Abstract
Accurate chromosome segregation depends on the kinetochore, the complex of proteins that link microtubules to centromeric DNA1. The budding yeast kinetochore consists of more than 80 proteins assembled on a 125bp region of DNA1. We studied the assembly and function of kinetochore components by fusing individual kinetochore proteins to the lactose repressor (LacI) and testing their ability to improve the segregation of a plasmid carrying tandem repeats of the lactose operator (LacO). Targeting Ask1, a member of the Dam1-DASH microtubule-binding complex, creates a synthetic kinetochore that performs many functions of a natural kinetochore: it can replace an endogenous kinetochore on a chromosome, biorient sister kinetochores at metaphase of mitosis, segregate sister chromatids, and repair errors in chromosome attachment. We show the synthetic kinetochore’s functions do not depend on the DNA-binding components of the natural kinetochore but do require other kinetochore proteins. We conclude that tethering a single kinetochore protein to DNA triggers the assembly of the complex structure that directs mitotic chromosome segregation.
Keywords: kinetochore, mitosis, chromosome segregation, Dam1 complex, DASH complex
The kinetochore ensures accurate chromosome segregation by attaching chromosomes to the microtubules of the spindle. The kinetochore is built on centromeric DNA and in mitosis, the two kinetochores of replicated sister chromatids attach to microtubules from opposite spindle poles1. The opposing forces on the two kinetochores are resisted by cohesin molecules that encircle the two sister chromatids2, creating tension at the kinetochores and stabilizing their binding to microtubules. If both kinetochores attach to the same pole, they sense the reduced tension and release their microtubules, activating the spindle checkpoint, delaying anaphase, and allowing another attempt at proper attachment. At the onset of anaphase, cohesin is cleaved and sister chromatids separate from one another, allowing their kinetochores to travel to the spindle poles by following the ends of depolymerizing microtubules1.
The budding yeast centromere is 125 base pairs long, assembles one microtubule-binding site, and recruits more than 80 proteins. Many of these kinetochore proteins are conserved among eukaryotes3 and assemble into biochemically and genetically defined complexes. The highly conserved KMN network includes the Mtw1 and Ndc80 complexes, but the DNA-binding CBF3 complex and microtubule-binding Dam1-DASH complex are less conserved1.
To understand how the kinetochore assembles, we asked if we could build a synthetic kinetochore in budding yeast. We recruited a lactose repressor (LacI)-kinetochore protein fusion to a plasmid that lacked a centromere but carried multiple tandem repeats of the lactose operator (LacO) (Figure 1A) and monitored the segregation of this plasmid. Without a centromere, plasmids prefer to segregate to the mother cell at division and are lost at a high frequency4; forming a synthetic kinetochore should dramatically reduce the rate at which the test plasmid is lost5 (Figure 1B).
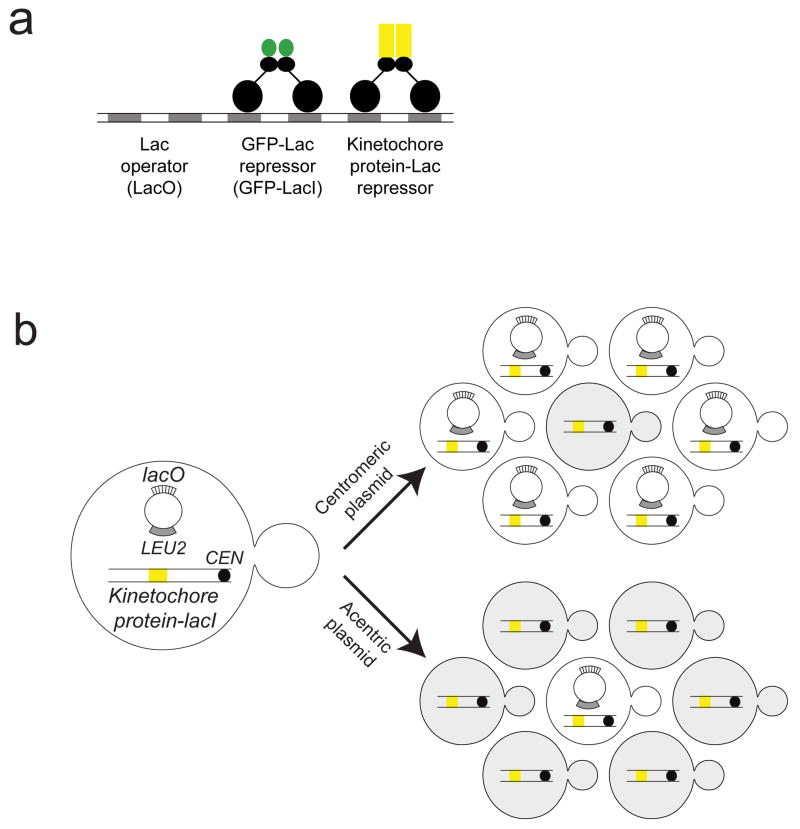
Figure 1. Assay for a synthetic kinetochore.
