Abstract
Resistance to anoikis, the subtype of apoptosis triggered by lack of adhesion, contributes to malignant transformation and the development of metastasis. Although several lines of evidence suggest that p53 plays a critical role in anoikis, the pathway(s) that connect cell detachment to p53 remain undefined. Here we identify the serine/threonine kinase SIK1 (salt-inducible kinase 1) as a regulator of p53-dependent anoikis through the use of a kinome-wide loss-of-function screen. Inactivation of SIK1 compromised p53 function in anoikis and permitted cells to grow in an anchorage-independent manner. In vivo, SIK1 loss facilitated metastatic spread and survival of disseminated cells as micrometastases in lungs. The presence of functional SIK1 was required for LKB1 activity in regulating p53-dependent anoikis and suppressing anchorage-independent growth, Matrigel invasion, and metastatic potential. In human cancers, decreased SIK1 expression closely correlated with development of distal metastases in breast cancers from three independent cohorts. Together, these findings indicate that SIK1 links LKB1 to p53-dependent anoikis and suppresses metastasis.
INTRODUCTION
Metastasis is the primary cause of cancer death and treatment failure in cancer patients. Although metastasis has been considered a late event in tumor progression, recent studies suggest that it may also occur early in the course of cancer development (1-4). One extreme example of this involves the class of metastatic cancers with unknown primary lesions (CUP), which are likely to result from early metastasis (5).
Adhesion-mediated signaling regulates adherent cell growth and differentiation when cells are in the appropriate tissue context. Loss of adhesion induces anoikis, a specific type of apoptosis that plays a key role in removing cells that have become displaced from their proper location and in thus maintaining tissue and organ homeostasis (6-9). To successfully metastasize, cancer cells must acquire the ability to resist anoikis and survive after detachment from their matrix contacts (10). Various oncogenes have been implicated in rendering cells resistant to anoikis (7), including activated phosphoinositide-3-kinase (PI3K) and its downstream effector Akt [also known as Protein Kinase B (PKB)], activated focal adhesion kinase (FAK), and the neurotrophin receptor TrkB (11-14).
The tumor suppressor p53 plays a central role in coordinating responses to stresses induced by a wide array of stimuli. p53 protein in cells is maintained at basal amounts under normal conditions. In response to genotoxic stresses, such as ultraviolet (UV) or γ-irradiation, p53 abundance rises substantially and triggers either cell cycle arrest or apoptosis (15). However, different stimuli elicit different degrees of p53 response. For example, in response to glucose starvation, adenosine monophosphate (AMP)-activated protein kinase- (AMPK) elicits only a moderate increase in p53 protein, which is nonetheless sufficient to elicit cell cycle arrest (16, 17).
One of the upstream regulators that activates AMPK is the kinase LKB1 (18). LKB1 was initially identified as a gene for which germ line mutations are associated with familial Peutz-Jeghers syndrome (PJS), a condition characterized by the development of hamartomatous polyps in the gastrointestinal tract and enhanced predisposition for tumors (19). In breast carcinoma, loss of LKB1 protein correlates with poor outcome (20) (21). Somatic mutations of LKB1 are frequently found in lung adenocarcinomas and squamous cell carcinomas (22). More recently, ablation of LKB1 in genetically engineered mice implicated LKB1 as a critical barrier to pulmonary metastasis (23). LKB1 is a tumor suppressor kinase implicated in the regulation of multiple biological pathways including those governing angiogenesis, cell cycle arrest, cell polarity, and energy metabolism (24). Notably, LKB1 is also the master upstream regulator of at least 12 other AMPK-related kinases (18), suggesting that it may contribute to tumorigenesis and metastasis through mechanisms other than AMPK regulation.
As a major apoptosis regulator, p53 also plays a critical role in anoikis and metastasis. p53-dependent anoikis has been demonstrated in many cell types (8), including epithelial cells (25-28). Inactivation of p53 promotes metastasis in a number of mouse tumor models. For example, loss of p53 function induces metastases in PyMT- (mouse polyoma virus middle T antigen) driven hepatocellular carcinoma (29) and in K-Ras-driven lung cancers (23). Moreover, combined somatic loss of E-cadherin and p53, but not that of E-cadherin alone, induces resistance to anoikis and the development of invasive skin and breast carcinomas (30). However, the signaling pathway(s) that regulate p53-dependent anoikis and control metastasis is largely unknown. To identify regulators of p53-mediated anoikis, we performed a kinome-wide, RNAi-based loss-of-function screen to identify kinases that regulate anchorage-independent (AI) growth. Here, we report that SIK1 (salt-inducible kinase 1) is an essential upstream regulator of p53-mediated anoikis that links the tumor suppressors LKB1 and p53, acts to suppress metastasis in experimental models, and is correlated with clinical outcome in human cancer.
RESULTS
A loss-of-function screen identifies SIK1, a kinase that suppresses the AI growth of HMECs
We showed previously that activation of the PI3K signaling pathway can transform human telomerase reverse transcriptase (hTERT) catalytic subunit-immortalized human mammary epithelial cells (tHMECs) (31, 32). Notably, in this experimental model, even though the tHMECs have lost expression of the gene encoding cyclin-dependent kinase 4 inhibitor A (p16INK4a) and show increased abundance of endogenous c-Myc (33-35), inactivation of p53 is required in combination with PI3K activation to overcome anoikis and permit growth of colonies on soft agar, a form of anchorage independent (AI) growth (31, 32). In these and related studies, functional inactivation of p53 was accomplished through expression of the SV40 large T antigen (LT), or of a dominantly interfering mutant of p53 (p53DD), or by suppressing p53 expression with short hairpin RNAs (shRNAs)(31, 32, 36).
To identify kinases that regulate p53 activity when cells undergo AI-growth, we employed this same experimental model together with lentiviral delivery of a shRNA library targeting the human kinome (Fig. 1A). A pool of lentiviruses containing ~2500 shRNAs targeting 488 human kinases, as well as p53, was generated and introduced into tHMEC cells expressing a constitutively activated tumor mutant allele of PIK3CA, PIK3CA-H1047R , referred to as tHMEC-P cells. After 4 weeks, AI colonies were picked, and vector-specific primers were used to identify the integrated shRNAs harbored by the cells derived from these colonies. As expected, we found that the p53-specific shRNA (shTP53) scored multiple times in each screen, validating this approach. We also identified a number of kinases that scored multiple times (Table S1), some of which have already been implicated in regulating p53 function, apoptosis, or both (37-41). Only one candidate gene targeted by multiple shRNAs scored more frequently than did p53 (Table S1). This gene encodes the serine/threonine kinase SIK1 (also known as salt-inducible kinase1, SIK, SNF1LK, or MSK), a member of the AMPK-related kinase family (18).
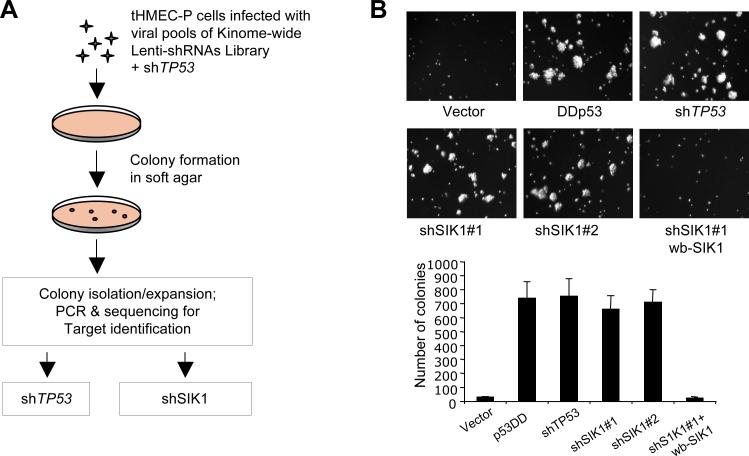
Fig. 1. Identification of SIK1 as a suppressor of AI-growth in HMECs.
(A) Experimental outline for the kinome-wide shRNA screen that identified SIK1 as a suppressor of AI-growth of tHMEC-P cells. (B) AI colony formation of tHMEC-P cells expressing various cDNAs or shRNAs as indicated, and cells expressing shSIK1#1 with add-back of a wobble SIK1 (wb-SIK1). The number of colonies is per 60 mm plate. Results are shown as the mean ± SD for 3 independent experiments.
