SUMMARY
The bacterial transposon Tn7 directs transposition into actively replicating DNA by a mechanism involving the transposon-encoded protein TnsE. Here we show that TnsE physically and functionally interacts with the processivity factor of the DNA replication machinery in vivo and in vitro. Our work establishes an in vitro TnsABC+E transposition reaction reconstituted from purified proteins and target DNA structures. Using the in vitro reaction we confirm that the processivity factor specifically reorders TnsE-mediated transposition events on target DNAs in a way that matches the bias with active DNA replication in vivo. The TnsE interaction with an essential and conserved component of the replication machinery and a DNA structure reveals a new mechanism by which Tn7, and probably other elements, select target-sites associated with DNA replication.
Keywords: beta sliding clamp, transposition, processivity factor, in vitro transposition
INTRODUCTION
Transposons are genetic elements that are capable of moving from one location to another within a cell. The bacterial transposon Tn7 and its relatives are abundantly distributed amongst bacteria in a wide variety of medical and environmental settings (Parks and Peters, 2007; Parks and Peters, 2009). Tn7 has served as a model system for transposition, especially for the understanding of transposon target-site selection (reviewed in Peters and Craig, 2001b, and Craig et al., 2002). Target-site selection is the process by which transposons assess new DNA molecules for potential insertion. While most transposable elements possess a weak target DNA sequence preference that guides target-site selection, Tn7 utilizes two distinct target-site selection pathways. In one pathway a sequence-specific DNA binding protein directs transposition into a single site within the bacterial chromosome, and in the other a separate protein recognizes a process associated with DNA replication. These two target selection pathways optimize vertical and horizontal transmission of the transposable element respectively (Craig, 2002; Parks and Peters, 2009).
Tn7 encodes five genes whose products conduct transposition (Craig et al., 2002). TnsA and TnsB comprise the transposase that catalyzes the DNA breakage and joining reactions at the transposon ends to mobilize the element. TnsC is an AAA regulator protein that activates the transposase when an appropriate target DNA has been found (Stellwagen and Craig, 1998). TnsD and TnsE identify target DNAs and signal TnsABC to activate transposition (Craig, 2002). Target-site selection is a prerequisite for activation of transposition with Tn7; transposon excision and insertion does not occur until an appropriate target has been identified. TnsD recognizes a specific site, called its attachment site or attTn7, by binding to a highly conserved DNA sequence within the 3’ end of the glmS gene. The TnsE protein recognizes an incompletely defined feature associated with discontinuous DNA replication (Peters and Craig, 2001a) that is over-represented or especially accessible in mobile plasmids, called conjugal plasmids, as they enter a new host cell (Wilkins and Lanka, 1993; Wolkow et al., 1996).
While TnsE-mediated transposition preferentially occurs into mobile plasmids undergoing conjugal DNA replication, at a lower frequency, the TnsABC+E machinery also recognizes sites within the bacterial chromosome with a preference for the region where DNA replication terminates and regions proximal to DNA double-strand breaks (Peters and Craig, 2000; Shi et al., 2008). The orientation of the transposon ends following TnsE-mediated transposition indicates that discontinuously replicated DNA is in some way recognized by TnsE (Peters and Craig, 2001a; Wolkow et al., 1996). As mobile plasmids enter a new host cell, they replicate in a single direction by a discontinuous process, like lagging-strand DNA synthesis (Wilkins and Lanka, 1993). In both mobile plasmids and in the chromosome, transposition events occur in a single orientation correlating with the direction of replication progression (Peters and Craig, 2001a; Peters and Craig, 2001b; Wolkow et al., 1996). It has been shown that TnsE is a DNA binding protein that preferentially binds to DNA structures that present a free 3’-recessed end (Peters and Craig, 2001a). Given that TnsD relies in part on additional host factors in activating transposition (Sharpe and Craig, 1998), it is conceivable that host factors associated with discontinuous DNA replication allow the selection of targets during TnsE-mediated transposition.
An intriguing host factor candidate that could allow the TnsABC+E transposition machinery to target lagging-strand DNA synthesis is the DNA replication processivity factor. Processivity factors are essential clamp proteins that encircle DNA and serve as a mobile platform, linking proteins to DNA (Johnson and O'Donnell, 2005; Warbrick, 2000). Interestingly, the inactive pogo element, found in Drosophila, encodes a transposase that has been shown to interact with the processivity factor (Warbrick et al., 1998). Because the element is no longer active, no functional link between this interaction and transposition has yet been established (Warbrick, 2000). It therefore remains unclear if the interaction was important in the original element. An interaction with the processivity factor could possibly regulate transposition with DNA replication and repair, or could be involved in target-site selection. We reasoned that TnsE might use an interaction with the processivity factor to direct transposition into certain forms of DNA replication.
The processivity factor, β clamp in Bacteria and PCNA in Eukaryotes and Archaea, is enriched on discontinuously replicating DNA (reviewed in Johnson and O'Donnell, 2005, and references therein). β and PCNA have been shown to interact with many proteins involved in DNA repair, Okazaki fragment maturation, and regulation of the cell cycle (Johnson and O'Donnell, 2005; Warbrick, 2000). The processivity factor binding motif found in proteins that interact with β and PCNA fits into a hydrophobic cleft found in the clamp proteins (Dalrymple et al., 2001; Jeruzalmi et al., 2001a; Johnson and O'Donnell, 2005; Warbrick, 2000). Competition for this common region of the clamp appears to play a role in coordination of proteins involved in DNA metabolism (Lopez de Saro et al., 2004).
In this work we reveal and characterize an interaction between TnsE and the β processivity factor, and reconstitute the TnsABC+E transposition pathway using purified components. The in vitro reaction confirms that two factors, interaction with a DNA structure and with the processivity factor account for the bias of TnsABC+E transposition with active DNA replication. In vitro and in vivo analysis of the TnsE-β interaction explains how Tn7 targets DNA replication without negatively affecting the cell. These findings likely reveal a general strategy used by other unrelated transposons for directing transposition to DNA replication intermediates.
RESULTS
TnsE interacts with the processivity factor
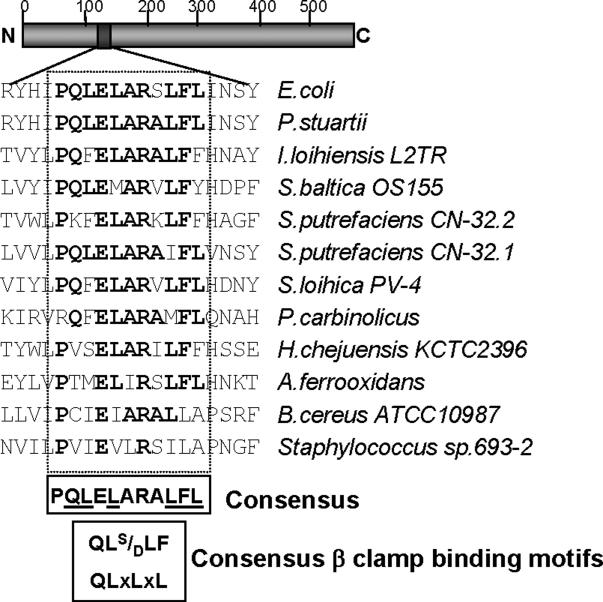
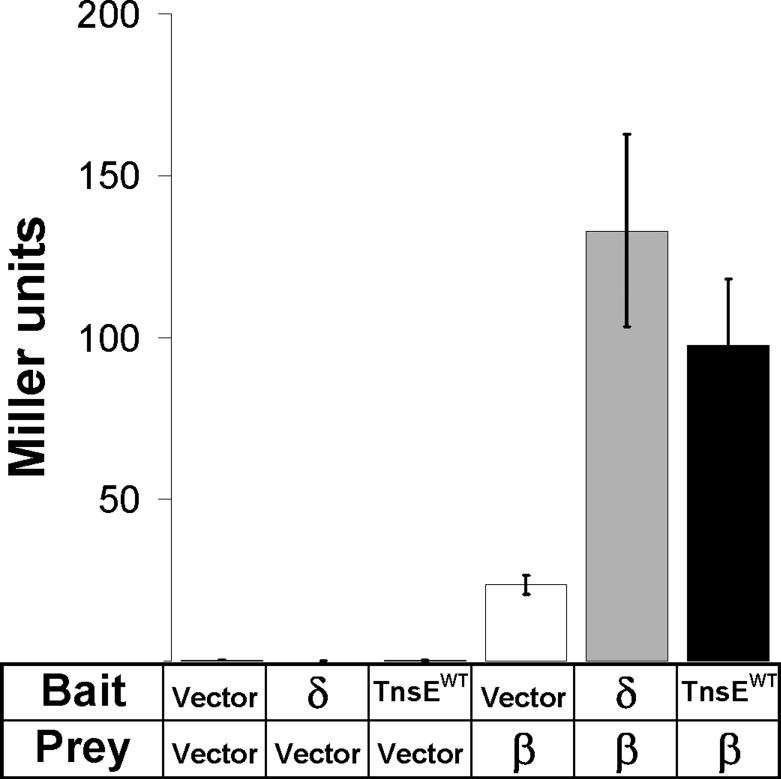
In an effort to understand how TnsE identifies a target DNA we looked for conserved motifs within the amino acid sequence of TnsE. We found a sequence that shows a modest resemblance to the consensus processivity factor binding motifs found in bacterial host proteins (QL(S/D)LF (Dalrymple et al., 2001) and QLxLxL (Wijffels et al., 2004))(Figure 1). Analysis of amino acid alignments of all known and predicted tnsE gene products (Parks and Peters, 2009) using the ClustalW algorithm (Thompson et al., 1994) revealed a highly conserved sequence PQLELARALFL (Figure 1). We hypothesized that TnsE recognizes lagging-strand DNA synthesis through an interaction with the processivity factor. We first tested for the TnsE-β interaction using TnsE and β protein derivatives fused to the yeast GAL4 transcription activation and DNA binding domains, respectively (Fields and Song, 1989). The presence and extent of the interaction in the two-hybrid assay was monitored by determining the β-galactosidase (β-gal) activity in a reporter strain containing the lacZ gene under control of a GAL4 promoter (Liachko and Tye, 2005; Miller, 1992). We also included a positive control for the β interaction, the δ subunit of the clamp-loader, which has been shown to interact with β in multiple assays (Johnson and O'Donnell, 2005). The yeast two-hybrid assay indicated that TnsE and β do indeed interact (Figure 2).
