Abstract
We created a visualization tool called Circos to facilitate the identification and analysis of similarities and differences arising from comparisons of genomes. Our tool is effective in displaying variation in genome structure and, generally, any other kind of positional relationships between genomic intervals. Such data are routinely produced by sequence alignments, hybridization arrays, genome mapping, and genotyping studies. Circos uses a circular ideogram layout to facilitate the display of relationships between pairs of positions by the use of ribbons, which encode the position, size, and orientation of related genomic elements. Circos is capable of displaying data as scatter, line, and histogram plots, heat maps, tiles, connectors, and text. Bitmap or vector images can be created from GFF-style data inputs and hierarchical configuration files, which can be easily generated by automated tools, making Circos suitable for rapid deployment in data analysis and reporting pipelines.
The continuing advances in speed, quality, and affordability of whole-genome analysis, including genome sequencing, have transitioned the comparative genomics field from the realm of comparing reference sequence assemblies to comparing assemblies of individual genomes. Whereas interspecies analysis leverages information about one species to further the understanding of biological mechanisms in another, comparative methods are now used to discover differences between individuals and the extent to which these differences affect response to the environment, such as susceptibility to disease and responsiveness to therapy.
Our growing ability to collect enormous amounts of sequence information to support such studies is arguably outpacing the rate at which we devise new methods to store, process, analyze, and visualize these data. Any new approaches in data modeling and analysis need to be accompanied with corresponding innovations in the visualization of these data. To mitigate the inherent difficulties in detecting, filtering, and classifying patterns within large data sets, we require instructive and clear visualizations that (1) adapt to the density and dynamic range of the data, (2) maintain complexity and detail in the data, and (3) scale well without sacrificing clarity and specificity.
The application of a germane data representation and its corresponding visualization to a domain-specific problem has historically improved the effectiveness of not only the presentation of the data, but also its analysis and dissemination. In some cases, the benefit of a new approach has altered how these data are perceived and investigated. Examples of this include the application of tree maps to show distribution of disk usage on a file system (Johnson and Shneiderman 1991) and hierarchical biological data (McConnell et al. 2002); directed graphs to depict networks, pathways, and phylogenetic information (Darwin 1859; Ciccarelli et al. 2006; Letunic and Bork 2007); and clustered heat maps to visualize array and expression data (Sneath 1957; Eisen et al. 1998). These approaches exemplify the virtues of an effective visualization: clarity, a high data-to-ink ratio (Tufte 1992), and favorable scaling characteristics. They have been widely adopted because they addressed pressing visualization problems within a domain where data sets were previously opaque to effective visual inspection.
Presently, a pressing visualization problem lies in the domain of comparative genomics and specifically in the comparative genomics of individuals. We need to establish a visual paradigm for displaying relationships between genomes in order to leverage the large amounts of sequence data that have been collected and to expand the power of the field of personal genomics. Previous efforts to visualize positional relationships applied linearly arranged ideograms, connected by lines, to represent rearrangements (Dicks 2000; Kozik et al. 2002; Yang et al. 2003; Choudhuri et al. 2004; Engels et al. 2006; Lee et al. 2006; Jakubowska et al. 2007; Kuenne et al. 2007; Sinha and Meller 2007). One approach uses encoding in HSL (hue, saturation, lightness) color space to perform three-way comparisons (Baran et al. 2007). The methods embodied in these approaches are effective for illustrating local alignments between similar sequences. However, the shortcoming of the linear layout becomes apparent in representations that associate many ideograms with numerous relationships (e.g., Fig. 2c in Lee et al. 2006). In such figures, multitudes of lines transgress unrelated ideograms and make patterns very difficult to discern. To mitigate this, color maps are used (e.g., Fig. 2 in Sinha and Meller 2007) as an effective way to represent large syntenic blocks. Although color maps address the problem of overburdened visualizations by mapping a position pair onto a position and a color, they reduce the texture and richness of the data. Circularly arranged ideograms are prevalent in visualizations of microbial genomes, which are circular (Gibson and Smith 2003; Sato and Ehira 2003; Kerkhoven et al. 2004; Stothard and Wishart 2005; Pritchard et al. 2006; Ghai and Chakraborty 2007). At least one report combined paired-position data with a circular layout to show relationships between genomic positions (specifically, pathways) (Ekdahl and Sonnhammer 2004) and hinted at the benefit of adopting the circular layout for application to structural data.
