Abstract
Background
Geocoding, the process of assigning each case a set of coordinates that closely approximates its true location, is an important component of spatial epidemiological studies. The failure to accurately geocode cases adversely affects the validity and strength of conclusions drawn from the analysis. We investigated whether there were differences among geographic locations and demographic classes in the ability to successfully geocode West Nile virus (WNV) cases in South Dakota. We successfully geocoded 1,354 cases (80.8%) to their street address locations and assigned all 1,676 cases to ZIP code tabulation areas (ZCTAs). Using spatial scan statistics, significant clusters of non-geocoded cases were identified in central and western South Dakota. Geocoding success rates were lower in areas of low population density and on Indian reservations than in other portions of the state. Geocoding success rates were lower for Native Americans than for other races. Spatial epidemiological studies should consider the potential biases that may result from excluding non-geocoded cases, particularly in rural portions of the Great Plains that contain large Native American populations.
Keywords: geocoding, cluster detection, West Nile virus, rural populations, Native Americans populations
Introduction
The use of geographic information systems (GIS) and spatial statistics is increasing in the fields of public health research and epidemiology (Beale et al., 2008). Many studies use GIS and spatial statistics to help understand the risk factors that are associated with disease incidence. For example, epidemiological cancer studies frequently investigate the associations of environmental exposures and geographical variables with cancer (Oliver et al. 2005b). Studies of infectious diseases also use GIS and spatial statistics to investigate the influences of climate, land cover, and other environmental variables that affect habitat suitability for the vectors and the hosts (Ostfeld et al. 2005). To carry out this type of spatial analysis, it is often desirable to have a precise spatial location for each human disease case. Geocoding, the process of assigning each case a spatial coordinate that closely approximates its true location, is therefore an important component of spatial epidemiological studies. Disease cases can be geocoded to points (e.g., latitude and longitude) or to polygons (e.g., U.S. Census tracts or ZIP code tabulation areas). In this study, we use the term geocoding to refer specifically to address matching, the process of determining the spatial coordinates of the point that represents the residential street address of each disease case.
There are two main types of error that occur when geocoding: (1) inaccuracy of the geocoded location and (2) inability to geocode all the desired locations. Obtaining accurate geocoded locations is essential because inaccurate locations adversely affect the validity and strength of conclusions drawn from the analysis (Mazumdar et al., 2008). Not having the ability to geocode all desired locations result in missing data points that must be excluded from spatial analyses. For example, 95% of human cases were successfully geocoded and included in an analysis of West Nile virus cases in Chicago and Detroit (Ruiz et al. 2007), whereas only 86% of cases were successfully geocoded in a study of breast cancer encompassing the entire state of Connecticut (Gregorio et al. 1999). Excluding non-geocoded data can affect the analysis in various ways, from reducing the statistical power of the spatial analysis to producing a selection or geographic bias, which may result in non-random spatial clustering of missing data (Vach et al., 1997; Oliver et al., 2005a; Zimmerman et al., 2008).
There are several reasons for the failure of geocoding: incorrect addresses in the case record file, misspelled words or improper abbreviations of street names; missing street segments in the reference file; and the use of rural routes and post office box numbers (Zimmerman et al. 2008; McElory 2003). A geographic selection bias occurs when there is a non-random pattern of the non-geocoded case data. This bias can result in the detection of disease clusters in particular subgroups of the population while decreasing the power to detect disease clusters in other subgroups. In a study done in Virginia, prostate cancer incidence clusters identified at a county level differed significantly depending on whether all the cases or only those cases that were geocoded to a census tract were used (Oliver et al., 2005a; 2005b). By investigating the factors that influence this selection bias in geocoding disease incidence, we can evaluate whether the non-geocoded cases are spatially clustered, whether the patterns of geocoding success are associated with environmental variables, and whether the inability to geocode is associated with particular subsets of the population.
