Abstract
De novo sequencing is a spectrum analysis approach for mass spectrometry data to discover post-translational modifications in proteins; however, such an approach is still in its infancy and is still not widely applied to proteomic practices due to its limited reliability. In this work, we describe a de novo sequencing approach for the discovery of protein modifications based on identification of the proteome UStags. The de novo information was obtained from Fourier-transform tandem mass spectrometry data for peptides and polypeptides from a yeast lysate, and the de novo sequences obtained were selected based on filter levels designed to provide a limited yet high quality subset of UStags. The DNA-predicted database protein sequences were then compared to the UStags, and the differences observed across or in the UStags (i.e., the UStags’ prefix and suffix sequences and the UStags themselves) were used to infer possible sequence modifications. With this de novo–UStag approach, we uncovered some unexpected variances within several yeast protein sequences due to amino acid mutations and/or multiple modifications to the predicted protein sequences. To determine false discovery rates, two random (false) databases were independently used for sequence matching, and ∼3% false discovery rates were estimated for the de novo–UStag approach. The factors affecting the reliability (e.g., existence of de novo sequencing noise residues and redundant sequences) and the sensitivity of the approach were investigated and described. The combined de novo–UStag approach complements the UStag method previously reported by enabling the discovery of new protein modifications.
The UStag method for unambiguous peptide and polypeptide identification has recently been demonstrated for the analysis of enzymatically (e.g., tryptic) digested cell lysates1 and for the determination of natural intracellular proteolysis (degradation) of proteins2 using accurate Fourier-transform tandem mass spectrometry (FT-MS/MS) data. Sequences are determined to be UStags when the accurately measured consecutive fragments reveal these sequences to be unique in the genome for single proteins. The UStags reported1,2 are assigned for the candidates that have the top closest spectral similarities to the MS/MS measurement (e.g., candidates ranked from Sequest). Advantage of such a database search–UStag approach is that it produces sequence identities with extremely low false discovery rates for peptides/polypeptides having a large range of lengths and with various amino acid termini.1,2 Also, this approach is capable of identifying unknown or unexpected changes, deviations, and errors from the predicted protein sequences.1 However, the amino acid changes, deviations, and errors either on the UStag’s prefix (i.e., the part of sequence prior to a UStag in the sequencing direction) or within the UStag itself may initially be missed since the b and y ion fragmentation patterns may not match at all the genome sequence used initially to provide a “close match” candidate list in the database search–UStag process. De novo sequencing is able to measure mass differences and consider modifications due to the mass change regardless of what the genome sequencing dictates and thus can enhance and complement the UStag approach to reveal modifications missed from the database search–UStag approach.
Several de novo algorithms have been developed, and Pevtsov et al.3 evaluated five de novo algorithms (AUDENS, Lutefisk, NovoHMM, PepNovo, and PEAKS) using a limited size data set containing peptides digested from model proteins. Probably because of the limited ability to correctly identify peptides (e.g., only ∼30% of all peptides were identified without errors as claimed by Frank et al.4), most de novo peptide identification approaches have not been widely applied for the identification of proteome proteins and their modifications. Efforts to improve the reliability of de novo sequencing have been made. These include the utilization of accurate mass measurements (e.g., using Orbitrap spectrometers),4,5 complementary methods for peptide fragmentations,5 and the introduction or use of more advanced statistical scoring systems.6–9 Direct use of currently available de novo tools is fraught with difficulties oftentimes because these algorithms typically only work when applied to small peptides with low charge states (e.g., ≤3) since they have been primarily developed for lowresolution MS/MS. Additionally, complex sequence variances, including the unexpected/unknown variances, involving multiple modifications on a single peptide/polypeptide sequence that deviate from the predicted genome sequence, are difficult to identify with high confidence using current sequence analysis methods. Therefore, there lies a need to develop reliable tools for the confident identification of these modified peptides/ polypeptides without limitation of the modification complexities (e.g., any combination of modifications listed in UNIMOD (http://www.unimod.org/).
In this work, we developed a de novo–UStag approach for the identification of protein post-translational modifications (PTMs). On the basis of the assignments of UStags, the protein sequences were specified and the variances of the specified protein sequences from their genome predictions due to various reasons including amino acid mutations or database errors and complex multiple PTMs on the protein sequences were confidently determined. The zero-charge state spectra were used for the de novo sequencing. A moderate sized data set that contained ∼30 000 Orbitrap FT-MS/MS spectra was used for the development, examination, and evaluation of this de novo–UStag approach. The data set was obtained from a yeast Saccharomyces cerevisiae lysate without the addition of extracellular enzymes for digestion (i.e., the sample only contained polypeptides and peptides generated by the intracellular proteases). The most intense (or highly abundant) peaks that had isotopic envelopes were used for de novo sequencing, and the factors that influenced the reliability of the de novo sequencing were examined. The UStags were conservatively assigned from de novo sequences with the removal of de novo noise residues. The results from various examinations shown in this work were also informative to develop reliable and sensitive de novo sequencing methods for FT-MS/ MS proteomic data sets.
METHODS
Description of the FT-MS/MS Data Set Used in This Study
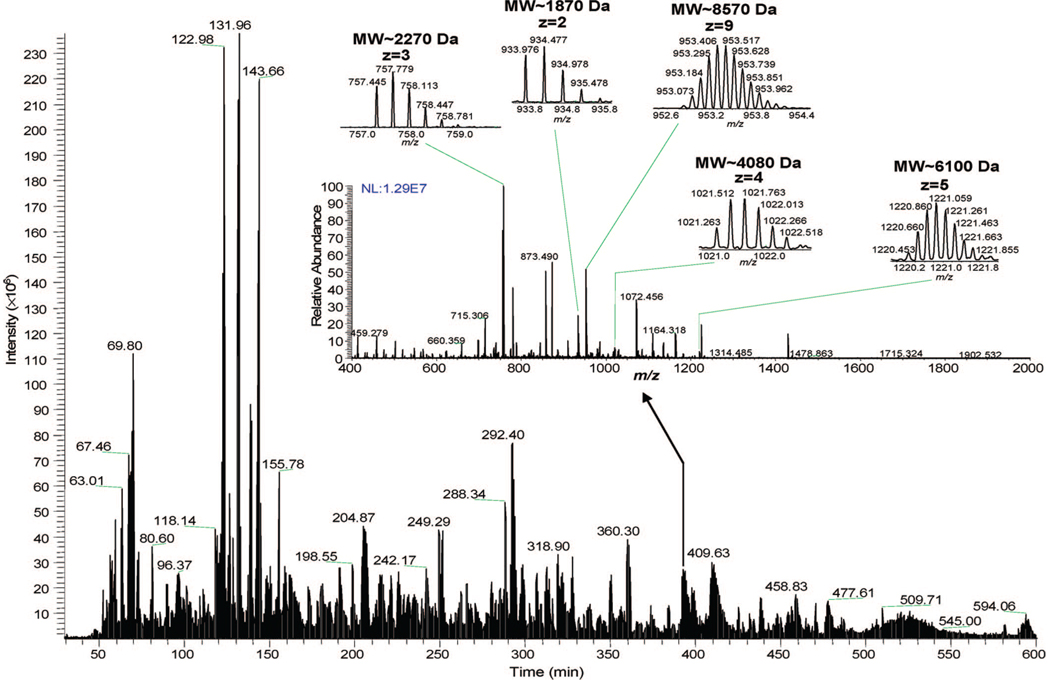
The FT-MS/MS data set used in this work was that used for a previous study on the intracellular proteolytic degradation of yeast proteins.2 Briefly, a lysate was quickly extracted from yeast Saccharomyces cerevisiae cells with the use of pressure cycling technology and in the presence of a protease inhibitor cocktail (Roche, Indianapolis, IN). The lysate obtained was directly separated on a home-built high-efficiency capillary LC system10 that was coupled online to a LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific., San Jose, CA). A total of 30 760 FT-MS/MS spectra and 6 156 precursor FT-MS spectra were acquired during the 600-min LC separation on a long reversed-phase column (120-cm length × 100-µm i.d. capillary packed with 3-µm C4-silica). Both FT-MS and MS/MS spectra were collected at a resolution of 30K; the FT-MS/MS employed an isolation window of 3 m/z units. Figure 1 shows the base peak chromatogram obtained from this experiment, showing examples of various sizes of peptides and polypeptides from a precursor MS spectrum.
Figure 1.
The LC–Orbitrap MS/MS base peak chromatogram of a yeast lysate used as the test sample in this work. The experiment was completed in 600 min; a mass spectrum from a precursor was inserted to show the various sizes of molecular species in the test sample. Experimental conditions and the collected data sets are described in the text.
De-Isotoping of FT-MS/MS Spectra and de Novo Sequencing
Deisotoping of high-resolution FT-MS and MS/MS raw file was accomplished with use of an in-house developed software ICR2LS (http://ncrr.pnl.gov/software/) with parameters previously described.2 The neutral monoisotopic masses generated from de-isotoping were used for de novo sequencing. The de novo function was recursive in nature with the objective to extend the constructed sequence with additional residue per each call. The logic of this function is described in Table 1 in the Supporting Information. The de novo sequence in this context was the sequence of triplets (fragment mass, charge state, residue) ordered on the fragment mass so that the difference between two neighboring fragment masses can fit within the specified precision to the mass of an amino acid (AA) residue. A mass error tolerance of 0.005 u was used for sequencing, which allowed for the resolution of all individual AA masses except for isobaric ones (e.g., I/L, N/GG, and Q/GA/AG). No gaps of > 1 amino acid were allowed. The output of de novo sequencing was a list that contained all sequences read from the FT-MS/MS spectra, which was used for the next steps in our de novo–UStag approach as described below.