Figure 1.
Alignment of TnsE homologs reveals a putative β clamp binding motif. A representation of the ∼538 amino acid TnsE protein found in E. coli is shown with the amino (N) and carboxy (C) termini indicated. An alignment of TnsE homologs encompassing the putative β clamp binding motif is presented with the putative motif boxed. The consensus sequence of the region between residues 121−131 containing the putative β clamp binding motif are shown in bold. Underlined residues correspond to positions where alanine substitutions were made in the TnsEβMA mutants. The consensus β clamp binding motifs found in bacterial host proteins (QL[S/D]LF (Dalrymple et al., 2001), QLxLxL (Wijffels et al., 2004) are given for reference.
Figure 2.
The TnsE-β interaction can be detected using a yeast two-hybrid assay. For the assay the bacterial proteins were fused to either the yeast DNA binding domain (Bait) or the yeast transcription activation domain (Prey)(Liachko and Tye, 2005). The β-fusion alone displays a slight auto-activation effect, while TnsE and the positive control, the δ subunit of the clamp loader, display interaction signals significantly above background. Interaction was measured by Miller assay and is reported in Miller Units (Miller, 1992). Error bars indicate standard error of the mean (n=4).
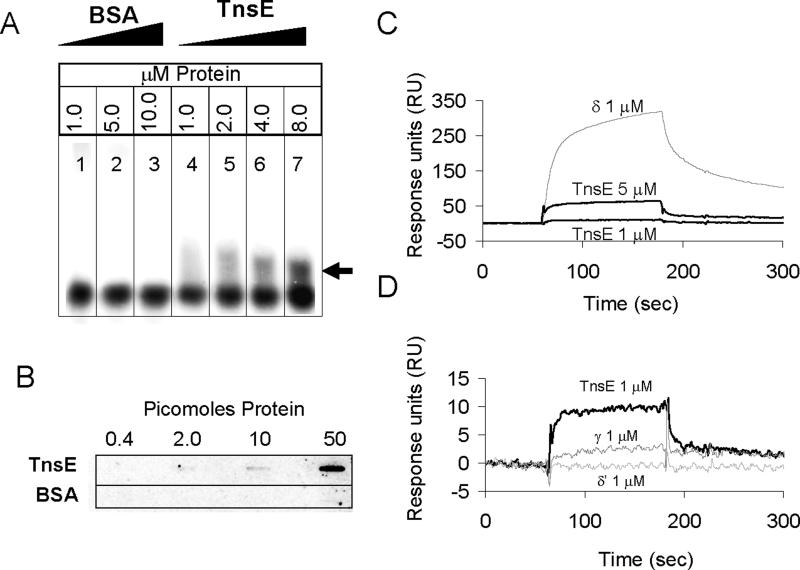
To confirm the interaction between TnsE and β in vitro, we purified a modified β protein that could be labeled by the addition of 32P phosphate by PKA enzyme (32P-β)(Kelman et al., 1995a; Kelman et al., 1995b). We tested the TnsE-β interaction using a protein mobility shift assay (Lopez de Saro et al., 2006; Lopez de Saro and O'Donnell, 2001). As a negative control we tested increasing concentrations of BSA, which is similar TnsE in size and charge (69.2 KDa, pI 5.8, for BSA versus 61.2 KDa, pI 5.7, for TnsE), and found no shifted product (Figure 3.A., lanes 1−3). A shift in the electrophoretic mobility of 32P-β upon addition of increasing concentrations of TnsE demonstrated that TnsE and β do form a complex (Figure 3.A., lanes 4−7). We used a far western blot technique to further confirm the in vitro interaction (Einarson et al., 2007). Serial dilutions of TnsE and BSA were spotted on a membrane in triplicate using a slot blot apparatus (Amersham Bioscience). The membrane was then probed with 32P-β. Consistent with the band shift assay, we found that TnsE retained 32P-β, indicating interaction, while equivalent concentrations of BSA did not (Figure 3.B.).
Figure 3. TnsE and β clamp interact in three distinct in vitro assays.
A. Protein mobility shift assays confirm the TnsE-β interaction in vitro. 32P-labeled β monomer (10 nM) (32P-β alone in lane 1) is unaffected by the addition of up to 10 μM BSA (lanes 1− 3) in a 4% native polyacrylamide gel, but produces a shifted product (black arrow) with the addition of TnsEwt (lanes 4−7).
B. Far western blots show that TnsE retains 32P signal when probed with 32P-labeled β clamp.
C. Surface Plasmon Resonance experiments reveal interaction between β and TnsE and enable comparisons to be drawn between this interaction and the well-characterized clamp loader interactions. Reference subtracted traces are shown for 1 μM δ (positive interaction control), 5 μM TnsE and 1μM TnsE. The KD of the TnsE-β interaction is calculated to be ∼2.44 μM.
D. Comparison between TnsE-β interaction and γ-β and δ’-β interactions provide further reference for the strength of the TnsE-β interaction. Reference subtracted sensograms for 1 μM γ (as monomer), 1 μM δ’, and 1 μM TnsE are shown.
For a more thorough and quantitative analysis of the TnsE-β interaction we utilized a Biacore instrument that is capable of monitoring relatively weak protein-protein interactions by surface plasmon resonance (SPR). For these experiments we immobilized purified β protein in varying concentrations by cross-linking them to the surface of a CM-5 SPR chip (GE Healthcare). Various concentrations of test proteins were then flowed over the surface of the chip to monitor any interaction between proteins, followed by dissociation under the flow of buffer alone (Figure 3.C. and 3.D.). We determined the KD of the TnsE-β interaction to be ∼2.44 μM (EXPERIMENTAL PROCEDURES). For comparison, we tested the β interaction with 1 μM (as monomers) of three different components of the minimal clamp-loader complex (the γ-complex, γ3δδ’), providing us with a pre-established set of standard proteins with varying dissociation constants (Leu and O'Donnell, 2001). We found that the TnsE-β interaction is intermediate between the primary β contacting subunit δ (Figure 3.C.) and the γ subunit (Figure 3.D.); however, direct comparison with the γ subunit is complicated by the fact purified γ forms tetramers in solution (Dallmann and McHenry, 1995). As a negative control we tested for interaction with δ’ and found no interaction, similar to previously reported results (Leu and O'Donnell, 2001). To confirm that our calculations were comparable to previously reported values, we calculated the KD of the δ-β interaction. Under the conditions used here we determine the KD for the δ-β interaction to be ∼0.06 μM, similar to the ∼0.03 μM value reported by Leu and O'Donnell (Leu and O'Donnell, 2001)(EXPERIMENTAL PROCEDURES).
