Abstract
BCOR (BCL6 co-repressor) represses gene transcription by interacting with BCL-6 1, 2. BCOR mutation is responsible for oculo-facio-cardio-dental (OFCD) syndrome, characterized by canine teeth with extremely long roots, congenital cataracts, craniofacial defects and congenital heart disease3–5. Here we show that BCOR mutation increased osteo/dentinogenic potentials of mesenchymal stem cells (MSCs) isolated from an OFCD patient, providing a molecular explanation for abnormal root growth. AP-2α was identified as a repressive target of BCOR, and BCOR mutation resulted in abnormal activation of AP-2α. Gain- and loss-of-function assays suggested that AP-2α was a key factor that mediated increased osteo/dentinogenic capacity of MSCs. Moreover, we found that BCOR maintained tissue homeostasis and gene silencing by epigenetic mechanisms. BCOR mutation increased histone H3K4/36 methylation in MSCs, thereby reactivating transcription of silenced target genes. In summary, by studying a rare human genetic disease, we unravel an epigenetic mechanism for control of human adult stem cell function.
Oculofacialcardiodental syndrome (OFCD) is a rare genetic disorder characterized by teeth with extremely long roots (radiculomegaly), and craniofacial, eye and cardiac abnormalities3–7. OFCD is inherited in an X-linked dominant pattern in heterozygous females, and males with OFCD cannot survive due to embryonic lethality. Frequent eye anomalies include congenital cataracts and microphthalmia. Facial deformities include long narrow face, high nasal bridge, and cleft palate. Congenital cardiac abnormalities include septal defect and mitral valve defect. Among the many dental defects reported in OFCD patients, enlarged roots of canine teeth is the most consistent and typical finding3–7. The roots mainly consist of dentin, a bone-like mineralized tissue that serves to anchor the tooth in alveolar bone. After tooth eruption, root formation is well-synchronized with alveolar bone growth and virtually stops growing at certain ages8–11. On the contrary, in OFCD patients, the roots of mandibular incisors and canines can grow continuously until they reach the lower border of the mandible; the roots of maxillary incisors and canines can extend to the cortical plate of the orbit. Thus, canine radiculomegaly has been considered to be an important sign for OFCD diagnosis3–7.
Genetic studies have found that mutations in BCL6 co-repressor (BCOR) are responsible for OFCD syndrome3. The most common mutations in BCOR are truncation and frameshift mutations, resulting in premature termination of the protein with deletion of the C-terminal domain3–5. BCOR was originally identified as a co-repressor of the transcription repressor BCL-61. Chromosomal translocations of BCL-6 are common genomic alterations in Non-Hodgkin’s B-cell lymphomas12. BCOR has been found to interact with histone deacetylase (HDAC), demethylase and H2A ubiquitin ligase, suggesting that BCOR may mediate repression through chromatin modification1, 13, 14. Mouse Bcor has been found to be expressed in tooth primordium, eye, neural tube and branchial arches which correlate with tissues affected in OFCD patients15. Genetic studies, through deleting Bcor in embryonic stem cells, suggest that Bcor plays an important role in early mouse embryonic development16.
Mesenchymal stem cells (MSCs) were originally isolated from bone marrow and are multipotent since they can differentiate into a variety of cell types including osteoblasts, chondrocytes, myocytes and adipocytes. Growing evidence indicates that MSCs are also present in non-bone marrow tissues17–21. Recently, a new population of MSCs has been isolated from dental and craniofacial tissues based on their stem cell properties11, 22, 23. These cells are multipotent, osteo/dentinogenic and capable of self-renewal. When transplanted into immunocompromised mice, they generated bone/dentin-like mineralized tissue and were capable of repairing dental and craniofacial defects11, 24. Characterization of MSCs from root apical papilla strongly suggests that these cells are responsible for root dentin formation and root growth. Since OFCD patients have enlarged and continuously growing root, BCOR mutation may have an intrinsic effect on the proliferation and function of MSCs from root apical papilla. To test our hypothesis, we isolated MSCs from the root apical papilla of an OFCD patient and examined their stem cell properties. While BCOR mutation did not affect stem cell marker expression, BCOR mutation resulted in enhanced osteo/dentinogenic potential of MSCs by inducing AP-2α. Mechanistically, we found that BCOR mutation increased histone H3 K4/36 methylation and reduced binding of BCL-6 to the AP-2α promoter, thereby resulting in a loss of BCL-6/BCOR repressive function. Our results provide a molecular explanation for the abnormal root growth of OFCD syndrome.
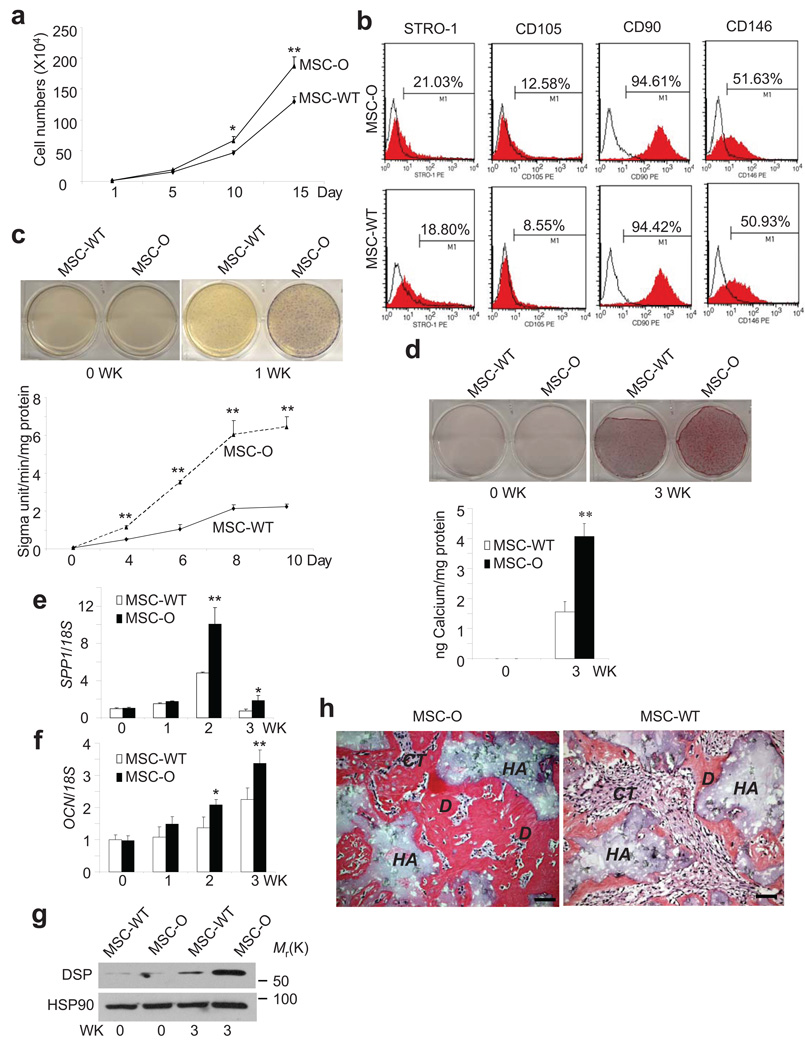
