Abstract
Mutations in the transmembrane protease, serine 3 (TMPRSS3) gene, encoding a transmembrane serine protease, cause autosomal recessive deafness childhood (DFNB8) or congenital onset (DFNB10). TMPRSS3 mutations have been mainly identified in patients from Asian and Mediterranean countries and seem to be a rare finding in the Northern European population so far. The identification of two novel pathogenic TMPRSS3 mutations (c.646C→T − R216C; c.916G→A − A306T) is described in four affected siblings of German origin with postlingual hearing loss, treated by bilateral cochlear implantation with good results. Although TMPRSS3 mutations are supposed to be a rare cause of autosomal recessive hearing loss, in families with postlingual disease onset TMPRSS3 is the most favourable candidate gene after exclusion of GJB2 mutations.
Autosomal recessive non‐syndromic deafness is the most common type of inherited hearing impairment, with an account of approximately 80% cases. Over 50 autosomal recessive non‐syndromic hearing loss loci have been mapped, but to date only about half of the genes have been isolated (hereditary hearing loss homepage: http://webh01.ua.ac.be/hhh/). Among these, connexin 26 (GJB2) is responsible for a large number of cases (30–60%).1,2,3 The typical phenotype of autosomal recessive non‐syndromic hearing loss is profound prelingual.
By contrast, postlingual forms seem to be either autosomal dominant or maternally inherited because of mitochondrial mutations. Autosomal recessive hearing loss with childhood onset is a rare clinical finding.4 Exceptions are caused by mutations in the transmembrane protease, serine 3 (TMPRSS3) gene on chromosome 21q22,5,6 which can lead to autosomal recessive prelingual (DFNB10) as well as to postlingual deafness (DFNB8).7TMPRSS3 encodes a transmembrane serine protease, which is inter alia expressed in the organ of Corti, the stria vascularis and in the spiral ganglion, and activates the epithelial sodium channel.8,9 Although the mutations of TMPRSS3 have been identified in patients with hearing loss from Asian and Mediterranean countries, in the Caucasian population the proportion of patients with hearing loss caused by TMPRSS3 mutations seem to be small, with an estimated frequency of <1%.10,11
In this study, we describe the detection of two new missense mutations (R216C and A306T) in TMPRSS3 in a family of German origin.
Patients and methods
Patients
Our family included four affected siblings, two females and two males, with postulated autosomal recessive postlingual non‐syndromic hearing loss as well as six non‐affected brothers and sisters (fig 1A). The non‐affected parents were unrelated, and of German origin. The first onset of hearing loss was reported at an age of 6 years, with progression to deafness by about 20 years in all affected individuals. In the affected siblings, GJB2 mutations were first excluded. As controls, we screened 100 normal hearing probands of German origin.
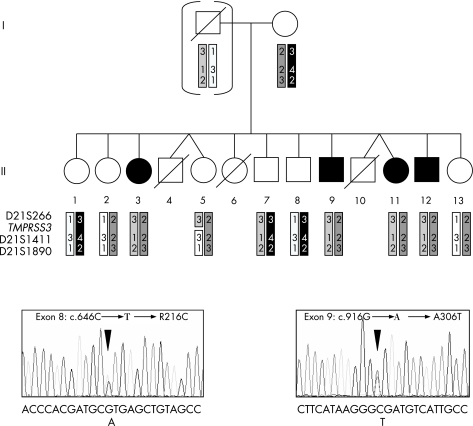
Figure 1 (A) Family pedigree and results of linkage analysis with polymorphic microsatellite markers spanning the transmembrane protease, serine 3 (TMPRSS3) locus on chromosome 21q22 (paternal DNA was not available, therefore the paternal haplotypes were delineated from the typing results in his children). (B) Direct sequencing of exons 8 and 9 of TMPRSS3 in patients II.3 and II.9 showed compound heterozygosity of two new missense mutations (sequencing was performed using the reverse primers).
DNA studies
Linkage analysis
Blood samples from 11 family members were obtained after informed consent was given. Genomic DNA was isolated from blood samples using standard protocols. For haplotype analysis, polymorphic microsatellite markers spanning the TMPRSS3 locus were used (centromere–D21S266–TMPRSS3–D21S1411–D21S1890–telomere). The PCR products were run on an ABI 3130 automatic sequencer (Applied Biosystems, Darmstadt, Germany).