A) Representation of binding of the Lac repressor (LacI) to the Lac operator (LacO). A dimer of LacI binds to palindromic LacO sequences. In our assays, we express a GFP-LacI fusion protein, a kinetochore protein-LacI fusion, or both (so that we can visualize the synthetic kinetochore). B) A diagram of the assay to determine if tethering individual proteins to DNA forms a synthetic kinetochore. Cells express a chromosomally integrated gene that fuses a kinetochore protein to LacI and the fusion protein is recruited to a plasmid that carries an array of tandem repeats of LacO and confers the ability to synthesize leucine, but lacks a centromere (acentric). The cells are grown in the absence of selection and the fraction containing the plasmid is measured. If the tethered protein confers kinetochore function, the plasmid will segregate to many of the daughter cells (like the control centromeric plasmid); if not, only a small fraction of the cells retain the plasmid (like the control acentric plasmid).
Our initial assay measured the fraction of cells that contain the plasmid after growth in medium that does not select for the plasmid. Under these conditions, only 20% of cells contain a centromere-less plasmid, compared to 80% when the plasmid carries a centromere (Figure 1B). We screened 17 kinetochore protein-LacI fusions (Supplementary Table 1). Only one, the fusion between the C-terminus of Ask1 and the N-terminus of LacI, enhanced the plasmid’s segregation: 65% of the cells carried the LacO-containing plasmid. We got similar results with plasmids containing 8 or 256 copies of LacO (Supplementary Table 2).
Ask1 is an essential protein, which is a component of the 10-member Dam1-DASH complex1, 3, whose association with the kinetochore depends on microtubules6–8. The N-terminal half of the protein enhances plasmid segregation and complements a deletion of ASK1 (Supplemental Figure 1) and localizes to spindle microtubules indistinguishably from full-length Ask1. This correlation suggests that Ask1-LacI recruits the microtubule-binding activity of the Dam1-DASH complex to the LacO array. Fusions of other members of the Dam1-DASH complex to LacI did not enhance plasmid segregation (Supplementary Table 1, 2) even though several could replace the endogenous, essential gene. The observation by Kiermaier et al. that a different fusion to Dam1 could rescue plasmid segregation suggests that some of our fusions are only partially functional9.
The Ask1-LacI fusion nucleates a “synthetic kinetochore” on the LacO array that can replace the natural kinetochore and direct chromosome segregation. Chromosomes with two centromeres (dicentrics) are mitotically unstable10, prompting us to ask if chromosomes carrying a natural and a synthetic kinetochore were lost frequently. In diploid cells, the wild type version of chromosome III is lost at a rate of 1 to 2 × 10−6/cell/generation, but adding the synthetic kinetochore to this chromosome increased the loss rate 275 fold to 5.5 × 10−4 showing that the synthetic kinetochore can interfere with a natural kinetochore’s ability to direct chromosome segregation (Figure 2A).
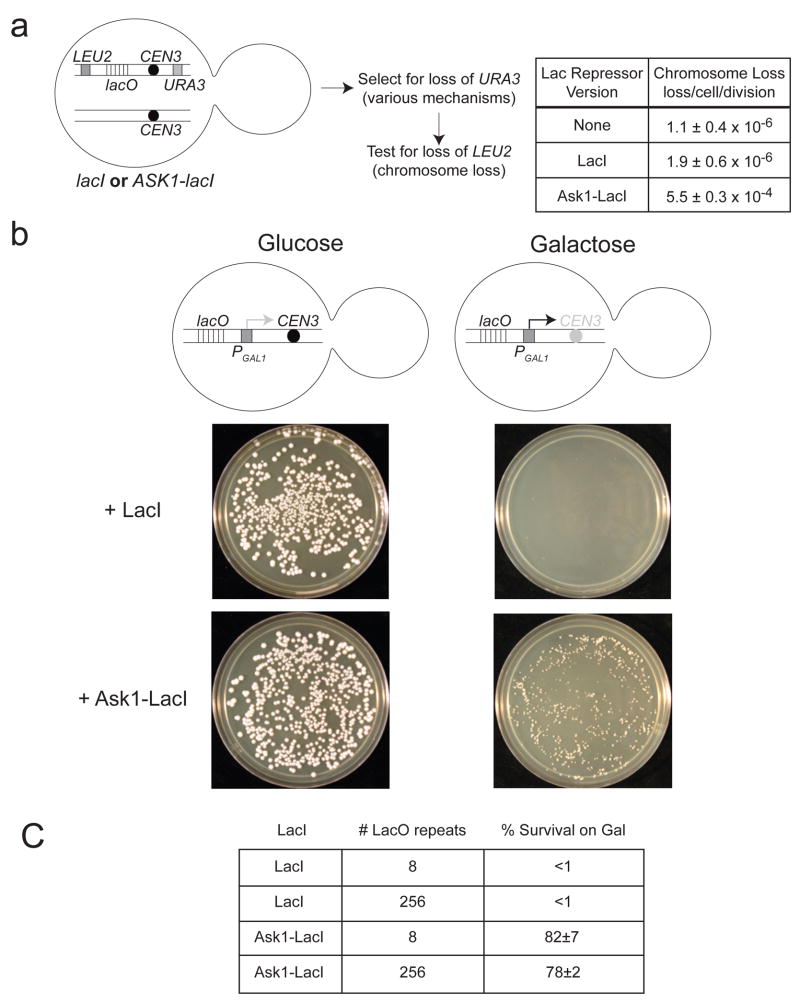
Figure 2. The synthetic kinetochore can replace a natural kinetochore.
