Abstract
BACKGROUND
The childhood onset of idiopathic cardiac hypertrophy that occurs without a family history of cardiomyopathy can portend a poor prognosis. Despite morphologic similarities to genetic cardiomyopathies of adulthood, the contribution of genetics to childhood-onset hypertrophy is unknown.
METHODS
We assessed the family and medical histories of 84 children (63 boys and 21 girls) with idiopathic cardiac hypertrophy diagnosed before 15 years of age (mean [±SD] age, 6.99±6.12 years). We sequenced eight genes: MYH7, MYBPC3, TNNT2, TNNI3, TPM1, MYL3, MYL2, and ACTC. These genes encode sarcomere proteins that, when mutated, cause adult-onset cardiomyopathies. We also sequenced PRKAG2 and LAMP2, which encode metabolic proteins; mutations in these genes can cause early-onset ventricular hypertrophy.
RESULTS
We identified mutations in 25 of 51 affected children without family histories of cardiomyopathy and in 21 of 33 affected children with familial cardiomyopathy. Among 11 of the 25 children with presumed sporadic disease, 4 carried new mutations and 7 inherited the mutations. Mutations occurred predominantly (in >75% of the children) in MYH7 and MYBPC3; significantly more MYBPC3 missense mutations were detected than occur in adult-onset cardiomyopathy (P<0.005). Neither hypertrophic severity nor contractile function correlated with familial or genetic status. Cardiac transplantation and sudden death were more prevalent among mutation-positive than among mutation-negative children; implantable cardioverter–defibrillators were more frequent (P=0.007) in children with family histories that were positive for the mutation.
CONCLUSIONS
Genetic causes account for about half of presumed sporadic cases and nearly two thirds of familial cases of childhood-onset hypertrophy. Childhood-onset hypertrophy should prompt genetic analyses and family evaluations.
The Diagnosis of Childhood Cardiomyopathies can be prompted by abnormal physical findings that occur without symptoms or by life-threatening events, including sudden death, which is the presenting manifestation in 3.5% of affected children.1–4 Despite sophisticated medical management, rates of death and cardiac transplantation among children with symptomatic childhood-onset cardiomyopathy approach 40%.2,3 The early age at diagnosis and the striking differences in morbidity and mortality that distinguish childhood cardiomyopathies from adult-onset cardiomyopathies have been interpreted to indicate distinct causes of these pathologic conditions.5,6
Adult-onset hypertrophic cardiomyopathy is a prevalent genetic condition caused by inherited or new mutations in genes that encode sarcomere proteins, including cardiac β-myosin heavy chain (MYH7), cardiac myosin-binding protein C (MYBPC3), cardiac troponin T (TNNT2), cardiac troponin I (TNNI3), essential myosin light chain (MYL3), regulatory myosin light chain (MYL2), α-tropomyosin (TPM1), cardiac actin (ACTC), and titin (TTN).7 Less commonly, hypertrophic cardiomyopathy is caused by mutations in other genes.8,9 The range of ages at clinical diagnosis of hypertrophic cardiomyopathy is broad; however, manifestations before 14 years of age are atypical, even in children with an inherited gene mutation.10
Mutations affecting the γ2 regulatory subunit of AMP-activated protein kinase (PRKAG2) (which regulates substrate use for energy production) cause early-onset left ventricular hypertrophy with arrhythmias11 and, more rarely, fatal infantile cardiac glycogenosis.12 Mutations in the gene encoding lysosome-associated membrane protein 2 (LAMP2), which is located on the X chromosome, cause massive left ventricular hypertrophy in boys in whom systemic manifestations (the Danon disease) may also develop.13 Because LAMP2 mutations are usually clinically silent in female carriers, affected boys appear to have sporadic rather than inherited disease.
To determine whether some childhood-onset left ventricular hypertrophies share a genetic cause with hypertrophic cardiomyopathy, we sequenced genes encoding eight sarcomere proteins, PRKAG2, and LAMP2 in 84 children with isolated, idiopathic left ventricular hypertrophy diagnosed before 15 years of age.
METHODS
SUBJECTS
Studies were performed in accordance with institutional guidelines and approved by the ethics committees of Brigham and Women’s Hospital and Baylor College of Medicine. Written informed consent was provided by the parents and age-appropriate assent was provided by the children. Subjects were participants in the Pediatric Cardiomyopathy Registry of the National Heart, Lung, and Blood Institute or were identified through referrals to investigators. All subjects had isolated, unexplained left ventricular hypertrophy (defined as a wall thickness >2 SD above the normal-population mean for body-surface area) that was diagnosed at or before 15 years of age. Subjects with extracardiac disease (suggesting the Danon disease, glycogen storage disease [types I through VI], Diamond–Blackfan anemia, muscular dystrophies, skeletal myopathies, or mitochondrial diseases) or secondary causes of hypertrophy (e.g., congenital malformations, hypertension, or drug exposure) were excluded. Family histories were obtained from parents and, when possible, medical records were reviewed. The ancestry of subjects and controls was determined by parental report or self-report with the use of a list of options (see Table 1 of the Supplementary Appendix, available with the full text of this article at www.nejm.org).
DNA SEQUENCING AND CONFIRMATION OF SEQUENCE VARIANTS
DNA was extracted from whole blood.13,14 Exons of MYH7, MYBPC3, TNNT2, TNNI3, TPM1, MYL2, MYL3, ACTC, PRKAG2, and LAMP2 were amplified from genomic DNA with the use of previously described primers and methods.11,13,15 After poly-merase-chain-reaction (PCR) purification with the use of a QIAquick purification kit (Qiagen), DNA was sequenced with a dye-terminator cycle-sequencing system (ABI PRISM, Applied Biosystems).