Table 1.
Examples of de Novo Sequences and Their Database Correspondentsa
| scan | de novo sequence | length | yeast sequence | length | description |
|---|---|---|---|---|---|
| 20004 | FAVGPLEKGELLELSAGGGG | 20 | GGGASLELLEGKELPGVAF | 19 | Rand_G |
| FAVGPLEKGELLELSQGGG | 19 | SLELLEGKELPGVAF | 15 | Rand_G, Q/AG | |
| 22092 | DTVKLIEDFNNVGQQNE | 17 | ENQQGVNNFDEILKVTD | 17 | correct sequence |
| 6304 | KAVTATDGGGIIVTNG | 16 | GNTVIIGGGDTATVAK | 16 | correct sequence |
| KAVTATDGGGIIVTNY | 16 | NTVIIGGGDTATVAK | 15 | Rand_Y | |
| KAVTATDGNIIVTNG | 15 | GDTATVAK | 8 | N/GG | |
| 19275 | ELLELSAGGGTSVHSIK | 17 | KISHVSTGGGASLELLE | 17 | correct sequence |
| ELLEISQGGTSVHSLK | 16 | SHVSTGG | 7 | I/L | |
| ELLEISQGGTSVHSLK | 16 | QSIELLE | 7 | I/L | |
| ELIEISQGGTSVHSIK | 16 | KISHVSTGG | 9 | Q/AG | |
| 8606 | GGGTSVHSIKDTVGYK | 16 | KYGVTDKISHVSTGGG | 16 | correct sequence |
| GNTSVHSIKDTVGYK | 15 | KYGVTDKISHVST | 13 | N/GG | |
| NGTSVHSIKDTVGYK | 15 | KYGVTDKISHVST | 13 | N/GG | |
| 6125 | NVVVIGHVDSGKSTT | 15 | NVVVIGHVDSGKSTT | 15 | correct sequence |
| NVVVGLHVDSGKSTT | 15 | GLHVDS | 6 | IG/GL | |
| NVVVGLHVDSGKSM | 14 | GLHVDS | 6 | IG/GL, Rand_M | |
| NVVLVGHVDSGKSTT | 15 | DVHGVL | 6 | VI/LV | |
| NVVLVGHVDSGKSM | 14 | DVHGVL | 14 | VI/LV, Rand_M | |
| NVVVAVHVDSGKSTT | 15 | VVAVH | 5 | IG/AV | |
| NVVVAVHVDSGKSM | 14 | VVAVH | 5 | IG/AV, Rand_M | |
| 13554 | QGKLEVPGYVDIVKT | 15 | QGKLEVPGYVDIVKT | 15 | correct sequence |
| QGKLEVPGYVVEVKT | 15 | QGKLEVPGYV | 5 | VE/DI | |
| QGKIEVPGYVVEVKT | 15 | GYVVE | 5 | VE/DI, I/L | |
| 19600 | KTGVIVGEDVHNLFT | 15 | KTGVIVGEDVHNLFT | 15 | correct sequence |
| KGTVIVGEDVHN | 12 | VIVGEDVHN | 9 | TG/GT | |
| 8865 | HVSTGGGASLELLEG | 15 | HVSTGGGASLELLEG | 15 | correct sequence |
| GVSTGGGASLELLE | 14 | VSTGGGASLELLE | 13 | Rand_G | |
| 14274 | VVVKEVQDGLIEKL | 14 | LKEILGDQVEKVVV | 14 | correct sequence |
| 6117 | NKQTSNIKNTVANL | 14 | NKQTSNIKNTVANL | 14 | correct sequence |
| 9486 | LLKEKKVYPDVLYT | 14 | LLKEKKVYPDVLYT | 14 | correct sequence |
| 3228 | DSLADAAAKSPTEK | 14 | KETPSKAAADALSD | 14 | correct sequence |
| 17298 | TGVIVGEDVHNLE | 13 | TGVIVGEDVHNL | 12 | Rand_E |
| 12912 | QWGAPIGEKDTVG | 13 | VTDKEGIPAGWQ | 12 | Rand_G |
Only long sequences containing 13-AA-residues obtained from de novo sequencing were listed. The database sequences are from ftp://genome ftp.stanford.edu/pub/yeast/data_download/sequence/GenBank/.
The random construction of sequences from de novo (i.e., the false discovery rate of de novo sequencing) was examined with two false (incorrect) databases. These false databases included a Shewanella oneidensis database that contained 4 897 protein entries11 and a scrambled yeast Saccharomyces cerevisiae database that randomly rearranged the AA residues for each protein entry from the original database (ftp://genome-ftp.stanford.edu/pub/yeast/data_download/sequence/GenBank/).
Uniqueness Inspection of de Novo Sequences
De novo sequences that contained ≥5-AA residues were matched to the yeast database for the inspection of their uniqueness (note: no unique sequences existed for <5-residue yeast sequences1). The matches were accepted for any ≥5-residue subsequence/sequence regardless if the entire de novo sequence matched the predicted genome sequence(s). When a subsequence/sequence was found only from a single database protein, the sequence/subsequence was then examined using the residue replacement filter (RRF) function1 to remove the ambiguities generated from the isobaric segments. The uniqueness was further examined for the subsequences obtained by removing one AA residue from either end of the matched unique sequences or subsequences, and those that passed all these examinations were assigned as the de novo-sequenced UStags. Removal of one residue along with an examination of uniqueness for the resultant subsequences allowed for the determination of reliable UStags even with the existence of de novo sequencing noise residues (details will be shown in the Results section below).
Determination of Protein Sequence Modifications
The UStags determined from the de novo sequences/subsequences were aligned with the specific database sequences according to the direction and location of the de novo sequences or subsequences. If the masses of the precursor, fragmental ions that were composed of a UStag, and UStag prefix and suffix sequences were completely in agreement with those of the database sequences (mass tolerances, 0.005 u for sequencing of amino acids and 10 ppm for measurement of precursor and fragment locations), the peptide/polypeptide sequences were considered as not modified. If any of these masses deviated from those of the database sequences, the peptide/polypeptide sequence was examined further for possible modification(s). The mass shifts of the UStag prefix and suffix sequences were searched against a modification list (http://www.unimod.org/) with consideration of prefix and suffix amino acids. When the mass shifts could only be explainable by the combination of multiple modifications, manual inspection of the spectra were needed for confirmation.
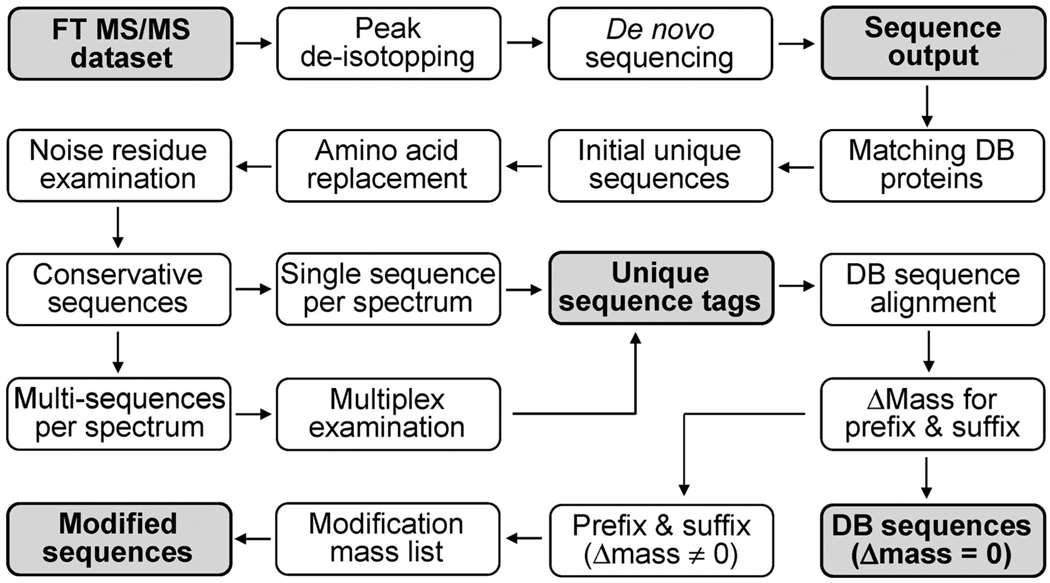
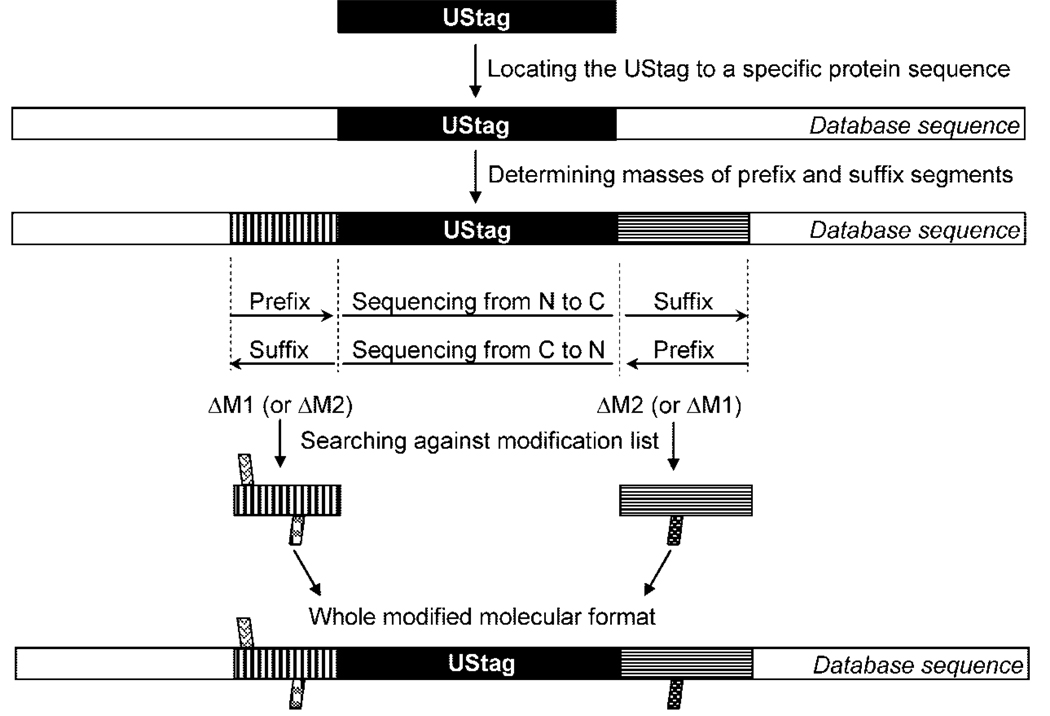
Figure 2 shows a flowchart of the overall procedures used for the de novo–UStag approach developed in this work. Some steps will be detailed below in combination with the discussion of the results.