To address the challenge in displaying large volumes of genomic rearrangement data, we have developed Circos, which applies the circular ideogram layout to display relationships between genomic intervals. This approach builds on the established use of circular maps in which concept relationships extracted from text are displayed (Zytkow and Rauch 1999). Circos's initial application was to visualize end-sequence profiling (Volik et al. 2003) and fingerprint profiling (Krzywinski et al. 2007) of cancer genomes.
Although Circos is general and useable in any data domain, features have been added to mitigate inherent difficulties in visualizing large-scale multi-sample genomic data. Specifically, to address the issue of sparseness, the scale on each ideogram can be independently adjusted (both locally and globally) to attenuate or increase the visibility of a region. To accommodate visualizations that focus on regions of interest, axis breaks can be used to map chromosomes onto any number of ideograms, which themselves can be drawn in any order or orientation. To help demonstrate patterns in the data, data points can have their value and format characteristics altered by flexible rules. Finally, to help integrate Circos into analysis pipelines, image creation is controlled by flat-text configuration files, which can be created and adjusted automatically.
Circos is a mature software package and has been used to display genomic data (Constantine 2007; Jaillon et al. 2007; Campbell et al. 2008; Hampton et al. 2009; Meyer et al. 2009), in online genomics resources (Forbes et al. 2008), and mainstream periodicals and newspapers (Constantine 2007; Duncan 2007; Ostrander 2007; Zimmer 2008), as well as to visualize data from other fields of study (Corum and Hossain 2007).
Results
Platform and configuration
Circos is a command-line application written in Perl, and thus is easily deployable on any system for which Perl is available (http://www.cpan.org/ports/). Inputs are GFF-style data files and Apache-like configuration files—both can be easily generated by automated tools. Configuration is modular, and parameter blocks can be reused by importing them from multiple files. Each data track can contain a rule block that filters and formats data elements based on position, value, or previous formatting. Output images can be created in PNG (8 or 24 bit) or SVG formats. Configuration and data files required to create versions of Figures 1–8 are available as Supplemental material.
Applications
To illustrate how Circos's functionality can be applied to comparative and personal genomics, we present a series of image archetypes (Figs. 1–4, see foldout) that are being used as part of a multi-patient whole-genome analysis (data not shown; see Methods) of genomic rearrangements in follicular lymphoma, a common B-cell malignancy. These images range in resolution from whole-genome (3 Gb), to a fingerprint-map contig (10 Mb), to a single bacterial artificial chromosome (BAC) clone (100 kb), and finally to a sequence contig (10 kb). A second image series (Figs. 5–7) demonstrates how axis breaks and ideogram magnification can be used to zoom in on regions of interest. Batch image generation is demonstrated in Figure 8 (see foldout), which comprises 39 images that illustrate synteny between the dog and human genomes. The configuration and data files to create each figure are available in the Supplemental material.
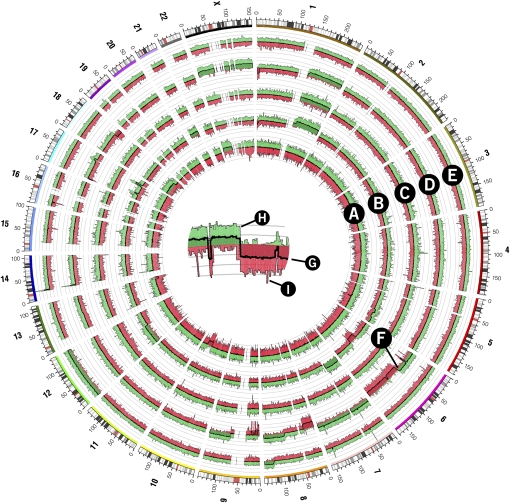
Figure 5.
Copy number whole-genome profiles of five follicular lymphoma tumor samples generated from the Affymetrix Mapping 500K array. Samples are represented in each of the five histograms in tracks A–E. A crop of histogram region F is shown in the center of the figure to demonstrate the structure of each histogram track. The central thick line (G) represents the average probe value across 250 adjacent probes. The area between this line and y = 0 is filled with green or red depending on whether the average value is positive (i.e., increased copy number value) or negative (decreased copy number value), respectively. Variability within each 250-probe set is shown in histogram components H and I, which show the maximum and minimum of three-probe average values within the set, respectively. The area under the maximum and minimum traces is filled with a lighter green or red, respectively.
