Abstract
Mutations in the GJB2 gene are a major cause of non‐syndromic recessive hearing loss in many countries. In a significant fraction of patients, only monoallelic GJB2 mutations known to be either recessive or of unclear pathogenicity are identified. This paper reports a novel GJB2 mutation, −3438C→T, found in the basal promoter of the gene, in trans with V84M, in a patient with profound hearing impairment. This novel mutation can abolish the basal promoter activity of GJB2. These results highlight the importance of extending the mutational screening to regions outside the coding region of GJB2.
Keywords: GJB2, connexin 26, Cx26, promoter, hearing loss
Hereditary hearing loss is a genetically heterogeneous disorder with 85 loci and 39 nuclear disease genes reported to date.1 Most of the cases of genetic hearing loss are non‐syndromic, and of these, most are autosomal recessive. In some populations, up to 50% of cases of prelingual, non‐syndromic sensorineural hearing loss (NSSHL) are due to mutations in a single gene, GJB2,2 which codes for connexin 26 protein (Cx26). Therefore, GJB2 is normally the first gene to be tested in patients with hearing loss. Mutational screening performed to date has usually focused on the coding region. Few studies have been conducted on the non‐coding exon 1 of GJB2, and even fewer on the promoter region of this gene. As a result of the GJB2 screening performed to date, the majority of patients in various populations have been reported with only one GJB2 mutation, either recessive or of unclear pathogenicity. Some of these cases were elucidated upon screening for the two common GJB6 deletions.3,4,5 The remaining cases possibly have pathogenic mutations yet to be found in the promoter region or other non‐coding regions of GJB2. Notably, pathogenic mutations have been identified in the 5′ untranslated region (UTR) and promoter region of GJB1 (Cx32),6,7,8 another gene of the connexin family. In this paper, we report the genetic identification and functional analysis of the first GJB2 promoter mutation.
Methods
Subjects and audiometry
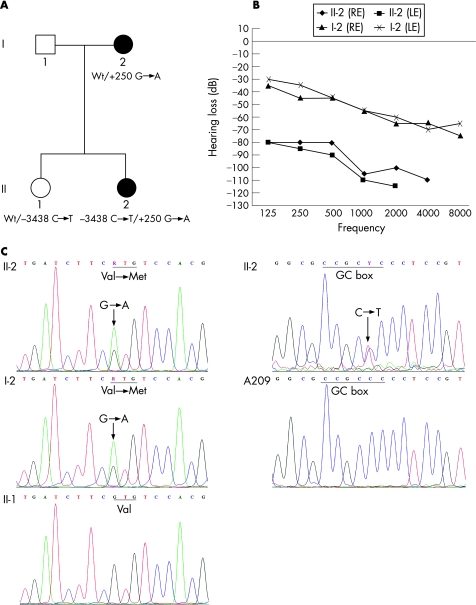
We studied a Portuguese family with NSSHL: the 35‐year old‐mother and her two daughters (fig 1A). The mother (I‐2) and the 12‐year proband (II‐2), had the hearing evaluated in both ears using pure‐tone audiometry. Both presented with bilateral sensorineural hearing loss, but while the mother was only moderately affected, the proband had profound hearing impairment (fig 1B). The other child (II‐1) had normal hearing. No biological or audiological data was available for the father (I‐1).
Figure 1 (A) Pedigree of the family here analysed. Black symbols, affected family members; GJB2 genotypes are displayed below the patients' symbols. (B) Audiograms from right ear (RE) and left ear (LE) of the affected members (II‐2 and I‐2), at the ages of 12 and 35 years, respectively. Patient II‐2 has bilateral profound hearing loss, and patient I‐2 has bilateral moderate hearing impairment. The hearing ability of both patients decreases across the frequencies tested, being most compromised at the high frequencies. (C) Electrophoreograms showing a heterozygous nucleotide change, +250 G→A, resulting in the V84M mutation, in I‐2 and II‐2; this substitution is not present in II‐1, Patient II‐2 is also heterozygous for the −3438C→T mutation in the basal promoter region, changing the GC box sequence from CCGCCC to CCGCTC. This alteration is not present in the control patient, A209.