Figure 2.
A flowchart showing the procedures used for the de novo–UStag approach developed in this work. Each step in the flowchart is described in the Methods and Results sections of the text.
RESULTS
Isotopic Envelopes Measured from Orbitrap Mass Spectrometry for Peptides and Polypeptides
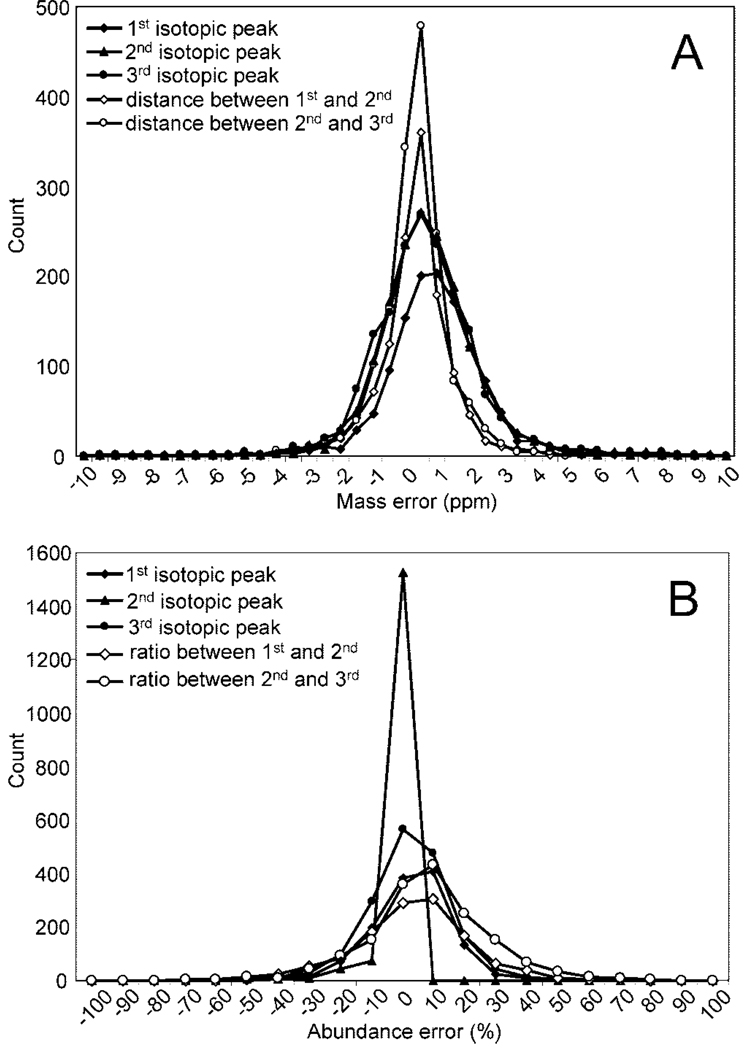
In our de novo sequencing, isotopic envelopes of the FT-MS/MS and precursor MS spectra were determined according to the peak m/z location and abundance with a reported algorithm12 that had been adopted into the ICR2LS software (http://ncrr.pnl.gov/software/). The algorithm may be reliable to describe the theoretical distribution;12 however, experimental measurements may vary from the theoretical distribution due to stochastic limitations on the mass accuracy and isotopic peak intensities. We examined the isotopic distributions measured from an Orbitrap mass spectrometer with ∼1 100 peptides identified confidently with the UStags approach from the proteomic sample.2 These peptides have molecular masses ranging from ∼600–8 000 u (the mass distribution for these peptides is given in Supporting Information Figure 1), and the examination results are shown in Figure 3. Deviations between those calculated according to peptide composition and measured experimentally were typically within 5 ppm (Figure 3A) for the isotopic peaks examined (i.e., the three most abundant isotopic peaks of each peptide) and were slightly smaller for the difference of adjacent isotopic peaks. These deviations were ascribed to spectrometric accuracy.1 Deviations of both isotopic peak abundances and their ratios were in a range of 30–40% (Figure 3B) but significantly smaller (e.g., ∼10%) for the peptides’ second isotopic peak. Higher intensities are typically observed for the second isotopic peak in comparison to the other peaks, which would explain the smaller abundances deviations observed. For peptide fragments, we anticipate situations similar to those observed above for peptide molecules.
Figure 3.
Examination of peptide isotopic distributions measured from the Orbitrap MS for the proteomic sample tested in this work. The first three isotopic peaks were used for this examination; the variances for (A) isotopic peak location and (B) abundance were calculated by [(experimental value – theoretical value)/theoretical value] where theoretical values were calculated according to the peptide composition.
De Novo Sequences from the Proteomic FT-MS/MS Data Set
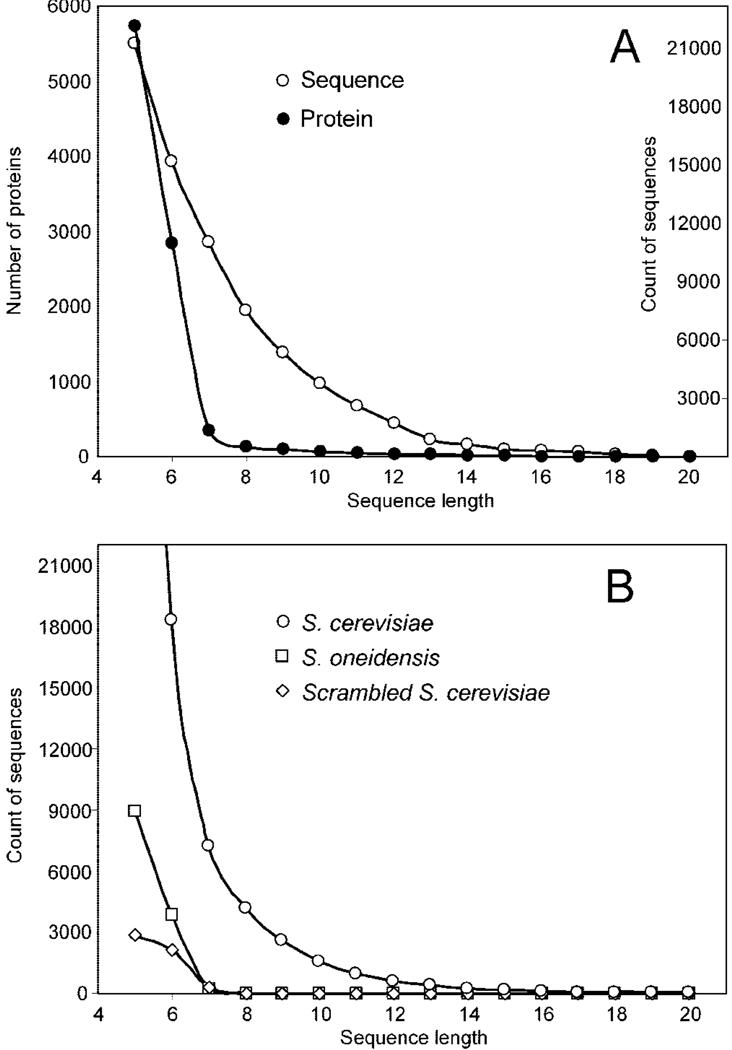
From de novo sequencing of the 30 760 FT-MS/MS spectra in the data set, we obtained 21 252 counts of ≥5-AA-residue sequences (note: one sequence may yield multiple counts as it can be obtained from different scans of the same or different peptides). These sequence counts generated 69 366 counts with ≥5-residue subsequences matching one or multiple database proteins (note: one de novo sequence can generate multiple subsequences that match different database proteins, see below). These matched sequences or subsequences corresponded to 5 733 of the total 5 889 yeast database proteins. Similarly, 18 412 counts of de novo sequences were located in 2 848 yeast database proteins with ≥6 residues. This level of proteome coverage was ∼10-fold higher than those obtained from the previous database search–UStag approach.2 Random matching was examined by searching these de novo sequences against the false databases (see Methods section). The searching results (Figure 4B) showed that ∼20% of the ≥6-residue de novo sequences could be matched to proteins with six residues in each false database, indicating that the de novo sequencing-constructed 5–6-residue sequences had poor specificity to proteome proteins despite the fact that the sequencing was for the intense fragments (i.e., those having isotopic peaks) measured with accurate FT-MS/MS (e.g., <0.005 u mass errors for sequencing). The numbers of sequence counts and the related yeast proteins were greatly reduced with an increase of sequence length (Figure 4A). The magnitude of counts became small as the sequence length increased to ≥7 residues, while the random matches reduced to ∼4% and ∼0.3% with an increase of sequence length of ≥7 and ≥8 residues, respectively. If sequence length is used as a filter for de novo outputs, matching 7–8-database residues should be a reasonable cutoff threshold for achievement of an acceptable false discovery rate.