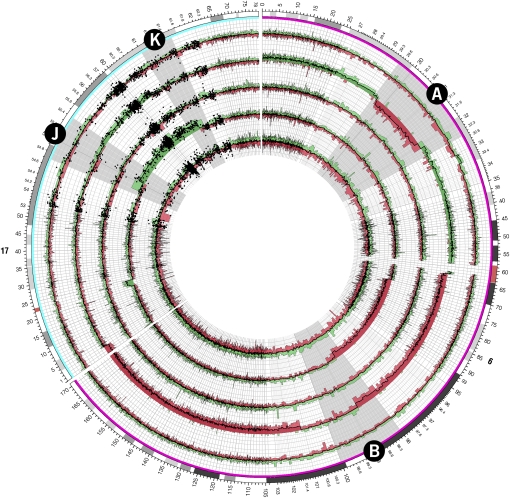
Figure 7.
Copy number profiles for chromosomes 6 and 17 of five follicular lymphoma tumor samples generated from the Affymetrix Mapping 500K array. Regions of interest A and B on chromosomes 6 and J and K on chromosomes 17 (corresponding to similarly labeled zoomed ideograms in Fig. 6) are magnified by a continuous scale expansion in these regions. Individual probe values are shown as a scatterplot in the vicinity of regions J and K.
Whole-genome view of genomic rearrangements
Figure 1 shows a whole-genome visual representation of genomic rearrangements in multiple genomes with the aim to identify breakpoint clusters and recurrent alterations. Presented are structural data derived from primary tumor samples from five patients diagnosed with follicular lymphoma. For each sample, the figure shows large-scale rearrangements, density of small-scale events, and copy number profile (see Methods). For example, at 130 Mb on chromosome 1, each sample shows large deletions and inversions as well as translocations that involve chromosomes 4, 16, and 17. This region also marks the beginning of significantly increased copy number in three samples (middle three rings in track F) that continues to the end of the 1q arm.
The resolution of a whole-genome view precludes the display of individual small-scale events at their native scale. To overcome this, scatter, histogram, and text tracks can be used as density plots, such as in Figure 1, track E, where glyph size is proportional to the number of small-scale events in each 5-Mb region.
A whole-genome view such as this can act as a departure point for in-depth investigation. Presently, we focus on the recurrent t(14;18) translocation between BCL2 and IGH gene regions, typical of follicular lymphoma (Yunis et al. 1982), and use Figure 1 as the initial image in a series that demonstrates the use of Circos at higher magnification. Using image maps, Figure 1 can be made interactive online, with elements being clickable for displaying features or their annotation in greater depth.
Identifying BACs spanning rearrangement breakpoints
A BAC-based fingerprint map was generated for each primary tumor sample, and each BAC was annotated with fingerprint-based alignments to the human reference sequence (see Methods). In this manner, the fingerprint map, which typically comprises thousands of contigs, was used to identify large-scale structural changes in the tumor genomes. Figure 2 demonstrates three such large-scale changes at the magnification level of a single fingerprint map contig (1–10 Mb). Each contig (Fig. 2, track A) was selected from the map of a different sample (patients 10, 13, and 21) and contains at least two BAC clones whose alignments (Fig. 2, track F) indicate a rearrangement breakpoint captured within the BACs.
At this magnification, the correspondence between the order of BACs in the map contig and on the sequence assembly can be demonstrated clearly and used to identify structural changes in the tumor genomes. For example, the fingerprint-based alignments map BACs 163K07 and 252O11 from a contig of patient 10 to two regions of chromosome 4 (97.1–98.1 Mb, 127.5–128.5 Mb), indicating that these BACs capture a rearrangement. Moreover, alignments from the second half of this contig are inverted in their progression, relative to clone order, suggesting that the rearrangement is an inversion. The classical t(14;18) translocation is exemplified by the alignments of BACs 175B19 and 278H11 from a contig of patient 13. The rearrangement in the map contig from patient 21 presents in a more complex fashion and is likely due to either an inversion or deletion.
The magnification of ideograms in Figure 2 is sufficiently high to allow for the display (track G) of small-scale events (indels or single nucleotide polymorphisms), which previously appeared in a density track (Fig. 1, track E). The layout in Figure 2 permits identification of any correlation between these events and rearrangements. Since these small-scale events are still much smaller than their associated glyph, the glyphs are magnified and scaled proportionally to the size of the event using data remap rules in the configuration of track G.
Views such as Figure 2 competently show the structural correspondence between representations of two genomes, such as those of a tumor sample and a reference. Other juxtapositions of this kind are common, such as two representations of the same genome (e.g., physical map and sequence assembly, to cross-validate their construction), or a representation of two closely related genomes (e.g., a physical map of one bacterial strain and sequence assembly of another, to study interstrain variation).