MSCs were isolated from apical papilla of an OFCD patient undergoing surgical root apex removal due to radiculomegaly. This patient had a single nucleotide deletion, c.2613delC, resulting in a frameshift mutation with premature stop codon, p.F871Lfs8X4. Like many of the mutations found in other OFCD patients, this frameshift mutation led to the deletion of the BCOR C-terminus (approximately 800 amino acid deletion). MSCs isolated from the OFCD patient (MSC-O) proliferated at a faster rate in vitro than wild type MSCs isolated from a healthy human subject (MSC-WT; Fig. 1a; Supplementary Fig. S1a). Of note, the expression levels of BCOR mRNA in MSC-WT and MSC-O cells were similar (Supplementary Fig. S1b,c). Since radiculomegaly indicated that root was heavily mineralized, increased proliferation could not fully account for the phenotype of OFCD. Thus, we further examined whether BCOR mutation intrinsically affected MSC function. As shown in Fig. 1b, FACS profiling showed that MSC-O expressed stem cell markers of MSCs similar to those of MSC-WT cells11, 20, 21. Root dentin is a specialized mineralized tissue like bones8, 10, 11. Since OFCD patients display abnormal root formation, we tested whether MSC-O cells had increased osteo/dentinogenic potentials. Both MSC-O and MSC-WT cells were treated with differentiation-inducing media containing dexamethasone, ascorbic acid and β-glycerophosphate as described previously11. To minimize the effect of cell growth on differentiation, both MSC-O and MSC-WT cells were plated in a confluent condition. As shown in Fig. 1c, shortly upon induction, alkaline phosphatase (ALP) activity, an early marker for osteo/dentinogenic differentiation, was more strongly induced in MSC-O cells as compared with MSC-WT cells. Three weeks after induction, Alizarin Red staining revealed that calcium deposition or mineralization was also significantly higher in MSC-O cells than in MSC-WT cells (Fig. 1d). Consistently, Real-time RT-PCR found that bone/dentin extracellular matrix genes, including OCN and SPP1, were more strongly induced in MSC-O cells as compared with MSC-WT cells (Fig. 1e,f). Dental sialoprotein (DSP) is an extracellular matrix protein highly expressed in dentin relative to other tissues8, 10, 25. We found that the induction of DSP was significantly higher in MSC-O cells than in MSC-WT cells (Fig. 1g). Finally, our in vivo transplantation also demonstrated that MSC-O cells generated more bone/dentin-like mineralized tissues than MSC-WT cells (Fig. 1h).
Figure 1. BCOR mutation results in enhanced osteo/dentinogenic potentials of MSCs from an OFCD patient.
a,BCOR mutation promoted MSC proliferation. Values are mean ± s.d for triplicate samples from a representative experiment. Student’s t test was performed to determine statistical significance. *P < 0.05; **P < 0.01. b, BCOR mutation did not affect the expression of stem cell surface markers by flow cytometry. Cells were sorted on a FACSCalibur flow cytometer and analyzed using Cell Quest software (BD Bioscience). c, BCOR mutation resulted in enhanced ALP activity in MSCs. Values are mean ± s.d for triplicate samples from a representative experiment. Student’s t test was performed to determine statistical significance. **P < 0.01. d, BCOR mutation resulted in enhanced mineralization in MSC-O cells. Values are mean ± s.d for triplicate samples from a representative experiment. Student’s t test was performed to determine statistical significance. **P < 0.01. e,f, BCOR mutation resulted in enhanced expression of OCN and SPP1 in MSC-O cells. The expressions of both OCN and SPP1 were examined by Real-time RT-PCR. Values are mean ± s.d for triplicate samples from a representative experiment. Student’s t test was performed to determine statistical significance. **P < 0.01. g, BCOR mutation resulted in enhanced DSP expression in MSC-O cells. DSP expression was examined by Western blot analysis. HSP90 was used as an internal control. Uncropped images of the blots are shown in the Supplementary information. h, BCOR mutation resulted in enhanced mineralized tissue formation in vivo. Both MSC-O and MSC-WT cells were transplanted into SCID mice for 8 weeks. D, bone/dentin-like tissues; HA, hydroxyapatite tricalcium carrier; CT, connective tissues. Scale bar, 100 µm.
Since OFCD is an X-linked dominant syndrome in heterozygous females, the initial MSC-O cells from the patient may be a mixed population expressing wild type or mutant BCOR mRNA due to X-inactivation. Thus, it is important to determine whether enhanced osteo/dentinogenic differentiation in passaged MSC-O cells is mainly due to cells expressing mutant BCOR mRNA. Total RNA from MSC-O cells was extracted and RT-RCR was performed using the specific primers which were from different exons and spanned the mutation site of BCOR. The PCR products were subcloned into a TA clone vector and sequenced. We found that the majority of clones (52 out of 60) expressed mutant BCOR (Supplementary Fig. S1d), suggesting that the mutant cells outgrew the wild-type cells in expanded MSC-O cell cultures. To further confirm our results, we also used shRNA to knock-down BCOR in MSC-WT cells. The knock-down of BCOR expression also significantly enhanced osteo/dentinogenic differentiation of MSC-WT cells (Supplementary Fig. S2). Moreover, the knock-down of BCOR in MSCs isolated from the dental pulp, also known as dental pulp stem cells (DPSCs)26, potently enhanced osteo/dentinogenic differentiation (Supplementary Fig. S3).