In this paper, we investigated whether there was a selection bias in the ability to geocode West Nile virus (WNV) cases in South Dakota by comparing the spatial patterns of geocoded and non-geocoded cases at a ZIP code tabulation area (ZCTA) level, examining the influences of population density and Indian reservations on geocoding success at a ZCTA level, and comparing the demographic characteristics of geocoded and non-geocoded cases at an individual level. WNV is a vector-borne pathogen that has affected much of the world. It is an arthropod-transmitted virus, or arbovirus, that is maintained in an enzootic cycle with birds as the primary reservoir hosts and mosquitoes as the primary vectors. WNV was first discovered in Uganda in 1937 and has spread throughout the globe reaching New York City in 1999 (Hayes et al., 2005). Within the next three years, WNV was carried westward reaching South Dakota and the Rocky Mountains in 2002. The Great Plains region has consistently high annual WNV incidence rates compared to the rest of the United States. South Dakota was found to have the highest state-level cumulative incidence of WNV of 32.2 per 100,000 from 2002–2006 (Lindsey et al., 2008). From 2002–2007 South Dakota has had a total of 1,676 human cases of WNV, including 295 cases of WNV neuroinvasive disease and 26 WNV-associated deaths. The sustained high incidence of WNV is the Northern Great Plains is related to climatic and land use patterns that are believed to provide a highly suitable ecological niche for Culex tarsalis, a particularly efficient mosquito vector (Wimberly et al. 2008).
South Dakota, which has an average population density of 9.9 per mile2, encompasses a wide range of population densities and a variety of habitats for the vectors and hosts of WNV. South Dakota’s lowest population density at the block group level is 0.01 per mile2, while the highest population density is 133 per mile2. Also, the landscape across South Dakota changes significantly from east to west; shifting from agricultural landscapes to grasslands and prairies to the rugged Badlands and pine forests of the Black Hills. In addition, South Dakota contains seven Indian Reservations comprising approximately 20% of the state’s land mass and about 8.3% of the South Dakota’s population is Native American (U.S. Census Bureau 2008). Although a previous study demonstrated that failure to geocode occurs more frequently in rural than in urban areas (Oliver et al. 2005), no studies to date have explored the implications of incomplete geocoding in the rural landscape of the northern Great Plains, or in regions with large Native American populations.
To examine the issue of geocoding bias and its potential implications for spatial analyses, we addressed the following hypotheses. (1) spatial clustering exists among non-geocoded cases at the ZCTA level; (2) geocoding success is lower for rural ZCTAs versus non-rural ZCTAs (based on three classes of population density); (3) geocoding success is lower in ZCTAs located within Indian reservations versus non-reservation ZCTAs; (4) geocoding success at the individual level varies with factors such as race, age, sex, and clinical diagnosis, and (5) there will be an increase in geocoding success over time due to the continuing improvements in data collection and surveillance efforts.
Methods
Geocoding of WNV Cases
There were 1,676 WNV disease cases for 2002–2007, from which 1,263 cases (75.4%) were geocoded by the South Dakota Department of Health (SDDOH) using a two-stage geocoding method (Table 1). The geocoding function in ArcGIS 9.2 was used with the South Dakota One Call roads layer as the street reference data to locate the approximate position of each physical street address along the length of a street segment containing address ranges. For those addresses that had a matched score of 80 or above, ArcGIS automatically created a feature for that attribute. For those that did not match, an outside search engine, Live Search Maps (http://maps.live.com/default.aspx), was used to help approximate the location of the physical address, export the spatial coordinates, and match them interactively in ArcMap. As a third step, we were able to use a variety of additional information sources, including web mapping software (yahoo.maps.com) to geocode an additional 91 (5.4%) WNV cases. Three hundred and twenty-two (19.2%) WNV cases were un-geocodable; while 1,354 (80.8%) WNV case locations were successfully geocoded. All geocoded coordinates were converted into a custom South Dakota coordinate system based on the Lambert Conformal Conic projection.
Table 1.
Summary of geocoding success rates.