Identifying sequence contigs containing breakpoints
As part of our whole-genome analysis of the structure of follicular lymphomas, we used short-read Illumina technology to fully sequence a set of BAC clones that capture putative rearrangements. In Figure 3, we show the sequence assembly of nine such BACs (from patients 6, 8, 10, 11, 12, 13, 16, 24, and 25) that capture the t(14;18) translocation to illustrate the fact that this rearrangement's breakpoint position is variable, as previously reported (Cleary and Sklar 1985; Bakhshi et al. 1987; Marculescu et al. 2002).
In Figure 3, sequence contigs that capture the t(14;18) translocation can be easily identified, as can be the coverage of adjacent sequence by neighboring sequence contigs. These breakpoint contigs are highlighted in red and have their alignment ribbons drawn at higher opacity (Fig. 3, track E). The application of transparency to image elements allows layering of data, such as ribbons, or data tracks, such as histograms. The tile track (Fig. 3, tracks B and D) is used to represent the alignments (track D) of BAC sequence contigs (track B) on the reference sequence assembly (track C).
Exploring breakpoint structure
Sequence contigs that were found to span the breakpoint (Fig. 3, track E) are shown at higher magnification in Figure 4. This figure demonstrates the precise structure of alignments within the breakpoint cluster on 14q32 and 18q21, vis-à-vis the exon structure of BCL2 and IGH in these regions. In this figure, sequence contig ideograms are interspersed with reference assembly ideograms to better separate the ribbon groups to chromosomes 14 and 18.
Circos can draw ideograms in any order and orientation. For example, in Figure 4, the scale of sequence contigs progresses counterclockwise, while the scale of reference sequence ideograms progresses clockwise. Precise tick mark and tick label control is possible, including placement of the tick ring and formatting of tick labels. In Figure 4, to maintain relevant precision and avoid long labels, the tick labels of reference sequence ideograms are abbreviated to their last three digits (e.g., the position 58,910 kb is shown as 910 kb), which are the only digits that change in the labels across the image.
Local and global scale transformation
A unique aspect of Circos is its ability to adjust the global magnification for each ideogram and, furthermore, to smoothly vary the magnification within a region. This kind of local scale adjustment is effective to emphasize fine structure of data in a region while preserving context.
Figure 5 depicts one kind of dense data set that benefits from examination at various length scales. This figure shows whole-genome copy number profiles of five lymphoma samples generated using the Affymetrix Mapping 500K array. Although there are several large regions in which copy number values are consistently altered, most of the statistically significant variation in the figure occurs over short distances, which cannot be effectively shown at this resolution. Moreover, very few array probe values depart sufficiently from the average to be meaningful, making regions of interest in the data set sparse.
Figure 5 suggests an abrupt change in copy number at ∼60 Mb of chromosome 6 (track F) and spikes in copy number increase on chromosome 17, but details of these features cannot be discerned at this scale. To explore these regions in detail, Figure 6 uses breakout ideograms at higher magnification. Here, the global scale for each ideogram is adjusted independently (from 5× to 40× magnification) to show the fine structure in the data. By imposing a minimum distance between adjacent tick marks and their labels, Circos automatically renders tick mark labels for smaller intervals in zoomed regions (Fig. 6, tracks C, D, J, K). Using rules that toggle the visibility of data elements, individual array probe values are superimposed on the average histogram tracks in the region of Figure 6, tracks J and K.
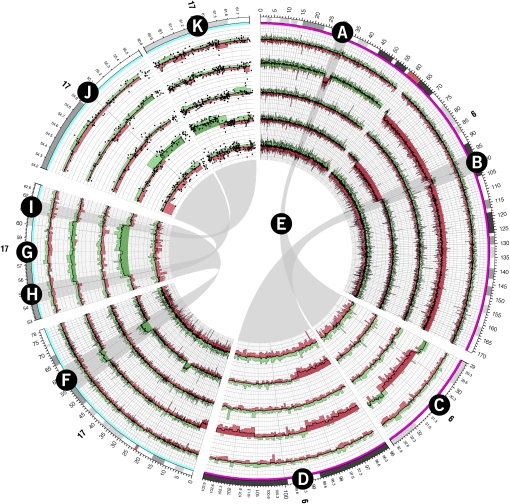
Figure 6.