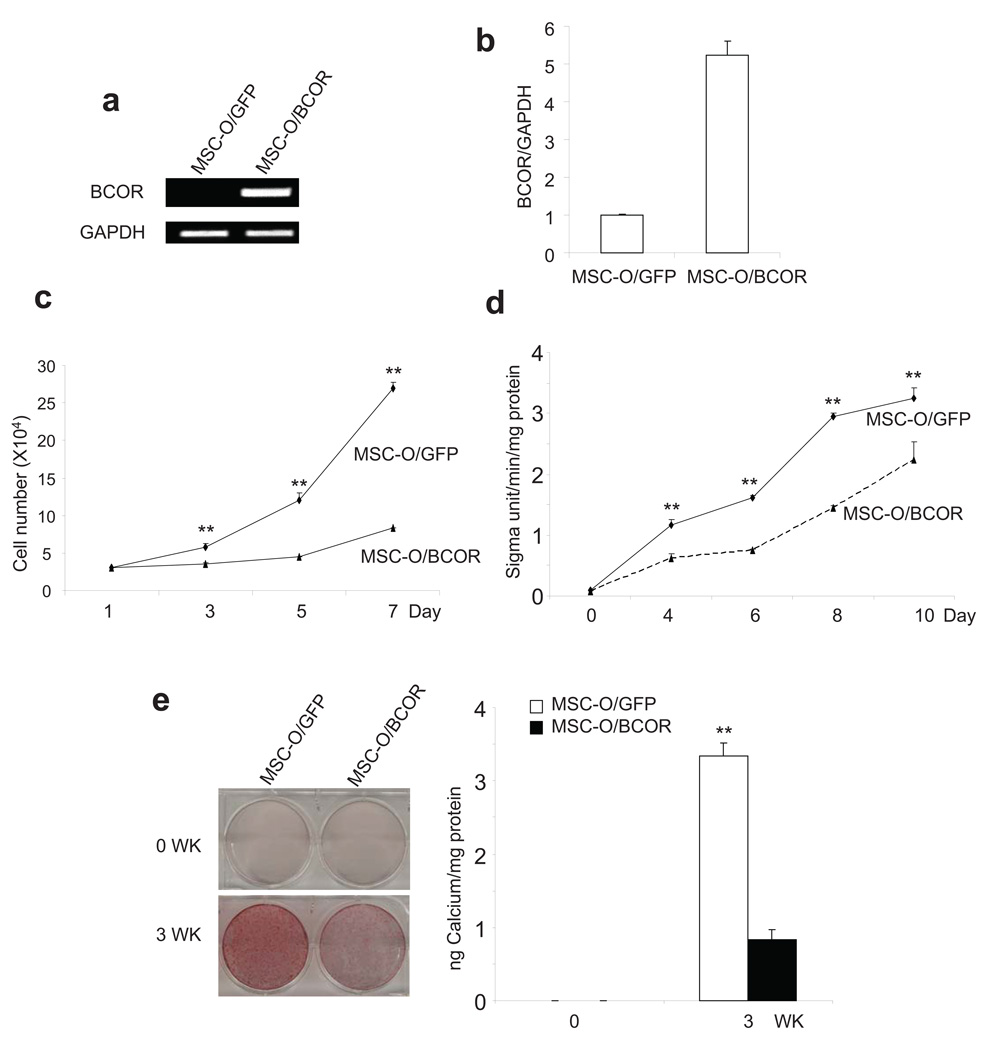
To determine whether enhanced osteo/dentinogenic potentials associated with MSC-O cells were directly due to BCOR mutation, we tested whether the restoration of wild type BCOR (isoform C) in MSC-O could inhibit osteo/dentinogenic potentials of MSC-O cells. To rule out clonal variation, MSC-O cells were transduced with retroviruses expressing Flag-BCOR and control vector. As shown in Fig. 2a, using specific primers to detect Flag-BCOR, RT-PCR confirmed that wild type BCOR was stably expressed in MSC-O (MSC-O/BCOR) cells, but not in control cells (MSC-O/GFP). Real-time RT-PCR showed a 5-fold increase in BCOR mRNA expression in MSC-O/BCOR cells compared to MSC-O/GFP cells (Fig. 2b), using primers which detect both endogenous BCOR and ectopic Flag-BCOR. The over-expression of wild type BCOR significantly inhibited the proliferation of MSC-O cells (Fig. 2c). Restoration of wild type BCOR in MSC-O cells strongly inhibited ALP activity upon induction of differentiation (Fig. 2d). Consistently, mineralization was potently inhibited in MSC-O/BCOR cells compared with MSC-O/GFP cells as determined by Alizarin Red staining (Fig. 2e).
Figure 2. The restoration of wild type BCOR in MSC-O cells inhibited cell differentiation and proliferation.
a, Over-expression of BCOR in MSC-O cells. Wild type Flag-BCOR was ectopically expressed in MSC-O cells as determined by RT-PCR using specific primers for Flag-BCOR. GAPDH was used as an internal control. b, BCOR over-expression was determined by Real-time RT-PCR. Real-time RT-PCR was performed using primers which detected endogenous BCOR and ectopic Flag-BCOR. c, Over-expression of BCOR inhibited MSC-O cell proliferation. Values are mean ± s.d for triplicate samples from a representative experiment. Student’s t test was performed to determine statistical significance. **P < 0.01. d. Over-expression of BCOR inhibited ALP activity in MSC-O cells. Values are mean ± s.d for triplicate samples from a representative experiment. Student’s t test was performed to determine statistical significance. **P < 0.01. e, Over-expression of BCOR inhibited mineralization in MSC-O cells. Values are mean ± s.d for triplicate samples from a representative experiment. Student’s t test was performed to determine statistical significance. **P < 0.01.
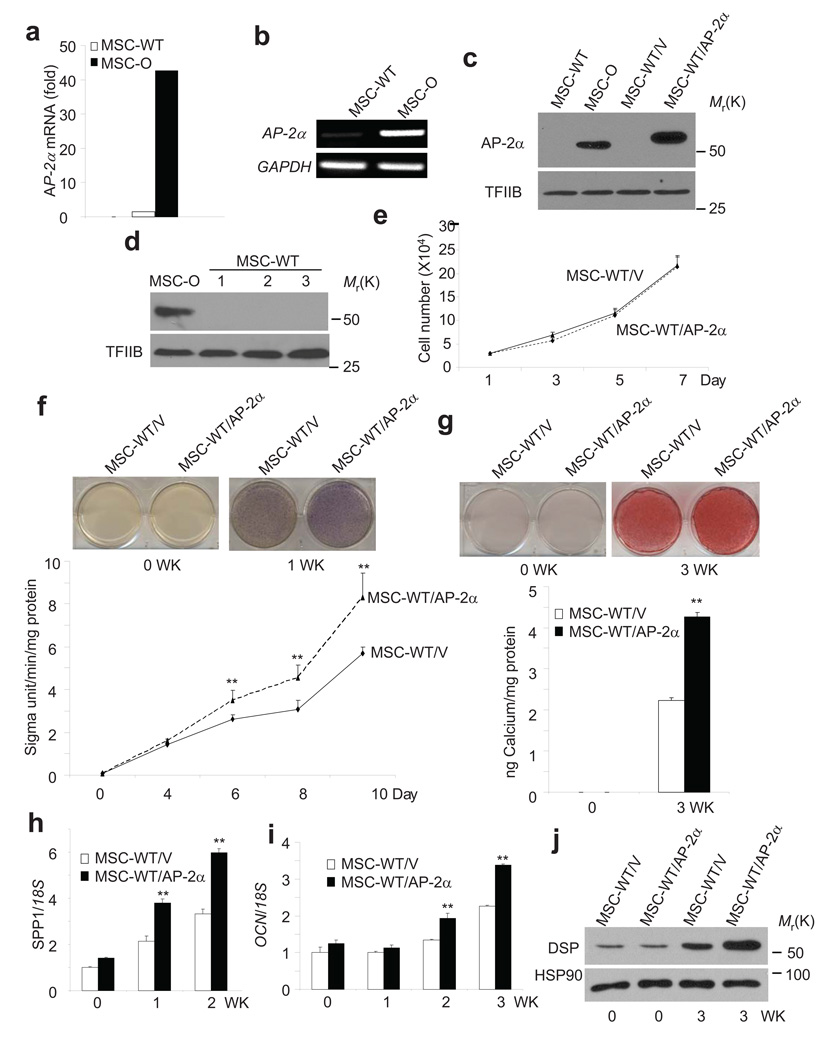
Our studies suggest that BCOR mutation has intrinsic effects on MSC differentiation capacity. However, because BCOR mutation increases cell proliferation, the enhancement of osteo/dentinogenic potentials of MSCs may be an indirect effect. To resolve this crucial issue, we performed gene expression profiling to identify the BCOR target genes that might be associated with osteo/dentinogenic potentials using Affymetrix Human Genome U133 Plus 2.0 Array. Together with this information, gain- and loss-of-function experiments would help to determine whether BCOR-regulated target genes were crucial for enhanced osteo/dentinogenic potentials, thereby providing direct evidence to verify the functional role of BCOR mutation in osteo/dentinogenic capacity. Importantly, microarray revealed that the transcription factor AP-2α was the second highest differentially expressed gene in MSC-O cells as compared to MSC-WT cells (Fig. 3a). Other highly expressed genes in MSC-O cells (>5-fold) relative to MSC-WT cells were listed in Supplementary Table 1. Previously, genetic studies found that AP-2α was associated with craniofacial development and the knock-out of AP-2α caused craniofacial and skeletal defects27, 28. RT-PCR confirmed that AP-2α was strongly expressed in MSC-O cells, but not in MSC-WT cells (Fig. 3b). Consistently, Western blot analysis found that AP-2α was highly expressed in MSC-O cells, but only was barely detected in MSC-WT cells (Fig. 3c). Furthermore, we found that AP-2α was not expressed in MSCs from three different healthy human subjects, indicating that the difference was unlikely due to individual variation (Fig. 3d).