A) The presence of a synthetic kinetochore interferes with normal chromosome segregation. The cartoon shows the relevant features of the genotype of a diploid strain carrying one normal copy of chromosome III and one that carried both the natural and synthetic kinetochore. We measured the loss frequency of this chromosome in cells where the synthetic kinetochore is active (due to expression of the Ask1-LacI fusion) and those where it is not (cells without LacI and cells expressing unfused LacI). We first looked for loss of URA3 and then looked for those that had also lost LEU2 to determine that the entire chromosome was lost. The quoted range represents the 95% confidence interval. B) The synthetic kinetochore can substitute for a natural kinetochore. Cartoon of a conditional centromere. Galactose-induced transcription from the GAL1 promoter inactivates CEN3, rendering the natural kinetochore functional on glucose but inactive on galactose. Only cells expressing the Ask1-LacI fusion form colonies on galactose plates. Pictures were taken after 3 days of growth on glucose plates and 5 days of growth on galactose plates at 30°C. C) 8 or 256 repeats of the LacO array give similar results. Plating experiments were performed on strains with 8 or 256 repeats of the LacO array. Approximately 500 cells were plated to YPGlu or YPGal. The percent of cells that survived on YPGal compared to YPGlu was calculated and the average taken from 3 independent experiments where 4 plates were counted per experiment ± s.d.
We made the natural centromere repressible by placing the GAL1 promoter upstream of the centromere; transcription from a strong promoter towards the centromere inactivates the kinetochore11. Adding galactose activates the GAL1 promoter, disrupting the natural kinetochore, which is fully functional in glucose-grown cells (Figure 2B).
We made haploid cells with the synthetic kinetochore on chromosome III, 23kb from the galactose-repressed CEN3. In the absence of Ask1-LacI, these cells form colonies on glucose, but less than 1% form colonies on galactose because they cannot tolerate the loss of chromosome III (Figure 2B). With Ask1 bound to the chromosome, 78 ± 2% of the cells form colonies on galactose plates. This experiment shows that the synthetic kinetochore can segregate a chromosome, although less well than a natural kinetochore, since the colonies are smaller than those of wildtype cells.
To ask how many copies of Ask1 allow chromosome segregation, we repeated this assay with a LacO array containing only 8 repeats. We found that 8 and 256 repeats gave similar results (Figure 2C). Since LacO binds a dimer of LacI, no more than 16 copies of Ask1 can be recruited to the smaller array, similar to the number of Ask1 molecules in the natural kinetochore12.
We followed chromosome segregation by expressing a green fluorescent protein-LacI fusion (GFP-LacI) that binds the LacO array13. This fusion reveals the tagged chromosomes as GFP dots but has no effect on the ability of Ask1-LacI to direct their segregation (data not shown). We monitored chromosome segregation within a single cell cycle by releasing cells from G1, with or without Ask1-LacI, and allowing them to arrest in anaphase (due to the cdc15-2 mutation). In glucose-grown cells, the natural centromere segregated the chromosomes to opposite ends of the spindle in 99±1% of the cells (Figure 3A). With the natural centromere turned off, and without Ask1-LacI, the chromosome tends to stay in the mother cell; only 27±9% segregated their sister chromatids into mother and bud, but 71±9% of cells contained two GFP dots in the mother. With the synthetic kinetochore on, and the natural centromere off, 74±8% of cells segregated sister chromatids properly between mother and bud. This experiment demonstrates that the synthetic kinetochore can direct ordered chromosome segregation, although this segregation is not as faithful as a natural kinetochore.
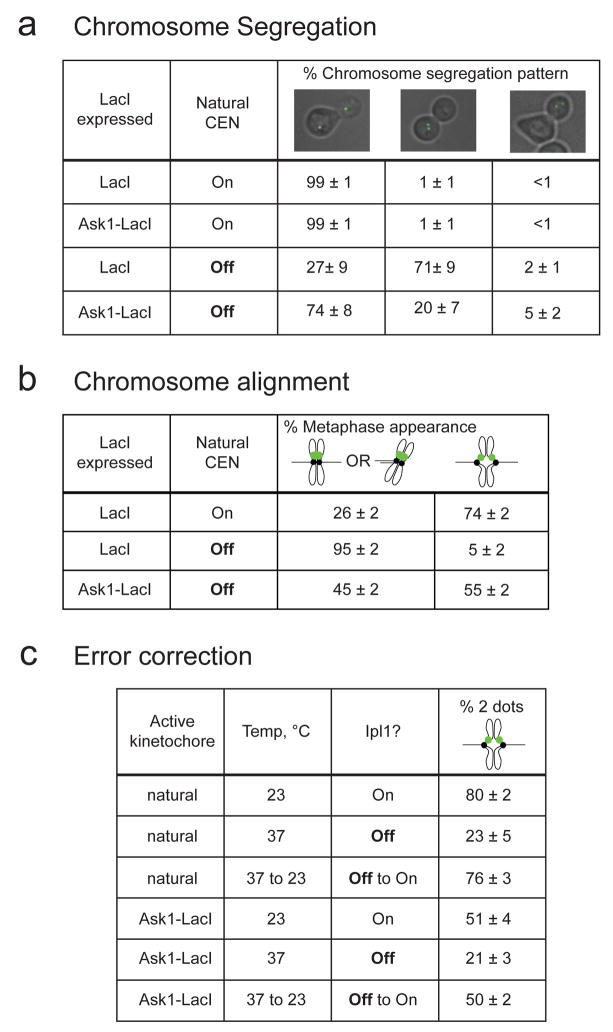
Figure 3. The synthetic kinetochore can align and segregate chromosomes and correct attachment errors.
