Abstract
Transcription factors of the basic helix-loop-helix (bHLH) family are critical regulators of muscle cell differentiation. For example, Myod drives skeletal muscle differentiation, and Hand2 potentiates cardiac muscle differentiation. Understanding how these bHLH factors regulate distinct transcriptional targets in a temporally and spatially controlled manner is critical for understanding their activity in cellular differentiation. We previously showed that Pbx homeodomain proteins modulate the activity of Myod to promote the differentiation of fast-twitch skeletal muscle. Here, we test the hypothesis that Pbx proteins are also necessary for cardiac muscle differentiation through interacting with Hand2. We show that Pbx proteins are required for the activation of cardiac muscle differentiation in zebrafish embryos. Loss of Pbx activity leads to delay of myocardial differentiation and subsequent defective cardiac morphogenesis, similar to reduced Hand2 activity. Genetic interaction experiments support the hypothesis that Pbx proteins modulate the activity of Hand2 in myocardial differentiation. Furthermore, we show that Pbx proteins directly bind the promoter of the myocardial differentiation gene myl7 in vitro, supporting a direct role for Pbx proteins in promoting cardiac muscle differentiation. Our findings demonstrate new roles for Pbx proteins in vertebrate cardiac development and also provide new insight into connections between the transcriptional regulation of skeletal and cardiac muscle differentiation programs.
Keywords: pbx, hand2, zebrafish, heart development, muscle differentiation
Introduction
Development of the heart involves the complex orchestration of cellular specification, differentiation, and morphogenesis. Even slight disruptions of embryonic cardiac development can lead to congenital heart disease (CHD) in humans. Insight into the genetic basis of CHD has come from the identification of human mutations associated with CHD and also from model organism studies of cardiac development (Clark et al., 2006; Olson, 2006; Pierpont et al., 2007). Several genes encoding transcription factors comprise a conserved regulatory network with major roles in heart development, including the factors Nkx2–5, MEF2, GATA, Tbx and Hand1/2 (Clark et al., 2006; Olson, 2006). These factors control cardiac cell fates and morphogenesis by regulating expression of downstream genes such as those encoding contractile proteins. Although mutations in these transcription factor genes have been associated with CHD in humans, there is often phenotypic heterogeneity associated with single-gene defects, as seen for NKX2–5 and TBX5 (Srivastava and Olson, 2000; Clark et al., 2006). This heterogeneity points to potential roles for modifier loci in affecting the phenotypic severity of CHD. A challenge now is to understand how additional factors might modify the transcriptional programs controlled by the major heart factors.
Skeletal muscle has served as a model for understanding general principles guiding the transcriptional control of cellular differentiation. Vertebrate skeletal muscle development is coordinated by a family of four basic helix-loop-helix (bHLH) myogenic regulatory factors (MRFs): Myod, Myf5, MRF4 and Myogenin (Pownall et al., 2002; Tapscott, 2005). These factors, in particular Myod, are sufficient to convert non-muscle cells into skeletal muscle (Weintraub et al., 1991; Tapscott, 2005). Myod directly activates the expression of additional transcription factors, including Myogenin, and acts in a feed-forward mechanism in cooperation with those factors to directly activate a skeletal muscle differentiation gene expression program (Tapscott, 2005; Cao et al., 2006). Recent studies have shown that Myod and another bHLH protein, Hand, are both critical for striated bodywall muscle formation in C. elegans (Fukushige et al., 2006), whereas Hand proteins regulate development of cardiac muscle, but not skeletal muscle, in flies and chordates (Olson, 2006). The conserved roles for bHLH proteins in regulating muscle development suggest that there are fundamental core mechanisms for generating contractile cell types (Fukushige et al., 2006).
Because the MRFs directly regulate the expression of many genes necessary for skeletal muscle differentiation, understanding how these bHLH proteins regulate distinct transcriptional targets in a temporally and spatially controlled manner has been critical for understanding their activity. One mechanism recently proposed is the use of “molecular beacons”, or cofactors that bind the promoter of specific transcriptional targets and act as signals to attract a MRF to that promoter (Berkes et al., 2004). Pbx homeodomain proteins appear to function as such factors. Pbx proteins are TALE (Three Amino acid Loop Extension)-class homeodomain proteins that are best characterized as transcriptional cofactors for Hox proteins (Moens and Selleri, 2006). However, Pbx proteins interact with factors other than Hox proteins and have been shown to cooperatively bind DNA with MRFs, including Myod (Knoepfler et al., 1999; Berkes et al., 2004). Pbx proteins bind directly to promoters of certain Myod transcriptional targets, such as Myogenin, and they bind the Myogenin promoter prior to Myod in mammalian myoblast cell lines (Berkes et al., 2004). Thus, Pbx appears to mark a subset of Myod target genes for transcriptional activation by Myod.
We recently tested the requirements for Pbx in interacting with Myod in zebrafish skeletal muscle development (Maves et al., 2007). Zebrafish have five pbx genes, but only pbx2 and pbx4 are expressed during early developmental stages, and both can be readily knocked down to generate Pbx-null embryos (Waskiewicz et al., 2002; Maves et al., 2007). We found that Pbx proteins are indeed required for Myod to activate myogenin expression and are necessary for Myod to activate expression of fast-twitch skeletal muscle differentiation genes (Maves et al., 2007). Pbx and Myod also exhibit genetic interactions in the regulation of myogenin expression and fast-muscle differentiation (Maves et al., 2007).
Here, we wanted to further test the hypothesis that Pbx proteins regulate cellular differentiation by modulating bHLH factor activity, in particular by testing whether Pbx proteins interact with Hand2 in cardiac muscle differentiation. Although Hand proteins are critical regulators of heart development in zebrafish and mice, the mechanisms of Hand function in the heart are not well understood (Srivastava et al., 1997; Firulli et al., 1998; Riley et al., 1998; Yelon et al., 2000; Firulli, 2003). Pbx gene deficiencies in mice were recently shown to cause cardiac defects that resemble human CHDs (Chang et al., 2008; Stankunas et al., 2008), but early roles of Pbx genes in myocardial differentiation have not yet been addressed. Here we show that Pbx proteins are indeed required for cardiac muscle differentiation in zebrafish. Our results suggest that Pbx proteins regulate myocardial differentiation and morphogenesis by interacting with Hand2. These connections between Pbx and bHLH factors support the idea of a core developmental program regulating the transcriptional control of contractile muscle cell differentiation.
Materials and methods
Zebrafish Stocks
Zebrafish (Danio rerio) were raised and staged as previously described (Westerfield, 1995). Time (hpf) refers to hours post-fertilization at 28.5°C. In some cases, embryos were raised for periods at room temperature, about 25°C. The wild-type stock used was AB. hand2s6 embryos were generously provided by Zayra Garavito-Aguilar and Debbie Yelon, and this allele has been previously described (Yelon et al., 2000)
Morpholino and mRNA injections
pbx2; pbx4 morpholino cocktails were previously described (Maves et al., 2007). hand2 translation-blocking morpholino, generously provided by Debbie Yelon, was injected at 9 ng per embryo at the 1-cell stage (sequence: 5′-CCTCCAACTAAACTCATGGCGACAG-3′). We somite-stage-matched sibling control and MO-treated embryos when collecting embryos for staining or for RNA harvesting. For the experiments of Fig. 5, control MO (pbx2-MO3 mismatch control; Maves et al., 2007), was used to balance the total amounts of MO injected.
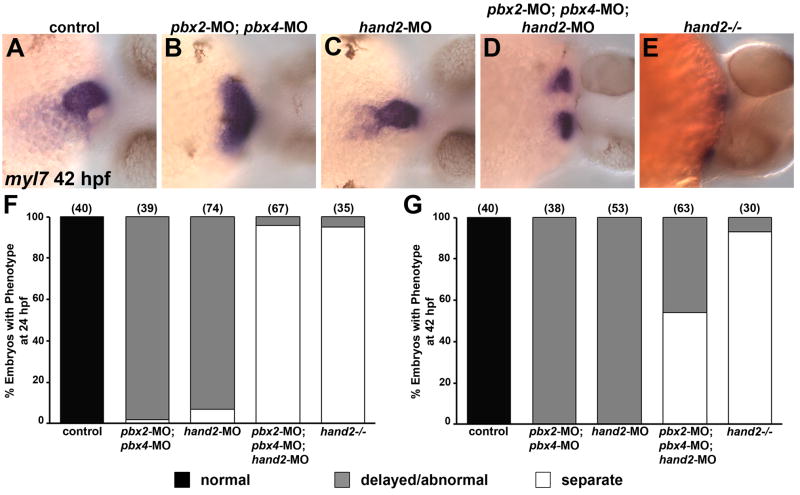
Fig. 5.
