtered, mappable read counts for each subject library ranged from 1.5 M to 3.8 M in run 1, and from 2.3 M to 4.8 M in run 2 (Supplemental Table 1).
We evaluated our improved padlock method with four metrics: sensitivity, uniformity, reproducibility, and the accuracy of the genotypes.
Sensitivity: The sensitivity of our multiplexed capture, which is related to multiplexity, can be assessed by the fraction of loci that are covered by at least one mapped read. With 2–3 million mapped reads, the sensitivity ranged from 90.8% to 94.0% (Fig. 3A; Supplemental Fig. 3). Each library offered the potential to resequence 537,770 Target10 base positions for each subject, and yielded an average of 346,749 (64.5%) Target10 base position allele determinations per library (replicates uncombined, Supplemental Table 1) due to residual probe circularization limitations, sequencing limitations, and filters used to ensure genotyping accuracy.
Uniformity: For both sequencing runs over all samples, 54.4%–56.5% of all captured targets had coverage levels within a 10-fold range, and 87.2%–92.7% had coverages within a 100-fold range (Fig. 3A; Supplemental Fig. 4).
Reproducibility: We conducted a technical replicate of our capturing experiment for HapMap sample NA10835 using our improved protocols. Read counts between the NA10835 replicates were correlated at >0.98 within sequencing runs (Fig. 3B; Supplemental Fig. 6), and at >0.95 across sequencing runs (see Supplemental Text). High reproducibility across the genotypes determined from the NA10835 replicates was also observed within the individual sequencing runs (>99.8% agreement in genotypes and >0.92 Spearman's rank correlation between genotype scores; see Supplemental Text).
Accuracy: Details are described in depth in following sections. Furthermore, to identify characteristics that may lead to further improvement of capture efficiency, we analyzed the relationship between coverage (i.e., the number of reads per site) and sequence features inherent in the probe design (Supplemental Fig. 7).
Figure 3.
Improved performance of padlock technology. (A) Uniformity of target sites. For each sample, log-normalized coverage levels from sequencing of padlock probe reaction products were computed for each captured target as the log10 of the number of target-mapped, filtered reads divided by the total number of mapped, filtered reads from the reaction. Targets were then ranked for each sample from highest to lowest numbers of mapped, filtered reads and plotted. Except at the extremes, curves exhibit a gradually decreasing slope, indicating that a large number of targets have coverage levels within two orders of magnitude. The plot above depicts sequencing run 1; sequencing run 2 is very similar (Supplemental Fig. 4). For both sequencing runs, overall samples, 54.4%–56.5% of all captured targets had coverage levels within a 10-fold range, and 87.2%–92.7% had coverage within a 100-fold range. (B) Reproducibility of padlock capture. Scatter plot of read coverage of the technical replicate libraries sequenced for NA10835. Pearson correlation coefficients (R) between read counts are provided for all 53,777 target sites (all), all target sites for which one of the replicates has nonzero coverage (one ≠0), and all for which both replicates have nonzero coverage (both ≠0). All Pearson correlation coefficients are >98.1%. The scatter plot is presented on a log–log scale and therefore only contains points corresponding to targets in the “both ≠0” set. The plot above depicts sequencing run 1; sequencing run 2 is very similar (Supplemental Fig. 6). For details on sequencing runs and read mapping and filtering, see text and Supplemental Text.
Genotype determination and spurious sequencing artifact
We determined genotypes for Target10 sites using an algorithm that makes use of Illumina base call quality scores, base-specific base call error rates, and Bayesian prior probabilities for the likelihood of a locus being a site of variation (see Methods and Supplemental Text). The algorithm delivers not only a genotype call for each site but also a genotype score indicating the confidence of the call. We generally used a genotype score of 5 as a cutoff, which represents a 1 × 10−5 chance that the genotype was called in error given the algorithm's error model. We optimized genotype calling accuracy using four measures, including the heterozygosity of Target10 loci by read position, CpG polymorphic allele fractions by read position, genotype call concordance with independent SNP data for the subjects, and profiles of dbSNP and non-dbSNP sites of variation (findings below and in Supplemental Text). Using this latter measure in our initial work with run 1, we found a consistently and apparently anomalously high degree of sharing of some heterozygous genotypes across subjects. Using conventional Sanger sequencing, we sequenced five dbSNP and 10 non-dbSNP highly shared heterozygous sites in all six subjects. The highly shared dbSNP sites were all validated, but none of the non-dbSNP sites were correctly called in any of the six subjects. Ultimately, we resequenced the same libraries used for run 1 but found that the anomaly persisted with run 2. However, the anomalously shared loci were distinct in the two runs, and the anomaly disappeared when we considered only those loci that were called with the same genotype in both runs (the “intersection” of the runs) (See Supplemental Text for extensive discussion and Supplemental Fig. 15 for examples). Since the libraries and computational processing were identical in all other respects, these observations suggest that incorrect reads for small subsets of probes may be generated within the sequencing process itself by an as yet unidentified mechanism. On this basis, we performed all analysis of genotypes based on the intersection, which comprises 442,937 Target10 loci genotyped in at least one subject sample.