| 2002 | 2003 | 2004 | 2005 | 2006 | 2007 | Total | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | (%) | N | (%) | N | (%) | N | (%) | N | (%) | N | (%) | N | (%) | |
| Total WNV Cases | 37 | 2.2 | 1039 | 62.0 | 51 | 3.0 | 229 | 13.7 | 113 | 6.7 | 207 | 12.4 | 1676 | 100.0 |
| Geocoded | 26 | 70.3 | 790 | 76.0 | 46 | 90.2 | 211 | 92.1 | 102 | 90.3 | 179 | 86.5 | 1354 | 80.8 |
| South Dakota One Call | 13 | 50.0 | 411 | 52.0 | 15 | 32.6 | 84 | 39.8 | 44 | 43.1 | 68 | 38.0 | 635 | 46.9 |
| Live Search Maps | 9 | 34.6 | 334 | 42.3 | 29 | 63.0 | 117 | 55.5 | 43 | 42.2 | 96 | 53.6 | 628 | 46.4 |
| Interactively | 4 | 15.4 | 45 | 5.7 | 2 | 4.4 | 10 | 4.7 | 15 | 14.7 | 15 | 8.4 | 91 | 6.7 |
| Not Gecoded | 11 | 29.7 | 249 | 24.0 | 5 | 9.8 | 18 | 7.9 | 11 | 9.7 | 28 | 13.5 | 322 | 19.2 |
Data on human WNV disease occurrence from 2002 to 2007 were also summarized at the ZIP code level. ZIP codes were mapped using the South Dakota ZIP Code Tabulation Area (ZCTA) map produced by the U.S. Census Bureau. The 2000 population data for each ZCTA were also obtained from the U.S. Census Bureau. ZIP codes are developed by the U.S. Postal Service and changes are made frequently. ZIP codes can be split, discontinued, added, or expanded. This continual change in ZIP codes makes it difficult for their boundaries to be mapped precisely. Therefore, ZCTAs were developed by the U.S. Census Bureau as spatial units that meet requests by data users for statistical data by ZIP Code areas (Grubesic and Matisziw, 2006). Because ZCTA’s are aggregated Census blocks they do not precisely match the actual ZIP codes used by the U.S. Postal service, and some addresses are located in different ZCTA’s relative to their ZIP code.
The successfully geocoded WNV cases were combined in ArcGIS 9.2 and exported as a shapefile. The spatial join tool in ArcGIS was used to associate the geocoded WNV cases with their respective ZCTA by identifying the WNV cases that fell within the boundary of each ZCTA. The attribute table of the geocoded points thus contained the year, ZCTA, ZIP code, county, age, sex, race and clinical diagnosis for each case. For the unsuccessfully geocoded WNV cases, the ZIP codes from the original case record were used to assign a ZCTA to each case. Four unsuccessfully geocoded WNV case addresses were manually located and linked their respective ZCTAs using city and county information because of unreadable ZIP Codes. These datasets were then aggregated to the ZCTA level to provide counts of the total numbers of geocoded and non-geocoded WNV cases for each ZCTA.
Analysis
The spatial scan statistic was used for cluster analyses at the ZCTA level (Kulldorff, 1997). This method used a variable-sized circular window to test for possible clusters with varying incidence and geocoding success rates. The center of the window was positioned on the centroid of each ZCTA, and a range of window sizes and shapes were examined. A purely spatial Poisson model was used to search for the most likely clusters of high rates using a maximum spatial cluster size of 5% of the total population. A test of statistical significance was carried out using a Monte Carlo simulation of 9,999 random datasets generated under the null hypothesis. The analysis based on geocoding success rates used the total number of non-geocoded cases as the case variable and the total number of cases as the population. This method identified zones where the proportion of non-geocoded cases was higher than expected. Cluster analysis was carried out using the SaTScan™ software package (Kulldorff, 2006).
To investigate possible geocoding bias in rural areas, all of the WNV cases from 2002–2007 were classified into three groups according to the population density of their ZCTA. The population density was computed using the ZCTA population and area data from the 2000 U.S. Census of Population and Housing. The low group contained WNV cases which occurred in ZCTAs with a population density of 0.0–0.50 per mile2. The medium group contained WNV cases which occurred in ZCTAs with a population density of 0.51–1.00 per. mile2. The high group contained WNV cases that occurred in ZCTAs with a population density of 1.01–133.20 per mile2.
To investigate geocoding success rates on Indian reservations, all WNV cases from 2002–2007 were separated into reservation and non-reservation groups. There are seven reservations in South Dakota—Yankton, Sisseton-Wahpeton, Cheyenne, Crow Creek, Lower Brule, Pine Ridge and Rosebud. Standing Rock, Cheyenne, Pine Ridge, Rosebud, and Yankton reservations were defined by county boundaries (Corson, Ziebach, Dewey Shannon, Jackson, Todd, and Charles Mix). For the Sisseton-Wahpeton, Crow Creek, and Lower Brule reservations all geocodable cases in the counties encompassing these reservations were mapped and classified as reservation or non-reservation using the case coordinates and a map of the reservation boundaries. For the non-geocodable cases, city and ZIP code were used to determine whether or not they were located on a reservation.