Pbx and Hand2 genetically interact in cardiac morphogenesis. (A–E) RNA in situ expression of myl7 at 42 hpf in (A) control, (B) pbx2-MO; pbx4-MO, (C) hand2-MO, (D) pbx2-MO; pbx4-MO; hand2-MO, or (E) hand2−/− embryos. Embryos are shown in frontal views, dorsal towards the right. (F–G) Quantification of phenotypes observed using myl7 expression at (F) 24 hpf or (G) 42 hpf. Normal phenotypic classes (black) correspond to tube (24 hpf) or looped (42 hpf) patterns depicted in Fig. 4D and Fig. 5A, respectively. Delayed or abnormal phenotypic classes (gray) correspond to images shown for pbx2-MO; pbx4-MO and hand2-MO embryos in Fig. 4D (24 hpf) or Fig. 5B–C (42 hpf). The separate, or cardia bifida, phenotypic classes (white) correspond to two myl7 domains and resemble those described for pbx2-MO; pbx4-MO and hand2-MO embryos in Fig. 4A or for pbx2-MO; pbx4-MO; hand2-MO and hand2−/− embryos in Fig. 5D–E. Numbers in parentheses denote numbers of embryos. hand2−/−embryos were derived from clutches that contained about 25% mutant embryos; their hand2+/+ and +/− siblings showed normal myl7 expression.
pCS2-hand2 was made by using RT-PCR to amplify full-length zebrafish hand2, which was cloned into the XhoI site of pCS2. mRNA was synthesized using the mMessage mMachine kit (Ambion). About 200–300 pg of hand2 mRNA was injected per embryo at the 1-cell stage.
RNA in situ hybridization
RNA in situ hybridizations were performed as previously described (Maves et al., 2007). The following cDNA probes were used: tnnt2 (Sehnert et al., 2002); ttna (EST wu:fb96b11); cmlc1 (Reiter et al., 1999); vmhc (Yelon et al., 1999); aldh1a2 (Begemann et al., 2001); myl7 (Yelon et al., 1999); lft1 (EST cb73, Thisse et al., 2001); hand2 (Yelon et al., 2000); nkx2.5 (Lee et al., 1996); scl (Schoenebeck et al., 2007); gata4 (Reiter et al., 1999); myod (Weinberg et al., 1996); krox-20 (egr2b-Zebrafish Information Network; Oxtoby and Jowett, 1993); amhc (Berdougo et al., 2003); nppa (Berdougo et al., 2003).
Embryos were photographed using a Zeiss Axioplan2 microscope and Zeiss AxioCam MRc digital camera or a Zeiss Stemi SV6 and Nikon CoolPix 4500 digital camera. Images were assembled using Adobe Photoshop.
cDNA microarray analysis
pbx2-MO; pbx4-MO embryos as well as their control siblings (pbx2-MO3 mismatch control) were collected at the 10s or 18s stage from three independent sets of injections. A subset of embryos from each set of injections were used for in situ hybridization to confirm somite staging (using myod) and loss of krox-20 expression in pbx2-MO; pbx4-MO embryos (see Figures 2 and 3 for examples). Total RNA harvesting, cRNA labeling and hybridization to Affymetrix Zebrafish Genome arrays, and statistical analyses of the gene expression profiles of control versus pbx2-MO; pbx4-MO were performed as previously described (Maves et al., 2007). We used the cut-offs of >1.5-fold change and a False Discovery Rate of <0.05 to identify genes whose expression is significantly dependent on Pbx function. Array data are available at NCBI GEO (www.ncbi.nlm.nih.gov/geo/), accession GSE8428.
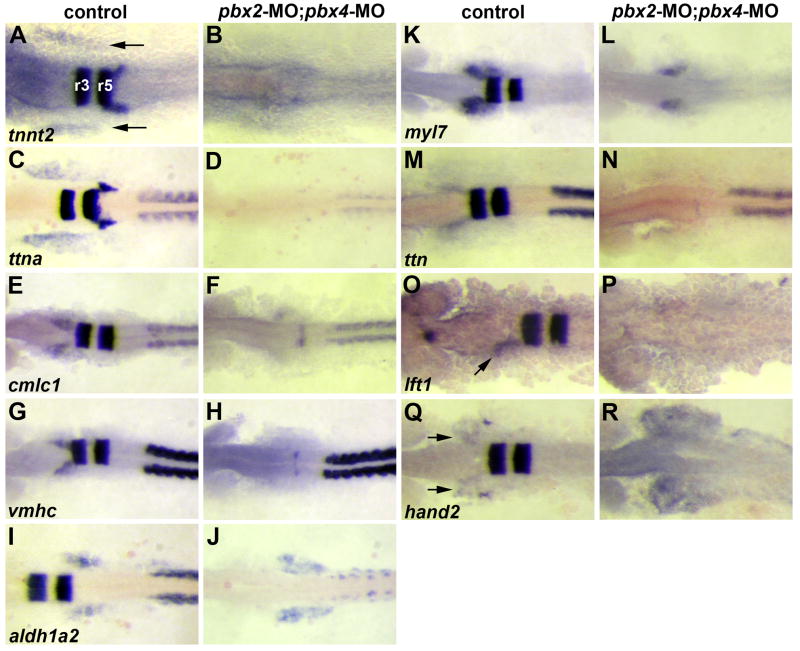
Fig. 2.
Validation of microarray-identified Pbx-dependent heart genes. (A–R) RNA in situ expression of genes from Table 1 in (A,C,E,G,I,K,M,O,Q) wild-type control or (B,D,F,H,J,L,N,P,R) pbx2-MO; pbx4-MO embryos. (A–D) are at 10 somite stage (10s). (E–N, Q–R) are at 18s. (O–P) are at 20s to achieve more robust expression of lft1 in controls. krox-20, labeled in hindbrain rhombomeres r3 and r5 in (A), is included in all in situs to control for loss of Pbx. The wild-type heart primordium expression domains of tnnt2, ttna, cmlc1, vmhc, myl7, lft1 and hand2 are just anterior-lateral to r3 krox-20 expression (arrows in A,O,Q). n≥10 for each marker in control or pbx2-MO; pbx4-MO. Embryos are shown in dorsal view, anterior towards the left.
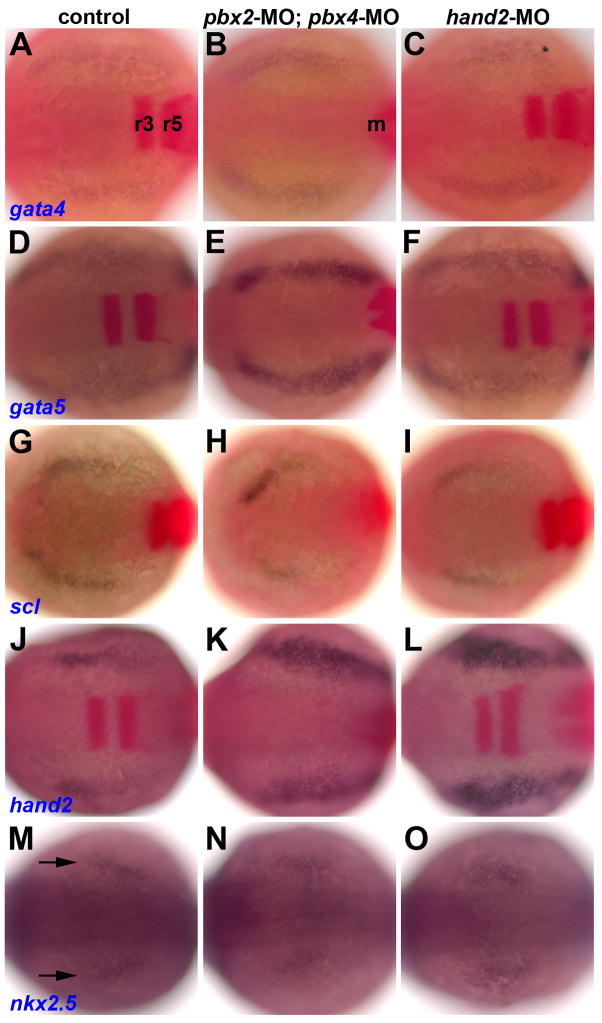
Fig. 3.
