Abstract
The trace metal copper is essential for a variety of biological processes, but extremely toxic when present in excessive amounts. Therefore, concentrations of this metal in the body are kept under tight control. Central regulators of cellular copper metabolism are the copper‐transporting P‐type ATPases ATP7A and ATP7B. Mutations in ATP7A or ATP7B disrupt the homeostatic copper balance, resulting in copper deficiency (Menkes disease) or copper overload (Wilson disease), respectively. ATP7A and ATP7B exert their functions in copper transport through a variety of interdependent mechanisms and regulatory events, including their catalytic ATPase activity, copper‐induced trafficking, post‐translational modifications and protein–protein interactions. This paper reviews the extensive efforts that have been undertaken over the past few years to dissect and characterise these mechanisms, and how these are affected in Menkes and Wilson disease. As both disorders are characterised by an extensive clinical heterogeneity, we will discus how the underlying genetic defects correlate with the molecular functions of ATP7A and ATP7B and with the clinical expression of these disorders.
Keywords: copper, Menkes disease, ATP7A, Wilson disease, ATP7B
Many trace elements require a delicate homeostatic balance to ensure that the needs for normal cellular processes are met, but at the same time, toxicity due to excessive accumulation of these elements needs to be prevented. Copper is an excellent example of such a trace element. It is required for numerous cellular processes, including mitochondrial respiration, antioxidant defence, neurotransmitter synthesis, connective tissue formation, pigmentation, peptide amidation and iron metabolism (table 1).1 However, in amounts that exceed cellular needs, copper is highly toxic, owing to its potential to facilitate the production of reactive oxygen species by means of Fenton chemistry.2 Refined mechanisms have evolved to regulate intake, excretion and the cellular distribution of copper (Box 1). The importance of these regulatory mechanisms is underlined by several hereditary human disorders of copper homeostasis. These disorders can broadly be divided into two classes; (1) diseases associated with copper deficiency (Menkes disease (MD), OMIM 309400;3 and occipital horn syndrome (OHS), OMIM 304150), and (2) diseases associated with copper excess (Wilson disease (WD), OMIM 277900;4 Indian childhood cirrhosis (ICC), OMIM 215600;5 endemic Tyrolean infantile cirrhosis (ETIC), OMIM 215600;6 and idiopathic copper toxicosis (ICT); OMIM 2156007,8). The clinical expression of several of these disorders is highly heterogeneous. In this review, we discuss both the genetics and molecular–functional defects underlying MD and WD, the genotype–phenotype correlations of these disorders and how such correlations might be explained by the molecular–functional defects. Additional information of interest to the reader, but not essential to the scope of this review, is provided in the text boxes.
Table 1 Functions of copper‐dependent enzymes.
| Enzyme | Function | Consequences of deficiency | ||
|---|---|---|---|---|
| Caeruloplasmin | Iron and copper transport | Decreased circulating copper levels, iron deficiency | ||
| Cytochrome C oxidase | Mitochondrial respiration | Hypothermia, muscle weakness | ||
| Dopamine β‐hydroxylase | Catecholamine production | Hypothermia, neurological defects | ||
| Lysyl oxidase | Connective tissue formation | Laxity of skin and joints | ||
| Peptidylglycine α‐amidating mono‐oxygenase | Peptide amidation | Neuroendocrine defects | ||
| Superoxide dismutase | Antioxidant defence | Diminished protection against oxidative stress | ||
| Tyrosinase | Pigment formation | Hypopigmentation of hair and skin |
Copper‐transporting ATPases and human disease
Diseases associated with copper deficiency
MD is an X‐linked recessive disorder characterised by a general copper deficiency.3,9 The incidence of the disease is estimated to range between 1:40 000 and 1:350 000.10,11,12 Clinical features of MD are a direct consequence of dysfunction of several copper‐dependent enzymes (cuproenzymes; table 1), secondary to an inability to load these enzymes with copper. Based on the symptoms, two forms of MD have been described; classic MD and mild MD, a less severe form. The clinical features of classic MD typically comprise neurological defects (severe mental retardation, neurodegeneration, seizures), growth retardation, hypothermia, laxity of skin and joints, hypopigmentation, and peculiar “kinky” or “steely” hair.13,14,15 Patients present at 2–3 months of age, and owing to the severity of the disorder, death usually occurs by 3 years of age. Patients with mild MD have a longer lifespan, and in these patients the neurological defects in particular are less profound.16,17,18 OHS (also known as X‐linked cutis laxa, or Ehlers–Danlos syndrome type IX) is allelic to MD and its symptoms generally overlap, with the most notable exception being that neurological abnormalities are far less severe or even absent in OHS. Patients with OHS present with slightly subnormal intelligence and seizures are generally absent. Connective tissue abnormalities represent the predominant features of OHS. Characteristic is the formation of occipital exostoses resulting from calcification of the trapezius and sternocleidomastoid muscles at their attachments to the occipital bone.13,19 Treatment of MD consists of copper replacement therapy in which the following fundamental issues should be taken into consideration: (1) the block in intestinal absorption of copper must be bypassed, (2) patients must be identified and treatment started as early in life as possible, (3) circulating copper must be delivered to the brain, and (4) copper must be available within cells for cuproenzyme biosynthesis.20,21 The only currently available treatment option consists of administration of copper–histidine, a naturally occurring copper–amino acid complex in serum.22,23 Although copper replacement therapy with copper histidine results in significant improvement in some patients, leading to an increased lifespan, it has not been uniformly beneficial and the prognosis of patients with classic MD remains inevitably poor. The age at which treatment is started and the severity of the disease seem to be among the main determinants for the outcome of this treatment.20,24
Both MD and OHS are caused by mutations in the ATP7A gene,18,25,26,27 which encodes a highly conserved copper translocating P(1B)‐type ATPase with orthologues present in eukaryotes, prokaryotes and archaea.28ATP7A mRNA is expressed in a wide range of human tissues, but its expression is notably low and sometimes even undetectable in the adult liver.25,27 The ATP7A polypeptide contains several conserved domains required for ATPase function and copper binding (Box 2). It has been estimated that one‐third of MD cases arise from de novo mutations.13 Over 200 MD‐causing mutations have been identified, among which small deletions/insertions, nonsense mutations, missense mutations, and splice site mutations are represented with equal frequency.29,30 Small deletions/insertions and nonsense mutations are found throughout the whole gene. In contrast, missense mutations are almost exclusively distributed between the first transmembrane domain and the stop codon. The relative lack of MD‐causing or OHS‐causing missense mutations within the six metal‐binding sites (MBSs) located in the amino terminal tail suggests that functional redundancy exists among these six MBSs. Splice‐site mutations are mostly clustered between exons 6 and 8, which encode a region just upstream of the first transmembrane domain, and between exons 21 and 22, a region encoding the last transmembrane domain. Interestingly, splice site mutations appear to be over‐represented in patients with OHS.29,30
Several mouse models for MD and OHS have been described, collectively known as mottled mice.31 The mottled mouse phenotypes show a similar variability, as seen between MD and OHS patients. The dappled subtype displays the most severe phenotype; affected mice usually die during prenatal development. In contrast, blotchy mice display a milder phenotype, reminiscent of OHS. Both dappled and blotchy, as well as other mottled subtypes, are caused by mutations in Atp7a, the murine orthologue of ATP7A.32,33,34,35,36,37,38,39,40,41,42 Transgenic expression of ATP7A in mottled mice of the brindled subtype rescues the phenotype and partially restores the copper balance.43 Calamity, a zebrafish mutant defective in the orthologue of ATP7A (atp7a), has recently been bred.44 Calamity zebrafish display a general copper deficiency phenotype and form an excellent model to study the function of atp7a in the developing zebrafish.
Diseases associated with copper excess
WD is an autosomal recessive disorder characterised by defective copper excretion, with an estimated incidence between 1:30 000 and 1:100 000.45 Clinical features of WD result from toxic accumulation of copper, primarily in the liver and the brain, and therefore may include hepatic abnormalities (cirrhosis and chronic hepatitis, culminating in progressive liver failure), neurological defects (parkinsonian features, seizures), and psychiatric symptoms (personality changes, depression, psychosis).45,46
In some severe cases, patients present with fulminant liver failure. A characteristic feature often found in patients with WD is the Kayser–Fleischer ring, a deposition of copper in the Descemet membrane visible as a gold‐brown ring around the periphery of the cornea. In addition, serum levels of the cuproenzyme caeruloplasmin are often found greatly reduced in patients with WD as a result of rapid degradation of the copper‐free form of caeruloplasmin formed in patients with WD.47,48 This type of caeruloplasmin deficiency is distinct from acaeruloplasminaemia, a disorder of iron metabolism caused by mutations in the caeruloplasmin gene.49 The presentation of WD is strongly heterogeneous, even among patients with the same mutations. Differences have been found in age at presentation, severity of the disease and the predominance of hepatic versus neurological symptoms.48 Treatment of WD focuses on two aspects: (1) copper excretion from the body must be promoted, and (2) copper absorption from the diet must be reduced.45 The first aspect is best accomplished by copper chelation therapy using penicillamine,50 trientine50 or ammonium tetrathiomolybdate.51,52 The latter remains under clinical trial, but seems particularly promising as, when taken with meals, this compound also prevents absorption of copper from the diet. Dietary copper absorption is also efficiently inhibited by zinc ingestion and by omitting copper‐rich dietary components. Controversy exists as to which approach constitutes the more effective treatment regimen, and proper randomised control studies to clarify this issue are lacking. In some cases, the preferred method includes initial copper chelation followed by zinc therapy to prevent remission of high copper concentration in the liver. In general, these treatment options are very effective; however, if treatment is ineffective, or in circumstances of fulminant liver failure, liver transplantation provides an effective cure.
The gene mutated in WD, ATP7B, encodes a copper‐transporting P‐type ATPase that is highly homologous to ATP7A.53,54,55,56 The expression pattern of ATP7B is strikingly different to that of ATP7A, as ATP7B is most abundantly expressed in the liver. Other tissues that express ATP7B include kidney, brain, lung, heart, mammary gland and placenta.53,56,57,58 Almost 300 mutations in ATP7B that are associated with WD development have been described.59 In contrast to MD, relatively few small insertions/deletions and splice site mutations have been identified in patients with WD; in fact, almost 60% of all identified WD‐causing mutations are missense mutations. The distribution of these mutations over ATP7B is similar to the distribution of MD‐causing missense mutations in ATP7A.30 Although most WD‐causing mutations are rare and only reported in single families, some are more common and account for a large portion of WD cases. The most prevalent mutations are H1069Q in Europe and North America, and R778L in southeast Asia.60
ICC, ETIC and ICT form a second class of copper‐overload disorders that are distinct from WD.61,62 Most patients with ICC, ETIC and ICT die at an early age due to liver failure as a consequence of decompensated liver cirrhosis. Neurological defects are not found in ICC, ETIC or ICT. Phenotypic expression of ICC, ETIC and some cases of ICT appears to be associated with both an excessive copper intake and an underlying genetic defect.5,6,8,63 Attempts to identify the genetic causes for these disorders have remained unsuccessful, although several candidate genes, including ATP7B, have been excluded.61,62
Several animal models of copper overload diseases have been described.64 Long–Evans cinnamon (LEC) rats and toxic‐milk mice suffer from abnormalities in the Atp7b gene, making these valid spontaneous models for WD.65,66,67,68,69,70 In addition, an engineered Atp7b knockout mouse had recently been generated.71 The hepatic abnormalities seen in both the LEC rat and the toxic‐milk mouse closely resemble those found in WD, but neurological defects have only been found in the Atp7b knockout mouse, but not in the LEC rat nor in the toxic‐milk mouse.64,71 Another interesting animal model characterised by hepatic copper overload is copper toxicosis (CT) in Bedlington terriers .72 Pathophysiologically, CT is similar to WD, although neurological defects have not been found and serum caeruloplasmin concentrations are normal. Furthermore, linkage analysis has excluded Atp7b as a candidate gene for CT, suggesting that CT is more likely to be a model for ICC, ETIC or ICT.73,74 Using positional cloning approaches, a deletion in COMMD1 (formerly MURR1), resulting in complete absence of the COMMD1 protein, was identified as a genetic cause for CT.75,76,77 This observation initially led to the suggestion of COMMD1 as a candidate gene for human copper overload disorders. However, no disease‐causing mutations in COMMD1 have been detected in several cohorts of patients with WD, ICC, ETIC or ICT61,78,79,80,81,82 Possibly, other genetic causes for CT in Bedlington terriers exist, as no mutation in COMMD1 could be detected in several pedigrees with affected dogs.83,84 Nine homologues of COMMD1 have recently been reported, which should be prioritised in the search for alternative genetic causes of CT.85
Functions of copper‐transporting ATPases
The physiological functions of ATP7A and ATP7B can largely be deduced from the observed phenotypes in MD and WD, respectively. In MD, copper transfer across the mucosal barrier is impaired, whereas in WD hepatic excretion of copper into the bile is reduced, suggesting that both proteins are rate limiting for cellular copper export. Biochemical and clinical observations in patients with MD or WD suggested an additional role for ATP7A and ATP7B in cuproenzyme biosynthesis, as markedly reduced copper incorporation of several cuproenzymes has been found in MD and WD.47,86,87,88,89,90
Regulation of cellular copper export
Direct evidence for a copper export function of ATP7A and ATP7B came from cell culture studies showing that absence of ATP7A in MD‐derived patient fibroblasts resulted in cellular copper accumulation and increased copper retention, despite normal copper uptake rates.91 This copper retention phenotype could be corrected by expression of either ATP7A or ATP7B.92 Consistent with these observations, overexpression of ATP7A or ATP7B in a variety of cell lines results in a decrease in copper accumulation and retention, and in an increased tolerance to elevated environmental copper levels.93,94,95 Direct proof for the actual copper translocation function of these proteins came from studies showing that overexpression of ATP7A or ATP7B resulted in increased translocation of 64Cu into isolated membrane vesicles, which has also been shown using purified ATP7A reconstituted in soybean asolectin liposomes.96,97,98 Taken together, these data suggest that both ATP7A and ATP7B function as ATP‐dependent copper export pumps. The difference in phenotype between MD and WD is mainly determined by differences in tissue distribution and cellular localisation of ATP7A and ATP7B, as discussed below. Unfortunately, the effects of MD‐causing and WD‐causing mutations on the copper transport activity of ATP7A and ATP7B have only been determined using complementation assays of ΔCcc2 yeast. Although these yeast studies have clearly shown the inability of mutated ATP7A and ATP7B proteins to mediate copper delivery to the secretory pathway, no studies have yet been performed to directly elucidate the effects of disease‐associated mutations on the transport activity of ATP7A and ATP7B. Significant efforts are currently being made to develop several new tools that allow detection of changes in intracellular copper concentrations and bioavailability.99,100,101,102 We have developed an MRE–luciferase reporter, which is a sensitive and robust tool to assess the ability of ATP7A and ATP7B to exort copper from the cytosol, and appears useful to assess the effects of MD‐causing and WD‐causing mutations on cytosolic bioavailable copper (van den Berghe and Klomp, unpublished observations).
Regulation of cuproenzyme biosynthesis
Loss of function of several cuproenzymes is a characteristic feature of both MD and WD. Using fibroblasts isolated from patients with MD or mottled mice, it has been shown that ATP7A deficiency directly results in reduced activities of the copper‐dependent enzymes lysyl oxidase, tyrosinase, cytochrome C oxidase, extracellular superoxide dismutase and peptidylglycine α‐amidating mono‐oxygenase.86,87,88,89,90 Although not consistently found among all patients, WD is often associated with a dramatic reduction in serum caeruloplasmin levels as a result of rapid degradation of the copper‐free form of caeruloplasmin secreted from the hepatocytes of patients with WD.47,48
In yeast, the orthologue of ATP7A and ATP7B, Ccc2p, serves a similar function. Ccc2p is required for incorporation of copper into the caeruloplasmin homologue Fet3p, a ferrireductase enabling growth of yeast under iron‐deficient conditions.103 Incorporation of copper into Fet3p takes places within the Golgi apparatus, where Ccc2p resides under basal culturing conditions.104 Expression of both ATP7A and ATP7B rescues the growth phenotype of Ccc2 knockout yeast on iron‐deficient media, making this an excellent model to study copper‐transporting, ATPase‐dependent cuproenzyme biosynthesis.105,106 Using this Ccc2 knockout complementation assay, effects of WD‐causing and MD‐causing mutations on translocation of copper into the trans‐Golgi network (TGN) lumen and subsequent incorporation of copper in cuproenzymes can be determined. This approach has been undertaken by several investigators, and indicates that multiple MD‐causing and WD‐causing mutations diminish or even completely abrogate the ability of ATP7A or ATP7B to rescue Fet3p biosynthesis or the Ccc2 knockout growth phenotype (tables 2 and 3). Taken together, these observations imply that ATP7A and ATP7B are critical for both proper delivery of copper to the TGN and subsequent cuproenzyme biosynthesis, and that mutations in ATP7A and ATP7B that perturb this process account for many of the clinical symptoms of MD and WD (table 1).