DNA sequence variants were confirmed by means of restriction-enzyme digestion of PCR-amplified fragments with the use of oligonucleotide primers (Table 2 of the Supplementary Appendix). Previously unreported nonsynonymous sequence variants (Table 1, and Table 3 of the Supplementary Appendix) were assessed with the use of the described procedures in DNA samples obtained from 180 unrelated persons who were matched by ancestral origin to the subjects.
Table 1.
Clinical and Genetic Profiles of Patients with Childhood-Onset Left Ventricular Hypertrophy.*
| Variable | Sporadic Disease | Familial Disease | ||||
|---|---|---|---|---|---|---|
| Total | Mutation- Positive |
Mutation- Negative |
Total | Mutation- Positive |
Mutation- Negative |
|
| Probands (no.) | 51 | 25 | 26 | 33 | 21 | 12 |
| Male sex (no.) | 39 | 21 | 18 | 24 | 14 | 10 |
| Age at onset (yr) | 6.04±6.2 | 6.52±6.6 | 5.28±5.6 | 10.5±5.8 | 10.3±5.0 | 10.4±6.7 |
| Maximum LVWT (cm)† | 1.79±1.06 | 1.92±1.16 | 1.59±0.87 | 1.98±0.91 | 2.15±0.93 | 1.68±0.83 |
| Fractional shortening (%)† | 45±13 | 45±9.3 | 45±18 | 41±4 | 42±3 | 39±5 |
| Sudden death from cardiac causes (no.) |
3 | 3 | 0 | 0 | 0 | 0 |
| ICD (no.) | 2‡ | 2‡ | 0 | 8 | 5 | 3 |
| Cardiac transplantation (no.) | 4§ | 3§ | 1 | 1 | 1 | 0 |
Plus–minus values are means ±SD. ICD denotes implantable cardioverter–defibrillator, and LVWT left ventricular wall thickness.
There were no significant differences in the maximum LVWT (before and after adjustment for age, body-surface area, or both) or fractional shortening among groups.
One child died suddenly from cardiac causes.
One child had an ICD, underwent cardiac transplantation, and died suddenly from cardiac causes.
STATISTICAL ANALYSIS
Descriptive data for continuous clinical values were expressed as means ±SD or discrete values (numbers or percentages). Differences in clinical characteristics between groups were assessed with the use of Proc GLM software for continuous variables (SAS Institute) or the Proc Logistic procedure for discrete variables (SAS Institute).16 Distributions of types of sarcomere-protein gene mutations in groups were compared with the use of two-tailed exact P values, with a P value of less than 0.05 considered to indicate statistical significance.
RESULTS
We studied 84 unrelated children (63 boys and 21 girls) with unexplained left ventricular hypertrophy diagnosed at or before 15 years of age (mean age, 6.99±6.12 years) (Table 1). Hypertrophy was determined by means of echocardiography (Fig. 1) and defined as a mean left ventricular wall thickness of at least 2 SD above the normal value, stratified according to body-mass index or age.1
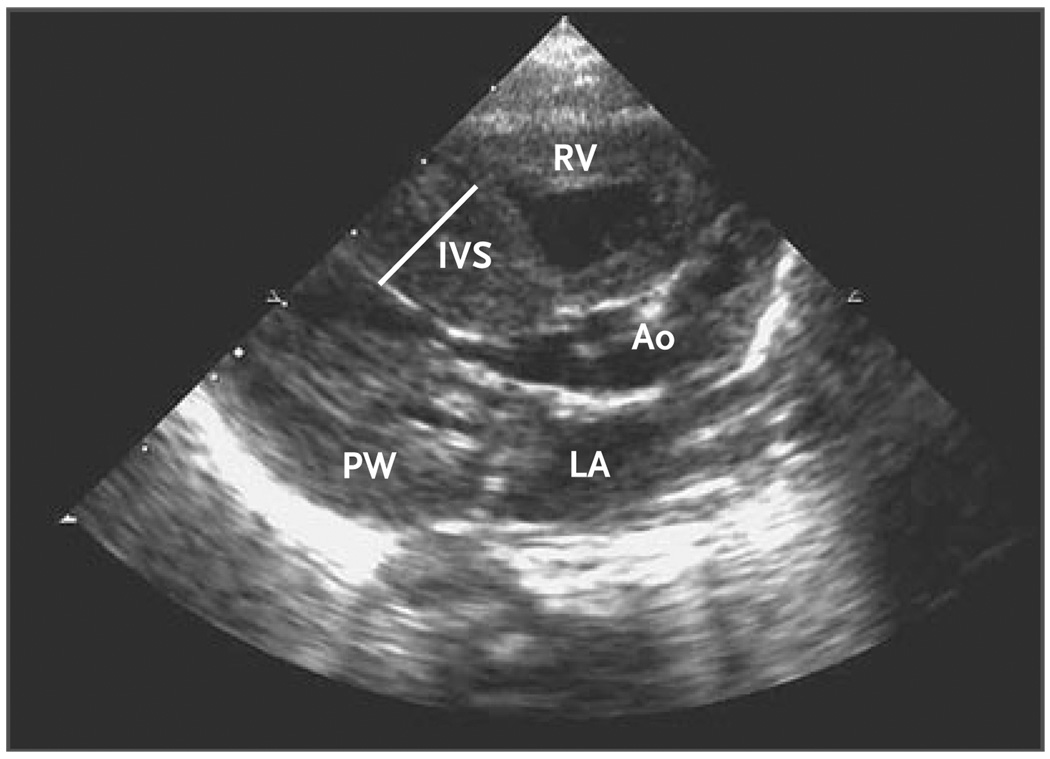
Figure 1. Echocardiogram Showing Unexplained Asymmetric Left Ventricular Hypertrophy Diagnosed in the First Week of Life during Evaluation of a Boy for Aspiration Pneumonia.