Early heart specification is normal in pbx2-MO; pbx4-MO and hand2-MO embryos. (A–O) RNA in situ expression at the 7–9 somite stage of (A–C) gata4, (D–F) gata5, (G–I) scl, (J–L) hand2, and (M–O) nkx2.5, all in blue, in (A,D,G,J,M) control, (B,E,H,K,N) pbx2-MO; pbx4-MO, or (C,F,I,L,O) hand2-MO embryos. Also included in (A–L), in red, are krox-20, labeled in (A) in r3 and r5 in the hindbrain, and myod, labeled m in (B) and used to confirm somite staging, but is not visible here in all embryos. Arrows in (M) mark nkx2.5 expression. n≥13 for each marker in control, pbx2-MO; pbx4-MO, or hand2-MO. Embryos are shown in dorsal view, anterior towards the left.
EMSA
Electrophoretic mobility shift assays were performed as previously described (Berkes et al., 2004). pbx4 and meis3 pCS2 expression constructs were previously described (Pöpperl et al., 2000; Vlachakis et al., 2000).
Results
Pbx2 and Pbx4 are required for normal heart function
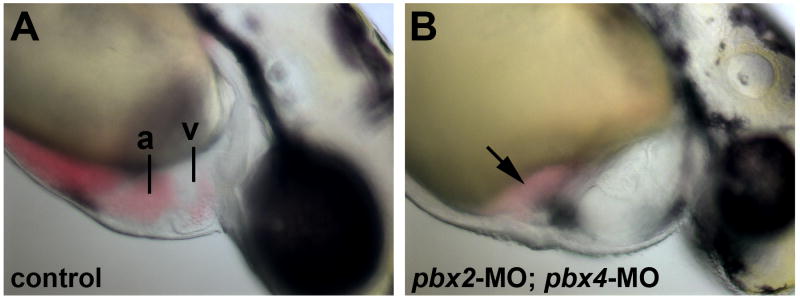
To address the role of Pbx proteins in zebrafish heart development, we used anti-sense morpholino oligos (MOs), injected at the 1-cell stage, to knock down the maternal and zygotic expression of both Pbx2 and Pbx4 (Waskiewicz et al., 2002). We previously demonstrated that these combined pbx2 and pbx4 morpholinos specifically and effectively knock down all Pbx protein at least up to 24 hours post fertilization (hpf) (Maves et al., 2007). In wild-type zebrafish embryos, the two heart chambers, the atrium and ventricle, are distinct and functioning by 48 hpf (Glickman and Yelon, 2002; Fig. 1A). In pbx2-MO; pbx4-MO embryos, a weakly contractile heart forms and appears morphologically abnormal, accompanied by mild pericardial edema (30/30 embryos exhibited a similar phenotype). Heart function appears disrupted, with blood pooling by the atrium (Fig. 1B). We confirmed the defect in heart function by assessing the heartbeat at 48 hpf. Wild-type control embryos had 156±18 beats/min (n=10) and pbx2-MO; pbx4-MO embryos had 86±16 beats/min (n=9). Although it is not clear from these live observations whether abnormal heart size or shape causes the functional defects, our finding that Pbx proteins are required for normal heart function is consistent with previous studies in zebrafish (Pöpperl et al., 2000) and mice (Chang et al., 2008; Stankunas et al., 2008).
Fig. 1.
Defective heart development in pbx2-MO; pbx4-MO embryos. (A–B) Lateral views of live (A) wild-type and (B) pbx2-MO; pbx4-MO embryos at 48 hpf. pbx2-MO; pbx4-MO embryos show mild pericardial edema and blood pooled near the atrium (arrow). Anterior is to the right. a, atrium. v, ventricle.
Microarray analysis identifies Pbx-dependent heart gene expression
To investigate the underlying requirements for Pbx proteins in heart development, we used Affymetrix expression microarrays to compare gene expression in pbx2-MO; pbx4-MO embryos with control siblings (injected with pbx2-MO mismatch control), as previously described (Maves et al., 2007). Two stages were examined: 10 somites/14 hpf, when early heart specification occurs, and 18 somites/18 hpf, when early heart differentiation occurs (Yelon et al., 1999; Glickman and Yelon, 2002). Microarray experiments on total RNA from whole embryos identified Pbx-dependent genes in all tissues (Maves et al., 2007). From the gene lists generated from the arrays, we identified the genes that are expressed in wild-type heart precursors, according to the publicly available RNA in situ database (Thisse and Thisse, 2005), or that are known to regulate vertebrate heart development (Table 1). For each of these heart genes, we performed RNA in situ hybridization or quantitative RT-PCR to confirm that these genes indeed show altered expression in pbx2-MO; pbx4-MO embryos (Table 1; Fig. 2). Embryos were co-labeled with krox-20, a Pbx-dependent hindbrain gene, to confirm full Pbx knock-down (Waskiewicz et al., 2002; Maves et al., 2007; Fig. 2). In pbx2-MO; pbx4-MO embryos, tnnt2, ttna, cmlc1, vmhc, myl7, and lft1 all show reduced heart primordium expression, whereas bon, sox32, and hand2 show increased expression, consistent with our array results (Table 1; Fig. 2). aldh1a2 shows reduced somite and eye expression in pbx2-MO; pbx4-MO embryos, consistent with our array results (Table 1; Figs. 2I–J; Maves et al., 2007), but shows expanded lateral plate mesoderm expression (Figs. 2I–J). These array analyses and validations thus define the genomic expression requirements for Pbx proteins at early and intermediate stages in heart development and support the hypothesis that Pbx proteins are required for proper heart development.
Table 1.
Microarray-identified Pbx-dependent heart genes
| Gene Symbol | Affymetrix ID number | Fold change at 10s (controlMO: pbx2-MO; pbx4-MO | Wild-type expression at 10s | RNA in situ expression in pbx2-MO; pbx4- MO at 10s | RNA in situ expression in hand2−/− or hand2- MO at 10s |
|---|---|---|---|---|---|
| tnnt2 | Dr.14260.1.S1_a_at | 1.55 | Heart primordium | Reduced | Reduced |
| ttna | Dr.4681.1.A1_at | 1.53 | Heart primordium; somites | Reduced (1) | Not affected |
| bon | Dr.8084.1.S1_at | −1.87 | Not expressed | 10X increase over control (qRT-PCR) | Similar to control (qRT-PCR) |
| sox32 | Dr.12573.1.S1_at | −1.88 | Endoderm | 3X increase over control (qRT-PCR) | Similar to control (qRT-PCR) |
| Gene Symbol | Affymetrix ID number | Fold change at 18s (controlMO: pbx2-MO; pbx4-MO | Wild-type expression at 18s | RNA in situ expression in pbx2-MO; pbx4- MO at 18s | RNA in situ expression in hand2−/− or hand2- MO at 18s |
| cmlc1 | Dr.8379.1.S1_at | 3.62 | Heart; somites | Heart domain reduced | Heart domain reduced |
| vmhc | Dr.10607.1.A1_at | 3.20 | Heart; somites | Heart domain reduced (1) | Reduced (2) |
| aldh1a2 | Dr.5206.1.S1_at | 2.26 | Lateral plate mesoderm (LPM); somites; eye | LPM domain increased; eye and somite domains reduced; increased in tail bud (1) | Not affected |
| myl7 | Dr.642.1.S1_at | 1.90 | Heart | Reduced | Reduced (2) |
| ttna | Dr.4681.1.A1_at | 1.85 | Heart; somites | Reduced (1) | Heart domain reduced |
| lft1 | Dr.8208.1.S1_at | 1.60 | Heart; forebrain; notochord | Reduced in heart and forebrain | Reduced in heart |
| hand2 | Dr.8328.1.S1_at | −1.70 | Heart; ventral mesoderm | Increased | Reduced in hand2−/− (2); Increased in hand2-MO |
| sox32 | Dr.12573.1.S1_at | −2.05 | Not expressed | 5X increase over control (qRT-PCR) | Similar to control (qRT-PCR) |
Of the Pbx-dependent heart genes (Table 1), most (tnnt2, ttna, cmlc1, vmhc, and myl7) are involved in cardiac contractile function and can be categorized as myocardial differentiation genes. In particular, cardiac troponin T2 (tnnt2), titin (ttna), cardiac myosin light chain-1 (cmlc1), ventricular myosin heavy chain (vmhc), and myosin light polypeptide 7 (myl7) have all been shown to be necessary for heart muscle differentiation and function in zebrafish (Sehnert et al., 2002; Xu et al., 2002; Auman et al., 2007; Chen et al., 2008). Therefore, the disrupted heart function in pbx2-MO; pbx4-MO embryos is likely due to defective myocardial differentiation.