We confirmed that two of the SIK1-specific shRNAs (shSIK1#1 and shSIK1#2) that scored in the screen decreased SIK1 mRNA and protein abundance (fig. S1). tHMEC-P cells that expressed either of these two shSIK1s formed AI colonies as robustly as cells harboring an shRNA targeting p53 (shTP53) or the dominant interfering mutant p53DD (43) (Fig. 1B). To eliminate the possibility that the observed effects were due to off-target effects of RNAi, we generated a retroviral expression vector encoding a “wobble” mutant of SIK1 that is insensitive to silencing by shSIK1#1. Expression of this wobble SIK1 in tHMEC-P harboring shSIK1#1 inhibited AI growth (Fig. 1B), suggesting that the induced AI growth specifically derived from SIK1 loss.
SIK1 catalytic activity is required for p53 phosphorylation in response to loss of adhesion
Because p53 abundance increases when cells detach from the extracellular matrix (ECM) (28, 44), we investigated the effect of SIK1 loss on p53 abundance when HMECs were shifted to suspension culture. The abundance of endogenous p53 increased moderately in cells expressing a control shRNA that targeted luciferase (shLuc) (Fig. 2A), consistent with previous findings in MCF7 and squamous cancer cells (28, 44). In contrast, induction of p53 was not observed when tHMECs expressing shSIK1 were shifted to suspension culture (Fig. 2A). To explore the relationship between SIK1 and p53 abundance in cells undergoing AI growth, we examined the abundance of p53 in tHMECs derived from AI colonies resulting from expression of shSIK1s. Strikingly, tHMEC-P-shSIK1 cells derived from AI colonies showed greatly diminished amounts of p53 protein, comparable to those of cells carrying shTP53, whereas expression of the wobble SIK1 restored p53 abundance in these AI colony-derived cells to amounts comparable to those in cells grown under attached conditions (Fig. 2B). Moreover, the p53 response to DNA-damaging agent adriamycin remained intact in SIK1 deficient cells (Fig. 2B). These data suggest that loss of SIK1 renders cells resistant to anoikis by attenuating the increase in p53 abundance that takes place in response to cell detachment. Notably, p53 mRNA abundance was not altered in cells by shSIK1 (fig. S2), suggesting that p53 is modulated post-translationally.
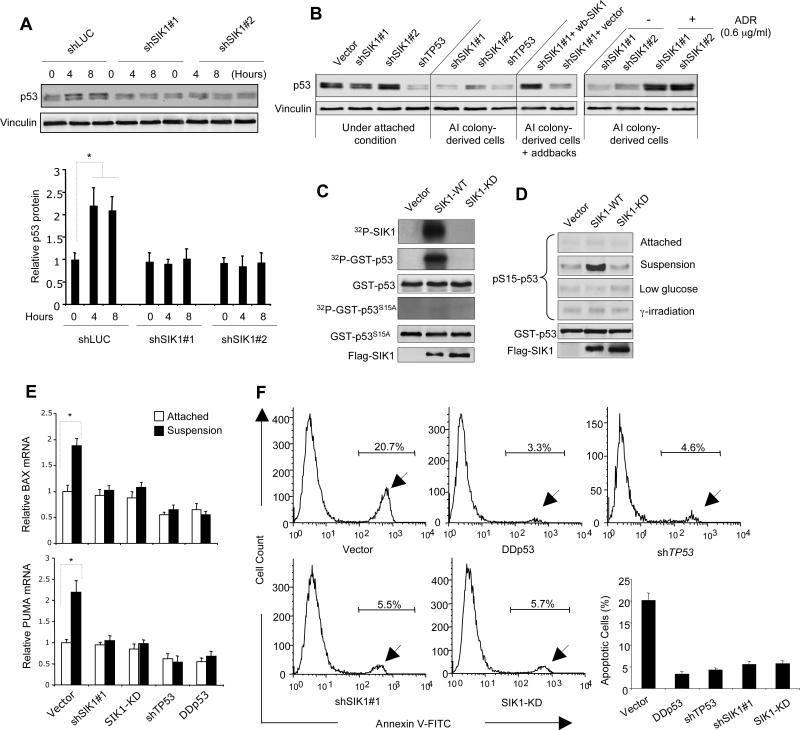
Fig. 2. SIK1 is required for p53-dependent anoikis.
(A) p53 protein abundance in HMECs expressing shSIK1s or shLuc cultured under attached or single cell suspension conditions for 4 or 8 hours, were examined by immunoblot analysis. Vinculin used as a loading control. Results are shown as the mean ± SD for 3 independent experiments. * p <0.01 (t-test) (B) Immunoblot analysis of p53 abundance in various tHMEC-P cell lines as indicated, as well as in colony-derived tHMEC-P-shSIK1 cells treated with adriamycin (ADR) (4.5 hr). wb-SIK1 (wobble SIK1), or vector, was expressed in AI colony-derived tHMEC-P-shSIK1#1 cells as indicated. (C) Kinase assays were used to detect auto-phosphorylation of SIK1 and phosphorylation of GST-p53 or GST-p53 S15A by Flag-tagged SIK1-WT or SIK1-KD immunoprecipitates. Radioactive signals were detected by phosphorimaging. (D) Phosphorylation of p53 at Ser15 was determined by immunoblot following kinase assay using Flag-tagged SIK1 immunocomplexes (WT or KD) from U2OS cells subjected to different conditions. (E) PUMA and BAX expression in tHMEC-P cells expressing shSIK1#1, SIK1-KD, shTP53, or p53DD cultured attached or in suspension for 16 hours was determined by real-time RT-PCR. Mean ± SD for 3 independent experiments are shown. * p <0.01 (t-test) (F) Inactivation of SIK1 renders cells resistant to anoikis. tHMEC-P cells were cultured in suspension for 2 days, and the percentage of apoptotic cells expressing various cDNAs or shRNAs was determined by Annexin-V staining. Mean ± SD for 3 independent experiments are shown. * p <0.001 (t-test).
Phosphorylation of p53 in response to several distinct types of cell stress promotes p53 stabilization (45). To investigate the possibility that the kinase activity of SIK1 may be involved in regulating p53 through phosphorylation, we constructed Flag-epitope-tagged versions of wild-type (WT) SIK1 and of a catalytically inactive mutant (SIK1-KD) in which lysine 56 is substituted with methionine (K56M) (46) and expressed them in U2OS human osteosarcoma cells. We first performed an in vitro SIK1 kinase assay using SIK1 immune complexes isolated from cells cultured in suspension, with a glutathione-S-transferase (GST)-p53 fusion protein containing the amino-terminus of human p53 (amino acids 1-105) (47) as a substrate. Both auto-phosphorylation and phosphorylation of the p53 fragment (Fig. 2C) were apparent in assays using immune complexes containing WT SIK1 but not those containing the SIK1-KD mutant. Several types of cellular stress stimulate p53 activation through phosphorylation of serine 15 (S15) (48). For instance, the ATM (ataxia telangiectasia mutated)-ATR (ataxia telangiectasia and Rad3 related)-CHK kinases perform this task in response to DNA damage. More germane here, AMPK regulates phosphorylation of S15 in response to low glucose (16, 17, 49), raising the possibility that SIK 1 is involved in the phosphorylation of this site when cells lose adhesion. Indeed, a form of the GST-p53 substrate containing a serine to alanine mutation at 15 (S15A) was not phosphorylated (Fig 2C), suggesting that Ser15 is targeted by the SIK1 immune complex. To test this hypothesis further, we expressed Flag-epitope-tagged versions of WT or SIK1-KD in U2OS cells and performed additional kinase assays followed by immunoblot analysis with antibody that specifically recognizes p53 phosphorylated at Ser15 (p-S15 p53). Immune complexes isolated from cells expressing WT SIK1, but not the SIK1-KD mutant, phosphorylated p53 at S15, when isolated from cells growing under suspension conditions (Fig. 2D). In contrast, immune complexes of WT SIK1 isolated from cells subjected to γ-irradiation or glucose deprivation failed to phosphorylate p53 at S15 (Fig. 2D). Because the p53 Ser15 motif does not match the optimal consensus motif for SIK1 phosphorylation, it is possible that a second kinase associated with the SIK1 immune complex may directly phosphorylate p53. However, our findings clearly suggest that SIK1 kinase activity is necessary for p53 phosphorylation in response to loss of adhesion.
SIK1 is required for the induction of p53-dependent anoikis
We next sought to determine whether SIK1 is required for the functional activation of p53 in apoptosis induced by cell detachment. Specifically, we asked whether transcriptional targets of p53 were upregulated in anoikis by assessing changes in the expression of the p53 target genes, BAX and PUMA (Fig. 2E). Using quantitative reverse transcription polymerase chain reaction (RT-PCR) analysis, we found increased BAX and PUMA expression in control cells grown in suspension (Fig. 2E). In contrast, we did not find increased BAX or PUMA mRNA in cells deficient in p53 or SIK1 activity (Fig. 2E), indicating that SIK1 is involved in the regulation of p53-dependent anoikis.