Genotype validation
Of all dbSNP loci annotated to be in our Target10 regions (5059), we verified Target10 location matches of 5008 by aligning Target10 reference sequences against dbSNP sequences with stringent parameters (Supplemental Text; Supplemental Table 2). The number of these dbSNP loci that could be genotyped in an individual subject depended strongly on coverage (R2 = 0.885; Supplemental Fig. 8). When we compared the bases actually found at these sites with previously observed alleles annotated for them in dbSNP, we found consistency rates >99.5% for all subjects, with statistically significantly greater consistency for dbSNP loci annotated as having been validated compared with dbSNP loci annotated with “unknown” validation (Supplemental Fig. 9). Additionally, among these sites, 2025 had been genotyped by the HapMap project for NA10835 (Frazer et al. 2007) and 217–245 had been genotyped independently on the Affymetrix SNP 500K platform for the other five Personal Genome Project subjects. Between 98.5% and 100% of these independently assessed genotypes matched our Target10 genotypes (Supplemental Table 3). For those genotypes assessed as heterozygous by the independent data source, the fraction of Target10 genotypes that did not match the independent genotypes was 0%–3%. These results confirm the accuracy of our padlock capture library sequencing and computational methods for genotyping.
Candidate novel SNPs
We identified 489 loci within the Target10 regions that were heterozygous in at least one subject and were not among our 5008 location-verified dbSNP loci. There were also 13 loci that were homozygous in each subject but for which the homozygous genotypes differed. These 502 loci represent candidate novel SNPs (Supplemental Table 4). Three hundred fifty-four of the heterozygous sites (71%) and eight of the 13 homozygous but differing loci (62%) are at the target CpG positions (26 and 27). Subject PGP10 is heterozygous at 215 non-dbSNP positions, a much higher number than the other subjects, which range from 65 to 87, depending on coverage (Supplemental Fig. 8). Taking the coverage relationship into account, PGP10 exhibits about three times the number of non-dbSNP heterozygous loci than the other subjects would have at PGP10's coverage level. PGP10 also exhibits less sharing of heterozygous genotypes compared with the other subjects (Supplemental Text; Supplemental Fig. 11). This and other findings below may relate to PGP10's African-American (AA) ancestry versus the European-American (EA) ancestry of the other five subjects, as the higher degree of genetic variation in AA populations may present SNPs uncommon in EA populations.
Biases in target capture
Target capture by padlock probes involves different enzyme behavior at the ligation junction than at other gap positions, while Illumina sequence quality can vary with read position (Supplemental Fig. 5). Both phenomena can potentially introduce biases into sequence data by read position. To assess for the presence of such effects, we analyzed heterozygosity (nucleotide diversity) by Target10 read position, and also compared the frequencies of CpG polymorphisms measured on an allele basis between target and nontarget positions. We found evidence of biases toward increased polymorphism rates in target CpG positions, especially in position 27 (see Discussion and Supplemental material). However, the size of the bias is small compared with the magnitude of the CpG polymorphism overall. For instance, the variation in heterozygosity between positions 26 and 27 is only ∼1.25-fold compared with a ninefold change between these positions and 28–35 (Supplemental Table 5), while the difference between CpG polymorphic allele fractions across all positions is ∼1.31-fold compared with a 12.5-fold difference between CpG and non-CpG allele fractions (see Supplemental material). Heterozygosity is expected to be higher in positions 26–27 than 28–35 because of the high mutability of CpG dinucleotides, while positions 28–35 should be close to overall chromosome 21 averages. In fact, the average heterozygosity in positions 28–35 is 0.00067, slightly higher than the 0.00052 value found for chromosome 21 by the HapMap project (Sachidanandam et al. 2001). A higher value for positions 28–35 is expected because Target10 regions are often in CpG islands, so that these positions have a high frequency of CpGs compared with chromosome 21 as a whole, even though they are not CpG target positions (see Supplemental material). Meanwhile, the overall 12.5-fold difference between CpG polymorphic allele fractions and non-CpG allele fractions is close to estimates of CpG versus non-CpG mutation rates (see below).
CpG variation rates and forces influencing CpG and non-CpG variation
To estimate the impact of CpG context on mutation rate, we determined the ancestral state of each SNP using the chimpanzee genome. Comparison of densities of CpG and non-CpG SNPs in our padlock probes suggests that the rate of transitions originated at CpG sites is 13.7 times higher than the rate of non-CpG transitions. This estimate is in good agreement with previous studies (Kondrashov 2003; Hwang and Green 2004; Schmidt et al. 2008). This is a slight underestimate of the impact of CpGs on mutation rate because mutations in the chimpanzee lineage reduce the estimate of the number of CpG dinucleotides in the ancestral sequence.
Our estimates are unlikely to be biased toward various genomic features as CpG sites surveyed were chosen to be a large representative subset of all chromosome 21 CpG sites irrespective of known annotations. Correspondingly, <5% of CpG sites surveyed were located in known protein-coding regions, and only 20 SNPs there were detected.
