Summary
An alternative fabrication method is presented for production of masters for single- or multilayer polymeric microfluidic devices in a standard laboratory environment, precluding the need for a cleanroom. This toner transfer masking (TTM) method utilizes an office laser printer to generate a toner pattern which is thermally transferred to a metal master to serve as a mask for etching. With master fabrication times as little as one hour (depending on channel depth) using commercially-available equipment and supplies, this approach should make microfluidic technology more widely accessible to the non-expert—even the non-scientist. The cost of fabrication consumables was estimated to be < $1 per master, over an order of magnitude decrease in consumable costs compared to standard photolithography. In addition, the use of chemical etching allows accurate control over the height of raised features (i.e., channel depths), allowing the flexibility to fabricate multiple depths on a single master with little added time. Resultant devices are shown capable of pneumatic valving, three-dimensional channel formation (using layer-connecting vias), droplet fluidics, and cell imaging and staining. The multiple-depth capabilities of the method are proven useful for cellular analysis by fabrication of handheld, disposable devices used for trapping and imaging of live murine pancreatic islets. The precise fluidic control provided by the microfluidic platform allows subsequent fixing and staining of these cells without significant movement, thus spatial correlation of imaging and staining is attainable—even with rare alpha cells that constitute only ∼10% of the islet cells.
Keywords: toner, lithography, membrane, valve, cell imaging, islets of Langerhans, droplet fluidics
Introduction
Fluidic manipulation and imaging of cellular systems has traditionally been carried out using simple glass slides, polymer dishes, multi-well plates, or flow cells. With these tools, the introduction of stimulants, inhibitors, or staining agents is accomplished by bulk addition to the cellular media. Microfluidic devices have emerged as alternative tools for handling and imaging cells 1-8. Polydimethylsiloxane (PDMS) devices are well-suited for imaging due to their transparency in the visible spectrum 9, 10, and have been used for various purposes such as imaging of pancreatic islets 2, 4 and staining of cell cultures 7, 8. The gas permeability of PDMS also provides a facile route for maintaining O2 or CO2, levels in long-term cell cultures. Finally, the small fluidic volumes of these devices are typically in the nL range (10-9 L). This is well-matched to the volumes of the cellular systems, and thus provides a novel platform for the analysis of single cell contents 6 or secretions 1. Volumetric reduction also results in significant decreases in reagent costs 11, 12, which is particularly important for expensive reagents such as antibodies.
Microfluidic technology should provide a plethora of novel and useful tools to biologists and cellular imaging scientists. Unfortunately in practice, there exists reluctance in implementing these devices as routine tools, and several authors have alluded to fabrication constraints as a likely cause 13-16. Standard fabrication of polymeric microfluidic devices requires a regularly-maintained cleanroom facility with specialized lighting for working with UV-sensitive materials 9, 10. This requirement alone is a major roadblock for many research groups. Furthermore, much of the equipment and materials needed for photolithography are expensive. On the other hand, the most commonly used device substrate, PDMS, is relatively inexpensive. Therefore, an alternative fabrication method—one that removes the necessity for a cleanroom and expensive reagents—would be advantageous and could render microfluidic technology more accessible to the non-expert.
Several alternative fabrication methods have been developed in recent years to address these problems. In keeping with the rapid and low-cost criteria described above, promising methods for microchip fabrication have been adapted from home-built electronics techniques 17, in which researchers have used standard office printers to generate masters 14, 18-20, channel walls 15, or etchant masks 16 for microfluidic devices. Coltro et al. used toner from a laser printer directly as the microchannel walls for microchip electrophoresis 15, then later adapted the technique by thermally transferring the toner to a glass substrate to serve as an etchant mask 16. Since glass has been well-characterized as a substrate in microfluidics, the latter technique is promising. Conversely, fabrication of glass devices requires etching with hydrofluoric acid (which must be handled very carefully) and subsequent thermal bonding (which is time consuming and inefficient). The approach, therefore, requires special equipment or safety measures which are not typically available in a standard laboratory. Bao et al. 19 utilized laser-printed toner as a positive-relief master for PDMS channel fabrication, applying their system to electrophoresis as well; and Backhouse and coworkers 20 used a wax printer to create similar master structures. Although the latter two methods provide a rapid and inexpensive alternative for PDMS microchip fabrication, they rely on the toner layer thickness (∼10 μm) to define the channel depth, limiting their use in many applications.
In the current work, we describe a technique that is essentially a hybrid between the toner-based etchant masking used for home-built electronics 17 and the highly successful cleanroom-based technique of multi-layer soft lithography developed by the Whitesides 9, 10 and Quake 21, 22 groups. Laser-printed toner is thermally transferred to a brass substrate, which is subsequently etched to form masters for PDMS devices. The approach, referred to herein as toner transfer masking (TTM), provides more flexibility and controllability in channel depth than the previous toner-based methods 14, 18-20. Turnaround times for master fabrication can be as little as one hour or less (depending on channel depth), allowing rapid iterations of design, fabrication, and testing. Since the TTM technique does not require cleanroom facilities and utilizes a standard office laser printer with mostly over-the-counter materials, fabrication costs are reduced by over an order of magnitude compared to photolithography. TTM is thus the first alternative to cleanroom fabrication that not only provides the benefits of speed and cost reduction (estimated < $1 per master, see Supplementary Information), but also provides accurate control of multiple channel depths on the same master. These devices are proven capable of elastomeric valving, three dimensional channel formation (using layer-connecting vias), and droplet fluidics. To prove their utility for manipulation and analysis of cellular systems, the devices are also used for trapping, imaging, and staining of live murine pancreatic islets. Since cell movement during flow is minimized within the device 4, images of stained islets could be spatially correlated with intracellular calcium oscillation data without performing tedious islet flattening techniques 23 that require overnight culture on extracellular matrix. These results demonstrate that a rapid and inexpensive method for fabricating microfluidic devices can provide novel tools for cellular imaging while simultaneously reducing reagent costs and analysis time.