To further examine the role of SIK1 in anoikis, we plated tHMEC-P cells in suspension on plates coated with a layer of agar. In the absence of cell adhesion, the percentage of apoptotic cells after 2 days of suspension culture, as measured by annexin V-FITC staining followed by flow cytometric analysis, was reduced in tHMEC-P cells expressing a dominant-interfering p53 mutant (p53DD), shTP53, shSIK1#1, or SIK1-KD compared to that in cells infected with the control vector (Fig. 2F). Interestingly, the SIK1-KD mutant acted in a dominant-interfering fashion, suppressing anoikis in cells undergoing loss of attachment. Using an AI colony formation assay, we confirmed that expression of SIK1-KD but not WT SIK1 permitted robust colony formation of tHMEC-P cells in soft agar (fig. S3), consistent with the effects seen with the shSIK1s (Fig.1B). Together, these studies suggest that loss of SIK1 activity rendered cells resistant to anoikis.
Loss of SIK1 facilitates metastatic spread in the absence of primary lesions
tHMECs carrying the oncogenes PIK3CA and H-Ras, referred to as tHMEC-PR cells, only form tumors in immunodeficient mice if p53 is also inactivated (31). To see whether inactivation of SIK1 can substitute for loss of p53 function in tumor formation in vivo, we re-engineered tHMEC-PR cells to express shSIK1 or the SIK1-KD mutant instead of p53DD or shTP53, and implanted these cells in mammary fat pads. Whereas tHMEC-PR cells expressing p53DD or shTP53 formed tumors in all cases within 2 months (Fig. 3A), inactivation of SIK1 did not lead to efficient primary tumor formation; we noted only one tumor, which formed after a long latency (4 months), in each of the mouse groups injected with cells expressing shSIK1 or SIK1-KD of Experiment A (Fig. 3A). The remaining mice, which had been injected with HMEC deficient in SIK1 activity and had remained free of primary tumors, were sacrificed at the end of 6 months and subjected to necropsy and histopathological analysis.
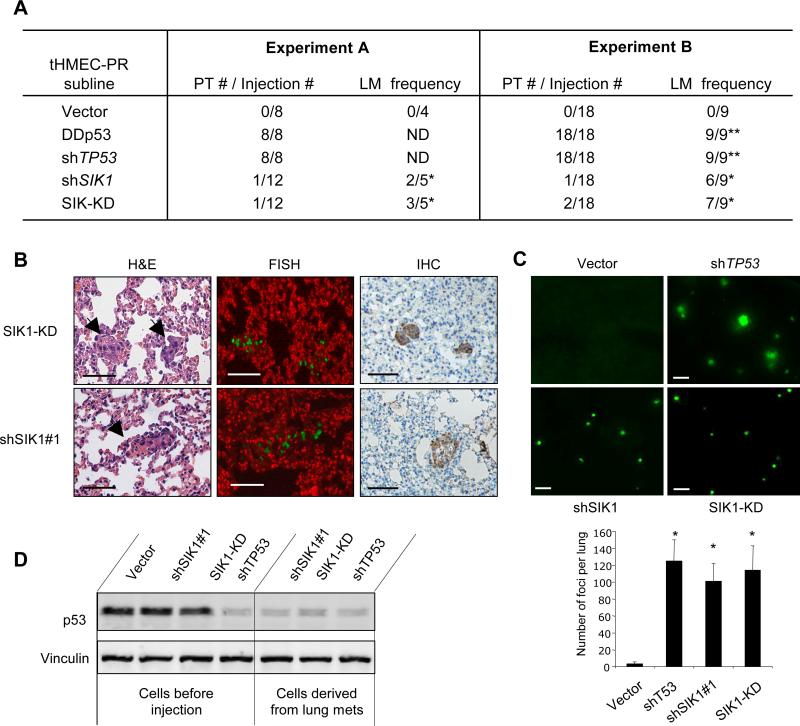
Fig. 3. Loss of SIK1 contributes to metastasis.
(A) Orthotopic xenograft assay. 2 × 106 cells from the indicated population were resuspended in a final volume of 120 μl and injected into each inguinal mammary fat pad of athymic mice. When the primary tumor size reached 1.8cm, mice were sacrificed (Experiment A) or subjected to surgical removal of the tumors (Experiment B). After 6 months, all of experimental mice were sacrificed and subjected to histopathological examination of the lungs. PT, primary tumor; LM, lung metastasis; ND, not determined. * micrometastases; ** macrometasteses. (B) Representative images for lung micrometastases by H&E staining (left panels), genomic FISH with mouse Cot1 DNA (red) and human Cot1 DNA (green) (middle panels), and IHC with anti-human cytokeratin (right panels). Upper and lower panels show pulmonary micrometastases from mice injected with tHMEC-P cells expressing SIK1-KD or shSIK1 as indicated. Scale bars, 50 mm. (C) Experimental metastasis assay. Representative fluorescence images of lungs of mice injected through tail veins with GFP-labeled tHMEC-P cells expressing various constructs. Number of foci in each lung was scored under fluorescence microscope and data are shown as mean ± S.E.M. (n=6-8), * p <0.001 (t-test). Scale bars, 0.5 mm. (D) Western analysis of p53 abundance in tHMEC-PR cells expressing shTP53, shSIK1#1, or SIK1-KD maintained in attached culture or derived from lung metastases in NOD/SCID mice.
Surprisingly, although we found no primary tumors, we observed small metastatic lesions in the lungs of the mice injected with cells expressing shSIK1 or the SIK1 KD mutant (Fig. 3A and B). To confirm that these microscopic pulmonary lesions had indeed originated from the injected HMECs, we performed genomic fluorescent in situ hybridization (FISH) using a human Cot1 probe (green) (Fig. 3B, middle panels) and immunohistochemical analysis (IHC) with anti-human cytokeratin (Fig. 3B, right panels) on paraffin sections prepared from the same lung samples, and found that the cells from these lesions were of human origin. In a separate experiment with larger cohorts of animals, we found that all mice injected with cells deficient in p53 function developed primary tumors as well as macroscopic metastases in lungs (Fig. 3A, Experiment B). In contrast, of the mice injected with cells expressing either shSIK1 or SIK-KD, most had microscopic metastases in their lungs but no primary tumors (Fig. 3A, Experiment B).
Loss of SIK1 permits formation of micrometastases
To compare the effects of SIK1 with those of p53 in suppressing metastasis, we employed a tail vein assay for experimental metastasis. Specifically, we injected green fluorescent protein (GFP)-labeled tHMEC-PR cells harboring shTP53, shSIK1, or SIK1-KD into the tail veins of non-obese diabetic/severe combined immunodeficiency (NOD/SCID) mice, and then isolated the lungs of these animals to assess the number of metastatic foci six weeks after injection. Notably, cells deficient in either p53 or SIK1 activity formed a similar number of foci in the lungs (Fig. 3C). Consistent with the observations obtained from the orthotopic xenograft study described above (Fig. 3A), we found that cells lacking p53 developed larger nodules than those expressing shSIK1 or SIK1-KD, which formed only microscopic foci (Fig. 3C).
To determine if the micrometastases formed by cells lacking SIK1 activity could develop into large nodules after a longer period of time, two additional cohorts of mice injected with tHMEC-PR cells expressing shSIK1 or SIK1-KD were housed for 16 weeks before their lungs were isolated for examination. Indeed, a few large nodules were observed in the lungs of most of the mice injected with cells expressing shSIK1 or SIK1-KD (Table S2). Strikingly, we found greatly reduced amounts of p53 protein in nodules derived from SIK1- deficient cells compared to that in comparable cells that had not been inoculated into animals (Fig. 3D), reminiscent of the reduced p53 abundance in colony-derived cells lacking SIK1 (Fig. 2B). These observations suggest that inactivation of SIK1 attenuates the increase in p53 activity in response to loss of adhesion and allows cells to escape anoikis and survive as micrometastases, but is not sufficient to allow cells to propagate into large tumors in vivo. The prolonged survival afforded by SIK1 loss may provide a selective advantage to these cells by allowing them to acquire additional mutations (involving p53 or other genes) necessary to allow growth of these micrometastases into sizable tumors.