In comparison with intergenic regions, CpG transition density per site was 15% lower in intronic regions and 3.2 times lower in coding regions. As expected within coding regions, SNP density per site was higher at fourfold degenerate sites than at nondegenerate sites. We observed one nonsense CpG transition (arginine to stop codon). It corresponded to the known rs4148974 SNP present in a facultative exon of the NADH dehydrogenase (ubiquinone) flavoprotein 3 gene (NDUFV3).
We also applied these data to ongoing analyses into the mechanisms of mutation rate and the basis of mutation rate heterogeneity. Mutation rate in mammals is known to be heterogeneous at a megabase scale (Wolfe et al. 1989; Smith and Lercher 2002; Gaffney and Keightley 2005). Various factors may contribute to mutation rate heterogeneity. Local CpG methylation level would affect the rate of CpG mutations and would leave the rate of non-CpG mutations unaffected. Fidelity of DNA replication and efficiency of the mismatch repair (MMR) system would primarily impact non-CpG mutation rate because mutations arising from deamination of methylcytosines escape the MMR system. Other factors, such as DNA exposure to damage and efficiency of base excision repair (BER) would impact both CpG and non-CpG mutation rate. Thus, comparison of CpG and non-CpG SNP densities along the chromosome may be informative about relative contributions of forces influencing mutation rate.
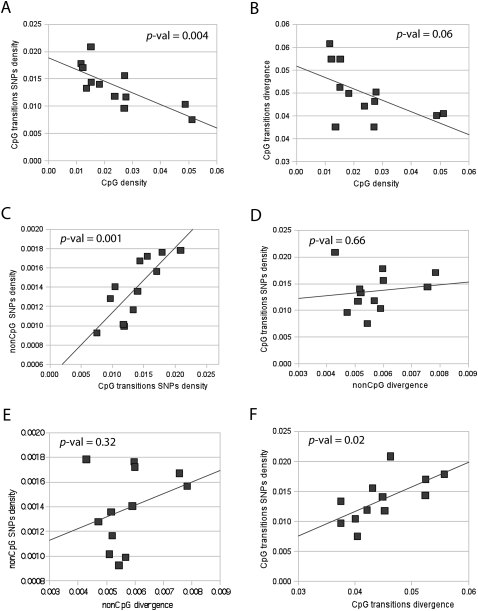
To analyze changes in the variation rate along the chromosome 21, we pooled CpG transitions and all non-CpG variants into 3-Mb windows. We observed a negative correlation between CpG content (fraction of CpG dinucleotides in a window) and the CpG polymorphism density per CpG site (Fig. 4A). Divergence with chimpanzee shows a similar but weaker effect (Fig. 4B). This suggests that the CpG mutation rate is heterogeneous and CpG dinucleotides are preserved in regions of lower mutation rate. An alternative (although less likely) explanation may be provided by natural selection maintaining hypermutable contexts.
Figure 4.
Correlations between polymorphism, interspecies divergence, and CpG content. We analyzed divergence in the chimpanzee lineage after divergence from human using orangutan as an outgroup in order to compensate for bias due to padlock probe design based on the presence of CpGs in the human sequence. SNP densities were calculated as normalized densities per site of a specific type. To calculate the density of CpG SNPs, we divided the total number of the observed CpG polymorphisms in the region by the combined length of all surveyed CpG nucleotides in the region. Correspondingly, to calculate the density of non-CpG SNPs, we divided the total number of observed non-CpG polymorphisms in the region by the combined length of all surveyed non-CpG nucleotides in the region. (A) Correlation between densities of SNPs originated as transitions in CpG sites and fraction of CpGs in the region. (B) Correlation between substitutions due to CpG transitions in the chimpanzee lineage after divergence with humans and fraction of CpGs in the region in the human genome. (C) Correlation between densities of SNPs originated as transitions in CpG sites and non-CpG SNP density. (D) Correlation between densities of SNPs originated as transitions in CpG sites with non-CpG divergence in the chimpanzee lineage after split with humans. (E) Correlation between non-CpG SNP density with non-CpG divergence in the chimpanzee lineage after split with humans. (F) Correlation between densities of SNPs originated as transitions in CpG sites with divergence in the chimpanzee lineage due to CpG transitions.