Pbx requirements resemble those for Hand2 in early myocardial differentiation
The reduced vmhc and myl7 expression in pbx2-MO; pbx4-MO embryos resembles that seen in zebrafish hand2 mutants, and Hand2 is required for myocardial differentiation in zebrafish (Yelon et al., 2000; Schoenebeck et al., 2007). We therefore asked whether the Pbx-dependent genes identified by our array analyses are also Hand2-dependent. We used a morpholino against hand2 to knock down hand2 function. We confirmed that the morpholino knock-down caused reduced myl7 and vmhc expression, similar to hand2−/− embryos (Table 1; Yelon et al., 2000), and we also confirmed that myl7 expression defects in hand2-MO embryos were rescued by hand2 mRNA (Supplemental Fig. 1). We find that, indeed, most of the Pbx-dependent genes, in particular the myocardial differentiation genes, are also Hand2-dependent (Table 1). In addition, hand2 expression itself is regulated by both Pbx and Hand2, as hand2 expression increases in pbx2-MO; pbx4-MO embryos and in hand2-MO embryos (Table 1; Figs. 2Q-R). Zebrafish hand2−/− embryos show reduced hand2 expression because the mutant alleles directly affect hand2 transcription (Yelon et al., 2000), whereas the hand2 MO targets Hand2 translation. Although we do not know the levels of Hand2 protein expression in hand2-MO or pbx2-MO; pbx4-MO embryos, we interpret the increased hand2 expression in hand2-MO and in pbx2-MO; pbx4-MO embryos as indicative of decreased Hand2 activity. Thus, Pbx-deficient embryos resemble Hand2-deficient embryos in two respects: they both have similar requirements for myocardial differentiation gene expression, and they both regulate hand2 expression.
We observe, however, two notable differences between Pbx and Hand2 regulation of the microarray-identified genes. First, aldh1a2 expression in the lateral plate mesoderm increases in pbx2-MO; pbx4-MO embryos but is not affected in hand2-MO embryos (Table 1; Figs. 2I–J). Second, the endodermal genes bon and sox32 show increased expression in pbx2-MO; pbx4-MO embryos but are not affected in hand2-MO embryos (Table 1). We address these differences in the Discussion, and, nevertheless, we conclude that our microarray analysis and validations reveal similar requirements for Pbx and Hand2 in early myocardial differentiation.
Early myocardial specification appears normal in Pbx-deficient embryos
In order to address the origins of the myocardial differentiation defects in Pbx-deficient embryos, we investigated early myocardial specification in pbx2-MO; pbx4-MO embryos. An early fate map of zebrafish cardiac progenitors at the 6–9 somite stage (Schoenebeck et al., 2007) has defined markers of anterior lateral plate mesoderm (ALPM; gata4, gata5, tbx20), rostral ALPM (presumptive blood and vasculature; scl), caudal ALPM (hand2), and medial-caudal ALPM (nkx2.5). These markers exhibit normal expression in hand2−/− embryos, showing that hand2 is not needed for early cardiac specification (Schoenebeck et al., 2007). We analyzed expression of these fate-map markers in pbx2-MO; pbx4-MO embryos and hand2-MO embryos at 7–9 somites (Fig. 3). The hand2 expression domain is expanded in both pbx2-MO; pbx4-MO and hand2-MO embryos at 7 somites (Figs. 3J–3L), similar to the hand2 upregulation at 18 somites (Figs. 2Q–R) and further supporting the correspondence between loss of Pbx and Hand2 activities. Expression of the other early cardiac fate-map markers appears largely normal in pbx2-MO; pbx4-MO embryos or in hand2-MO embryos (Fig. 3 and data not shown), although scl expression may be subtly reduced in pbx2-MO; pbx4-MO embryos (Fig. 3G–3H). Thus, early cardiac progenitors and myocardial specification appear intact in the absence of Pbx. Furthermore, aside from hand2, expression of these early cardiac fate-map markers was not identified as altered in our pbx2-MO; pbx4-MO microarray experiment. This analysis shows that Pbx, like Hand2, is not required for establishing early domains of specific heart precursors but rather for subsequent myocardial differentiation.
Pbx-deficient and hand2-MO embryos exhibit similar cardiac differentiation and morphogenesis defects
We next wanted to investigate the extent of the myocardial differentiation defects in pbx2-MO; pbx4-MO embryos compared to hand2-MO embryos. Subsequent to the activation of myocardial differentiation genes at around the 13–18 somite stage (15.5–18 hours post fertilization, hpf; Yelon et al., 1999), the bilateral myocardial precursors migrate to the midline, merge to form the cardiac cone, and then, by 26 hpf, elongate to form the heart tube (Yelon and Glickman, 2002). Myocardial differentiation appears to be required for myocardial morphogenesis because zebrafish embryos with defective myocardial differentiation have defects in myocardial morphogenesis (Yelon et al., 1999; Reiter et al., 1999; Yelon et al., 2000). However, zebrafish loci that are required for myocardial differentiation can exhibit very different requirements for myocardial morphogenesis. For example, zebrafish hand2 mutants show reduced myocardial precursors with defective midline migration and subsequent cardia bifida, whereas nkx-deficient embryos have normal midline migration but then fail to properly elongate the heart tube (Yelon et al., 2000; Trinh et al., 2005; Targoff et al., 2008). Because zebrafish pbx2-MO; pbx4-MO embryos have early myocardial differentiation defects, we wanted to determine whether they also have a characteristic myocardial morphogenesis defect. We therefore examined the expression of myl7, vmhc and amhc at three time points of myocardial morphogenesis: the 21 somite-stage (21s), when the myocardial precursors have merged to form the cardiac cone (Fig. 4A), 25 hpf, when the heart tube has elongated (Fig. 4D), and 42 hpf, when the heart tube has looped such that the atrium lies to the right of the ventricle (Fig. 4G). In control embryos, myl7 labels all myocardial precursors, vmhc labels ventricular precursors, and amhc labels atrial precursors (Berdougo et al., 2003). We find that myl7- and vmhc-expressing heart precursors continue to be reduced and show delayed midline migration at 21s in both pbx2-MO; pbx4-MO embryos and hand2-MO embryos, while amhc expression is absent (Fig. 4A-C). At 25 hpf, myocardial precursors in pbx2-MO; pbx4-MO and hand2-MO embryos have merged abnormally at the midline and have failed to elongate into the heart tube (Fig. 4D–F). At 42 hpf, cardiac morphogenesis appears similarly abnormal in both pbx2-MO; pbx4-MO and hand2-MO embryos: there appear to be separate vmhc-expressing and amhc-expressing chambers, however their shapes are abnormal, appear somewhat smaller than controls, and have not looped properly (Fig. 4G–I). These analyses reveal that the hand2-MO phenotype does not fully correspond to the hand2 null mutant phenotype at 25 hpf and beyond, as most hand2-MO embryos show delayed midline fusion of heart precursors rather than cardia bifida, or separate lateral hearts, at 25 hpf and 42 hpf (Yelon et al., 2000; also see Fig. 5). However, this suggests that the heart morphogenesis defects in fully Pbx-deficient embryos correspond to reduced Hand2 function. We also analyzed differentiated hearts of pbx2-MO; pbx4-MO and hand2-MO embryos compared to control embryos, using dual labeling of the atrium-specific antibody S46 with the whole heart marker MF20. We see similar results as the in situ analysis at 42 hpf, in that both chambers are reduced and similar morphogenesis defects occur in pbx2-MO; pbx4-MO and hand2-MO embryos (data not shown). Taken together, our analyses show that pbx2-MO; pbx4-MO embryos have both myocardial differentiation and morphogenesis defects that closely resemble those seen in embryos with reduced Hand2 function.
Fig. 4.