Activation of SIK1 restores p53 abundance and anoikis to LKB1-deficient cells
The LKB1 tumor suppressor provides a critical barrier to cancer metastasis (21, 23, 50) and functions in part through the regulation of p53 (51, 52). Suppression of LKB1 expression with LKB1-specific shRNAs (shLKB1) in tHMEC-P cells allowed these cells to form AI colonies (Fig. 4A). Because a previous study reported that LKB1-null cells display decreased basal p53 abundance following serial passage in culture (51), we also assessed p53 abundance in anchorage-independent colonies derived from tHMEC-P-shLKB1 cells. A decrease in basal p53 abundance was apparent in LKB1-deficient colonies as compared to comparable cells cultured under attached conditions (Fig. 4B), similar to the decrease in p53 seen in AI colony-derived tHMECs-P-shSIK1 cells (Fig. 2B).
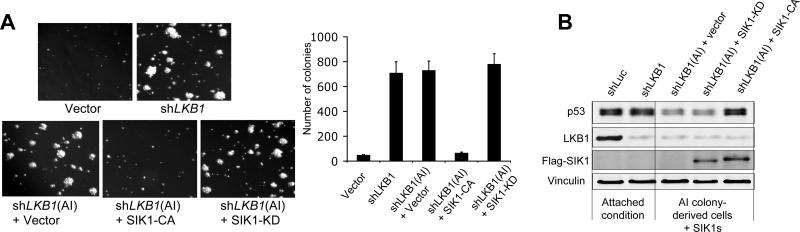
Fig. 4. SIK1 activation suppresses AI growth of LKB1-deficient HMECs.
(A) Colony formation assays of tHMEC-P cells expressing shLKB1 or shLuc. SIK1-CA, but not SIK1-KD, suppressed AI growth of AI-derived tHMEC-P-shLKB1 cells. Mean ± SD for 3 independent experiments are shown. (B) Western blot analysis of p53 and LKB1 abundance in tHMEC-P cells expressing shLKB1 grown in attached culture or AI-derived tHMEC-P-shLKB1 cells expressing SIK1-KD or -CA as indicated. Flag-tagged SIK1 (KD or CA) HMECs was probed with anti-Flag antibody.
Because LKB1 is a key upstream activating kinase for SIK1 (18), we asked whether SIK1 links LKB1 to p53-dependent anoikis. We derived cells from tHMEC-P-shLKB1 colonies and stably introduced a constitutively activated form of SIK1, the T-loop mutant T172E (18) (SIK1-CA) or, as a negative control, the SIK1-KD mutant. Expression of SIK1-CA, but not SIK1-KD, restored p53 to normal amounts in these AI-derived LKB1-deficient cells and abolished their ability to form AI colonies (Fig. 4A and B), suggesting that the LKB1-SIK1 axis is involved in regulation of p53 abundance and apoptosis with loss of adhesion.
SIK1 couples LKB1 to p53-dependent suppression of metastasis
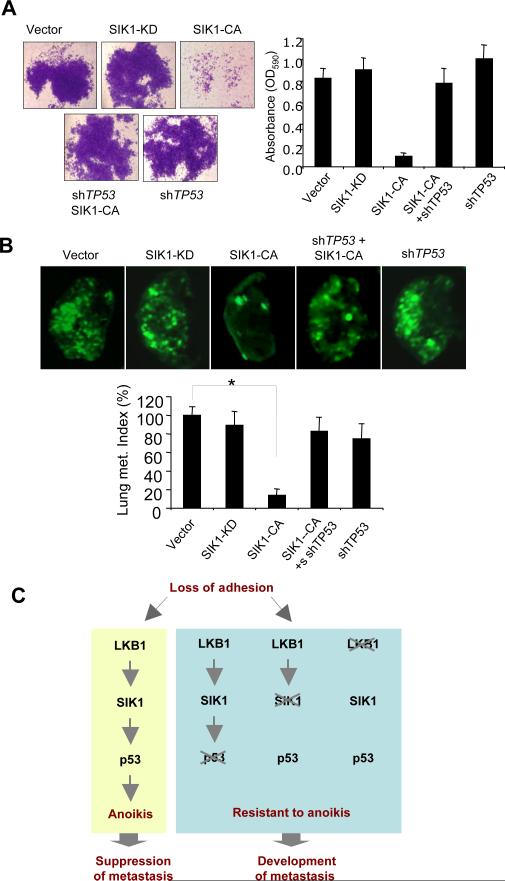
To further examine whether SIK1 mediates LKB1 signals leading to p53-dependent suppression of tumor metastasis, we selected an A549 lung cancer cell line that lacks LKB1 expression but retains wild-type p53 to perform reconstitution/disruption experiments. Reconstitution of A549 cells with LKB1-WT inhibits their AI growth and metastatic potential (23). To determine whether activation of SIK1 in A549 cells had a similar effect, we introduced SIK1-CA and SIK1-KD into A549 cells and subjected them to AI growth, Matrigel invasion, and experimental metastasis assays. Expression of SIK1-CA in these LKB1-null cells markedly suppressed Matrigel invasion and pulmonary metastasis, whereas SIK1-KD expression had no effect (Fig. 5A and B). To further assess whether p53 is required for SIK1 function, we introduced shTP53 into A549 cells expressing SIK1-CA. Notably, removal of p53 abolished the effect of SIK1-CA on suppression of invasiveness and metastatic potential (Fig. 5A and B). Together, our findings establish the connections among LKB1, SIK1, and the p53 pathway in the regulation of cancer invasiveness and metastasis (Fig. 5C).
Fig. 5. SIK1 links LKB1 to p53-dependent suppression of metastasis.
(A) A549 cells expressing various versions of SIK1 with or without co-expression of shTP53 were subjected to Matrigel invasion assay. Mean ± SD for 3 independent experiments. (B) GFP-labeled A549 cells expressing various constructs as indicated were injected into tail veins of NOD/SCID mice and lung metastasis was assessed 6 weeks post injection. Representative fluorescence images of mouse lungs are shown. Lung metastasis burden was shown as indices pooled with each group of mice expressed as % (± SEM) over controls (n=6-9), * p < 0.001 (t-test). (C) Summary of LKB1-SIK1 pathway that mediates p53-dependent anoikis and suppresses metastasis.
Loss of SIK1 correlates with poor clinical outcome in breast cancers
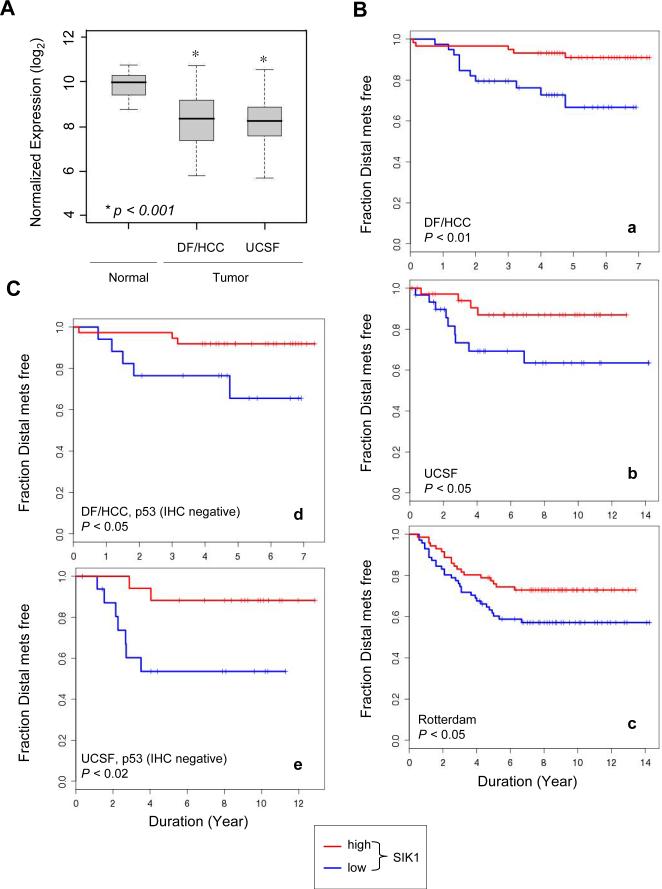
Although a small number of somatic mutations in SIK1 have been identified in gliomas, melanomas, and lung cancers (53), we found that SIK1 expression was significantly lower in primary breast tumors than in normal breast tissues in data from two independent cohorts (Fig. 6A) (54, 55). To explore whether SIK1 expression correlates with clinical outcome, we analyzed SIK1 expression profiles in tumor samples from three independent cohorts of breast cancer patients at the Dana-Farber/Harvard Cancer Center (DF/HCC), USCF and Rotterdam cancer centers with up to 7, 14, and 14 years of clinical follow-up, respectively (55-57). In all three cohorts, we found that low SIK1 expression was associated with a significantly shorter interval before the appearance of distal metastases (Fig. 6B, panels a, b, and c). We further examined subsets of breast cancers from the DF/HCC and UCSF cohorts (the Rotterdam database lacks p53 status) that were negative for p53 immunohistochemical (IHC) staining, which usually identifies tumors harboring wild-type (WT) p53 (55, 58). We found that low SIK1 expression in both subsets of p53-negative patients showed close correlation with higher distal recurrence rate (Fig. 6C, panels d and e). In addition, we found an ovarian cancer cohort with a significant correlation of lower SIK1 abundance with higher malignant grade that is associated with higher metastatic potential (fig. S4), suggesting that low SIK1 expression may be associated with progression in other cancer types.