The observed heterogeneity of CpG variation rate (and consequently the heterogeneity of the CpG content) along the chromosome could parallel the overall pattern of variation, or it could be CpG-specific. We find that CpG and non-CpG SNP densities are highly correlated (Fig. 4C). However, this correlation is expected to be strong because many factors unrelated to mutation rates impact both CpG and non-CpG variation. These factors include variation in coalescent times influenced by historic changes in human population size and population structure (Marth et al. 2003), background selection (Charlesworth et al. 1993), hitchhiking effect (Smith and Haigh 1974), and biased gene conversion (Webster and Smith 2004). To exclude the effect of these population factors, we analyzed the dependency of the CpG polymorphism rate on species divergence in CpG and non-CpG sites. SNP densities in both CpG and non-CpG sites do not display a statistically significant correlation with non-CpG divergence in the chimpanzee lineage in our data set (Fig. 4D,E). This correlation would reflect the influence of mutation rate components common to CpG and non-CpG mutations, such as exposure of DNA to damage and the efficiency of the BER system (Stamatoyannopoulos et al. 2009). It may also reflect the effect of natural selection. Contributions of these forces to the megabase-scale heterogeneity of the human polymorphism rate appear limited, at least in our sparse data set. The density of SNPs originated in CpG sites shows a stronger significant correlation with CpG divergence in the chimpanzee lineage (Fig. 4F). This suggests that local CpG mutation rate is primarily influenced by factors specific to CpG dinucleotides. We hypothesize that heterogeneity of the methylation rate along the chromosome may underlie the heterogeneity of the CpG mutation rate.
A more precise analysis would be possible with larger targeted CpG data sets than these relatively sparse chromosome 21-based data. However, the results above were confirmed using species divergence and population variation data in the genomic regions included in phase I of the ENCODE project (data not shown). In these regions, correlation of SNP densities in both CpG and non-CpG sites with non-CpG divergence appears statistically significant, but still remains much weaker than correlation between diversity and divergence in CpG sites.
Discussion
The cost of DNA sequencing has been continuously dropping in the past several years toward the goal of sequencing a human genome at $1000. However, regardless of the ever-lowering cost of DNA sequencing, it is always more cost-effective to selectively sequence regions of interest, thus allowing more samples to be analyzed. Until the cost of the target capture overwhelms the sequencing cost, targeted sequencing remains a viable and highly demanded approach in genetic studies and diagnosis.
In this work, we improved the padlock-based capturing method significantly by refining the probe design algorithm and probe synthesis protocol, extending the hybridization time, increasing the amount of the probes, and tuning the dNTP concentration. We applied this improved technology to amplify 53,777 regions of 40 bp containing CpGs across human chromosome 21. A uniform gap size was chosen for the simplicity of sequencing library construction; new protocols developed herein have also successfully applied to probe sets with various target sizes, such as the exon set (JB Li, K Zhang, and GM Church, unpubl.).
The capture efficiency has increased >10,000-fold compared with our previous report (Porreca et al. 2007), where <20% of 55,000 exons were amplified and most of the heterozygous SNPs were erroneously called as homozygous. We are now able to observe over 50,000 sites of 53,777 desired targets with ∼3 million reads from a single lane of the Illumina Genome Analyzer. The uniformity of different targets was also significantly improved; ∼90% and ∼55% of all targets were within a 100- and 10-fold range of each other, respectively. The accuracy of genotyping calls was estimated to be 98.5% for NA10835 in comparison with HapMap data, and 99.2%–100% accurate for the other subjects with PGP Affymetrix 500K SNP data. The modified experimental protocol mainly accounted for the significant improvement. In addition, these metrics were achieved with a series of constraints in the probe design. Lastly, steady improvement of our oligonucleotide synthesis technology (Agilent) enabled us to start with roughly equal amounts of probe precursors (Fig. 1B).
Compared with the microarray-based “on surface” hybridization approach to enrich targets (Albert et al. 2007; Hodges et al. 2007; Okou et al. 2007), our significantly improved padlock-based “in solution” technology has led to (1) efficient hybridization as >1% of genomic copies successfully form circles at each locus, (2) a requirement of ∼20-fold less input genomic DNA, and (3) better scalability of the reactions, particularly when the sample size is large. Although a recently developed method based on “in solution” hybridization has solved these problems (Gnirke et al. 2009), the close to 100% specificity of the padlock approach is unmatched since two linked probing sequences, rather than one, need to be precisely hybridized to form circles (Porreca et al. 2007). However, padlock-based target capture technology is subject to biases in circle formation due to the enzymatic requirements of the reaction. With the new protocols developed in this work, the uniformity of distribution among different target regions has been significantly improved. For example, the fraction of sites within 100-fold in abundance has increased from 16% to ∼90%.
The uniformity could be further improved based on what we learned in this work. First, selection of extension and ligation arms could be more flexible in a wider window flanking the target. This would make it possible to design probes for difficult regions or probes that better satisfy the design criteria. Additionally, we observed that the first base on the ligation arm of the padlock probe (proximal end to the target gap) contributed to the success of circle formation, and the G+C content of targets led to amplification bias (Supplemental Fig. 7). These pitfalls could be avoided or alleviated by the flexibility in probe design. In addition, longer padlock probes may further improve the uniformity as demonstrated in a recent smaller scale study (Krishnakumar et al. 2008). Lastly, due to the fact that the abundance of reads per site spans a three-log magnitude range and this difference in abundance is systematic rather than random (Fig. 3), the probes could be divided into three or more subsets so that the sites would fall into a 10-fold range in each subset (Deng et al. 2009).