Table 2 Effects of MD‐causing missense mutations on function and regulation of ATP7A .
| Mutation | Cuproenzyme biosynthesis | Catalytic ATPase activity | Localisation | Post‐translational modifications | Protein–protein interactions | ||||
|---|---|---|---|---|---|---|---|---|---|
| A629P | Reduced rescue ΔCcc2 yeast106 | ||||||||
| S637L | Reduced rescue ΔCcc2 yeast205 | ||||||||
| R844H | No signal observed, protein absent in patient fibroblasts258 | ||||||||
| G860V | No signal observed, protein absent in patient fibroblasts258 | ||||||||
| L873R | No rescue ΔCcc2 yeast114 | Increased formation of acylphosphate intermediate114 | Cell periphery and plasma membrane,114 no copper response114 | ||||||
| G876R | No signal observed, protein absent in patient fibroblasts258 | ||||||||
| Q924R | Partial in cell periphery258 | ||||||||
| C1000R | No tyrosinase activity142 | Normal, TGN,114,258 no copper response114 | |||||||
| A1007V | Normal, TGN258 | ||||||||
| G1015D | Normal, TGN258 | ||||||||
| G1019D | Reduced rescue ΔCcc2 yeast106 | Partial ER mislocalisation,146 copper response present146 | Impaired glycosylation146 | ||||||
| D1044G | Normal, TGN258 | ||||||||
| K1282E | Normal, TGN258 | ||||||||
| G1300E | Normal, TGN258 | ||||||||
| G1302V | Partial in cell periphery258 | ||||||||
| N1304S | Reduced rescue ΔCcc2 yeast259 | ||||||||
| N1304K | Normal, TGN258 | ||||||||
| D1305A | Normal, TGN258 | ||||||||
| G1315R | Normal, TGN258 | ||||||||
| A1325V | No signal observed, protein absent in patient fibroblasts258 | ||||||||
| A1362D | Reduced rescue ΔCcc2 yeast205 | ||||||||
| A1362V | Normal, TGN, no copper response141 | ||||||||
| G1369R | No signal observed, protein absent in patient fibroblasts258 | ||||||||
| S1397F | Normal, TGN258 |
ER, endoplasmic reticulum; TGN, trans‐Golgi network.
Table 3 Effects of WD‐causing missense mutations on function and regulation of ATP7B.
| Mutation: | Cuproenzyme biosynthesis | Catalytic ATPase activity | Localisation | Post‐translational modifications | Protein–protein interactions | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| G85V | Extensive ER mislocalisation149 | Decreased interaction with ATOX1176 | ||||||||
| Increased interaction with COMMD1149 | ||||||||||
| L492S | Decreased interaction with ATOX1176 | |||||||||
| Increased interaction with COMMD1149 | ||||||||||
| G591D | Extensive ER mislocalisation149 | Absence of copper‐induced phosphorylation152 | Decreased interaction with ATOX1176 | |||||||
| Increased interaction with COMMD1149 | ||||||||||
| A604P | Increased interaction with COMMD1149 | |||||||||
| R616W | Normal, TGN126 | |||||||||
| G710S | Normal, TGN126 | |||||||||
| P760L | Normal, TGN126 | |||||||||
| D765N | Normal rescue ΔCcc2 yeast143 | Partial ER mislocalisation126,127 | ||||||||
| Reduced copper response127 | ||||||||||
| M769V | Normal rescue ΔCcc2 yeast143 | Normal, TGN126,127 | ||||||||
| Copper response unaffected127 | ||||||||||
| L776V | Normal rescue ΔCcc2 yeast143 | Partial ER mislocalisation127 | ||||||||
| Reduced copper response127 | ||||||||||
| R778Q | Reduced rescue ΔCcc2 yeast143 | |||||||||
| R778L | Reduced rescue ΔCcc2 yeast143 | Extensive ER mislocalisation127 | ||||||||
| Copper response absent127 | ||||||||||
| W779X | Cytoplasmic clusters126 | |||||||||
| Copper response absent126 | ||||||||||
| G943S | Normal rescue ΔCcc2 yeast143 | Normal, TGN127 | ||||||||
| Copper response absent127 | ||||||||||
| R969Q | Normal, TGN126 | |||||||||
| T997M | No rescue ΔCcc2 yeast143 | |||||||||
| V995A | Normal rescue ΔCcc2 yeast143 | |||||||||
| P992L | Reduced rescue ΔCcc2 yeast143 | Normal, TGN126 | ||||||||
| E1064A | Absence of ATP binding117 | |||||||||
| H1069Q | No rescue ΔCcc2 yeast105 | Absence of ATP binding,117 Absence of ATP hydrolysis activity214 | Extensive ER mislocalisation,126,148 mislocalisation to aggrosomes144 | |||||||
| Reduced rescue ΔCcc2 yeast260 | ||||||||||
| R1115H | Slightly decreased ATP binding117 | |||||||||
| N1270S | Reduced ATP hydrolysis activity214 | Normal, TGN126 | ||||||||
| Normal, endosome144 |
ER, endoplasmic reticulum; TGN, trans‐Golgi network.
Molecular mechanisms of ATP7A and ATP7B function and the effects of disease‐causing mutations
ATP7A and ATP7B exert their functions in copper transport through a variety of interdependent mechanisms and regulatory events. These include catalytic ATPase activity, copper‐dependent trafficking, post‐translational modifications and protein–protein interactions. In recent years, extensive efforts have been undertaken to dissect and characterise these mechanisms, which will be discussed in this section in the context of the pathophysiology of MD and WD. As explained below, these mechanisms are highly interdependent, and specific MD‐causing or WD‐causing mutations can exert effects on multiple levels. Therefore, care should be taken in the interpretation of how such effects relate to the pathogenesis of MD and WD.
ATPase catalytic cycle
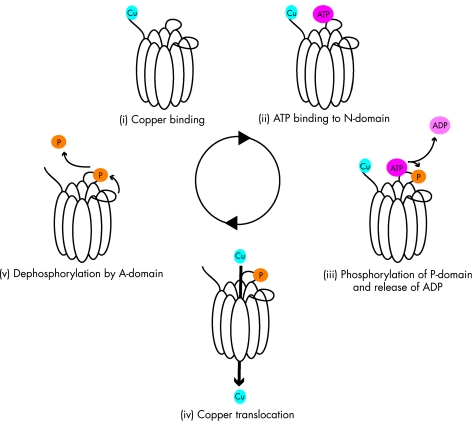
The ion translocation cycle by P‐type ATPases is believed to occur through a general cycling model involving several discrete stages in which ATP hydrolysis drives translocation of the target ion (schematically depicted in fig 1).107 These stages are: (i) binding of the target ion, (ii) binding of ATP to the nucleotide‐binding (N)‐domain, (iii) ATP hydrolysis and phosphorylation of the phosphorylation (P)‐domain, (iv) translocation of the target ion, and (v) dephosphorylation of the P‐domain by the actuator (A)‐domain.
Figure 1 Schematic representation of the general ATPase catalytic cycle. Copper translocation by ATP7A and ATP7B is believed to occur through a general cycling model involving several discrete stages. These stages include (i) binding of the target ion, (ii) binding of ATP to the N‐domain, (iii) ATP hydrolysis and phosphorylation of the P‐domain, (iv) translocation of the target ion, and (v) dephosphorylation of the P‐domain by the A‐domain.
This model suggests that copper plays a key regulatory role in the catalytic cycle of ATP7A and ATP7B. ATP7A and ATP7B contain a number of putative copper‐binding sites, of which the amino terminal MBSs are the best characterised.108,109 The amino terminal tail containing these MBSs interacts with the N‐domain of ATP7B, leading to inhibition of ATP binding.110 This interaction is inhibited by copper, providing a potential mechanism for copper‐regulated availability of the N‐domain to bind ATP.110 Consistent with this hypothesis, both ATP hydrolysis and formation of the acylphosphate intermediate (step (3) in the model) are also dependent on copper.98,111,112,113,114 The effect of copper on acylphosphate intermediate formation is cooperative, suggesting that the six amino terminal MBSs have a regulatory role in the formation of the acylphosphate intermediate.98,112,113 Indeed, MBSs 5 and 6 are required for the cooperative effect of copper on acylphosphate intermediate formation.112 However, mutation of all six amino terminal MBSs only mildly affects the rate and extent of catalytic phosphorylation of ATP7A, raising the possibility that other copper binding sites, such as the conserved intramembranous CPC motif, play an important role in the regulation of this process.111
The next steps in the model predict that copper translocation by ATP7A and ATP7B is dependent on ATP binding and hydrolysis. Indeed, translocation of copper into membrane vesicles isolated from rat liver, and in vitro copper translocation activities of ATP7A and ATP7B, are ATP‐dependent.96,97,98,115,116 Using purified recombinant protein fragments, ATP binding to both the N‐domain and P‐domain of ATP7B has been found.110,117,118 Molecular modelling analysis supports this experimental observation.119 The exact binding site for ATP in the N‐domain has not yet been identified, but analysis of the solution structure of the N‐domain of ATP7B has implicated a number of residues in this process, including H1069, G1099, G1101, I1102, G1149 and N1150.118 Interestingly, H1069, G1099, G1101 and I1102 are all residues found mutated in WD, and H1069Q represents the most frequent WD‐causing mutation in Europe and North America.59 Consistent with a role for this residue in ATP binding, the H1069Q mutation results in almost complete absence of ATP binding to ATP7B.117 However, within the N‐domain, >40 different WD‐causing mutations have been reported, indicating that more residues might be involved in either direct ATP binding or coordination.59 This possibility is supported by experiments showing that the WD‐causing E1064A mutation results in complete absence of ATP binding to the N‐domain of ATP7B.117
Molecular modelling analysis suggests that after initial ATP binding to the N‐domain of ATP7B, conformational changes take place that bring the ATP binding site within the N‐domain in close proximity to the P‐domain.119 This could potentially promote ATP binding and phosphorylation of the P‐domain, the third step in the catalytic cycle model.119 ATP binding in the P‐domain of ATP7B has been suggested to take place in the vicinity of D1027.119 This residue is part of the DKTG motif, which is highly conserved in all P‐type ATPases, and is presumed to be the target of phosphorylation by the γ‐phosphate of ATP.107 Mutation of this aspartic acid residue in either ATP7A or ATP7B completely prevents formation of an acylphosphate intermediate, thereby supporting this hypothesis.111,113,114 Furthermore, this mutation in ATP7A is associated with a complete loss of copper translocation activity, the next step in the general model.111
Although copper translocation can be measured using isolated membrane vesicles from cells expressing ATP7A or ATP7B, or using purified ATP7A or ATP7B reconstituted in soybean asolectin liposomes, studies have yet to be performed to characterise the effects of MD‐causing and WD‐causing mutations on this particular step in the cycling model.96,98 Furthermore, no studies have yet been performed to unravel the role of the intramembranous copper‐binding CPC motif, which is presumed to play an essential role in this step. However, it has been shown that intact MBSs in the amino terminal region of ATP7A are required for proper translocation of 64Cu in isolated membrane vesicles.120
Dephosphorylation of the aspartic acid residue is mediated by intrinsic phosphatase activity, in which the A‐domain plays a key role. The signals that induce this phosphatase activity, and the mechanisms behind it, have not yet been elucidated, but an essential role has been ascribed to the conserved TGE motif. Mutation of this motif in ATP7A results in hyperphosphorylation of the protein.114 Although the TGE motif per se has not been reported to be mutated in MD or WD, adjacent residues in ATP7A and ATP7B are known sites of MD‐causing and WD‐causing mutations.59,114 In fact, the MD‐causing L873R mutation, two amino acids upstream of the TGE motif, results in hyperphosphorylation of ATP7A.114
Copper‐dependent localisation of ATP7A and ATP7B
Under basal conditions, ATP7A and ATP7B are localised within the TGN (fig 2).105,121,122 This localisation is consistent with their function in cuproenzyme biosynthesis, as several cuproenzymes are synthesised within the secretory pathway. Some controversy exists about the localisation of ATP7B, as it has also been suggested that this protein resides in an endosomal compartment.123,124 In addition, a smaller isoform of ATP7B exists, which has been localised to mitochondria.125 However, the current general agreement is that ATP7B is localised to the TGN, as this has been confirmed by several independent groups, including at ultrastructural resolution in human liver biopsies.105,126,127,128,129,130,131,132,133
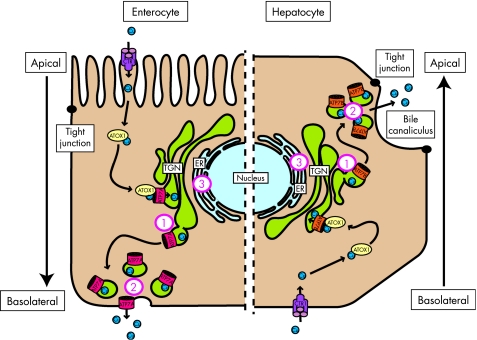
Figure 2 Schematic representation of copper‐induced relocalisation of ATP7A and ATP7B. Left side depicts an enterocyte, and right side represents and hepatocyte. In both cells, copper enters through the copper transporter 1 (CTR1), and is then distributed via the copper chaperone ATOX1 to ATP7A or ATP7B residing in the TGN. After a rise in copper concentrations, ATP7A and ATP7B relocalise from the TGN to the cell periphery, and in the case of ATP7A also the plasma membrane, to facilitate excretion of copper. The main difference in these two copper transport pathways lies in the direction. In the enterocyte, ATP7A facilitates excretion of copper into the bloodstream at the basolateral side, whereas in the hepatocyte copper is excreted at the apical side into the bile. The numbers indicate localisation defects of ATP7A and ATP7B due to MD‐causing and WD‐causing mutations, respectively: (1) lack of copper responsiveness, resulting in constitutive localisation at the TGN, (2) constitutive localisation at the cell periphery, and (3) mislocalisation at the ER, presumably due to misfolding.
A key mechanism in the regulation of the copper export function of ATP7A and ATP7B became apparent from studies showing that the subcellular distribution of both proteins was sensitive to the concentration of copper to which the cell was exposed (fig 2, Box 3). In response to raised copper levels, ATP7A reversibly relocalises to a peripheral vesicular compartment and to the plasma membrane.122,134 In polarised cells, and in intestinal tissue sections, ATP7A specifically localises towards the basolateral membrane upon copper exposure, consistent with its function in transferring copper across the intestinal barrier.135,136,137,138 ATP7A overexpressed in mouse liver tissue also localises at the hepatocyte basolateral membrane.139 Specific targeting of ATP7A to the basolateral membrane appears to be mediated through a putative PDZ binding motif present in the carboxy terminal tail of ATP7A. Deletion of this motif results in targeting of ATP7A to the apical membrane in response to raised copper levels.136 ATP7B undergoes a similar copper‐induced relocalisation to a peripheral vesicular compartment. Although ATP7B has not been unequivocally detected at the plasma membrane of polarised cells, the idea that the transporter rapidly recycles between the peripheral vesicular compartment and the plasma membrane cannot be excluded.105,140 In polarised hepatocytic cell lines, exposure to high copper concentrations results in localisation of ATP7B in the proximity of the apical vacuoles, a structure reminiscent of the bile canaliculus.46,128,130,133,140 This observation is consistent with its proposed function in excretion of copper from the hepatocyte via the bile. Small amounts of ATP7B localised in the proximity of the bile canaliculus have also been detected in human liver tissues.130 Signals mediating the specific targeting of ATP7B towards the apical region seem to be present within the first 63 amino acids, a region that is not present in ATP7A.133 Taken together, these data indicate that differences in the trafficking destinations of ATP7A and ATP7B are not caused by general cell‐type specific differences in regulation of polarised membrane protein localisation, but rather are determined by intrinsic signals present in their amino acid sequences. The difference in the directionality of copper‐induced relocalisation of ATP7A and ATP7B illustrates why two distinct, but very similar, copper export proteins are required to ensure proper copper uptake and excretion in higher organisms. In addition, this difference, resulting in distinct physiological functions of these homologous proteins in maintaining whole‐body copper homeostasis, explains the opposed copper transport defects seen in MD and WD.