Fig. 6. SIK1 correlation with clinical outcome in human cancer.
(A) SIK1 expression in normal breast or tumor samples from the DF/HCC or the UCSF cohorts is shown in boxplots. The mean for each group is indicated by the black center line, the first and third quantiles (the inter-quantile range, IQR) are indicated by the edges of the grey area. The extreme values (within 1.5 times the IQR from the upper or lower quantile) are indicated by the ends of the lines extending from the IQR. The difference in log2 expression ratios between normal and tumor samples was tested with Welch's t-test, p < 0.001. (B) and (C) Kaplan-Meier plots were used to assess the association of the levels of SIK1 transcripts with development of distal metastases. Samples were grouped as described in Materials and Methods. a, the DF/HCC cohort of breast cancer patients (n=133). b, the UCSF cohort of breast cancer patients (n=112). c, the Rotterdam cohort of breast cancer patients (n= 286). d, the subset of DF/HCC breast cancer patients whose tumors are negative for p53 IHC staining (n=76). e, the subset of UCSF breast cancer patients whose tumors are negative for p53 IHC staining (n=57).
DISCUSSION
SIK1 and p53
Using a loss-of-function screening approach, we identified SIK1 as a kinase that when suppressed substitutes for p53 inactivation to permit immortalized HMECs expressing PIK3CA-H1047R to grow anchorage-independently. Although the ability to form AI colonies usually correlates with tumorigenic potential, our study demonstrated that, whereas loss of p53 allowed the formation of both primary and metastatic tumors, loss of SIK1 facilitated the metastatic spread of cells, allowing them to lodge in distant tissues as micrometastases, but did not promote frank tumorigenesis. Our findings suggest that, because p53 performs several tumor suppressing functions and only one of these depends on SIK1, micrometastases formed due to SIK1 loss would still need to lose p53 or other p53 functions to grow into clinically manifest tumors.
SIK1 is a link between LKB1 and p53
Recent studies have demonstrated that loss of LKB1 is associated with invasiveness and metastasis in breast, lung, and endometrial adenocarcinomas (23, 31, 32, 50). However, the mechanism by which LKB1 contributes to the suppression of tumor formation and metastases development has remained elusive. Notably, LKB1 regulates the AMPK family of 13 kinases (18). Among these, AMPKa2 is the best characterized kinase, acting downstream of LKB1 to regulate mTOR signaling (59, 60). Jones et al. found that AMPKa2 is required for p53-dependent growth arrest in response to low glucose (16). However, it is not clear how AMPK links LKB1 to tumor suppression.
Several studies have associated LKB1 function and p53 activity (51, 52), but the mechanistic link between these two tumor suppressors remains elusive. Herein, using biochemical, genetic, and functional analyses both in cell culture and experimental animal models, we have shown that SIK1 acts as an important link that couples LKB1 to p53-mediated anoikis and suppression of metastasis. Recent studies have shown that SIK1 is also involved in other cellular activities, such as regulation of active sodium transport in response to sodium stress (61), CREB-(cAMP response element-binding protein) mediated phosphorylation of TORC2 (transducer of regulated CREB activity 2) and class II histone deacetylases (HDACs) (42, 62), and regulation of transforming growth factor β (TGFβ) signaling (63). However, it is not clear how these other activities of SIK1 correlate with the role described in the current study. In aggregate, our findings establish the LKB1-SIK1-p53 axis as a potential clinically important pathway in the development of metastatic disease.
SIK1 and metastatic cancers
It is widely believed that metastasis is a late event in cancer progression, and that metastases are derived from primary tumors that become invasive. However, recent studies have challenged this notion, showing that metastasis can occur early in the course of tumor development, even in the absence of detectable primary tumors (2-4). Indeed, approximately 10% of all cancer patients present with CUP at the time of diagnosis (5, 64). In some of these cases, a primary tumor becomes apparent with further study, particularly with the implementation of more sensitive imaging technology. However, in approximately 3% of all cancer patients, the primary site is never identified (64, 65).
Although it has been postulated that some primary tumors may regress completely and leave their metastases behind, our finding provides an alternative explanation for this longstanding clinical mystery. Our observations suggest that some precancerous cells, such as the SIK1-deficient cells examined here, escape anoikis to disseminate and reside in distant tissues and organs before primary tumor formation. The prolonged survival of these precancerous cells increases the risk for them to acquire additional oncogenic alternations that promote the development of malignant metastatic lesions. If the precancerous cells residing at secondary sites acquire additional “hit(s)” and progress into malignancies before the cells at the primary site develop into a tumor, CUP would occur. Consistent with this hypothesis, an epidemiological survey of more than 12,000 breast cancer patients indicated that metastatic spread had likely initiated several years prior to clinical diagnosis of primary cancer (66). Thus, metastases can develop before, after, or parallel to the primary tumor. Indeed, recent studies also found that certain genes are involved in primary tumor development whereas others promote metastasis (67, 68), indicating that metastasis can be regulated independently from tumor formation. Advancing our understanding of the genetic mechanisms underlying metastasis and identifying predictive markers for this process should prove important for both treatment and prevention of cancer.
Materials and Methods
Kinome-wide shRNA screen
tHMEC-P cells were infected with a lentiviral shRNA library with viral titer ~1.0X107 infectious particles/ml constructed in pLK0.1 consisting of ~2500 shRNAs directed against ~488 human kinases (provided by the RNAi Consortium, Broad Institute, Cambridge, MA) (69). After infection, cells were briefly selected with puromycin and then subjected to a colony formation assay. Genomic DNA from AI colonies was prepared for PCR amplification using the pLKO forward CAAGGCTGTTAGAGAGATAATTGGA and reverse GACGTGAAGAATGTGCGAGA primers. The PCR-amplified products were sequenced to identify the integrated shRNAs.
Plasmid constructs and cell culture
The cDNA clone of SIK1 (Origene) was subcloned into pWzl-Blast by PCR with primers containing Flag epitope-encoding sequence to generate Flag-tagged SIK1. The SIK1 mutant alleles, KD (K56M) and CA (T182E), were generated by site-directed mutagenesis (Stratagene). All of these constructs were verified by sequence analysis. tHMEC lines stably expressing shSIK1s, shLKB1, or various alleles of SIK1 were generated by viral infection. Culture conditions of tHMECs were as described previously (32).
Preparation of Immune complexes and in vitro SIK1 kinase assay
U2OS cells were transfected with 10μg of SIK1 plasmid (SIK1 WT or KD) or control plasmid. At 24 hr post transfection, cells were subjected to conditions for suspension culture, low glucose (0.5 mM Glucose) or γ-irradiation (5Gy), and 18 hrs later they were harvested and lysed in RIPA (Radio Immuno Precipitation Assay) buffer as described above and the debris was cleared by centrifugation. The supernatant fraction was incubated with anti-Flag M2 agarose beads (Sigma) for 2 hr. The beads were washed and used in a kinase reaction containing 50mM Tris HCl pH7.5, 5 mM MgCl2, 1mM NaF, 10mM β-glycerolphosphate, 1mM DTT, 0.2 mM ATP with 0.5μCi/μl (γ-32P) ATP (for hot reaction) or 100μM ATP (for cold reaction) and 1 μg of recombinant GST-p53 fusion proteins containing the first 105 amino acids of human p53 (GST-p53) or (GST-p53S15A) as substrates. GST-p53 proteins were purified from E. coli cultures with GST-Sepharose beads (Amersham Biosciences).(16)
Immunoblotting and antibodies
Cells were lysed in RIPA buffer (Pierce) supplemented with protease and phosphatase inhibitors as described (32). Cell lysates were cleared by centrifugation and proteins were resolved by SDS-PAGE and transferred to nitrocellulose. Blots were probed using the following antibodies: anti-SIK1 (Abgent), anti-p53 (DO-1) (Santa Cruz biotechnology), anti-LKB1 (Upstate biotechnology), anti-Flag and anti-vinculin (Sigma) and visualized by using Odyssey Scanner (Li-Cor, Lincoln, NE) as described (31).