Our improved padlock-based capture technology can be extended to a variety of applications. For example, one can efficiently capture genomic regions, such as SNPs, exons, and contiguous regions containing susceptible mutations from gene mapping studies. Such tools are greatly demanded in projects including personal genomes, cancer genomes, and genome-wide association follow-up studies. In addition, the cDNA derived from RNA can also be targeted to quantitatively measure allele frequencies in gene expression (Zhang et al. 2009) and to identify RNA editing events (Li et al. 2009). Because of the superior specificity of the padlocks, we recently applied them to bisulfite converted human genomes to profile cytosine methylation (Ball et al. 2009; Deng et al. 2009).
We showcased the utility of data obtained using our padlock probes by accurately estimating the CpG mutation rate as at least 13.7 times the non-CpG mutation rate in humans, using the chimpanzee genome to identify ancestral CpGs. A simple quantitative analysis of the data obtained using padlock probes suggests that CpG polymorphism density is highly variable along the chromosome. Comparison of human polymorphism and species divergence in CpG versus non-CpG sites demonstrates that this heterogeneity is primarily due to factors specific to CpG dinucleotides. This suggests that variation in methylation rate may be an important determinant of local mutation rate in humans. This analysis does not exclude the alternative hypothesis that hypermutable CpG contexts are preserved by purifying selection.
We believe that the inexpensive and rapid analysis of CpG mutations has a potential for a number of applications in human genetics. First, genetic diagnostics of autosomal dominant genetic disorders and discovery of genes involved in developmental defects requires detection of de novo mutation events. Highly accurate sequencing of the complete genome or even “exome” is likely to remain prohibitively expensive in coming years. At the same time, a much less expensive screening of CpG mutations will have more than one-third of a chance to find the relevant de novo sequence change at a small fraction of the cost (Cooper and Youssoufian 1988; Cooper and Krawczak 1993). Therefore, CpG screening can be used as an efficient first pass for detecting de novo mutations. Second, rare alleles involved in human complex phenotypes can be detected by systematic resequencing of phenotyped populations. CpG screening may provide a more efficient study design because it will detect a large fraction of rare alleles at a small fraction of the cost. We expect to carry this study design forward in larger phenotyped populations such as those from Genome-Wide Association Studies and the Personal Genome Project.
Methods
Selection of 53,777 CpG loci across human chr21
Human genomic sequences (hg18) were downloaded from the UCSC Genome Browser (http://genome.ucsc.edu). The total number of CpGs on human chr21 is 371,971. We started with 223,058 nonoverlapping probes that could possibly cover the maximum number of CpGs on human chromosome 21, and 101,822 of them were nonrepetitive, which made probe design possible. We designed 53,777 probes that can be accommodated by the capacity of oligonucleotide synthesis on a single microarray, with the following stringent considerations: (1) the CGs did not fall into the repetitive regions; (2) the Tm of the extension arm ranges 50°C–58°C, with 53°C being optimal; the Tm of the ligation arm ranges 53°C–61°C, with 58°C being optimal; (3) the length of extension and ligation arms ranges from 17 to 25 nt; (4) the arms were compared against a pre-computed occurrence table of all 12-mers in the human genome. The sum of the 12-mer occurrences in the arms was normalized by the arm length. Only arms with a normalized 12-mer count less than 2000 were retained; (5) the pair of extension and ligation arms separated by a gap was BLASTed against the human genome. When both arms matched a second location in the genome, the probe was discarded; and (6) the GC content of both arms was 30%–70%.
Improved padlock capturing protocol
Generation of padlock probes
Using a programmable microarray (Agilent Technologies), we synthesized 53,777 oligos (150-mers), cleaved them off the microarray, and collected them in a single Eppendorf tube. Each of the oligo species is ∼0.2 fmol, totaling ∼10 pmol of oligos synthesized on one array. The sequence of the 150-mer oligo is ATCAAGCCGAAGACAGTGTT[ligation_arm][random_ligation]TCTCTGCTGCTTCAGCTTCCCAGTCGTGGTACATACGAGCGATATCCGACGGTAGTGTAC[random_extension][extension_arm]GATCCAGGAAATTCGCGCTA. Random sequences may be added to extend ligation and extension arms to 25 bases for uniform probe size.
A 100-μL PCR reaction was assembled with 90 μL of Platinum Taq supermix (Invitrogen), 50 pmol each of forward primer, A*T*C*AAGCCGAAGACAGTGT/deoxyUridine/ (* denotes phosphothiorate bond), and reverse primer, /5phos/TAGCGCGAATTTCCTGGATC (both from IDT), 0.5× SYBR Green (Invitrogen), and 10–100 fmol of template. The quantitative PCR program was 5 min at 95°C; nine to 15 cycles of 30 sec at 95°C, 1 min at 58°C, and 1 min at 72°C; and 5 min at 72°C. We first used 1% (∼100 fmol) of oligos provided by Agilent in a single 100-μL reaction with nine cycles. Approximately 10 fmol of the purified PCR product was used as template in each of the 5 × 96 100-μL reactions with 12–15 cycles in the second round of PCR. The PCR product was purified with a QIAquick PCR purification kit (Qiagen).