Copy number profiles for chromosomes 6 and 17 of five follicular lymphoma tumor samples generated from the Affymetrix Mapping 500K array. Probe values were averaged across 20 adjacent probes. Several regions showing large copy number changes are shown on ideograms with an expanded scale. The structure of the histogram track is the same as used in Figure 5. Regions A and B on chromosomes 6 are shown at 10× magnification on ideograms C and D, respectively, with ribbons E connecting these regions with their zoomed ideograms. Similarly, region F on chromosome 17 is shown at 5× magnification as ideogram G. Regions H and I on ideogram G are shown at 40× magnification on ideograms J and K, respectively. Individual probe values are shown as scatterplots on ideograms J and K.
Whereas Figure 6 used breakout ideograms to render regions of chromosomes 6 and 17 in greater detail, Figure 7 accomplishes the same task by a continuous magnification increase in the regions of interest. Regions A and B on chromosome 6 are smoothly zoomed to 10× magnification, and regions J and K on chromosomes 17 are zoomed to 20×. With this approach, the profile of individual probe values in a region of interest can be shown while keeping the rest of the data in view.
Whole-genome syntenic profile
Sequence similarity profiles between two genomes are complex and difficult to visualize. By grouping adjacent regions of similarity into larger syntenic blocks, the data can be distilled into a visual form that is both coherent and interpretable (see Methods). Figure 8 illustrates how synteny between two genomes can be shown at a scale of 250 kb. Each panel in the figure represents the synteny between a single dog chromosome and the entire human reference sequence. By representing these blocks as transparent ribbons, it is possible to indicate multiple similarity targets for a given stretch of dog sequence (e.g., dog chromosome 31, which shows similarity to both human chromosomes 3 and 21), rather than a single consensus target.
Run-time rules
Through rule blocks in the configuration file, Circos allows for control over visibility and format of every data element based on its position, value, or format characteristics. Run-time rules permit changing the appearance of a figure within regions of interest (or ranges of data values), without needing to provide a new data set. Rules can simplify the task of identifying patterns in data by applying formatting that contrasts a subset of the data to the baseline.
Rules facilitate batch generation of a panel of images such as Figure 8. Each image in the panel was generated from the same configuration file, and execution varied only in command-line parameters that specified the identity of the dog chromosome and the scale at which it should be shown. In this figure, rules were used to color ribbons based on human chromosome target and to hide ribbons with ends smaller than 250 kb to limit the complexity of the figure. Rule blocks were used in nearly every figure, such as in Figure 3 to extend alignment curves to ticks for BACs with translocations, in Figure 5 to color parts of the histogram based on the sign of the probe value, and in Figure 6 to limit the display of probe values to breakout ideograms of chromosomes 17.
Discussion
Circos has been used to visualize data from the field of genomics, generate images for book and magazine covers, and even to provide scientific context to a David Cronenberg cinema artbook (De Gaetano 2008). It can generate images that are clear and informative to the investigator and attractive and compelling to the general public.
The core strengths of Circos are twofold. First, Circos provides an effective and scalable means to illustrate relationships between genomic positions. The comparison of intervals such as sequences and genome assemblies is commonplace, and Circos fills a need to visualize information in this data domain. Second, Circos is designed to allow flexible and easy rearrangement of elements in the image. While the circular framework of ideograms forms the foundation, the extent to which data tracks and their content can be visualized remains to be explored by the imagination of the investigator. There is an extensive online set of tutorials (presently there are about 80 tutorials), each with a thorough discussion of a specific feature and with sample images, configuration files, and data. Each tutorial provides a set of recipes that can be used as a departure point in generating visualizations of common data sets.
The flexibility of layout and formatting of graphical elements allows the creation of diverse visualizations in various data domains. For example, Circos can be effectively used to graphically represent tabular data. In this application, the concept of ideograms is subverted. Here, ideograms do not represent regions of chromosomes but individual rows or columns of a table, and a ribbon, instead of a structural relationship, represents the value of a cell for a given row and column.
A recurring challenge with genomic data is their sparseness and the small size of features relative to the supporting scale. For example, a rearrangement data set may be a list of small deletions, sized on the order of 1 kb. Features of this size cannot be drawn to scale on an ideogram, requiring the use of a density plot (Fig. 1, track E). However, by using run-time rules, it is possible to automatically resize these small features to a size that is discernable (Fig. 2, track G). It is equally challenging to effectively represent sparse or nonuniformly distributed data, which inherently do not make effective use of the space within a figure. An example of these kinds of data is epigenomic methylation state information, which is sampled at large but nonuniformly distributed positions in the genome (Eckhardt et al. 2006). Circos was applied to visualize these data in Zimmer (2008), using a connector track (also used in Fig. 3, track G) to map nonuniformly distributed genomic primer positions at which methylation values were measured with uniformly distributed stacked histograms that relate the extent of methylation.