The putative β clamp binding motif of TnsE is involved in binding β
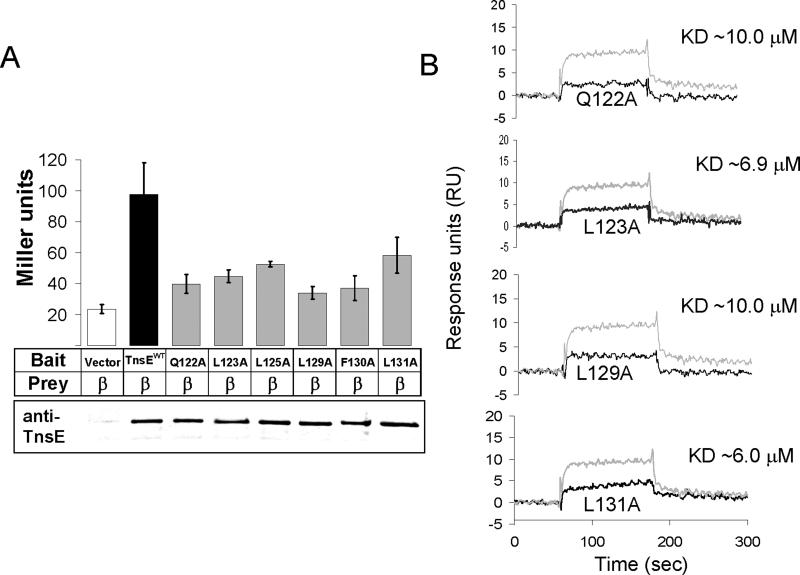
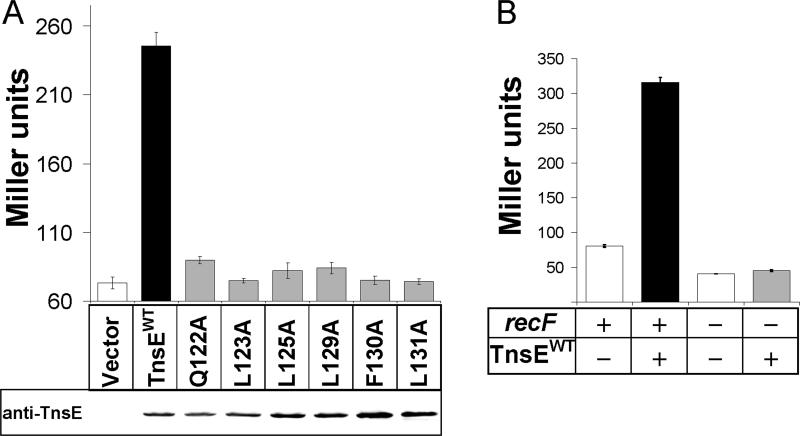
To test if the region of TnsE that resembles the β clamp binding motif is important for binding β, we constructed a set of tnsE mutants replacing specific amino acids within this region with alanines (collectively referred to as tnsEβMA). Amino acids were chosen for mutation based on their presumed relationship to the consensus β clamp binding and their conservative among TnsE proteins (Figure 1). The tnsEβMAmutations were first tested in the yeast two-hybrid assay described above. Each alanine substitution resulted in decreased β-gal activity in the reporter strain, supporting the idea that the conserved region is important for interaction with β and possibly comprises a region that interacts with the hydrophobic pocket of the β clamp (Figure 4.A.). Western blots did not indicate changes in stability or expression resulting from the TnsEβMA mutations that would account for these observations (Figure 4.A.).
Figure 4.
A. Quantification of the yeast two-hybrid assay reveals a defect in the β interaction with TnsE proteins with alanine substitutions in the putative β interaction motif. The alanine substitutions in the putative β clamp binding motif consistently reduce TnsE-β interaction levels. Interaction was measured by Miller assay and is reported in Miller Units (Miller, 1992). Western blots using anti-TnsE antibodies are given below the bar graph. Error bars indicate standard error of the mean (n=4).
B. Surface Plasmon Resonance results for purified TnsEβMA mutants confirm a loss in interaction between TnsE and β in vitro. All four TnsEβMA mutants tested show a decrease in β interaction. Reference subtracted results from 1 μM of each of the TnsEβMA mutant proteins tested are shown (black lines) overlaid with results from 1 μM TnsEWT (gray lines) for comparison.
To confirm that the TnsEβMA mutants are weaker in interaction with β in vitro we purified a subset of the TnsEβMA proteins. Using the SPR system we compared interaction between β and 1 μM of each purified TnsE allele. In all cases, the SPR sensograms indicated weaker interaction between each of the mutant TnsE alleles and β (Figure 4.B.) and loosely corresponded to the relative loss in interaction observed in the yeast two-hybrid assay. The β interaction with the mutant TnsE proteins varied in KD from ∼6.0 to ∼10.0 μM, as analyzed by the BIAevaluation software (Biacore, 1997). The SPR results and the yeast two-hybrid data support the view that the conserved region of TnsE is involved in the interaction between TnsE and β.
Interaction with the processivity factor is essential for transposition in vivo
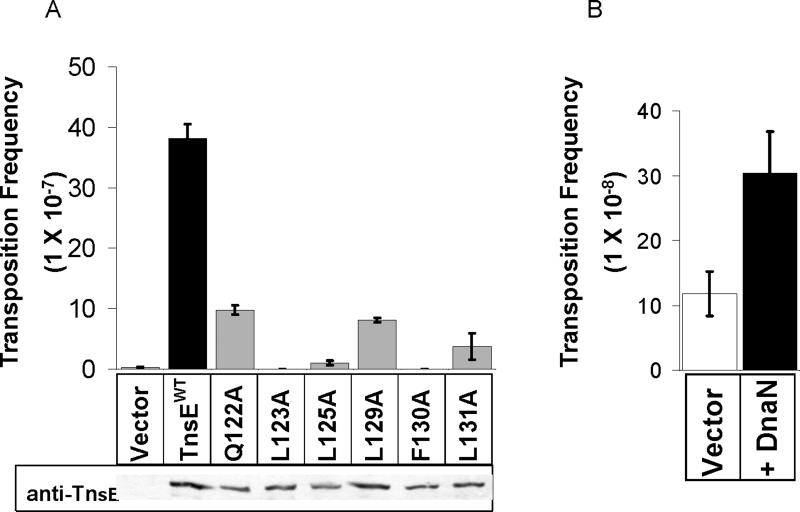
If binding to the β clamp is required to activate transposition, we expected to see a decrease in transposition with the tnsEβMA mutants, due to their attenuated β clamp binding ability. Interaction with the clamp via the interaction motif is required for activity of DNA polymerases (Johnson and O'Donnell, 2005), although these protein-protein interactions include more extensive surface interactions beyond the conserved motif (Bunting et al., 2003). Using an in vivo transposition assay (McKown et al., 1988), we found that the alanine mutations abolished or significantly reduced (p<0.005, two-tailed unequal variance t-test) the frequency of TnsABC+E transposition (Figure 5.A.). Western blots show that the stability and expression of these mutants could not account for such sharp reduction or complete loss of transposition (Figure 5.A.). We found no negative effects with the tnsEβMAmutants on other Tn7 transposition pathways (data not shown). These results indicate that the decreases we observed in transposition frequency are consistent with decreased β clamp binding ability, supporting the view that activation of transposition via the TnsE pathway is dependent on binding to the β clamp. Similarly, others have shown that mutant host proteins with attenuated β clamp binding ability display decreased or abolished activity in vivo (Beuning et al., 2006; Johnson and O'Donnell, 2005; Simmons et al., 2008; Sutton, 2004). In vivo, TnsE must compete with many proteins for binding to the clamp (Johnson and O'Donnell, 2005), which may explain some discrepancies between in vitro TnsEWe hypothesized that if TnsE-β interaction and in vivo TnsE activity.
Figure 5. An in vivo transposition assay reveals a defect in the ability of TnsEβMA mutants to activate transposition, and an increase in transposition frequency when β is over-expressed.
A. Transposition frequency is reduced significantly below wild-type with each of the six TnsEβMA mutations. Transposition was monitored in cells expressing TnsABC and wild type TnsE or a mutant TnsE containing an alanine substitution at one of six positions in the putative β clamp interacting motif (see text for details). A western blot using an anti-TnsE antibody is displayed below the graph. Transposition assays were conducted in recA− cells containing tns genes on plasmids using a lambda delivery vector (McKown et al., 1988). Error bars indicate the standard error of the mean (n=3).
B. Over-expression of β results in a significant increase in TnsE-mediated transposition. Transposition was monitored in cells expressing TnsABC+E with a plasmid expressing β or an empty vector. Error bars indicate standard error of the mean (n=3).
We hypothesized that if TnsE relies on the β clamp for target recognition we might increase the likelihood of TnsE recognizing a target by increasing the concentration of β in the cell. We performed a transposition assay with moderate over-expression of β and found that the transposition frequency increased, compared to the empty vector control (Figure 5.B.). We found this effect to be specific to the TnsABC+E transposition pathway, as there was no significant change in the TnsABC+D transposition frequency when β was over-expressed (3.3 (+/−0.9) ×10−5 transposition events per donor molecule with wild type levels of β, and 3.6 (+/−0.6) × 10−5 with β over-expressed). The responsiveness of TnsABC+E transposition to the concentration of β is consistent with a dependence on interaction with β for target-site selection and subsequent activation of transposition.