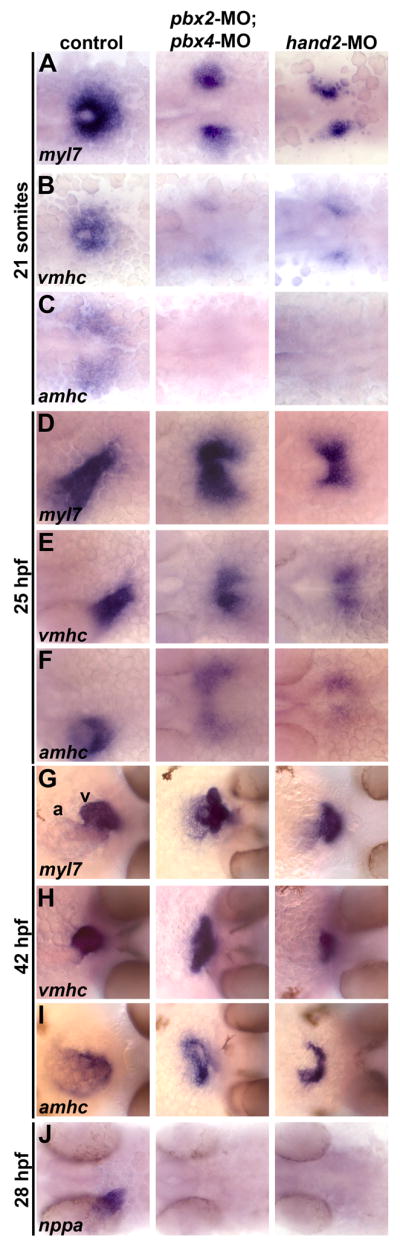
Pbx-deficient and hand2-MO embryos exhibit similar cardiac differentiation and morphogenesis defects. (A–J) RNA in situ expression of (A,D,G) myl7, (B,E,H) vmhc, (C,F,I) amhc, and (J) nppa at (A–C) 21 somites, (D–F) 25 hours post fertilization (hpf), (G–I) 42 hpf, and (J) 28 hpf. In control embryos shown, myl7 labels all myocardial precursors, vmhc labels ventricular precursors, and amhc labels atrial precursors (Berdougo et al., 2003). Ventricle (v) and atrium (a) are labeled in (G). n≥20 for each marker in control, pbx2-MO; pbx4-MO, or hand2-MO. (A–F) Embryos are shown in dorsal view, anterior towards the left. (G-I) Embryos are shown in frontal views, dorsal towards the right.
To further investigate and compare the myocardial differentiation defects in pbx2-MO; pbx4-MO and hand2-MO embryos, we examined expression of natriuretic peptide precursor A (nppa), a myocardial differentiation gene that is regulated by Hand2 in mice (Thattaliyath et al., 2002). nppa expression in the zebrafish heart initiates around 24 hpf (data not shown). By 28 hpf, nppa fails to be activated in either pbx2-MO; pbx4-MO or hand2-MO embryos (Fig. 4J). Even by 42 hpf, nppa expression remains significantly downregulated in pbx2-MO; pbx4-MO or hand2-MO embryos (data not shown). These findings thus further corroborate the correspondence between the requirements for Pbx and Hand2 in myocardial differentiation.
Pbx and Hand2 genetically interact in cardiac morphogenesis
In two respects, our results strongly suggest that Pbx interacts with Hand2 in myocardial differentiation. First, most of the Pbx-regulated heart genes identified in our microarray analysis are also Hand2-regulated (Table 1), and second, the myocardial differentiation and morphogenesis phenotype of pbx2-MO; pbx4-MO embryos closely resembles that of zebrafish hand2 knock-down embryos (Fig. 4). We therefore wanted to determine whether Pbx and Hand2 exhibit genetic interactions in myocardial differentiation by simultaneously knocking down the functions of both factors using combinations of pbx2; pbx4 and hand2 morpholinos. We envisioned three possible results for pbx2-MO; pbx4-MO; hand2-MO embryos: 1. They may show a more severe phenotype than hand2 null embryos. This result may occur if Pbx and Hand2 act additively and independently to regulate myocardial differentiation. 2. They may show a less severe phenotype than pbx2-MO; pbx4-MO embryos. Because hand2 expression is upregulated in pbx2-MO; pbx4-MO embryos (Fig. 2), blocking hand2 function in pbx2-MO; pbx4-MO embryos may act to rescue the pbx2-MO; pbx4-MO phenotype. 3. They may resemble hand2 null embryos, which exhibit cardia bifida at 25 hpf and 42 hpf and are more severe than pbx2-MO; pbx4-MO embryos or hand2-MO embryos (Yelon et al., 2000; Fig. 5E). This result may occur if Pbx provides no further function in heart development than facilitating Hand2 function. Whereas knock down of either Pbx or Hand2 function led to delayed and abnormal heart morphogenesis (Figs. 5B–C, 5F–G), as described above for Figure 4, when we knocked down both Pbx and Hand2 function, we observed that, at both 24 hpf and 42 hpf, most pbx2-MO; pbx4-MO; hand2-MO embryos exhibit cardia bifida (Figs. 5D, 5F–G). With simultaneous Pbx and Hand2 knock-down, we did not observe any evidence for a more severe heart phenotype or loss of myl7 or vmhc expression than that observed in hand2 null embryos, nor did we observe any evidence that Hand2 knock-down rescued the Pbx knock-down (Fig. 5 and data not shown). The simultaneous Pbx and Hand2 knock-down thus corresponds to the hand2 null phenotype, supporting an interaction between Pbx and Hand2 in promoting myocardial differentiation and morphogenesis.
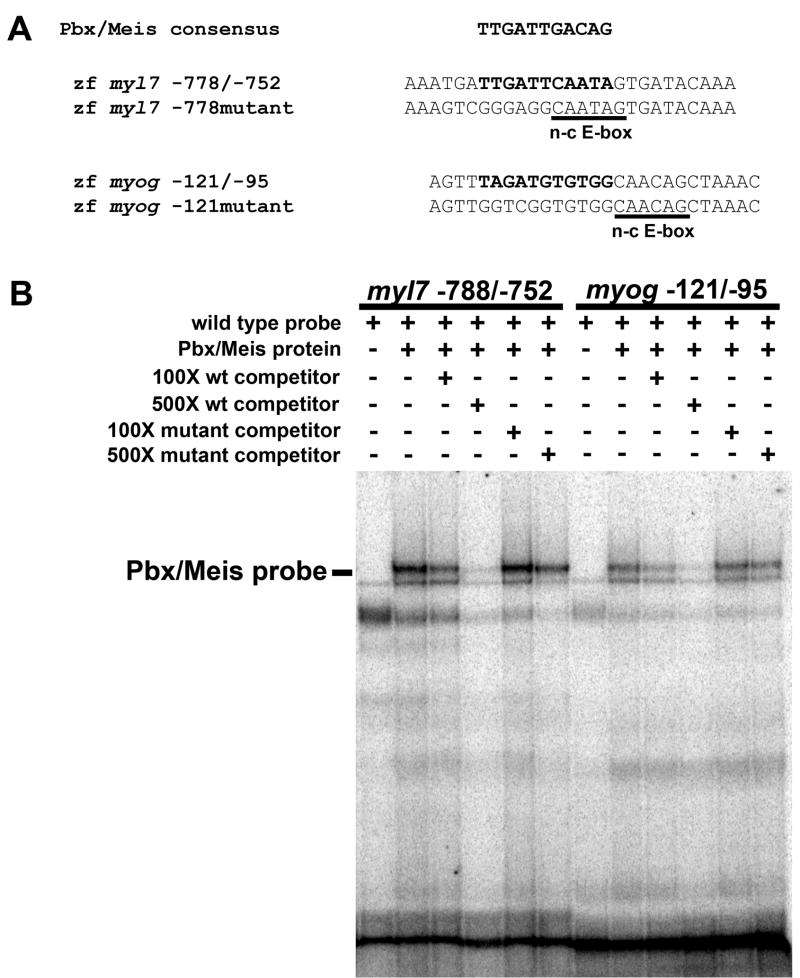
Pbx proteins bind to the myl7 promoter in vitro
We next wanted to determine whether Pbx directly regulates expression of cardiac muscle differentiation genes. Because Pbx recruits Myod to the Myogenin promoter (Berkes et al., 2004), Pbx may also recruit Hand2 to myocardial differentiation gene promoters. The zebrafish myl7 promoter has been partially characterized through gfp reporter analyses in transgenic embryos (Huang et al., 2003). A fragment containing 870bp of 5′ upstream regulatory sequence mimics endogenous myl7 expression (Huang et al., 2003). Using the binding-site prediction program Match1.0, we identified a potential Pbx binding site at −778bp that appears to more closely resemble the Pbx/Meis consensus binding site than the conserved Pbx/Meis site in the myogenin promoter (Fig. 6A; Berkes et al., 2004). We tested whether this site is capable of binding Pbx4, together with cofactor Meis3, through gel-shift assays (EMSA), similar to how we have assayed Pbx/Meis binding in previous studies (Berkes et al., 2004; Maves et al., 2007). We find that Pbx4/Meis3 binding to the −778/−752bp oligo is inhibited by unlabeled wild-type oligo but not by an oligo containing a mutated Pbx binding site (Fig. 6B), showing that Pbx proteins can bind directly to the myl7 promoter in vitro. As a control, we observe similar results with an oligo containing the myogenin Pbx/Meis site (Fig. 6B). Because there is a non-canonical E-box sequence adjacent to the Pbx binding site in the myl7 promoter, similar to the non-canonical E-box adjacent to the Pbx site in the myogenin promoter (Fig. 6A; Berkes et al., 2004), we wanted to address whether Hand2 protein interacts with Pbx at the myl7 promoter. We performed additional gel shift assays but were unable to observe any binding of Hand2 protein or Hand2 combined with E12 protein to the −778/−752 oligo, nor were we able to observe any cooperative binding of Hand2/E12 with Pbx/Meis to the −778/−752 oligo (data not shown). Thus, the mechanism by which Pbx and Hand2 proteins interact at myocardial differentiation gene promoters remains to be determined. Although we do not yet know whether Pbx proteins bind myl7 or other myocardial promoters in vivo, our EMSA results strongly support a direct role for Pbx regulation of myocardial differentiation gene expression.