AI colony-formation assay
For AI colony formation assays, 5 × 104 cells were plated into each 60-mm plate with a bottom layer of 0.6% agar and a top layer of 0.3% agar. For library screening, 5 × 105 cells were seeded into each 100-mm plate. Colonies were counted after 4 weeks and analyzed as described (32) or isolated for expansion or analysis (70).
Suspension culture and Anoikis assay
Cells were plated at a density of 105 cells/well in 6-well plates coated with a layer of 1% agar. Two days later, the apoptotic cell population was determined using FITC-conjugated Annexin-V (BD Biosciences) and FACS analysis as described (30).
Orthotopic xenograft assay and histological analysis
Six- to eight-week-old Balb/c nude mice (CanN.Cg-Foxn1nu Crl, Charles River Laboratories) were γ-irradiated with one dose of 400 rad and subjected to mammary fat pad injections with HMECs as described previously (31). The mice were monitored routinely for tumor growth and were either sacrificed (Experiment A) or resected (Experiment B) when the primary tumor reached 1.8cm in diameter or after 6 months. 6 months post injection, any remaining mice were sacrificed and fixed in 10% buffered formalin for histopathological examination of mammary, brain, liver, lung, and ovary tissues at DF/HCC Rodent Histopathology Core. IHC analysis was performed with anti-human cytokeratin clone AE1/AE3 (Dako North America Inc.) at DF/HCC Specialized HistoPathology Core.
Experimental metastasis assay
GFP-labeled HMEC (1×106 re-suspended in PBS) or A549 cells (5×105 re-suspended in PBS) were injected into the lateral tail vein of NOD/SCID mice (Charles River Laboratories). Mice were sacrificed 6 or 16 weeks post injection and the lungs were dissected for fluorescence microscopic and histopathological examination. The lung metastasis burden was analyzed by scanning the fluorescence intensity of all lobes of each lung using KODAK Image Station 4000MM Imaging System. The total intensity of green fluorescence over each lung area was computed by Kodak Molecular Imaging software (version 4.5.0b6 SE) to obtain mean values of fluorescence intensity. The mean fluorescence intensity of the lungs from mice bearing vector-expressing cells was used as a control (defined as 100%). The lung metastasis index for each mouse was calculated as percentage of the mean fluorescence intensity over the control.
Matrigel invasion assay
Assays were performed in Boyden chambers with 8 μm pore filter inserts for 24-well plates (BD Bioscience). A549 cells (0.5 × 105) in 200 μl of serum-free medium were added to the upper chamber with the filters pre-coated with matrigel (BD Bioscience). The lower chamber was filled with 300 μl of medium containing 10% FBS as attractant. After 48 hours incubation at 37°C, cells on the underside of the filter were fixed and stained with 0.5% crystal violet. Cells that remained in the gel or attached to the upper side of the filter were removed with cotton swabs. Cells on the underside of the filter were examined by light microscopy and subsequently extracted with 10% citric acid and the optical density was measured at 590 nm by spectrophotometer for quantitation of cell number.
Genomic Fluorescence in Situ Hybridization (FISH)
Paraffin sections of lung tissues were assayed for the presence of mouse and human cells using FISH with species-specific genomic probes as described (71). Briefly, sections were denatured and hybridized with probes against SpectrumGreen-dUTP-labeled human Cot-1 DNA (Invitrogen) and SpectrumRed-dUTP-labeled mouse Cot-1 DNA (Invitrogen). Fluorescence-labeled probes were generated by nick translation (Abott Molecular).
Real-time RT-PCR
Total RNA was purified using the RNeasy kit (Qiagen). 2 micrograms of total purified RNA was subjected to a reverse transcriptase reaction according to high capacity cDNA reverse transcription kit (Applied Biosystems). cDNA corresponding to approximately 10 ng of starting RNA was used for each of four replicates for quantitative PCR. Human PUMA, BAX and GAPDH (as an endogenous control) were amplified with commercially designed Taqman gene expression assays (Applied Biosystems) and the Taqman universal PCR master mix (Applied Biosystems). Quantitative expression data were acquired and analyzed using a 7300 Real-Time PCR System (Applied Biosystems).
Statistical analysis
SIK1 gene expression and p53 status (determined by IHC analysis) of the UCSF cohort of breast cancer samples were retrieved from the website http://caarraydb.nci.nih.gov/caarray (55). In this data set of Affmetrix U133, SIK1 transcript units in tumors vary from 51 to 1506 (mean 378) with no normal reference available. However, SIK1 transcript units in another breast cancer data set of U133 platform (NCBI GEO database accession number GSE3744) (54, 56) showed comparable values, ranging from 74 to 1042 (mean 326) in tumors, and 278 to 2669 (mean 1154) in normal human breast tissues. All stage II-III (n=117) samples, including a subset of p53 wild-type samples (n=57), were assigned into two groups according to SIK1 transcript levels: low (≤ 200) and high (≥ 400). Stage IV samples and three samples with unknown clinical stage information were excluded from analysis. Information on SIK1 gene expression for the Rotterdam cohort of breast cancer samples (n=286) that were lymph node negative at presentation were retrieved from Rotterdam cohort (57). The mean of transcript units was 1040, with no normal reference available. Samples were assigned into two groups: SIK1-low (cutoff-bottom 25%) and SIK1-high (cutoff-top 25%). Distal metastases recurrence curves were derived from the Kaplan-Meier method and the degree of difference among groups was analyzed by log-rank test. R package version 2.4.1 was used for statistical computing.
Supplementary Material
Footnotes
One sentence summary: SIK1 loss suppresses anoikis and promotes metastasis in the absence of primary tumors.
Editor's Summary
The tumor suppressor p53 promotes anoikis, the apoptotic death of cells that lose adhesion and become detached from their appropriate location, thereby suppressing cancer metastasis. Here, Cheng et al. show that the serine/threonine kinase SIK1 (salt-inducible kinase 1) regulates p53-dependent anoikis. Loss of SIK1 facilitated the anchorage-independent growth of cultured cells and promoted the development of micrometastases in the absence of primary tumors in immunodeficient mice. Moreover, decreased SIK1 expression correlated with appearance of distal metastases in human breast cancer. The authors thus propose that SIK1, by promoting anoikis, prevents the dissemination of precancerous cells and thereby plays a critical role in inhibiting metastasis.