To investigate factors such as sex, race and clinical diagnosis, each factor was converted into a categorical variable. Race was categorized into three groups (White, Native American and Other) due to the low numbers of other races. Clinical diagnosis was categorized into encephalitis/meningitis or WNV fever. A contingency table was then made for each biological factor against the geocoded and non-geocoded WNV cases. Age was analyzed as a continuous variable. Mean ages were computed for both the geocoded and non-geocoded cases.
The dataset was separated into two groups, epidemic years (2002–2003) and endemic years (2004–2007). An epidemic is defined as an increase in the prevalence of a disease over baseline rates; therefore 2002 and 2003 were considered epidemic years because of the appearance of the disease in 2002 and the exceptionally high WNV incidence in 2003. 2004 to 2007 were considered endemic years due to the lower numbers of WNV cases during these years.
Chi-square analyses were used to evaluate the association of geocoding success with the different risk factors: population density group (N=1,675), and reservation group (N=1,676 at the ZCTA level; and sex (N=1,673), race (N=1,655), and clinical diagnosis (N=1,675) at the individual level. Student’s t-test was used to determine whether the mean ages (N=1,676) differed between geocoded and non-geocoded WNV cases at the individual level. The number of cases in each analysis varied due to missing data. A log linear model was used to determine whether population density (low, medium, high), race (Native American, White, Other), or reservation (vs. non-reservation), were independently associated with the ability to geocode. Statistical analysis was carried out using JMP (SAS Institute, Cary, NC, US, version 7.0).
Results
We successfully geocoded 1,354 cases (80.8%). A high percent of the WNV cases that were not successfully geocoded were rural routes or PO boxes (60%, 192/322). The remaining non-geocodable WNV cases were individuals with no address on record or addresses that could not be found (40%, 128/322).
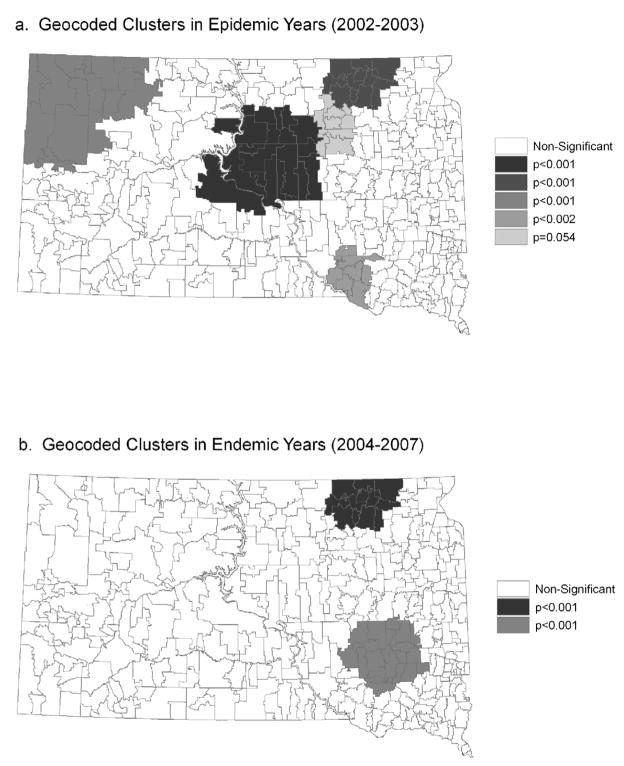
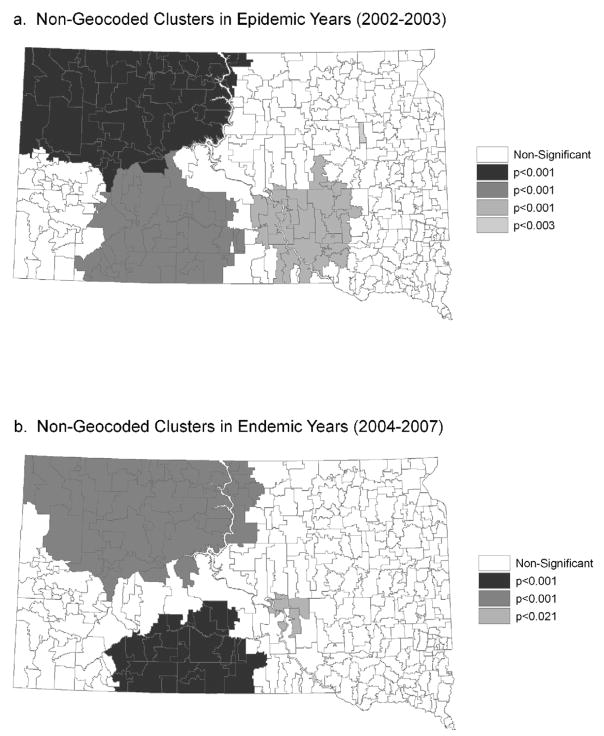
For the geocoded cases, there was a consistent cluster of high WNV rates in northeastern South Dakota, along the James River in both the epidemic and endemic years (Figure 1). During the epidemic years there was also a pattern of clusters following the James River that was concentrated more in the northern part of the state. Clusters also existed in central and northwestern South Dakota during the epidemic years. For the non-geocoded cases, there were four significant clusters containing a large proportion of South Dakota reservations and rural areas in the epidemic years 2002 and 2003 (Figure 2). These clusters encompassed ZCTAs where the proportion of cases that were not successfully geocoded was higher than expected. In the endemic years, 2004 to 2007, there were three significant clusters containing the three major SD reservations. Except for the northwestern portion of the state during the epidemic years, the overlap between clusters of geocoded and non-geocoded cases was relatively small. As the population density of the ZCTAs decreased, the probability of geocoding success also decreased (Table 2). The percent of WNV cases that were successfully geocoded was significantly lower in reservations than in non reservations (Table 2).