The interventricular septum (IVS) measured 12 mm, and the posterior wall (PW) measured 7 mm. The ejection fraction was 75%, and the calculated left ventricular mass was 41 g. Genetic analyses revealed a previously unreported missense mutation, MYBPC3 Arg495Gly; other amino acid substitutions at this codon are known causes of hypertrophic cardiomyopathy. Ao denotes aorta, LA left atrium, and RV right ventricle.
The ages of the children at diagnosis ranged from 2 days to 15 years. Initial clinical evaluations were prompted by abnormal physical findings (e.g., murmur or cardiomegaly) identified during routine examinations or by symptoms that were nonspecific (e.g., irritability or lack of feeding in an infant) or suggestive of heart disease (e.g., chest pain). Three children died suddenly from cardiac causes, and in one child this was the presenting manifestation. Over a 5-year follow-up period, 10 children received implantable cardioverter–defibrillators and 5 children underwent cardiac transplantation.
Family histories were positive for cardiomyopathy in 33 children but negative in 51 children who were presumed to have sporadic disease. There were no significant differences with regard to sex distribution, age at presentation, maximum left ventricular wall thickness, contractile function, sudden death, or cardiac transplantation between the group of children with familial disease and the group of children with presumed sporadic cardiomyopathy (Table 1). However, significantly more children with a family history of cardiomyopathy received implantable cardioverter–defibrillators (8 of 33 children, vs. 2 of 51 children with no family history; P = 0.007).
Nucleotide sequences encoding eight sarcomere-protein genes (MYH7, MYBPC3, TNNT2, TNNI3, TPM1, MYL3, MYL2, and ACTC) and two metabolic genes (PRKAG2 and LAMP2) were determined in each proband. Among children with presumed sporadic cardiomyopathy, 112 sequence variants were identified (Fig. 1 of the Supplementary Appendix). Seventy-nine synonymous variants or intronic polymorphisms distant from conserved splicing sequences were not predicted to alter the protein structure. Six nonsynonymous MYBPC3 variants (Val158Met, Ser236Gly, Arg326Gln, Val-896Met, Arg1002Trp, and Gln1233Ter) are known polymorphisms.15,17,18 These 85 variants were not studied further.
Among the remaining 27 sequence variants, 3 (MYBPC3 IVS31+2t→g, MYL3 IVS5+6c→t, and TNNT2 IVS14+6c→t) were predicted to alter RNA splice sites or conserved flanking sequences. To assess the effect of these three variants on splicing, we carried out in vitro cell-based assays (Fig. 2 of the Supplementary Appendix).19,20 Only one variant, MYBPC3 IVS31+2t→g, disrupted RNA splicing in a way that would be predicted to alter protein structure (Fig. 2 of the Supplementary Appendix), and it is thus similar to an established MYBPC3 splice-site mutation that causes familial hypertrophic cardiomyopathy.21 We therefore evaluated MYBPC3 IVS31+2t→g, but not MYL3 IVS5+6c→t or TNNT2 IVS14+6c→t, along with the other 24 variants predicted to affect protein structure and function.
We observed 25 unique, nonsynonymous sequence variants in MYH7, MYBPC3, TNNT2, TNNI3, ACTC, and PRKAG2 (Table 2, and Fig. 1 of the Supplementary Appendix) in children with presumed sporadic cardiomyopathy. We did not observe associations between variant residues in the three other genes associated with hypertrophic cardiomyopathy — TPM1, MYL2, and MYL3 — and childhood-onset left ventricular hypertrophy. Each nonsynonymous sequence variant was independently confirmed by restriction-enzyme digestion of PCR-amplified sequences. Fourteen of the 25 variants (Table 2) have been reported to cause familial hypertrophic cardiomyopathy15,18,22–33; 12 of these variants also encode missense residues, 1 is predicted to delete an amino acid, and 1 is predicted to prematurely terminate protein translation.
Table 2.
Gene Mutations in Patients with Childhood-Onset Cardiac Hypertrophy and Presumed Sporadic Cardiomyopathy.*
| Gene | Mutation | Sequence | Consequence | Charge | Previously Reported Hypertrophic Cardiomyopathy Mutations |
|---|---|---|---|---|---|
| MYH7 | Lys146Asn | G→T | Missense | −1 | Yes |
| MYH7 | Val606Met | G→A | Missense | 0 | Yes |
| MYH7 | Arg663His | G→A | Missense | 0 | Yes |
| MYH7 | Arg719Gln | G→A | Missense | −1 | Yes |
| MYH7 | Val763Met | G→A | Missense | 0 | No |
| MYH7 | Arg787Cys | C→T | Missense | −1 | No |
| MYH7 | Leu908Val | C→G | Missense | 0 | Yes |
| MYH7 | Glu924Lys | G→A | Missense | +2 | Yes |
| MYH7 | Leu1414Met | C→A | Missense | 0 | No |
| MYBPC3 | Gly278Glu | G→A | Missense | −1 | Yes |
| MYBPC3 | Gly490Arg | G→A | Missense | +1 | Yes |
| MYBPC3 | Arg495Gly | C→G | Missense | −1 | No |
| MYBPC3 | Arg502Trp† | C→T | Missense | −1 | Yes |
| MYBPC3 | Arg502Gln | G→A | Missense | −1 | Yes |
| MYBPC3 | Asp605Asn | G→A | Missense | + 1 | Yes |
| MYBPC3 | Arg943ter | C→T | Truncation | NA | Yes |
| MYBPC3 | Thr1028Ser | C→G | Missense | 0 | No |
| MYBPC3 | IVS31+2t→g‡ | t→g | Truncation | NA | No |
| MYBPC3 | Gly1248Arg | G→a | Missense | + 1 | No |
| TNNT2 | Arg92Gln | G→A | Missense | −1 | Yes |
| TNNT2 | Glu96del | Δ GAG | Codon 96 deleted | + 1 | No |
| TNNI3 | Lys178del | Δ AAG | Codon 178 deleted | −1 | Yes |
| ACTC | His90Tyr | C→T | Missense | −1 | No |
| ACTC | Arg97Cys | C→T | Missense | −1 | No |
| PRKAG2 | His530Arg | A→G | Missense | 0 | No |
Mutations are denoted by normally encoded amino acid residue number, substituted amino acid or termination signal (ter), or altered splice signal (intervening sequence). Sequence refers to nucleotide substitution, and consequence refers to the mutational effects on protein. Charge is the altered charge by the mutant amino acid change. NA denotes not available. Three children had compound mutations: MYH7 Arg663His and MYH7 Val763Met, MYBPC3 Thr1028Ser and MYBPC3 IVS31+2t→g, and MYH7 Arg787Cys and ACTC Arg97Cys.