Results
Toner Transfer Masking (TTM)
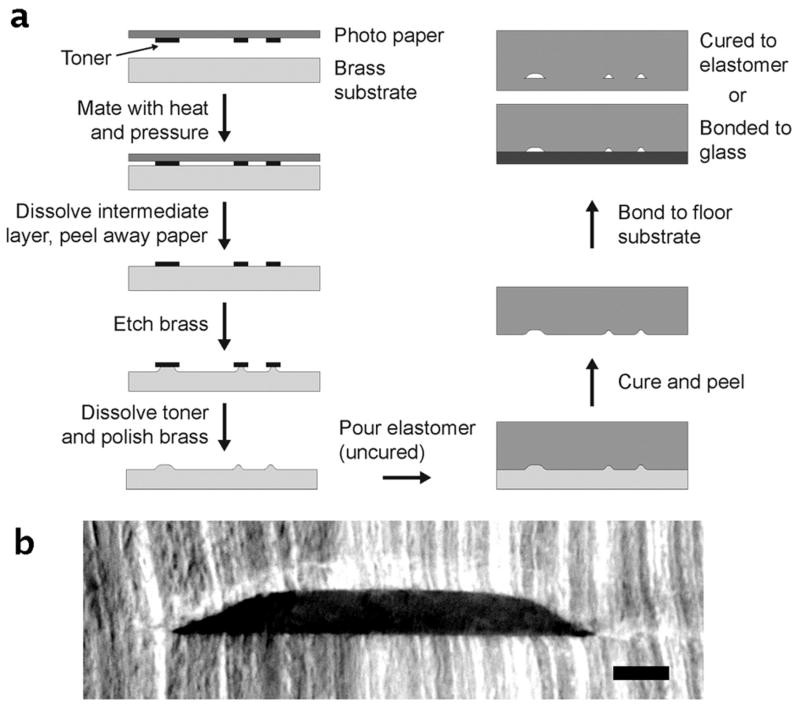
A graphical summary of the TTM method is shown in Fig. 1a. The technique begins with an ordinary office laser printer and can be carried out in a standard laboratory environment (no cleanroom required). In this work, a 1200 dpi, single tone laser printer (HP LaserJet 4350n) is used to print patterns onto glossy photographic paper (Photo Basic Gloss, Staples) that is designed for inkjet printers. This type of paper is typically coated with a water-soluble layer of starch, with or without imbedded polymer particles for controlling ink absorption 24. Consequently, the hydrophobic, laser-printed toner particles can be printed onto the starch layer of the inkjet paper, and the toner can be transferred to another substrate (brass in this work) by simply applying heat and pressure, dissolving the starch layer in water, and finally peeling away the paper (Fig. 1a). The exposed regions of the brass substrate are then etched to the desired depth, and the toner is removed using acetone. It is important to note that these former two steps of etching and toner removal can be repeated to give multi-depth devices. The brass substrate is then polished using a commercially available metal polish (Brasso®), and the resultant substrate is then ready to serve as a master for polymeric devices. In this work, polydimethylsiloxane (PDMS) is poured over the master, cured, peeled away, and bonded to a floor substrate (PDMS or glass). Further details of this fabrication process are included below.
Figure 1.
Toner transfer masking (TTM) process for rapid, inexpensive, and cleanroom-free fabrication of microfluidic devices. (a) Illustration of the simple fabrication process in which masters are etched from brass then used to mold poly(dimethylsiloxane) (PDMS) microfluidic chips. The turnaround time for master fabrication (∼1-2 h) as well as device molding (∼1 h), dominated by the brass etching and polymer curing times, is typically around 2-3 hours. (b) An optical image of a sliced cross-section of a PDMS device, fabricated by TTM, reveals the channel cross-section to be trapezoidal, with tapered side walls that are faintly sigmoidal. Scale bar is 100 μm.
The TTM technique was used to etch brass masters for casting PDMS microfluidic devices. Commercially available brass strips (or shim) of 0.813 mm (0.032 inch) thickness (Small Parts, Inc.; Miramar, FL, USA) was used as the substrate for masters. These brass strips could be cut to the appropriate size by dicing with a band saw or even by cutting with a pair of shears, and the working surfaces were then sanded evenly using sandpaper to improve the toner adhesion. As depicted in Fig. 1a, the toner was first transferred to the brass sheet using heat and pressure, which was carried out using a clothing iron on the highest heat setting, requiring an average of 10 min per transfer. The adhered pieces were immersed in room temperature water to dissolve the intermediate layer, and the paper was peeled away after approximately 10 min. The brass was then etched in a 20% solution (% w/v) of ammonium persulfate (APS, (NH4)2S2O8), which is commercially available at electronics suppliers. This solution was found to etch the brass substrate at an average dissolution rate of 0.428 ± 0.047 mg cm-2 min-1 by simply weighing the brass sheet on a fine balance in increments as etching was carried out over a 90 min period. Moreover, the data revealed that the mass removed during etching was directly proportional to the etch depth, with a linear correlation coefficient of 0.99993 and a y-intercept of -0.09534 ± 0.10819 μm, essentially zero (Supplementary Fig. S1). This allowed the brass weight to be used as a simple and accurate indicator of microfluidic channel depth via Equation 1,
| (1) |
where dch represents the microfluidic channel depth after molding, detch is the etch depth of the brass master, Δm is the measured change in brass mass after etching, Aexp is the area of exposed brass (calculated from original mask pattern), and ρbrass is the density of the brass sheet (measured as 8500 mg cm-3). Equation 1 was applied for fabrication of single- and multi-depth masters. For example, with a dissolution rate of 0.428 mg cm-2 min-1 and a typical Aexp of 6.45 cm2, etching for only one hour would provide a master for polymeric channels of 30.3 μm depth, corresponding to 166 mg of removed brass. A cross-section of a typical channel is shown in Fig. 1b, in which a PDMS device was sliced with a razor blade then imaged using a wide-field microscope. The image reveals that the cross-section of the channel could be approximated as a rounded trapezoid, with tapered side walls that are faintly sigmoidal (80 μm deep channel). This cross-section is an important factor to consider when designing patterns for the TTM fabrication, and it has recently been proposed that this very cross-section is optimal for low-pressure microfluidic valving 25.