Figure 3. BCOR mutation increases AP-2α expression in MSC-O cells.
a, Gene expression profile revealed that AP-2α was highly expressed in MSC-O cells. b, AP-2α was highly expressed in MSC-O cells compared with MSC-WT cells. AP-2α expression was determined by RT-PCR. GAPDH was used as an internal control. c, AP-2α was highly expressed in MSC-O cells compared with MSC-WT cells as determined by Western blot analysis. Uncropped images of the blots are shown in the Supplementary information. d, AP-2α was not detected in normal MSCs cells from three different normal human subjects. Uncropped images of the blots are shown in the Supplementary information. e, Over-expression of AP-2α did not affect MSC proliferation. Values are mean ± s.d for triplicate samples from a representative experiment. f, Over-expression of AP-2α increased ALP activity in MSC cells. Values are mean ± s.d for triplicate samples from a representative experiment. Student’s t test was performed to determine statistical significance. **P < 0.01. g, Over-expression of AP-2α increased mineralization in MSCs. Values are mean ± s.d for triplicate samples from a representative experiment. Student’s t test was performed to determine statistical significance. **P < 0.01. h, Over-expression of AP-2α enhanced SPP1 expression in MSCs. SPP1 was determined by Real-time RT-PCR. Values are mean ± s.d for triplicate samples from a representative experiment. Student’s t test was performed to determine statistical significance. **P < 0.01. i, Over-expression of AP-2α enhanced OCN expression. OCN was determined by Real-time RT-PCR. Values are mean ± s.d for triplicate samples from a representative experiment. Student’s t test was performed to determine statistical significance. **P < 0.01. j, Over-expression of AP-2α enhanced DSP expression in MSC cells. DSP expression was determined by Western blot analysis. HSP90 was used as an internal control. Uncropped images of the blots are shown in the Supplementary information.
To determine whether AP-2α was a key mediator for enhanced osteo/dentinogenic potentials resulting from BCOR mutation, we first tested whether the over-expression of AP-2α potentiated differentiation capacity of MSC-WT cells. To avoid clonal variation, MSC-WT cells were transduced with retroviruses expressing Flag-AP-2α or control vector. Western blot analysis confirmed that AP-2α was expressed (Fig. 3c). Over-expression of AP-2α did not affect MSC proliferation (Fig. 3e). As shown in Fig. 3f, g, we found that over-expression of AP-2α significantly enhanced ALP activity and mineralization upon induction. Real-time RT-PCR and Western blot analysis also revealed that the over-expression of AP-2α enhanced expressions of OCN, SPP1, and DSP (Fig. 3h, i, j).
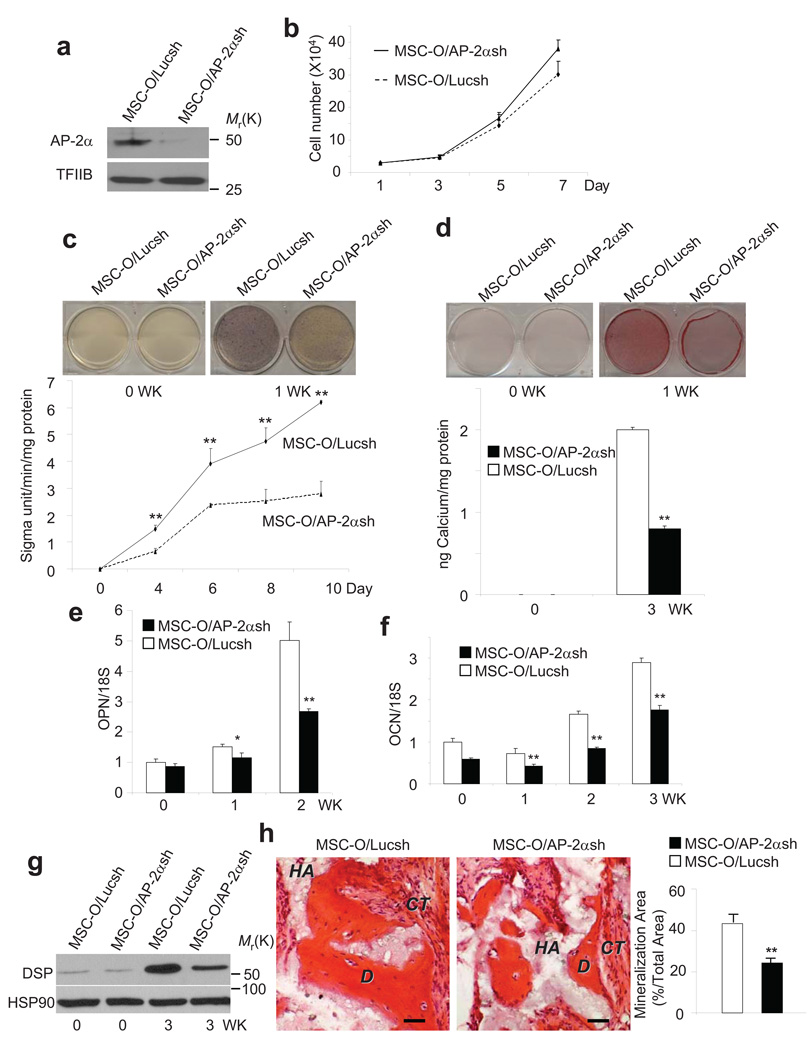
To further confirm that AP-2α was responsible for enhanced osteo/dentinogenic potentials of MSCs, we utilized small hairpin RNA (shRNA) to knock-down AP-2α expression in MSC-O cells. Western blot analysis confirmed that approximately 90% of AP-2α in MSC-O cells were knocked down by retroviruses expressing shRNA against AP-2α (Fig. 4a). The knock-down of AP-2α did not significantly change cell proliferation (Fig. 4b), but it significantly reduced ALP activity and mineralization in MSC-O cells (Fig. 4c,d). Consistently, the knock-down of AP-2α decreased expressions of OCN, SPP1 and DSP (Fig. 4e–g). We transplanted both MSC-O cells expressing AP-2α shRNA (MSC-O/AP-2αshRNA) and MSC-O expressing luciferase shRNA (MSC-O/LucshRNA) into nude mice. As shown in Fig. 4h, the knock-down of AP-2α also significantly reduced bone/dentin-like tissue formation in vivo.