Sixteen tubes of 100-μL reactions with 1× λ exonuclease buffer, 5.6 μg of PCR products, and 25 units of λ exonuclease were incubated for 1 h at 37°C, then 15 min at 75°C. The reaction was purified with a QIAquick PCR purification kit, eluted with 400 μL of dH2O, and quantified with a Nanodrop (Thermo Scientific) at 45 ng/μL. Eight tubes of 60-μL reactions with 2.25 μg of λ exonuclease-treated single-stranded DNA, 1× DpnII reaction buffer (NEB), and 200 pmol of Guide_DpnII (GCGCGAATTTCCTGGATCNN) were denatured for 5 min at 95°C, ramped to 60°C at 0.1°C/sec, and incubated for 10 min at 60°C and 1 min at 37°C. For each of the 60-μL reactions, we then added 50 units of DpnII (NEB) and five units of USER (NEB), and incubated the reaction for 3 h at 37°C. The post-processing 110-mer padlock probes were size selected on 6% denaturing acrylmide gels (Invitrogen) and eluted in 100 μL of dH2O. The total concentration of the 110-mer padlock probes was estimated to be 16 ng/μL (441 nM) by a denaturing acrylmide gel.
Hybridization
The genomic DNAs of the HapMap sample, NA10835, and five Personal Genome Project (PGP) samples (NA20431, NA21070, NA21660, NA21781, and NA21833) were obtained from Coriell. In a 15-μL reaction, 1× Ampligase buffer (Epicentre), 500 ng (0.25 amol) of genomic DNA, and 48 ng (1.32 pmol) of probes were mixed (each probe to gDNA molar ratio = 100:1; numbers change accordingly for other ratios), denatured for 10 min at 95°C, ramped at 0.1°C/sec to 60°C, and then hybridized for 24 h at 60°C. We then added 2 μL of gap filling and sealing mix (5.4 μM dNTPs [100×, numbers change accordingly for 1×, 10×, 1000×, and 10,000×], two units of Taq Stoffel fragment [Applied Biosystems], and 2.5 units of Ampligase [Epicentre] in Ampligase storage buffer [Epicentre]), and incubated the reaction for 15 min, 1 h, 1 d, 2 d, or 5 d at 60°C. We also tried cycling the reaction: after 1 d at 60°C, we applied 10 cycles of 2 min at 95°C followed by 2 h at 60°C. To remove the linear DNA, we lowered the incubation temperature to 37°C, immediately added 2 μL of Exonuclease I (20 units/μL) and 2 μL of Exonuclease III (200 units/μL) (both from USB), and incubated the reaction for 2 h at 37°C followed by 5 min at 94°C.
Amplification of captured circles
The circles were amplified by two 100-μL PCR reactions with 50 μL of 2× iQ SYBR Green supermix (Bio-Rad), 10 μL of circle template (from above), and 40 pmol each of forward (CAAGCAGAAGACGGCATACGAGCGATATCCGACGGTAGTGTAC) and reverse (AATGATACGGCGACCACCGACACTAACACGACTGGGAAGCTGAAGCAGCAG) primers (IDT). The PCR program was 3 min at 96°C; three cycles of 30 sec at 95°C, 30 sec at 60°C, and 30 sec at 72°C; and 10 cycles of 30 sec at 95°C, 1 min at 72°C, and 5 min at 72°C. The desired PCR products were gel purified and quantified. For each sample, 10–20 fmol of DNA was sequenced by both Illumina Genome Analyzer version 1 and updated version 2 with the custom primer (CACTAACACGACTGGGAAGCTGAAGCAGCAGAGA).
Illumina sequence processing, genotyping, and SNP identification
Illumina reads were mapped to target sequences by aligning them along their full 35-bp lengths to the 35 bp of human genome reference sequence corresponding to each of the 53,777 targets using in-house built dynamic programming software. Only reads with at most one mismatch or gap compared with a target, and no alignment better than two mismatches away from distinct target sequences, were accepted.
Genotypes were computed using an in-house-developed algorithm that takes into account base-specific base calling error rates determined from mapped reads, base call quality scores, and prior probabilities for genotypes. The algorithm computes a probability P(genotype = xy | Illumina base calls and qualities) for all 10 possible genotypes xy, calls the genotype xy with the highest probability, and assigns the value −log10(1 − P) as a confidence score. This score measures the probability of a miscall given the algorithm's error model. Additional details are provided in the Supplemental Text.
SNPs were identified as loci that were found to be heterozygous in at least one subject, or as homozygous in all subjects that could be genotyped for the locus with at least one subject having a different homozygous genotype than the others. Variants were associated with known dbSNP loci by downloading all dbSNP entries with moltype “single” and locType “exact” in the chromosome 21 ranges occupied by all 53,777 padlock probe Target10 regions by using the UCSC Genome Browser (http://genome.ucsc.edu), and then verifying that the location of the dbSNP SNP mapped precisely to the candidate SNP position (build 129). Additional details on SNP matching can be found in the Supplemental Text.