References and Notes
- 1.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt-Kittler O, Ragg T, Daskalakis A, Granzow M, Ahr A, Blankenstein TJ, Kaufmann M, Diebold J, Arnholdt H, Muller P, Bischoff J, Harich D, Schlimok G, Riethmuller G, Eils R, Klein CA. From latent disseminated cells to overt metastasis: genetic analysis of systemic breast cancer progression. Proc Natl Acad Sci U S A. 2003;100:7737–7742. doi: 10.1073/pnas.1331931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Husemann Y, Geigl JB, Schubert F, Musiani P, Meyer M, Burghart E, Forni G, Eils R, Fehm T, Riethmuller G, Klein CA. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Podsypanina K, Du YC, Jechlinger M, Beverly LJ, Hambardzumyan D, Varmus H. Seeding and propagation of untransformed mouse mammary cells in the lung. Science. 2008;321:1841–1844. doi: 10.1126/science.1161621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van de Wouw AJ, Jansen RL, Speel EJ, Hillen HF. The unknown biology of the unknown primary tumour: a literature review. Ann Oncol. 2003;14:191–196. doi: 10.1093/annonc/mdg068. [DOI] [PubMed] [Google Scholar]
- 6.Ruoslahti E, Reed JC. Anchorage dependence, integrins, and apoptosis. Cell. 1994;77:477–478. doi: 10.1016/0092-8674(94)90209-7. [DOI] [PubMed] [Google Scholar]
- 7.Frisch SM, Screaton RA. Anoikis mechanisms. Curr Opin Cell Biol. 2001;13:555–562. doi: 10.1016/s0955-0674(00)00251-9. [DOI] [PubMed] [Google Scholar]
- 8.Grossmann J. Molecular mechanisms of “detachment-induced apoptosis--Anoikis”. Apoptosis. 2002;7:247–260. doi: 10.1023/a:1015312119693. [DOI] [PubMed] [Google Scholar]
- 9.Gilmore AP. Anoikis. Cell Death Differ. 2005;12(Suppl 2):1473–1477. doi: 10.1038/sj.cdd.4401723. [DOI] [PubMed] [Google Scholar]
- 10.Mehlen P, Puisieux A. Metastasis: a question of life or death. Nat Rev Cancer. 2006;6:449–458. doi: 10.1038/nrc1886. [DOI] [PubMed] [Google Scholar]
- 11.van Nimwegen MJ, van de Water B. Focal adhesion kinase: a potential target in cancer therapy. Biochem Pharmacol. 2007;73:597–609. doi: 10.1016/j.bcp.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Shaw LM, Rabinovitz I, Wang HH, Toker A, Mercurio AM. Activation of phosphoinositide 3-OH kinase by the alpha6beta4 integrin promotes carcinoma invasion. Cell. 1997;91:949–960. doi: 10.1016/s0092-8674(00)80486-9. [DOI] [PubMed] [Google Scholar]
- 13.Dufour G, Demers MJ, Gagne D, Dydensborg AB, Teller IC, Bouchard V, Degongre I, Beaulieu JF, Cheng JQ, Fujita N, Tsuruo T, Vallee K, Vachon PH. Human intestinal epithelial cell survival and anoikis. Differentiation state-distinct regulation and roles of protein kinase B/Akt isoforms. J Biol Chem. 2004;279:44113–44122. doi: 10.1074/jbc.M405323200. [DOI] [PubMed] [Google Scholar]
- 14.Douma S, Van Laar T, Zevenhoven J, Meuwissen R, Van Garderen E, Peeper DS. Suppression of anoikis and induction of metastasis by the neurotrophic receptor TrkB. Nature. 2004;430:1034–1039. doi: 10.1038/nature02765. [DOI] [PubMed] [Google Scholar]
- 15.Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 16.Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 17.Lee CH, Inoki K, Karbowniczek M, Petroulakis E, Sonenberg N, Henske EP, Guan KL. Constitutive mTOR activation in TSC mutants sensitizes cells to energy starvation and genomic damage via p53. EMBO J. 2007;26:4812–4823. doi: 10.1038/sj.emboj.7601900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lizcano JM, Goransson O, Toth R, Deak M, Morrice NA, Boudeau J, Hawley SA, Udd L, Makela TP, Hardie DG, Alessi DR. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. Embo J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, Bignell G, Warren W, Aminoff M, Hoglund P, Jarvinen H, Kristo P, Pelin K, Ridanpaa M, Salovaara R, Toro T, Bodmer W, Olschwang S, Olsen AS, Stratton MR, de la Chapelle A, Aaltonen LA. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 20.Shen Z, Wen XF, Lan F, Shen ZZ, Shao ZM. The tumor suppressor gene LKB1 is associated with prognosis in human breast carcinoma. Clin Cancer Res. 2002;8:2085–2090. [PubMed] [Google Scholar]
- 21.Zhuang ZG, Di GH, Shen ZZ, Ding J, Shao ZM. Enhanced expression of LKB1 in breast cancer cells attenuates angiogenesis, invasion, and metastatic potential. Mol Cancer Res. 2006;4:843–849. doi: 10.1158/1541-7786.MCR-06-0118. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez-Cespedes M, Parrella P, Esteller M, Nomoto S, Trink B, Engles JM, Westra WH, Herman JG, Sidransky D. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 2002;62:3659–3662. [PubMed] [Google Scholar]
- 23.Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, Torrice C, Wu MC, Shimamura T, Perera SA, Liang MC, Cai D, Naumov GN, Bao L, Contreras CM, Li D, Chen L, Krishnamurthy J, Koivunen J, Chirieac LR, Padera RF, Bronson RT, Lindeman NI, Christiani DC, Lin X, Shapiro GI, Janne PA, Johnson BE, Meyerson M, Kwiatkowski DJ, Castrillon DH, Bardeesy N, Sharpless NE, Wong KK. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 24.Alessi DR, Sakamoto K, Bayascas JR. LKB1-dependent signaling pathways. Annu Rev Biochem. 2006;75:137–163. doi: 10.1146/annurev.biochem.75.103004.142702. [DOI] [PubMed] [Google Scholar]
- 25.Ilic D, Almeida EA, Schlaepfer DD, Dazin P, Aizawa S, Damsky CH. Extracellular matrix survival signals transduced by focal adhesion kinase suppress p53-mediated apoptosis. J Cell Biol. 1998;143:547–560. doi: 10.1083/jcb.143.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bachelder RE, Ribick MJ, Marchetti A, Falcioni R, Soddu S, Davis KR, Mercurio AM. p53 inhibits alpha 6 beta 4 integrin survival signaling by promoting the caspase 3-dependent cleavage of AKT/PKB. J Cell Biol. 1999;147:1063–1072. doi: 10.1083/jcb.147.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vitale M, Di Matola T, Bifulco M, Casamassima A, Fenzi G, Rossi G. Apoptosis induced by denied adhesion to extracellular matrix (anoikis) in thyroid epithelial cells is p53 dependent but fails to correlate with modulation of p53 expression. FEBS Lett. 1999;462:57–60. doi: 10.1016/s0014-5793(99)01512-4. [DOI] [PubMed] [Google Scholar]
- 28.Ravid D, Maor S, Werner H, Liscovitch M. Caveolin-1 inhibits cell detachment-induced p53 activation and anoikis by upregulation of insulin-like growth factor-I receptors and signaling. Oncogene. 2005;24:1338–1347. doi: 10.1038/sj.onc.1208337. [DOI] [PubMed] [Google Scholar]
- 29.Lewis BC, Klimstra DS, Socci ND, Xu S, Koutcher JA, Varmus HE. The absence of p53 promotes metastasis in a novel somatic mouse model for hepatocellular carcinoma. Mol Cell Biol. 2005;25:1228–1237. doi: 10.1128/MCB.25.4.1228-1237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Derksen PW, Liu X, Saridin F, van der Gulden H, Zevenhoven J, Evers B, van Beijnum JR, Griffioen AW, Vink J, Krimpenfort P, Peterse JL, Cardiff RD, Berns A, Jonkers J. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell. 2006;10:437–449. doi: 10.1016/j.ccr.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 31.Zhao JJ, Liu Z, Wang L, Shin E, Loda MF, Roberts TM. The oncogenic properties of mutant p110alpha and p110beta phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc Natl Acad Sci U S A. 2005;102:18443–18448. doi: 10.1073/pnas.0508988102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao JJ, Gjoerup OV, Subramanian RR, Cheng Y, Chen W, Roberts TM, Hahn WC. Human mammary epithelial cell transformation through the activation of phosphatidylinositol 3-kinase. Cancer Cell. 2003;3:483–495. doi: 10.1016/s1535-6108(03)00088-6. [DOI] [PubMed] [Google Scholar]
- 33.Romanov SR, Kozakiewicz BK, Holst CR, Stampfer MR, Haupt LM, Tlsty TD. Normal human mammary epithelial cells spontaneously escape senescence and acquire genomic changes. Nature. 2001;409:633–637. doi: 10.1038/35054579. [DOI] [PubMed] [Google Scholar]
- 34.Wang J, Hannon GJ, Beach DH. Risky immortalization by telomerase. Nature. 2000;405:755–756. doi: 10.1038/35015674. [DOI] [PubMed] [Google Scholar]