This mutation was identified in three children.
This mutation was identified in two children.
Eleven sequence variants were not previously reported: MYH7 Val763Met, MYH7 Arg787Cys, MYH7 Leu1414Met, MYBPC3 Arg495Gly, MYBPC3 Thr1028Ser, MYBPC3 IVS31+2t→g, MYBPC3 Gly1248Arg, TNNT2 Glu96del, ACTC His90Tyr, ACTC Arg97Cys, and PRKAG2 His530Arg. Nine of these previously unreported variants caused missense changes and altered amino acid residues that are highly conserved during mammalian evolution (Fig. 3 of the Supplementary Appendix), one variant deleted a highly conserved glutamic acid residue in TNNT2, and one variant altered MYBPC3 splicing (detailed above). Each of these 11 previously unreported variants was absent in 180 unrelated persons matched by ancestral origin to the subjects (data not shown) and in more than 1000 chromosomes of unaffected persons (unpublished data). Furthermore, sequence analyses of these eight sarcomere-protein genes in 34 persons without left ventricular hypertrophy revealed no variants.34 On the basis of a chi-square analysis, the probability that 25 nonsynonymous sequence variants occurred by chance in 51 cases of pediatric-onset left ventricular hypertrophy is less than 1 in 500,000. We conclude that each of these previously unreported variants, like the 14 mutations known to cause hypertrophic cardiomyopathy, caused left ventricular hypertrophy in these children.
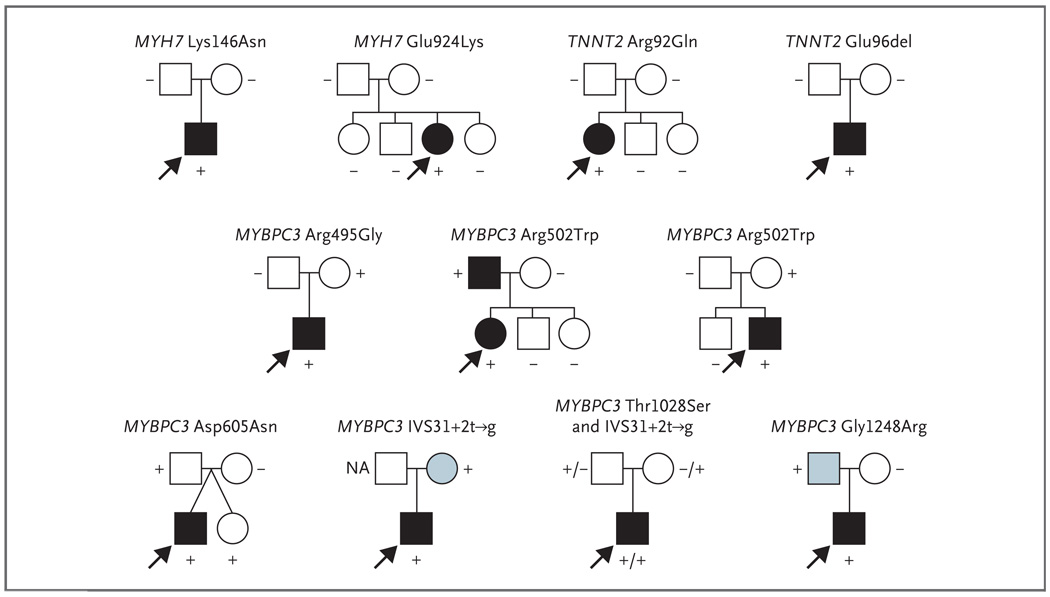
Three of the 25 children with presumed sporadic cardiomyopathy who had mutations (12%) had two different sarcomere-gene mutations: MYH7 Arg663His and Val763Met; MYH7 Arg787Cys and ACTC Arg97Cys; and MYBPC3 Thr1028Ser and MYBPC3 IVS31+2t→g. We observed the MYBPC3 IVS31+2t→g mutation in isolation in another child; others27 have reported MYH7 Arg663His in isolation as the cause of hypertrophic cardiomyopathy. To determine whether sporadic left ventricular hypertrophy in the children reflected new mutational events, we genotyped the parents of 11 probands (Fig. 2) and confirmed parental relationships using 22 polymorphic microsatellite markers (data not shown). Four mutations (MYH7 Lys146Asn, MYH7 Glu924Lys, TNNT2 Arg92Gln, and TNNT2 Glu96del) were absent from both parents, confirming that new mutations accounted for sporadic cardiomyopathy in these children. In contrast, mutations found in seven children were also present in the parents, indicating that these mutations were inherited (Fig. 2). Clinical studies revealed cardiomyopathy in one parent, were not performed in two parents, and showed no abnormalities in five parents, two of whom carried one of the two MYBPC3 mutations carried by their compound heterozygous (Thr1028Ser and IVS31+2t→g) affected child.