Fig. 6.
Pbx proteins bind the myl7 promoter in vitro. (A) Pbx/Meis consensus binding site sequence (Berkes et al., 2004) aligned relative to putative Pbx binding site sequences (highlighted in bold) in the zebrafish myl7 promoter and myogenin (myog) promoter. The non-canonical E-box (n-c E-box) sites are underlined. Also shown are sequences of mutated Pbx binding sites. Sequences represent oligos used in EMSA. (B) Pbx4 and Meis3 proteins were generated by in vitro translation using a reticulocyte lysate and subjected to EMSA with the specified myl7 or myog probes. Pbx/Meis-bound probe band is indicated. The non-specific bands were observed in multiple gel shifts. Addition of unlabeled myl7 −778/−752 probe or myog −121/−95 probe decreases binding of Pbx/Meis, whereas addition of mutant unlabeled probes does not.
Discussion
In this work, we test the hypothesis that Pbx proteins regulate cellular differentiation by modulating bHLH factor activity, in particular by testing whether Pbx proteins interact with Hand2 in cardiac muscle differentiation. Our data reveal that Pbx proteins in zebrafish are necessary for proper heart development and function by regulating gene expression required for myocardial differentiation. Our results show that Pbx proteins have similar requirements to Hand2 in myocardial differentiation and morphogenesis and, furthermore, demonstrate a genetic interaction between Pbx and Hand2. Thus, our findings underscore the critical roles of Pbx proteins in promoting muscle cell differentiation and suggest that Pbx proteins may modulate bHLH factor activity more broadly than previously appreciated.
Our results demonstrate that Pbx and Hand2 are required for the expression of similar cardiac muscle differentiation genes and show similar requirements for cardiac morphogenesis. However, we observed two notable differences between Pbx and Hand2 regulation of the microarray-identified Pbx-dependent genes. First, aldh1a2 expression in the lateral plate mesoderm increases in pbx2-MO; pbx4-MO embryos but is not affected in hand2-MO embryos (Table 1; Fig. 2). aldh1a2 encodes an enzyme responsible for the synthesis of retinoic acid. Because retinoic acid normally restricts the number of cardiac progenitor cells (Keegan et al., 2005), increased aldh1a2 expression in pbx2-MO; pbx4-MO embryos could lead to reduced myocardial differentiation. However, the increased aldh1a2 expression in pbx2-MO; pbx4-MO embryos occurs during somitogenesis, while retinoic acid acts on cardiac progenitors during gastrulation (Keegan et al., 2005). Second, the endodermal genes bon and sox32 show increased expression in pbx2-MO; pbx4-MO embryos but are not affected in hand2-MO embryos (Table 1). In zebrafish, loss of bon, which encodes a Mix homeodomain factor, or sox32 function leads to delayed cardiac morphogenesis and cardia bifida (Kikuchi et al., 2000; Alexander et al., 1999). Thus, the connection between increased endodermal marker expression in pbx2-MO; pbx4-MO embryos and cardiac differentiation and morphogenesis is not immediately clear. In spite of these differences between Pbx and Hand2 gene regulation, we conclude that our microarray analysis and validations reveal similar requirements for Pbx and Hand2 in early myocardial differentiation and suggest that they act together in a common pathway.
In addition to Hand2, other transcription factors could potentially interact with Pbx proteins in heart development. GATA and Nkx family factors are expressed in myocardial precursors and play critical roles in early myocardial differentiation and morphogenesis (Olson, 2006; Schoenebeck and Yelon, 2007). However, zebrafish gata5 mutants have early defects in gata4 and nkx2.5 expression and thus have early myocardial specification defects that are not observed in Pbx-null embryos (Reiter et al., 1999; Fig. 3). Zebrafish Nkx-deficient embryos have heart tube extension defects but do not exhibit early myocardial differentiation defects (Targoff et al., 2008). Pbx proteins are well-known cofactors for Hox proteins (Moens and Selleri, 2006), so Pbx proteins may regulate heart development through these factors. The full roles for Hox proteins in cardiac development are not known, but, in zebrafish, Hoxb5b negatively regulates cardiomyocyte numbers (Waxman et al., 2008). If Pbx acts with Hoxb5b in this role, the hoxb5b knock-down phenotype (increased amhc-expressing cardiomyocytes; Waxman et al., 2008) is not consistent with the reduced amhc expression in Pbx-null embryos (Fig. 4). Thus, although Pbx may function with additional factors in myocardial differentiation, the requirements for Pbx most closely correspond with those for Hand2, underscoring the interaction between these two factors.
The advantage of using zebrafish in these studies is that we characterized the Pbx-null embryonic heart phenotype by knocking down both Pbx2 and Pbx4. In zebrafish, pbx4 likely plays a major role relative to pbx2 during early heart development. Zebrafish zygotic pbx4 mutant embryos have been reported to have impaired circulation and pericardial edema at 5 days post fertilization (Pöpperl et al., 2000), and we find that pbx4−/− embryos have reduced myl7 and vmhc expression and cardiac morphogenesis defects (L.M., unpublished data), although not as severe as pbx2-MO; pbx4-MO embryos. Pbx gene deficiencies in mice were recently shown to cause cardiac defects that resemble human CHDs (Chang et al., 2008; Stankunas et al., 2008). In particular, loss of specific Pbx genes led to distinct cardiac outflow tract defects, with Pbx1 playing a major role (Stankunas et al., 2008). The role of Pbx in cardiac outflow tract development likely reflects Pbx function within cardiac neural crest cells (Chang et al., 2008). These studies did not address whether early myocardial differentiation is affected in Pbx-deficient mice, thus, it is not yet clear whether Pbx proteins have conserved roles in zebrafish and mammalian heart development. In order to gain full insight into the potential roles of Pbx in CHD in humans, it will be necessary to characterize the requirements for Pbx genes in early mouse heart development, in particular to test whether Pbx genes are required for early myocardial differentiation. The ability to generate mouse embryos deficient for multiple alleles of Pbx1, Pbx2, and Pbx3 (Capellini et al., 2006; Stankunas et al., 2008) will greatly contribute to analyzing Pbx requirements in early heart development.
Our zebrafish studies suggest that Pbx genes may interact with Hand1 or Hand2 in mammalian heart development, but this is not yet explored. Pbx genes and Hand2 both function in limb development in zebrafish (Pöpperl et al., 2000; Yelon et al., 2000) and in mice (Charite et al., 2000; Capellini et al., 2006). However, whereas Pbx and Hand2 act together in the zebrafish heart, in the mouse limb they appear to function in parallel pathways (Capellini et al., 2006), and Hand2 expression is not dependent on Pbx in the mouse limb (Capellini et al., 2006) as it is in the zebrafish heart (this work). Future examinations of Pbx and Hand2 functions in the heart or limb would benefit from complementary approaches in zebrafish and mouse.
Our studies reveal several similarities between the roles of Pbx in cardiac muscle development and in skeletal muscle development (this work and Maves et al., 2007). In both muscle systems: 1. Pbx is needed for muscle differentiation and contractile gene expression, 2. Pbx is needed for proper initiation of muscle gene activation, because Pbx-dependent genes eventually turn on, 3. Pbx genetically interacts with a bHLH factor, Myod or Hand2, 4. Expression of the bHLH genes myod or hand2 increases (while their activity decreases) in Pbx-null embryos, and 5. Pbx-null embryos have similar, but not as severe, differentiation phenotypes compared to complete loss of Myod or Hand2. These similarities strongly suggest that Pbx may have comparable interactions with Hand2 in cardiac development as Pbx does with Myod in skeletal muscle.
These connections between Pbx and bHLH factors support the idea of a core developmental program regulating the transcriptional control of contractile muscle cell differentiation, in which Pbx proteins facilitate the ability of bHLH factors to recognize and activate a set of myosins and other genes needed for differentiation. We speculate that Pbx/bHLH interactions could function even more broadly in cellular differentiation, in contexts such as neuronal differentiation and pancreas development, where Pbx and the NeuroD family of bHLH factors both play important roles (Dutta et al., 2001; Waskiewicz et al., 2002; Westerman et al., 2003).