Illumina reads for both runs 1 and 2 are available from the NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession number SRA007914. All candidate novel SNPs identified in this study, and also all known SNP loci for which a heterozygous genotype or two disagreeing homozygous genotypes could be found among the six subjects, were submitted to dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) where they were assigned accession numbers ss120032891– ss120035117. dbSNP was also provided with individual subject genotypes for all of the known dbSNP and candidate novel SNP loci identified in this study. All dbSNP submissions used handle “CHURCH_CG54K.”
Genotyping accuracy determination
Accuracy was determined primarily by comparing genotypes determined for all subjects with independently available SNP genotype data for subsets of loci that we could identify as assayed in both our and the independent studies. For NA10835, we used 2025 SNP loci genotyped by the HapMap project downloaded from http://ftp.hapmap.org/genotypes/latest/forward/non-redundant/. For the other five subjects, we used 217–246 SNPs that were assayed by Affymetrix SNP 500K arrays for the Personal Genome Project (http://personalgenomes.org). See Supplemental Text for additional details.
Heterozygosity and CpG polymorphic allele fraction determinations
Heterozygosity was computed as the number of heterozygous genotypes divided by the total number of genotypes. CpG polymorphic allele fractions were computed by counting the total number of alleles that could be determined in subject genotypes that correspond to CpGs in the hg18 human genome reference sequence in which Cs appeared as Ts (for CpG→TpG variations), or where Gs appeared as As (for CpG→CpA variations), and dividing by the total numbers of alleles of any sort determined for these CpG positions. Non-CpG polymorphic allele fractions were computed similarly. For additional details, see Supplemental Text.
Calculations of CpG and non-CpG divergence
We used orangutan as an outgroup to determine which nucleotide differences between the human and chimpanzee genomes occurred in the chimpanzee lineage. The human–orangutan alignment (hg18 vs. ponAbe2) was downloaded from the UCSC Genome Browser website (http://genome.ucsc.edu). CpG transition diversity was estimated by dividing the total number of CpG sites conserved between human and orangutan but harboring a transition in chimpanzee sequence by the total number of CpG sites conserved between human and orangutan and alignable with the chimpanzee genome. Non-CpG transitions and transversions were combined in a single class of non-CpG substitutions and their density was calculated in a similar way.
Calculation of CpG and non-CpG SNP densities
CpG transitions SNP density was calculated by dividing the number of reliably detected transitions in CpG nucleotides (with genotype scores of 5 or higher) by the total number of CpG sites among the first 10 probe's positions detected with a genotype score of 5. Non-CpG transitions and transversions were combined in a single class of non-CpG SNPs and their density was calculated in a similar way.
Statistical analysis of correlations
Statistical significance of the observed correlations was analyzed by three methods: (1) by construction of linear regression models with the subsequent application of an F-test, (2) by Spearman's rank-order correlation test, and (3) by Kendall's rank correlation test. All three methods produced very similar P-values.
Acknowledgments
We thank Jay Shendure, Gregory Porreca, and Joseph Chou for input in the padlock technology development; Uri Laserson, Madeleine Price Ball, Michael Chou, Alon Keinan, and Heidi Rehm for discussion and/or critical reading of the manuscript; and the Harvard Research Institute Technology Group for computing resources. Funding came from NHGRI Center of Excellence in Genome Sciences and NHLBI targeted sequencing grants.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to the NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA007914.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.092213.109.