Figure 2. Family Pedigrees of 11 Patients with Childhood-Onset Idiopathic Cardiac Hypertrophy and No Family History of Cardiomyopathy.
Mutations identified in patients (arrows) were studied in family members, and their clinical status was ascertained. The plus symbol denotes the presence of the mutation, the minus symbol the absence of the mutation, and NA mutation status not available. Four mutations that were not detected in parents were new, and seven mutations were inherited from one parent. Circles denote female family members, squares male family members, solid symbols clinically affected family members, open symbols clinically unaffected members, and blue symbols family members with unknown clinical status.
To determine the parental origins of chromosomes for new mutations, closely linked polymorphisms were analyzed. Haplotype analyses were not informative in two subjects (with mutations MYH7 Lys146Asn and TNNT2 Glu96del). Haplotype analyses indicated that the new mutation MYH7 Glu924Lys arose on a maternal chromosome (Fig. 4A of the Supplementary Appendix), and the new mutation TNNT2 Arg92Gln arose on a paternal chromosome (Fig. 4B of the Supplementary Appendix).
We also sequenced genes in 33 patients who had childhood-onset cardiac hypertrophy and family histories of cardiomyopathy but who had not undergone previous genetic studies. Twenty-two nonsynonymous sequence variants (Table 3 of the Supplementary Appendix) were identified in 21 patients (64%); each variant altered a sarcomere-protein gene: MYH7 (in 9 patients), MYBPC3 (in 7 patients), TNNT2 (in 2 patients), TNNI3 (in 1 patient), TPM1 (in 2 patients), and MYL3 (in 1 patient). No variants were found in ACTC or MYL2. Fifteen sequence variants were previously reported as cardiomyopathy mutations (Table 3 of the Supplementary Appendix). The remaining seven variants (MYH7 Glu903Gly, MYH7 Ser1836Leu, MYBPC3 Ile154Thr, MYBPC3 Asp605del, MYBPC3 Ser858Asn, TMP1 Ser215Leu, and MYL3 Met173Val) were absent from 180 unrelated, ancestrally matched persons (data not shown) and from more than 1000 chromosomes of unaffected persons (unpublished data). Each of these new variants altered amino acid residues that are highly conserved throughout mammalian evolution (Fig. 5 of the Supplementary Appendix), predicting that the missense residue alters protein structure.
Among mutation-positive children with familial disease, three (14%) had compound mutations (MYBPC3 Arg502Trp and MYBPC3 Ser858Asn; MYBPC3 Arg495Gln and TNNI3 Arg141Gln; and MYBPC3 Ile154Thr and MYBPC3 Asp605del).
DISCUSSION
Genetic studies of childhood-onset cardiomyopathy characterized by unexplained left ventricular hypertrophy in the absence of systemic disease revealed a gene mutation in 46 of 84 patients (55%). Rates of mutation detection were similar among children with familial cardiomyopathy (64%; 95% confidence interval [CI], 45 to 80) and among children with presumed sporadic cardiomyopathy (49%; 95% CI, 36 to 62). We suggest that unexplained hypertrophy that presents during childhood should prompt evaluation for a potential genetic cause of the cardiac changes.
All but 1 of the 46 mutations identified occurred in a sarcomere-protein gene. Thus, despite considerable clinical differences between childhood-onset hypertrophy and hypertrophic cardiomyopathy in adulthood, our data define an etiologic relationship between these pathologic conditions.
We considered whether the disease gene, mutation type, or mutation number accounted for the early clinical presentation, poor prognosis, or male predilection observed in this study cohort. More than 75% of the children who had mutation-positive hypertrophy with a family history (16 of 21 children) or without a family history (20 of 25 children) had at least one mutation in MYH7 (encoding β-myosin heavy chain) or MYBPC3 (encoding myosin-binding protein C). Mutations in these genes that encode thick filaments also account for the majority of cases of familial hypertrophic cardiomyopathy, elderly-onset hypertrophic cardiomyopathy (onset after 50 years of age), and left ventricular hypertrophy in participants of the Framingham Heart Study (Table 3).
Table 3.
Distribution of Sarcomere-Protein Gene Mutations in Different Populations.
| Variable | Sarcomere Mutations* |
Familial Hypertrophic Cardiomyopathy† |
Elderly-Onset Left Ventricular Hypertrophy‡ |
Childhood-Onset Left Ventricular Hypertrophy § |
|
|---|---|---|---|---|---|
| Familial | Sporadic | ||||
| Gene (%) | |||||
| MYBPC3 | 34.4 | 24.1 | 10.7 | 21.2 | 25.5¶ |
| MYH7 | 41.2 | 23.4 | 1.6 | 33.3 | 17.6¶ |
| TNNT2 | 8.0 | 4.2 | 0 | 6.1 | 3.9 |
| TNNI3 | 7.4 | 4.9 | 2.5 | 3.0 | 2.0 |
| TPM1 | 2.9 | 0.4 | 0 | 6.1 | 0 |
| MYL3 | 1.2 | 0.5 | 0 | 3.0 | 0 |
| MYL2 | 3.1 | 1.8 | 0 | 0 | 0 |
| ACTC | 1.7 | 0 | 0 | 0 | 3.9 |
| Mutation detection (%) ‖ | NA | 59.3 | 16.4 | 63.6 | 49.0 |
| Probands (no.) | NA | 551 | 122 | 33 | 51 |
This category refers to the percentage of mutations that have been detected in each sarcomere-protein gene. Data are from the CardioGenomics Project Web site15 and unpublished data. NA denotes not available.