Figure 1.
Geographic clusters of geocoded WNV cases identified using the spatial scan statistic at the ZCTA level. Only the cases that were successfully geocoded at the street address level were included in the analysis. (a) Epidemic years (2002–2003) and (b) Endemic years (2004–2007).
Figure 2.
Geographic clusters of non-geocoded WNV cases identified using the spatial scan statistic at the ZCTA level. (a) Epidemic years (2002–2003) and (b) Endemic years (2004–2007).
Table 2.
Selected characteristics of geocoded and non-goecoded populations.
| Geocoded population (N = 1354) |
Non-Geocoded population (N = 322) |
p-Values* |
|||
|---|---|---|---|---|---|
| Covariate | N | % | N | % | |
| Sex | 0.130 | ||||
| Female | 588 | 79.1 | 155 | 20.9 | |
| Male (Unknown, N=3) |
763 | 82.0 | 167 | 18.0 | |
| Race | <0.001 | ||||
| White | 1286 | 85.7 | 215 | 14.3 | |
| Native American | 44 | 29.9 | 103 | 70.1 | |
| Other (Unknown, N=21) |
6 | 85.7 | 1 | 14.3 | |
| Clinical Diagonsis | 0.370 | ||||
| WNV Fever | 1097 | 80.4 | 268 | 19.6 | |
| Encephalitis / Meningitis (Unknown, N=1) |
256 | 82.6 | 54 | 17.4 | |
| Location | <0.001 | ||||
| Reservation | 68 | 38.6 | 108 | 61.4 | |
| Non-Reservation (Unknown, N=0) |
1286 | 85.7 | 214 | 14.3 | |
| Population Density | <0.001 | ||||
| Low (0.0–0.50/mi2) | 595 | 69.2 | 265 | 30.8 | |
| Group Medium (0.51–1.0/mi2) | 166 | 84.7 | 30 | 15.3 | |
| Group High (1.1–133.0/mi2) (Unknown, N=1) |
592 | 95.6 | 27 | 4.36 | |
| Time | <0.001 | ||||
| Epidemic | 816 | 75.8 | 260 | 24.2 | |
| Endemic | 538 | 89.7 | 62 | 10.3 | |
p-values based on chi-square analyses
Overall, there were few differences in the individual-level covariates between the geocoded and non-geocoded populations (Table 2). Race was the only covariate that was significantly different between geocoded and non-geocoded cases (p< 0.001). Native Americans were the second largest population in South Dakota (8.3% of the SD population), yet 32% of the non-geocoded cases were Native American, whereas only 3.3% of the geocoded cases were Native American. This result supports the finding of a lower ability to geocode individuals on reservations. Age, sex, and type of disease (encephalitis/meningitis vs. WNV fever) were not associated with the ability to geocode. When categorizing the WNV cases into groups based on time (epidemic vs. endemic years), there was a significant difference in the proportion of cases that could be geocoded (Table 2) with a larger percent of cases not being geocoded during the epidemic years. In a multivariate analysis including population density, race, and reservation, we found both population density and race to be associated with the geocoding success rate (Table 3). This finding indicated a significant independent effect of both of these factors.