Figure 4.
Counts of various lengths of de novo sequences and their matches to different types of database sequences. (A) Counts of total de novo sequences obtained form the tested data set and the number of yeast proteins containing the various lengths of de novo sequences or subsequences. (B) Counts of de novo sequences that contain various length sequences or subsequences of correct yeast S. cerevisiae and false S. oneidensis and scrambled yeast S. cerevisiae database sequences.
Noise Residues and Redundant Sequences in de Novo Sequencing
It was common to observe multiple de novo sequences from a single FT-MS/MS spectrum, and these multiple sequences were constructed mainly from the same ions. Table 1 lists some ≥13-residue sequences outputted from the de novo sequencing and their corresponding database sequence matches. Four different cases were observed from these examples. First, de novo sequences were completely matched to the database sequences (see those labeled as “correct sequence”), and these types of sequences seem able to be directly applied to the protein identification. Second, new de novo sequences were generated due to isobaric AA segments (e.g., I/L, N/GG, Q/AG/GA), which were expected for mass sequencing and which could be removed with the RRF.1 Third, new sequences were generated due to the reverse order placement of two to three adjacent residues (e.g., VI/LV, TG/GT) and small isobaric sequence segments (e.g., IG/AV, VE/DI). These new sequences could match the database sequences with five to six residues. Finally, one or two nondatabase-predicted residues were sequenced prior to and/or after the predicted sequences (see those labeled with Rand_X).
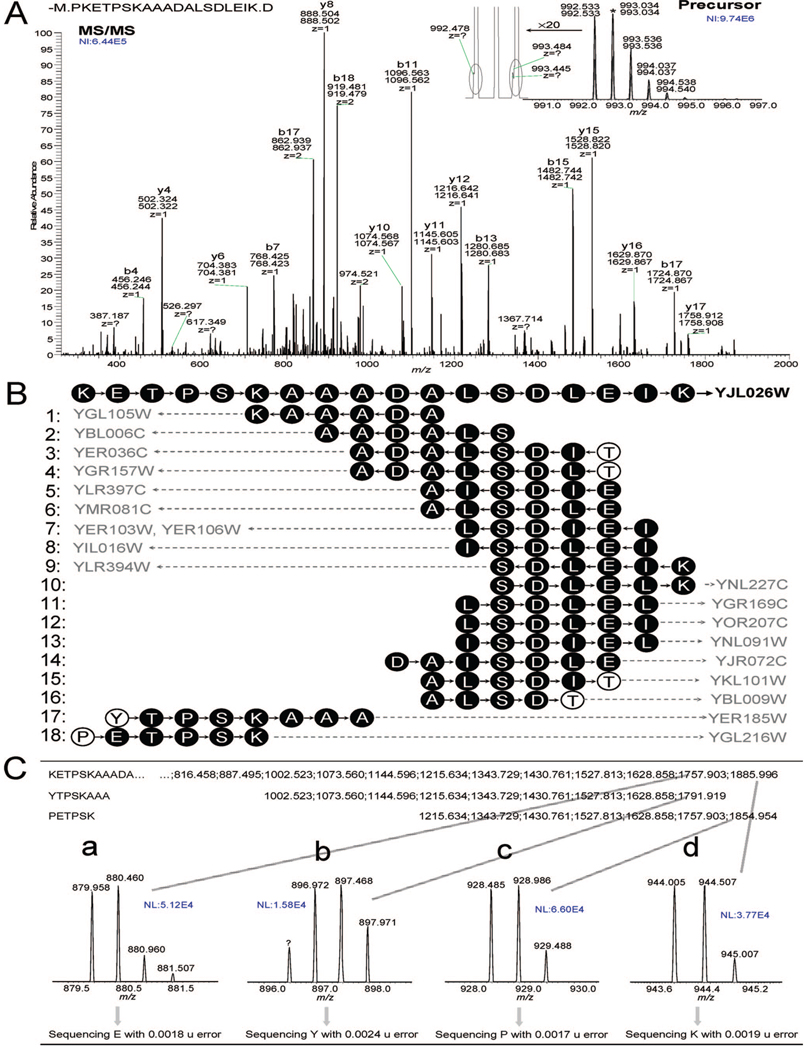
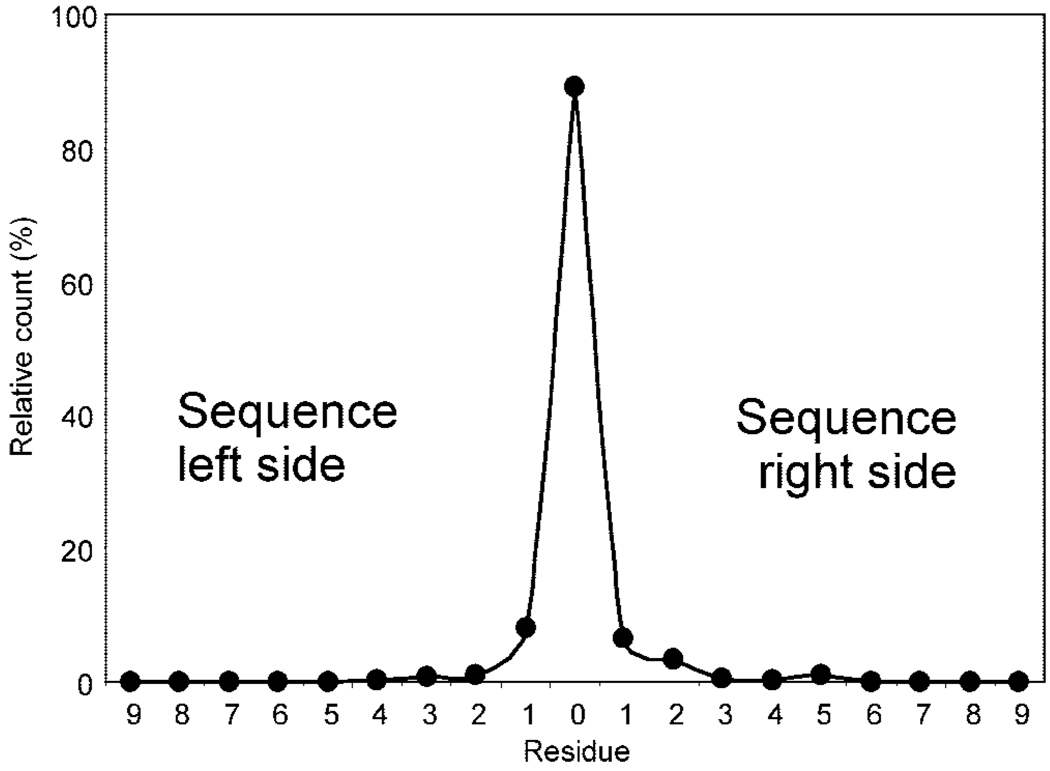
We next inspected the FT-MS/MS spectra and their precursors to examine sources possibly responsible for the generation of the multiple sequences. Figure 5 shows an example where de novo sequencing generated 19 sequences related to various database proteins from a single spectrum. Peptide PKETPSKAAADALS-DLEIK of yeast ribonucleoside-diphosphate reductase was unambiguously assigned to its FT-MS/MS spectrum (Figure 5A) using three types of information including the de novo sequence KETPSKAAADALSDLEIK, the correct location of b and y ions that covered all intense peaks (labeled in Figure 5A), and the agreement between the molecular mass and precursor (inserted in Figure 5A). However, the de novo sequencing also outputted 18 other sequences that matched to different yeast proteins (Figure 5B). These 18 sequences only explained a small portion of the whole spectrum (e.g., front, middle, or last portion of the spectrum, see Figure 5B), and the resultant proteins assigned were believed to be incorrect. Of the 18 sequences, 12 sequences (i.e., 1, 2, 5–14) were fully located in the assigned peptide in the same or different directions, and these de novo redundant sequences could be automatically removed with the implementation of the UStag uniqueness constraint. The other six sequences (i.e., 3, 4, 15–18) were generated due to the detection of one new residue (i.e., Thr for 3, 4, 15, 16; Tyr for 17; Pro for 18). Fragment ions for the generation of new residues were examined (Figure 5C), and no significant differences between these ions and those generated by the identified peptide were found in the isotopic pattern, sequencing accuracy, and peak intensity. This suggests that none of these parameters could be directly used as a filter to remove the new residues. A possible mix-up of b and y fragments cannot explain the ions that were generated by new residues even with consideration of all modifications listed in UNIMOD (e.g., b16-piperidination with a mass error of −0.007 u was the most possible interfering species for Figure 5C,b and y16-biotinylation with a mass error of 0.013 u for Figure 5C,c; these mass errors were significantly larger than those produced by the mass spectrometer, e.g., <0.005 u for sequencing). Similarly, no other types of fragmentation ions (i.e., a, c, x, or z ions) could explain these ions. Examination of the precursor ion revealed the existence of low intensity species (see species with m/z 992.478, 993.445, 993.484 inserted in Figure 5A) in the m/z zone of 993.032 ± 1.5 u where peaks were isolated for fragmentation (see Methods section). These minor contaminants may probably be responsible for the ions of Figure 5C,b,c and their resultant new residues, which we refer to as the de novo noise. A similar situation was found for the reversed order of the adjacent residues and isobaric small sequence segments (not shown). Figure 6 illustrates the distribution of the nondatabase-predicted residues at two sides of the ≥5 database-matching residues in the ≥7-residue sequences obtained from de novo sequencing of the whole MS/MS data set tested in this study. Approximately 90% of the ≥7-residue de novo sequences were fully located in the database prediction, while the other ∼6% with one nonpredicted residue and ∼4% with ≥2 nonpredicted residues were not located due to the reversed order residues of the adjacent residues (described above).