TnsABC+E transposition reaction can be reconstituted in vitro
To confirm the molecular components required for targeting active DNA replication in vivo we established an in vitro system for TnsABC+E transposition using purified components. While in vitro systems for two other Tn7 transposition pathways exist (the TnsD pathway (Bainton et al., 1993) and an untargeted mutant core machinery pathway termed TnsABC* (Stellwagen and Craig, 2001)), there have been no reports of the reconstitution of the TnsE pathway. We took advantage of previous analyses of TnsE DNA binding that indicated that the protein shows a strong preference for DNA structures containing 3'-recessed ends (Peters and Craig, 2001a), a structure more commonly found on the lagging-strand than the leading-strand (Johnson and O'Donnell, 2005). For our in vitro assay, we constructed DNA-target molecules that contain a 20 bp single-stranded gap in the duplex DNA (EXPERIMENTAL PROCEDURES). Transposition was monitored by transforming the deproteinated reaction products into highly competent E.coli (DH5α) . The donor plasmid, which contains the Tn7 element, possesses a conditional origin of replication that will not replicate in E. coli that do not express the π protein (Shafferman et al., 1982). Therefore, chloramphenicol resistant colonies will only result if the Tn7 element (carrying the chloramphenicol resistance cassette) moves into the target plasmid during the in vitro transposition reaction.
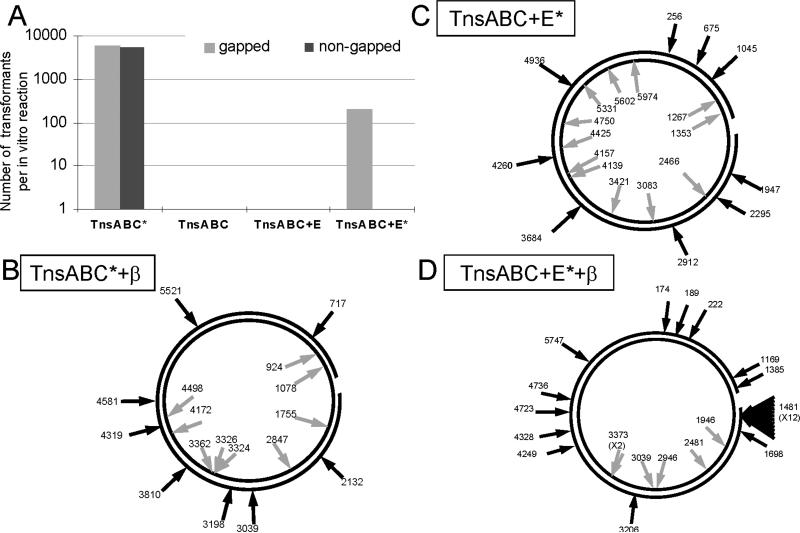
An initial test to determine if the gapped plasmid could be used as a target and recovered following in vitro transposition was necessary. The cell must repair multiple gaps in the DNA, one created intentionally as described above and two created by the transposition reaction (Craig, 2002). To ensure that we could recover the plasmids, we carried out in vitro transposition reactions using purified components of the TnsABC core transposition machinery alone containing a mutant form of TnsC (TnsCA225V or TnsC*) which is still sensitive to targeting signals, but does not require the TnsD or TnsE proteins in vivo or in vitro (Stellwagen and Craig, 1997). We found that we could readily monitor TnsABC* transposition using this new assay, and the 20 bp gap that was constructed into these DNAs could be repaired by the host following transformation (Figure 6.A., data not shown). There was no apparent difference in the recovery of gapped versus non-gapped DNAs in the transformation assay (Figure 6.A.). As expected, the wild-type TnsABC control yielded no detectable transposition events (Figure 6.A.)
Figure 6. An in vitro TnsABC+E transposition assay shows that DNA structures are important for TnsE-mediated transposition, and that the β clamp is required to direct the orientation of transposon ends.
A. A representative bar graph representing the number of transposon insertions recovered per in vitro reaction shows that the presence of a single-stranded gap in the target DNA substrate is essential for activation of TnsE-mediated transposition, but shows no effect on the untargeted TnsABC* pathway.
B. Transposition via the TnsABC* pathway into the β-loaded gapped substrate indicates that there are no indirect affects of β or gapped DNA on the core transposition machinery that could account for the results observed in TnsABC+E reactions. Black circles represent the target DNA, with a gap in one circle representing the location of the ssDNA gap. Black arrows on the outside of the DNA represent the left-to right orientation of the transposon ends with respect to the free 3’ end of the gapped DNA, while gray arrows inside the circles represent the opposite (right-to-left) orientation. The position of each insertion is given next to the arrow. The ssDNA gap resides between positions 1415−1435.
C. In vitro reactions containing TnsABC+E* and gapped DNA alone. Transposition events mapped in the target DNA appear to be randomly distributed, and with no particular orientation of transposon ends.
D. Reactions containing TnsABC+E* and β clamp loaded onto the gapped DNA substrate display a rearrangement of transposon insertions. Many insertions were found at a single base pair junction 66 bases from the free 3’ end of the ssDNA gap (position 1481), and occurred in a single left-to-right orientation at that site. Other insertions were found throughout the target DNA, almost all in the left-to-right orientation.
We assayed transposition with purified wild-type TnsABC+E proteins using gapped and ungapped DNA substrates. We also monitored transposition using hyperactive mutants of TnsE (TnsE*) that were isolated in previous work, all of which were shown to strictly obey the same target site preference for DNA replication and the orientation bias with DNA replication found with the wild type protein in vivo (Peters and Craig, 2001a). The mutant proteins allow a ∼300 to ∼1000-fold increase in transposition when compared to the wild type protein in vivo (Peters and Craig, 2001a). We found that TnsABC+E transposition could be reconstituted in vitro, but that this process required the use of a gapped DNA substrate and could only be detected using hyperactive TnsE mutants (Figure 6.A. and data not shown). In multiple trials of this experiment no transposition events were ever detected unless the target DNA contained the single-strand gap (Figure 6.A.). The dependence on increased-activity mutant TnsEs could indicate that the signal is simply too low with the wild type protein to be detected in our current assay, or that the mutant proteins no longer require some factor needed by the wild type protein. Sequencing of DNA adjacent to both of the transposon ends, in plasmids isolated from chloramphenicol resistant colonies, confirmed that transposition had occurred as indicated by the presence of the characteristic 5 bp duplication found with Tn7 transposition (Craig, 2002). As expected we found that transposition with TnsABC* was random across the gapped DNA substrates with respect to position and orientation (see below, Figure 6.B., data not shown). Interestingly, we found that in vitro transposition with TnsABC+E* also appeared to occur randomly in the gapped DNA substrate (Figure 6.C.). This was somewhat surprising given the absolute bias with DNA replication found in vivo, but was consistent with the separation of activation and targeting found in other Tn7 in vitro reactions (See DISCUSSION)(Rao et al., 2000).
To determine the effect of β in TnsE-mediated transposition in vitro, the β clamp was loaded onto the DNA structure using a purified minimal clamp-loader system (γ3δδ’). The gapped structure presents a site in the DNA that can be used to load β onto the DNA substrate (Leu et al., 2000). Clamps are expected to be loaded onto the gapped DNA with a single orientation that is dependent on which strand of DNA contains a free 3’ end (Johnson and O'Donnell, 2005), and may be preferentially retained at the 3’ end through interactions with the DNA (Georgescu et al., 2008). We loaded the β-clamp onto the DNA substrates using a 20-fold molar excess of β with respect to DNA, then purified the β-loaded structures away from the clamp-loader components and free β proteins (EXPERIMENTAL PROCEDURES) (Leu et al., 2000). We did not detect significant increases in TnsE-mediated transposition using the β-loaded gapped DNA structures (data not shown). However, upon mapping the location and orientation of transposon insertions, we found a dramatically different pattern compare Figures than that exhibited by the gapped-DNA substrate without β (compare Figures 6.C. and 6.D. We found that 80% of the insertion events occurring in the β-loaded substrate were in a single orientation, and that 40% of them were found at a single base pair junction (at position 1481), proximal to the location of the single-stranded DNA gap (66 bp from the 3’ end). These insertions were isolated from separate transformations, ensuring that they are independent transposition events and not siblings. The presence of a single site where many insertions were recovered is reminiscent of TnsD-mediated insertion into attTn7 (Craig, 2002)) and the influence of triplex DNA structures on TnsABC* transposition (Rao and Craig, 2001) (see DISCUSSION). When we monitored TnsABC* transposition on the same β-loaded gapped DNA substrates we found no change in the distribution or orientation of insertion events from what was found in gapped substrates without β; the insertions were found in both orientations without any preference for any position in the plasmid (Figure 6.B.). Significantly, the TnsABC+E* insertions are consistent with in vivo data with respect to the orientation of transposon end alignment with the 3’ end of the nascent lagging-strand. While β may not be necessary to activate transposition in vitro, its presence on DNA is required to recapitulate the exact targeting activity observed with TnsE-mediated transposition in vivo (Peters and Craig, 2001a).