Table 3.
Multicontingency table of reservation, population density and race by ability to geocode.
| Population Densityab | Racea | Geocoded |
||
|---|---|---|---|---|
| Yes | No | |||
| Non-Reservation | Low | Native American | 8 | 17 |
| White | 528 | 154 | ||
| Other | 3 | 2 | ||
| Medium | Native American | 6 | 7 | |
| White | 146 | 7 | ||
| Other | 3 | 1 | ||
| High | Native American | 15 | 2 | |
| White | 561 | 24 | ||
| Other | 15 | 0 | ||
| Reservation | Low | Native American | 11 | 63 |
| White | 42 | 28 | ||
| Other | 3 | 1 | ||
| Medium | Native American | 3 | 13 | |
| White | 8 | 2 | ||
| Other | 0 | 0 | ||
| High | Native American | 1 | 1 | |
| White | 0 | 0 | ||
| Other | 0 | 0 | ||
Both race and population density were significantly associated with the ability to geocode (both, p<0.001).
Population density/mi2: Low 0.00–0.50; Medium 0.51–1.00; High 1.10–133.00
Discussion
One of the main reasons for using GIS and spatial statistics in epidemiological research is to investigate and understand the geographic patterns of disease (Beale et al., 2008). The advantages and disadvantages of analyzing geocoded data have been important issues in the spatial epidemiology literature. If precisely geocoded epidemiological case data is available, investigations of spatial clustering can reveal the underlying disease pattern. Based on these observed patterns, researchers can then begin to investigate the environmental factors that influence the geographic pattern of disease. However, as shown in this study and previous studies (Gregorio et al. 1999; Oliver et al., 2005a; Gilboa et al., 2006; Zimmerman et al., 2008), the results of spatial analysis may not accurately represent the pattern of disease when the geocoding success rate is less than 100%. If geocoding success is higher in certain geographic areas or in particular segments of the population, then the results of statistical analyses may reflect the pattern of geocoding success rather than the true pattern of disease. Our findings demonstrate that patterns of geocoding success in South Dakota are indeed spatially clustered at the ZCTA level, and that clusters of geocoded cases detected at the ZCTA level are generally located in portions on the state where geocoding success rates are relatively high.
We hypothesized that population density would have a positive influence on success rates for geocoding WNV cases. The results from the population density analysis supported this hypothesis, showing that cases in areas of low density have the lowest probability of being successfully geocoded. The inability to geocode rural routes and PO boxes is one of the most important causes of low geocoding success rates in rural areas (Hurley et al., 2003; Zimmerman et al. 2008). Thus, states like South Dakota that have a large rural population are likely to have lower geocoding success rates than other states where a greater proportion of the population lives in suburban and urban areas. A major implication of this bias is that we may fail to detect real WNV clusters that exist in areas where geocoding success rates are low (Oliver et al. 2005a). In South Dakota, this problem is likely to particularly acute in the sparsely populated western portion of the state. Analyses of WNV virus may be particularly sensitive to the resulting selection bias because incidence rates are highest in rural populations (Wimberly et al., 2008).
Race was shown to be a significant factor influencing geocoding success rates at the individual level. The Native American population is the second largest race (8.3%) in South Dakota and Indian reservations occupy approximately 40% of the state’s rural land area. Thus, the rural effects discussed previously are one possible explanation for low geocoding success among Native Americans. However, the results of the log-linear model indicated that race also had an effect on geocoding success that was independent of population density. This selection bias could lead to an underestimate of disease incidence in geographical areas with a large Native American population in analyses that are based solely on the successfully geocoded cases. Similar selection biases resulting from variability in geocoding success have been reported in other studies. For example, Gilboa et al. (2006) found that geocoding success rates were lower for Latinos than non-Latinos in a study of air quality and birth defects in Texas. Gregorio et al. (1999) found that geocoding success rates were higher for non-whites than whites in a study of breast cancer in Connecticut.
A limitation of our study was that the analyses of spatial clustering and geographic associations of non-geocoded cases could only be carried out at the ZCTA level. Therefore, although we were able to identify particular areas within South Dakota that have low rates of geocoding success, we were not able to assess the impacts of spatially variability in geocoding success on spatial analyses conducted on the point locations of individual cases. Such an analysis would require a complete and accurate census of all residence locations, such as the dataset developed for spatial analysis of geocoding failure in rural Iowa (Zimmerman et al., 2008). It was not feasible to develop such a dataset for our retrospective study of WNV cases in South Dakota. However, the ZCTA-level analyses presented here still provide valuable information on statewide patterns of geocoding success that can be used to select areas for more detailed follow-up studies, and can help target efforts to increase the rates of geocoding success for future disease surveillance efforts.