Several MD‐causing and WD‐causing mutations are associated with defects in copper‐induced relocalisation of ATP7A and ATP7B respectively, suggesting that this is a key event that precedes cellular copper excretion by ATP7A and ATP7B. Therefore, such defects might possibly be associated with the pathogenesis of MD and WD. In general, MD‐causing and WD‐causing mutations can result in three types of localisation defects (tables 2 and 3, fig 3). The first type displays a normal steady‐state localisation of the protein within the TGN, but responsiveness to copper is lost.114,127,141,142 One mutation of interest that has been shown to result in such a defect is the MD‐causing C1000R mutation in ATP7A, which disrupts the putative copper‐binding CPC motif, suggesting that in this case impaired copper binding causes the lack of copper‐induced trafficking to the cell periphery.114 This type of localisation defect could potentially explain some of the biochemical symptoms seen in patients with MD or WD. For example, although the G943S mutation in ATP7B prohibits copper‐induced trafficking of ATP7B, this mutation still permits cuproenzyme biosynthesis in the ΔCcc2 complementation assay.127,143 Although this correlation does not exist for all mutations causing this type of localisation defect, it could explain the normal caeruloplasmin production found in some patients with WD. Unfortunately, serum caeruloplasmin levels have only been described for one patient who was compound heterozygous for the G943S mutation, and this was indeed found to be within normal range.127 Although this type of localisation defect might still permit cuproenzyme biosynthesis in MD, resulting in alleviation of the clinical phenotype, this might not always be obvious in patients, owing to an impairment in copper translocation across the mucosal barrier. More likely, this type of localisation defect could allow for the beneficial effects of copper replacement therapy in these patients. The second type of localisation defects is on the opposite side of the spectrum. Several MD‐causing and WD‐causing mutations result in constitutive peripheral localisation of ATP7A or ATP7B, respectively.114,126,141 Of interest is the MD‐causing L873R mutation in ATP7A, targeting a residue located adjacent to the conserved TGE motif. In this case, a constitutively peripheral localisation of ATP7A as a result of this mutation has been associated with hyperphosphorylation of ATP7A, suggesting that copper‐induced relocalisation is dependent on the ATPase catalytic cycle (further discussed in Box 3).114 The third type of localisation defect is probably the most common and clinically important. MD‐causing and WD‐causing mutations result in mislocalisation, or possibly retention, of ATP7A and ATP7B within the endoplasmic reticulum.126,127,140,144,145,146,147,148,149 Strikingly, this defect has been found for the two most common WD‐causing mutations, H1069Q and R778L.126,127,148 Some controversy exists about the localisation of the H1069Q variant, as this has also been reported to localise to aggresomes.144 ER mislocalisation of proteins is often due to misfolding and associated with proteasomal degradation.150 A well‐known example of this process in human disease development is the ER‐associated degradation of the ΔF508 CFTR mutant in cystic fibrosis.151 In analogy, the H1069Q mutation in ATP7B indeed results in an increased proteolysis rate.144,148 Whether the increased proteolysis resulting from the H1069Q mutation is due to defects in the conformation of ATP7B remains unclear, but the solution structure of the N‐domain of ATP7B containing the H1069Q mutation shows no folding defects.118
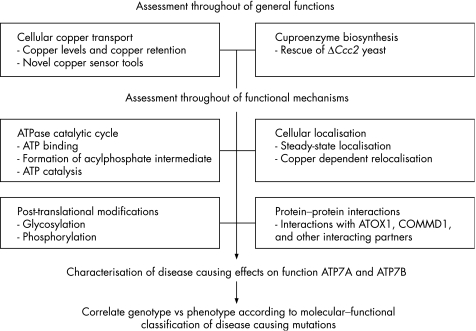
Figure 3 Proposed flow chart to analyse MD‐causing and WD‐causing mutations in ATP7A and ATP7B and to correlate the molecular defects to disease phenotype. MD‐causing and WD‐causing mutations can have profound effects on the functions of ATP7A and ATP7B. Thorough analysis of the functional and mechanistic defects resulting from these mutations will enhance understanding of MD and WD pathophysiology and the clinical heterogeneity seen in patients.
Post‐translational modifications
Most efforts in investigating putative post‐translational modifications of ATP7A and ATP7B have focused on the formation of the acylphosphate intermediate during the catalytic cycle. However, recent studies have led authors to suggest that both ATP7A and ATP7B are subject to basal and copper‐induced phosphorylation that is distinct from the formation of the acylphosphate intermediate.152,153 Both basal and copper‐induced phosphorylation could still be observed with a catalytically inactive mutant of ATP7B, indicating that these types of phosphorylation are indeed distinct from the formation of the acylphosphate intermediate, and that specific kinases are required.154 Copper‐induced phosphorylation of ATP7B is partially inhibited by an inhibitor of casein kinase II.152 Phosphoamino acid analysis indicates that both ATP7A and ATP7B are phosphorylated on serine residues.153,155 In addition to phosphorylation, modification of ATP7A with N‐linked glycan chains has also been found.121 Although little is known about the functional effects of ATP7A glycosylation, it is interesting that this type of modification is probably specific to ATP7A, as the amino acid sequence of ATP7B does not contain any consensus glycosylation sites at relevant extracytoplasmic domains.
Taken together, studies on post‐translational modifications of ATP7A and ATP7B are still in their initial phases, but could shed important new light on our understanding of the regulation of these copper‐transporting ATPases. The importance of these modifications is emphasised by the absence of copper‐induced phosphorylation of ATP7B due to the WD‐causing G591D mutation, and absence of ATP7A glycosylation, due to the MD‐causing G1019D mutation (tables 1, 2).146,152
Protein–protein interactions
Protein–protein interactions form an essential mechanism through which many proteins exert their functions. Mapping of the protein–protein interactome provides a valuable framework for elucidating the functional organisation of the human proteome, and consequently to understanding of the molecular pathology of human disease.156 In recent years, significant process has been made in unravelling the ATP7A and ATP7B interactome (Box 4). In the following, we elaborate on two of the interacting partners of ATP7A and ATP7B that can be directly linked to the development of copper homeostasis disorders.
Owing to its toxic potential, concentrations of free copper inside the cell are extremely low. In fact, it has been estimated that yeast maintains a concentration of <1 free copper ion per cell.157 As a consequence of such low free copper concentrations, donor proteins are required for the delivery of substrate copper to copper‐transporting ATPases. Originally isolated as a suppressor of oxidative toxicity in yeast, Atx1p (and its human orthologue ATOX1) was shown to be required for copper‐transporting ATPase‐mediated cuproenzyme biosynthesis158,159,160 Subsequently, it was demonstrated that ATOX1 interacts with both ATP7A and ATP7B.161,162 This interaction is conserved in yeast and bacteria, illustrating its importance.163,164,165,166 An MxCxxC‐containing MBS, homologous to those in ATP7A and ATP7B, is present in ATOX1, through which it has been shown to bind copper.163,167 The interaction of ATOX1 with ATP7A or ATP7B is copper‐dependent and requires intact MBSs of both ATOX1 and ATP7A or ATP7B.161,162,163,168,169,170,171 These data suggest that ATOX1 delivers copper to ATP7A and ATP7B, which in fact has been demonstrated in vitro.172,173,174,175 Atox1 knockout mice display a phenotype similar to that seen in patients with MD including symptoms such as growth failure, skin laxity, hypopigmentation and seizures.160 Consistent with a copper excretion defect caused by defective copper delivery to ATP7A, cultured fibroblasts isolated from Atox1 knockout mice exhibit an increase in copper retention and content.160,176 A time‐dependent and dose‐dependent impairment of copper‐induced relocalisation of ATP7A has also been found in these cells.176 One possible defect underlying the development of MD and WD could thus be an impairment of the interaction of ATOX1 with ATP7A or ATP7B respectively. Indeed, several mutations in the amino terminal tail of ATP7B prevent its interaction with ATOX1.161 Two of these mutations, G85V and G591D, affect highly conserved glycine residues in the proximity of the MxCxxC core sequence in MBS 1 and 6 respectively, indicating an important role for this conserved residue in coordination of the ATOX1–ATP7B interaction.161 However, these mutations also result in mislocalisation of ATP7B to the ER, which might also underlie the loss of interaction of ATP7B with ATOX1 caused by these mutations.149
Recent studies have shown that COMMD1, the protein defective in CT, interacts with ATP7B and that this interaction is mediated by the amino terminal tail of ATP7B.177 Transient knock‐down of COMMD1 in HEK293 cells results in increased cellular copper levels.178 These data suggest that COMMD1 and ATP7B cooperate in the excretion of copper from the hepatocyte, and that absence of the interaction between these proteins underlies the pathophysiology of CT in Bedlington terriers with the COMMD1 deletion. Strikingly, COMMD1 also interacts with ATP7A, indicating that COMMD1 has a role in general copper homeostasis that is not restricted to the liver (de Bie et al., unpublished observation). COMMD1 has also recently been implicated in several other cellular processes, including the nuclear factor (NF)‐κB and HIF1 signalling pathways.85,179,180,181,182,183,184 In these two pathways, COMMD1 is thought to exert its regulatory role by regulating the proteasomal degradation of key components of these pathways.179,181,182,185 With this in mind, COMMD1 might also regulate the proteasomal degradation of ATP7A and ATP7B, thus regulating copper homeostasis. Other examples exist in which cuproenzymes or copper transport proteins, such as the copper chaperone for superoxide dismutase 1 (CCS) and hephaestin, are regulated by means of proteasomal degradation in response to altering copper levels.186,187 The WD‐causing mutations G85V, L492S, G591D, and A604P result in increased binding of ATP7B to COMMD1, suggesting that disruption of this interaction could underlie the development of WD in some cases.149 The G85V and G591D mutations have also been associated with ER mislocalisation and increased degradation of ATP7B, supporting the hypothesis that COMMD1 facilitates the degradation of ATP7B.149 Recently, 10 homologues of COMMD1 that are characterised by a conserved domain have been described, and are also involved in regulation of NF‐κB signalling.85,184 It would be interesting to investigate if these homologues of COMMD1 also have a functional role in copper homeostasis, possibly through interactions with ATP7B or ATP7A.
Genotype–phenotype correlations in the development of MD and WD
Proper characterisation of the defects in MD and WD is required to resolve the clinical heterogeneity that is seen in both disorders. Either environmental or genetic variations might underlie this clinical heterogeneity. For example, development of ICC, ETIC and ICT is associated with a high copper intake.5,6,8,63 Although such correlation has not been found for WD, it is possible that high copper intake might exacerbate the symptoms, whereas dietary components having low copper but high zinc concentrations might have a beneficial effect. Genetic variations in genes other than ATP7A and ATP7B might modulate the clinical expression of MD and WD. In various other disorders with a mendelian mode of inheritance, it has been shown that modifier genes can modulate penetrance, dominance modification, expressivity and pleiotrophy.188 This is exemplified by genetic modification of the severity and expressivity of cystic fibrosis by polymorphisms in various genes, including mannose‐binding lectin and transforming growth factor β (reviewed by Cutting189 and Boyle190). COMMD1 has been proposed as a modifier gene for the clinical presentation of WD, as heterozygosity for a silent missense mutation in COMMD1 was found to be possibly associated with an earlier onset of the disorder in patients with known ATP7B mutations.80 In other cohorts of patients with WD, no association between variations in COMMD1 and the clinical expression of WD was found.78,81,82 However, these cohorts were not classified to include patients with WD with identical mutations in ATP7B, which should be performed to reliably assess the possible role of COMMD1 as a modifier gene for the clinical presentation of WD. Variations genes coding for prion protein and apolipoprotein E have been proposed to modify the presentation of WD, although for the latter this has not been consistently found.191,192,193,194
Notwithstanding the role modifier genes and environmental factors play, it is evident from the functional data discussed above that different mutations in ATP7A or ATP7B can potentially result in different functional effects. Therefore, it appears likely that also the type of mutation, or the residue(s) affected, modulate the clinical expression of MD or WD. For example, functional redundancy among the six amino terminal MBSs explains the relatively low amount of MD‐causing or WD‐causing missense mutations within the amino terminal tails of ATP7A and ATP7B. In addition, it has recently been found that translation reinitiation after a deletion in the amino terminal coding region of the ATP7A transcript produces a truncated protein containing only the fifth and sixth MBSs. Interestingly, this truncated protein was still partially functional, and the patient in which this phenomenon was found displayed a remarkably mild MD phenotype.195 A suggestive correlation between the severity of the phenotype and the extent to which missense mutations impair the function of Atp7a was recently found in three types of Mottled mice.196
Many studies have focused on establishing genotype–phenotype correlations in both MD and WD. Severe mutations such as nonsense and frameshift mutations causing insertions/deletions are generally believed to completely disrupt protein function, and would therefore be expected to yield a more severe clinical phenotype. Indeed, for WD it has been shown that severe mutations lead to an earlier onset of the disease, but a correlation with the type of presentation (neurological vs. hepatic) has not been found.197,198,199,200 Similarly, it has been suggested that severe mutations in ATP7A result in classic MD, whereas mild MD is caused by mutations that still permit residual activity of ATP7A.201 On the other hand, OHS is often associated with splice site mutations. Although these mutations are predicted to dramatically affect ATP7A mRNA splicing, expression of the normal transcript is often still detectable, indicating that minor amounts of normally spliced ATP7A expressed in these patients are sufficient to allow for an alleviation of the phenotype.18,40,202,203,204 In addition, it was suggested in a recent case report that, in two brothers with MD carrying the same mutation in ATP7A, differences in severity of the disease correlated with the amount of ATP7A expressed.205
Genotype–phenotype correlation analyses for missense mutations are less clear. Approximately 30% of all cases with MD result from de novo mutations, as a result of which mutations are often very rare, thus prohibiting proper genotype–phenotype correlation analysis using large numbers of patients. Unfortunately, such studies in WD are also hampered by the large number of mutations detected; most mutations are very rare, and therefore most patients with WD are compound heterozygous. In a large number of studies, no genotype–phenotype correlation could be found. As most of these studies relied on small patient cohorts, combining data from several independent studies in a meta‐analysis could be useful to shed more clarity on this issue, as has been the case for the common H1069Q mutation.206 In several independent cohorts, a correlation between ATP7B H1069Q homozygosity and a neurological presentation of WD has been found.206,207,208,209,210,211 In addition, this genotype has been associated with a significantly later age at onset.198,199,206,208,209,210,211,212,213,214,215 In several other studies, one or both of these observations could not be statistically confirmed.198,207,212,213,214,215,216,217,218,219,220 However, a meta‐analysis of all genetic studies before 2004 devoted to H1069Q genotype and phenotype indicated that overall, H1069Q homozygosity is indeed associated with a late neurological presentation of WD.206 A similar correlation with a later age at onset in WD has been found for patients homozygous for the R969Q mutation.199 In some cohorts, it seems that homozygosity for the R778L mutation correlates with an early, hepatic presentation of WD.221,222 It has been suggested that alternative splicing of ATP7B in the brain, resulting in absence of exon 8 (harbouring the R778 residue), underlies this suggested correlation.222,223 However, here also, studies could not confirm the correlation of the homozygous R778L genotype with hepatic presentation, although a tendency towards this observation was sometimes present.224,225,226
Conclusion
Mutations in the structurally and functionally highly homologous copper‐transporting ATPases ATP7A and ATP7B underlie both copper deficiency and overload diseases. Tremendous progress has been made in the characterisation of the function of these proteins, and how this is impaired in MD and WD. From the data presented in tables 2 and 3, it is evident that different mutations in ATP7A or ATP7B result in a variety of defects in the molecular functions of these proteins. Consistent with this notion, to some extent a correlation of the genotype with the heterogeneous phenotypes has been found in these disorders. However, it is also very likely that some of the different mutations in ATP7A or ATP7B result in overlapping functional impairments. This overlap could form a potential bias in genotype–phenotype correlation studies, in which correlations might be missed due to such an overlap in test and control patients. To overcome this potential bias, genotype–phenotype correlations should be performed using patient groups classified primarily using the functional effects of the disease‐causing mutations. This approach would also permit the performance of correlation studies with relatively mild or rare, mutations. At present, this approach is unfortunately not yet possible, as only a limited number of MD‐causing or WD‐associated variants of ATP7A and ATP7B have been functionally characterised (tables 2 and 3). Furthermore, a thorough characterisation of all molecular mechanisms that participate in the copper transport functions of ATP7A and ATP7B would be required, as these mechanisms are highly interdependent (as schematically suggested in fig 3). Such characterisation of mutations in ATP7A and ATP7B should be an important focus of future studies on the functional genetics of MD and WD, and will result in valuable insights into the molecular pathogenesis of MD and WD.