A) The synthetic kinetochore directs chromosome segregation. To monitor the segregation of sister chromatids, cells whose chromosome III carried both the LacO array and the conditional centromere (see Fig. 2B) were grown in either glucose (CEN3 ON) or galactose (CEN3 OFF) and arrested in anaphase. The synthetic kinetochore is active in cells expressing Ask1-LacI and inactive in those expressing LacI. All cells are expressing GFP-LacI and thus the sister chromatids are marked with GFP dots at the LacO array. Cells were classified into three categories: GFP dots separated with one in mother and one in the bud, two GFP dots in the mother, and two GFP dots in the daughter. Values in this figure represent the mean percent of cells ± s.d. from 3 independent experiments where at least 200 cells per experiment were counted. B) The synthetic kinetochore aligns chromosomes on the metaphase spindle. To determine if sister chromatids are able to biorient at metaphase, cells containing the LacO array and the conditional centromere were grown in either glucose or galactose, arrested in metaphase, and analyzed for transient separation of sister chromatids. If sister chromatids are transiently separated, two GFP dots are detected. If sister chromatids are bioriented but not separated or are mono-oriented, only one GFP dot is detected. The synthetic kinetochore is active in cells expressing Ask1-LacI and inactive in those expressing LacI. C). The synthetic kinetochore corrects errors in initial chromosome alignment. To verify that Ipl1 is needed for correct alignment of the synthetic kinetochore, cells carrying the temperature sensitive ipl1-321 mutant were arrested in G1 and then allowed to proceed to a metaphase arrest at either 23 °C (Ipl1 on), or 37 °C (Ipl1 off). To measure error correction, cells that had gone from G1 to the metaphase arrest at 37 °C were returned to 23 °C, restoring the activity of Ipl1. All assays with the synthetic kinetochore were conducted after inactivating the natural centromere and cells were scored as having one or two GFP-LacI dots.
Natural kinetochores biorient at metaphase; the sister kinetochores attach to opposite spindle poles and are pulled about 0.5μm apart from each other1. This separation can be seen by placing a GFP tag 1.8kb from the centromere and arresting the cells in metaphase (by depleting Cdc20, the activator of the anaphase promoting complex (APC))14–16; with the natural centromere active, 74±2% of metaphase-arrested cells contained two separate GFP dots (Figure 3B), whereas the synthetic kinetochore was separated in 55±2% of cells when the natural centromere was off. Without a synthetic kinetochore and with the natural centromere off, only 5±2% of cells contained two GFP dots. We conclude that the synthetic kinetochore can biorient at metaphase.
In some cells, both sister kinetochores initially attach to the same pole of the spindle. Left uncorrected, this error leads to one daughter with two copies of the chromosome and one with none. The kinetochore detects and corrects this error: the protein kinase Ipl1 (the yeast Aurora B homolog) induces kinetochores that are not under tension to release their microtubules17–20.
We asked if the synthetic kinetochore could detect and correct orientation errors. Cells carrying the ipl1-321 mutation detect and correct errors at 23 °C but not 37 °C17. When the mutant went from G1 to metaphase at 23°C, a natural kinetochore was visibly bioriented in 80±2% of the cells (Figure 3C). But if the cells went from G1 to metaphase at 37°C only 23±5% of the cells contained two GFP dots. These errors could be corrected; shifting the metaphase-arrested cells from 37 °C to 23 °C, raised the fraction of cells containing two GFP dots to 76±3%.
We showed that the synthetic kinetochore can also correct errors in attachment (Figure 3C). Cells carrying ipl1-321 and a chromosome with both the galactose-repressible (PGAL1-CEN3) and the synthetic kinetochore were arrested in G1, exposed to galactose to disrupt the formation of the natural kinetochore and allowed to progress from G1 to a metaphase arrest at different temperatures. When Ipl1 was active (23 °C), 51±4% of the synthetic kinetochores were visibly bi-oriented. In contrast, only 21±3% of cells that had reached metaphase at 37 °C contained two GFP dots. When these arrested cells were transferred to 23 °C this fraction rose to 50±2%, showing that the synthetic kinetochore can both sense and correct orientation errors.
Finally, we determined which other kinetochore proteins were required to produce a synthetic kinetochore. We obtained cells with temperature-sensitive mutations in a variety of kinetochore proteins3 and verified that they formed spindles (demonstrating that they had microtubules) and prevented natural kinetochores from biorienting at 37 °C. We asked whether the corresponding proteins were needed to align the synthetic kinetochores on the spindle: if the protein plays no role, the kinetochores will biorient and resolve into two GFP dots at 37 °C, but mutations that affect the synthetic kinetochore will prevent biorientation.
The synthetic kinetochore requires proteins in several different kinetochore complexes. The Ndc80 complex may have the kinetochore’s principal microtubule binding activity1, 21 and the Mtw1 complex appears to regulate this connection14. We tested mutations in three components of the Ndc80 complex (Ndc80, Nuf2, Spc24) and three of the Mtw1 complex (Nsl1, Dsn1 and Mtw1) and found that all six proteins were needed for biorientation (Figure 4A).