Figure 4. AP-2α is a key mediator for enhancing osteo/dentinogenic potentials of MSCs by BCOR mutation.
a, The knock-down of AP-2α in MSC-O cells. MSC-O/Lucsh, MSC-O cells expressing luciferase shRNA; MSC-O/AP-2αsh, MSC-O cells expressing AP-2α shRNA. Uncropped images of the blots are shown in the Supplementary information. b, The depletion of AP-2α in MSC-O cells did not significantly affect cell proliferation. Values are mean ± s.d for triplicate samples from a representative experiment. c, The knock-down of AP-2α reduced ALP activity in MSC-O cells. Values are mean ± s.d for triplicate samples from a representative experiment. Student’s t test was performed to determine statistical significance. **P < 0.01. d, The knock-down of AP-2α reduced mineralization in MSC-O cells. Values are mean ± s.d for triplicate samples from a representative experiment. Student’s t test was performed to determine statistical significance. **P < 0.01. e, The knock-down of AP-2α decreased SPP1 in MSC-O cells as determined by Real-time RT-PCR. Values are mean ± s.d for triplicate samples from a representative experiment. Student’s t test was performed to determine statistical significance. *P < 0.05; **P < 0.01. f, The knock-down of AP-2α decreased OCN in MSC-O cells as determined by Real-time RT-PCR. Values are mean ± s.d for triplicate samples from a representative experiment. Student’s t test was performed to determine statistical significance. **P < 0.01. g, The knock-down of AP-2;α decreased DSP expression in MSC-O cells. Uncropped images of the blots are shown in the Supplementary information. h, The knock-down of AP-2α reduced mineralized tissue formation in vivo. Both MSC-O/AP-2α shRNA and MSC-O/Luc shRNA cells were transplanted subcutaneously into the dorsal surface of 10-wk old immunocompromised beige mice. Values are mean ± s.d, n = 5. Student’s t test was performed to determine statistical significance. **P < 0.01. Scale bar, 100 µm.
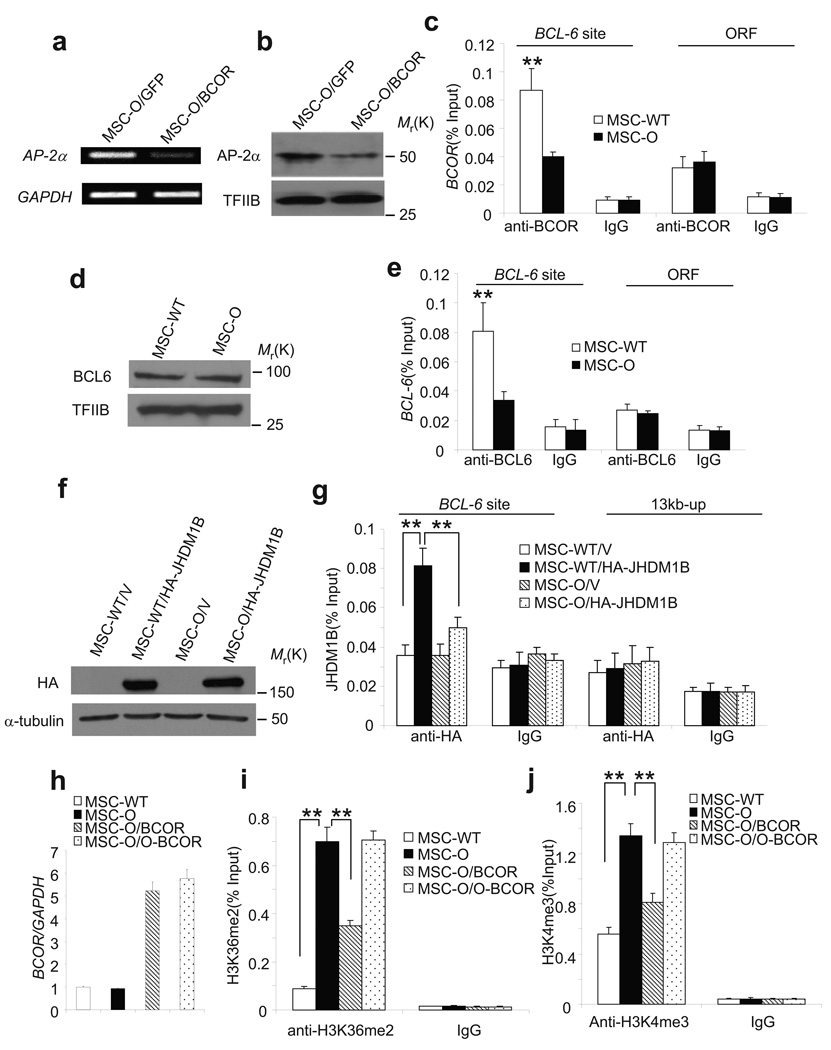
We found that over-expression of BCOR suppressed AP-2α expression in MSC-O cells, as determined by RT-PCR and Western blot analysis (Fig. 5a,b). Conversely, the knock-down of BCOR in MSC-WT cells increased AP-2α expression (Supplementary Fig. S2b). These results further confirm that BCOR controls AP-2α expression. We then examined whether BCOR was associated with the AP-2α promoter in MSC cells. A BCL6-binding site (TTTAGGAA), which is located 1439 bp upstream of the transcription start site, was found on the AP-2α promoter. Chromatin co-immunoprecipitation (ChIP) assays revealed that BCOR was present at the BCL6-binding site of AP-2α in MSC-WT cells, but not in MSC-O cells (Fig. 5c). Of note, anti-BCOR antibodies could not recognize the mutant BCOR proteins in MSC-O cells. As a negative control, BCOR was not detected in a region located in the open reading frame (ORF) of AP-2α. Interestingly, although BCL-6 was expressed at similar levels (Fig. 5d), BCL-6 binding to the AP-2α promoter was reduced in MSC-O cells compared with MSC-WT cells (Fig. 5e). Recent studies have shown that the BCOR complex contains polycomb group proteins and JHDM1B/FBXL10 demethylase13, 14. While the functional role of these molecules in the BCOR complex is not clear, their findings suggest that BCOR may utilize an epigenetic mechanism to direct gene silencing. Abnormal histone methylation due to BCOR mutation might affect BCL-6 binding to the AP-2α promoter. JHDM1B is a histone demethylase that has been shown to demethylate trimethylated lysine 4 or dimethylated 36 on histone 3 (H3K4me3 or H3K36me2)29, 30. In general, methylation at H3K4 and H3K36 is associated with transcriptional activation 31, 32. Since the BCOR C-terminus is required to interact with these molecules, we hypothesized that BCOR mutation in OFCD might impair the recruitment of JHDM1B to chromatin. However, despite our repeated efforts, available anti-JHDM1B antibodies did not work for our ChIP assays. To overcome this problem, we retrovirally expressed HA-JHDM1B in MSC-WT and MSC-O cells (Fig. 5f) and performed ChIP assays using anti-HA antibodies. As shown in Fig. 5g, HA-JHDM1B on the AP-2α promoter in MSC-WT cells was significantly higher than that in MSC-O cells. Moreover, we directly examined whether BCOR mutation affected H3K4/36 methylation of the AP-2α promoter in MSCs. ChIP assays revealed that H3K36me2 on the AP-2α promoter in MSC-O cells was 7-fold higher than in MSC-WT cells (Fig. 5i). H3K4me3 on the AP-2α promoter in MSC-O cells was also significantly increased (Fig. 5j). To further confirm our results, we over-expressed both wild type and OFCD-mutant forms of BCOR (O-BCOR) in MSC-O cells (Fig. 5h). ChIP assays showed that the restoration of wild type-BCOR significantly reduced H3K4me3 and H3K36me2 in MSC-O cells (Fig. 5i, j). On the contrary, over-expression of O-BCOR had no effects on H3K4/36 methylation in MSC-O cells. Of note, a recent study showed that JHDM1B is an H3K36me2-specific demethylase. H3K4me3 changes might be indirect33. To determine whether the BCOR/JHDM1B complex played a role in the inhibition of MSC functions, we utilized shRNA to knock-down JHDM1B (Supplementary Fig. S4a) in MSC-WT cells. The depletion of JHDM1B resulted in the induction of AP-2α expression (Supplementary Fig. S4b). Moreover, the depletion of JHDM1B enhanced osteo/dentinogenic differentiation of MSC-WT cells (Supplementary Fig. S4c–f). Finally, since the BCOR complex is associated with ubiquitylation of histone H2A, we performed ChIP assays to determine whether BCOR mutation affects ubiquintylation of H2A. ChIP assays revealed that ubiquintylation of H2A was significantly reduced in MSC-O cells as compared with MSC-WT cells (Supplementary Fig. S5).