This category refers to the percentage of unrelated probands (persons younger than 50 years of age with a diagnosis of familial hypertrophic cardiomyopathy) with a mutation in each sarcomere-protein gene. Data are from the CardioGenomics Project Web site,15 Richard et al.,26 and Morita et al.35
This category refers to the percentage of unrelated probands (persons older than 50 years of age with a diagnosis of hypertrophic cardiomyopathy or unexplained left ventricular hypertrophy and without a family history of disease) with a mutation in each sarcomere-protein gene. Data are from Morita et al.,34 Anan et al.,36 and Niimura et al.37
This category refers to the percentage of unrelated probands (children in the familial and sporadic cohorts with a diagnosis of unexplained left ventricular hypertrophy) with a mutation in each sarcomere-protein gene.
The frequency of MYBPC3 and MYH7 in the sporadic pediatric cohort was significantly higher than the frequency in the elderly cohort with unexplained left ventricular hypertrophy (P<0.001) and was not significantly different from the frequency in the cohort with familial hypertrophic cardiomyopathy.
Mutation detection refers to the percentage of persons in each cohort who had a sarcomere-protein gene mutation or, in this study, a PRKAG2 mutation.
The two types of MYBPC3 mutations are those that are predicted to result in the substitution of one amino acid for another (also called missense mutations) and truncation mutations, which are predicted to foreshorten the protein. A total of 13 of the 17 MYBPC3 mutations found in patients with childhood-onset hypertrophy were missense mutations (8 in patients with presumed sporadic disease and 5 in patients with familial disease), whereas among 162 previously reported MYBPC3 mutations,15 only 64 were missense mutations (P = 0.005); the remainder encoded truncations.
This high proportion of MYBPC3 missense mutations in childhood-onset hypertrophy may imply greater functional consequences of MYBPC3 missense mutations than of truncation mutations. The variable stability of different mutant messenger RNAs, mutant proteins, or both might influence clinical expression. This model is supported by the observation that only small amounts of truncated myosin-binding protein C polypeptides, if any, are incorporated into the sarcomere.38 Alternatively, earlier clinical expression of MYBPC3 missense mutations than of truncation mutation may result from distinct effects on the biophysical properties of myosin-binding protein C.
Virtually all known MYH7 mutations, including those that we report here, are missense mutations. Background genotypes, lifestyle, and other factors probably contribute to the age at which different MYH7 mutations and other sarcomere-protein genes become clinically manifest. The strikingly unequal sex distribution of patients in this study provides support for the influence of background genotype on clinical expression. Although dominant defects in genes are equally transmitted to male and female offspring, boys predominated among the children with mutations (35 boys vs. 11 girls; P = 0.002) (Table 1). Unequal sex distribution was also observed among patients with presumed sporadic cardiomyopathy (39 boys and 12 girls; P< 0.001), those with cardiomyopathy of unknown cause (18 boys and 8 girls with presumed sporadic cardiomyopathy and 10 boys and 2 girls with familial cardiomyopathy), and subjects enrolled in the Pediatric Cardiomyopathy Registry of the National Heart, Lung, and Blood Institute and the Australian Cardiomyopathy Registry.1,2 Understanding the biology that underpins a heightened clinical expression of cardiomyopathy in young boys will be important.
We considered whether genetic status correlated with other important clinical characteristics of childhood-onset cardiomyopathy. No significant differences in the severity of left ventricular hypertrophy or contractile function were observed between children with and those without a mutation (Table 1). Among children with sporadic disease, seven serious adverse events (sudden death from cardiac causes or cardiac transplantation) occurred in six children (approximately 10%), which is consistent with the reported high morbidity and mortality in childhood-onset hypertrophy. Five of these six children had single gene mutations: MYH7 (in two children), MYBPC3 (in two children), and TNNT2 (in one child). Although the study size and number of adverse events were too small to achieve significant differences between children with and those without mutations, the prognosis for children who had mutations with sporadic disease tended to be poorer. There were adverse events in 24% of mutation-positive children and in 4% of mutation-negative children.
As compared with children with presumed sporadic disease, significantly more children with a family history of cardiomyopathy received implantable cardioverter–defibrillators (P = 0.007). These devices may have prevented sudden death in children with familial disease, whereas three sudden deaths occurred in children with presumed sporadic disease. Because a positive family history is a clinical surrogate for a genetic condition, we speculate that gene-based diagnosis in childhood-onset hypertrophy may improve management strategies that reduce the risk of sudden death.
An unexpected observation from our genetic studies was that some children with presumed sporadic cardiomyopathy had an inherited mutation. Among the 11 probands whose parents were available for genotyping, only 4 had truly sporadic disease (resulting from new mutations). The remainder had inherited an MYBPC3 mutation (Fig. 2). The significant overrepresentation of MYBPC3 mutations in patients without hypertrophy (P = 0.001) is consistent with previous studies31 that show left ventricular hypertrophy in only 55% of MYPBC3 mutation carriers before 40 years of age. Our clinical evaluations of the parents genotyped in this study revealed only one parent with cardiomyopathy and clinically significant arrhythmias. Given the lifetime penetrance of sarcomere-gene mutations and the increased potential for coexisting conditions (particularly atrial fibrillation, which increases in frequency with disease duration), we recommend clinical evaluations or genetic studies in all first-degree family members of children with unexplained left ventricular hypertrophy.
In conclusion, approximately half of all cases of childhood-onset isolated cardiac hypertrophy are caused by a mutation of genes that are routinely screened in adults with unexplained left ventricular hypertrophy. Analyses of genes encoding some sarcomere-protein genes, PRKAG2, and LAMP2 (the latter only if warranted by clinical findings) can precisely define the cause and help to identify family members at risk. The application of management strategies developed for hypertrophic cardiomyopathy may improve outcomes in children with sarcomere-gene mutations.