Figure 5.
An example showing noise residues and redundant sequences observed for de novo sequencing of the proteomic LC-FT-MS/MS data set. (A) The FT-MS/MS and its precursor MS spectra were assigned to the peptide PKETPSKAAADALSDLEIK of yeast ribonucleotidediphosphate reductase; the assignment was achieved with the database search–UStag approach, and the abundant peaks in the MS/MS spectrum were labeled fragments of the assigned peptide. (B) The redundant sequences generated from de novo sequencing and corresponding database protein matching. (C) Spectral evidence showing the ions adopted for correct sequencing and noise residues.
Figure 6.
Distribution of noise residues from de novo sequencing. The de novo sequences having ≥7 residues were used for this examination; the number of nondatabase-predicted residues prior to (left side) and after (right side) the database sequence matched were counted by calculation of their frequencies (%).
UStag Assignment from de Novo Sequences
De novo sequences containing ≥7-database-matching residues found in our data sets were considered reliable sequences representing proteome proteins (Figure 4B). Protein assignment from these sequences, however, was not necessarily unambiguous due to the existence of the same (or homologous) sequence portions from different proteins of a given proteome.1 This situation was similar to that for some small peptides, i.e., some peptides could be correctly identified but could not be used for protein identification because they were not unique to a specific protein. UStags was designed for sequence-based protein identification1 and therefore it can solve these issues by addressing whether or not the sequence identified is unique. Use of UStags for de novo sequencing also enables the automatic removal of redundant sequences such as the new sequences generated from the reverse order of sequences and isobaric segments (e.g., 12 redundant sequences in Figure 5B). The existence of noise residues (Figure 5 and Figure 6) observed for de novo sequencing of accurate FT-MS/ MS data arouses a new concern for de novo-sequenced UStags, that is, a nonunique sequence can occasionally become unique by the pickup of a noise residue (e.g., sequence 17 in Figure 5B). In order to solve this uncertainty factor, we added a new requirement for de novo-sequenced UStags: the UStags sequenced from de novo are required to remain unique after removal of one residue from either side of the sequences and the multiple UStags from de novo are allowed for a single spectrum only when they are constructed from different series of fragmental ions. With these requirements, we filtered the all de novo sequences and obtained 390 UStags.
Random false discovery rates for the de novo-sequenced UStags were examined. Using the requirements described above, we obtained 23 and 28 unique sequences (listed in Table 2) from two false databases, respectively. Thirteen of the sequences from each false database were coincidently actual sequences also found in the correct database. With the exclusion of these same sequences, the de novo-sequenced UStags were evaluated to have ∼2.5% [i.e., (23–13)/390] and 3.8% [i.e., (28–13)/390] false matching rates examined with the two different false databases, respectively. Among the false matching sequences, 7 and 8 sequences from the two false databases, respectively, were located in the correct database except for the pickup of one noise residue; and the other 3 and 7 sequences from the two false databases, respectively, included the reverse of isobaric segments (e.g., EK/ KE, QD/DAG). These suggest that the false hits examined for the de novo–UStag approach with use of the false databases were not random ones but instead resulted from the existence of the same short sequences (e.g., with ∼5 residues) in both the correct and false databases.
Table 2.
De Novo Sequences That Uniquely Match False Databases and Their Homologous Counterparts in the Correct Databasea
|
S. oneidensis unique sequences |
S. cerevisiae sequences |
|||||
|---|---|---|---|---|---|---|
| scan | sequence | prot and seq location | length | sequence | prot and seq location | length |
| 5372 | TTFAEDI | SO_1871.110.202.7.1 | 7 | LEAEDEAFTT | YER003C.412.18.10.0 | 10 |
| 5942 | ASSGSSG | SO_3669.237.461.7.0 | 7 | ASSGSSG | YML100W.230.869.7.0 | 7 |
| 6304 | LGGGDTA | SO_1324.291.178.7.1 | 7 | GNTVIIGGGDTATVAK | YCR012W.364.53.16.1 | 16 |
| 6765 | AKVTLEP | SO_0256.24.306.7.1 | 7 | KVTLEP | YFR029W.451.228.6.1 | 6 |
| 6866 | PGAGKGTQA | SO_2018.9.206.9.1 | 9 | IGPPGAGKGTQA | YDR226W.12.211.12.1 | 12 |
| 7063 | TVPAYFND | SO_1126.140.500.8.1 | 8 | VVTVPAYFND | YAL005C.141.502.10.1 | 10 |
| 8222 | KTLVGVG | SO_3392.296.60.7.0 | 7 | GVGVIT | YDL144C.11.346.6.1 | 6 |
| 9092 | GKSAESL | SO_3527.119.201.7.0 | 7 | ADIISEASKGK | YGR240C.839.149.11.1 | 11 |
| 10072 | GFVTHVL | SO_4528.131.139.7.1 | 7 | IVHTVFTS | YDR012W.34.329.8.0 | 8 |
| 10214 | GLLKYVD | SO_1325.781.702.7.1 | 7 | GIIKYVD | YCR073C.1170.162.7.1 | 7 |
| 12505 | ITKNVLL | SO_4249.7.403.7.1 | 7 | ITKNVLL | YJR001W.454.149.7.1 | 7 |
| 12576 | KEKLQER | SO_0704.362.184.7.1 | 7 | NSYEKEKLQERLAKL | YLR259C.380.193.15.1 | 15 |
| 13168 | AVGIDLGTT | SO_2268.21.600.9.1 | 9 | KAVGIDLGTT | YAL005C.3.640.10.1 | 10 |
| 15505 | GLVEKVVS | SO_2268.407.214.8.0 | 8 | LEVKVV | YAL035W.635.368.6.0 | 7 |
| 15510 | SVKEVVL | SO_1142.888.187.7.1 | 7 | VVEKVS | YDL133W.426.12.6.0 | 6 |
| 17958 | ITMGEEI | SO_1251.24.60.7.1 | 7 | VTLTMGE | YLR034C.361.113.7.1 | 7 |
| 18174 | VGSTGNA | SO_4420.121.50.7.0 | 7 | GSTGN | YML007W.402.249.5.0 | 5 |
| 20839 | LAVSVKA | SO_0354.53.264.7.0 | 7 | KVSVA | YBR086C.845.102.5.1 | 5 |
| 21372 | YGAALTD | SO_0578.352.449.7.0 | 7 | ETAPVIDTLAAGY | YGL103W.97.53.13.1 | 13 |
| 23212 | DIIIAVDI | SO_0428.224.513.8.0 | 8 | VAIIID | YHR197W.124.640.6.1 | 11 |
| 24421 | VADESIT | SO_2978.17.383.7.0 | 7 | VADESLT | YLL050C.8.136.7.0 | 7 |
| 24546 | LEAIDALE | SO_2255.940.225.8.1 | 8 | LLEAIDAIEQ | YPR080W.226.233.10.1 | 10 |
| 26826 | VTGIGGI | SO_4383.16.405.7.0 | 7 | IGGIGTV | YBR118W.254.205.7.1 | 7 |
|
S. oneidensis unique sequences |
S. cerevisiae sequences |
|||||
|---|---|---|---|---|---|---|
| scan | sequence | prot and seq location | length | sequence | prot and seq location | length |
| 4923 | QSEEIDT | s_YGR193C.225.186.7.0 | 7 | TDIEESQIQTNYD | YKR059W.6.390.13.1 | 13 |