Eight unrelated patients, presenting with different degrees of hearing loss, and with only a monoallelic GJB2 variant previously identified, were also part of this study. Six of them have one controversial GJB2 variant (M34T, R127H or G160S), and the other two patients harbour one recessive GBJ2 allele (35delG). None of these eight patients has either of the two GJB6 deletions, del(GJB6‐D13S1830) and del(GJB6‐D13S1854).
The control population used comprised 70 non‐related, hearing people from the Portuguese general population.
Informed consent was obtained from each participant before collecting blood for DNA extraction.
Screening of the GJB2 coding region and receptor splice site
The coding region and receptor splice site of GJB2 gene were analysed by single‐strand conformational polymorphism (SSCP) in both affected members of the family (I‐2 and II‐2). Sequencing of all the GJB2 coding region and receptor splice site was performed to identify the sequence variation detected by SSCP and to exclude other possible mutations in that genomic region. Before analysis of GJB2 basal promoter, both I‐2 and II‐2 were screened for the common GJB6 deletions, del(GJB6‐D13S1830) and del(GJB6‐D13S1854), following the method used by del Castillo et al.4
Screening of the GJB2 donor splice site, exon 1 and basal promoter
For the three members of the family under study, the 70 Portuguese controls and the eight patients with only a monoallelic GJB2 variant, a GJB2 region of 1009 bp, comprising the donor splice site, exon 1 and part of the 5′ upstream sequence, was amplified by PCR using the forward primer 5′‐CgTTCgTTCggATTggTgAg‐3′ and the reverse primer 5′‐CAgAAACgCCCgCTCCAgAA‐3′. The PCR products from the proband and one control patient were sequenced using the internal forward primer 5′‐ggCTCAAAggAACTAggAgATCg‐3′, with the same reverse primer as that used for amplification. These primers, used for sequencing, delimit a 539 bp GJB2 region (from nucleotides −3573 to −3034 relative to the initiator methionine), which includes the 128 bp basal promoter, exon 1 and the donor splice site.
The presence of the −3438C→T mutation identified in the proband was investigated in her mother and sister, and in the 70 Portuguese controls, by restriction analysis of the PCR products with the enzyme BsrBI (New England Biolabs, Quinta do Paizinho, Carnaxide, Portugal). The eight patients who had only a monoallelic GJB2 variant were assessed by sequencing for the presence of this mutation or other mutations in the donor splice site, exon 1 or basal promoter. The basal promoter, exon 1 and donor splice site of GJB2 gene can be found in GenBank (accession number U43932.1).
Reporter‐gene assay for assessment of wild‐type and mutant GJB2 basal promoter activity
A fragment of the promoter region from GJB2 alleles containing either a cytosine or a thymine at position −3438 relative to the initiator methionine was amplified by PCR using the forward primer 5′‐ATACAgAgCTCACAgAggACAACgACCACAg‐3′, designed to contain a SacI site, and the reverse primer 5′ TTAgTACCATggAgggCCgCAACACCTgTC‐3′, designed to contain an NcoI site. The PCR products were cloned into the vector pCR2.1‐topo using a commercial kit (TOPO TA pCR 2.1, TOPO Cloning Kit (ref. 45‐064); Invitrogen, Prat de Llobregat, Barcelona, Spain). The SacI–NcoI fragment was excised by double digestion with the enzymes SacI and NcoI, and was then directionally cloned into pGL3‐basic vector (Promega, Lisbon, Portugal) using its SacI and NcoI sites. A fragment upstream of the basal promoter and downstream of the SacI site was then removed by digestion with SacI and XmaI followed by self‐ligation of the remaining plasmid. The resulting constructs contained, therefore, a fragment of the GJB2 promoter region ranging from nucleotides −3348 to −3490, which includes the entire basal promoter, cloned upstream of the firefly luciferase coding sequence. The constructs termed −3438C‐pGL3 (wild‐type sequence) and −3438T‐pGL3 (containing the mutation −3438C→T) were assessed by sequencing and a diagnostic digestion with BsrBI to confirm sequence integrity. HEK‐293 or Caco‐2 cells were transfected with equimolar amounts of pRL‐TK vector (Promega) and −3438C‐pGL3, −3438T‐pGL3 or pGL3‐basic, using a commercial transfection reagent (FuGENE 6.0; Roche, Basel, Switzerland). The transfected cells were lysed 48h post‐transfection. Luminescence of firefly luciferase (coded by pGL3‐basic vector and by the pGL3 constructs) and Renilla luciferase (coded by pRL‐TK vector) was assessed (Dual‐Luciferase® Reporter Assay System; Promega) and a multilayer counter plate reader (Victor(2) 1420‐002; Perkin Elmer Wallack, Norton, Ohio, USA). The reporter gene assay was performed five times on HEK‐293 cells and four times on Caco‐2 cells. The firefly luciferase luminescence values were normalised against the Renilla luciferase luminescence values for each sample in each experiment. For each construct the average value of normalised luciferase luminescence was calculated and plotted in a histogram and analysed using the t test and Microsoft Excel software.