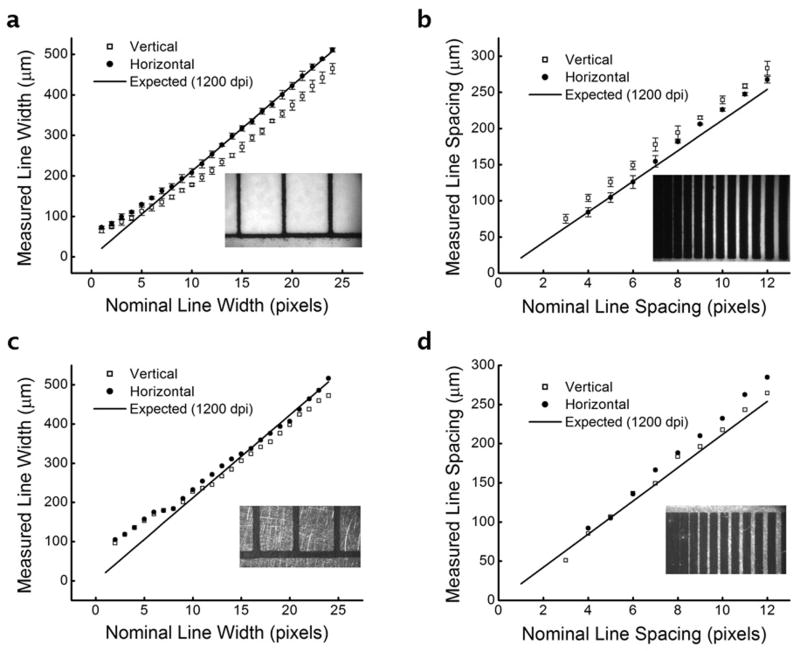
Next, the resolution of the TTM method for master fabrication was investigated. A resolution test pattern (Supplementary Fig. S2; 1- to 24-pixel line widths, 1- to 12-pixel line spacing, vertical and horizontal) was designed and printed in triplicate, and a wide-field microscope was used to collect digital images of the printed patterns on the paper substrate. Image analysis algorithms were written using ImageJ 26 to rapidly quantify the line widths or line spacing. As shown in Fig. 2a, the line widths (data points) of laser-printed toner on inkjet paper (glossy photographic paper) correlated well with the expected line widths (solid line) based on a resolution of 1200 dpi (21.2 μm pixel-1). Interestingly, between 12 and 24 pixels of nominal width, the vertical line widths (open squares) were offset from the expected values by 50.6 ± 9.1 μm, while the horizontal line widths (filled circles) were essentially equal to the expected values (differences of 0.0 ± 7.6 μm). This result suggested that the printer had a slightly rectangular aspect ratio of transfer. Below about 10-12 nominal pixels, both the vertical and horizontal line widths began to converge and deviate slightly from expected behavior (with decreasing width). A similar result was seen with measured line spacing (Fig. 2b), where horizontal line spacing was consistent with expected values and vertical line spacing was slightly offset. Below 3-4 nominal pixels, the lines of toner began to merge and adhere to each other, resulting in gaps and inconsistent spacing. The line widths and spacing of thermally transferred toner on brass was also measured (Fig. 2c-d), and the results were similar to the toner printed on paper. The aspect ratio difference seemed to be reduced upon thermal transfer. A possible explanation for this effect is that the printer deposits equal volumes of toner for each pixel, which could reduce aspect ratio differences upon application of heat and pressure. Despite the slightly rectangular aspect ratio and the deviation at lower line widths, the results shown in Fig. 2a-d indicated that the printed and transferred line widths and spacing followed a consistent, predictable pattern dependent upon the characteristics of the printer. Using this data, along with empirical observations through design iterations, it was concluded that the technique was suitable for line widths and spacing ≥ 100 μm (≥ 5 pixels at 1200 dpi). Improvements in resolution should be possible using laser printers with higher than 1200 dpi resolution, at the expense of increases in overall cost.
Figure 2.
Toner line widths printed onto photographic paper (vertical and horizontal), then transferred to brass substrates, were measured using wide-field microscopy and image analysis (example image inset). (a) The horizontal line widths (filled circles) correlated well with the expected values of a 1200 dpi printer (solid line), while the vertical line widths (open squares) revealed a slightly rectangular pixel aspect ratio of the printer. (b) Toner line spacing was measured in a similar fashion, confirming the rectangular pixel aspect ratio. (c) Line widths and (d) spacing after thermal transfer to brass substrates. Insets show typical images from each analysis. Error bars represent standard deviations about mean values.
Fabrication of Multilayer PDMS Devices
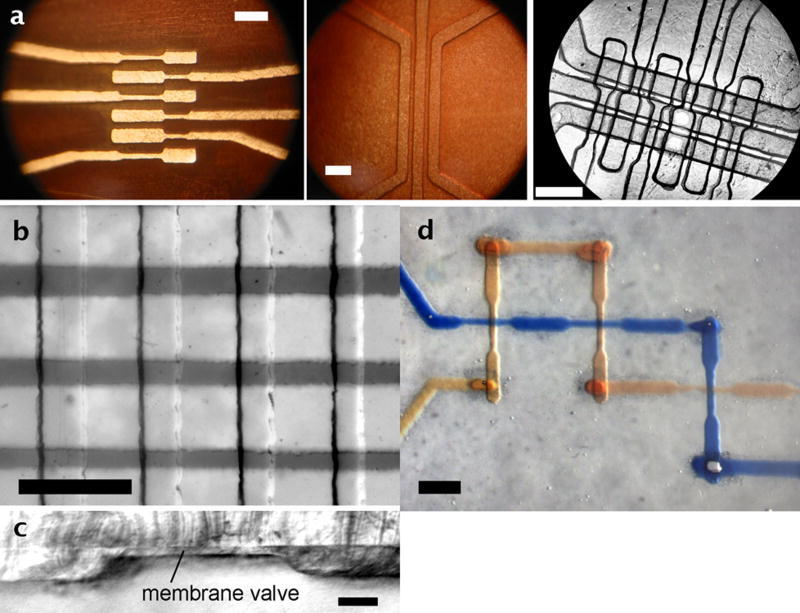
In order to evaluate the flexibility of the TTM master fabrication for microfluidic device production, several multilayer assembly schemes were developed. Owing to the widespread applicability of PDMS devices made using multilayer soft lithography (MSL) methods 9, 10, 21, 22, these devices were made using a modified version of MSL with the TTM-fabricated masters. Again, this method did not require the use of cleanroom facilities. Furthermore, the MSL could be carried out without the use of a spin-coater. In order to achieve accurate and controllable membrane thicknesses, thin sheets of brass shim (thicknesses of 25.4 μm, 38.1 μm, or 50.8 μm {0.01, 0.015, 0.02 in}; Small Parts, Inc.; Miramar, FL, USA) were used as spacer layers 27 between the un-etched portions of the brass master and a transparency film covered by a sheet of glass. Using a modification of the method developed by Beebe and coworkers 27, PDMS was poured over the pneumatic layer master, 25.4-μm brass shim spacers were placed accordingly, and the master/transparency/glass assembly was clamped tightly and heated to PDMS curing temperature (70 °C, 1-2 h). Next, a thick (5-mm) layer of PDMS was cured over the fluidic master. The fluidic and pneumatic layers could be joined by either partial curing and annealing 21 or by plasma oxidation and adhering, and these joined layers were sealed to a glass slide by plasma oxidation and adhering. Fig. 3a shows images of brass pneumatic (left) and fluidic (middle) masters fabricated by TTM along with the resultant device (right). Fig. 3b shows another assembled device with crossing channels for valving, with air in the pneumatic channels (lower) and dye solution in the fluidic channels (upper). As shown by the sliced cross-section in Fig. 3c, the PDMS membrane separating the pneumatic channels (shown) and the fluidic channels (not shown) was 26.6 ± 0.7 μm (±σ) in thickness, a value similar to the 25.4-μm thickness of the brass shim, as expected. The pneumatic actuation channel below the membrane was 74.0 ± 1.1 μm in depth and ∼560 μm in width. Analyses of other sliced sections showed a membrane thickness variation of <4% relative standard deviation (RSD), indicating that the TTM method could be reliably coupled with MSL to produce arrays of microfluidic valves. Without any use of cleanroom facilities, the valve structures shown here were fabricated with a density of approximately 4 valves mm-2.