Box 1 Copper homeostasis
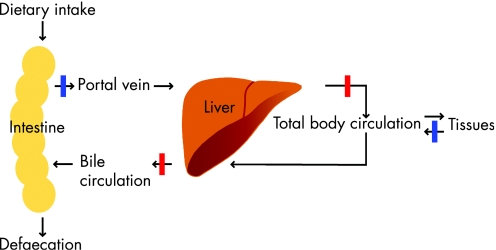
Copper is taken up from the diet in enterocytes and subsequently effluxed to the liver through the portal vein. This step is blocked in MD and OHS (blue bar in fig 4). From the liver, copper is distributed to the general circulation to provide tissues with required copper. Excretion of the majority (98%) of copper from the body is mediated by biliary export, indicating that the liver plays a central role in the regulation of body copper homeostasis.227 Copper excretion from the liver is defective in WD, ICC, ETIC and ICT (red bars in fig 4), resulting in accumulation of excess copper in the liver. To ensure copper homeostasis on the cellular level, refined mechanisms to regulate copper uptake, distribution and excretion have evolved. Copper import in the cell takes place via the copper transporters 1 and 2.102,228,229 After uptake, distribution of intracellular copper is facilitated by a group of proteins called copper chaperones, which function to deliver free copper to its sites of utilisation. Several human copper chaperones have been described; CCS delivers copper to SOD1 in the cytosol and mitochondria,230,231 COX17 delivers copper to the cytochrome C oxidase complex in mitochondria,232,233 and ATOX1 delivers copper to the copper‐transporting ATPases ATP7A and ATP7B in the trans‐Golgi complex.158,159 These copper‐transporting ATPases play an essential role in the export of copper from the cell. Dysfunction of these proteins underlies the development of MD, OHS and WD, and forms the topic of this review.
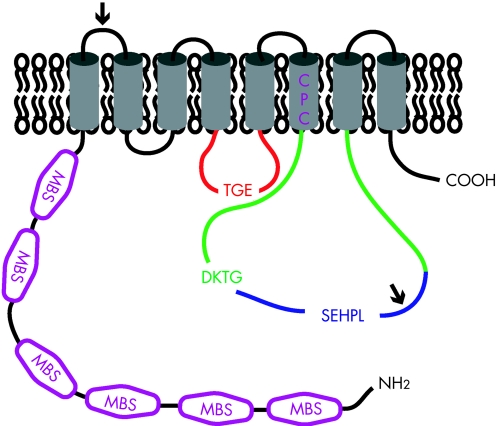
Box 2 Structure of copper‐transporting ATPases
ATP7A and ATP7B are highly homologous and both belong to the heavy metal‐transporting P(1B)‐type ATPases. The basic topology of ATP7A and ATP7B is depicted in figure 5. The polypeptide sequences of ATP7A and ATP7B are 54% identical, however two sequence inserts are present in ATP7A that are not present in ATP7B (the position of these sequences are marked with an arrow). Several conserved motifs are present in both ATP7A and ATP7B that are characteristic for the P‐type ATPase protein family.107 These motifs are required for ATP catalysis and include the nucleotide binding domain (N‐domain; depicted in blue), the phosphorylation domain (P‐domain; depicted in green) and the actuator domain (A‐domain; depicted in red). Highly conserved signature residues are present in these motifs; SEHPL in the N‐domain, DKTG in the P‐domain, and TGE in the A‐domain. The specific functions of these domains are discussed in the text. Within the amino terminal tail, six MBSs; depicted in purple) are present, each containing the core sequence MxCxxC. These MBSs bind Cu(I) in a stoichiometry of one atom of Cu(I) per MBS.108,109 Structural analysis of separate MBSs of ATP7A or ATP7B has shown a conserved βαββαβ folding structure.234,235,236,237,238,239,240 The sequence and structure of these MBSs are highly conserved in evolution and to other proteins, such as the copper chaperone ATOX1.28 These amino‐terminal MBSs in ATP7A and ATP7B are required for several aspects of their function, including copper translocation, incorporation of copper in cuproenzymes, ATPase activity, localisation and trafficking, and protein–protein interactions.106,111,112,114,120,131,136,162,168,171,241,242,243 Strikingly, however, bacterial and yeast orthologues of ATP7A and ATP7B only contain one or two MBSs, indicating that the six MBSs in ATP7A and ATP7B might be partially redundant. In general, it is believed that P‐type ATPases contain binding sites for their substrate ion within their transmembrane regions to facilitate transfer of these ions across the membrane barrier.107 Trans‐membrane domain 6 contains a highly conserved CPC motif (depicted in purple) that is characteristic for heavy metal transporting P‐type ATPases.244,245 Peptides containing this motif have been shown to bind copper, indicating that the CPC motif could indeed be involved in the actual transfer of copper across the membrane.240
Figure 4 Schematic representation of the physiology of copper homeostasis.
Figure 5 Schematic representation of the topology of ATP7A and ATP7B.
Box 3 Signals and mechanisms mediating copper‐induced trafficking of ATP7A and ATP7B
Signals and mechanisms underlying the localisation and copper‐induced trafficking of ATP7A and ATP7B have been the focus of many studies. Steady‐state TGN localisation of ATP7A has been shown to be mediated by a putative TGN‐targeting signal within transmembrane domain 3.147 Although transmembrane region 3 of ATP7B shows a high degree of conservation with that of ATP7A, it is unknown if it serves the same TGN‐targeting function. Copper‐induced relocalisation of ATP7A and ATP7B appears to be initiated by direct binding of copper to ATP7A and ATP7B. Deletion or mutation of all amino terminal MBS or the putative copper‐binding CPC sequence results in an inability to induce the copper‐dependent relocalisation.114,131,136,242,243 Strikingly, however, the presence of only one intact amino terminal MBS is sufficient for normal copper‐induced relocalisation.131,242,243,246,247 This observation indicates that the six different MBSs have redundant functions, at least in copper‐induced relocalisation of ATP7A and ATP7B. However, some studies have attributed a particularly important role to MBSs 5 and 6 for copper‐induced relocalisation, suggesting that these specific MBSs serve a different function from the others.131,242,246 In addition to copper binding, phosphorylation status plays an important role in the regulation of trafficking of ATP7A and ATP7B. Mutation of the aspartic acid residue in the DKTG motif, which prohibits the formation of an acylphosphate intermediate, abolishes copper‐induced trafficking of ATP7A and ATP7B from the TGN to the cell periphery.114,140 Conversely, hyperphosphorylation of ATP7A or ATP7B induced by mutation of the TGE motif results in a constitutive peripheral localisation.114 A combination of mutations of the TGE motif, the six amino terminal MBSs and the CPC motif in ATP7B also shows constitutive peripheral localisation.247 This fascinating observation can be explained in two ways. It is possible that other metal‐binding sites are present in ATP7B through which relocalisation can be induced by copper. Alternatively, ATP7B constitutively cycles between the TGN and the cell periphery, and copper binding serves as a retention signal, inhibiting retrograde trafficking to the TGN. Retrograde trafficking of ATP7A from the plasma membrane back to the TGN requires a dileucine motif within the carboxyterminal tail.134,248,249 Although this motif might serve as a classic clathrin‐mediated endocytosis targeting motif, other mechanisms play a role in the internalisation of ATP7A from the plasma membrane.250,251 A trileucine motif is present at the same position of ATP7B, and although ATP7B does not reach the plasma membrane, mutation of this motif also results in a constitutive peripheral localisation of ATP7B.140
These studies have provided valuable insights into the mechanisms behind copper‐induced trafficking of ATP7A and ATP7B. One of the most important remaining questions is how exactly copper is exported from the cell after this event has occurred, and this should be the focus of future studies.
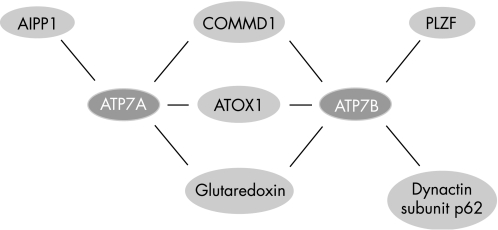
Box 4 The ATP7A and ATP7B interactome
Identification of protein–protein interactions is a powerful tool to understand protein function and how perturbation of protein function results in human disease. Over the past few years several interacting partners for ATP7A and ATP7B have been reported, resulting in a rapid elaboration of the ATP7A and ATP7B interactome (figure 6).177,252,253,254,255 The best characterised interacting partner of ATP7A and ATP7B is ATOX1. Through its role to deliver copper to ATP7A and ATP7B, ATOX1 plays an essential role in the function of ATP7A and ATP7B, which is further discussed in the text. However, additional proteins might be involved in the delivery of copper to ATP7A and ATP7B. Among these is glutaredoxin, which interacts with both ATP7A and ATP7B in a copper‐dependent manner, and has been hypothesised to regulate copper binding by ATP7A and ATP7B.252 In addition, copper delivery to ATP7A and ATP7B might be regulated by the immunophillin FKBP52. This protein interacts with ATOX1 in a copper‐dependent manner, and overexpression of FKBP52 results in increased cellular copper efflux.256 The exact mechanisms through which FKBP52 attenuates its role in the copper‐excretion pathway need to be further characterised. COMMD1, the protein defective in CT, interacts with both ATP7A and ATP7B149,177 (de Bie et al, unpublished observations). The function of these interactions remains to be characterised, but the phenotype resulting from absence of COMMD1 in CT suggests that COMMD1 and ATP7B cooperate to facilitate billiary copper excretion (further discussed in the text). Another recently identified interacting partner for ATP7A is AIPP1.255 This PDZ domain‐containing protein binds to the carboxy terminal tail of ATP7A, where a putative PDZ binding motif is present. As this motif is required for targeting of ATP7A to the basolateral membrane, it has been suggested that AIPP1 has a regulatory role in the copper‐induced trafficking of ATP7A.136,255 Through its amino terminal tail, ATP7B interacts with the dynactin subunit p62 in a copper‐dependent manner.253 As the dynactin protein complex is involved in membrane vesicle movement along microtubules, this interaction suggests that the p62 dynactin subunit facilitates copper‐induced trafficking of ATP7B.257 Further identification and characterisation of novel interacting partners of ATP7A and ATP7B will also shed new light on previously unanticipated functions of these proteins, as evidenced by the recently observed interaction between ATP7B and a promyelocytic leukemia zinc finger protein (PLZF) isoform. Characterisation of the interaction between the PLZF isoform and ATP7B suggested that ATP7B attenuates activation of the ERK signaling pathway.254 Further investigation is needed to determine the implications of this observation.
Figure 6 The ATP7A and ATP7B interactome.
Acknowledgements
We thank members of the Leo Klomp and Cisca Wijmenga laboratories for helpful discussions. The work in these two laboratories is funded by the Netherlands Organization for Scientific Research (Zon‐MW, grant 40‐00812‐98‐03106), the Dutch Digestive Diseases Foundation (MLDS, grant WS 02‐34), the Wilhelmina Children's Hospital (WKZ) Fund (grant 901‐04‐219). The authors declare they have no competing interests.
Abbreviations
A‐domain - actuator domain
AIPP1 - ATPase interacting PDZ protein 1
CCS - copper chaperone for superoxide dismutase 1
CT - copper toxicosis in Bedlington terriers
CTR1 - copper transporter 1
ER - endoplasmic reticulum
ETIC - endemic Tyrolean infantile cirrhosis
ICC - Indian childhood cirrhosis
ICT - idiopathic copper toxicosis
LEC - Long–Evans cinnamon
MBS - metal‐binding site
MD - Menkes disease
N‐domain - nucleotide‐binding domain
NF - nuclear factor
OHS - occipital horn syndrome
OMIM - Online Mendelian Inheritance in Man
P‐domain - phosphorylation domain
PLZF - promyelocytic leukemia zinc finger protein
SOD1 - superoxide dismutase 1
TGN - trans‐Golgi network
WD - Wilson disease
Footnotes
Competing interests: None declared.
References
- 1.Culotta V C, Gitlin J D. Disorders of copper transport. In: Scriver CR, Beaudet AL, Sly WS, et al eds. The metabolic and molecular bases of inherited disease. New York: McGraw‐Hill, 200123105–3126. [Google Scholar]
- 2.Stohs S J, Bagchi D. Oxidative mechanisms in the toxicity of metal ions. Free Radic Biol Med 199518321–336. [DOI] [PubMed] [Google Scholar]
- 3.Menkes J H, Alter M, Steigleder G K, Weakley D R, Sung J H. A sex‐linked recessive disorder with retardation of growth, peculiar hair, and focal cerebral and cerebellar degeneration. Pediatrics 196229764–779. [PubMed] [Google Scholar]
- 4.Wilson S A K. Progressive lenticular degeneration: A familial nervous disease associated with cirrhosis of the liver. Brain 191234295–509. [DOI] [PubMed] [Google Scholar]
- 5.Tanner M S. Role of copper in Indian childhood cirrhosis. Am J Clin Nutr 199867(Suppl)1074–81S. [DOI] [PubMed] [Google Scholar]
- 6.Muller T, Feichtinger H, Berger H, Muller W. Endemic Tyrolean infantile cirrhosis: an ecogenetic disorder. Lancet 1996347877–880. [DOI] [PubMed] [Google Scholar]
- 7.Muller T, Muller W, Feichtinger H. Idiopathic copper toxicosis. Am J Clin Nutr 199867(Suppl)1082–6S. [DOI] [PubMed] [Google Scholar]
- 8.Scheinberg I H, Sternlieb I. Wilson disease and idiopathic copper toxicosis. Am J Clin Nutr 199663842–5S. [DOI] [PubMed] [Google Scholar]
- 9.Danks D M, Campbell P E, Walker‐Smith J, Stevens B J, Gillespie J M, Blomfield J, Turner B. Menkes' kinky‐hair syndrome. Lancet 197211100–1102. [DOI] [PubMed] [Google Scholar]
- 10.Tonnesen T, Kleijer W J, Horn N. Incidence of Menkes disease. Hum Genet 199186408–410. [DOI] [PubMed] [Google Scholar]
- 11.Gu Y H, Kodama H, Shiga K, Nakata S, Yanagawa Y, Ozawa H. A survey of Japanese patients with Menkes disease from 1990 to 2003: incidence and early signs before typical symptomatic onset, pointing the way to earlier diagnosis. J Inherit Metab Dis 200528473–478. [DOI] [PubMed] [Google Scholar]
- 12.Danks D M, Scriver C R, Beaudet A L, Sly W M. Disorders of Copper transport. In: et al eds. The metabolic and molecular basis of inherited disease. New York: McGraw‐Hill, 19952211–2235.
- 13.Kaler S G. Metabolic and molecular bases of Menkes disease and occipital horn syndrome. Pediatr Dev Pathol 1998185–98. [DOI] [PubMed] [Google Scholar]
- 14.Mercer J F. Menkes syndrome and animal models. Am J Clin Nutr 199867(Suppl)1022–8S. [DOI] [PubMed] [Google Scholar]
- 15.Tumer Z, Horn N. Menkes disease: underlying genetic defect and new diagnostic possibilities. J Inherit Metab Dis 199821604–612. [DOI] [PubMed] [Google Scholar]
- 16.Procopis P, Camakaris J, Danks D M. A mild form of Menkes steely hair syndrome. J Pediatr 19819897–99. [DOI] [PubMed] [Google Scholar]
- 17.Danks D M. The mild form of Menkes disease: progress report on the original case. Am J Med Genet 198830859–864. [DOI] [PubMed] [Google Scholar]
- 18.Kaler S G, Gallo L K, Proud V K, Percy A K, Mark Y, Segal N A, Goldstein D S, Holmes C S, Gahl W A. Occipital horn syndrome and a mild Menkes phenotype associated with splice site mutations at the MNK locus. Nat Genet 19948195–202. [DOI] [PubMed] [Google Scholar]
- 19.Proud V K, Mussell H G, Kaler S G, Young D W, Percy A K. Distinctive Menkes disease variant with occipital horns: delineation of natural history and clinical phenotype. Am J Med Genet 19966544–51. [DOI] [PubMed] [Google Scholar]
- 20.Kaler S G. Diagnosis and therapy of Menkes syndrome, a genetic form of copper deficiency. Am J Clin Nutr 199867(Suppl)1029–34S. [DOI] [PubMed] [Google Scholar]
- 21.Sheela S R, Latha M, Liu P, Lem K, Kaler S G. Copper‐replacement treatment for symptomatic Menkes disease: ethical considerations. Clin Genet 200568278–283. [DOI] [PubMed] [Google Scholar]
- 22.Sarkar B, Lingertat‐Walsh K, Clarke J T. Copper‐histidine therapy for Menkes disease. J Pediatr 199312828–830. [DOI] [PubMed] [Google Scholar]
- 23.Sarkar B, Kruck T P A. Copper‐amino acids complexes in human serum. In: Peisach J, Aisen P, Blumberg WE, eds. The biochemistry of copper. New York: Academic Press, 1966183–196.