Dye‐transfer assay for assessment of Wt‐Cx26 and V84M‐Cx26 function
Gap‐junctional communication in Wt‐Cx26 and V84M‐Cx26‐expressing cells was assessed by intercellular spread of lucifer yellow (LY) and neurobiotin (NBN). LY and NBN were delivered to HeLa cells using the whole‐cell configuration of the patch‐clamp technique. HeLa cells were transiently transfected with 500 ng Wt‐Cx26‐enhanced green fluorescent protein (EGFP) cDNA or V84M‐Cx26‐EGFP cDNA, using a commercial transfection reagent (Lipofectamine LTX; Invitrogen) according to the manufacturer's recommendations. Transfected HeLa cells grown on 9 mm coverslips were transferred to a recording chamber 24 hours after transfection. Cells in which significant EGFP‐positive gap junction plaques were identified were selected for dye transfer. Following 5‐minute whole‐cell recordings, cells were fixed in 4% paraformaldehyde for 20 minutes and washed in phosphate‐buffered saline. LY and NBN were detected and imaged as previously described.9
Results
Analysis of the coding region of the GJB2 gene revealed that the proband and her mother were heterozygous for the V84M mutation (fig 1C). No other coding GJB2 variants were found. The V84M mutation has been reported previously to be associated with profound hearing loss.10,11 In the latter study, V84M was in trans with the 35delG mutation or M34T, suggesting that the V84M mutation is a recessive GJB2 mutation associated with profound hearing loss. As the proband analysed in our study was heterozygous for V84M and had profound hearing impairment, we hypothesised that she could yet possess either one of the two common GJB6 deletions, del(GJB6‐D13S1830) and del(GJB6‐D13S1854), or a non‐coding GJB2 mutation. Therefore, we tested the proband and her mother for the presence of del(GJB6‐D13S1830) or del(GJB6‐D13S1854). The deletions were not detected in either patient. We then sequenced the GJB2 donor splice site, exon 1 and basal promoter of the proband. A previously unreported substitution, −3438C→T, was found in the heterozygous state within the basal promoter. Restriction analysis with the enzyme BsrBI revealed that the proband's normal‐hearing sister also had one allele −3438C→T, but their mother, who had moderate hearing impairment, did not (data not shown). This variation was not found in the 70 hearing individuals from the Portuguese general population. Neither this nor other mutations were found in the donor splice site, exon 1 or basal promoter of seven of the eight monoallelic patients analysed. However, one monoallelic patient, heterozygous for the controversial R127H variant, was shown to be heterozygous for the donor splice‐site recessive mutation IVS1+1 G→A.