Methods
Whole-genome structural data of follicular lymphomas
The multi-patient data set and corresponding in-depth analysis of the structure of the lymphoma genomes will be presented in detail elsewhere. Presently, we focus on illustrating how Circos can be used to interrogate these and similar data, and we include a brief snapshot of the data set to orient the reader in interpreting the visualizations.
Whole-genome structural data (data not shown) from primary tumor samples from patients diagnosed with follicular lymphoma were used to generate Figures 1–7. A BAC library was created from each tumor sample (average insert size of libraries ranged between 130 and 200 kb) and subjected to restriction-digest fingerprinting (Marra et al. 1997; Schein et al. 2004; Mathewson et al. 2007) using an EcoRI/EcoRV double digest to a depth of fivefold to sixfold. Rearrangements (translocations, inversions, deletions) were identified from alignments of fingerprinted BACs onto the human reference sequence (Krzywinski et al. 2007). Copy number changes in the samples were identified using the Affymetrix Mapping 500K array. Individual BACs identified to capture rearrangements were subject to short-read Illumina sequencing and assembled with ABySS (Simpson et al. 2009).
Generation of synteny bundles between dog and human genomes
Sequence similarity data relating the dog (UCSC, canFam2, May 2005) and human (UCSC, hg18, Mar 2006) genomes were downloaded from the UCSC Genome Browser (http://www.genome.ucsc.edu) from the chainCanFam2 table (track: Dog Chain; track group: Comparative Genomics). This data set comprises about 2.16 million gapped alignments between the two assemblies. Alignment bundles were built up from individual alignments by a scheme (implemented by bundlelinks, a utility tool in the circos-tools distribution) that grouped alignments into sets. For a given alignment set, (1) all alignments related the same pair of dog and human chromosomes, (2) any alignment was no further than 250 kb away from its nearest neighbor in the set, and (3) the number of alignments in the set was at least three. Each set is represented in Figure 8 as a ribbon whose ends represent the extent of the set alignments on the dog and human chromosomes. Figure 8 shows sets that spanned at least 250 kb on both human and dog chromosomes.
Utility tools
Several utility tools are bundled with Circos to help analyze, filter, and format data. filterlinks parses a link file and selects only those links that pass positional criteria. orderchr applies simulated annealing to a link data set to generate an ideogram order that minimizes (or maximizes) the number of links that cross in the image. bundlelinks is used to identify links that are corroborated by other adjacent links (used for Fig. 8). binlinks is used to generate density tracks, suitable for scatter/line/histogram tracks, based on the number of links within a sliding window. tableviewer is a collection of tools that is used to parse tabular data and generate data and configuration files for visualizing tables with Circos.
Acknowledgments
M.K. thanks the many individuals who sent invaluable suggestions, comments, and bug reports (specifically, Perseus Missirlis, Gordon Robertson, Martin Rijlaarsdam, and Stefan Conrady), as well as Art Directors who helped Circos enter the public fray (David Constantine, New York Times; Jonathan Corum, New York Times; John Grimwade, Conde Nast; Domenico de Gaetano, Volumina; Derek Bacchus, Pearson Science; Barbara Aulicino, American Scientist; Nikki Greenwood, Seed Magazine). We thank the members of the GSC mapping group for preparing the raw data presented in this work, especially Matthew Field and Andrew Mungall for helpful discussions. M.A.M. and S.J.J. are scholars of the Michael Smith Foundation for Health Research. Development of Circos was supported by Genome Canada and Genome British Columbia, and the National Cancer Institute/Terry Fox Foundation.
Footnotes
[Supplemental material is available online at http://www.genome.org. Circos is licensed under GPL and available at http://mkweb.bcgsc.ca/circos. An interactive online version of Circos designed to visualize tabular data is available at http://mkweb.bcgsc.ca/circos/tableviewer.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.092759.109.