TnsE is capable of disrupting normal coordination of host protein interaction with the processivity factor
What are the consequences to the host cell when a foreign protein binds to the processivity factor? Since TnsE is able to bind the β clamp, we reasoned that over-expression of TnsE might interrupt the coordination of host proteins that normally bind to the clamp. To test this idea we monitored SOS induction following considerable over-expression of TnsE. The SOS response is a regulatory network that is normally repressed until DNA damage occurs. Persistently stalled, blocked, or collapsed replication forks are known to trigger the SOS response (Kuzminov, 1995). Constitutive induction of the SOS response is a phenotype associated with some mutant β clamp proteins (Maul et al., 2007; Sutton, 2004). We found that very high expression levels of TnsE do induce the SOS response (Figure 7.A.). Moreover, we found that the ability of TnsE to induce SOS is dependent on the putative β clamp interaction motif. Each of the tnsEβMA mutants significantly reduced (p<0.0005, two-tailed unequal variance t-test) the level of SOS induction (Figure 7.A.). Similar to the constitutive SOS phenotype observed in cells expressing a defective β protein (Flores et al., 2005), elimination of the RecFOR system suppressed SOS induction in cells over-expressing TnsE (Figure 7.B.). This result indicates that over-expression of TnsE leads to single-stranded gaps in DNA, not double-stranded DNA breaks (Kuzminov, 1995). Wild-type and moderately over-expressed levels of TnsE (from wild-type and Plac promoters, respectively) do not lead to the induction of the SOS response (data not shown), and TnsE-mediated transposition is not dependent on SOS induction since all in vivo transposition assays in this study were carried out in recA− cells. These experiments and the results from our analysis using SPR indicate that TnsE has evolved to minimize interference with normal DNA metabolism while maintaining the ability to interact with the highly conserved and essential processivity factor for use in targeting transposition.
Figure 7. Over-expression of TnsE results in activation of the SOS DNA damage response.
A. Activation of SOS is consistent with interference with normal traffic on the β clamp. Cells that over-express wild type TnsE activate the SOS response. Strains expressing the TnsEβMA mutant proteins show a significant reduction in the SOS response compared to the wild type protein, likely due to a compromised interaction with β. Western blot results for membranes probed with anti-TnsE that are displayed below the graph. SOS was monitored by Miller assay and in a reporter strain containing a sulA::lac fusion and is reported in Miller units (Miller, 1992; Sutton, 2004). Error bars indicate standard error of the mean (n=3).
B. SOS induction provoked by TnsE over-expression is abolished in the recF-background. RecF is essential for the RecFOR pathway of RecA loading onto single-stranded DNA gaps. Error bars indicate standard error of the mean (n=3).
DISCUSSION
Our results indicate that there are two critical features recognized by TnsE during DNA replication: a specific DNA structure and the processivity clamp on DNA. While structural components are needed for activating TnsE-mediated transposition in vitro, the presence of the β clamp processivity factor is essential for specifically redirecting TnsE-mediated transposition events in a manor that recapitulates the profile found in vivo with DNA replication. We conclude that Tn7 likely evolved the capacity to interact with the processivity factor as a way of recognizing discontinuous DNA replication displayed by mobile plasmids.
Tn7 participates in multiple transposition pathways that have been established in vitro (Bainton et al., 1993; Stellwagen, 2001). These systems provide unique perspectives on the relationship between activation and target-site selection of transposons, and highlight the importance of target DNA structure and interaction with host factors for Tn7 transposition. Multiple mutant TnsC alleles can allow Tn7 transposition that does not require either the TnsD or TnsE protein for activation (Stellwagen and Craig, 1997). TnsABC* transposition events appear to occur randomly in vitro and in vivo (Biery et al., 2000; Peters and Craig, 2000; Seringhaus et al., 2006; Stellwagen and Craig, 1997)(Figure 6.B. and data not shown). Rao and Craig showed that random TnsABC* transposition events can be redirected in vitro within target DNA molecules to a hotspot adjacent to a pyrimidine triplex (Rao and Craig, 2001; Rao et al., 2000). While triplex forming DNAs cannot activate the wild type protein, they are sufficient to redirect active TnsABC* complexes to a specific hotspot in target DNAs. This work strongly suggested that a structure induced in target DNAs is an important component of Tn7 target recognition and that the activation and targeting signals are separable. It is also thought that the primary role of the TnsD protein in targeting transposition events into the attTn7 site involves the creation of a distortion in the DNA adjacent to its binding site in the glmS gene (Kuduvalli et al., 2001).
While activation of TnsABC+E transposition requires a structure in the target DNA, a separate signal is required to target transposition events, similar to what was found in the TnsABC* pathway with triplex DNA target structures (Rao and Craig, 2001). In the in vitro transposition experiments, clamps were loaded onto target DNAs in a single orientation, which was determined by the strand of DNA that contained the gap (Johnson and O'Donnell, 2005). Transposition events received by the clamp-loaded DNA substrates were predominantly in a single orientation and at a site in the DNA near where clamps are expected to transiently reside (Figure 6.D.)(Georgescu et al., 2008).
The dependence of TnsABC+E transposition on interaction with β and a DNA structure allows Tn7 to modulate the activity of this transposition pathway depending on the replication status of the target DNA. Given that β clamps and single-stranded DNA gaps accumulate on the DNA strand that is replicated discontinuously (Johnson and O'Donnell, 2005), the TnsE-β interaction and DNA structure specific activation helps explain how TnsE directs transposon insertion to the conjugal DNA replication process. The nature of conjugal DNA replication may leave β clamps especially vulnerable to TnsE interaction. Conjugal plasmids replicate by a discontinuous process that resembles lagging-strand DNA synthesis that occurs during chromosomal replication (Wilkins and Lanka, 1993). While processivity clamps are expected to be enriched both on the chromosome and on conjugal plasmids (Johnson and O'Donnell, 2005), a notable difference is that discontinuous conjugal replication is not physically coupled to continuous replication within the same replisome as it is in chromosomal replication (Wilkins and Lanka, 1993). The protein complex that is present at a standard DNA replication fork may limit the exposure of the β clamp and gapped DNA structures more effectively than the uncoupled DNA replication found in conjugal replication. Our data suggest that TnsE has evolved to not interrupt the exchange of proteins on clamps during normal DNA replication when expressed at moderate concentrations (Figures 5, 7, and data not shown). The distribution of TnsE-mediated insertions in the chromosome (Peters and Craig, 2001a) is likely explained by the ability of TnsE to interact with β clamps that are still topologically linked to DNA, but not actively involved in chromosomal DNA replication. For example, in cells lacking mobile plasmids TnsE-mediated insertions occur in the region where DNA replication terminates and sites proximal to repaired DNA double-strand breaks (Peters and Craig, 2000; Shi et al., 2008).
The processivity factor appears to play a pivotal role in the coordination of activity at the replication fork. TnsE likely binds to the same face of the clamp as MutS, Ligase, Pol III and others (Johnson and O'Donnell, 2005; Lopez de Saro and O'Donnell, 2001; Simmons et al., 2008). The presence and importance of β clamp binding motif in TnsE suggests that interaction with the clamp occurs at least in part through the same hydrophobic pocket on the C-terminal face that appears to be involved in coordination of protein-protein interactions. The orientation bias and location of transposition events we observed in vitro were consistent with interaction with the C-terminal face of the clamp, which is expected to preferentially reside at the 3’ junction of the gap in the target DNA (Georgescu et al., 2008)(Figure 6.D.). Clamps are also free to slide about the DNA (Johnson and O'Donnell, 2005; Laurence et al., 2008), which may explain insertions that occurred into other parts of the target DNA, yet in an orientation that appears to be dictated by the clamp. The SOS induction phenotype observed with TnsE over-expression further supports the notion that TnsE interacts with the clamp on the same face as host proteins involved in DNA replication and repair (Figure 7). The TnsABC+E transposition complex may use a similar mechanism for detecting strand polarity as has been suggested for mismatch repair systems (MMR) in Eukaryotes and in some Prokaryotes. The ability to interact with only one face of the processivity factor has been suggested to allow strand discrimination in MMR so that newly-replicated DNA containing errors can be selectively removed (Jiricny, 2006; Simmons et al., 2008). Based on our in vitro transposition experiments, interaction with the β clamp directs the activity of TnsE in a similarly directional manner (Figure 6.D.), resulting in the orientation bias with replication that we observe in vivo with TnsE-mediated insertions (Peters and Craig, 2001b).