Figure 3.
Multilayer PDMS devices were fabricated using toner transfer masking (TTM) and multilayer soft lithography (MLS). (a) Wide-field images of two brass masters and the final device made with micro-pneumatic valves using MLS (left: pneumatic master, middle: fluidic master, right: final device). Image shows the fourth valve from left during actuation. (b) Image of assembled crossing PDMS valve structures, with air in the pneumatic channels (lower) and dye solution in the fluidic channels (upper). (c) Image of a sliced cross-section of a PDMS valve from (b), showing the membrane thickness of 26.6 ± 0.7 μm (±σ) above a pneumatic actuation channel of 74.0 ± 1.1 μm depth and ∼560 μm width. (d) Image of crossing channels in a three-dimensional channel network, including vias with average volume of 2.5 ± 0.6 nL, made using TTM and MLS. Scale bars are 1 mm in (a), (b), and (d); 100 μm in (c).
Fig. 3d shows that the TTM method also has the flexibility to fabricate three-dimensional channel patterns connected by through-membrane vias 28. This process required a modified version of the TTM method, with three patterns to be printed on photo paper: a lower layer pattern, a layer-connecting via pattern, and an upper layer pattern (patterns and modified method included in Supplementary Information and Fig. S3). This method resulted in vias of minimal volume, i.e. with similar cross-section to the channels themselves, and no leakage was observed between crossing channels. Using microscopy and image analysis, via volumes were measured as 2.5 ± 0.6 nL (n = 5 vias), allowing minimal dead-volume in solution transfer between layers. More importantly, the ability to fabricate both microfluidic valves and vias at low cost, without cleanroom facilities, greatly expands the design flexibility of the TTM method.
Droplet Microfluidics
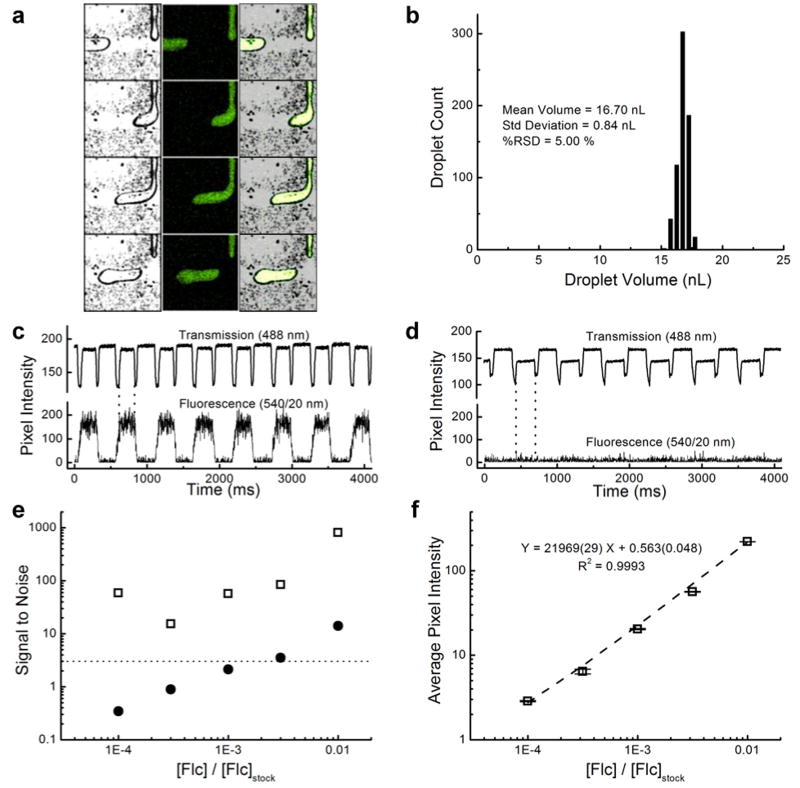
Droplet microfluidics, a relatively new subset of the microfluidics field 29-31, has been shown to be highly effective in applications such as millisecond kinetics measurements 30, fluidic computations 31, and high-throughput single copy DNA amplification 32. This novel technology has great potential due to its ability to passively segregate picoliter to nanoliter volumes of reagents into well-defined, monodisperse droplets that can be thought of as miniature chemical or biological reactors. Droplet formation is highly dependent upon the surface properties and the dimensions of the microfluidic channels 33. To test the flexibility of the TTM method, masters were designed and fabricated for nanoliter droplet formation at a microfluidic T-junction 33, and PDMS devices were made from these masters. As shown in Fig. 4, the devices were capable of formation of monodisperse populations of fluorescein-containing aqueous droplets in silicone oil. Fig. 4a shows a montage image taken from a confocal transmission and fluorescence video of droplet formation, with the transmitted images shown on the left, the fluorescence images in the middle, and the combined images on the right. Fig. 4b depicts the distribution of droplet volumes, with a mean value of 16.70 ± 0.84 nL (± σ), corresponding to an RSD of 5.0 %.
Figure 4.
Aqueous-in-oil droplet formation at a microfluidic T-junction. (a) Confocal transmission (left), fluorescence (middle), and combined (right) images as acquired during fluorescein droplet formation in silicone oil. (b) Droplets were monodisperse, with an average volume of 16.70 ± 0.84 nL (± σ). (c) Correlation of transmission and fluorescence intensities allowed spatial and temporal lock-in-detection due to the ‘sample chopping’ effect of the droplets, even at (d) fluorescein concentrations below the LOD. (e) Signal-to-noise enhancements up to ∼800-fold were possible using lock-in spatial filtering (open squares), compared to unprocessed data (filled circles). Three of the five concentrations were undetectable (below the 3σ dotted line) until the data was processed. (f) Processed signal was linear over several orders of magnitude of concentration. Error bars represent standard deviations about mean values.