References
- Bakhshi A, Wright JJ, Graninger W, Seto M, Owens J, Cossman J, Jensen JP, Goldman P, Korsmeyer SJ. Mechanism of the t(14;18) chromosomal translocation: Structural analysis of both derivative 14 and 18 reciprocal partners. Proc Natl Acad Sci. 1987;84:2396–2400. doi: 10.1073/pnas.84.8.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran R, Robert M, Suematsu M, Soga T, Tomita M. Visualization of three-way comparisons of omics data. BMC Bioinformatics. 2007;8:72. doi: 10.1186/1471-2105-8-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri JV, Schleiermacher C, Kurtz S, Giegerich R. GenAlyzer: Interactive visualization of sequence similarities between entire genomes. Bioinformatics. 2004;20:1964–1965. doi: 10.1093/bioinformatics/bth161. [DOI] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- Cleary ML, Sklar J. Nucleotide sequence of a t(14;18) chromosomal breakpoint in follicular lymphoma and demonstration of a breakpoint-cluster region near a transcriptionally active locus on chromosome 18. Proc Natl Acad Sci. 1985;82:7439–7443. doi: 10.1073/pnas.82.21.7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantine D. Close-ups of the genome, species by species by species. New York Times. 2007:F4. January 23, [Google Scholar]
- Corum J, Hossain F. Naming names. New York Times. 2007:41. December 16, [Google Scholar]
- Darwin C. On the origin of species by means of natural selection. John Murray; London, UK: 1859. [Google Scholar]
- De Gaetano D. Chromosomes. Volumina; Torino, Italy: 2008. [Google Scholar]
- Dicks J. Graphical tools for comparative genome analysis. Yeast. 2000;17:6–15. doi: 10.1002/(SICI)1097-0061(200004)17:1<6::AID-YEA15>3.0.CO;2-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan DE. Welcome to the future. Conde Nast Portfolio. 2007;November:192–197. 220–222. [Google Scholar]
- Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekdahl S, Sonnhammer EL. ChromoWheel: A new spin on eukaryotic chromosome visualization. Bioinformatics. 2004;20:576–577. doi: 10.1093/bioinformatics/btg448. [DOI] [PubMed] [Google Scholar]
- Engels R, Yu T, Burge C, Mesirov JP, DeCaprio D, Galagan JE. Combo: A whole genome comparative browser. Bioinformatics. 2006;22:1782–1783. doi: 10.1093/bioinformatics/btl193. [DOI] [PubMed] [Google Scholar]
- Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, Menzies A, Teague JW, Futreal PA, Stratton MR. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr Protoc Hum Genet. 2008;57:10.11.1–10.11.26. doi: 10.1002/0471142905.hg1011s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghai R, Chakraborty T. Comparative microbial genome visualization using GenomeViz. Methods Mol Biol. 2007;395:97–108. doi: 10.1007/978-1-59745-514-5_6. [DOI] [PubMed] [Google Scholar]
- Gibson R, Smith DR. Genome visualization made fast and simple. Bioinformatics. 2003;19:1449–1450. doi: 10.1093/bioinformatics/btg152. [DOI] [PubMed] [Google Scholar]
- Hampton OA, Den Hollander P, Miller CA, Delgado DA, Li J, Coarfa C, Harris RA, Richards S, Scherer SE, Muzny DM, et al. A sequence-level map of chromosomal breakpoints in the MCF-7 breast cancer cell line yields insights into the evolution of a cancer genome. Genome Res. 2009;19:167–177. doi: 10.1101/gr.080259.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Jakubowska J, Hunt E, Chalmers M, McBride M, Dominiczak AF. VisGenome: Visualization of single and comparative genome representations. Bioinformatics. 2007;23:2641–2642. doi: 10.1093/bioinformatics/btm394. [DOI] [PubMed] [Google Scholar]
- Johnson B, Shneiderman B. Tree-Maps: A space-filling approach to the visualization of hierarchical information structures. IEEE Visualization—Proceedings of the 2nd Conference on Visualization '91; 1991. [DOI] [Google Scholar]
- Kerkhoven R, van Enckevort FH, Boekhorst J, Molenaar D, Siezen RJ. Visualization for genomics: The Microbial Genome Viewer. Bioinformatics. 2004;20:1812–1814. doi: 10.1093/bioinformatics/bth159. [DOI] [PubMed] [Google Scholar]