Mutation analysis
All coding exons and the exon–intron boundaries of the TMPRSS3 gene were amplified by PCR using standard protocols (primer sequences were designed by using the Primer3 software: http://frodo.wi.mit.edu/cgi‐bin/primer3/primer3_www.cgi). PCR products were sequenced using the Big Dye Terminator Cycle Ready Reaction Kit V.1.0 (Applied Biosystems) on an ABI 3130 sequencer. The initial analysis included one affected person and the mother. When a sequence variant was identified, direct sequencing was performed in all family members to verify its segregation with the phenotype.
To allow rapid genotyping of controls, we developed PCR‐based restriction fragment length polymorphism assays for the identified mutations R216C and A306T. In both assays, 5 µl of the PCR product was digested with 5 U of NsiI (R216C mutation) and 5 U of MslI (A306T mutation), according to the manufacturer's instructions (New England Biolabs, Frankfurt, Germany). Digestion products were electrophoresed on 3% agarose gels.
Results and discussion
In a large German family with four siblings affected by postlingual non‐syndromic hearing loss, we searched for TMPRSS3 mutations as this gene is known to cause autosomal recessive hearing loss of postlingual onset.
Firstly, the segregation of three highly polymorphic microsatellite markers flanking TMPRSS3 was studied for linkage with the phenotype. We reconstructed the haplotypes of all available family members (fig 1A). One recombination was found (person II.5) between the two tested markers D21S266 and D21S1411, but direct sequencing showed that the recombination must have occurred telomeric from the coding sequence of TMPRSS3. However, these microsatellite typing results were consistent with linkage to the TMPRSS3 locus.
By direct sequencing of the exons and intron–exon boundaries of TMPRSS3, we identified compound heterozygosity of two new mutations cosegregating in the four affected family members (fig 1B).
The mutation in exon 8 (c.646C→T) results in an arginine to cysteine substitution at codon 216 (R216C) and affects the same codon as the known pathogenic mutation c.647G→T (R216L).9 The second mutation was found in exon 9 (c.916G→A) and leads to an alanine to threonine substitution (A306T). Both missense mutations change evolutionary highly conserved amino acids. They were not found in 200 control chromosomes of German origin. Analysis of the mutant TMPRSS3 harbouring a R216L mutation showed that it failed to undergo proteolytic cleavage and could not activate the epithelial sodium channel.9
Apart from the two new identified pathogenic mutations, we detected two known polymorphisms cosegregating with the maternal pathogenic mutation (intron 7: IVS7‐3ins(TA), rs2839500; exon 8: c.757A→G, p.I253V, rs284372665).
Owing to the large number of known genes associated with hearing loss, the identification of the underlying genetic cause of autosomal recessive non‐syndromic hearing loss is difficult in cases where GJB2 mutations have been excluded. Although TMPRSS3 mutations account only for a small number of Caucasian patients with hereditary hearing loss,10,11 we think that in the cases of postlingual disease onset and possible autosomal recessive inheritance, TMPRSS3 is the most favourable candidate gene. The detection of compound heterozygosity for mutations in TMPRSS3 in our family impressively illustrates that testing of rare disease genes is worthwhile also in cases without evidence for consanguinity.
Data access
GenBank: AB038157.
Acknowledgements
We thank the family who participated in this study.
Abbreviations
TMPRSS3 - transmembrane protease, serine 3 gene
Footnotes
Competing interests: None declared.
Informed consent has been obtained from patients for publication of their details in this paper.