Notably, droplet edges could be easily detected using the transmitted signal of the 488 nm laser line. As a result, this system allowed the use of spatial and temporal lock-in detection. These droplets could be used as ‘sample choppers’ to greatly improve the fluorescence limit of detection. Serial dilutions of fluorescein were passed through the aqueous portion of the device, and the images were processed using an in-house written, spatial lock-in detection algorithm (Matlab). Example traces of single-point detection are shown at high (Fig. 4c) and low (Fig. 4d) fluorescein concentrations. Using this type of linear detection, low concentrations of fluorescein could not be distinguished from the signals in droplets containing only buffer solution. However, by applying the lock-in detection algorithm to the data, signal-to-noise ratios (S/N) could be greatly enhanced. Fig. 4e compares the single-point detection S/N values (filled circles) with those of the lock-in processed values (open squares). The dotted line represents the 3σ cutoff for reliably discriminating between signal and reference droplets (buffer only), based on the standard deviation of the background signal, σ. Three of the five concentrations tested were not detectable (< 3σ) until processing with the lock-in algorithm, at which point the S/N values were significantly enhanced (up to ∼800-fold). The processed signal was linear over several orders of magnitude of concentration (Fig. 4f). To the knowledge of the authors, this combination of droplet fluidics with spatial and temporal lock-in detection is novel. To accomplish this, the TTM technique provided a valuable tool for rapid iterations of device design, fabrication, and testing.
Cell Trapping, Imaging, and Staining
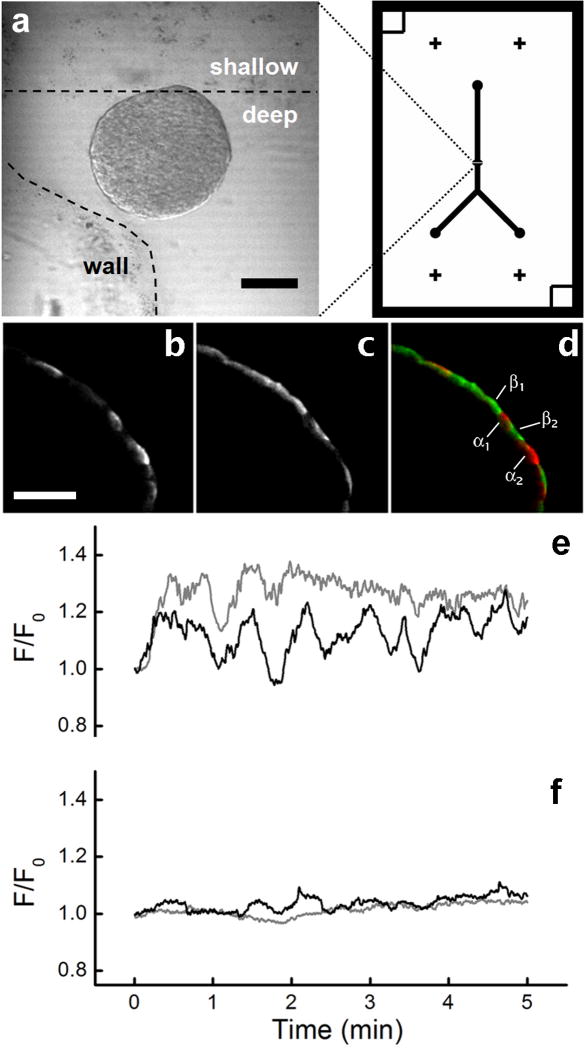
Finally, the TTM method was evaluated for master fabrication with the applications of cell trapping, stimulation, imaging, and staining. Masters were fabricated based on a previous design used from trapping and imaging islets of Langerhans 4, in which a shallow wall trap (weir structure) is used to trap islets while maintaining an open path for fluid flow within the device. PDMS devices were fabricated as shown in Fig. 1 and permanently sealed to a glass coverslip using plasma oxidation and adhering. A key advantage of this device design is that the islet can be gently held in a stationary position for hours of flow time by the coverslip, ceiling, and wall trap of the device. This permits time-lapse imaging of similar regions after many different treatments without perturbing the intracellular metabolism within the islet 4. In the present work, islets were loaded onto the device (Fig. 5a), and oscillations of a calcium indicator, fluo-4, were imaged during glucose stimulation to determine the dynamics of intra-cellular free calcium (Fig. 5e-f). The device then allowed immunostaining followed by subsequent imaging of this staining (Fig. 5b-d). Since the islet remained stationary within the device, intracellular free calcium oscillations could be correlated with immunostaining, permitting reliable identification of intracellular metabolism in rare pancreatic α-cells (∼10% of islet). With this technique, it was possible to show that α-cells within the islet were electrically active at 2.0 mM glucose, while β-cells were essentially inactive (Fig. 5e-f). Notably, the microfluidic platform allowed these experiments—including islet loading, imaging, staining, and follow-up imaging—to be accomplished in only one day, consuming an order of magnitude less of expensive antibody reagents compared to standard staining experiments. Alternative methods require tedious islet flattening techniques 23 that require overnight culture on extracellular matrix, and the staining is typically an overnight step as well.
Figure 5.
Trapping of islets of Langerhans for imaging. (a) Confocal transmission (488 nm) image of an islet after loading and trapping at the weir region of a device fabricated by multi-depth TTM (deep = ∼100 μm, shallow = ∼25 μm). The dashed line outlines the deep channel region. The device design, with a printed toner area of 25.4 mm (1.00 in) width by 38.1 mm (1.50 in) height, is shown on the right. Scale bar is 100 μm. Peripheral immunostaining of (b) glucagon- and (c) insulin-specific islet cells was accomplished in < 4 h and could be correlated with intracellular free calcium oscillations. (d) The merged image shows very little co-localization of staining, allowing labeling of peripheral islet cells as either α- (glucagon+, red) or β-cells (insulin+, green). (e) Calcium oscillations that spatially correlated with glucagon+ cells indicated α-cell activity at 2 mM glucose (α1, black trace; α2, gray trace), while (f) insulin+ β-cells were essentially silent (β1, black trace; β2, gray trace). Scale bars are 100 μm.