The results of this study caution that statewide spatial analyses based only on successfully geocoded cases have the potential to yield spurious results. For example, such an analysis might erroneously conclude that Native Americans have lower risk of WNV than other races, and might fail to detect clusters of high WNV incidence that occur in rural areas or on Indian reservations. Various approaches can be used to minimize these biases. In this study, 100% of the WNV cases were successfully assigned to ZCTAs, and area-based spatial analysis using ZCTAs can therefore be used without the possibility of spatial bias from variable geocoding success rates. Although many area-based spatial epidemiological studies have traditionally used counties as analysis units, the use of ZCTAs for spatial epidemiology has recently been encouraged because ZCTAs are smaller than counties and therefore have the potential to reveal finer-scale patterns of disease incidence (Eisen and Eisen, 2007). However, a limitation of this approach is that the mapped ZCTA boundaries developed by the U.S. Census Bureau do not precisely match the spatial boundaries of the ZIP codes that are utilized by the U.S. Postal Service, resulting in an unknown level of error in assigning disease cases to ZCTAs (Grubesic and Matisziw, 2006; Krieger 2002). Furthermore, the arbitrary boundaries of ZCTAs are not necessarily the most relevant spatial units to use for the analysis of WNV or other diseases.
When geocoded coordinates are available for individual disease cases, spatial analysis methods can be applied that do not require the study area to be subdivided into ZCTAs, counties, or other arbitrary areal units. Detailed measurements of environmental risk factors can be taken in the vicinity of the geocoded cases, rather than assuming that risk is homogeneous across ZCTAs or counties. However, this type of localized analysis is only valid if most individuals can be assumed to have contracted the disease either at or near their home address. Furthermore, imprecision in the geocoding process introduces error into these coordinates which can affect the outcome of spatial analyses (Mazumdar et al., 2008).
For WNV in South Dakota, analyses of the effects of land cover and land use on the pattern of individual disease cases offer an opportunity to examine environmental risk factors at a relatively fine spatial resolution and highlight neighborhoods that are at a particularly high risk for WNV. Key environmental factors of interest include various types of land use (e.g., cropland, pasture, development), irrigation, and distance from ponds, wetlands, sloughs, and other potential breeding sites. At the present time, however, this type of analysis should be restricted to sub-regions of the state where geocoding success rates are relatively high. In the future, additional research should be conducted to assess the impacts of low geocoding success on spatial analyses conducted at the individual case level, and strategies need to be developed that will help to increase the rates of geocoding success on Indian reservations and in rural areas.
Acknowledgments
Christine Wey was supported by a Joseph F. Nelson Research Mentorship Award from South Dakota State University. Financial support for this research was provided by National Institutes of Health, National Institute of Allergy and Infectious Diseases (grant R01-AI079411). Aki Michimi and an anonymous reviewer provided helpful comments on earlier drafts of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beale L, Abellan JJ, Hodgson S, Jarup L. Methodologic issues and approaches to spatial epidemiology. Environmental Health Perspectives. 2008;116:1105–1110. doi: 10.1289/ehp.10816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen L, Eisen RJ. Need for improved methods to collect and present spatial epidemiologic data for vector borne diseases. Emerging Infectious Diseases. 2007;13:1816–1820. doi: 10.3201/eid1312.070211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa SM, Mendola P, Olshan AF, Harness C, Loomis D, Langlois PH, Savitz DA, Herring AM. Comparison of residential geocoding methods in population-based study of air quality and birth defects. Environmental Research. 2006;6:259–262. doi: 10.1016/j.envres.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Gregorio DI, Cromley E, Mrozinski R, Walsh SJ. Subject loss in spatial analysis of breast cancer. Health & Place. 1999;5:173–177. doi: 10.1016/s1353-8292(99)00004-0. [DOI] [PubMed] [Google Scholar]