In skeletal muscle, Pbx and Myod exhibit both genetic as well as biochemical interactions and cooperatively bind in a complex together on the Myogenin promoter, which contains Pbx and Myod binding sites (Berkes et al., 2004; Maves et al., 2007). By binding to Myod target gene promoters, Pbx is thought to act as a signal for Myod to recognize and activate those promoters (Berkes et al., 2004). While we were able to demonstrate that Pbx proteins can bind the myocardial myl7 promoter in vitro (Fig. 6), we were unable to demonstrate that Hand2 can bind the myl7 promoter, either in the absence or presence of Pbx. Several studies have revealed that Hand proteins may activate promoters independently of DNA binding by physically interacting with transcriptional complexes (Dai et al., 2002; McFadden et al., 2002; Thattaliyath et al., 2002; Morin et al., 2005). Also, Liu et al. (Liu et al., 2009) have recently shown that mice lacking the DNA-binding domain of Hand2 have normal early heart development. Whether Pbx/Myod and Pbx/Hand2 interactions share common biochemical mechanisms remains to be determined.
Supplementary Material
Supplemental Fig. 1. hand2 RNA rescues myl7 expression in hand2-MO embryos. (A–D) RNA in situ expression of myl7 at the 20s stage in embryos injected with (A) control gfp mRNA, (B) hand2 mRNA, (C), hand2-MO and gfp mRNA, or (D) hand2-MO and hand2 mRNA. Embryos are shown in dorsal view, anterior towards the left. (A) In 20s control embryos, heart precursors form a cone on the embryonic midline. (B) Embryos injected with hand2 mRNA have about wild-type numbers of myl7-expressing cells but heart morphogenesis and cone formation is slightly impaired. (C) Embryos injected with hand2-MO have reduced lateral myl7 domains with defective midline migration. (D) hand2 mRNA largely rescues the hand2-MO phenotype, although most embryos show some impaired morphogenesis. (E) Quantification of the rescue data. Numbers in parentheses denote numbers of embryos. The phenotypic classes correspond to the images shown in panels A–C.
Acknowledgments
We thank Zayra Garavito-Aguilar, Vladimir Korzh, Didier Stainier, Andrew Waskiewicz and Debbie Yelon for generously providing reagents. The Zebrafish International Resource Center (supported by grants RR12546 and RR15402-01 from the NIH) provided cDNA clones. We thank Andrew Waskiewicz and Yi Cao for their initial contributions to the array experiments. We also thank Sean Rhodes for technical assistance and Debbie Yelon and members of the Moens and Tapscott labs for advice. C.B.M. is an investigator with the Howard Hughes Medical Institute. This work was supported by NIH AR45113 (S.J.T.) and a Research Development Grant from the Muscular Dystrophy Association (L.M.).
Footnotes
Contributions
AT and LM carried out the experiments. SJT and LM conceived the experiments. CBM provided the zebrafish facility. LM analyzed the data and wrote the paper. All authors have approved the final version of this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander J, Rothenberg M, Henry GL, Stainier DY. casanova plays an early and essential role in endoderm formation in zebrafish. Dev Biol. 1999;215:343–357. doi: 10.1006/dbio.1999.9441. [DOI] [PubMed] [Google Scholar]
- Auman HJ, Coleman H, Riley HE, Olale F, Tsai HJ, Yelon D. Functional modulation of cardiac form through regionally confined cell shape changes. PLoS Biol. 2007;5:e53. doi: 10.1371/journal.pbio.0050053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader D, Masaki T, Fischman DA. Immunochemical analysis of myosin heavy chain during avian myogenesis in vivo and in vitro. J Cell Biol. 1982;95:763–770. doi: 10.1083/jcb.95.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann G, Schilling TF, Rauch GJ, Geisler R, Ingham PW. The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development. 2001;127:2189–2199. doi: 10.1242/dev.128.16.3081. [DOI] [PubMed] [Google Scholar]
- Berdougo E, Coleman H, Lee DH, Stainier DYR, Yelon D. Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development. 2003;130:6121–6129. doi: 10.1242/dev.00838. [DOI] [PubMed] [Google Scholar]
- Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell. 2004;14:465–477. doi: 10.1016/s1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
- Cao Y, Kumar RM, Penn BH, Berkes CA, Kooperberg C, Boyer LA, Young RA, Tapscott SJ. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J. 2006;25:502–511. doi: 10.1038/sj.emboj.7600958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capellini TD, Di Giacomo G, Salsi V, Brendolan A, Ferretti E, Srivastava D, Zappavigna V, Selleri L. Pbx1/Pbx2 requirement for distal limb patterning is mediated by the hierarchical control of Hox gene spatial distribution and Shh expression. Development. 2006;133:2263–2273. doi: 10.1242/dev.02395. [DOI] [PubMed] [Google Scholar]
- Chang CP, Stankunas K, Shang C, Kao SC, Twu KY, Cleary ML. Pbx1 functions in distinct regulatory networks to pattern the great arteries and cardiac outflow tract. Development. 2008;135:3577–3586. doi: 10.1242/dev.022350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charite J, McFadden DG, Olson EN. The bHLH transcription factor dHand controls Sonic hedgehog expression and establishment of the zone of polarizing activity during limb development. Development. 2000;127:2461–2470. doi: 10.1242/dev.127.11.2461. [DOI] [PubMed] [Google Scholar]
- Chen Z, Huang W, Dahme T, Rottbauer W, Ackerman MJ, Xu X. Depletion of zebrafish essential and regulatory myosin light chains reduces cardiac function through distinct mechanisms. Cardiovasc Res. 2008;79:97–108. doi: 10.1093/cvr/cvn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KL, Yutzey KE, Benson DW. Transcription factors and congenital heart defects. Annu Rev Physiol. 2006;68:97–121. doi: 10.1146/annurev.physiol.68.040104.113828. [DOI] [PubMed] [Google Scholar]
- Dai YS, Cserjesi P, Markham BE, Molkentin JD. The transcription factors GATA4 and dHAND physically interact to synergistically activate cardiac gene expression through a p30-dependent mechanism. J Biol Chem. 2002;277:24390–24398. doi: 10.1074/jbc.M202490200. [DOI] [PubMed] [Google Scholar]
- Dutta S, Gannon M, Peers B, Wright C, Bonner-Weir S, Montminy M. PDX:PBX complexes are required for normal proliferation of pancreatic cells during development. Proc Natl Acad Sci USA. 2001;98:1065–1070. doi: 10.1073/pnas.031561298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli AB, McFadden DG, Lin Q, Srivastava D, Olson EN. Heart and extra-embryonic mesoderm defects in mouse embryos lacking the bHLH transcription factor Hand1. Nature Genetics. 1998;18:266–270. doi: 10.1038/ng0398-266. [DOI] [PubMed] [Google Scholar]
- Firulli AB. A HANDful of questions: the molecular biology of the heart and neural crest derivatives (HAND)-subclass of basic helix-loop-helix transcription factors. Gene. 2003;312:27–40. doi: 10.1016/s0378-1119(03)00669-3. [DOI] [PubMed] [Google Scholar]
- Fukushige T, Brodigan TM, Schriefer LA, Waterston RH, Krause M. Defining the transcriptional redundancy of early bodywall muscle development in C. elegans: evidence for a unified theory of animal muscle development. Genes Dev. 2006;20:3395–3406. doi: 10.1101/gad.1481706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman NS, Yelon D. Cardiac development in zebrafish: coordination of form and function. Sem Cell Dev Biology. 2002;13:507–513. doi: 10.1016/s1084952102001040. [DOI] [PubMed] [Google Scholar]
- Huang CJ, Tu CT, Hsiao CD, Hsieh FJ, Tsai HJ. Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev Dyn. 2003;228:30–40. doi: 10.1002/dvdy.10356. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Trinh LA, Reiter JF, Alexander J, Yelon D, Stainier DY. The zebrafish bonnie and clyde gene encodes a Mix family homeodomain protein that regulates the generation of endodermal precursors. Genes Dev. 2000;14:1279–1289. [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Bergstrom DA, Uetsuki T, Dac-Korytko I, Sun YH, Wright WE, Tapscott SJ, Kamps MP. A conserved motif N-terminal to the DNA-binding domains of myogenic bHLH transcription factors mediates cooperative DNA binding with Pbx-Meis1/Prep1. Nucleic Acids Res. 1999;27:3752–3761. doi: 10.1093/nar/27.18.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Xu Q, Breitbart RE. A new tinman-related gene, nkx2.7, anticipates the expression of nkx2.5 and nkx2.3 in zebrafish heart and pharyngeal endoderm. Dev Biol. 1996;180:722–731. doi: 10.1006/dbio.1996.0341. [DOI] [PubMed] [Google Scholar]