Figure 5. BCOR represses AP-2α transcription by epigenetic mechanisms.
a, Over-expression of BCOR suppressed AP-2α expression in MSC-O cells as determined by RT-PCR. b, Over-expression of BCOR suppressed AP-2α expression in MSC-O cells as determined by Western blot analysis. Uncropped images of the blots are shown in the Supplementary information. c, BCOR was not detected in the AP-2α promoter in MSC-O cells. Chromatin and DNA complexes were immunoprecipitated with anti-BCOR antibodies. All error bars represent s.d. (n=3). Student’s t test was performed to determine statistical significance. **P < 0.01. d, BCL-6 expression in MSC-WT and MSC-O cells was examined by Western blot analysis. Uncropped images of the blots are shown in the Supplementary information. e, BCOR mutation impaired BCL-6 binding to the AP-2α promoter. ChIP assays were performed with anti-BCL-6 antibodies or control IgG. The error bars represent s.d. (n=3). Student’s t test was performed to determine statistical significance. **P < 0.01. f, Over-expression of JHDM1B in MSC-WT and MSC-O cells. Cells were transduced with retroviruses expressing HA-JHDM1B or control empty vector. Uncropped images of the blots are shown in the Supplementary information. g, BCOR mutation failed to recruit JHDM1B to the AP-2α promoter. ChIP assays were performed with anti-HA antibodies or control IgG. The error bars represent s.d. (n=3). Student’s t test was performed to determine statistical significance. **P < 0.01. h, Over-expression of BCOR or O-BCOR in MSC-O cells. The error bars represent s.d. (n=3). i, BCOR mutation resulted in increased histone H3K36 methylation in the AP-2α promoter. ChIP assays were performed with anti-H3K36me2 antibodies or control IgG. The error bars represent s.d. (n=3). **P < 0.01. h, BCOR mutation resulted in increased histone H3K4 methylation in the AP-2α promoter. ChIP assays were performed with anti-H3K4me3 antibodies or control IgG. The error bars represent s.d. (n=3). **P < 0.01.
Our studies provide a possible explanation for dental and craniofacial defects of OFCD patients. We showed that BCOR mutation led to the upregulation of AP-2α in MSCs and promoted osteo/dentinogenesis. Mechanistically, BCOR plays a critical role in development and maintenance of homeostasis via epigenetic modification of histone methylation. In normal conditions, BCOR interacts with JHDM1B and represses gene transcription by inhibiting H3K36/4 methylation on the target gene promoter in MSCs. In OFCD patients, the BCOR mutation fails to recruit JHDM1B to the target gene promoter, resulting in increased H3K36/4 methylation and transcription activation of silenced gene in MSCs. Supporting this conclusion, the depletion of JHDM1B in MSCs also induces AP-2α expression and enhances osteo/dentinogenic differentiation of MSCs. Based on analysis of our microarray results, the BCOR complex may repress a large number of genes in MSCs. In addition to AP-2α, it is possible that other genes may also play a role in dental and craniofacial defects of OFCD patients. For example, PAX3, which is associated with craniofacial development, is also activated in MSC-O cells. In addition to craniofacial defects, cataracts are the most frequent ocular phenotype of OFCD patients3, 4. Interestingly, transgenic mice over-expressing AP-2α developed cataracts34, further supporting the notion that AP-2α plays a role in the pathogenesis of OFCD. MSCs are also involved in cardiac development and formation. In future studies, it will be interesting to examine how BCOR mutation affects heart development and whether AP-2α plays a role in congenial heart defects associated with OFCD patients. In summary, by studying a rare human genetic disease, we identified the BCOR complex as a novel negative regulator of osteo/dentiogenic capacity of MSCs.
METHODS
Cell Cultures and Viral Infection
Tissues were obtained under approved guidelines set by the University of California San Francisco IRB with informed patient consent. Cells were grown in a humidified 5% CO2 incubator at 37°C in DMEM alpha modified Eagle’s medium (Invitrogen) supplemented with 15% fetal bovine serum (FBS; Invitrogen). The full-length AP-2α and JHDM1B mRNAs from MSC-O cells were amplified by RT-PCR and subsequently subcloned into pQCXIP retroviral vector (BD Biosciences). pCLMFG Flag-BCOR/C IRES eGFP plasmids were kindly provided by Dr. Vivian Bardwell at the University of Minnesota. Viral packaging was prepared as described previously35. For viral infection, MSCs were plated overnight and then infected with retroviruses in the presence of polybrene (6 µg/ml, Sigma-Aldrich) for 6 hr. The target sequences for shRNA were: AP-2α, 5’-TCCAGGAAGATCTTTAAGA-3’; BCOR, 5’-GATGGCTTCAGTGCTATAT-3’; JHDM1B, 5’-GAGTCAAGACGTAGAATAA-3’ and luciferase, 5’-GTGCGTTGCTAGTACCAAC-3’. The shRNA was subcloned into a pSIREN retroviral vector (BD Bioscience) and retrovirus packaging was performed as described previously35.