Interaction with the processivity factor may constitute a general mechanism for targeting transposition into actively replicating DNA. The transposase of the inactive pogo element, found in Drosophila, has been shown to bind to the DNA replication processivity factor (PCNA), but the function of this interaction remains a mystery (Warbrick, 2000; Warbrick et al., 1998). Transposases of other inactive transposons that are abundant in humans (tigger elements, estimated to be present at ∼3000 copies) and in Arabidopsis (lemi1 elements) also possess putative PCNA binding motifs (Warbrick, 2000; Warbrick et al., 1998). Because none of these elements are active, determination of the functional relevance of their interaction with the processivity factor is not possible. We suggest that TnsE has evolved that ability to identify the β clamp as a mechanism for targeting processing events found during the mobilization of plasmids. A wide range of transposable elements may use a similar mechanism to target DNA replication and/or DNA repair. While mechanistically very different from Tn7, the transposon Tn917 displays target selection profile that resembles that of Tn7 transposition with the TnsABC+E pathway in the chromosome (Garsin et al., 2004). The single polypeptide transposase of Tn917 contains an amino acid sequence (QLCLAR) that resembles the β clamp binding motif described in this work (Figure 1). In plants, the transposase of the Ac element has been shown to be stimulated by active DNA replication (Chen et al., 1992) and contains the sequence QKRIVGFF (A. Parks and J. Peters, unpublished observation), similar to many previously reported PCNA interaction motifs, or PIP-boxes, with the consensus sequence QxxIxxFF (Warbrick, 2000).
For Tn7, the interaction with the processivity factor appears to be primarily used to activate transposition directed into mobilized plasmids, providing Tn7 with a means of moving to a new host. Since Tn7-like elements are found in a wide variety of hosts (Parks and Peters, 2009), TnsE-mediated transposition shows promise as a new tool for probing the mechanisms and evolution of genetic processes involving processivity factors.
EXPERIMENTAL PROCEDURES
Plasmids and Strains
All E. coli strains were constructed using P1 transduction according to standard genetic techniques (Table S1) (Peters, 2007). All primers were purchased from Integrated DNA Technologies (Table S2). Plasmid construction was accomplished by established methods and detailed in Table S3 (Sambrook et al., 1989). Site directed mutagenesis of the putative β clamp binding motif within tnsE was carried out using PCR and sub-cloned into the various expression vectors (Table S2). The bottom strand primers are shown in Table S3 to indicate the specific mutations. Yeast two-hybrid vectors were constructed using the Gateway system (Invitrogen) essentially as described in the manufacturers recommendations and elsewhere (Liachko and Tye, 2005). pGAP plasmid was constructed from a pGEM-T cloning vector containing the attTn7 locus (pGEM-attTn7) by insertion of a fragment containing two recognition sites (20 bp apart from each other) for the nicking enzyme Nb. BbvCI (NEB).
Computational analysis
The tnsE genes identified in previous work (Parks and Peters, 2009) were aligned using the ClustalW algorithm (Thompson et al., 1994) through the online Jalview server (Clamp et al., 2004)(Table S4). A consensus sequence was generated using the Jalview software (Clamp et al., 2004).
Yeast two-hybrid assay
Yeast strain EGY40[pSH18−34] (Golemis, 2002) was co-transformed with pBTMgw and pGADgw derivatives and selected on SC-Leu-Trp plates. Overnight yeast cultures of each isolate were grown in 5 ml SC–Trp–Leu dropout media (Sunrise Science Products) to an OD600 ∼1. 1ml of cells were spun down and resuspended in Z-buffer. Miller assays were performed as described by Amberg et. al. (Amberg et al., 2006)
Protein purification and labeling
His-6-tagged TnsE proteins were purified as described (Peters and Craig, 2001a). TnsA, TnsB, TnsC, and TnsCA225V were purified as described in (Bordi et al., 2008). A modified β protein that could be labeled with 32P phosphate was purified using the previously described method and vector (Kelman et al., 1995b). β was labeled with 32P as described using αAMP Protein Kinase (PKA) purchased from NEB (Kelman et al., 1995a).
The δ, δ’, and γ proteins were each purified using the IMPACT system from NEB according to manufacturers recommendations. Cleaved proteins were eluted and dialyzed in storage buffer (20 mM Tris (pH 7.5), 100 mM KCl, 20% glycerol, 0.5 mM EDTA, 1mM DTT).
The minimal clamp-loader γ complex (γ3δδ’) (Jeruzalmi et al., 2001b) was assembled from individually purified proteins in clamp-loading buffer (20 mM Tris-HCl (pH 7.5), 4% glycerol, 8mM MgCl2, 1mM ATP, 2mM DTT, 0.1 mM EDTA) and further purified away from monomers by size exclusion chromatography on a Superdex G-200 column (GE Healthcare). Fractions were assayed for clamp loading activity as described in (Leu et al., 2000) using nicked pGAP plasmid and 32P-β and the most active fractions were used in experiments requiring β-loaded DNA.
Preparation of target DNA substrates
The gapped substrate was made as described in (Wang and Hays, 2001). Briefly, supercoiled pGAP plasmid containing two Nb. BvcCI nicking sites separated by 20 bp was digested with Nb. BbvCI enzyme (NEB). A competitor DNA oligo (JEP 348) complementary to the sequence flanked by the nick was then added in 50-fold molar excess and incubated at 85°C for 10 minutes to displace the 20 nucleotide fragment. After reannealing, gapped plasmids were purified away from the 20-mer duplex DNA and the excess single-stranded oligomers using Amicon unitra-4 (MWCO 100 kDa) centrifugal filter (Millipore).
The β-clamp was loaded onto gapped or nicked pGAP plasmid as described in (Leu et al., 2000) by incubation of minimal clamp-loader (1 pmol), β (25 pmol as β dimer), and gapped pGAP plasmid (1.25 pmol) in clamp-loading buffer (20 mM Tris-HCl (pH 7.5), 4% glycerol, 8mM MgCl2, 1mM ATP, 2mM DTT, 0.1 mM EDTA). The β-loaded DNA was purified away from clamp-loader and free β-clamp by size exclusion chromatography on a 2 ml 4% agarose bead column (MP biomedicals) in gel-filtration buffer (20 mM Tris-HCl (pH 7.5), 4% glycerol, 100 μg/ml BSA , 2mM DTT). 200-μl fractions were collected and two peak fractions containing β-loaded DNA were pooled and used for in vitro transposition assays.
Protein Gel Mobility Shift Assays
Native polyacrylamide gel electrophoresis assays using 32P-β were performed as described (Lopez de Saro and O'Donnell, 2001). Each 15 μl reaction was composed of 25 mM Tris-Cl (pH 7.5), 0.5 mM EDTA, 10% glycerol, 50 μg/ml bovine serum albumin, 100 mM KCl, 5 mM DTT, 10 nM 32P-β, and TnsE or BSA as indicated in the figure legends. Samples were incubated at 37 °C for 5 min, then 10 μl of the reaction was run on a 4% native polyacrylamide gel (4% acrylamide:bisacrylamide 29:1, 0.5 × TBE buffer, 5% glycerol). Electrophoresis was performed in 0.5 × TBE buffer (45 mM Tris borate, 45 mM boric acid, 2.5 mM EDTA) at 70 V for 8 hours (23°C). Gels soaked in 20% ethanol and 5% glycerol for 10 minutes then dried overnight. 32P-β was detected using a PhosphorImager (Molecular Dynamics). Images were quanitifed using ImageJ software (Rasband, 1997), and the KD was calculated by plotting the area of shifted product peaks (32P signal) vs. TnsE concentration. KD is equal to the concentration of TnsE at which ½ predicted maximal 32P-signal in the shifted product is achieved.
Far Western Blot
A Hybond-ECL nitrocellulose membrane was prepared according to manufacturers recommendations (Amersham Biosciences). Proteins were bound to the wet membrane in the quantities noted in the figure legend using a slot blot apparatus (Amersham Biosciences). The membrane was then blocked overnight at 25°C, in blocking buffer containing 20 mM HEPES (pH 7.5), 50 mM KCl, 10 mM MgCl2, 1 mM DTT, 0.1% Triton X-100, 1% BSA. The membrane was probed as described previously (Einarson et al., 2007). An interaction buffer composed of 20 mM HEPES (pH 7.5), 50 mM KCl, 10 mM MgCl2, 1 mM DTT, 0.1% Triton X-100, 5% glycerol, 1% BSA, and containing 5 nM 32P-β was used to probe the membrane at 4°C for 3.5 hours. Following interaction, the membrane was washed four times with PBS (10 mM Na-phosphate, pH7.2, 0.9% NaCl, 0.2% Triton X-100), and twice with PBS augmented with 100 mM KCl. The membrane was dried for 30 min at 42°C and 32P-β was detected using a PhosphorImager (Molecular Dynamics).