- Liu N, Barbosa AC, Chapman SL, Bezprozvannaya S, Qi X, Richardson JA, Yanagisawa H, Olson EN. DNA binding-dependent and –independent functions of the Hand2 transcription factor during mouse embryogenesis. Development. 2009;136:933–942. doi: 10.1242/dev.034025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maves L, Waskiewicz AJ, Paul B, Cao Y, Tyler A, Moens CB, Tapscott SJ. Pbx homeodomain proteins direct Myod activity to promote fast-muscle differentiation. Development. 2007;134:3371–3382. doi: 10.1242/dev.003905. [DOI] [PubMed] [Google Scholar]
- McFadden DG, McAnally J, Richardson JA, Charite J, Olson EN. Misexpression of dHAND induces ectopic digits in the developing limb bud in the absence of direct DNA binding. Development. 2002;129:3077–3088. doi: 10.1242/dev.129.13.3077. [DOI] [PubMed] [Google Scholar]
- Miller JB, Crow MT, Stockdale FE. Slow and fast myosin heavy chain content defines three types of myotubes in early muscle cell cultures. J Cell Biol. 1985;101:1643–1650. doi: 10.1083/jcb.101.5.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Morin S, Pozzulo G, Robitaille L, Cross J, Nemer M. MEF2-dependent recruitment of the HAND1 transcription factor results in synergistic activation of target promoters. J Biol Chem. 2005;280:32272–32278. doi: 10.1074/jbc.M507640200. [DOI] [PubMed] [Google Scholar]
- Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxtoby E, Jowett T. Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res. 1993;21:1087–1095. doi: 10.1093/nar/21.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierpont ME, Basson CT, Benson DW, Jr, Gelb BD, Giglia TM, Goldmuntz E, McGee G, Sable CA, Srivastava D, Webb CL. Genetic basis for congenital heart defects: current knowledge. Circulation. 2007;115:3015–3038. doi: 10.1161/CIRCULATIONAHA.106.183056. [DOI] [PubMed] [Google Scholar]
- Pöpperl H, Rikhof H, Chang H, Haffter P, Kimmel CB, Moens CB. Lazarus is a novel pbx gene that globally mediates hox gene function in zebrafish. Mol Cell. 2000;6:255–267. doi: 10.1016/s1097-2765(00)00027-7. [DOI] [PubMed] [Google Scholar]
- Pownall ME, Gustafsson MK, Emerson CP., Jr Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu Rev Cell Dev Biol. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- Reiter JF, Alexander J, Rodaway A, Yelon D, Patient R, Holder N, Stainier DY. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 1999;13:2983–2995. doi: 10.1101/gad.13.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley P, Anson-Cartwright L, Cross JC. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nature Genetics. 1998;18:271–275. doi: 10.1038/ng0398-271. [DOI] [PubMed] [Google Scholar]
- Schoenebeck JJ, Keegan BR, Yelon D. Vessel and blood specification override cardiac potential in anterior mesoderm. Dev Cell. 2007;13:254–267. doi: 10.1016/j.devcel.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenebeck JJ, Yelon D. Illuminating cardiac development: Advances in imaging add new dimensions to the utility of zebrafish genetics. Semin Cell Dev Biol. 2007;18:27–35. doi: 10.1016/j.semcdb.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnert AJ, Huq A, Weinstein BM, Walker C, Fishman M, Stainier DY. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat Genet. 2002;31:106–110. doi: 10.1038/ng875. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nature Genetics. 1997;16:154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Olson EN. A genetic blueprint for cardiac development. Nature. 2000;407:221–226. doi: 10.1038/35025190. [DOI] [PubMed] [Google Scholar]
- Stankunas K, Shang C, Twu KY, Kao SC, Jenkins NA, Copeland NG, Sanyal M, Selleri L, Cleary ML, Chang CP. Pbx/Meis deficiencies demonstrate multigenetic origins of congenital heart disease. Circ Res. 2008;103:702–709. doi: 10.1161/CIRCRESAHA.108.175489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- Targoff KL, Schell T, Yelon D. Nkx genes regulate heart tube extension and exert differential effects on ventricular and atrial cell number. Dev Biol. 2008;322:314–321. doi: 10.1016/j.ydbio.2008.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thattaliyath BD, Firulli BA, Firulli AB. The basic-helix-loop-helix transcription factor HAND2 directly regulates transcription of the Atrial Naturetic Peptide gene. J Mol Cell Cardiol. 2002;34:1325–1344. doi: 10.1006/jmcc.2002.2085. [DOI] [PubMed] [Google Scholar]
- Thisse B, Pflumio S, Fürthauer M, Loppin B, Heyer V, Degrave A, Woehl R, Lux A, Steffan T, Charbonnier XQ, Thisse C. Expression of the zebrafish genome during embryogenesis. ZFIN Direct Data Submission. 2001 ( http://zfin.org)
- Thisse C, Thisse B. High throughput expression analysis of ZF-Models Consortium clones. ZFIN Direct Data Submission. 2005 ( http://zfin.org)
- Trinh LA, Yelon D, Stainier DYR. Hand2 regulates epithelial formation during myocardial differentiation. Curr Biol. 2005;15:441–446. doi: 10.1016/j.cub.2004.12.083. [DOI] [PubMed] [Google Scholar]
- Vlachakis N, Ellstron DR, Sagerstrom CG. A novel pbx family member expressed during early zebrafish embryogenesis forms trimeric complexes with Meis3 and Hoxb1b. Dev Dyn. 2000;217:3227–3239. doi: 10.1002/(SICI)1097-0177(200001)217:1<109::AID-DVDY10>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Waskiewicz AJ, Rikhof HA, Moens CB. Eliminating zebrafish pbx proteins reveals a hindbrain ground state. Dev Cell. 2002;3:723–733. doi: 10.1016/s1534-5807(02)00319-2. [DOI] [PubMed] [Google Scholar]
- Waxman JS, Keegan BR, Roberts RW, Poss KD, Yelon D. Hoxb5b acts downstream of retinoic acid signaling in the forelimb field to restrict heart field potential in zebrafish. Dev Cell. 2008;15:923–934. doi: 10.1016/j.devcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg ES, Allende ML, Kelly CS, Abdelhamid A, Murakami T, Andermann P, Doerre OG, Grunwald DJ, Riggleman B. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development. 1996;122:271–280. doi: 10.1242/dev.122.1.271. [DOI] [PubMed] [Google Scholar]
- Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, Blackwell TK, Turner D, Rupp R, Hollenberg S, et al. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- Westerman BA, Murre C, Oudejans CBM. The cellular Pax-Hox-Helix connection. Biochim Biophys Acta. 2003;1629:1–7. doi: 10.1016/j.bbaexp.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Xu X, Meiler SE, Zhong TP, Mohideen M, Crossley DA, Burggren WW, Fishman MC. Cardiomyopathy in zebrafish due to mutation in an alternatively spliced exon of titin. Nat Genet. 2002;30:205–209. doi: 10.1038/ng816. [DOI] [PubMed] [Google Scholar]
- Yelon D, Horne SA, Stainier DYR. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev Biol. 1999;214:23–37. doi: 10.1006/dbio.1999.9406. [DOI] [PubMed] [Google Scholar]
- Yelon D, Ticho B, Halpern ME, Ruvinsky I, Ho RK, Silver LM, Stainier DYR. The bHLH transcription factor Hand2 plays parallel roles in zebrafish heart and pectoral fin development. Development. 2000;127:2573–2582. doi: 10.1242/dev.127.12.2573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1. hand2 RNA rescues myl7 expression in hand2-MO embryos. (A–D) RNA in situ expression of myl7 at the 20s stage in embryos injected with (A) control gfp mRNA, (B) hand2 mRNA, (C), hand2-MO and gfp mRNA, or (D) hand2-MO and hand2 mRNA. Embryos are shown in dorsal view, anterior towards the left. (A) In 20s control embryos, heart precursors form a cone on the embryonic midline. (B) Embryos injected with hand2 mRNA have about wild-type numbers of myl7-expressing cells but heart morphogenesis and cone formation is slightly impaired. (C) Embryos injected with hand2-MO have reduced lateral myl7 domains with defective midline migration. (D) hand2 mRNA largely rescues the hand2-MO phenotype, although most embryos show some impaired morphogenesis. (E) Quantification of the rescue data. Numbers in parentheses denote numbers of embryos. The phenotypic classes correspond to the images shown in panels A–C.