Supplementary Material
Acknowledgments
Supported by grants from the Howard Hughes Medical Institute (to Dr. C.E. Seidman), the National Heart, Lung, and Blood Institute (to Drs. J.G. Seidman and Towbin), the National Football League Charities Foundation (to Drs. J.G. Seidman and C.E. Seidman), and the Children’s Cardiomyopathy Foundation (to Dr. Towbin).
We thank Josh Gorham, Steven DePalma, and Daniel Herman for excellent technical assistance and statistical support.
REFERENCES
- 1.Lipshultz SE, Sleeper LA, Towbin JA, et al. The incidence of pediatric cardiomyopathy in two regions of the United States. N Engl J Med. 2003;348:1647–1655. doi: 10.1056/NEJMoa021715. [DOI] [PubMed] [Google Scholar]
- 2.Nugent AW, Daubeney PE, Chondros P, et al. The epidemiology of childhood cardiomyopathy in Australia. N Engl J Med. 2003;348:1639–1646. doi: 10.1056/NEJMoa021737. [DOI] [PubMed] [Google Scholar]
- 3.Nugent AW, Daubeney PE, Chondros P, et al. Clinical features and outcomes of childhood hypertrophic cardiomyopathy: results from a national population-based study. Circulation. 2005;112:1332–1338. doi: 10.1161/CIRCULATIONAHA.104.530303. [DOI] [PubMed] [Google Scholar]
- 4.Colan SD, Lipshultz SE, Lowe AM, et al. Epidemiology and cause-specific outcome of hypertrophic cardiomyopathy in children: findings from the Pediatric Cardiomyopathy Registry. Circulation. 2007;115:773–781. doi: 10.1161/CIRCULATIONAHA.106.621185. [DOI] [PubMed] [Google Scholar]
- 5.Yetman AT, McCrindle BW. Management of pediatric hypertrophic cardiomyopathy. Curr Opin Cardiol. 2005;20:80–83. doi: 10.1097/01.hco.0000153452.45341.36. [DOI] [PubMed] [Google Scholar]
- 6.Maron BJ. Hypertrophic cardiomyopathy in childhood. Pediatr Clin North Am. 2004;51:1305–1346. doi: 10.1016/j.pcl.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 7.Seidman JG, Seidman C. The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell. 2001;104:557–567. doi: 10.1016/s0092-8674(01)00242-2. [DOI] [PubMed] [Google Scholar]
- 8.Lind JM, Chiu C, Semsarian C. Genetic basis of hypertrophic cardiomyopathy. Expert Rev Cardiovasc Ther. 2006;4:927–934. doi: 10.1586/14779072.4.6.927. [DOI] [PubMed] [Google Scholar]
- 9.Osio A, Tan L, Chen SN, et al. Myozenin 2 is a novel gene for human hypertrophic cardiomyopathy. Circ Res. 2007;100:766–768. doi: 10.1161/01.RES.0000263008.66799.aa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maron BJ, McKenna WJ, Danielson GK, et al. American College of Cardiology/European Society of Cardiology clinical expert consensus document on hypertrophic cardiomyopathy: a report of the American College of Cardiology Foundation Task Force on Clinical Expert Consensus Documents and the European Society of Cardiology Committee for Practice Guidelines. J Am Coll Cardiol. 2003;42:1687–1713. doi: 10.1016/s0735-1097(03)00941-0. [DOI] [PubMed] [Google Scholar]
- 11.Arad M, Benson DW, Perez-Atayde AR, et al. Constitutively active AMP kinase mutations cause glycogen storage disease mimicking hypertrophic cardiomyopathy. J Clin Invest. 2002;109:357–362. doi: 10.1172/JCI14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burwinkel B, Scott JW, Bührer C, et al. Fatal congenital heart glycogenosis caused by a recurrent activating R531Q mutation in the gamma 2-subunit of AMP-activated protein kinase (PRKAG2), not by phosphorylase kinase deficiency. Am J Hum Genet. 2005;76:1034–1049. doi: 10.1086/430840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arad M, Maron BJ, Gorham JM, et al. Glycogen storage diseases presenting as hypertrophic cardiomyopathy. N Engl J Med. 2005;352:362–372. doi: 10.1056/NEJMoa033349. [DOI] [PubMed] [Google Scholar]
- 14.Roberts AE, Hult B, Rehm HL, et al. The PTPN11 gene is not implicated in nonsyndromic hypertrophic cardiomyopathy. Am J Med Genet A. 2005;132:333–334. doi: 10.1002/ajmg.a.30405. [DOI] [PubMed] [Google Scholar]
- 15.CardioGenomics Project Web site. Boston: NHLBI Program for Genomic Applications, Harvard Medical School; 2007. [Accessed April 7, 2008]. The program in genomics applications. (at http://cardiogenomics.med.harvard.edu.) [Google Scholar]
- 16.SAS-STAT user’s guide: release 8.1. Cary, NC: SAS Institute; 2000. [Google Scholar]
- 17.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Driest SL, Jaeger MA, Ommen SR, et al. Comprehensive analysis of the beta-myosin heavy chain gene in 389 unrelated patients with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44:602–610. doi: 10.1016/j.jacc.2004.04.039. [DOI] [PubMed] [Google Scholar]
- 19.Church DM, Stotler CJ, Rutter JL, Murrell JR, Trofatter JA, Buckler AJ. Isolation of genes from complex sources of mammalian genomic DNA using exon amplification. Nat Genet. 1994;6:98–105. doi: 10.1038/ng0194-98. [DOI] [PubMed] [Google Scholar]