Surface Plasmon Resonance
Surface plasmon resonance experiments were carried out essentially as reported in (Leu and O'Donnell, 2001) using a Biacore 2000 instrument (GE healthcare). All proteins used in these experiments were further purified by gel filtration using a Superdex G-200 column (GE healthcare) immediately prior to these experiments and eluted directly in running buffer consisting of 10 mM HEPES (pH 7.4), 150 mM NaCl, 3.4 mM EDTA, and 0.005% Tween 20. The β protein was diluted 1:10 into 100mM sodium acetate (pH 5.5), then immobilized at a flow rate of 5 μl/min on a CM-5 chip using N-hydroxysulfosuccinimide (NHS) and 1-Ethyl-3-[3-dimethylaminopropyl]carbodiimide (EDC) cross-linking to final amounts of 1900, 6300, and 9600 response units (RU) (GE Healthcare). 1M Ethanolamine-HCl was used to block all remaining reactive groups. A reference cell containing no protein was also prepared for subtraction of non-specific interaction signal between proteins and the chip. All Interaction experiments were carried out at a flow rate of 10 μl/min. Kinetic calculations were carried out using the BIAevaluation software (Biacore, 1997) and KD values were confirmed by plotting the RU at equilibrium versus protein concentration, and calculating the protein concentration at which ½ the maximum RU has been reached.
In vivo Transposition Assay
Transposition assays were conducted in the strain NLC51 containing tns genes encoded on plasmids as described (McKown et al., 1988). A miniTn7 element containing a KamR cassette was introduced using a defective lambda phage (γKK1) that cannot integrate or replicate in the NLC51 background strains used in this assay. Transposition frequency was obtained by dividing the number of Kanamycin resistant colonies by the number of infectious γKK1 phage used in the assay.
In vitro transposition assay
In vitro transposition reactions contained 26 mM HEPES (pH 7.6), 15 mM Tris (pH 7.6), 4% glycerol, 50 μg/ml bovine serum albumin, 2 mM ATP (pH 7.0), 1.5 mM dithiothreitol, 100 μg/ml tRNA, 0.05 mM EDTA, 8.3 mM NaCl, 9.4 mM KCl, 0.06 mM MgCl2,15 mM magnesium acetate, 0.2 nM of the donor plasmid pGPS2.1 (NEB), 3.2 nM of recipient plasmid, 12.5 nM TnsA, 3 nM TnsB, 5 nM TnsCA225V or TnsCwt, and 3.6 nM of TnsE in a final volume of 100 μl. All of the reaction mixture components except TnsA, TnsB, magnesium acetate, and the donor DNA were preincubated for 20 min at 30°C. The final reaction mixture components were then added, and the reaction was allowed to proceed for an additional 45 min at 30°C. The reaction was stopped by phenol:chloroform extraction and the DNA was ethanol precipitated and resuspended in 40 μl H2O. The DNA was then assayed for transposition by transforming E. coli (DH5α) cells and selecting for chloramphenicol resistance. Plasmids were isolated from chloramphenicol resistant colonies, and the position of the transposition was determined using a primer specific to the left end of the Tn7 element (Table S3). About 50% of the insertions were sequenced from both ends to ensure that real transposition occurred. Inclusion of sequences from separate transformation pools insured that siblings were not confused with independent insertions.
SOS induction assay
AP330 or its derivatives containing pBAD24 derivatives were grown overnight in LB media containing 0.2% glucose and ampicillin (100 μg/ml) at 37°C. Overnight cultures were diluted 1:100 in fresh LB with 0.2% arabinose and ampicillin then grown two hours at 37°C. β galactosidase activity was measured by the Miller assay as described (Miller, 1992).
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grant R01 GM069508. We thank Mark Sutton and Bik Tye for strains and plasmids and helpful discussions. We also thank Eric Alani, Bik Tye, Jeff Roberts, and the members of the Peters laboratory for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Amberg DC, Burke DJ, Strathern JN. Assay of {beta}-Galactosidase in Yeast: Permeabilized Cell Assay. Cold Spring Harbor Protocols 2006. 2006 doi: 10.1101/pdb.prot4158. 10.1101/pdb.prot4158. pdb.prot4158- [DOI] [PubMed] [Google Scholar]
- Bainton RJ, Kubo KM, Feng J-N, Craig NL. Tn7 transposition: target DNA recognition is mediated by multiple Tn7-encoded proteins in a purified in vitro system. Cell. 1993;72:931–943. doi: 10.1016/0092-8674(93)90581-a. [DOI] [PubMed] [Google Scholar]
- Beuning PJ, Sawicka D, Barsky D, Walker GC. Two processivity clamp interactions differentially alter the dual activities of UmuC. Mol Microbiol. 2006;59:460–474. doi: 10.1111/j.1365-2958.2005.04959.x. [DOI] [PubMed] [Google Scholar]
- Biacore I. BIAevaluation software handbook, version 3.0. Biacore (Inc); Uppsala, Sweden: 1997. [Google Scholar]
- Biery MC, Steward F, Stellwagen AE, Raleigh EA, Craig NL. A simple in vitro Tn7-based transposition system with low target site selectivity for genome and gene analysis. Nucleic Acids Res. 2000;28:1067–1077. doi: 10.1093/nar/28.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordi C, Butcher BG, Shi Q, Hachmann AB, Peters JE, Helmann JD. In vitro mutagenesis of Bacillus subtilis by using a modified Tn7 transposon with an outward-facing inducible promoter. Appl Environ Microbiol. 2008;74:3419–3425. doi: 10.1128/AEM.00476-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting KA, Roe SM, Pearl LH. Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the beta-clamp. Embo J. 2003;22:5883–5892. doi: 10.1093/emboj/cdg568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Greenblatt IM, Dellaporta SL. Molecular analysis of Ac transposition and DNA replication. Genetics. 1992;130:665–676. doi: 10.1093/genetics/130.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Craig N, Craigie R, Gellert M, Lambowitz A. Mobile DNA II. ASM Press; Washington, D.C.: 2002. [Google Scholar]
- Craig NL. Tn7. In: Craig Nancy L, Craigie R, Gellert M, Lambowitz-Alan M, editors. Mobile DNA II. ASM Press; Washington, DC: 2002. pp. 423–456. [Google Scholar]
- Dallmann HG, McHenry CS. DnaX complex of Escherichia coli DNA polymerase III holoenzyme. Physical characterization of the DnaX subunits and complexes. J Biol Chem. 1995;270:29563–29569. [PubMed] [Google Scholar]
- Dalrymple BP, Kongsuwan K, Wijffels G, Dixon NE, Jennings PA. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc Natl Acad Sci U S A. 2001;98:11627–11632. doi: 10.1073/pnas.191384398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einarson MB, Pugacheva EN, Orlinick JR. Far Western: Probing Membranes. Cold Spring Harbor Protocols 2007. 2007 doi: 10.1101/pdb.prot4759. 10.1101/pdb.prot4759. pdb.prot4759- [DOI] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Flores MJ, Sanchez N, Michel B. A fork-clearing role for UvrD. Mol Microbiol. 2005;57:1664–1675. doi: 10.1111/j.1365-2958.2005.04753.x. [DOI] [PubMed] [Google Scholar]
- Garsin DA, Urbach J, Huguet-Tapia JC, Peters JE, Ausubel FM. Construction of a Enterococcus faecalis Tn917-mediated-gene-disruption library offers insight into Tn917 insertion patterns. Journal of Bacteriology. 2004;186:7280–7289. doi: 10.1128/JB.186.21.7280-7289.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu RE, Kim SS, Yurieva O, Kuriyan J, Kong XP, O'Donnell M. Structure of a sliding clamp on DNA. Cell. 2008;132:43–54. doi: 10.1016/j.cell.2007.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis E. Protein-Protein Interactions. CSHL Press; Cold Spring Harbor NY: 2002. [Google Scholar]
- Jeruzalmi D, O'Donnell M, Kuriyan J. Crystal structure of the processivity clamp loader gamma (gamma) complex of E. coli DNA polymerase III. Cell. 2001a;106:429–441. doi: 10.1016/s0092-8674(01)00463-9. [DOI] [PubMed] [Google Scholar]
- Jeruzalmi D, Yurieva O, Zhao Y, Young M, Stewart J, Hingorani M, O'Donnell M, Kuriyan J. Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell. 2001b;106:417–428. [PubMed] [Google Scholar]
- Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- Johnson A, O'Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- Kelman Z, Naktinis V, O'Donnell M. Radiolabeling of proteins for biochemical studies. Methods Enzymol. 1995a;262:430–442. doi: 10.1016/0076-6879(95)62034-6. [DOI] [PubMed] [Google Scholar]