- 24.Olivares J L, Bueno I, Gallati S, Ramos F J. Late‐onset treatment in Menkes disease: is there a correlation between genotype and response to therapy? Clin Genet 200669363–366. [DOI] [PubMed] [Google Scholar]
- 25.Chelly J, Tumer Z, Tonnesen T, Petterson A, Ishikawa‐Brush Y, Tommerup N, Horn N, Monaco A P. Isolation of a candidate gene for Menkes disease that encodes a potential heavy metal binding protein. Nat Genet 1993314–19. [DOI] [PubMed] [Google Scholar]
- 26.Mercer J F, Livingston J, Hall B, Paynter J A, Begy C, Chandrasekharappa S, Lockhart P, Grimes A, Bhave M, Siemieniak D.et al Isolation of a partial candidate gene for Menkes disease by positional cloning. Nat Genet 1993320–25. [DOI] [PubMed] [Google Scholar]
- 27.Vulpe C, Levinson B, Whitney S, Packman S, Gitschier J. Isolation of a candidate gene for Menkes disease and evidence that it encodes a copper‐transporting ATPase. Nat Genet 199337–13. [DOI] [PubMed] [Google Scholar]
- 28.Arnesano F, Banci L, Bertini I, Ciofi‐Baffoni S, Molteni E, Huffman D L, O'Halloran T V. Metallochaperones and metal‐transporting ATPases: a comparative analysis of sequences and structures. Genome Res 200212255–271. [DOI] [PubMed] [Google Scholar]
- 29.Tumer Z, Moller L B, Horn N. Mutation spectrum of ATP7A, the gene defective in Menkes disease. Adv Exp Med Biol 199944883–95. [DOI] [PubMed] [Google Scholar]
- 30.Hsi G, Cox D W. A comparison of the mutation spectra of Menkes disease and Wilson disease. Hum Genet 2004114165–172. [DOI] [PubMed] [Google Scholar]
- 31.Mercer J F, Ambrosini L, Horton S, Gazeas S, Grimes A. Animal models of Menkes disease. Adv Exp Med Biol 199944897–108. [DOI] [PubMed] [Google Scholar]
- 32.Mercer J F, Grimes A, Ambrosini L, Lockhart P, Paynter J A, Dierick H, Glover T W. Mutations in the murine homologue of the Menkes gene in dappled and blotchy mice. Nat Genet 19946374–378. [DOI] [PubMed] [Google Scholar]
- 33.Levinson B, Vulpe C, Elder B, Martin C, Verley F, Packman S, Gitschier J. The mottled gene is the mouse homologue of the Menkes disease gene. Nat Genet 19946369–373. [DOI] [PubMed] [Google Scholar]
- 34.Cecchi C, Biasotto M, Tosi M, Avner P. The mottled mouse as a model for human Menkes disease: identification of mutations in the Atp7a gene. Hum Mol Genet 19976425–433. [DOI] [PubMed] [Google Scholar]
- 35.Grimes A, Hearn C J, Lockhart P, Newgreen D F, Mercer J F. Molecular basis of the brindled mouse mutant (Mo(br)): a murine model of Menkes disease. Hum Mol Genet 199761037–1042. [DOI] [PubMed] [Google Scholar]
- 36.Murata Y, Kodama H, Abe T, Ishida N, Nishimura M, Levinson B, Gitschier J, Packman S. Mutation analysis and expression of the mottled gene in the macular mouse model of Menkes disease. Pediatr Res 199742436–442. [DOI] [PubMed] [Google Scholar]
- 37.Levinson B, Packman S, Gitschier J. Deletion of the promoter region in the Atp7a gene of the mottled dappled mouse. Nat Genet 199716224–225. [DOI] [PubMed] [Google Scholar]
- 38.Mori M, Nishimura M. A serine‐to‐proline mutation in the copper‐transporting P‐type ATPase gene of the macular mouse. Mamm Genome 19978407–410. [DOI] [PubMed] [Google Scholar]
- 39.Reed V, Boyd Y. Mutation analysis provides additional proof that mottled is the mouse homologue of Menkes' disease. Hum Mol Genet 19976417–423. [DOI] [PubMed] [Google Scholar]
- 40.Das S, Levinson B, Vulpe C, Whitney S, Gitschier J, Packman S. Similar splicing mutations of the Menkes/mottled copper‐transporting ATPase gene in occipital horn syndrome and the blotchy mouse. Am J Hum Genet 199556570–576. [PMC free article] [PubMed] [Google Scholar]
- 41.George A M, Reed V, Glenister P, Chelly J, Tumer Z, Horn N, Monaco A P, Boyd Y. Analysis of Mnk, the murine homologue of the locus for Menkes disease, in normal and mottled (Mo) mice. Genomics 19942227–35. [DOI] [PubMed] [Google Scholar]
- 42.Mototani Y, Miyoshi I, Okamura T, Moriya T, Meng Y, Yuan Pei X, Kameo S, Kasai N. Phenotypic and genetic characterization of the Atp7a(Mo‐Tohm) mottled mouse: a new murine model of Menkes disease. Genomics 200687191–199. [DOI] [PubMed] [Google Scholar]
- 43.Llanos R M, Ke B X, Wright M, Deal Y, Monty F, Kramer D R, Mercer J F. Correction of a mouse model of Menkes disease by the human Menkes gene. Biochim Biophys Acta 20061762485–493. [DOI] [PubMed] [Google Scholar]
- 44.Mendelsohn B A, Yin C, Johnson S L, Wilm T P, Solnica‐Krezel L, Gitlin J D. Atp7a determines a hierarchy of copper metabolism essential for notochord development. Cell Metab 20064155–162. [DOI] [PubMed] [Google Scholar]
- 45.Ala A, Walker A P, Ashkan K, Dooley J S, Schilsky M L. Wilson's disease. Lancet 2007369397–408. [DOI] [PubMed] [Google Scholar]
- 46.Gitlin J D. Wilson disease. Gastroenterology 20031251868–1877. [DOI] [PubMed] [Google Scholar]
- 47.Scheinberg I H, Gitlin D. Deficiency of ceruloplasmin in patients with hepatolenticular degeneration (Wilson's disease). Science 1952116484–485. [DOI] [PubMed] [Google Scholar]
- 48.Riordan S M, Williams R. The Wilson's disease gene and phenotypic diversity. J Hepatol 200134165–171. [DOI] [PubMed] [Google Scholar]
- 49.Harris Z L, Klomp L W, Gitlin J D. Aceruloplasminemia: an inherited neurodegenerative disease with impairment of iron homeostasis. Am J Clin Nutr 199867(Suppl)972–7S. [DOI] [PubMed] [Google Scholar]
- 50.Walshe J M. Wilson's disease; new oral therapy. Lancet 195627025–26. [DOI] [PubMed] [Google Scholar]
- 51.Brewer G J, Dick R D, Yuzbasiyan‐Gurkin V, Tankanow R, Young A B, Kluin K J. Initial therapy of patients with Wilson's disease with tetrathiomolybdate. Arch Neurol 19914842–47. [DOI] [PubMed] [Google Scholar]
- 52.Brewer G J, Dick R D, Johnson V, Wang Y, Yuzbasiyan‐Gurkan V, Kluin K, Fink J K, Aisen A. Treatment of Wilson's disease with ammonium tetrathiomolybdate. I. Initial therapy in 17 neurologically affected patients. Arch Neurol 199451545–554. [DOI] [PubMed] [Google Scholar]
- 53.Bull P C, Thomas G R, Rommens J M, Forbes J R, Cox D W. The Wilson disease gene is a putative copper transporting P‐type ATPase similar to the Menkes gene. Nat Genet 19935327–337. [DOI] [PubMed] [Google Scholar]
- 54.Tanzi R E, Petrukhin K, Chernov I, Pellequer J L, Wasco W, Ross B, Romano D M, Parano E, Pavone L, Brzustowicz L M. The Wilson disease gene is a copper transporting ATPase with homology to the Menkes disease gene. Nat Genet 19935344–350. [DOI] [PubMed] [Google Scholar]
- 55.Yamaguchi Y, Heiny M E, Gitlin J D. Isolation and characterization of a human liver cDNA as a candidate gene for Wilson disease. Biochem Biophys Res Commun 1993197271–277. [DOI] [PubMed] [Google Scholar]
- 56.Petrukhin K, Fischer S G, Pirastu M, Tanzi R E, Chernov I, Devoto M, Brzustowicz L M, Cayanis E, Vitale E, Russo J J. Mapping, cloning and genetic characterization of the region containing the Wilson disease gene. Nat Genet 19935338–343. [DOI] [PubMed] [Google Scholar]
- 57.Michalczyk A A, Rieger J, Allen K J, Mercer J F, Ackland M L. Defective localization of the Wilson disease protein (ATP7B) in the mammary gland of the toxic milk mouse and the effects of copper supplementation. Biochem J 2000352565–571. [PMC free article] [PubMed] [Google Scholar]
- 58.Kuo Y M, Gitschier J, Packman S. Developmental expression of the mouse mottled and toxic milk genes suggests distinct functions for the Menkes and Wilson disease copper transporters. Hum Mol Genet 199761043–1049. [DOI] [PubMed] [Google Scholar]
- 59.Kenny S, Cox D W. Wilson disease mutation database. http://www.medicalgenetics.med.ualberta.ca/wilson/index.php
- 60.Ferenci P. Regional distribution of mutations of the ATP7B gene in patients with Wilson disease: impact on genetic testing. Hum Genet 2006120151–159. [DOI] [PubMed] [Google Scholar]
- 61.Muller T, van de Sluis B, Zhernakova A, van Binsbergen E, Janecke A R, Bavdekar A, Pandit A, Weirich‐Schwaiger H, Witt H, Ellemunter H, Deutsch J, Denk H, Muller W, Sternlieb I, Tanner M S, Wijmenga C. The canine copper toxicosis gene MURR1 does not cause non‐Wilsonian hepatic copper toxicosis. J Hepatol 200338164–168. [DOI] [PubMed] [Google Scholar]
- 62.Wijmenga C, Muller T, Murli I S, Brunt T, Feichtinger H, Schonitzer D, Houwen R H, Muller W, Sandkuijl L A, Pearson P L. Endemic Tyrolean infantile cirrhosis is not an allelic variant of Wilson's disease. Eur J Hum Genet 19986624–628. [DOI] [PubMed] [Google Scholar]
- 63.Muller T, Schafer H, Rodeck B, Haupt G, Koch H, Bosse H, Welling P, Lange H, Krech R, Feist D, Muhlendahl K E, Bramswig J, Feichtinger H, Muller W. Familial clustering of infantile cirrhosis in Northern Germany: A clue to the etiology of idiopathic copper toxicosis. J Pediatr 1999135189–196. [DOI] [PubMed] [Google Scholar]
- 64.Fuentealba I C, Aburto E M. Animal models of copper‐associated liver disease. Comp Hepatol 200325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamaguchi Y, Heiny M E, Shimizu N, Aoki T, Gitlin J D. Expression of the Wilson disease gene is deficient in the Long‐Evans Cinnamon rat. Biochem J 19943011–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu J, Forbes J R, Chen H S, Cox D W. The LEC rat has a deletion in the copper transporting ATPase gene homologous to the Wilson disease gene. Nat Genet 1994541–545. [DOI] [PubMed]
- 67.Muramatsu Y, Yamada T, Moralejo D H, Cai Y, Xin X, Miwa Y, Izumi K, Matsumoto K. The rat homologue of the Wilson's disease gene was partially deleted at the 3′ end of its protein‐coding region in Long‐Evans Cinnamon mutant rats. Res Commun Mol Pathol Pharmacol 199589421–424. [PubMed] [Google Scholar]
- 68.Ono T, Fukumoto R, Takada S, Nagao T, Yoshida M C. Responsible gene for hepatitis of the LEC rat (hts) is the homolog of the human Wilson's disease (WD) gene. Transplant Proc 1995271545. [PubMed] [Google Scholar]
- 69.Ono T, Fukumoto R, Kondoh Y, Yoshida M C. Deletion of the Wilson's disease gene in hereditary hepatitis LEC rats. Jpn J Genet 19957025–33. [DOI] [PubMed] [Google Scholar]
- 70.Theophilos M B, Cox D W, Mercer J F. The toxic milk mouse is a murine model of Wilson disease. Hum Mol Genet 199651619–1624. [DOI] [PubMed] [Google Scholar]
- 71.Buiakova O I, Xu J, Lutsenko S, Zeitlin S, Das K, Das S, Ross B M, Mekios C, Scheinberg I H, Gilliam T C. Null mutation of the murine ATP7B (Wilson disease) gene results in intracellular copper accumulation and late‐onset hepatic nodular transformation. Hum Mol Genet 199981665–1671. [DOI] [PubMed] [Google Scholar]
- 72.Hardy R M, Stevens J B, Stowe C M. Chronic progressive hepatitis in Bedlington terriers associated with elevated liver copper concentrations. Minnesota Veterinarian 19751513–24. [Google Scholar]
- 73.Dagenais S L, Guevara‐Fujita M, Loechel R, Burgess A C, Miller D E, Yuzbasiyan‐Gurkan V, Brewer G J, Glover T W. The canine copper toxicosis locus is not syntenic with ATP7B or ATX1 and maps to a region showing homology to human 2p21. Mamm Genome 199910753–756. [DOI] [PubMed] [Google Scholar]
- 74.Van de Sluis B, Breen M, Nanji M, van Wolferen M, de Jong P, Binns M M, Pearson P L, Kuipers J, Rothuizen J, Cox D W, Wijmenga C, van Oost B A. Genetic mapping of the copper toxicosis locus in Bedlington terriers to dog chromosome 10, in a region syntenic to human chromosome region 2p13–p16. Hum Mol Genet 19998501–507. [DOI] [PubMed] [Google Scholar]
- 75.Klomp A E, van de Sluis B, Klomp L W, Wijmenga C. The ubiquitously expressed MURR1 protein is absent in canine copper toxicosis. J Hepatol 200339703–709. [DOI] [PubMed] [Google Scholar]
- 76.Van de Sluis B, Rothuizen J, Pearson P L, van Oost B A, Wijmenga C. Identification of a new copper metabolism gene by positional cloning in a purebred dog population. Hum Mol Genet 200211165–173. [DOI] [PubMed] [Google Scholar]
- 77.Forman O P, Boursnell M E, Dunmore B J, Stendall N, van den Sluis B, Fretwell N, Jones C, Wijmenga C, Rothuizen J, van Oost B A, Holmes N G, Binns M M, Jones P. Characterization of the COMMD1 (MURR1) mutation causing copper toxicosis in Bedlington terriers. Anim Genet 200536497–501. [DOI] [PubMed] [Google Scholar]
- 78.Wu Z Y, Zhao G X, Chen W J, Wang N, Wan B, Lin M T, Murong S X, Yu L. Mutation analysis of 218 Chinese patients with Wilson disease revealed no correlation between the canine copper toxicosis gene MURR1 and Wilson disease. J Mol Med 20061–5. [DOI] [PubMed]
- 79.Coronado V A, Bonneville J A, Nazer H, Roberts E A, Cox D W. COMMD1 (MURR1) as a candidate in patients with copper storage disease of undefined etiology. Clin Genet 200568548–551. [DOI] [PubMed] [Google Scholar]
- 80.Stuehler B, Reichert J, Stremmel W, Schaefer M. Analysis of the human homologue of the canine copper toxicosis gene MURR1 in Wilson disease patients. J Mol Med 200482629–634. [DOI] [PubMed] [Google Scholar]
- 81.Weiss K H, Merle U, Schaefer M, Ferenci P, Fullekrug J, Stremmel W. Copper toxicosis gene MURR1 is not changed in Wilson disease patients with normal blood ceruloplasmin levels. World J Gastroenterol 2006122239–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lovicu M, Dessi V, Lepori M B, Zappu A, Zancan L, Giacchino R, Marazzi M G, Iorio R, Vegnente A, Vajro P, Maggiore G, Marcellini M, Barbera C, Kostic V, Farci A M, Solinas A, Altuntas B, Yuce A, Kocak N, Tsezou A, De Virgiliis S, Cao A, Loudianos G. The canine copper toxicosis gene MURR1 is not implicated in the pathogenesis of Wilson disease. J Gastroenterol 200641582–587. [DOI] [PubMed] [Google Scholar]
- 83.Hyun C, Lavulo L T, Filippich L J. Evaluation of haplotypes associated with copper toxicosis in Bedlington terriers in Australia. Am J Vet Res 2004651573–1579. [DOI] [PubMed] [Google Scholar]
- 84.Coronado V A, Damaraju D, Kohijoki R, Cox D W. New haplotypes in the Bedlington terrier indicate complexity in copper toxicosis. Mamm Genome 200314483–491. [DOI] [PubMed] [Google Scholar]
- 85.Burstein E, Hoberg J E, Wilkinson A S, Rumble J M, Csomos R A, Komarck C M, Maine G N, Wilkinson J C, Mayo M W, Duckett C S. COMMD proteins, a novel family of structural and functional homologs of MURR1. J Biol Chem 200528022222–22232. [DOI] [PubMed] [Google Scholar]