- 20.Probst V, Kyndt F, Potet F, et al. Haploinsufficiency in combination with aging causes SCN5A-linked hereditary Lenègre disease. J Am Coll Cardiol. 2003;41:643–652. doi: 10.1016/s0735-1097(02)02864-4. [DOI] [PubMed] [Google Scholar]
- 21.Watkins H, Conner D, Thierfelder L, et al. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat Genet. 1995;11:434–437. doi: 10.1038/ng1295-434. [DOI] [PubMed] [Google Scholar]
- 22.Ingles J, Doolan A, Chiu C, Seidman J, Seidman C, Semsarian C. Compound and double mutations in patients with hypertrophic cardiomyopathy: implications for genetic testing and counselling. J Med Genet. 2005;42(10):e59. doi: 10.1136/jmg.2005.033886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watkins H, Rosenzweig A, Hwang DS, et al. Characteristics and prognostic implications of myosin missense mutations in familial hypertrophic cardiomyopathy. N Engl J Med. 1992;326:1108–1114. doi: 10.1056/NEJM199204233261703. [DOI] [PubMed] [Google Scholar]
- 24.Fananapazir L, Epstein ND. Genotype-phenotype correlations in hypertrophic cardiomyopathy: insights provided by comparisons of kindreds with distinct and identical beta-myosin heavy chain gene mutations. Circulation. 1994;89:22–32. doi: 10.1161/01.cir.89.1.22. [DOI] [PubMed] [Google Scholar]
- 25.Van Driest SL, Ackerman MJ, Ommen SR, et al. Prevalence and severity of “benign” mutations in the beta-myosin heavy chain, cardiac troponin T, and alpha-tropomyosin genes in hypertrophic cardiomyopathy. Circulation. 2002;106:3085–3090. doi: 10.1161/01.cir.0000042675.59901.14. [DOI] [PubMed] [Google Scholar]
- 26.Richard P, Charron P, Carrier L, et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 27.Gruver EJ, Fatkin D, Dodds GA, et al. Familial hypertrophic cardiomyopathy and atrial fibrillation caused by Arg663His beta-cardiac myosin heavy chain mutation. Am J Cardiol. 1999;83(12A):13H–18H. doi: 10.1016/s0002-9149(99)00251-9. [DOI] [PubMed] [Google Scholar]
- 28.Consevage MW, Salada GC, Baylen BG, Ladda RL, Rogan PK. A new missense mutation, Arg719Gln, in the beta-cardiac heavy chain myosin gene of patients with familial hypertrophic cardiomyopathy. Hum Mol Genet. 1994;3:1025–1026. doi: 10.1093/hmg/3.6.1025. [DOI] [PubMed] [Google Scholar]
- 29.Epstein ND, Cohn GM, Cyran F, Fananapazir L. Differences in clinical expression of hypertrophic cardiomyopathy associated with two distinct mutations in the beta-myosin heavy chain gene: a 908Leu→Val mutation and a 403Arg→Gln mutation. Circulation. 1992;86:345–352. doi: 10.1161/01.cir.86.2.345. [DOI] [PubMed] [Google Scholar]
- 30.Van Driest SL, Vasile VC, Ommen SR, et al. Myosin binding protein C mutations and compound heterozygosity in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44:1903–1910. doi: 10.1016/j.jacc.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 31.Niimura H, Bachinski LL, Sangwatan-aroj S, et al. Mutations in the gene for cardiac myosin-binding protein C and late-onset familial hypertrophic cardiomyopathy. N Engl J Med. 1998;338:1248–1257. doi: 10.1056/NEJM199804303381802. [DOI] [PubMed] [Google Scholar]
- 32.Alders M, Jongbloed R, Deelen W, et al. The 2373insG mutation in the MYBPC3 gene is a founder mutation, which accounts for nearly one-fourth of the HCM cases in the Netherlands. Eur Heart J. 2003;24:1848–1853. doi: 10.1016/s0195-668x(03)00466-4. [DOI] [PubMed] [Google Scholar]
- 33.Watkins H, McKenna WJ, Thierfelder L, et al. Mutations in the genes for cardiac troponin T and alpha-tropomyosin in hypertrophic cardiomyopathy. N Engl J Med. 1995;332:1058–1064. doi: 10.1056/NEJM199504203321603. [DOI] [PubMed] [Google Scholar]
- 34.Morita H, Larson MG, Barr SC, et al. Single-gene mutations and increased left ventricular wall thickness in the community: the Framingham Heart Study. Circulation. 2006;113:2697–2705. doi: 10.1161/CIRCULATIONAHA.105.593558. [DOI] [PubMed] [Google Scholar]
- 35.Morita H, DePalma SR, Arad M, et al. Molecular epidemiology of hypertrophic cardiomyopathy. Cold Spring Harb Symp Quant Biol. 2002;67:383–388. doi: 10.1101/sqb.2002.67.383. [DOI] [PubMed] [Google Scholar]
- 36.Anan R, Niimura H, Takenaka T, Hamasaki S, Tei C. Mutations in the genes for sarcomeric proteins in Japanese patients with onset sporadic hypertrophic cardiomyopathy after age 40 years. Am J Cardiol. 2007;99:1750–1754. doi: 10.1016/j.amjcard.2007.01.066. [DOI] [PubMed] [Google Scholar]
- 37.Niimura H, Patton KK, McKenna WJ, et al. Sarcomere protein gene mutations in hypertrophic cardiomyopathy of the elderly. Circulation. 2002;105:446–451. doi: 10.1161/hc0402.102990. [DOI] [PubMed] [Google Scholar]
- 38.Flavigny J, Souchet M, Sébillon P, et al. COOH-terminal truncated cardiac myosin-binding protein C mutants resulting from familial hypertrophic cardiomyopathy mutations exhibit altered expression and/or incorporation in fetal rat cardiomyocytes. J Mol Biol. 1999;294:443–456. doi: 10.1006/jmbi.1999.3276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.