- Grubesic TH, Matisziw TC. On the use of ZIP codes and ZIP code tabulation areas (ZCTAs) for the spatial analysis of epidemiological data. International Journal of Health Geographics. 2006;5:58. doi: 10.1186/1476-072X-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes EB, Komar N, Nasci RS, Montgomery SP, O’Leary DR, Campbell GL. Epidemiology and transmission dynamics of West Nile Virus disease. Emerging Infectious Diseases. 2005;11:1167–1173. doi: 10.3201/eid1108.050289a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley SE, Saunders TM, Nivas R, Hertz A, Reynolds P. Post office box addresses: a challenge for geographic information system-based studies. Epidemiology. 2003;14:386–391. doi: 10.1097/01.EDE.0000073161.66729.89. [DOI] [PubMed] [Google Scholar]
- Krieger N, Waterman PD, Chen JT, Soobader MJ, Subramanian SV, Carson R. ZIP code caveat: Bias due to spatiotemporal mismatches between ZIP codes and U.S. census-defined geographic areas -- The Public Health Disparities Geocoding Project. American Journal of Public Health. 2002;92:1100–1102. doi: 10.2105/ajph.92.7.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger N, Chen JT, Waterman PD, Soobader MJ, Subramanian SV, Carson R. Geocoding and monitoring of U.S. socioeconomic inequalities in mortality and cancer incidence: Does the choice of area-based measure and geographic level matter? The Public Health Disparities Geocoding Project. American Journal of Epidemiology. 2002;156:471–482. doi: 10.1093/aje/kwf068. [DOI] [PubMed] [Google Scholar]
- Kulldorff M. SaTScanTM v7.0.3: Software for the spatial and space-time scan statistics. Information Management Services, Inc; 2006. [Google Scholar]
- Kulldorff M. A spatial scan statistic. Communications in Statistics: Theory and Methods. 1997;26:1481–1496. doi: 10.1080/03610927708831932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey N, Kuhn S, Campbell GL, Hayes E. West Nile Virus: Neuroinvasive Disease Incidence in the United States, 2002–2006. Vector-Borne and Zoonotic Diseases. 2008;8:35–40. doi: 10.1089/vbz.2007.0137. [DOI] [PubMed] [Google Scholar]
- Mazumdar S, Rushton G, Smith BJ, Zimmerman DL, Donham KJ. Geocoding accuracy and the recovery of relationships between environmental exposures and health. International Journal of Health Geographics. 2008;7:13. doi: 10.1186/1476-072X-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy JA, Remington PL, Trentham-Dietz A, Robert SA, Newcomb PA. Geocoding addresses from a large population-based study: lessons learned. Epidemiology. 2003;14:399–407. doi: 10.1097/01.EDE.0000073160.79633.c1. [DOI] [PubMed] [Google Scholar]
- Oliver MN, Matthews KA, Siadaty M, Hauck FR, Pickle LW. Geographic bias related to geocoding in epidemiologic studies. International Journal of Health Geographics. 2005a;4:29. doi: 10.1186/1476-072X-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver MN, Smith E, Siadaty M, Hauck F, Pickle RLW. A spatial analysis of prostate cancer incidence and race in Virginia, 1990–1999. American Journal of Preventive Medicine. 2005b;30:S67–76. doi: 10.1016/j.amepre.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Ostfeld R, Glass G, Keesing F. Spatial epidemiology: an emerging (or re-emerging) discipline. Trends in Ecology and Evolution. 2005;6:228–236. doi: 10.1016/j.tree.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Ruiz MO, Walker ED, Foster ES, Haramis LD, Kitron UD. Association of West Nile virus illness and urban landscapes in Chicago and Detroit. International Journal of Health Geographics. 2007;6:10. doi: 10.1186/1476-072X-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Census Bureau. State & Country Quick Facts. 2008 ≪ http://quickfacts.census.gov/qfd/states/46000.html≫.
- Vach W. Some issues in estimating the effect of prognostic factors from incomplete covariate data. Statistics in Medicine. 1997;16:57–72. doi: 10.1002/(sici)1097-0258(19970115)16:1<57::aid-sim471>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Wimberly MC, Hildreth MB, Boyte SP, Lindquist E, Kightlinger L. Ecological niche of the 2003 West Nile virus epidemic in the northern Great Plains of the United States. PLOS One. 2008:e3744. doi: 10.1371/journal.pone.0003744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman DL, Fang X, Mazumdar S. Spatial clustering of the failure to geocode and its implications for the detection of disease clustering. Statistics in Medicine. 2008;27:4254–4266. doi: 10.1002/sim.3288. [DOI] [PubMed] [Google Scholar]