- 86.Royce P M, Camakaris J, Danks D M. Reduced lysyl oxidase activity in skin fibroblasts from patients with Menkes' syndrome. Biochem J 1980192579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Petris M J, Strausak D, Mercer J F. The Menkes copper transporter is required for the activation of tyrosinase. Hum Mol Genet 200092845–2851. [DOI] [PubMed] [Google Scholar]
- 88.Kodama H, Okabe I, Yanagisawa M, Kodama Y. Copper deficiency in the mitochondria of cultured skin fibroblasts from patients with Menkes syndrome. J Inherit Metab Dis 198912386–389. [DOI] [PubMed] [Google Scholar]
- 89.El Meskini R, Culotta V C, Mains R E, Eipper B A. Supplying copper to the cuproenzyme peptidylglycine alpha‐amidating monooxygenase. J Biol Chem 200327812278–12284. [DOI] [PubMed] [Google Scholar]
- 90.Qin Z, Itoh S, Jeney V, Ushio‐Fukai M, Fukai T. Essential role for the Menkes ATPase in activation of extracellular superoxide dismutase: implication for vascular oxidative stress. FASEB J 200620334–336. [DOI] [PubMed] [Google Scholar]
- 91.Chan W Y, Garnica A D, Rennert O M. Cell culture studies of Menkes kinky hair disease. Clin Chim Acta 197888495–507. [DOI] [PubMed] [Google Scholar]
- 92.La Fontaine S L, Firth S D, Camakaris J, Englezou A, Theophilos M B, Petris M J, Howie M, Lockhart P J, Greenough M, Brooks H, Reddel R R, Mercer J F. Correction of the copper transport defect of Menkes patient fibroblasts by expression of the Menkes and Wilson ATPases. J Biol Chem 199827331375–31380. [DOI] [PubMed] [Google Scholar]
- 93.Camakaris J, Petris M J, Bailey L, Shen P, Lockhart P, Glover T W, Barcroft C, Patton J, Mercer J F. Gene amplification of the Menkes (MNK; ATP7A) P‐type ATPase gene of CHO cells is associated with copper resistance and enhanced copper efflux. Hum Mol Genet 199542117–2123. [DOI] [PubMed] [Google Scholar]
- 94.La Fontaine S, Firth S D, Lockhart P J, Brooks H, Parton R G, Camakaris J, Mercer J F. Functional analysis and intracellular localization of the human menkes protein (MNK) stably expressed from a cDNA construct in Chinese hamster ovary cells (CHO‐K1). Hum Mol Genet 199871293–1300. [DOI] [PubMed] [Google Scholar]
- 95.Katano K, Safaei R, Samimi G, Holzer A, Rochdi M, Howell S B. The copper export pump ATP7B modulates the cellular pharmacology of carboplatin in ovarian carcinoma cells. Mol Pharmacol 200364466–473. [DOI] [PubMed] [Google Scholar]
- 96.Voskoboinik I, Brooks H, Smith S, Shen P, Camakaris J. ATP‐dependent copper transport by the Menkes protein in membrane vesicles isolated from cultured Chinese hamster ovary cells. FEBS Lett 1998435178–182. [DOI] [PubMed] [Google Scholar]
- 97.Voskoboinik I, Greenough M, La Fontaine S, Mercer J F, Camakaris J. Functional studies on the Wilson copper P‐type ATPase and toxic milk mouse mutant. Biochem Biophys Res Commun 2001281966–970. [DOI] [PubMed] [Google Scholar]
- 98.Hung Y H, Layton M J, Voskoboinik I, Mercer J F, Camakaris J. Purification and membrane reconstitution of catalytically active Menkes copper‐transporting P‐type ATPase (MNK; ATP7A). Biochem J 2007401569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.van Dongen E M, Dekkers L M, Spijker K, Meijer E W, Klomp L W, Merkx M. Ratiometric fluorescent sensor proteins with subnanomolar affinity for Zn(II) based on copper chaperone domains. J Am Chem Soc 200612810754–10762. [DOI] [PubMed] [Google Scholar]
- 100.Zeng L, Miller E W, Pralle A, Isacoff E Y, Chang C J. A selective turn‐on fluorescent sensor for imaging copper in living cells. J Am Chem Soc 200612810–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang L, McRae R, Henary M M, Patel R, Lai B, Vogt S, Fahrni C J. Imaging of the intracellular topography of copper with a fluorescent sensor and by synchrotron x‐ray fluorescence microscopy. Proc Natl Acad Sci U S A 200510211179–11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.van den Berghe P V, Folmer D E, Malingre H E, van Beurden E, Klomp A E, van de Sluis B, Merkx M, Berger R, Klomp L W. Human copper transporter 2 is localized in late endosomes and lysosomes and facilitates cellular copper uptake. Biochem J 200740749–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yuan D S, Stearman R, Dancis A, Dunn T, Beeler T, Klausner R D. The Menkes/Wilson disease gene homologue in yeast provides copper to a ceruloplasmin‐like oxidase required for iron uptake. Proc Natl Acad Sci U S A 199592632–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yuan D S, Dancis A, Klausner R D. Restriction of copper export in Saccharomyces cerevisiae to a late Golgi or post‐Golgi compartment in the secretory pathway. J Biol Chem 199727225787–25793. [DOI] [PubMed] [Google Scholar]
- 105.Hung I H, Suzuki M, Yamaguchi Y, Yuan D S, Klausner R D, Gitlin J D. Biochemical characterization of the Wilson disease protein and functional expression in the yeast Saccharomyces cerevisiae. J Biol Chem 199727221461–21466. [DOI] [PubMed] [Google Scholar]
- 106.Payne A S, Gitlin J D. Functional expression of the menkes disease protein reveals common biochemical mechanisms among the copper‐transporting P‐type ATPases. J Biol Chem 19982733765–3770. [DOI] [PubMed] [Google Scholar]
- 107.Kuhlbrandt W. Biology, structure and mechanism of P‐type ATPases. Nat Rev Mol Cell Biol 20045282–295. [DOI] [PubMed] [Google Scholar]
- 108.Lutsenko S, Petrukhin K, Cooper M J, Gilliam C T, Kaplan J H. N‐terminal domains of human copper‐transporting adenosine triphosphatases (the Wilson's and Menkes disease proteins) bind copper selectively in vivo and in vitro with stoichiometry of one copper per metal‐binding repeat. J Biol Chem 199727218939–18944. [DOI] [PubMed] [Google Scholar]
- 109.DiDonato M, Narindrasorasak S, Forbes J R, Cox D W, Sarkar B. Expression, purification, and metal binding properties of the N‐terminal domain from the wilson disease putative copper‐transporting ATPase (ATP7B). J Biol Chem 199727233279–33282. [DOI] [PubMed] [Google Scholar]
- 110.Tsivkovskii R, MacArthur B C, Lutsenko S. The Lys1010‐Lys1325 fragment of the Wilson's disease protein binds nucleotides and interacts with the N‐terminal domain of this protein in a copper‐dependent manner. J Biol Chem 20012762234–2242. [DOI] [PubMed] [Google Scholar]
- 111.Voskoboinik I, Mar J, Strausak D, Camakaris J. The regulation of catalytic activity of the Menkes copper‐translocating P‐type ATPase. Role of high affinity copper‐binding sites. J Biol Chem 200127628620–28627. [DOI] [PubMed] [Google Scholar]
- 112.Huster D, Lutsenko S. The distinct roles of the N‐terminal copper‐binding sites in regulation of catalytic activity of the Wilson's disease protein. J Biol Chem 200327832212–32218. [DOI] [PubMed] [Google Scholar]
- 113.Tsivkovskii R, Eisses J F, Kaplan J H, Lutsenko S. Functional properties of the copper‐transporting ATPase ATP7B (the Wilson's disease protein) expressed in insect cells. J Biol Chem 2002277976–983. [DOI] [PubMed] [Google Scholar]
- 114.Petris M J, Voskoboinik I, Cater M, Smith K, Kim B E, Llanos R M, Strausak D, Camakaris J, Mercer J F. Copper‐regulated trafficking of the Menkes disease copper ATPase is associated with formation of a phosphorylated catalytic intermediate. J Biol Chem 200227746736–46742. [DOI] [PubMed] [Google Scholar]
- 115.Dijkstra M, In ‘t Veld G, van den Berg G J, Muller M, Kuipers F, Vonk R J. Adenosine triphosphate‐dependent copper transport in isolated rat liver plasma membranes. J Clin Invest 199595412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bingham M J, Burchell A, McArdle H J. Identification of an ATP‐dependent copper transport system in endoplasmic reticulum vesicles isolated from rat liver. J Physiol 1995482583–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Morgan C T, Tsivkovskii R, Kosinsky Y A, Efremov R G, Lutsenko S. The distinct functional properties of the nucleotide‐binding domain of ATP7B, the human copper‐transporting ATPase: analysis of the Wilson disease mutations E1064A, H1069Q, R1151H, and C1104F. J Biol Chem 200427936363–36371. [DOI] [PubMed] [Google Scholar]
- 118.Dmitriev O, Tsivkovskii R, Abildgaard F, Morgan C T, Markley J L, Lutsenko S. Solution structure of the N‐domain of Wilson disease protein: distinct nucleotide‐binding environment and effects of disease mutations. Proc Natl Acad Sci U S A 20061035302–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Efremov R G, Kosinsky Y A, Nolde D E, Tsivkovskii R, Arseniev A S, Lutsenko S. Molecular modelling of the nucleotide‐binding domain of Wilson's disease protein: location of the ATP‐binding site, domain dynamics and potential effects of the major disease mutations. Biochem J 2004382293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Voskoboinik I, Strausak D, Greenough M, Brooks H, Petris M, Smith S, Mercer J F, Camakaris J. Functional analysis of the N‐terminal CXXC metal‐binding motifs in the human menkes copper‐transporting P‐type ATPase expressed in cultured mammalian cells. J Biol Chem 199927422008–22012. [DOI] [PubMed] [Google Scholar]
- 121.Yamaguchi Y, Heiny M E, Suzuki M, Gitlin J D. Biochemical characterization and intracellular localization of the Menkes disease protein. Proc Natl Acad Sci U S A 19969314030–14035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Petris M J, Mercer J F, Culvenor J G, Lockhart P, Gleeson P A, Camakaris J. Ligand‐regulated transport of the Menkes copper P‐type ATPase efflux pump from the Golgi apparatus to the plasma membrane: a novel mechanism of regulated trafficking. Embo J 1996156084–95 [PMC free article] [PubMed] [Google Scholar]
- 123.Harada M, Sakisaka S, Terada K, Kimura R, Kawaguchi T, Koga H, Taniguchi E, Sasatomi K, Miura N, Suganuma T, Fujita H, Furuta K, Tanikawa K, Sugiyama T, Sata M. Role of ATP7B in biliary copper excretion in a human hepatoma cell line and normal rat hepatocytes. Gastroenterology 2000118921–928. [DOI] [PubMed] [Google Scholar]
- 124.Harada M, Kawaguchi T, Kumemura H, Terada K, Ninomiya H, Taniguchi E, Hanada S, Baba S, Maeyama M, Koga H, Ueno T, Furuta K, Suganuma T, Sugiyama T, Sata M. The Wilson disease protein ATP7B resides in the late endosomes with Rab7 and the Niemann‐Pick C1 protein. Am J Pathol 2005166499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lutsenko S, Cooper M J. Localization of the Wilson's disease protein product to mitochondria. Proc Natl Acad Sci U S A 1998956004–6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Huster D, Hoppert M, Lutsenko S, Zinke J, Lehmann C, Mossner J, Berr F, Caca K. Defective cellular localization of mutant ATP7B in Wilson's disease patients and hepatoma cell lines. Gastroenterology 2003124335–345. [DOI] [PubMed] [Google Scholar]
- 127.Forbes J R, Cox D W. Copper‐dependent trafficking of Wilson disease mutant ATP7B proteins. Hum Mol Genet 200091927–1935. [DOI] [PubMed] [Google Scholar]
- 128.Roelofsen H, Wolters H, Van Luyn M J, Miura N, Kuipers F, Vonk R J. Copper‐induced apical trafficking of ATP7B in polarized hepatoma cells provides a mechanism for biliary copper excretion. Gastroenterology 2000119782–793. [DOI] [PubMed] [Google Scholar]
- 129.Schaefer M, Roelofsen H, Wolters H, Hofmann W J, Muller M, Kuipers F, Stremmel W, Vonk R J. Localization of the Wilson's disease protein in human liver. Gastroenterology 19991171380–1385. [DOI] [PubMed] [Google Scholar]
- 130.Schaefer M, Hopkins R G, Failla M L, Gitlin J D. Hepatocyte‐specific localization and copper‐dependent trafficking of the Wilson's disease protein in the liver. Am J Physiol 1999276G639–G646. [DOI] [PubMed] [Google Scholar]
- 131.Mercer J F, Barnes N, Stevenson J, Strausak D, Llanos R M. Copper‐induced trafficking of the cU‐ATPases: a key mechanism for copper homeostasis. Biometals 200316175–184. [DOI] [PubMed] [Google Scholar]
- 132.La Fontaine S, Theophilos M B, Firth S D, Gould R, Parton R G, Mercer J F. Effect of the toxic milk mutation (tx) on the function and intracellular localization of Wnd, the murine homologue of the Wilson copper ATPase. Hum Mol Genet 200110361–370. [DOI] [PubMed] [Google Scholar]
- 133.Guo Y, Nyasae L, Braiterman L T, Hubbard A L. NH2‐terminal signals in ATP7B Cu‐ATPase mediate its Cu‐dependent anterograde traffic in polarized hepatic cells. Am J Physiol Gastrointest Liver Physiol 2005289G904–G916. [DOI] [PubMed] [Google Scholar]
- 134.Petris M J, Mercer J F. The Menkes protein (ATP7A; MNK) cycles via the plasma membrane both in basal and elevated extracellular copper using a C‐terminal di‐leucine endocytic signal. Hum Mol Genet 199982107–2115. [DOI] [PubMed] [Google Scholar]
- 135.Nyasae L, Bustos R, Braiterman L, Eipper B, Hubbard A. Dynamics of endogenous ATP7A (Menkes protein) in intestinal epithelial cells: copper‐dependent redistribution between two intracellular sites. Am J Physiol Gastrointest Liver Physiol 2007292G1181–G1194. [DOI] [PubMed] [Google Scholar]
- 136.Greenough M, Pase L, Voskoboinik I, Petris M J, O'Brien A W, Camakaris J. Signals regulating trafficking of Menkes (MNK; ATP7A) copper‐translocating P‐type ATPase in polarized MDCK cells. Am J Physiol Cell Physiol 2004287C1463–C1471. [DOI] [PubMed] [Google Scholar]
- 137.Monty J F, Llanos R M, Mercer J F, Kramer D R. Copper exposure induces trafficking of the menkes protein in intestinal epithelium of ATP7A transgenic mice. J Nutr 20051352762–2766. [DOI] [PubMed] [Google Scholar]
- 138.Ravia J J, Stephen R M, Ghishan F K, Collins J F. Menkes Copper ATPase (Atp7a) is a novel metal‐responsive gene in rat duodenum, and immunoreactive protein is present on brush‐border and basolateral membrane domains. J Biol Chem 200528036221–36227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ke B X, Llanos R M, Wright M, Deal Y, Mercer J F. Alteration of copper physiology in mice overexpressing the human Menkes protein ATP7A. Am J Physiol Regul Integr Comp Physiol 2006290R1460–R1467. [DOI] [PubMed] [Google Scholar]
- 140.Cater M A, La Fontaine S, Shield K, Deal Y, Mercer J F. ATP7B mediates vesicular sequestration of copper: insight into biliary copper excretion. Gastroenterology 2006130493–506. [DOI] [PubMed] [Google Scholar]