- 35.Utermark T, Schaffhausen BS, Roberts TM, Zhao JJ. The p110{alpha} Isoform of Phosphatidylinositol 3-Kinase Is Essential for Polyomavirus Middle T Antigen-Mediated Transformation. J Virol. 2007;81:7069–7076. doi: 10.1128/JVI.00115-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, Sjostrom SK, Garraway LA, Weremowicz S, Richardson AL, Greulich H, Stewart CJ, Mulvey LA, Shen RR, Ambrogio L, Hirozane-Kishikawa T, Hill DE, Vidal M, Meyerson M, Grenier JK, Hinkle G, Root DE, Roberts TM, Lander ES, Polyak K, Hahn WC. Integrative Genomic Approaches Identify IKBKE as a Breast Cancer Oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 37.Xia H, Qi H, Li Y, Pei J, Barton J, Blackstad M, Xu T, Tao W. LATS1 tumor suppressor regulates G2/M transition and apoptosis. Oncogene. 2002;21:1233–1241. doi: 10.1038/sj.onc.1205174. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Templeton DM. Cadmium activates CaMK-II and initiates CaMK-II-dependent apoptosis in mesangial cells. FEBS Lett. 2007;581:1481–1486. doi: 10.1016/j.febslet.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 39.Miyazaki T, Shen M, Fujikura D, Tosa N, Kim HR, Kon S, Uede T, Reed JC. Functional role of death-associated protein 3 (DAP3) in anoikis. J Biol Chem. 2004;279:44667–44672. doi: 10.1074/jbc.M408101200. [DOI] [PubMed] [Google Scholar]
- 40.Sanjo H, Kawai T, Akira S. DRAKs, novel serine/threonine kinases related to death-associated protein kinase that trigger apoptosis. J Biol Chem. 1998;273:29066–29071. doi: 10.1074/jbc.273.44.29066. [DOI] [PubMed] [Google Scholar]
- 41.Wang S, Guo M, Ouyang H, Li X, Cordon-Cardo C, Kurimasa A, Chen DJ, Fuks Z, Ling CC, Li GC. The catalytic subunit of DNA-dependent protein kinase selectively regulates p53-dependent apoptosis but not cell-cycle arrest. Proc Natl Acad Sci U S A. 2000;97:1584–1588. doi: 10.1073/pnas.97.4.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koo SH, Flechner L, Qi L, Zhang X, Screaton RA, Jeffries S, Hedrick S, Xu W, Boussouar F, Brindle P, Takemori H, Montminy M. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437:1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- 43.Shaulian E, Zauberman A, Ginsberg D, Oren M. Identification of a minimal transforming domain of p53: negative dominance through abrogation of sequence-specific DNA binding. Mol Cell Biol. 1992;12:5581–5592. doi: 10.1128/mcb.12.12.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Lu H, Dazin P, Kapila Y. Squamous cell carcinoma cell aggregates escape suspension-induced, p53-mediated anoikis: fibronectin and integrin alphav mediate survival signals through focal adhesion kinase. J Biol Chem. 2004;279:48342–48349. doi: 10.1074/jbc.M407953200. [DOI] [PubMed] [Google Scholar]
- 45.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 46.Doi J, Takemori H, Lin XZ, Horike N, Katoh Y, Okamoto M. Salt-inducible kinase represses cAMP-dependent protein kinase-mediated activation of human cholesterol side chain cleavage cytochrome P450 promoter through the CREB basic leucine zipper domain. J Biol Chem. 2002;277:15629–15637. doi: 10.1074/jbc.M109365200. [DOI] [PubMed] [Google Scholar]
- 47.Ariumi Y, Kaida A, Lin JY, Hirota M, Masui O, Yamaoka S, Taya Y, Shimotohno K. HTLV-1 tax oncoprotein represses the p53-mediated trans-activation function through coactivator CBP sequestration. Oncogene. 2000;19:1491–1499. doi: 10.1038/sj.onc.1203450. [DOI] [PubMed] [Google Scholar]
- 48.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 49.Imamura K, Ogura T, Kishimoto A, Kaminishi M, Esumi H. Cell cycle regulation via p53 phosphorylation by a 5'-AMP activated protein kinase activator, 5-aminoimidazole- 4-carboxamide-1-beta-D-ribofuranoside, in a human hepatocellular carcinoma cell line. Biochem Biophys Res Commun. 2001;287:562–567. doi: 10.1006/bbrc.2001.5627. [DOI] [PubMed] [Google Scholar]
- 50.Contreras CM, Gurumurthy S, Haynie JM, Shirley LJ, Akbay EA, Wingo SN, Schorge JO, Broaddus RR, Wong KK, Bardeesy N, Castrillon DH. Loss of Lkb1 provokes highly invasive endometrial adenocarcinomas. Cancer Res. 2008;68:759–766. doi: 10.1158/0008-5472.CAN-07-5014. [DOI] [PubMed] [Google Scholar]
- 51.Bardeesy N, Sinha M, Hezel AF, Signoretti S, Hathaway NA, Sharpless NE, Loda M, Carrasco DR, DePinho RA. Loss of the Lkb1 tumour suppressor provokes intestinal polyposis but resistance to transformation. Nature. 2002;419:162–167. doi: 10.1038/nature01045. [DOI] [PubMed] [Google Scholar]
- 52.Karuman P, Gozani O, Odze RD, Zhou XC, Zhu H, Shaw R, Brien TP, Bozzuto CD, Ooi D, Cantley LC, Yuan J. The Peutz-Jegher gene product LKB1 is a mediator of p53-dependent cell death. Mol Cell. 2001;7:1307–1319. doi: 10.1016/s1097-2765(01)00258-1. [DOI] [PubMed] [Google Scholar]
- 53.Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, Flanagan A, Teague J, Futreal PA, Stratton MR, Wooster R. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 55.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, Chen F, Feiler H, Tokuyasu T, Kingsley C, Dairkee S, Meng Z, Chew K, Pinkel D, Jain A, Ljung BM, Esserman L, Albertson DG, Waldman FM, Gray JW. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 56.Lu X, Wang ZC, Iglehart JD, Zhang X, Richardson AL. Predicting features of breast cancer with gene expression patterns. Breast Cancer Res Treat. 2008;108:191–201. doi: 10.1007/s10549-007-9596-6. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, Jatkoe T, Berns EM, Atkins D, Foekens JA. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 58.Davidoff AM, Humphrey PA, Iglehart JD, Marks JR. Genetic basis for p53 overexpression in human breast cancer. Proc Natl Acad Sci U S A. 1991;88:5006–5010. doi: 10.1073/pnas.88.11.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, Cantley LC. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;6:91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 60.Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci U S A. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sjostrom M, Stenstrom K, Eneling K, Zwiller J, Katz AI, Takemori H, Bertorello AM. SIK1 is part of a cell sodium-sensing network that regulates active sodium transport through a calcium-dependent process. Proc Natl Acad Sci U S A. 2007;104:16922–16927. doi: 10.1073/pnas.0706838104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berdeaux R, Goebel N, Banaszynski L, Takemori H, Wandless T, Shelton GD, Montminy M. SIK1 is a class II HDAC kinase that promotes survival of skeletal myocytes. Nat Med. 2007;13:597–603. doi: 10.1038/nm1573. [DOI] [PubMed] [Google Scholar]
- 63.Kowanetz M, Lonn P, Vanlandewijck M, Kowanetz K, Heldin CH, Moustakas A. TGFbeta induces SIK to negatively regulate type I receptor kinase signaling. J Cell Biol. 2008;182:655–662. doi: 10.1083/jcb.200804107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Briasoulis E, Pavlidis N. Cancer of Unknown Primary Origin. Oncologist. 1997;2:142–152. [PubMed] [Google Scholar]
- 65.Ismael G, de Azambuja E, Awada A. Molecular profiling of a tumor of unknown origin. N Engl J Med. 2006;355:1071–1072. doi: 10.1056/NEJMc061533. [DOI] [PubMed] [Google Scholar]
- 66.Engel J, Eckel R, Kerr J, Schmidt M, Furstenberger G, Richter R, Sauer H, Senn HJ, Holzel D. The process of metastasisation for breast cancer. Eur J Cancer. 2003;39:1794–1806. doi: 10.1016/s0959-8049(03)00422-2. [DOI] [PubMed] [Google Scholar]
- 67.Gupta GP, Nguyen DX, Chiang AC, Bos PD, Kim JY, Nadal C, Gomis RR, Manova-Todorova K, Massague J. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature. 2007;446:765–770. doi: 10.1038/nature05760. [DOI] [PubMed] [Google Scholar]
- 68.Nguyen DX, Massague J. Genetic determinants of cancer metastasis. Nat Rev Genet. 2007;8:341–352. doi: 10.1038/nrg2101. [DOI] [PubMed] [Google Scholar]
- 69.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 70.Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B, Zhao JJ, Roberts TM, Mandel G, Hannon GJ, Depinho RA, Chin L, Elledge SJ. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 71.Parrott JA, Nilsson E, Mosher R, Magrane G, Albertson D, Pinkel D, Gray JW, Skinner MK. Stromal-epithelial interactions in the progression of ovarian cancer: influence and source of tumor stromal cells. Mol Cell Endocrinol. 2001;175:29–39. doi: 10.1016/s0303-7207(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 72.We thank the Broad Institute RNAi Platform and David Root for advice and reagents. This work was supported in part by the National Institute of Health (CA089393 to JDI and CA134502 to JJZ), US DOD Breast Cancer Research Program (W81XWH-07-1-0408, WCH) and BC051565 (JJZ), the Tisch Family Fund (WCH), a Claudia Barr Award (JJZ), and a Woman's Cancer Program Award (JJZ). In compliance with Harvard Medical School guidelines, we disclose that WCH and JJZ are consultants for Novartis Pharmaceuticals, Inc.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.