Western blot analysis
Cells were lysed in RIPA buffer (10 mM Tris-HCL, 1 mM EDTA, 1% sodium dodecyl sulfate [SDS], 1% Nonidet P-40, 1: 100 proteinase inhibitor cocktail, 50 mM β-glycerophosphate, 50 mM sodium fluoride). The samples were separated on a 10% SDS polyacrylamide gel and transferred to PVDF membrane by a semi-dry transfer apparatus (Bio-Rad). The membranes were blotted with 5% milk for 2 hr and then incubated with primary antibodies overnight. The immunocomplexes were incubated with horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG (Promega) and visualized with SuperSignal reagents (Pierce). Primary antibodies were purchased from the following commercial sources: monoclonal antibodies against AP-2α, polyclonal antibodies against HSP90 and TFIIB (Santa Cruz, CA, USA); monoclonal antibodies against ubiquitinated H2A (Millipore); polyclonal antibodies against H3K4me3 (Abcam) and H3K36me2 (Upstate); polyclonal antibodies against dentin sialoprotein (DSP) (NIDCR/NIH, USA); polyclonal anti-BCL-6 antibodies (Cell Signaling); monoclonal antibodies against α-tubulin (Sigma-Aldrich).
ALP and Alizarin Red staining
MSCs were grown in mineralization-inducing media containing 100 µM ascorbic acid, 2 mM β-glycerophosphate and 10 nM dexamethasone. For ALP staining, after induction, cells were fixed with 70% ethanol and incubated with a solution of 0.25% naphthol AS-BI phosphate and 0.75% Fast Blue BB dissolved in 0.1 M Tris buffer (pH 9.3). ALP activity assay was performed with an ALP kit according to the manufacturer’s protocol (Sigma-Aldrich) and normalized based on protein concentrations. To assess mineralization, cells were induced for 2 to 3 weeks, fixed with 70% ethanol and stained with 2% Alizarin red (Sigma-Aldrich). To quantitatively determine calcium mineral density, Alizarin Red was destained with 10% cetylpyridinium chloride in 10 mM sodium phosphate for 30 minutes at room temperature. The concentration was determined by absorbance measurement at 562nm on a multiplate reader using a standard calcium curve prepared in the same solution. The final calcium levels in each group were normalized with the total protein concentrations prepared from duplicate plates20.
Reverse transcriptase-polymerase chain reaction (RT-PCR) and Real-time RT-PCR
Total RNA was isolated from MSCs using Trizol reagents (Invitrogen, Carlsbad, CA). The primers for AP-2α are: forward, 5’-CTCTCACCACCCGAGTGTCT-3’; reverse, 5’- GAGGTTGAAGTGGGTCAAGC-3’. The primers for BCOR are: forward, CTCAGGAGACCACCCAGTC-3’; reverse, 5’-CCCTGAGCCACAGATACTTG-3’. The primers for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) are: forward, 5’-GATCATCAGCAATGCCTCCT-3’; reverse, 5’-ACCTGGTGCTCAGTGTAGCC-3’. For RT-PCR, 2-µg aliquots of RNAs were synthesized using random hexamers and reverse transcriptase according to the manufacturer’s protocol (Invitrogen).
The real-time PCR reactions were performed using the QuantiTect SYBR Green PCR kit (Qiagen) and Icycler iQ Multi-color Real-time PCR Detection System. The primers for SPP1 are: forward, 5’-ATGATGGCCGAGGTGATAGT-3’; reverse, 5’-ACCATTCAACTCCTCGCTTT-3’. The primers for OCN are: forward, 5’-AGCAAAGGTGCAGCCTTTGT-3’; reverse, 5’- GCGCCTGGGTCTCTTCACT-3’. The primers for JHDM1B are: forward, 5’-ACTTGACCATACCAATGGCGGT-3’; reverse, 5’- AAGCTGGTCAGGATTGCCAGAA-3’. The primers for BCOR are: forward, 5’-CATAGTGCTTGTGGAACTCCG-3’; reverse, 5’- GGACACAGCTCTCCTGTTGC-3’. The primers for AP-2α are: forward, 5’- CTGCAGGGAGACGTAAAGC; reverse, 5’-GGCTAGGTGGACAGCTTCTC-3’. The primers for 18S rRNA are: forward, 5’-CGGCTACCACATCCAAGGAA-3’; reverse, 5’-GCTGGAATTACCGCGGCT-3’. The primers for GAPDH are: forward, 5’CGGCTACCACATCCAAGGAA-3’; reverse, 5’-AGCCACATCGCTCAGACACC-3’.
Human Affymetrix microarray
Total RNAs were extracted from MSC-WT and MSC-O cells with Trizol reagents and cleaned with an RNeasy kit (Qiagen). 5-µg aliquots of total RNA from each sample were transcribed to double-stranded complementary DNA (cDNA) using SuperScript II RT (Invitrogen) with an oligo-dT primer and then used to generate single stranded RNAs. The biotin-labeled RNAs were fragmented and hybridized with an Affymetrix Human Genome U133 Plus 2.0 Array. The arrays were scanned with the GeneArray scanner (Affymetrix). The one-step Tukey's Biweight Estimate was used to calculate signal intensity. Affymetrix® Microarray Suite (MAS) 5.0 was used for data analysis35.
ChIP assays
The assay was performed using a ChIP assay kit (Upstate) according to the manufacturer’s protocol. Polyclonal antibodies against BCOR were kindly provided by Dr. Vivian Bardwell at the University of Minnesota. Cells were incubated with 5 mM dimethyl 3,3’ dithiobispropionimidate-HCl (Pierce) solution for 10 min at room temperature before formaldehyde treatment. For each ChIP reaction, 2× 106 cells were used. All resulting precipitated DNA samples were quantified with Real-time PCR. Data were expressed as the percentage of input DNA. The BCL-6 binding site was detected in 1439 bp upstream of the AP-2α transcription start site. The surrounding region of the binding site was used for amplification. The primer sequences for the BCL-6-binding region of the AP-2α promoter are: forward, 5’GTGAGGGAATGCTCCAATCT-3’; reverse, 5’-CCTTTGATTCATCTGGGCTT. The primer sequences from ORF are: forward, 5’- CCTCGAAGTACAAGGTCACG-3’; reverse, 5’-GACACTCGGGTGGTGAGAG-3’. The primer sequences from 13kb up of AP-2α are: forward, 5’-CCGCCCTGTCTCTGGTACTTTC-3’; reverse, 5’-AGCACCTTCTATACAGCATTCG-3’.
Transplantation in nude mice
Approximately 4.0×106 of cells were mixed with 40 mg of hydroxyapatite/tricalcium phosphate (HA/TCP) ceramic particles (Zimmer) and then transplanted subcutaneously into the dorsal surface of 10-week-old immunocompromised beige mice as previously described11, 24. These procedures were performed in accordance with an approved animal protocol. 8 weeks after transplantation, the transplants were harvested, fixed with 10% formalin, decalcified with buffered 10% EDTA (pH 8.0), and then embedded in paraffin. Sections were deparaffinized, hydrated, and stained with H&E.