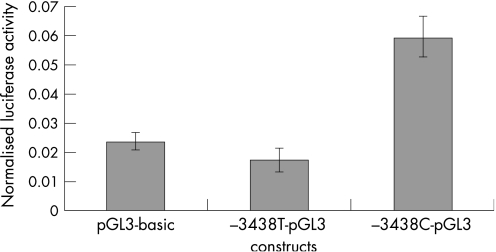
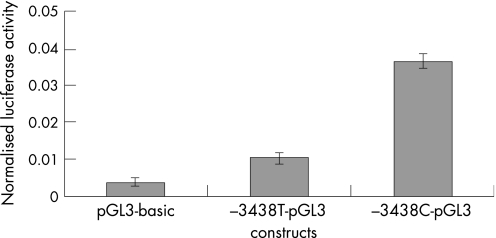
The novel −3438C→T mutation is localised 3438 nucleotides upstream of the initiator methionine, in the GC box at position −81 relative to the transcription start point (TSP). This GC box and another at position −93 relative to the TSP, regulate GJB2 basal transcription by interacting with the transcription factors Sp1 and Sp3, but the −81 box is the more important.12 Because the −3438C→T mutation disrupts this GC box, we considered it to be potentially pathogenic, and reporter‐gene assays were performed to confirm this. They revealed that the −3438C→T mutation abolishes the GJB2 basal promoter activity in HEK‐293 cells (fig 2), and greatly reduces it in Caco‐2 cells (fig 3). Figure 2 shows that the mean normalised value of firefly luciferase luminescence from cells transfected with the plasmid containing the wild‐type basal promoter (−3438C‐pGL3) was higher than and significantly different from the value obtained from cells transfected with the promoterless pGL3‐basic vector, which indicates promoter activity of the wild‐type basal promoter (p<0.01), as expected. In contrast, there was no significant difference of the mean normalised values of firefly luciferase luminescence between cells that were transfected either with the mutant basal promoter (−3438T‐pGL3) or with the pGL3‐basic vector (p = 0.29). This means that the mutation −3438C→T abolishes the activity of the GJB2 basal promoter in HEK‐293 cells. In Caco‐2 cells, the mutant GJB2 basal promoter showed some activity compared with the pGL3‐basic vector (p<0.001), but was much less active than the wild‐type basal promoter (p<<0.001) (fig 3).
Figure 2 Reporter‐gene assay in HEK‐293 cells.
Figure 3 Reporter gene assay in Caco‐2 cells.
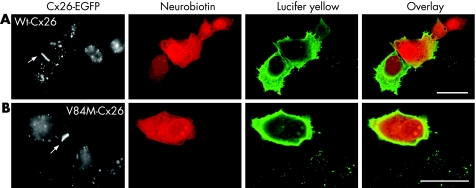
To confirm the suspected pathogenic effect of the V84M mutation we performed a dye‐transfer assay in HeLa cells. This experiment revealed clear differences between Wt‐Cx26 and V84M‐Cx26 function in the permeability of gap junctions to LY and NBN. In both groups of cells, there were detectable gap‐junction plaques between adjacent cells (fig 4). In all Wt‐Cx26 recordings (6/6) there was LY and NBN transfer to at least one adjacent cell (fig 4A). Conversely, in all V84M‐Cx26 recordings (8/8) there was no transfer of LY or NBN (fig 4B). The membrane capacitance (an approximation of the total continuous membrane area in contact with the recording pipette) was significantly larger in Wt‐Cx26 recordings (mean (SD) 54.8 (8.7) pF, n = 6) compared with V84M‐Cx26 recordings (29.6 (2.9) pF, n = 8, p<0.01; unpaired t test). These results suggest that cells expressing V84M‐Cx26 can form gap junction‐like aggregates at the plasma membrane, but this does not allow intercellular electrical or molecular coupling.
Figure 4 Dye‐transfer assay of connexin (Cx)26 function in Hela cells. (A) In cells expressing Wt‐Cx26, enhanced green fluorescent protein (EGFP)‐labelled gap junction plaques (arrow) were seen. Following whole‐cell dye injections, neurobiotin and lucifer yellow were seen to transfer to adjacent cells. (B) In cells expressing V84M‐Cx26, EGFP‐labelled gap junction plaques (arrow) were also apparent. There was no spread of neurobiotin or lucifer yellow in these cells. Scale bars, 20 μm.