| 6117 | KQTSNLK | s_YIL144W.612.80.7.0 | 7 | NKQTSNIKNTVANL | YPL106C.55.639.14.0 | 14 |
| 6661 | SEAKSGK | s_YMR229C.1306.424.7.1 | 7 | GSKAES | YGR082W.167.17.6.0 | 6 |
| 6842 | TGVLKPG | s_YKL215C.187.1100.7.0 | 7 | ETGVIKPG | YPR080W.266.193.8.0 | 8 |
| 8540 | IKQDLSTS | s_YOR023C.295.272.8.0 | 8 | ALKDAGLSTS | YJR045C.346.309.10.0 | 10 |
| 8557 | TGADKISD | s_YDR226W.90.133.8.1 | 8 | ALKDAGLST | YJR045C.346.309.9.0 | 9 |
| 8744 | KFKEEFE | s_YLR287C.238.118.7.0 | 7 | KFKEEFEKAQEINK | YDR002W.186.16.14.0 | 14 |
| 8863 | LKAGLVGL | s_YIL075C.87.859.8.0 | 8 | NLKAGIVGL | YBR025C.20.375.9.0 | 9 |
| 9012 | VKLIIATI | s_YDR545W.1437.360.8.1 | 8 | KIIIAT | YKR086W.633.439.6.1 | 6 |
| 9487 | NPVPPPL | s_YPL105C.687.163.7.0 | 7 | PVPPPL | YPL140C.86.421.6.0 | 6 |
| 10348 | GADLHKQ | s_YJR125C.270.139.7.1 | 7 | QKHIDAGA | YJR009C.107.226.8.0 | 8 |
| 10981 | VADNIEV | s_YPR032W.982.52.7.0 | 7 | ADNLE | YCR067C.834.232.5.0 | 5 |
| 11976 | LQGEVGT | s_YDR190C.69.395.7.0 | 7 | QGEVG | YFL049W.21.603.5.0 | 5 |
| 15505 | KVEVVLG | s_YDR093W.79.1534.7.1 | 7 | VEVVI | YPR032W.109.925.5.1 | 5 |
| 15541 | FSGKGID | s_YOR304W.841.280.7.1 | 7 | AAELGKGSFKY | YPR080W.46.413.11.0 | 11 |
| 16140 | IKKFEKE | s_YPL216W.895.208.7.1 | 7 | EKEFK | YNL304W.101.255.5.0 | 5 |
| 16356 | LKKQDFN | s_YKL129C.339.933.7.0 | 7 | FDQKK | YER125W.71.739.5.1 | 5 |
| 16696 | VKSSAQN | s_YDR356W.105.840.7.0 | 7 | VKSSAAGNT | YCR012W.358.59.9.0 | 9 |
| 17040 | SVKVDVL | s_YDR458C.586.78.7.1 | 7 | VDVKVV | YBR142W.52.722.6.0 | 6 |
| 17040 | VVVVEKVS | s_YKR084C.236.376.8.0 | 8 | QVEKVVVS | YPL240C.561.149.8.0 | 8 |
| 21948 | LENSLLVL | s_YHR099W.1843.1902.8.1 | 8 | AIENSLLVLD | YOR198C.291.180.10.1 | 10 |
| 22893 | KVAVGIST | s_YCR034W.49.299.8.0 | 8 | SLGVAV | YBR236C.228.209.6.1 | 6 |
| 23154 | IKEVLGHI | s_YOR361C.362.402.8.1 | 8 | IKEVLG | YGR234W.106.294.6.1 | 6 |
| 23310 | AAKVVKK | s_YGL105W.131.246.7.0 | 7 | KETTYDEIKKVVKAAAE | YJR009C.249.84.17.1 | 17 |
| 24434 | AAIASADLI | s_YMR080C.793.179.9.0 | 9 | AIASDAI | YER151C.352.561.7.0 | 7 |
| 25065 | DEVDNGI | s_YOR191W.1143.477.7.0 | 7 | PFDLLGNDVEDADVVVL | YLR150W.4.270.17.1 | 17 |
| 25065 | DNGILDF | s_YDR465C.400.13.7.0 | 7 | PFDLLGNDVEDADVVVL | YLR150W.4.270.17.1 | 17 |
| 25065 | VDADEVD | s_YDL126C.237.599.7.0 | 7 | PFDLLGNDVEDADVVVL | YLR150W.4.270.17.1 | 17 |
The sequences underlined are those matched to both a false (i.e., S. oneidensis or scrambled yeast S. cerevisiae) database and a correct (i.e., yeast S. cerevisiae) database.
Discovery of Protein PTMs from UStags
Figure 7 details the procedures used in the determination of protein PTMs from the de novo-sequenced UStags. Assignment of a UStag simultaneously locates the UStag position in a specific protein sequence. The UStag prefix mass was determined from the smallest fragment used to construct the UStag, while the suffix mass from the mass difference between the precursor MS and the largest UStag-constructing fragment. The difference in masses measured and predicted was then determined for the prefix (ΔM1) and suffix (ΔM2), respectively. A peptide or polypeptide was considered as not modified when ΔM1 and ΔM2 were within either 0.005 u or 10 ppm. Of the 390 UStags assigned, 293 could be located onto 336 different peptides and polypeptides within the mass tolerances (note: a UStag obtained from different scans might lead to the identification of different length peptides that originate from the same protein due to different prefixes or suffixes). The other 97 UStags had no database-predicted prefix or suffix sequence(s), making these UStags ideal to probe for PTMs.
Figure 7.
Determination of protein modifications from the de novo–UStag approach. The UStag was first located in a database protein specified by the UStag; the mass differences of the UStag’s prefix and suffix sequences were determined from the measurement and the database predictions for the specified protein; and the obtained mass differences were searched against the UNIMOD list to look for the protein modifications that may be responsible for the mass differences. For determination of unknown and multiple modifications, spectral manual inspection was needed at times for the examination of ions of modified fragments.
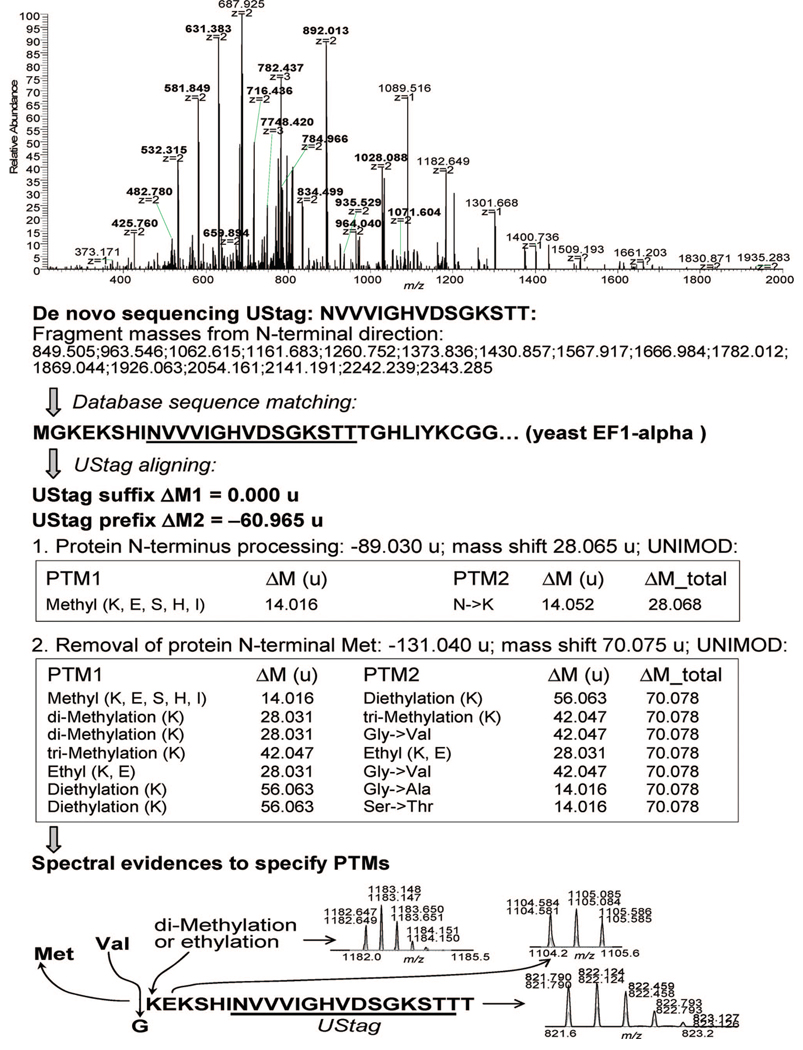
The prefix and suffix ΔM1 and ΔM2 of the 97 UStags were calculated and searched against the modification list (i.e., the UNIMOD list). Figure 8 illustrates an example of how the UStag’s ΔM1 and ΔM2 and the modification list were used to reveal complex PTMs on a single sequence. The UStag NVVVIGHVDS-GKSTT of yeast EF1-α protein was obtained from de novo sequencing. This UStag was located on Asn9–Thr23 amino acids of the protein and its suffix had a ΔM2 of 0.000 u, revealing no modification on the suffix. The prefix had a ΔM1 of −60.965 u from the database-predicted sequence. This ΔM1 value corresponded to either a 28.065 or 70.075 u discrepancy, depending on the protein N-terminus processing13 or not (i.e., removal of the protein N-terminal Met and acetylation on the new N-terminus would generate an additional shift of −89.030 u, or the single removal of Met would generate an additional shift of −131.065 u). The mass shift of 28.065 u can be explained by a combination of methylation and a substitution from Asn to Lys according to the UNIMOD searched with the mass tolerance of 0.005 u, but these modifications were excluded as there was no replaceable Asn in the suffix sequence GKEKSH. For the mass shift of 70.075 u, seven different combinations consisting of two modifications (listed in the figure) were matched with the precise mass shift. For this complex situation, inspection of the spectrum was used to find evidence for the confident assignment of the multiple modifications. The monoisotopic ion 1104.581 provided the evidence for the presence of fragment EKS…TTT, which narrowed the location of the modifications down to Lys3 and Gly2. Existence of the monoisotopic ion 1182.649 lead to the final determination of dimethylation (or ethylation) on the Lys3 and a substitution of Gly2 by Val. Taking into consideration of these modifications and amino acid substitution, the MS precursor ion was in agreement with the observed measurements (see the isotopic envelope, containing ion 821.790 in the figure).