Discussion and Conclusions
We have presented an alternative method for fabrication of microfluidic devices—referred to as toner-transfer masking (TTM)—that should make microfluidic tools more accessible to the non-expert. Several key advantages separate this method from typical photolithographic techniques. First, the metal masters are more robust than photoresist masters (e.g. SU-8 or AZ photoresists) on silicon wafers, thus they are more resistant to wear, and they can feasibly be reused indefinitely if care is taken. Second, turnaround times for master fabrication (as little as 1 h, depending on channel depth) are shown to be comparable to the PDMS curing time, thus providing a means for rapid iterations of design, fabrication, and testing. Third, since the technique does not require cleanroom facilities and utilizes a standard laser-jet printer with over-the-counter materials, fabrication consumable costs (estimated < $1 per master, see Supplementary Information) are reduced by over an order of magnitude compared to photolithography (estimated $13 per master). Supplies and equipment needed for this method can be cheaply and easily obtained through non-scientific commercial sources. This aspect alone should allow those with little or no expertise in microfluidic device fabrication to begin utilizing powerful microfluidic tools in their own research. Finally, the TTM technique allows accurate control of channel depths (see Equation 1), and the method can be easily extrapolated to fabrication of multi-depth devices (Fig. 5). These benefits, together, have not been achieved using other rapid fabrication techniques noted above 14, 15. In light of these advantages, it can be argued that the TTM method provides the best combination of fabrication flexibility, accessibility, speed, and cost reduction compared to any alternative to microfluidic device fabrication that has been reported to date.
In fact, the brass etching time could feasibly be extended to produce channels as deep as the brass sheet being etched, although this would eventually move the channel volumes outside the realm of microfluidics. Of course, for applications requiring line width resolution better than ≥ 100 μm, the TTM technique described here will not suffice (Fig. 2) without a higher-quality printer. As commonly performed in electronics fabrication, however, it should be possible to reduce the achievable channel width (and volume) by under-etching thin toner line widths.
Perhaps more important are the demonstrations that TTM is a flexible fabrication technique capable of producing elastomeric valving structures (Fig. 3b-c), three-dimensional patterns with through-membrane vias (Fig. 3d), droplet-generating devices (Fig. 4), and devices capable of cell trapping, stimulation, imaging, and staining (Fig. 5). These results were obtained without the use of any specialized equipment or photolithography rooms. The TTM method merely requires a standard office laser printer and a clothing iron to print toner onto photographic paper and transfer it to brass substrates. Although brass was characterized in this work, the method could be extrapolated to other etchable metals. Furthermore, since Soper and coworkers 34 have shown that milled brass masters provide excellent reproducibility for hot embossing of poly(methylmethacrylate) (PMMA) devices, the brass features produced in this work could be used as masters for various polymeric molding or hot embossing approaches. By comparison, the “Shrinky-Dink microfluidics” approach presented by Khine and coworkers 14, 35 allowed polystyrene sheets to be stacked and bonded to directly serve as the microfluidic channels in three dimensions, rather than creating a master for soft lithography. An advantage of their three-dimensional microfluidic devices is that they could be designed and fabricated to full functionality in a matter of minutes. It was possible to achieve variable height channels using this method, although the channel width, spacing, and depth resolutions were not characterized. However, it is important to note that, since the polystyrene is pre-stressed in the plane of the sheet (x-y plane), both of these methods achieve their reduction in lateral resolution at the expense of expanding the channel in the axial direction (z). Thus the volume of the shrunken channels will be essentially equal to the volume of the channels before shrinking. Moreover, for those who require reproducibility in channel depths or volumes, the toner shrinkage 14 and manual scribing 35 techniques may be limited.
Multilayer soft lithography 21 was achieved using TTM masters (Fig. 3). Interestingly, the rounded trapezoidal cross section shown here (Fig. 1b) has recently been proposed as the optimal geometry for elastomeric valving, requiring very low actuation pressures 25. Future work should be carried out to determine the required actuation pressures of the valves shown in Fig. 3b-c. Additionally, the three dimensional channels shown in this work were connected through vias of minimal dead volume (Fig. 3d), with an average volume of 2.5 ± 0.6 nL. By comparison to other rapid fabrication techniques, the manually-punched vias shown by Chen et al. 35 were approximately an order of magnitude larger in volume (2-3 μL), which is much larger than the total volume of typical microfluidic channel networks (100's of nL). Although these relatively large vias were shown capable of vortex-based mixing 35, the large dead volumes would be disadvantageous with respect to transit times and additional use of expensive reagents used for biological assays 36.
Finally, the TTM method was shown to provide a novel and rapid approach for reliable identification of intracellular metabolism of rare pancreatic α-cells within intact, live murine islets of Langerhans (Fig. 5). Handheld, disposable microfluidic devices were fabricated, and the devices were utilized for trapping islets, fluorescence imaging of oscillations in intracellular free calcium during glucose stimulation, and finally for alpha and beta cell-specific staining of these islets. Due to the superior fluidic control and cell manipulation provided by the microfluidic platform, these islets remained stationary throughout the imaging, stimulation, and staining procedures. Therefore, images of stained islets could be spatially correlated with the calcium oscillation data without performing tedious islet flattening techniques 23 that require overnight culture on extracellular matrix. Furthermore, the cost of expensive staining reagents, namely antibodies, could be reduced by an order of magnitude owing to the small volume of the device (∼1 μL total). These results demonstrate that a rapid and inexpensive method for fabricating microfluidic devices can provide novel tools for cellular imaging while simultaneously reducing reagent costs and analysis time.
Materials and Methods
Reagents
Polydimethylsiloxane (PDMS) precursors, Sylgard® 184 elastomer base and curing agent, were obtained from Dow Corning. NaCl, KCl, CaCl2, MgCl2, HEPES, Triton-X, bovine serum albumin (BSA), ammonium persulfate (APS), fluorescein, silicone oil AR-20, and D-glucose were purchased from Sigma-Aldrich (St. Louis, Missouri, United States). Guinea pig anti-insulin and rabbit anti-glulcagon were purchased from LINCO Research (St. Charles, Missouri, United States). Alexa 568 goat anti-guinea pig and Alexa 660 goat anti-rabbit were obtained from Invitrogen (Eugene, Oregon, United States). Fluo-4 AM dye, Gibco RPMI medium, phosphate buffered saline (PBS), and fetal bovine serum were also obtained from Invitrogen. Paraformaldehyde was obtained from Electron Microscopy Sciences (Hatfield, Pennsylvania, United States), and goat serum from the Jackson Laboratory (Bar Harbor, Maine, United States). The ferric chloride-based PCB etchant solution (Radio Shack, Inc.), Photo Basic Gloss paper (Staples, Inc.), and Brasso® metal polish (Ace Hardware, Inc.) were purchased from local suppliers.
Master Fabrication
The toner transfer masking (TTM) method was utilized to produce raised features in brass substrates (SmallParts, Inc.), which served as masters for PDMS devices. Details are described in the text and in Supplementary Information.