Discussion
Connexin 26, a transmembrane protein coded by the gene GJB2, and a component of gap junctions, is strongly expressed in the cochlea, both in epithelial and connective tissues, where it is believed not only to have a role in the recirculation of the ion K+,13 which is a crucial mechanism for the transduction of sound waves into nervous impulses and consequently for proper hearing function, but also in the permeability to signalling molecules and metabolites.14 The gene GJB2 is composed of two exons separated by an intron, and the coding region is entirely contained in exon 2. The basal promoter activity resides in the first 128 nucleotides upstream of the TSP and has two GC boxes, at positions −81 and −93 from the TSP, which are important for transcription.12
Most of the GJB2 sequence variations described to date are localised in the coding region, and only a few have been reported in non‐coding regions of the gene.15,16,17,18,19 Among the latter, there are two donor splice‐site recessive mutations.15,19 The rest of the non‐coding variations reported to date are localised upstream of the exon 1, in exon 1, the intron, the 5′ UTR of exon 2, or the 3′ UTR, but none of these have yet been proven to be pathogenic. One is the −493del10 deletion,16 located upstream of the basal promoter region, which occurs in most of the GJB2 alleles harbouring the M34T mutation. This deletion disrupts a MGF‐like sequence that is strongly homologous to the MGF binding site of the mouse β‐casein gene.20 It was found that GJB2 alleles containing both −493del10 and M34T were expressed in cultured keratinocytes,16 but it is not known whether the deletion altered the gene's normal expression and whether it has an effect in the cochlea.
We found a novel sequence variation in the gene GJB2 within the basal promoter, more specifically, in the −81 GC box. This novel mutation, −3438 C→T, was found in trans with the V84M mutation in a patient with profound hearing impariment, the proband of this study. Her mother, who had moderate hearing impairment, harboured V84M as the sole mutation in the coding region of GJB2, and tested negative for the −3438C→T mutation. The proband's normal‐hearing sister did not have the V84M, but was found to be heterozygous for the −3438C→T mutation. The V84M mutation has been reported in other cases of profound hearing loss,10,11 and one of these cases was a patient also harbouring the 35delG,11 which suggests that when V84M‐Cx26 is the only variant of Cx26 being produced, profound hearing loss can result. As the −81 GC box was found to be crucial for the transcription of the gene in both the MCF‐12A (human immortalised mammary epithelial) and RL95‐2 (human endometrial) cell lines,12 we hypothesised that the −3438 C→T mutation could cause loss of function of the GC box, impairing or abolishing the GJB2 transcription in the cochlea. In this way, the profound hearing loss of the proband analysed here could be due to expression of only the V84M‐Cx26 protein. To confirm that the GJB2 genotype −3438C→T/V84M could indeed be the cause of the profound hearing loss of the proband analysed in this study, we performed functional studies on both mutations.
The reporter‐gene assay, performed to assess the functionality of the basal promoter containing the −3438C→T mutation, revealed that this mutation abolishes the GJB2 basal promoter activity in the HEK‐293 cell line and greatly reduces it in the Caco‐2 cell line.
The dye‐transfer assay of V84M‐Cx26 gap‐junction properties revealed that this mutation results in communication‐incompetent cells. A recent study has suggested that homomeric expression of V84L‐Cx26 results in gap‐junction channels that have unitary channel conductance and LY permeability that is indistinguishable from that of Wt‐Cx26 channels.14 However, the basis of the hearing impairment caused by this mutation has been ascribed to impaired permeability to inositol trisphosphate. This would result in dysfunctional Ca2+ mobilisation in affected cochlear epithelial cells. In our study, HeLa cells expressing V84M‐Cx26 also formed morphologically normal gap‐junction plaques, but appeared to be uncoupled both electrically (based on membrane capacitance measurements), and metabolically (based on impaired LY and NBN transfer).
In conclusion, this study describes the first pathogenic, hearing loss‐related mutation impairing the basal promoter activity of the gene GJB2. It also shows that this or other basal promoter mutations, or even mutations in other non‐coding regions, may be present in some patients with hearing loss with only a monoallelic GJB2 recessive mutation. Thus, screening of the basal promoter and other non‐coding regions of the GJB2 gene, such as exon 1, UTRs and both splice sites, are important and necessary for improving genetic diagnosis regarding these monoallelic patients, and for subsequent genetic counselling. We also present the first functional evidence for the pathogenicity of the previously described V84M mutation.