Figure 8.
An example showing the determination of modification and amino acid substitution with the de novo–UStag approach. Removal of the N-terminal Met, Gly substitution by Val, and dimethylation (or ethylation) on Lys were determined for a single piece of the sequence (MGKEKSHINVVVIGHVDSGKSTTT) of a yeast protein. Explanation for the determination of these modifications is detailed in the text.
Using the procedure described above, we determined the PTMs for UStags that did not have a correct prefix and/or suffix and some results are listed in Table 3 (spectra are given in the Supporting Information). The de novo–UStag approach developed in this work enabled us to find the prefix-involved PTMs that were missed from the earlier reported UStag approach.2
Table 3.
Examples Showing the PTMs and Amino Acid Substitutions Identified from the de Novo-UStag Methoda
| scan | sequence description | sequence location | prefix ΔM | prefix modification | suffix ΔM | suffix modification |
|---|---|---|---|---|---|---|
| 4102 | [] [mseqlr] [QTFANAK] [] [kenrn] | YGL026C.7.7.0 | −89.033 | acetylation on Ser | 0.000 | none |
| 5152 | [] [matlhfvpqhe] [EEQVYSI] [S] [gkalk] | YMR235C.12.7.0 | −89.032 | acetylation on Ala | 0.005 | none |
| 5828 | [apsak] [a] [TAAKKAVVK] [gtngkkalkvr] [tsatf] | YOL127W.8.9.1 | −17.030 | y-H2O, Asn → Asp | 18.010 | H2O |
| 6482 | [m] [ppkkqve] [EKKVLLGR] [PGNNLKAGIV] [glanv] | YBR025C.9.8.0 | 28.029 | dimethyl on P or K | −0.002 | none |
| 6492 | [] [msdagrkgfg] [EKASEALK] [PDSQKSYAEQGKEYITDKADKVAGK] [vqped] | YFL014W.11.8.0 | −89.033 | acetylation on Ser | 0.003 | none |
| 8458 | [] [mseqlrqt] [FANAKKE] [NRNALVT] [fmtag] | YGL026C.9.7.0 | −89.032 | acetylation on Ser | 0.000 | none |
| 8846 | [m] [ppkkqveekkvllgrpgnn] [LKAGIVG] [LA] [nvgks] | YBR025C.21.7.0 | 28.028 | dimethyl on P or K | 0.000 | none |
| 8863 | [] [mppkkqve] [EKKVLLGR] [PGNNLKAGIVGLA] [nvgks] | YBR025C.9.8.0 | 28.027 | dimethyl on P or K | 0.001 | none |
| 9352 | [yssfl] [qketkddkpksil] [TDDMLFK] [a] [gvdvd] | YPL063W.57.7.0 | −17.032 | - H2O and Gln → Glu | 18.010 | H2O |
| 10614 | [] [mdkkkdll] [ENEQFLRI] [Q] [klnaa] | YNR019W 0.9.8.0 | 42.007 | acetylation on K | 0.004 | none |
| 13164 | [] [ms] [LSSKLSVQDLDL] [KDKRVFIR] [vdfnv] | YCR012W 0.3.12.1 | −0.007 | none | −89.024 | acetylation on Ser |
| 14527 | [] [mseqlrqt] [FANAKKE] [NRNALVTF] [mtagy] | YGL026C.9.7.0 | −89.032 | acetylation on Ser | 0.002 | none |
| 15493 | [kvagk] [vqpednkgvfqgvhdsaekgkdnaegqgesladqardymgaak] [SKLNDAVE][YVSGR] [vhgee] | YFL014W.87.8.1 | −0.003 | none | 16.006 | Phe → Tyr, or Ala → Ser, or oxid |
| 17414 | [vfasf] [irtandvltire] [VLGEQGKD] [VKIIVK] [ienqq] | YAL038W.227.8.0 | 0.978 | Asn → Asp | 0.003 | none |
| 18736 | [] [mseiqnk] [AETAAQD] [VQQKLEETKESLQNKGQEVK] [eqaea] | YOL109W.8.7.1 | −0.006 | none | −89.026 | aetylation on Ser |
| 19218 | [] [mseiqnka] [ETAAQDVQQKLEETKE] [SLQNK] [gqevk] | YOL109W.9.16.1 | −0.002 | none | −89.031 | aetylation on Ser |
| 21972 | [] [msd] [INEKLPELLQDAV] [LK] [asvpi] | YHR068W 0.4.13.1 | −0.001 | none | −89.026 | aetylation on Ser |
| 23730 | [] [msnpf] [DLLGNDVED] [ADVVVLPPKEIVKSNTSSK] [kadvp] | YLR150W 0.6.9.1 | −0.004 | none | −89.021 | aetylation on Ser |
| 24421 | [] [msrsgvav] [ADESLTAF] [NDLKLGKKYKFIL] [fglnd] | YLL050C.9.8.1 | −0.005 | none | −89.030 | aetylation on Ser |
| 25065 | [] [msn] [PFDLLGNDVEDADVVVL] [PPKEIVKSN] [tsskk] | YLR150W 0.4.17.1 | −0.004 | none | −89.020 | aetylation on Ser |
| 25345 | [] [ms] [NPFDLLG] [NDVEDADVVVLPPKEIVKS] [ntssk] | YLR150W 0.3.7.1 | −0.001 | none | −89.020 | aetylation on Ser |
For sequence description [xxx] [xxx] [XXX] [xxx] [xxx]: the square brackets from left to right encompass database sequences before the start of the peptide, UStag’s prefix or suffix sequence (depending on the de novo direction), the UStag, the UStag’s suffix or prefix sequence, and database sequence after the peptide; if a sequence has correct mass (filtered with 0.005 u for sequencing and 10 ppm for molecular mass), it is presented with capitalized letters. For sequence location, the letters and number(s) separated by periods are gene name, UStag start sites in protein sequence, UStag length, and de novo direction (0,N to C; 1,C to N).
Except for PTMs, we also found that some other nonaligned UStags (i.e., ΔM1 and ΔM2 values significantly larger than the measurement errors) resulted from artifacts during the sample preparation (e.g., oxidation on Met generated a mass shift of ∼15.995 u), mass spectrometry analysis (e.g., fragment water loss during MS/MS generated ΔM1 of −18.010 u and ΔM2 of +18.010 u), and de-isotoping errors (mass shifts of ∼1.000 u). Identification of these non-PTM artifacts are not detailed here.
Discovery of Protein Modifications/Nondatabase Sequences in the UStags
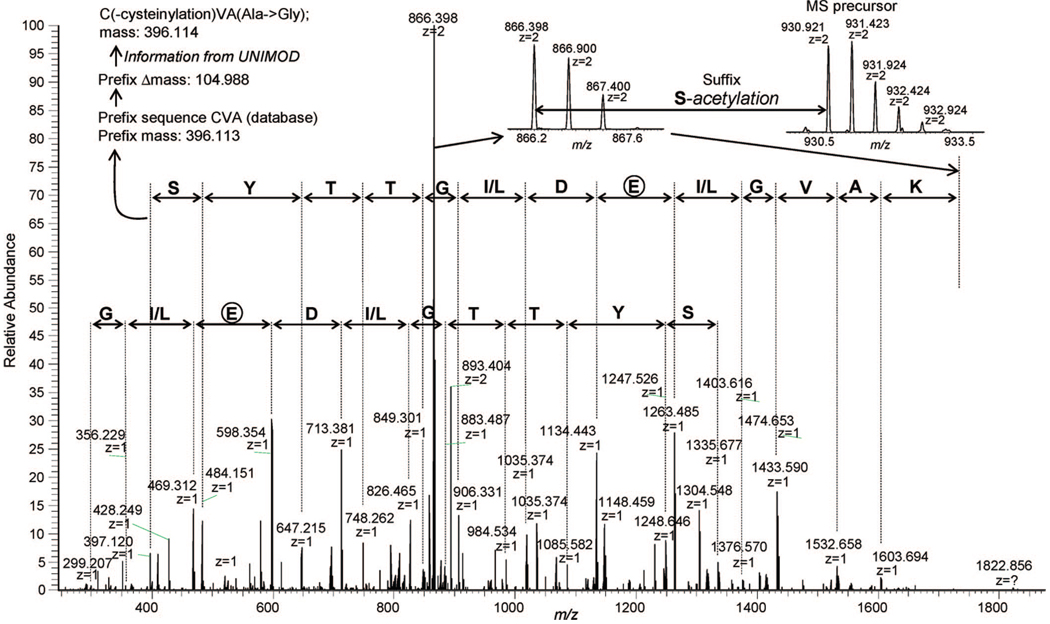
The UStags described above were typically obtained from ≥7-residue de novo sequences containing >5-database residues. We then examined the >7-residue de novo sequences that were not utilized for identifications. In a thorough inspection of all >7-residue de novo sequences obtained, the followings were found to be the reasons behind the rejection of these sequences for identification: not being unique sequences, the issue of which has been discussed in detail in the previous study;1 being redundant sequences resulted from isobaric segment replacement during de novo sequencing, as describe above for Table 1; being redundant sequences resulted from the reverse order of adjacent residues in the database sequence, as described for Table 1; being redundant sequences resulted from one or two de novo noise residues, as described for Table 1 and Figure 5 and Figure 6; being sequences filtered out during removal of one residue for the achievement of reliable UStags, as described above for UStag assignment. It was commonly observed that the sequences rejected for identification conformed to one or multiple reasons listed above. However, a set of sequences obtained from the de novo sequencing of one single spectrum was found with an exception to the above reasons, as only short pieces of the subsequences from the long de novo sequences matched the database sequences. Figure 9 shows the spectrum, and the set of de novo sequences [labeled with a format of YTTGI(L)DEI(L)G-VAK]. The entire sequence as presented did not match the database sequence. However, within it the sequence contained two subsequences KAVGI and DLGTTY that could recombine to form KAVGIDLGTTYS, a sequence located at Lys3–Ser14 of the yeast heat shock proteins SSA1, SSA2, and SSA4. The presence of the database-unpredicted Glu that divided the database sequence into two parts was confirmed with two directions of de novo sequences (see those labeled in Figure 9A), while additional PTMs including acetylation, cysteinylation, and substitution of Ala by Gly on the prefix and suffix of the sequence were determined according to the procedure described above. That is, the final peptide for interpretation of the spectrum was S(-acetyl)-KAVGIEDLGTTYSC(-cysteinyl)VA(A → G). This example demonstrates how the de novo–UStag approach enables the identification of complex PTMs through inspection of the inconsistencies between the de novo sequences and the database-predicted UStags.