- 141.Ambrosini L, Mercer J F. Defective copper‐induced trafficking and localization of the Menkes protein in patients with mild and copper‐treated classical Menkes disease. Hum Mol Genet 199981547–1555. [DOI] [PubMed] [Google Scholar]
- 142.Kim B E, Smith K, Petris M J. A copper treatable Menkes disease mutation associated with defective trafficking of a functional Menkes copper ATPase. J Med Genet 200340290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Forbes J R, Cox D W. Functional characterization of missense mutations in ATP7B: Wilson disease mutation or normal variant? Am J Hum Genet 1998631663–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Harada M, Sakisaka S, Terada K, Kimura R, Kawaguchi T, Koga H, Kim M, Taniguchi E, Hanada S, Suganuma T, Furuta K, Sugiyama T, Sata M. A mutation of the Wilson disease protein, ATP7B, is degraded in the proteasomes and forms protein aggregates. Gastroenterology 2001120967–974. [DOI] [PubMed] [Google Scholar]
- 145.Qi M, Byers P H. Constitutive skipping of alternatively spliced exon 10 in the ATP7A gene abolishes Golgi localization of the menkes protein and produces the occipital horn syndrome. Hum Mol Genet 19987465–469. [DOI] [PubMed] [Google Scholar]
- 146.Kim B E, Smith K, Meagher C K, Petris M J. A conditional mutation affecting localization of the Menkes disease copper ATPase. Suppression by copper supplementation. J Biol Chem 200227744079–44084. [DOI] [PubMed] [Google Scholar]
- 147.Francis M J, Jones E E, Levy E R, Ponnambalam S, Chelly J, Monaco A P. A Golgi localization signal identified in the Menkes recombinant protein. Hum Mol Genet 199871245–1252. [DOI] [PubMed] [Google Scholar]
- 148.Payne A S, Kelly E J, Gitlin J D. Functional expression of the Wilson disease protein reveals mislocalization and impaired copper‐dependent trafficking of the common H1069Q mutation. Proc Natl Acad Sci U S A 19989510854–10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.de Bie P, van de Sluis B, Burstein E, Berghe P V E, Muller P, Berger R, Gitlin J D, Wijmenga C, Klomp L W J. Distinct Wilson‐disease mutations in ATP7B are associated with enhanced binding to COMMD1 and reduced stability of ATP7B. Gastroenterology 200739863–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol 20057766–772. [DOI] [PubMed] [Google Scholar]
- 151.Cheng S H, Gregory R J, Marshall J, Paul S, Souza D W, White G A, O'Riordan C R, Smith A E. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell 199063827–834. [DOI] [PubMed] [Google Scholar]
- 152.Vanderwerf S M, Cooper M J, Stetsenko I V, Lutsenko S. Copper specifically regulates intracellular phosphorylation of the Wilson's disease protein, a human copper‐transporting ATPase. J Biol Chem 200127636289–36294. [DOI] [PubMed] [Google Scholar]
- 153.Voskoboinik I, Fernando R, Veldhuis N, Hannan K M, Marmy‐Conus N, Pearson R B, Camakaris J. Protein kinase‐dependent phosphorylation of the Menkes copper P‐type ATPase. Biochem Biophys Res Commun 2003303337–342. [DOI] [PubMed] [Google Scholar]
- 154.Lutsenko S, Petris M J. Function and regulation of the mammalian copper‐transporting ATPases: insights from biochemical and cell biological approaches. J Membr Biol 20031911–12. [DOI] [PubMed] [Google Scholar]
- 155.Vanderwerf S M, Lutsenko S. The Wilson's disease protein expressed in Sf9 cells is phosphorylated. Biochem Soc Trans 200230739–741. [DOI] [PubMed] [Google Scholar]
- 156.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck F H, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker E E. A human protein–protein interaction network: a resource for annotating the proteome. Cell 2005122957–968. [DOI] [PubMed] [Google Scholar]
- 157.Rae T D, Schmidt P J, Pufahl R A, Culotta V C, O'Halloran T V. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science 1999284805–808. [DOI] [PubMed] [Google Scholar]
- 158.Klomp L W, Lin S J, Yuan D S, Klausner R D, Culotta V C, Gitlin J D. Identification and functional expression of HAH1, a novel human gene involved in copper homeostasis. J Biol Chem 19972729221–9226. [DOI] [PubMed] [Google Scholar]
- 159.Lin S J, Pufahl R A, Dancis A, O'Halloran T V, Culotta V C. A role for the Saccharomyces cerevisiae ATX1 gene in copper trafficking and iron transport. J Biol Chem 19972729215–9220. [PubMed] [Google Scholar]
- 160.Hamza I, Faisst A, Prohaska J, Chen J, Gruss P, Gitlin J D. The metallochaperone Atox1 plays a critical role in perinatal copper homeostasis. Proc Natl Acad Sci U S A 2001986848–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hamza I, Schaefer M, Klomp L W, Gitlin J D. Interaction of the copper chaperone HAH1 with the Wilson disease protein is essential for copper homeostasis. Proc Natl Acad Sci U S A 19999613363–13368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Larin D, Mekios C, Das K, Ross B, Yang A S, Gilliam T C. Characterization of the interaction between the Wilson and Menkes disease proteins and the cytoplasmic copper chaperone, HAH1p. J Biol Chem 199927428497–28504. [DOI] [PubMed] [Google Scholar]
- 163.Pufahl R A, Singer C P, Peariso K L, Lin S J, Schmidt P J, Fahrni C J, Culotta V C, Penner‐Hahn J E, O'Halloran T V. Metal ion chaperone function of the soluble Cu(I) receptor Atx1. Science 1997278853–856. [DOI] [PubMed] [Google Scholar]
- 164.Multhaup G, Strausak D, Bissig K D, Solioz M. Interaction of the CopZ copper chaperone with the CopA copper ATPase of Enterococcus hirae assessed by surface plasmon resonance. Biochem Biophys Res Commun 2001288172–177. [DOI] [PubMed] [Google Scholar]
- 165.Banci L, Bertini I, Ciofi‐Baffoni S, Del Conte R, Gonnelli L. Understanding copper trafficking in bacteria: interaction between the copper transport protein CopZ and the N‐terminal domain of the copper ATPase CopA from Bacillus subtilis. Biochemistry 2003421939–1949. [DOI] [PubMed] [Google Scholar]
- 166.Radford D S, Kihlken M A, Borrelly G P, Harwood C R, Le Brun N E, Cavet J S. CopZ from Bacillus subtilis interacts in vivo with a copper exporting CPx‐type ATPase CopA. FEMS Microbiol Lett 2003220105–112. [DOI] [PubMed] [Google Scholar]
- 167.Hung I H, Casareno R L, Labesse G, Mathews F S, Gitlin J D. HAH1 is a copper‐binding protein with distinct amino acid residues mediating copper homeostasis and antioxidant defense. J Biol Chem 19982731749–1754. [DOI] [PubMed] [Google Scholar]
- 168.Strausak D, Howie M K, Firth S D, Schlicksupp A, Pipkorn R, Multhaup G, Mercer J F. Kinetic analysis of the interaction of the copper chaperone Atox1 with the metal binding sites of the Menkes protein. J Biol Chem 200327820821–20827. [DOI] [PubMed] [Google Scholar]
- 169.Portnoy M E, Rosenzweig A C, Rae T, Huffman D L, O'Halloran T V, Culotta V C. Structure‐function analyses of the ATX1 metallochaperone. J Biol Chem 199927415041–15045. [DOI] [PubMed] [Google Scholar]
- 170.Arnesano F, Banci L, Bertini I, Cantini F, Ciofi‐Baffoni S, Huffman D L, O'Halloran T V. Characterization of the binding interface between the copper chaperone Atx1 and the first cytosolic domain of Ccc2 ATPase. J Biol Chem 200127641365–41376. [DOI] [PubMed] [Google Scholar]
- 171.Van Dongen E M, Klomp L W, Merkx M. Copper‐dependent protein–protein interactions studied by yeast two‐hybrid analysis. Biochem Biophys Res Commun 2004323789–795. [DOI] [PubMed] [Google Scholar]
- 172.Yatsunyk L A, Rosenzweig A C. Cu(I) binding and transfer by the N terminus of the Wilson disease protein. J Biol Chem 20072828622–8631. [DOI] [PubMed] [Google Scholar]
- 173.Walker J M, Tsivkovskii R, Lutsenko S. Metallochaperone Atox1 transfers copper to the NH2‐terminal domain of the Wilson's disease protein and regulates its catalytic activity. J Biol Chem 200227727953–27959. [DOI] [PubMed] [Google Scholar]
- 174.Ralle M, Lutsenko S, Blackburn N J. Copper transfer to the N‐terminal domain of the Wilson disease protein (ATP7B): X‐ray absorption spectroscopy of reconstituted and chaperone‐loaded metal binding domains and their interaction with exogenous ligands. J Inorg Biochem 200498765–774. [DOI] [PubMed] [Google Scholar]
- 175.Walker J M, Huster D, Ralle M, Morgan C T, Blackburn N J, Lutsenko S. The N‐terminal metal‐binding site 2 of the Wilson's Disease Protein plays a key role in the transfer of copper from Atox1. J Biol Chem 200427915376–15384. [DOI] [PubMed] [Google Scholar]
- 176.Hamza I, Prohaska J, Gitlin J D. Essential role for Atox1 in the copper‐mediated intracellular trafficking of the Menkes ATPase. Proc Natl Acad Sci U S A 20031001215–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Tao T Y, Liu F, Klomp L, Wijmenga C, Gitlin J D. The copper toxicosis gene product Murr1 directly interacts with the Wilson disease protein. J Biol Chem 200327841593–41596. [DOI] [PubMed] [Google Scholar]
- 178.Burstein E, Ganesh L, Dick R D, van De Sluis B, Wilkinson J C, Klomp L W, Wijmenga C, Brewer G J, Nabel G J, Duckett C S. A novel role for XIAP in copper homeostasis through regulation of MURR1. Embo J 200423244–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ganesh L, Burstein E, Guha‐Niyogi A, Louder M K, Mascola J R, Klomp L W, Wijmenga C, Duckett C S, Nabel G J. The gene product Murr1 restricts HIV‐1 replication in resting CD4+ lymphocytes. Nature 2003426853–857. [DOI] [PubMed] [Google Scholar]
- 180.De Bie P, van de Sluis B, Klomp L, Wijmenga C. The Many Faces of the Copper Metabolism Protein MURR1/COMMD1. J Hered 200596803–811. [DOI] [PubMed] [Google Scholar]
- 181.Maine G N, Mao X, Komarck C M, Burstein E. COMMD1 promotes the ubiquitination of NF‐kappaB subunits through a cullin‐containing ubiquitin ligase. Embo J 200726436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.van de Sluis B, Muller P, Duran K, Chen A, Groot A J, Klomp L W, Liu P P, Wijmenga C. Increased activity of hypoxia‐inducible factor 1 is associated with early embryonic lethality in Commd1 null mice. Mol Cell Biol 2007274142–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Biasio W, Chang T, McIntosh C J, McDonald F J. Identification of Murr1 as a regulator of the human delta epithelial sodium channel. J Biol Chem 20042795429–5434. [DOI] [PubMed] [Google Scholar]
- 184.De Bie P, van de Sluis B, Burstein E, Duran K J, Berger R, Duckett C S, Wijmenga C, Klomp L W. Characterization of COMMD protein–protein interactions in NF‐kappaB signalling. Biochem J 200639863–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Greene W C. How resting T cells deMURR HIV infection. Nat Immunol 2004518–19. [DOI] [PubMed] [Google Scholar]
- 186.Bertinato J, L'Abbe M R. Copper modulates the degradation of copper chaperone for Cu,Zn superoxide dismutase by the 26 S proteosome. J Biol Chem 200327835071–35078. [DOI] [PubMed] [Google Scholar]
- 187.Nittis T, Gitlin J D. Role of copper in the proteosome‐mediated degradation of the multicopper oxidase hephaestin. J Biol Chem 200427925696–25702. [DOI] [PubMed] [Google Scholar]
- 188.Nadeau J H. Modifier genes in mice and humans. Nat Rev Genet 20012165–174. [DOI] [PubMed] [Google Scholar]
- 189.Cutting G R. Modifier genetics: cystic fibrosis. Annu Rev Genomics Hum Genet 20056237–260. [DOI] [PubMed] [Google Scholar]
- 190.Boyle M P. Strategies for identifying modifier genes in cystic fibrosis. Proc Am Thorac Soc 2007452–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Grubenbecher S, Stuve O, Hefter H, Korth C. Prion protein gene codon 129 modulates clinical course of neurological Wilson disease. Neuroreport 200617549–552. [DOI] [PubMed] [Google Scholar]
- 192.Merle U, Stremmel W, Gessner R. Influence of homozygosity for methionine at codon 129 of the human prion gene on the onset of neurological and hepatic symptoms in Wilson disease. Arch Neurol 200663982–985. [DOI] [PubMed] [Google Scholar]
- 193.Schiefermeier M, Kollegger H, Madl C, Polli C, Oder W, Kuhn H, Berr F, Ferenci P. The impact of apolipoprotein E genotypes on age at onset of symptoms and phenotypic expression in Wilson's disease. Brain 2000123585–590. [DOI] [PubMed] [Google Scholar]
- 194.Gu Y H, Kodama H, Du S L. Apolipoprotein E genotype analysis in Chinese Han ethnic children with Wilson's disease, with a concentration on those homozygous for R778L. Brain Dev 200527551–553. [DOI] [PubMed] [Google Scholar]
- 195.Paulsen M, Lund C, Akram Z, Winther J R, Horn N, Moller L B. Evidence that translation reinitiation leads to a partially functional Menkes protein containing two copper‐binding sites. Am J Hum Genet 200679214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kim B E, Petris M J. Phenotypic diversity of menkes disease in mottled mice is associated with defects in localization and trafficking of the ATP7A protein. J Med Genet. 2007; doi:10. 1136/jmg. 2007. 049627 [DOI] [PMC free article] [PubMed]
- 197.Gromadzka G, Schmidt H H, Genschel J, Bochow B, Rodo M, Tarnacka B, Litwin T, Chabik G, Czlonkowska A. Frameshift and nonsense mutations in the gene for ATPase7B are associated with severe impairment of copper metabolism and with an early clinical manifestation of Wilson's disease. Clin Genet 200568524–532. [DOI] [PubMed] [Google Scholar]
- 198.Thomas G R, Forbes J R, Roberts E A, Walshe J M, Cox D W. The Wilson disease gene: spectrum of mutations and their consequences. Nat Genet 19959210–217. [DOI] [PubMed] [Google Scholar]
- 199.Panagiotakaki E, Tzetis M, Manolaki N, Loudianos G, Papatheodorou A, Manesis E, Nousia‐Arvanitakis S, Syriopoulou V, Kanavakis E. Genotype–phenotype correlations for a wide spectrum of mutations in the Wilson disease gene (ATP7B). Am J Med Genet A 2004131168–173. [DOI] [PubMed] [Google Scholar]
- 200.Kusuda Y, Hamaguchi K, Mori T, Shin R, Seike M, Sakata T. Novel mutations of the ATP7B gene in Japanese patients with Wilson disease. J Hum Genet 20004586–91. [DOI] [PubMed] [Google Scholar]
- 201.Mercer J F. The molecular basis of copper‐transport diseases. Trends Mol Med 2001764–69. [DOI] [PubMed] [Google Scholar]