References
- 1.Zelante L, Gasparini P, Estivill X, Melchionda S, D'Agruma L, Govea N, Mila M, Monica M D, Lutfi J, Shohat M, Mansfield E, Delgrosso K, Rappaport E, Surrey S, Fortina P. Connexin26 mutations associated with the most common form of non‐syndromic neurosensory autosomal recessive deafness (DFNB1) in Mediterraneans. Hum Mol Genet 199761605–1609. [DOI] [PubMed] [Google Scholar]
- 2.Lench N, Houseman M, Newton V, Van Camp G, Mueller R. Connexin‐26 mutations in sporadic non‐syndromal sensorineural deafness. Lancet 1998351415. [DOI] [PubMed] [Google Scholar]
- 3.Gasparini P, Rabionet R, Barbujani G, Melchionda S, Petersen M, Brondum‐Nielsen K, Metspalu A, Oitmaa E, Pisano M, Fortina P, Zelante L, Estivill X. High carrier frequency of the 35delG deafness mutation in European populations. Genetic analysis consortium of GJB2 35delG. Eur J Hum Genet 2000819–23. [DOI] [PubMed] [Google Scholar]
- 4.Kalatzis V, Petit C. The fundamental and medical impacts of recent progress in research on hereditary hearing loss. Hum Mol Genet 199871589–1597. [DOI] [PubMed] [Google Scholar]
- 5.Veske A, Oehlmann R, Younus F, Mohyuddin A, Muller‐Myhsok B, Mehdi S Q, Gal A. Autosomal recessive non‐syndromic deafness locus (DFNB8) maps on chromosome 21q22 in a large consanguineous kindred from Pakistan. Hum Mol Genet 19965165–168. [DOI] [PubMed] [Google Scholar]
- 6.Bonne‐Tamir B, DeStefano A L, Briggs C E, Adair R, Franklyn B, Weiss S, Korostishevsky M, Frydman M, Baldwin C T, Farrer L A. Linkage of congenital recessive deafness (gene DFNB10) to chromosome 21q22.3. Am J Hum Genet 1996581254–1259. [PMC free article] [PubMed] [Google Scholar]
- 7.Scott H S, Kudoh J, Wattenhofer M, Shibuya K, Berry A, Chrast R, Guipponi M, Wang J, Kawasaki K, Asakawa S, Minoshima S, Younus F, Mehdi S Q, Radhakrishna U, Papasavvas M P, Gehrig C, Rossier C, Korostishevsky M, Gal A, Shimizu N, Bonne‐Tamir B, Antonarakis S E. Insertion of beta‐satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat Genet 20012759–63. [DOI] [PubMed] [Google Scholar]
- 8.Guipponi M, Vuagniaux G, Wattenhofer M, Shibuya K, Vazquez M, Dougherty L, Scamuffa N, Guida E, Okui M, Rossier C, Hancock M, Buchet K, Reymond A, Hummler E, Marzella P L, Kudoh J, Shimizu N, Scott H S, Antonarakis S E, Rossier B C. The transmembrane serine protease (TMPRSS3) mutated in deafness DFNB8/10 activates the epithelial sodium channel (ENaC) in vitro. Hum Mol Genet 2002112829–2836. [DOI] [PubMed] [Google Scholar]
- 9.Wattenhofer M, Sahin‐Calapoglu N, Andreasen D, Kalay E, Caylan R, Braillard B, Fowler‐Jaeger N, Reymond A, Rossier B C, Karaguzel A, Antonarakis S E. A novel TMPRSS3 missense mutation in a DFNB8/10 family prevents proteolytic activation of the protein. Hum Genet 2005117528–535. [DOI] [PubMed] [Google Scholar]
- 10.Wattenhofer M, Di Iorio M V, Rabionet R, Dougherty L, Pampanos A, Schwede T, Montserrat‐Sentis B, Arbones M L, Iliades T, Pasquadibisceglie A, D'Amelio M, Alwan S, Rossier C, Dahl H H, Petersen M B, Estivill X, Gasparini P, Scott H S, Antonarakis S E. Mutations in the TMPRSS3 gene are a rare cause of childhood nonsyndromic deafness in Caucasian patients. J Mol Med 200280124–131. [DOI] [PubMed] [Google Scholar]
- 11.Hutchin T, Coy N N, Conlon H, Telford E, Bromelow K, Blaydon D, Taylor G, Coghill E, Brown S, Trembath R, Liu X Z, Bitner‐Glindzicz M, Mueller R. Assessment of the genetic causes of recessive childhood non‐syndromic deafness in the UK—implications for genetic testing. Clin Genet 200568506–512. [DOI] [PubMed] [Google Scholar]