Accession numbers
The accession number for the microarray data is GSE15214.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Institute of Dental and Craniofacial Research Grants (R01DE1016513 and R01DE017684) and the Shapiro Family Charitable Foundation. We thank Dr. Vivian Bardwell for reagents.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
REFERENCES
- 1.Huynh KD, Fischle W, Verdin E, Bardwell VJ. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 2000;14:1810–1823. [PMC free article] [PubMed] [Google Scholar]
- 2.Ghetu AF, et al. Structure of a BCOR corepressor peptide in complex with the BCL6 BTB domain dimer. Mol Cell. 2008;29:384–391. doi: 10.1016/j.molcel.2007.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng D, et al. Oculofaciocardiodental and Lenz microphthalmia syndromes result from distinct classes of mutations in BCOR. Nat Genet. 2004;36:411–416. doi: 10.1038/ng1321. [DOI] [PubMed] [Google Scholar]
- 4.Oberoi S, Winder AE, Johnston J, Vargervik K, Slavotinek AM. Case reports of oculofaciocardiodental syndrome with unusual dental findings. Am J Med Genet A. 2005;136:275–277. doi: 10.1002/ajmg.a.30811. [DOI] [PubMed] [Google Scholar]
- 5.Horn D, et al. Novel mutations in BCOR in three patients with oculo-facio-cardio-dental syndrome, but none in Lenz microphthalmia syndrome. Eur J Hum Genet. 2005;13:563–569. doi: 10.1038/sj.ejhg.5201391. [DOI] [PubMed] [Google Scholar]
- 6.Hedera P, Gorski JL. Oculo-facio-cardio-dental syndrome: skewed X chromosome inactivation in mother and daughter suggest X-linked dominant Inheritance. Am J Med Genet A. 2003;123A:261–266. doi: 10.1002/ajmg.a.20444. [DOI] [PubMed] [Google Scholar]
- 7.Schulze BR, Horn D, Kobelt A, Tariverdian G, Stellzig A. Rare dental abnormalities seen in oculo-facio-cardio-dental (OFCD) syndrome: three new cases and review of nine patients. Am J Med Genet. 1999;82:429–435. doi: 10.1002/(sici)1096-8628(19990219)82:5<429::aid-ajmg13>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 8.Foster BL, Popowics TE, Fong HK, Somerman MJ. Advances in defining regulators of cementum development and periodontal regeneration. Curr Top Dev Biol. 2007;78:47–126. doi: 10.1016/S0070-2153(06)78003-6. [DOI] [PubMed] [Google Scholar]
- 9.Kim JW, Simmer JP. Hereditary dentin defects. J Dent Res. 2007;86:392–399. doi: 10.1177/154405910708600502. [DOI] [PubMed] [Google Scholar]
- 10.MacDougall M, Dong J, Acevedo AC. Molecular basis of human dentin diseases. Am J Med Genet A. 2006;140:2536–2546. doi: 10.1002/ajmg.a.31359. [DOI] [PubMed] [Google Scholar]
- 11.Sonoyama W, et al. Mesenchymal stem cell-mediated functional tooth regeneration in swine. PLoS ONE. 2006;1:e79. doi: 10.1371/journal.pone.0000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye BH, et al. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science. 1993;262:747–750. doi: 10.1126/science.8235596. [DOI] [PubMed] [Google Scholar]
- 13.Gearhart MD, Corcoran CM, Wamstad JA, Bardwell VJ. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol Cell Biol. 2006;26:6880–6889. doi: 10.1128/MCB.00630-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez C, et al. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics. 2007;6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Wamstad JA, Bardwell VJ. Characterization of Bcor expression in mouse development. Gene Expr Patterns. 2007;7:550–557. doi: 10.1016/j.modgep.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wamstad JA, Corcoran CM, Keating AM, Bardwell VJ. Role of the transcriptional corepressor Bcor in embryonic stem cell differentiation and early embryonic development. PLoS ONE. 2008;3:e2814. doi: 10.1371/journal.pone.0002814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jahagirdar BN, Verfaillie CM. Multipotent adult progenitor cell and stem cell plasticity. Stem Cell Rev. 2005;1:53–59. doi: 10.1385/SCR:1:1:053. [DOI] [PubMed] [Google Scholar]
- 18.Phinney DG, Prockop DJ. Concise review: mesenchymal stem/multipotent stromal cells: the state of transdifferentiation and modes of tissue repair--current views. Stem Cells. 2007;25:2896–2902. doi: 10.1634/stemcells.2007-0637. [DOI] [PubMed] [Google Scholar]
- 19.Scaffidi P, Misteli T. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat Cell Biol. 2008;10:452–459. doi: 10.1038/ncb1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi S, et al. Bone formation by human postnatal bone marrow stromal stem cells is enhanced by telomerase expression. Nat Biotechnol. 2002;20:587–591. doi: 10.1038/nbt0602-587. [DOI] [PubMed] [Google Scholar]
- 21.Shi S, Wang CY. Bone marrow stromal stem cells for repairing the skeleton. Biotechnol Genet Eng Rev. 2004;21:133–143. doi: 10.1080/02648725.2004.10648052. [DOI] [PubMed] [Google Scholar]
- 22.Seo BM, et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. 2004;364:149–155. doi: 10.1016/S0140-6736(04)16627-0. [DOI] [PubMed] [Google Scholar]
- 23.Arthur A, Rychkov G, Shi S, Koblar SA, Gronthos S. Adult Human Dental Pulp Stem Cells Differentiate Towards Functionally Active Neurons Under Appropriate Environmental Cues. Stem Cells. 2008 doi: 10.1634/stemcells.2007-0979. [DOI] [PubMed] [Google Scholar]
- 24.Chang J, et al. Noncanonical Wnt-4 signaling enhances bone regeneration of mesenchymal stem cells in craniofacial defects through activation of p38 MAPK. J Biol Chem. 2007;282:30938–30948. doi: 10.1074/jbc.M702391200. [DOI] [PubMed] [Google Scholar]
- 25.Butler WT, Brunn JC, Qin C. Dentin extracellular matrix (ECM) proteins: comparison to bone ECM and contribution to dynamics of dentinogenesis. Connect Tissue Res. 2003;44(Suppl 1):171–178. [PubMed] [Google Scholar]
- 26.Scheller EL, Chang J, Wang CY. Wnt/beta-catenin inhibits dental pulp stem cell differentiation. J Dent Res. 2008;87:126–130. doi: 10.1177/154405910808700206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schorle H, Meier P, Buchert M, Jaenisch R, Mitchell PJ. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature. 1996;381:235–238. doi: 10.1038/381235a0. [DOI] [PubMed] [Google Scholar]
- 28.Brewer S, Feng W, Huang J, Sullivan S, Williams T. Wnt1-Cre-mediated deletion of AP-2alpha causes multiple neural crest-related defects. Dev Biol. 2004;267:135–152. doi: 10.1016/j.ydbio.2003.10.039. [DOI] [PubMed] [Google Scholar]
- 29.Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature. 2007;450:309–313. doi: 10.1038/nature06255. [DOI] [PubMed] [Google Scholar]
- 30.Tsukada Y, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 31.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 32.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 33.He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nat Struct Mol Biol. 2008;15:1169–1175. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.West-Mays JA, Coyle BM, Piatigorsky J, Papagiotas S, Libby D. Ectopic expression of AP-2alpha transcription factor in the lens disrupts fiber cell differentiation. Dev Biol. 2002;245:13–27. doi: 10.1006/dbio.2002.0624. [DOI] [PubMed] [Google Scholar]
- 35.Park BK, et al. NF-kappaB in breast cancer cells promotes osteolytic bone metastasis by inducing osteoclastogenesis via GM-CSF. Nat Med. 2007;13:62–69. doi: 10.1038/nm1519. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.