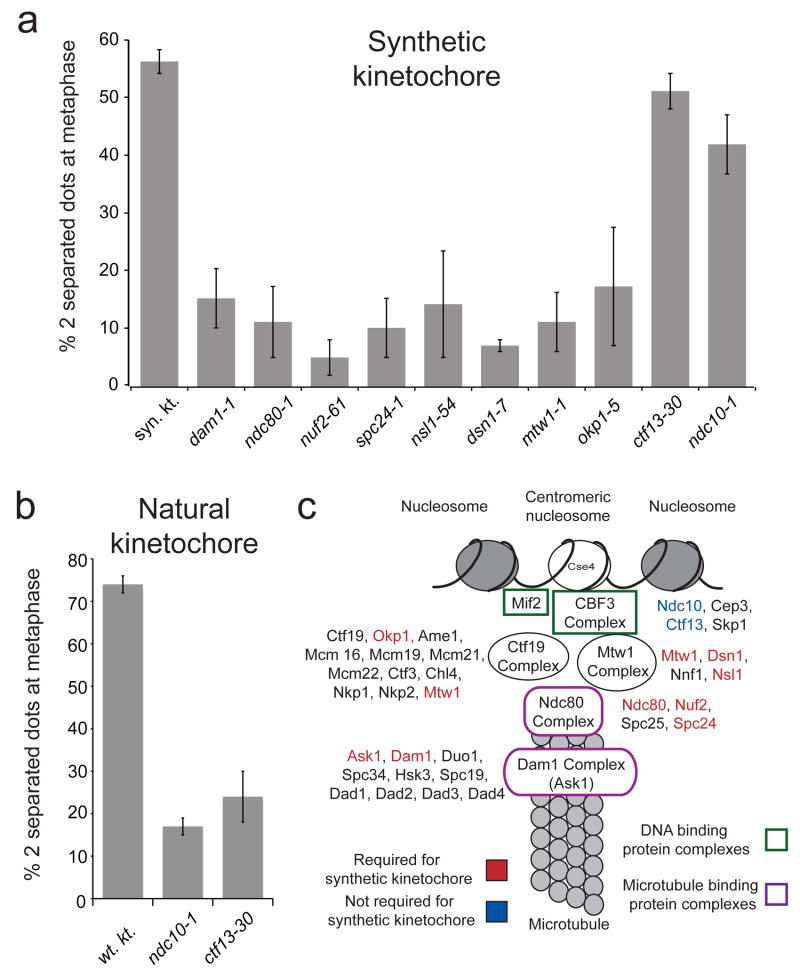
Figure 4. The synthetic kinetochore requires many components of natural kinetochores.
A) Dissection of the genetic requirements for the synthetic kinetochore. Cells carrying temperature-sensitive mutations in a variety of kinetochore proteins were analyzed for their ability to biorient a chromosome carrying a synthetic kinetochore at 37 °C. The control (Syn. Kt.) contains the synthetic kinetochore but does not have any kinetochore temperature-sensitive mutations. Graphs in part A and B show the mean values ± s.d. of 3 experiments where at least 200 cells per genotype were counted per experiment. The values for the percent with 2 separated dots are the following: syn. kt. 55 ± 2, dam1-1 15 ± 5, ndc80-1 11 ± 6, nuf2-61 5 ± 3, spc24-1 5 ± 3, nsl1-54 14 ± 9, dsn1-7 7 ± 1, mtw1-1 11 ± 5, okp1-5 17 ± 11, ctf13-30 50 ± 3, ndc10-1 41 ± 5. B) Mutants that allow the synthetic kinetochore to biorient prevent biorientation of natural kinetochores. A natural kinetochore was analyzed for its ability to biorient with either an ndc10-1 or ctf13-30 temperature sensitive mutation. The wildtype control (wt kt) does not have any kinetochore temperature-sensitive mutations. The values for the percent with 2 separated dots are the following: wt. kt. 74 ± 1, ndc10-1 18 ± 1, ctf13-30 24 ± 4. C) A cartoon showing the location of proteins whose absence keeps the synthetic kinetochore from biorienting (shown in red). Proteins that are dispensable for the synthetic kinetochore are shown in blue, and proteins listed in black were not tested. Components of the DNA-binding complex are outlined in green. Components of the microtubule-binding complex are outlined in purple.
We suspected that other members of the Dam1-DASH complex were needed at the synthetic kinetochore. Tests on Dam1 supported this idea; when Dam1 was inactivated, the synthetic kinetochore could no longer biorient. We tested one of the two essential members of the Ctf19 complex3 (Okp1) and found that it is also involved in biorientation of the synthetic kinetochore.
The proteins that bind to centromeric DNA are poorly conserved during evolution. We tested two of four components of CBF3 (Ndc10 and Ctf13), the essential DNA-binding complex of the yeast kinetochore; neither was needed to biorient the synthetic kinetochore (Figure 4A) but both mutations completely disrupt biorientation of natural kinetochores (Figure 4B). Our functional analysis suggests that the synthetic kinetochore recruits four complexes, including two that bind microtubules, but does not need the primary sequence-specific DNA binding activity of the natural kinetochore (Figure 4C).
Discussion
Taken together, our results lead to two surprising conclusions. First, recruiting multiple copies of a single protein, Ask1, produces a synthetic kinetochore that demonstrates many of a natural kinetochore’s functions. How close to a natural kinetochore is the synthetic kinetochore? The synthetic kinetochore aligns and segregates an entire chromosome, and corrects errors in the initial attachment to microtubules, properties that are completely absent from DNA molecules that lack centromeric DNA. Quantitatively, the synthetic kinetochore is inferior to the natural kinetochore in every assay. Deletions that remove 57 bps of CDEII, one of the three elements of the budding yeast centromere, behave as poorly as the synthetic kinetochore22, suggesting that precise control of chromatin structure optimizes the kinetochore’s activities23.