- 202.Ronce N, Moizard M P, Robb L, Toutain A, Villard L, Moraine C. A C2055T transition in exon 8 of the ATP7A gene is associated with exon skipping in an occipital horn syndrome family. Am J Hum Genet 199761233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Moller L B, Tumer Z, Lund C, Petersen C, Cole T, Hanusch R, Seidel J, Jensen L R, Horn N. Similar splice‐site mutations of the ATP7A gene lead to different phenotypes: classical Menkes disease or occipital horn syndrome. Am J Hum Genet 2000661211–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Gu Y H, Kodama H, Murata Y, Mochizuki D, Yanagawa Y, Ushijima H, Shiba T, Lee C C. ATP7A gene mutations in 16 patients with Menkes disease and a patient with occipital horn syndrome. Am J Med Genet 200199217–222. [DOI] [PubMed] [Google Scholar]
- 205.Donsante A, Tang J, Godwin S C, Holmes C S, Goldstein D S, Bassuk A, Kaler S G. Differences in ATP7A gene expression underlie intra‐familial variability in Menkes disease/occipital horn syndrome. J Med Genet 200744492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Stapelbroek J M, Bollen C W, van Amstel J K, van Erpecum K J, van Hattum J, van den Berg L H, Klomp L W, Houwen R H. The H1069Q mutation in ATP7B is associated with late and neurologic presentation in Wilson disease: results of a meta‐analysis. J Hepatol 200441758–763. [DOI] [PubMed] [Google Scholar]
- 207.Butler P, McIntyre N, Mistry P K. Molecular diagnosis of Wilson disease. Mol Genet Metab 200172223–230. [DOI] [PubMed] [Google Scholar]
- 208.Caca K, Ferenci P, Kuhn H J, Polli C, Willgerodt H, Kunath B, Hermann W, Mossner J, Berr F. High prevalence of the H1069Q mutation in East German patients with Wilson disease: rapid detection of mutations by limited sequencing and phenotype‐genotype analysis. J Hepatol 200135575–581. [DOI] [PubMed] [Google Scholar]
- 209.Firneisz G, Lakatos P L, Szalay F, Polli C, Glant T T, Ferenci P. Common mutations of ATP7B in Wilson disease patients from Hungary. Am J Med Genet 200210823–28. [DOI] [PubMed] [Google Scholar]
- 210.Houwen R H, Juyn J, Hoogenraad T U, Ploos van Amstel J K, Berger R. H714Q mutation in Wilson disease is associated with late, neurological presentation. J Med Genet 199532480–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Maier‐Dobersberger T, Ferenci P, Polli C, Balac P, Dienes H P, Kaserer K, Datz C, Vogel W, Gangl A. Detection of the His1069Gln mutation in Wilson disease by rapid polymerase chain reaction. Ann Intern Med 199712721–26. [DOI] [PubMed] [Google Scholar]
- 212.Waldenstrom E, Lagerkvist A, Dahlman T, Westermark K, Landegren U. Efficient detection of mutations in Wilson disease by manifold sequencing. Genomics 199637303–309. [DOI] [PubMed] [Google Scholar]
- 213.Czlonkowska A, Rodo M, Gajda J, Ploos van Amstel H K, Juyn J, Houwen R H. Very high frequency of the His1069Gln mutation in Polish Wilson disease patients. J Neurol 1997244591–592. [DOI] [PubMed] [Google Scholar]
- 214.Shah A B, Chernov I, Zhang H T, Ross B M, Das K, Lutsenko S, Parano E, Pavone L, Evgrafov O, Ivanova‐Smolenskaya I A, Anneren G, Westermark K, Urrutia F H, Penchaszadeh G K, Sternlieb I, Scheinberg I H, Gilliam T C, Petrukhin K. Identification and analysis of mutations in the Wilson disease gene (ATP7B): population frequencies, genotype–phenotype correlation, and functional analyses. Am J Hum Genet 199761317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Gromadzka G, Schmidt H H, Genschel J, Bochow B, Rodo M, Tarnacka B, Litwin T, Chabik G, Czlonkowska A. p.H1069Q mutation in ATP7B and biochemical parameters of copper metabolism and clinical manifestation of Wilson's disease. Mov Disord 200621245–248. [DOI] [PubMed] [Google Scholar]
- 216.Duc H H, Hefter H, Stremmel W, Castaneda‐Guillot C, Hernandez Hernandez A, Cox D W, Auburger G. His1069Gln and six novel Wilson disease mutations: analysis of relevance for early diagnosis and phenotype. Eur J Hum Genet 19986616–623. [DOI] [PubMed] [Google Scholar]
- 217.Ivanova‐Smolenskaya I A, Ovchinnikov I V, Karabanov A V, Deineko N L, Poleshchuk V V, Markova E D, Illarioshkin S N. The His1069Gln mutation in the ATP7B gene in Russian patients with Wilson disease. J Med Genet 199936174. [PMC free article] [PubMed] [Google Scholar]
- 218.Tarnacka B, Gromadzka G, Rodo M, Mierzejewski P, Czloonkowska A. Frequency of His1069Gln and Gly1267Lys mutations in Polish Wilson's disease population. Eur J Neurol 20007495–498. [DOI] [PubMed] [Google Scholar]
- 219.Loudianos G, Kostic V, Solinas P, Lovicu M, Dessi V, Svetel M, Major T, Cao A. Characterization of the molecular defect in the ATP7B gene in Wilson disease patients from Yugoslavia. Genet Test 20037107–112. [DOI] [PubMed] [Google Scholar]
- 220.Vrabelova S, Letocha O, Borsky M, Kozak L. Mutation analysis of the ATP7B gene and genotype/phenotype correlation in 227 patients with Wilson disease. Mol Genet Metab 200586277–285. [DOI] [PubMed] [Google Scholar]
- 221.Wu Z Y, Wang N, Lin M T, Fang L, Murong S X, Yu L. Mutation analysis and the correlation between genotype and phenotype of Arg778Leu mutation in chinese patients with Wilson disease. Arch Neurol 200158971–976. [DOI] [PubMed] [Google Scholar]
- 222.Yoo H W. Identification of novel mutations and the three most common mutations in the human ATP7B gene of Korean patients with Wilson disease. Genet Med 20024(Suppl)43S–8S. [DOI] [PubMed] [Google Scholar]
- 223.Petrukhin K, Lutsenko S, Chernov I, Ross B M, Kaplan J H, Gilliam T C. Characterization of the Wilson disease gene encoding a P‐type copper transporting ATPase: genomic organization, alternative splicing, and structure/function predictions. Hum Mol Genet 19941647–1656. [DOI] [PubMed]
- 224.Okada T, Shiono Y, Hayashi H, Satoh H, Sawada T, Suzuki A, Takeda Y, Yano M, Michitaka K, Onji M, Mabuchi H. Mutational analysis of ATP7B and genotype–phenotype correlation in Japanese with Wilson's disease. Hum Mutat 200015454–462. [DOI] [PubMed] [Google Scholar]
- 225.Liu X Q, Zhang Y F, Liu T T, Hsiao K J, Zhang J M, Gu X F, Bao K R, Yu L H, Wang M X. Correlation of ATP7B genotype with phenotype in Chinese patients with Wilson disease. World J Gastroenterol 200410590–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Gu Y H, Kodama H, Du S L, Gu Q J, Sun H J, Ushijima H. Mutation spectrum and polymorphisms in ATP7B identified on direct sequencing of all exons in Chinese Han and Hui ethnic patients with Wilson's disease. Clin Genet 200364(6)479–484. [DOI] [PubMed] [Google Scholar]
- 227.Wijmenga C, Klomp L W. Molecular regulation of copper excretion in the liver. Proc Nutr Soc 20046331–9 [DOI] [PubMed] [Google Scholar]
- 228.Moller L B, Petersen C, Lund C, Horn N. Characterization of the hCTR1 gene: genomic organization, functional expression, and identification of a highly homologous processed gene. Gene 200025713–22. [DOI] [PubMed] [Google Scholar]
- 229.Zhou B, Gitschier J. hCTR1: a human gene for copper uptake identified by complementation in yeast. Proc Natl Acad Sci U S A 199794(14)7481–7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Culotta V C, Klomp L W, Strain J, Casareno R L, Krems B, Gitlin J D. The copper chaperone for superoxide dismutase. J Biol Chem 199727223469–23472. [DOI] [PubMed] [Google Scholar]
- 231.Sturtz L A, Diekert K, Jensen L T, Lill R, Culotta V C. A fraction of yeast Cu, Zn‐superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria. A physiological role for SOD1 in guarding against mitochondrial oxidative damage. J Biol Chem 200127638084–38089. [DOI] [PubMed] [Google Scholar]
- 232.Glerum D M, Shtanko A, Tzagoloff A. Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J Biol Chem 199627114504–14509. [DOI] [PubMed] [Google Scholar]
- 233.Amaravadi R, Glerum D M, Tzagoloff A. Isolation of a cDNA encoding the human homolog of COX17, a yeast gene essential for mitochondrial copper recruitment. Hum Genet 199799329–333. [DOI] [PubMed] [Google Scholar]
- 234.Banci L, Bertini I, Del Conte R, D'Onofrio M, Rosato A. Solution structure and backbone dynamics of the Cu(I) and apo forms of the second metal‐binding domain of the Menkes protein ATP7A. Biochemistry 2004433396–3403. [DOI] [PubMed] [Google Scholar]
- 235.Banci L, Bertini I, Cantini F, DellaMalva N, Herrmann T, Rosato A, Wuthrich K. Solution structure and intermolecular interactions of the third metal‐binding domain of ATP7A, the Menkes disease protein. J Biol Chem 200628129141–29147. [DOI] [PubMed] [Google Scholar]
- 236.Banci L, Bertini I, Ciofi‐Baffoni S, Chasapis C T, Hadjiliadis N, Rosato A. An NMR study of the interaction between the human copper(I) chaperone and the second and fifth metal‐binding domains of the Menkes protein. Febs J 2005272865–871. [DOI] [PubMed] [Google Scholar]
- 237.DeSilva T M, Veglia G, Opella S J. Solution structures of the reduced and Cu(I) bound forms of the first metal binding sequence of ATP7A associated with Menkes disease. Proteins 2005611038–1049. [DOI] [PubMed] [Google Scholar]
- 238.Jones C E, Daly N L, Cobine P A, Craik D J, Dameron C T. Structure and metal binding studies of the second copper binding domain of the Menkes ATPase. J Struct Biol 2003143209–218. [DOI] [PubMed] [Google Scholar]
- 239.Jensen P Y, Bonander N, Karlsson B G, Horn N, Tumer Z, Farver O. Investigation of the copper binding sites in the Menkes disease protein, ATP7A. SSIEM Award. Society of the Study of Inborn Errors of Metabolism. J Inherit Metab Dis 199821195–198. [DOI] [PubMed] [Google Scholar]
- 240.Myari A, Hadjiliadis N, Fatemi N, Sarkar B. Copper(I) interaction with model peptides of WD6 and TM6 domains of Wilson ATPase: regulatory and mechanistic implications. J Inorg Biochem 2004981483–1494. [DOI] [PubMed] [Google Scholar]
- 241.Forbes J R, Hsi G, Cox D W. Role of the copper‐binding domain in the copper transport function of ATP7B, the P‐type ATPase defective in Wilson disease. J Biol Chem 199927412408–12413. [DOI] [PubMed] [Google Scholar]
- 242.Strausak D, La Fontaine S, Hill J, Firth S D, Lockhart P J, Mercer J F. The role of GMXCXXC metal binding sites in the copper‐induced redistribution of the Menkes protein. J Biol Chem 199927411170–11177. [DOI] [PubMed] [Google Scholar]
- 243.Goodyer I D, Jones E E, Monaco A P, Francis M J. Characterization of the Menkes protein copper‐binding domains and their role in copper‐induced protein relocalization. Hum Mol Genet 199981473–1478. [DOI] [PubMed] [Google Scholar]
- 244.Lutsenko S, Kaplan J H. Organization of P‐type ATPases: significance of structural diversity. Biochemistry 19953415607–15613. [DOI] [PubMed] [Google Scholar]
- 245.Solioz M, Vulpe C. CPx‐type ATPases: a class of P‐type ATPases that pump heavy metals. Trends Biochem Sci 199621237–241. [PubMed] [Google Scholar]
- 246.Cater M A, Forbes J, La Fontaine S, Cox D, Mercer J F. Intracellular trafficking of the human Wilson protein: the role of the six N‐terminal metal‐binding sites. Biochem J 2004380805–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Cater M A, La Fontaine S, Mercer J F. Copper binding to the N‐terminal metal‐binding sites or the CPC motif is not essential for copper‐induced trafficking of the human Wilson protein (ATP7B). Biochem J 2007401143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Petris M J, Camakaris J, Greenough M, LaFontaine S, Mercer J F. A C‐terminal di‐leucine is required for localization of the Menkes protein in the trans‐Golgi network. Hum Mol Genet 199872063–2071. [DOI] [PubMed] [Google Scholar]
- 249.Francis M J, Jones E E, Levy E R, Martin R L, Ponnambalam S, Monaco A P. Identification of a di‐leucine motif within the C terminus domain of the Menkes disease protein that mediates endocytosis from the plasma membrane. J Cell Sci 19991121721–32 [DOI] [PubMed] [Google Scholar]
- 250.Lane C, Petris M J, Benmerah A, Greenough M, Camakaris J. Studies on endocytic mechanisms of the menkes copper‐translocating P‐type ATPase (ATP7A; MNK). Endocytosis of the menkes protein. Biometals 20041787–98. [DOI] [PubMed] [Google Scholar]
- 251.Cobbold C, Coventry J, Ponnambalam S, Monaco A P. The Menkes disease ATPase (ATP7A) is internalized via a Rac1‐regulated, clathrin‐ and caveolae‐independent pathway. Hum Mol Genet 2003121523–1533. [DOI] [PubMed] [Google Scholar]
- 252.Lim C M, Cater M A, Mercer J F, La Fontaine S. Copper‐dependent interaction of glutaredoxin with the N termini of the copper‐ATPases (ATP7A and ATP7B) defective in Menkes and Wilson diseases. Biochem Biophys Res Commun 2006348428–436. [DOI] [PubMed] [Google Scholar]
- 253.Lim C M, Cater M A, Mercer J F, La Fontaine S. Copper‐dependent interaction of dynactin subunit p62 with the N terminus of ATP7B but not ATP7A. J Biol Chem 200628114006–14014. [DOI] [PubMed] [Google Scholar]
- 254.Ko J H, Son W, Bae G Y, Kang J H, Oh W, Yoo O J. A new hepatocytic isoform of PLZF lacking the BTB domain interacts with ATP7B, the Wilson disease protein, and positively regulates ERK signal transduction. J Cell Biochem 200699719–734. [DOI] [PubMed] [Google Scholar]
- 255.Stephenson S E, Dubach D, Lim C M, Mercer J F, La Fontaine S. A single PDZ domain protein interacts with the Menkes copper ATPase, ATP7A. A new protein implicated in copper homeostasis. J Biol Chem 200528033270–33279. [DOI] [PubMed] [Google Scholar]
- 256.Sanokawa‐Akakura R, Dai H, Akakura S, Weinstein D, Fajardo J E, Lang S E, Wadsworth S, Siekierka J, Birge R B. A novel role for the immunophilin FKBP52 in copper transport. J Biol Chem 200427927845–27848. [DOI] [PubMed] [Google Scholar]
- 257.Schroer T A. Dynactin. Annu Rev Cell Dev Biol 200420759–779. [DOI] [PubMed] [Google Scholar]
- 258.Moller L B, Bukrinsky J T, Molgaard A, Paulsen M, Lund C, Tumer Z, Larsen S, Horn N. Identification and analysis of 21 novel disease‐causing amino acid substitutions in the conserved part of ATP7A. Hum Mutat 20052684–93. [DOI] [PubMed] [Google Scholar]
- 259.Tang J, Robertson S, Lem K E, Godwin S C, Kaler S G. Functional copper transport explains neurologic sparing in occipital horn syndrome. Genet Med 20068711–718. [DOI] [PubMed] [Google Scholar]
- 260.Iida M, Terada K, Sambongi Y, Wakabayashi T, Miura N, Koyama K, Futai M, Sugiyama T. Analysis of functional domains of Wilson disease protein (ATP7B) in Saccharomyces cerevisiae. FEBS Lett 1998428281–285. [DOI] [PubMed] [Google Scholar]