Key points
We report identification of the first GJB2 mutation that lies in the basal promoter of the gene associated with recessive non‐syndromic hearing loss.
Reporter‐gene studies supported the deleterious nature of this promoter mutation.
As the −3438C→T mutation is in trans with V84M, we performed functional studies, which showed that V84M forms non‐functional channels in vitro.
Acknowledgements
We would like to thank the family for its contribution to the investigation, Dr Diana Blaydon for providing the pRL‐TK plasmid and for her help and advice regarding the functional studies and statistics, and Professor Alan Storey for providing the pGL3‐Basic vector. TM and H T were supported by the Portuguese Fundação para a Ciência e Tecnologia (Foundation for Science and Technology) grant SFRH/BD/19988/2004 and grant SSRH/BD/24575/2005, respectively. The study was also supported by a Wellcome Trust VIP award to TA and DPK. DJ is a Royal Society University Research Fellow.
Abbreviations
Cx - connexin
EGFP - enhanced green fluorescent protein
LY - lucifer yellow
NBN - neurobiotin
NSSHL - non‐syndromic sensorineural hearing loss
SSCP - single‐strand confomational polymorphism
TSP - transcription start point
UTR - untranslated region
Footnotes
Competing interests: None declared.
References
- 1.Petersen M B, Willems P J. Non‐syndromic, autosomal‐recessive deafness. Clin Genet 200669371–392. [DOI] [PubMed] [Google Scholar]
- 2.Kenneson A, Van Naarden Braun K, Boyle C. GJB2 (connexin 26) variants and nonsyndromic sensorineural hearing loss: a HuGE review. Genet Med 20024258–274. [DOI] [PubMed] [Google Scholar]
- 3.del Castillo I, Villamar M, Moreno‐Pelayo M A, del Castillo F J, Alvarez A, Telleria D, Menendez I, Moreno F. A deletion involving the connexin 30 gene in nonsyndromic hearing impairment. N Engl J Med 2002346243–249. [DOI] [PubMed] [Google Scholar]
- 4.del Castillo F J, Rodriguez‐Ballesteros M, Alvarez A, Hutchin T, Leonardi E, de Oliveira C A, Azaiez H, Brownstein Z, Avenarius M R, Marlin S, Pandya A, Shahin H, Siemering K R, Weil D, Wuyts W, Aguirre L A, Martin Y, Moreno‐Pelayo M A, Villamar M, Avraham K B, Dahl H H, Kanaan M, Nance W E, Petit C, Smith R J, Van Camp G, Sartorato E L, Murgia A, Moreno F, del Castillo I. A novel deletion involving the connexin‐30 gene, del(GJB6‐d13s1854), found in trans with mutations in the GJB2 gene (connexin‐26) in subjects with DFNB1 non‐syndromic hearing impairment. J Med Genet 200542588–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.del Castillo I, Moreno‐Pelayo M A, Del Castillo F J, Brownstein Z, Marlin S, Adina Q, Cockburn D J, Pandya A, Siemering K R, Chamberlin G P, Ballana E, Wuyts W, Maciel‐Guerra A T, Alvarez A, Villamar M, Shohat M, Abeliovich D, Dahl H H, Estivill X, Gasparini P, Hutchin T, Nance W E, Sartorato E L, Smith R J, Van Camp G, Avraham K B, Petit C, Moreno F. Prevalence and evolutionary origins of the del(GJB6‐D13S1830) mutation in the DFNB1 locus in hearing‐impaired subjects: a multicenter study. Am J Hum Genet 2003731452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houlden H, Girard M, Cockerell C, Ingram D, Wood N W, Goossens M, Walker R W, Reilly M M. Connexin 32 promoter P2 mutations: a mechanism of peripheral nerve dysfunction. Ann Neurol 200456730–734. [DOI] [PubMed] [Google Scholar]