The second conclusion is that Ask1 recruits at least two conserved kinetochore complexes even though Ask1 occurs in a complex not found outside fungi24. Because these complexes are conserved, we suggest that in higher eukaryotes, the other kinetochore complexes can also form associations without the centromere-binding proteins. In budding yeast, the Dam1-DASH complex associates with microtubules and is needed for poleward kinetochore movement and for chromosomes to switch from attaching to the side to the end of microtubules25. We infer the presence of other proteins because mutating them keeps Ask1 from forming a synthetic kinetochore. By this criterion, Ask1 recruits other subunits of the Dam1-DASH complex, and members of the Mtw1, Ctf19 and Ndc80 complexes. The N-terminal half of Ask1 is sufficient for this function. The C-terminus of this protein contains Cdk phosphorylation sites and regulates microtubule dynamics as cells enter anaphase26, 27, suggesting that Ask1’s different functions can be separated into different domains.
The synthetic kinetochore assembles without the centromere’s normal DNA binding proteins. This suggests that these proteins normally mark the site where a kinetochore should assemble, but that their interactions with other proteins and their effects on DNA structure are not essential for the fundamental activities of the kinetochore. The details of these missing interactions may explain the more accurate segregation of natural kinetochores23. We can exclude the possibility that Ask1 simply tethers the synthetic kinetochore to natural kinetochores; the ndc10-1 mutant, which disrupts every known aspect of kinetochore behavior8, 19, 28 (Figure 4B), has no effect on the synthetic kinetochore (Figure 4A). Our results offer two possible interpretations: i) interactions of the different kinetochore complexes on microtubules assemble a proto-kinetochore that is recruited to the chromosome by the Dam1-DASH complexes bound to the LacO array, ii) the multiple copies of the Dam1-DASH complex bound to the LacO array creates sites that recruit the other kinetochore protein complexes, independently of their interactions with microtubules.
Our findings support the view that the least conserved proteins of the kinetochore are those that connect it to DNA3. The identities of the proteins that bind centromeric DNA have changed during eukaryotic evolution whereas the core microtubule binding and central complexes have not. One explanation is that competition amongst variant centromere sequences has caused co-evolution of centromeric DNA and the proteins that bind it, whereas the conservation of microtubules requires conservation of the microtubule-binding activities of the kinetochore29.
We speculate that recruiting the microtubule binding activities of the kinetochore to novel DNA sequences has played important roles in evolution. One example is the appearance of a centromere at a new location on a chromosome. In humans, these neocentromeres form at regions that lack the repetitive α-satellite DNA found at normal human centromeres, yet neocentromeres bind kinetochore proteins, form heterochromatin, and segregate chromosomes in mitosis and meiosis30. Selfish DNA elements may have hijacked the microtubule binding activity of the kinetochore. One possible candidate is the yeast 2 μm circle, an endogenous plasmid that has been shown to associate with the spindle during mitosis, recruit Cse4, the centromere-specific histone variant, and whose accurate segregation depends on Ipl131, 32. Understanding the formation and function of synthetic kinetochores should thus provide insights into kinetochore assembly as well as the evolution of centromeres and kinetochores.
Methods
Yeast Strains, Techniques, and Media
All strains are isogenic with the W303 (ura3-1, ade2-1, his3-11, 15, leu2-3, 112, trp1-1, can1-100) background and are listed in Supplementary Table 3. Media, microbial and genetic techniques were essentially as described33.
Assay for a synthetic kinetochore
Strain Construction
To make Lac repressor (LacI) fusion proteins, the kinetochore proteins listed in Supplementary Table 1 were amplified without the stop codon by PCR from genomic DNA and ligated into a yeast integrating vector containing LacI driven by the HIS3 promoter (pAFS135)13. This vector was integrated into the HIS3 locus. This strain was then transformed with an ARS plasmid containing either 256 repeats (pSLB5) or 8 repeats (pDL10) of the LacO array and the LEU2 marker. As a control, GFP-LacI was integrated into the HIS3 locus and this strain was then transformed with the same ARS plasmids. Control strains containing a plasmid with a centromere and the LacO array were also made.
Plasmid stability assay
Cells were grown overnight in synthetic complete (SC) media lacking leucine and histidine (SC – HIS – LEU). They were diluted 1:50 and grown for 9 hours in YPD, doubling approximately 5 times. Approximately 500 cells were plated to SC – HIS – LEU and SC – HIS. Cells were counted and the % of cells containing the plasmid (# cells on SC – HIS – LEU/# cells on SC – HIS) was calculated. At least 5 plates were counted per experiment and each experiment was performed three times.
Complementation Test
To determine if constructs were functional, the kinetochore protein-LacI fusions were integrated into diploid yeast strains that were heterozygous for a replacement of the respective essential kinetochore gene with the kanMX4 marker. The diploid yeast was then sporulated using standard techniques33 and the spores were dissected. The fusion protein was considered functional if the fusion protein allowed yeast strains containing the knockout (kanMX4 or resistance to geneticin) to survive.