Figure 9.
De novo sequencing of a nondatabase-predicted protein sequence. The sequences YTTGI(L)DEI(L)GVAK were constructed in the same direction and S prior to Y was determined with manual inspection of low abundance ions; the sequences GI(L)EDI/LGTTYS were the subsequences of the above sequences constructed from the reverse direction. The Glu circled is the nondatabase-predicted residue, and its location was determined from both directions of de novo sequencing. Detailed explanations are given in the text.
DISCUSSION
The most attractive aspect of de novo sequencing is its potential to discover protein sequences that differ from database predictions due to amino acid modifications and mutations (or database errors). This work demonstrated this potential by revealing both unexpected and complex multiple modifications (e.g., see those shown in Figure 8 and Figure 9 and Table 3). The UStag identifications played a critical role in the facile and quick determination of such protein modifications from de novo sequencing by providing a list of accurate database protein sequences which unambiguously explain the spectra and in the end allows for extensive PTM searching. Additionally, being deemed a UStags in itself produces a high level of certainty and reliability for determination of modifications, which is of great concern when searching for modifications that are not previously known to exist on the proteins. From the de novo–UStag approach we did also detect new unexpected and complex modifications that could not be found from the previous database search–UStag approach2 because these modifications occurred on the UStag prefix sequences (see results listed in Table 3 and examples shown in Figure 8 and Figure 9) that resulted in the absence of precise y or b ions needed to identify UStags.
Theoretically, the amino acid sequences outputted from the de novo sequencing can be directly used to look for the UStags, correlate or align the database sequences, and compare the sequence difference(s) between the database predictions and experimental observations. However, the existence of redundant sequences and noise residues observed for the de novo sequences impairs the reliability of sequences outputted from de novo sequencing. This critical issue needs to be inspected further before de novo sequences can be reliably applied to study the protein sequence changes due to modifications. We addressed this issue with careful examinations of the de novo sequences and reported details of the examinations (see Figure 4 and Figure 6 and Table 1 and Table 2) for a proteomic data set. The short sequences (e.g., 5–6 residues) obtained from de novo sequencing were found to have limited usefulness to unambiguously specify (or identify) proteome proteins (Figure 4), although these sequences were determined using high-accuracy FT-MS/MS from abundant ions (i.e., those having isotopic peaks). The de novo sequencing commonly generated multiple sequences for individual MS/MS spectra (see examples shown in Figure 5), and these sequences were less likely ascribed to different molecular species as the sequences were constructed mainly from the same ions. Use of UStags to address the sequence uniqueness can remove the redundant sequences that were constructed from the same ions (Figure 5) but cannot remove the sequences that resulted from the same ions plus the noise residues. Thus far, we have no better approach to process the redundant sequences constructed with database sequences plus the de novo noise residue(s) than to simply cut off one terminal residue from either side of the sequences according to our examination results (Figure 5 and Figure 6). This simple cutoff step provided the UStag identifications from de novo sequencing with ∼97% accuracy (see the results described for Table 2).
The sensitivity of the de novo–UStag approach was affected by both selection of the ions for construction of the de novo sequences and manipulation of the resultant de novo sequences. In this work, we only adopted abundant fragment ions that had isotopic envelopes for de novo sequencing in order to develop a reliable de novo sequencing method for discovery of PTMs and, simultaneously, provide insights for factors affecting the de novo sequencing reliability. In combination with the subsequent processing step of de novo sequences (i.e., cutoff of one residue), the de novo–UStag approach described here had lower sensitivity than the database search–UStag approach previously reported,1,2 which led to an obvious reduction in the numbers of UStag and thus peptide identifications. For example, ∼400 peptides including both modified and unmodified ones were obtained from the de novo–UStag approach, in comparison to ∼1100 peptides identified from the database search–UStag approach where low-abundance ions were used for the calculation of the UStags.2 Use of the low-abundance ions for de novo sequencing to extend the sequence length can be a promising way to improve the de novo sequencing sensitivity, and utilization of such low abundance ions needs to be further studied to identify more UStags without a significant sacrifice in the identification reliability.
De novo sequencing requires the sample components to be analyzed by MS/MS be as pure as possible due to its weak tolerance for the noise residues occurring from contaminants (i.e., the ions from contaminants can occasionally generate new sequences when combining with those of the species to-be-analyzed). Narrowing the m/z zone or even selecting individual isotopic peaks for MS/MS fragmentation can purify the resultant spectra but greatly degrade the sensitivity. Improving the separation resolution prior to MS/MS analysis (e.g., with use of ultrahigh-pressure LC, sample fractionation prior to LC separation, and implementation of multidimensional separations) can help to achieve the same goal without a significant tradeoff in sensitivity.
ACKNOWLEDGMENT
This research was partially supported by The William R. Wiley Environmental Molecular Sciences Laboratory (EMSL) Intramural Research and Capability Development Program (Grant 22142), the U.S. Department of Energy (DOE) Office of Biological and Environmental Research, and the NIH National Center for Research Resources (Grant RR18522). Work was performed in the EMSL, a DOE national scientific user facility located on the campus of Pacific Northwest National Laboratory (PNNL) in Richland, Washington. PNNL is a multiprogram national laboratory operated by Battelle Memorial Institute for the DOE under Contract DE-AC05-76RLO-1830.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Shen Y, Tolić N, Hixson KK, Purvine SO, Pasă-Tolić L, Qian WJ, Adkins JN, Moore RJ, Smith RD. Anal. Chem. 2008;80:1871–1882. doi: 10.1021/ac702328x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen Y, Hixson KK, Tolić N, Purvine SO, Moore RJ, Smith RD. Anal. Chem. 2008;80:5819–5828. doi: 10.1021/ac800077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pevtsov S, Fedulova I, Mirzaei H, Buck C, Zhang X. J. Proteome Res. 2006;5:3018–3028. doi: 10.1021/pr060222h. [DOI] [PubMed] [Google Scholar]
- 4.Frank AM, Savitski AM, Nielsen ML, Zubarev RA, Pevzner PA. J. Proteome Res. 2007;6:114–123. doi: 10.1021/pr060271u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savitski MM, Nielsen ML, Kjeldsen F, Zubarev RA. J. Proteome Res. 2005;4:2348–2354. doi: 10.1021/pr050288x. [DOI] [PubMed] [Google Scholar]
- 6.Frank A, Pevzner P. Anal. Chem. 2005;77:964–973. doi: 10.1021/ac048788h. [DOI] [PubMed] [Google Scholar]
- 7.Bern M, Cai Y, Goldberg D. Anal. Chem. 2007;79:1393–1400. doi: 10.1021/ac0617013. [DOI] [PubMed] [Google Scholar]
- 8.Colinge J. Anal. Chem. 2007;79:7286–7290. doi: 10.1021/ac070488n. [DOI] [PubMed] [Google Scholar]
- 9.DiMaggio PA, Jr, Floudas CA, Lu B, Yates JR., III J. Proteome. Res. 2008;7:1584–1593. doi: 10.1021/pr700577z. [DOI] [PubMed] [Google Scholar]
- 10.Shen Y, Zhang R, Moore RJ, Kim J, Metz TO, Hixson KK, Zhao Z, Livesay EC, Udseth HR, Smith RD. Anal. Chem. 2005;77:3090–3100. doi: 10.1021/ac0483062. [DOI] [PubMed] [Google Scholar]
- 11.The total 4 897 database proteins were obtained by combination of 4 630 chromosomal proteins in Genbank, 148 megaplasmid proteins in Genbank, 24 excluded proteins found in TIGR database, and 104 proteins from predicted genes frameshifs and elimination of duplicates. Details can be found in Omics. 2004;8:239–254. [Google Scholar]
- 12.Horn DM, Zubarev RA, McLafferty FW. J. Am. Soc. Mass Spectrom. 2000;11:320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Chang YH. Proc. Natl. Acad. Sci. U.S.A. 1995;92:12357–12361. doi: 10.1073/pnas.92.26.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]