For purposes of clarity, the time required for each step from Fig. 1 has been included here. Sanding of working surfaces required approximately 1 min or less. Pattern transfer required approximately 10 min. Removal of the paper layer required about 10 min. Chemical etching time, which was typically the time-consuming step of the process, was dependent upon the required channel depth and exposed brass area (see Equation 1). After etching, substrate polishing (chemical and mechanical) required approximately 5-6 min. These steps require a maximum of only 27 min, excluding time of etching. If etching time is included, it can be estimated that masters for ∼20 μm deep channels could be fabricated completely within approximately one hour.
Microchip Fabrication
Various types of microfluidic devices were fabricated as described in the text using our TTM method. For single- and multi-layered channel patterns, appropriate ratios of Sylgard® 184 curing agent was mixed well with Sylgard® 184 elastomer base (Dow Corning), and the mixture was degassed under vacuum for 20-30 min. For single-layer devices, the degassed mixture was poured over a brass strip (or shim stock) master (SmallParts, Inc.), which was placed in a boat made of aluminum foil, and the boat was cured on a hot plate at 70 °C for 1-2 h. Multilayer devices were fabricated as described in the text and in Supplementary Information.
Image Acquisition and Analysis
Images of toner on paper and brass substrates were acquired using a TE300 Eclipse microscope (Nikon, Melville, New York, United States) with a side-mounted CoolSnap HQ camera (Roper Scientific, Tucson, Arizona, United States). Sliced cross-sections and whole microfluidic devices were imaged using an M2BIO microscope (Carl Zeiss, Thornwood, New York, United States) with a MicroPublisher RTV camera (QImaging, Tucson, Arizona, United States). These images were background-subtracted to correct for uneven illumination. Where appropriate, background images were taken in the absence of the device or cross-section. While imaging, illumination of the opaque brass substrates and the microfluidic devices/cross-sections was accomplished using an external fiber-optic illuminator. Droplet formation and confocal transmission images of pancreatic islets were imaged using an LSM 510 laser-scanning confocal microscope (Carl Zeiss) with a 10× 0.3 NA Plan-Neofluar objective, using a 488 nm argon ion laser for excitation and a 540/20 nm bandpass filter when detecting fluorescence emission. All other pancreatic islet images were collected with an LSM 5Live line-scanning confocal microscope (Carl Zeiss) with a 20× 0.8 NA Fluar objective. Intracellular calcium was imaged using a 488 nm diode laser for excitation and a 495 nm long-pass filter to detect fluorescence emission. Immunostained islets were imaged using 532 nm excitation, and 540-625 nm band-pass emission as well as 635 nm excitation, and 650 nm long-pass emission. All image analysis algorithms were written in-house using ImageJ 26 or Matlab.
Characterization of Printer Resolution
The characterization of line widths and spacing (Fig. 2) was carried out using a resolution test pattern. The pattern (refer to Fig. S1), which was designed in Adobe Illustrator then transferred to Adobe Photoshop (Adobe, Inc., San Jose, CA) and rasterized, included vertical and horizontal line widths between 1 and 24 pixels (21.2 to 508.0 μm) and spacing from 1 to 12 pixels (21.2 to 254.0 μm). The TE300 Eclipse optical microscope (Nikon) was used to acquire digital images of the lines printed on paper or transferred to the brass substrate. A 508-μm standard was imaged simultaneously with the first and last images to provide a baseline for quantitation of the line widths and spacing. Image analysis algorithms were written using ImageJ 26 to rapidly quantify the line widths or line spacing.
Islet isolation, on-chip calcium imaging, and immunostaining
Islets were isolated as described in 37, 38 and maintained in RPMI medium containing 10 % fetal bovine serum, 11 mM glucose at 37 °C under humidified 5 % CO2 for 24-48 h before imaging.
Isolated islets were stained with 4 μM Fluo-4 AM (Invitrogen) in imaging medium (125 mM NaCl, 5.7 mM KCl, 2.5 CaCl2, 1.2 mM MgCl2, 10 mM HEPES, 2 mM glucose, 0.1% BSA, pH = 7.4) at room temperature for 1-3 h prior to imaging of [Ca2+]i time course data. A single islet was loaded onto the microchip, which was held on a microscope stage in a humidified temperature controlled chamber, maintained at 37 °C. During imaging, each islet was perfused continually with imaging medium at a gravity driven flow rate of ∼3 μL min-1. Fluo-4 fluorescence was imaged on an LSM 5Live line-scanning confocal microscope (Zeiss) with a 20× 0.8NA Fluar Objective, using a 488 nm diode laser for excitation and a 495 nm long-pass filter to detect fluorescence emission.
Following acquisition of [Ca2+]i time course data, each islet was immunostained to identify alpha and beta cells. The islet was initially perfused for 10 minutes with 1× phosphate buffered saline (PBS) for washing, then with 4 % (w/v) paraformaldehyde for 20 min for fixation, both with a gravity driven flow rate of ∼3 μL min-1. The microchip was then transferred to an ice bath, where a syringe pump was used to deliver the remaining solutions through the device. Permeabilization buffer (1× PBS, 0.3 % Triton-X), blocking buffer (1× PBS, 0.15 % Triton-X, 5 % goat serum) and then equilibration buffer (1 × PBS, 1 % BSA, 0.2 % Triton-X) were all delivered, each for 15 minutes, at a flow rate of 1.3 μL min-1. Primary antibodies (guinea pig anti-insulin, 1:1000; rabbit anti-glucagon, 1:1000) in the equilibration buffer were delivered for 3 h at a flow rate of 0.3 μL min-1. Washing was then performed for 15 min before secondary antibodies (Alexa 568 goat anti-guinea pig, 1:1000; Alexa660 goat anti-rabbit, 1:1000) were delivered for 1 hour at a flow rate of 0.3 μL min-1.
A final wash was performed before imaging immunofluorescence on the same LSM 5Live microscope, using 532 nm excitation, 540-625 nm band-pass emission as well as 635 nm excitation, 650 nm long-pass emission.
Supplementary Material
Acknowledgments
Support for this work was provided by award numbers F32DK07964 (Easley), R01DK053434 (Piston), and P20GM072048 (Piston) from the National Institutes of Health. Support was also provided by the Department of Defense Medical Free-Electron Laser Program. The authors would like to thank the Vanderbilt Institute for Integrative Biosystem Research and Education (VIIBRE) for use of their profilometer.