- 7.Ionasescu V V, Searby C, Ionasescu R, Neuhaus I M, Werner R. Mutations of the noncoding region of the connexin32 gene in X‐linked dominant Charcot‐Marie‐Tooth neuropathy. Neurology 199647541–544. [DOI] [PubMed] [Google Scholar]
- 8.Wang H L, Wu T, Chang W T, Li A H, Chen M S, Wu C Y, Fang W. Point mutation associated with X‐linked dominant Charcot‐Marie‐Tooth disease impairs the P2 promoter activity of human connexin‐32 gene. Brain Res Mol Brain Res 200078146–153. [DOI] [PubMed] [Google Scholar]
- 9.Jagger D J, Forge A. Compartmentalized and signal‐selective gap junctional coupling in the hearing cochlea. J Neurosci 2006261260–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pandya A, Arnos K S, Xia X J, Welch K O, Blanton S H, Friedman T B, Garcia Sanchez G, Liu M X, Morell R, Nance W E. Frequency and distribution of GJB2 (connexin 26) and GJB6 (connexin 30) mutations in a large North American repository of deaf probands. Genet Med 20035295–303. [DOI] [PubMed] [Google Scholar]
- 11.Cheng X, Li L, Brashears S, Morlet T, Ng S S, Berlin C, Hood L, Keats B. Connexin 26 variants and auditory neuropathy/dys‐synchrony among children in schools for the deaf. Am J Med Genet A 200513913–18. [DOI] [PubMed] [Google Scholar]
- 12.Tu Z J, Kiang D T. Mapping and characterization of the basal promoter of the human connexin26 gene. Biochim Biophys Acta 19981443169–181. [DOI] [PubMed] [Google Scholar]
- 13.Kikuchi T, Kimar R S, Paul D L, Adams J C. Gap junctions in the rat cochlea: immunohistochemical and ultrastructural analysis. Anat Embryol 1995191101–118. [DOI] [PubMed] [Google Scholar]
- 14.Beltramello M, Piazza V, Bukauskas F F, Pozzan T, Mammano F. Impaired permeability to Ins(1,4,5)P3 in a mutant connexin underlies recessive hereditary deafness. Nat Cell Biol 2005763–69. [DOI] [PubMed] [Google Scholar]
- 15.Denoyelle F, Marlin S, Weil D, Moatti L, Chauvin P, Garabedian E N, Petit C. Clinical features of the prevalent form of childhood deafness, DFNB1, due to a connexin‐26 gene defect: implications for genetic counselling. Lancet 19993531298–1303. [DOI] [PubMed] [Google Scholar]
- 16.Houseman M J, Ellis L A, Pagnamenta A, Di W L, Rickard S, Osborn A H, Dahl H H, Taylor G R, Bitner‐Glindzicz M, Reardon W, Mueller R F, Kelsell D P. Genetic analysis of the connexin‐26 M34T variant: identification of genotype M34T/M34T segregating with mild‐moderate non‐syndromic sensorineural hearing loss. J Med Genet 20013820–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roux A F, Pallares‐Ruiz N, Vielle A, Faugere V, Templin C, Leprevost D, Artieres F, Lina G, Molinari N, Blanchet P, Mondain M, Claustres M. Molecular epidemiology of DFNB1 deafness in France. BMC Med Genet 200455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang H Y, Fang P, Ward P A, Schmitt E, Darilek S, Manolidis S, Oghalai J S, Roa B B, Alford R L. DNA sequence analysis of GJB2, encoding connexin 26: observations from a population of hearing impaired cases and variable carrier rates, complex genotypes, and ethnic stratification of alleles among controls. Am J Med Genet A 20061402401–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green G E, Scott D A, McDonald J M, Woodworth G G, Sheffield V C, Smith R J. Carrier rates in the midwestern United States for GJB2 mutations causing inherited deafness. JAMA 19992812211–2216. [DOI] [PubMed] [Google Scholar]
- 20.Kiang D T, Jin N, Tu Z J, Lin H H. Upstream genomic sequence of the human connexin26 gene. Gene 1997199165–171. [DOI] [PubMed] [Google Scholar]