- Kelman Z, Yao N, O'Donnell M. Escherichia coli expression vectors containing a protein kinase recognition motif, His6-tag and hemagglutinin epitope. Gene. 1995b;166:177–178. doi: 10.1016/0378-1119(95)00556-7. [DOI] [PubMed] [Google Scholar]
- Kuduvalli P, Rao JE, Craig NL. Target DNA structure plays a critical role in Tn7 transposition. EMBO J. 2001;20:924–932. doi: 10.1093/emboj/20.4.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A. Instability of inhibited replication forks in E. coli. Bioessays. 1995;17:733–741. doi: 10.1002/bies.950170810. [DOI] [PubMed] [Google Scholar]
- Laurence TA, Kwon Y, Johnson A, Hollars CW, O'Donnell M, Camarero JA, Barsky D. Motion of a DNA sliding clamp observed by single molecule fluorescence spectroscopy. J Biol Chem. 2008 doi: 10.1074/jbc.M800174200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu FP, Hingorani MM, Turner J, O'Donnell M. The delta subunit of DNA polymerase III holoenzyme serves as a sliding clamp unloader in Escherichia coli. J Biol Chem. 2000;275:34609–34618. doi: 10.1074/jbc.M005495200. [DOI] [PubMed] [Google Scholar]
- Leu FP, O'Donnell M. Interplay of clamp loader subunits in opening the beta sliding clamp of Escherichia coli DNA polymerase III holoenzyme. J Biol Chem. 2001;276:47185–47194. doi: 10.1074/jbc.M106780200. [DOI] [PubMed] [Google Scholar]
- Liachko I, Tye BK. Mcm10 is required for the maintenance of transcriptional silencing in Saccharomyces cerevisiae. Genetics. 2005;171:503–515. doi: 10.1534/genetics.105.042333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Saro F, Georgescu RE, Leu F, O'Donnell M. Protein trafficking on sliding clamps. Philos Trans R Soc Lond B Biol Sci. 2004;359:25–30. doi: 10.1098/rstb.2003.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Saro FJ, Marinus MG, Modrich P, O'Donnell M. The beta sliding clamp binds to multiple sites within MutL and MutS. J Biol Chem. 2006;281:14340–14349. doi: 10.1074/jbc.M601264200. [DOI] [PubMed] [Google Scholar]
- Lopez de Saro FJ, O'Donnell M. Interaction of the beta sliding clamp with MutS, ligase, and DNA polymerase I. Proc Natl Acad Sci U S A. 2001;98:8376–8380. doi: 10.1073/pnas.121009498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul RW, Ponticelli SK, Duzen JM, Sutton MD. Differential binding of Escherichia coli DNA polymerases to the beta-sliding clamp. Mol Microbiol. 2007;65:811–827. doi: 10.1111/j.1365-2958.2007.05828.x. [DOI] [PubMed] [Google Scholar]
- McKown RL, Orle KA, Chen T, Craig NL. Sequence requirements of Escherichia coli attTn7, a specific site of transposon Tn7 insertion. J Bacteriol. 1988;170:352–358. doi: 10.1128/jb.170.1.352-358.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. A short course in bacterial genetics. Cold Spring Harbor; New York: 1992. [Google Scholar]
- Parks AR, Peters JE. Transposon Tn7 is widespread in diverse bacteria and forms genomic islands. J Bacteriol. 2007;189:2170–2173. doi: 10.1128/JB.01536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AR, Peters JE. Tn7 elements: engendering diversity from chromosomes to episomes. Plasmid. 2009;61:1–14. doi: 10.1016/j.plasmid.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JE. Gene transfer - Gram-negative bacteria, Chapter 31. In: Reddy CA, Beveridge TJ, Breznak JA, Marzluf GA, Schmidt TM, S. L. R., editors. Methods for General and Molecular Microbiology. ASM Press; Washington D. C.: 2007. pp. 735–755. [Google Scholar]
- Peters JE, Craig NL. Tn7 transposes proximal to DNA double-strand breaks and into regions where chromosomal DNA replication terminates. Mol Cell. 2000;6:573–582. doi: 10.1016/s1097-2765(00)00056-3. [DOI] [PubMed] [Google Scholar]
- Peters JE, Craig NL. Tn7 recognizes target structures associated with DNA replication using the DNA binding protein TnsE. Genes & Dev. 2001a;15:737–747. doi: 10.1101/gad.870201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JE, Craig NL. Tn7: smarter than we thought. Nature Reviews/Molecular Cell Biology. 2001b;2:806–814. doi: 10.1038/35099006. [DOI] [PubMed] [Google Scholar]
- Rao JE, Craig NL. Selective recognition of pyrimidine motif triplexes by a protein encoded by the bacterial transposon Tn7. J Mol Biol. 2001;307:1161–1170. doi: 10.1006/jmbi.2001.4553. [DOI] [PubMed] [Google Scholar]
- Rao JE, Miller PS, Craig NL. Recognition of triple-helical DNA structures by transposon Tn7. Proc Natl Acad Sci USA. 2000;97:3936–3941. doi: 10.1073/pnas.080061497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband W. ImageJ. US National Institutes of Health; Bethesda, Maryland, USA: 1997. (Version 1.38x) [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd edn Cold Spring Harbor Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Seringhaus M, Kumar A, Hartigan J, Snyder M, Gerstein M. Genomic analysis of insertion behavior and target specificity of mini-Tn7 and Tn3 transposons in Saccharomyces cerevisiae. Nucleic Acids Res. 2006;34:e57. doi: 10.1093/nar/gkl184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafferman A, Kolter R, Stalker D, Helinski DR. Plasmid R6K DNA replication. III. Regulatory properties of the pi initiation protein. J Mol Biol. 1982;161:57–76. doi: 10.1016/0022-2836(82)90278-9. [DOI] [PubMed] [Google Scholar]
- Sharpe P, Craig NL. Host proteins can stimulate Tn7 transposition: a novel role for the ribosomal protein L29 and the acyl carrier protein. EMBO J. 1998;17:5822–5831. doi: 10.1093/emboj/17.19.5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Parks AR, Potter BD, Safir IJ, Luo Y, Forster BM, Peters JE. DNA damage differentially activates regional chromosomal loci for Tn7 transposition in Escherichia coli. Genetics. 2008;179:1237–1250. doi: 10.1534/genetics.108.088161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons LA, Davies BW, Grossman AD, Walker GC. Beta clamp directs localization of mismatch repair in Bacillus subtilis. Mol Cell. 2008;29:291–301. doi: 10.1016/j.molcel.2007.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen A, Craig NL. Gain-of-function mutations in TnsC, an ATP-dependent transposition protein which activates the bacterial transposon Tn7. Genetics. 1997;145:573–585. doi: 10.1093/genetics/145.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen A, Craig NL. Mobile DNA elements: controlling transposition with ATP-dependent molecular switches. TIBS. 1998;23:486–490. doi: 10.1016/s0968-0004(98)01325-5. [DOI] [PubMed] [Google Scholar]
- Stellwagen AE. Analysis of gain of function mutants of an ATP-dependent regulator of Tn7 transposition. J Mol Biol. 2001;305:633–642. doi: 10.1006/jmbi.2000.4317. [DOI] [PubMed] [Google Scholar]
- Stellwagen AE, Craig NL. Analysis of Gain of Function Mutants of an ATP-dependent Regulator of Tn7 Transposition. J Mol Biol. 2001;305:633–642. doi: 10.1006/jmbi.2000.4317. [DOI] [PubMed] [Google Scholar]
- Sutton MD. The Escherichia coli dnaN159 mutant displays altered DNA polymerase usage and chronic SOS induction. J Bacteriol. 2004;186:6738–6748. doi: 10.1128/JB.186.20.6738-6748.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hays JB. Simple and rapid preparation of gapped plasmid DNA for incorporation of oligomers containing specific DNA lesions. Mol Biotechnol. 2001;19:133–140. doi: 10.1385/MB:19:2:133. [DOI] [PubMed] [Google Scholar]
- Warbrick E. The puzzle of PCNA's many partners. Bioessays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Warbrick E, Heatherington W, Lane DP, Glover DM. PCNA binding proteins in Drosophila melanogaster : the analysis of a conserved PCNA binding domain. Nucleic Acids Res. 1998;26:3925–3932. doi: 10.1093/nar/26.17.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijffels G, Dalrymple BP, Prosselkov P, Kongsuwan K, Epa VC, Lilley PE, Jergic S, Buchardt J, Brown SE, Alewood PF, et al. Inhibition of protein interactions with the beta 2 sliding clamp of Escherichia coli DNA polymerase III by peptides from beta 2-binding proteins. Biochemistry. 2004;43:5661–5671. doi: 10.1021/bi036229j. [DOI] [PubMed] [Google Scholar]
- Wilkins B, Lanka E. DNA processing and replication during plasmid transfer between gram-negative bacteria. In: Clewell DB, editor. Bacterial Conjugation. Plenum Press; New York: 1993. [Google Scholar]
- Wolkow CA, DeBoy RT, Craig NL. Conjugating plasmids are preferred targets for Tn7. Genes & Dev. 1996;10:2145–2157. doi: 10.1101/gad.10.17.2145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.