References
- Akhras MS, Unemo M, Thiyagarajan S, Nyren P, Davis RW, Fire AZ, Pourmand N. Connector inversion probe technology: A powerful one-primer multiplex DNA amplification system for numerous scientific applications. PLoS One. 2007;2:e915. doi: 10.1371/journal.pone.0000915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Morgan MT, Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M. Human gene mutation. BIOS Scientific; Oxford, UK: 1993. [Google Scholar]
- Cooper DN, Youssoufian H. The CpG dinucleotide and human genetic disease. Hum Genet. 1988;78:151–155. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- Dahl F, Gullberg M, Stenberg J, Landegren U, Nilsson M. Multiplex amplification enabled by selective circularization of large sets of genomic DNA fragments. Nucleic Acids Res. 2005;33:e71. doi: 10.1093/nar/gni070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl F, Stenberg J, Fredriksson S, Welch K, Zhang M, Nilsson M, Bicknell D, Bodmer WF, Davis RW, Ji H. Multigene amplification and massively parallel sequencing for cancer mutation discovery. Proc Natl Acad Sci. 2007;104:9387–9392. doi: 10.1073/pnas.0702165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park IH, Yu J, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 2009;27:353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney DJ, Keightley PD. The scale of mutational variation in the murid genome. Genome Res. 2005;15:1086–1094. doi: 10.1101/gr.3895005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber K. Fixing the front end. Nat Biotechnol. 2008;26:1101–1104. doi: 10.1038/nbt1008-1101. [DOI] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardenbol P, Baner J, Jain M, Nilsson M, Namsaraev EA, Karlin-Neumann GA, Fakhrai-Rad H, Ronaghi M, Willis TD, Landegren U, et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat Biotechnol. 2003;21:673–678. doi: 10.1038/nbt821. [DOI] [PubMed] [Google Scholar]
- Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, Smith SW, Middle CM, Rodesch MJ, Albert TJ, Hannon GJ, et al. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- Hwang DG, Green P. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc Natl Acad Sci. 2004;101:13994–14001. doi: 10.1073/pnas.0404142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W, Foo JN, O'Roak BJ, Zhao H, Larson MG, Simon DB, Newton-Cheh C, State MW, Levy D, Lifton RP. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40:592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov AS. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Hum Mutat. 2003;21:12–27. doi: 10.1002/humu.10147. [DOI] [PubMed] [Google Scholar]
- Kondrashov FA, Ogurtsov AY, Kondrashov AS. Selection in favor of nucleotides G and C diversifies evolution rates and levels of polymorphism at mammalian synonymous sites. J Theor Biol. 2006;240:616–626. doi: 10.1016/j.jtbi.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Krishnakumar S, Zheng J, Wilhelmy J, Faham M, Mindrinos M, Davis R. A comprehensive assay for targeted multiplex amplification of human DNA sequences. Proc Natl Acad Sci. 2008;105:9296–9301. doi: 10.1073/pnas.0803240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Levanon EY, Yoon JK, Aach J, Xie B, LeProust E, Zhang K, Gao Y, Church GM. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- Marth G, Schuler G, Yeh R, Davenport R, Agarwala R, Church D, Wheelan S, Baker J, Ward M, Kholodov M, et al. Sequence variations in the public human genome data reflect a bottlenecked population history. Proc Natl Acad Sci. 2003;100:376–381. doi: 10.1073/pnas.222673099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okou DT, Steinberg KM, Middle C, Cutler DJ, Albert TJ, Zwick ME. Microarray-based genomic selection for high-throughput resequencing. Nat Methods. 2007;4:907–909. doi: 10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: New online mutation analysis and recommendations to users. Hum Mutat. 2002;19:607–614. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porreca GJ, Zhang K, Li JB, Xie B, Austin D, Vassallo SL, LeProust EM, Peck BJ, Emig CJ, Dahl F, et al. Multiplex amplification of large sets of human exons. Nat Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ, Willey DL, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- Schmidt S, Gerasimova A, Kondrashov FA, Adzuhbei IA, Kondrashov AS, Sunyaev S. Hypermutable non-synonymous sites are under stronger negative selection. PLoS Genet. 2008;4:e1000281. doi: 10.1371/journal.pgen.1000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Smith JM, Haigh J. The hitch-hiking effect of a favourable gene. Genet Res. 1974;23:23–35. [PubMed] [Google Scholar]
- Smith NG, Lercher MJ. Regional similarities in polymorphism in the human genome extend over many megabases. Trends Genet. 2002;18:281–283. doi: 10.1016/S0168-9525(02)02659-8. [DOI] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA, Adzhubei I, Thurman RE, Kryukov GV, Mirkin SM, Sunyaev SR. Human mutation rate associated with DNA replication timing. Nat Genet. 2009;41:393–395. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Ball E, Howells K, Phillips A, Mort M, Cooper DN. Human Gene Mutation Database: Towards a comprehensive central mutation database. J Med Genet. 2008;45:124–126. doi: 10.1136/jmg.2007.055210. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Kumar S. Neutral substitutions occur at a faster rate in exons than in noncoding DNA in primate genomes. Genome Res. 2003;13:838–844. doi: 10.1101/gr.1152803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sved J, Bird A. The expected equilibrium of the CpG dinucleotide in vertebrate genomes under a mutation model. Proc Natl Acad Sci. 1990;87:4692–4696. doi: 10.1073/pnas.87.12.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RY, Kuo KC, Gehrke CW, Huang LH, Ehrlich M. Heat- and alkali-induced deamination of 5-methylcytosine and cytosine residues in DNA. Biochim Biophys Acta. 1982;697:371–377. doi: 10.1016/0167-4781(82)90101-4. [DOI] [PubMed] [Google Scholar]
- Webster MT, Smith NG. Fixation biases affecting human SNPs. Trends Genet. 2004;20:122–126. doi: 10.1016/j.tig.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Sharp PM, Li WH. Mutation rates differ among regions of the mammalian genome. Nature. 1989;337:283–285. doi: 10.1038/337283a0. [DOI] [PubMed] [Google Scholar]
- Zhang K, Li JB, Gao Y, Egli D, Xie B, Deng J, Li Z, Lee JH, Aach J, LeProust E, et al. Digital RNA allelotyping reveals tissue-specific and allele-specific gene expression in human. Nat Methods. 2009;6:613–618. doi: 10.1038/nmeth.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]