Assay for Chromosome Loss
Strain Construction
A construct containing the 256 repeat LacO array (pAFS59) was integrated at the LEU2 locus of chromosome III. The URA3 gene was integrated on the other side of CEN3 at 116000 bp by targeting an integrating plasmid (pSLB71) containing URA3 to that location. This strain was transformed with pSLB57 (Ask1-LacI) or pAFS135 (GFP-LacI) and then mated to a wildtype haploid to make SLY769 and SLY768, respectively. A control strain, SLY767, with LacO:LEU2 but without LacI was also used. A conditional centromere was constructed by placing the GAL1 promoter in front of CEN3. The PCR integration34 plasmid pFA6A-TRP1-PGAL1 was targeted 100 bp upstream of CEN3 in a haploid strain containing LacO256:LEU2 or LacO8:LEU2. Primers are available upon request. These strains were then transformed with plasmids expressing either Ask1-LacI or (pSLB57) or GFP-LacI (pAFS135) and used in the haploid plating assays.
Assays for kinetochore function
Dicentric chromosome loss
To determine the rate of chromosome loss of the dicentric chromosome, the fluctuation assay was performed with minor variations from published methods35, 36. Cells were grown overnight in SC – LEU – HIS. The next evening, the cells were diluted 1:10,000 into SC with only 0.1% glucose and dispensed into 96 well dishes that were sealed. They were grown at 30°C overnight. The next day, some of the wells were counted using a Coulter Particle Counter (Beckman Coulter). The other wells were spotted onto plates containing 5-FOA and grown for 4 days at 30°C. Cells were counted and then replica plated to SC – LEU plates to determine the number of colonies that had lost both markers and thus all of chromosome III. The rate of chromosome loss was calculated using formulas described36. The calculation was made from 3 experiments where 45 cultures had been spotted for each genotype.
Plating Assay
SLY806, SLY807, DLY242, DLY245 strains with the conditional centromere and either 256 or 8 repeats of the LacO array at LEU2 with either Ask1-LacI or GFP-lacI were grown in SC – URA – LEU. Cells were counted and approximately 500 cells were plated to YPGlu or YPGal. The number of cells that survived on YPGal compared to YPGlu was calculated. At least 4 plates were counted per experiment and 3 independent experiments were performed. Photos represent 3 days of growth on YPD and 5 days of growth on YPGal at 30°C.
Chromosome Segregation
SLY849 and SLY850 strains with a conditional centromere, cdc15-2, GFP-lacI, 256 repeats of LacO at LEU2, and with or without Ask1-LacI were grown to early log phase in SC raffinose – URA – LEU – HIS at 25°C. 10 μg/ml of α-factor was added and cells were placed at 25°C for 2 hours to arrest them in G1. Cells were then resuspended in either YPGlu or YPGal + 10 μg/ml α-factor and placed at 37°C for 1 hour. Cells were washed 4 times at 37°C and released in YPGlu or YPGal. After they reached large-bud arrest at 37°C, they were fixed with paraformaldehyde and GFP dots were counted as described below. The experiment was repeated 4 times, counting at least 200 cells in each experiment.
Biorientation
The experiment was performed at 30°C with SLY834, SLY835 (conditional centromere, GFP-LacI, 256 repeats of LacO at LEU2, PMET3CDC20, with or without Ask1-LacI), and VBI313 (GFP-LacI, 256 repeats of LacO at CEN15, PMET3CDC20). Cells were grown to early log phase in SC raffinose –MET-HIS. They were arrested with 10 μg/ml of α-factor (SLY834, SLY835) or 1 μg/ml of α-factor (VBI313) for 2 hours. They were then resuspended in YPGal or YPGlu + α-factor for 2 hours. After washing 4 times, they were released into YPGlu or YPGal. Once the cells reached a large-budded arrest, they were fixed and GFP dots were counted. The experiment was repeated 4 times, counting at least 200 cells in each experiment.
Error Correction
SLY905 (conditional centromere, GFP-LacI, 256 repeats of LacO at LEU2, PMET3CDC20, Ask1-LacI, ipl1-321) and VBI312 (GFP-LacI, 256 repeats of LacO at CEN15, PMET3CDC20, ipl1-321) cells were grown to early log phase in SC raffinose –MET at 25°C. 1 μg/ml of α-factor + 0.2 mM copper sulfate was added and cells were placed at 25°C for 2 hours. Cultures were split and resuspended in YPGal + α-factor + copper sulfate and placed at either 23°C or 37°C for 1 hour. Cells were washed 4 times at the appropriate temperature and resuspended in YPGal. After they reached large-bud arrest at 23°C and 37°C, they were fixed with paraformaldehyde. A portion of the 37°C cultures was then placed at 23°C to allow for error correction for 2 hours. They were then fixed with paraformaldehyde and GFP dots were counted as described below. The experiment was repeated 4 times, counting at least 200 cells in each experiment.
Biorientation of kinetochore temperature-sensitive mutants
SLY969, 931, 892, 894, 899, 902, 909, 943, 934, 1000 cells (synthetic kinetochore with ctf13-30, ndc80-1, nuf2-61, nsl1-54, dsn1-7, mtw1-11, dam1-1, spc24-1, ndc10-1, and okp1-5 respectively) were grown to early log phase in SC raffinose – MET at 25°C. 10ug/ml of α-factor was added and cells were placed at 25°C for 2 hours. Cultures were resuspended in YPGal + 10ug/ml of α-factor and placed at 37°C for 1 hour. Cells were washed 4 times at 37°C and resuspended in YPGal. After they reached large-bud arrest, they were fixed with paraformaldehyde. The experiment was repeated 3 times, counting at least 200 cells in each experiment.