References
- 1.Roper MG, Shackman JG, Dahlgren GM, Kennedy RT. Anal Chem. 2003;75:4711–4717. doi: 10.1021/ac0346813. [DOI] [PubMed] [Google Scholar]
- 2.Rocheleau JV, Walker GM, Head WS, McGuinness OP, Piston DW. P Natl Acad Sci USA. 2004;101:12899–12903. doi: 10.1073/pnas.0405149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balagadde FK, You LC, Hansen CL, Arnold FH, Quake SR. Science. 2005;309:137–140. doi: 10.1126/science.1109173. [DOI] [PubMed] [Google Scholar]
- 4.Rocheleau JV, Remedi MS, Granada B, Head WS, Koster JC, Nichols CG, Piston DW. Plos Biol. 2006;4:221–227. doi: 10.1371/journal.pbio.0040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horsman KM, Barker SLR, Ferrance JP, Forrest KA, Koen KA, Landers JP. Anal Chem. 2005;77:742–749. doi: 10.1021/ac0486239. [DOI] [PubMed] [Google Scholar]
- 6.Huang B, Wu H, Bhaya D, Grossman A, Granier S, Kobilka BK, Zare RN. Science. 2007;315:81–4. doi: 10.1126/science.1133992. [DOI] [PubMed] [Google Scholar]
- 7.Gomez-Sjoberg R, Leyrat AA, Pirone DM, Chen CS, Quake SR. Anal Chem. 2007;79:8557–8563. doi: 10.1021/ac071311w. [DOI] [PubMed] [Google Scholar]
- 8.Park JW, Vahidi B, Taylor AM, Rhee SW, Jeon NL. Nat Protoc. 2006;1:2128–36. doi: 10.1038/nprot.2006.316. [DOI] [PubMed] [Google Scholar]
- 9.Duffy DC, McDonald JC, Schueller OJA, Whitesides GM. Anal Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 10.McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu HK, Schueller OJA, Whitesides GM. Electrophoresis. 2000;21:27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 11.Jin LJ, Ferrance J, Landers JP. Biotechniques. 2001;31:1332–+. doi: 10.2144/01316md01. [DOI] [PubMed] [Google Scholar]
- 12.Easley CJ, Karlinsey JM, Bienvenue JM, Legendre LA, Roper MG, Feldman SH, Hughes MA, Hewlett EL, Merkel TJ, Ferrance JP, Landers JP. P Natl Acad Sci USA. 2006;103:19272–19277. doi: 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peeni BA, Lee ML, Hawkins AR, Woolley AT. Electrophoresis. 2006;27:4888–4895. doi: 10.1002/elps.200600399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grimes A, Breslauer DN, Long M, Pegan J, Lee LP, Khine M. Lab Chip. 2008;8:170–172. doi: 10.1039/b711622e. [DOI] [PubMed] [Google Scholar]
- 15.Coltro WKT, da Silva JAF, da Silva HDT, Richter EM, Furlan R, Angnes L, do Lago CL, Mazo LH, Carrilho E. Electrophoresis. 2004;25:3832–3839. doi: 10.1002/elps.200406091. [DOI] [PubMed] [Google Scholar]
- 16.Coltro WKT, Piccin E, da Silva JAF, do Lago CL, Carrilho E. Lab Chip. 2007;7:931–934. doi: 10.1039/b702931d. [DOI] [PubMed] [Google Scholar]
- 17.Branson J, Naber J, Edelen G. Ieee T Educ. 2000;43:257–261. [Google Scholar]
- 18.Abdelgawad M, Wheeler AR. Adv Mater. 2007;19:133–137. [Google Scholar]
- 19.Bao N, Zhang Q, Xu JJ, Chen HY. J Chromatogr A. 2005;1089:270–5. doi: 10.1016/j.chroma.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Kaigala GV, Ho S, Penterman R, Backhouse CJ. Lab Chip. 2007;7:384–7. doi: 10.1039/b617764f. [DOI] [PubMed] [Google Scholar]
- 21.Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 22.Thorsen T, Maerkl SJ, Quake SR. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 23.Arkhammar POG, Terry BR, Kofod H, Thastrup O. Methods in Cell Science. 1998;19:255–268. [Google Scholar]
- 24.Moutinho IMT, Ferreira PJT, Figueiredo ML. Ind Eng Chem Res. 2007;46:6183–6188. [Google Scholar]
- 25.Pandolfi A, Ortiz M. J Micromech Microeng. 2007;17:1487–1493. [Google Scholar]
- 26.Abramoff MD, Magelhaes PJ, Ram SJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 27.Jo BH, Van Lerberghe LM, Motsegood KM, Beebe DJ. J MEMS. 2000;9:76–81. [Google Scholar]
- 28.Kartalov EP, Walker C, Taylor CR, Anderson WF, Scherer A. Proc Natl Acad Sci USA. 2006;103:12280–12284. doi: 10.1073/pnas.0602890103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thorsen T, Roberts RW, Arnold FH, Quake SR. Phys Rev Lett. 2001;86:4163–4166. doi: 10.1103/PhysRevLett.86.4163. [DOI] [PubMed] [Google Scholar]
- 30.Song H, Ismagilov RF. J Amer Chem Soc. 2003;125:14613–14619. doi: 10.1021/ja0354566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuerstman MJ, Garstecki P, Whitesides GM. Science. 2007;315:828–832. doi: 10.1126/science.1134514. [DOI] [PubMed] [Google Scholar]
- 32.Kumaresan P, Yang CJ, Cronier SA, Blazej RG, Mathies RA. Anal Chem. 2008;80:3522–3529. doi: 10.1021/ac800327d. [DOI] [PubMed] [Google Scholar]
- 33.Garstecki P, Fuerstman MJ, Stone HA, Whitesides GM. Lab Chip. 2006;6:437–446. doi: 10.1039/b510841a. [DOI] [PubMed] [Google Scholar]
- 34.Hupert ML, Guy WJ, Llopis SD, Shadpour H, Rani S, Nikitopoulos DE, Soper SA. Microfluid Nanofluid. 2007;3:1–11. [Google Scholar]
- 35.Chen CS, Breslauer DN, Luna JI, Grimes A, Chin WC, Lee LP, Khine M. Lab Chip. 2008;8:622–624. doi: 10.1039/b719029h. [DOI] [PubMed] [Google Scholar]
- 36.Stroock AD, Dertinger SKW, Ajdari A, Mezic I, Stone HA, Whitesides GM. Science. 2002;295:647–651. doi: 10.1126/science.1066238. [DOI] [PubMed] [Google Scholar]
- 37.Scharp DW, Kemp CB, Knight MJ, Ballinge WF, Lacy PE. Transplantation. 1973;16:686–689. doi: 10.1097/00007890-197312000-00028. [DOI] [PubMed] [Google Scholar]
- 38.Stefan Y, Meda P, Neufeld M, Orci L. J Clin Invest. 1987;80:175–183. doi: 10.1172/JCI113045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.