- Kozik A, Kochetkova E, Michelmore R. GenomePixelizer—a visualization program for comparative genomics within and between species. Bioinformatics. 2002;18:335–336. doi: 10.1093/bioinformatics/18.2.335. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Bosdet I, Mathewson C, Wye N, Brebner J, Chiu R, Corbett R, Field M, Lee D, Pugh T, et al. A BAC clone fingerprinting approach to the detection of human genome rearrangements. Genome Biol. 2007;8:R224. doi: 10.1186/gb-2007-8-10-r224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuenne CT, Ghai R, Chakraborty T, Hain T. GECO—linear visualization for comparative genomics. Bioinformatics. 2007;23:125–126. doi: 10.1093/bioinformatics/btl556. [DOI] [PubMed] [Google Scholar]
- Lee D, Choi JH, Dalkilic MM, Kim S. COMPAM: Visualization of combining pairwise alignments for multiple genomes. Bioinformatics. 2006;22:242–244. doi: 10.1093/bioinformatics/bti759. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Marculescu R, Le T, Bocskor S, Mitterbauer G, Chott A, Mannhalter C, Jaeger U, Nadel B. Alternative end-joining in follicular lymphomas' t(14;18) translocation. Leukemia. 2002;16:120–126. doi: 10.1038/sj.leu.2402324. [DOI] [PubMed] [Google Scholar]
- Marra MA, Kucaba TA, Dietrich NL, Green ED, Brownstein B, Wilson RK, McDonald KM, Hillier LW, McPherson JD, Waterston RH. High throughput fingerprint analysis of large-insert clones. Genome Res. 1997;7:1072–1084. doi: 10.1101/gr.7.11.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathewson CA, Schein JE, Marra MA. Large-scale BAC clone restriction digest fingerprinting. Curr Protoc Hum Genet. 2007:5.19.1–5.19.21. doi: 10.1002/0471142905.hg0519s53. [DOI] [PubMed] [Google Scholar]
- McConnell P, Johnson K, Lin S. Applications of Tree-Maps to hierarchical biological data. Bioinformatics. 2002;18:1278–1279. doi: 10.1093/bioinformatics/18.9.1278. [DOI] [PubMed] [Google Scholar]
- Meyer C, Kowarz E, Hofmann J, Renneville A, Zuna J, Trka J, Abdelali RB, Macintyre E, De Braekeleer E, De Braekeleer M, et al. New insights to the MLL recombinome of acute leukemias. Leukemia. 2009 doi: 10.1038/leu.2009.33. [DOI] [PubMed] [Google Scholar]
- Ostrander EA. Genetics and the shape of dogs. Am Sci. 2007;95:406–413. [Google Scholar]
- Pritchard L, White JA, Birch PR, Toth IK. GenomeDiagram: A python package for the visualization of large-scale genomic data. Bioinformatics. 2006;22:616–617. doi: 10.1093/bioinformatics/btk021. [DOI] [PubMed] [Google Scholar]
- Sato N, Ehira S. GenoMap, a circular genome data viewer. Bioinformatics. 2003;19:1583–1584. doi: 10.1093/bioinformatics/btg195. [DOI] [PubMed] [Google Scholar]
- Schein J, Kucaba T, Sekhon M, Smailus D, Waterston R, Marra M. High-throughput BAC fingerprinting. Methods Mol Biol. 2004;255:143–156. doi: 10.1385/1-59259-752-1:143. [DOI] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha AU, Meller J. Cinteny: Flexible analysis and visualization of synteny and genome rearrangements in multiple organisms. BMC Bioinformatics. 2007;8:82. doi: 10.1186/1471-2105-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneath PH. The application of computers to taxonomy. J Gen Microbiol. 1957;17:201–226. doi: 10.1099/00221287-17-1-201. [DOI] [PubMed] [Google Scholar]
- Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- Tufte E. Visual display of quantitative information. Graphics Press; Cheshire, CT: 1992. [Google Scholar]
- Volik S, Zhao S, Chin K, Brebner JH, Herndon DR, Tao Q, Kowbel D, Huang G, Lapuk A, Kuo WL, et al. End-sequence profiling: Sequence-based analysis of aberrant genomes. Proc Natl Acad Sci. 2003;100:7696–7701. doi: 10.1073/pnas.1232418100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Wang J, Yao ZJ, Jin Q, Shen Y, Chen R. GenomeComp: A visualization tool for microbial genome comparison. J Microbiol Methods. 2003;54:423–426. doi: 10.1016/s0167-7012(03)00094-0. [DOI] [PubMed] [Google Scholar]
- Yunis JJ, Oken MM, Kaplan ME, Ensrud KM, Howe RR, Theologides A. Distinctive chromosomal abnormalities in histologic subtypes of non-Hodgkin's lymphoma. N Engl J Med. 1982;307:1231–1236. doi: 10.1056/NEJM198211113072002. [DOI] [PubMed] [Google Scholar]
- Zimmer C. Now: The rest of the genome. New York Times. 2008:D1. November 11, [Google Scholar]
- Zytkow JM, Rauch J. Principles of data mining and knowledge discovery. Third European Conference on Principles and Practice of Knowledge Discovery in Databases, PKDD '99; Prague, Czech Republic: Springer; 1999. pp. 277–282. [Google Scholar]