Microscopy
To count the number of GFP dots, cells were fixed as follows: 0.9ml of culture was harvested, and 0.1ml of 4% paraformaldehyde in 0.1M potassium phosphate buffer, pH8.5 was added. The cells were incubated for 4 minutes at room temperature and were washed once with 1 ml of 0.1 M potassium phosphate buffer, pH 8.5, once with 1 ml of 1.2 M sorbitol 0.1 M potassium phosphate buffer, pH 8.5. A Nikon Eclipse E600 equipped with a 100X 1.4 NA lens (Nikon), GFP filter, a Cascade 512B digital camera (Photometrics) and MetaMorph software (Universal Imaging Corporation) was used to determine the number of GFP dots per cell by moving the focal plane through the sample and analyzing the live digital image on the computer screen. In all cases, at least 200 cells were counted per sample per experiment and each experiment repeated at least 3 times.
Supplementary Material
Acknowledgments
We thank Greg Lang, Joana Goncalves-Sa, John Koschwanez, Gregg Wildenberg, and Dai Tsuchiya for technical advice; Charles Asbury, Ted Salmon, Steve Elledge, Ajit Joglekar, Frank Solomon, Amy Rowat, and members of the Murray lab for critical reading of the manuscript; and Steve Elledge, John Kilmartin, and Sue Biggins for strains. This work was supported by a US National Institutes of Health (NIH) National Research Service Award fellowship to S.L. and an NIH grant to A.W.M. (GM043987).
References
- 1.Tanaka TU, Desai A. Kinetochore-microtubule interactions: the means to the end. Curr Opin Cell Biol. 2008;20:53–63. doi: 10.1016/j.ceb.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- 3.Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu Rev Biochem. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- 4.Murray AW, Szostak JW. Pedigree analysis of plasmid segregation in yeast. Cell. 1983;34:961–970. doi: 10.1016/0092-8674(83)90553-6. [DOI] [PubMed] [Google Scholar]
- 5.Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- 6.Cheeseman IM, et al. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J Cell Biol. 2001;155:1137–1145. doi: 10.1083/jcb.200109063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, et al. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 2002;16:183–197. doi: 10.1101/gad.959402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janke C, Ortiz J, Tanaka TU, Lechner J, Schiebel E. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. Embo J. 2002;21:181–193. doi: 10.1093/emboj/21.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiermaier E, Woehrer S, Peng Y, Mechtler K, Westermann SA. Dam1-based artificial kinetochore is sufficient to promote chromosome segregation in budding yeast. Nat Cell Biol. 2009 doi: 10.1038/ncb1924. in press. [DOI] [PubMed] [Google Scholar]
- 10.Haber JE, Thorburn PC. Healing of broken linear dicentric chromosomes in yeast. Genetics. 1984;106:207–226. doi: 10.1093/genetics/106.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill A, Bloom K. Genetic manipulation of centromere function. Mol Cell Biol. 1987;7:2397–2405. doi: 10.1128/mcb.7.7.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joglekar AP, Salmon ED, Bloom KS. Counting kinetochore protein numbers in budding yeast using genetically encoded fluorescent proteins. Methods Cell Biol. 2008;85:127–151. doi: 10.1016/S0091-679X(08)85007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- 14.Goshima G, Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- 15.He X, Asthana S, Sorger PK. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–775. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka T, Fuchs J, Loidl J, Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat Cell Biol. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- 17.Biggins S, et al. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He X, Rines DR, Espelin CW, Sorger PK. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 2001;106:195–206. doi: 10.1016/s0092-8674(01)00438-x. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka TU, et al. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 21.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 22.Gaudet A, Fitzgerald-Hayes M. Alterations in the adenine-plus-thymine-rich region of CEN3 affect centromere function in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:68–75. doi: 10.1128/mcb.7.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeh E, et al. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westermann S, et al. Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka K, Kitamura E, Kitamura Y, Tanaka TU. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J Cell Biol. 2007;178:269–281. doi: 10.1083/jcb.200702141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Elledge SJ. The DASH complex component Ask1 is a cell cycle-regulated Cdk substrate in Saccharomyces cerevisiae. Cell Cycle. 2003;2:143–148. [PubMed] [Google Scholar]
- 27.Higuchi T, Uhlmann F. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature. 2005;433:171–176. doi: 10.1038/nature03240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goh PY, Kilmartin JV. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 30.Marshall OJ, Chueh AC, Wong LH, Choo KH. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet. 2008;82:261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velmurugan S, Yang XM, Chan CS, Dobson M, Jayaram M. Partitioning of the 2-microm circle plasmid of Saccharomyces cerevisiae. Functional coordination with chromosome segregation and plasmid-encoded rep protein distribution. J Cell Biol. 2000;149:553–566. doi: 10.1083/jcb.149.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hajra S, Ghosh SK, Jayaram M. The centromere-specific histone variant Cse4p (CENP-A) is essential for functional chromatin architecture at the yeast 2-microm circle partitioning locus and promotes equal plasmid segregation. J Cell Biol. 2006;174:779–790. doi: 10.1083/jcb.200603042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherman F, Lawrence CW, Fink GR. Methods in yeast genetics. Cold Spring Harbor Laboratory Press; 1979. [Google Scholar]
- 34.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 35.Lang GI, Murray AW. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics. 2008;178:67–82. doi: 10.1534/genetics.107.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foster PL. Methods for determining spontaneous mutation rates. Methods Enzymol. 2006;409:195–213. doi: 10.1016/S0076-6879(05)09012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.