Abstract
The genus Cryptosporidium includes several species of intestinal protozoan parasites which multiply in intestinal epithelial cells. The impact of this infection on the transcriptome of cultured host cells was investigated using DNA microarray hybridizations. The expression of 14 genes found to be consistently up- or down-regulated in infected cell monolayers was validated with RT PCR. Using immunofluorescence we examined the expression of Protease Activated Receptor-2, which is encoded by one of the up-regulated genes. In infected cells this receptor localized to the host cell membrane which covers intracellular trophozoites and meronts. This observation indicates that the composition of the host cell membrane is affected by the developing trophozoite, a phenomenon which has not been described previously.
Keywords: Cryptosporidium parvum, human ileocecal epithelial cells, HCT-8, Protein Activated Receptor-2
1. Introduction
Cryptosporidiosis, a cosmopolitan diarrheal illness, is caused by several species of intestinal protozoa belonging to the genus Cryptosporidium. Cryptosporidium parvum primarily infects the epithelium of the small intestine, which typically results in acute, self-limited diarrhea. Immunodeficient individuals and young children are more vulnerable and may develop a persistent infection [1, 2]. Unlike infections caused by other pathogenic Apicomplexa, such as the parasite causing malaria, toxoplasmosis and coccidiosis, there is no reliable treatment against cryptosporidiosis. Understanding the interaction of the pathogen with the host is expected to facilitate the development of new therapies.
Several studies have documented morphological changes in host cells infected with C. parvum [3-5]. Studies of host cell invasion by C. parvum sporozoites using electron microscopy indicate that the microvilli are incorporated into the host membrane which covers the developing trophozoite [6]. Accumulation of host cell actin and actin-binding proteins at the host parasite interface was observed during parasite attachment and invasion, suggesting that C. parvum also induces the rearrangement of the host cell cytoskeleton [7, 8]. Host cell apoptosis has been demonstrated to occur in response to C. parvum infection in different cell lines [9-12]. Secretion of cytokines and chemokines by infected cells has also been described [13, 14].
Microarrays have been used to study protozoal infections, including those caused by Toxoplasma, Plasmodium, Eimeria, and Trypanosoma species, which all develop intracellularly. These studies have found that the expression of genes associated with cytokine and chemokine production is typically altered in response to infection [15-17]. The expression of host cell genes controlling cellular metabolism, cell adhesion, and DNA repair can also be affected, as is the expression of apoptosis related genes [18, 19]. Up-regulation of cell cycle related genes were identified in Toxoplasma gondii infected cell monolayers [15]. These data not only illustrate the complexity of the cellular processes modulated in response to intracellular protozoa, but also highlight commonalities in the transcriptional response to different intracellular pathogens.
Microarray analyses of the transcriptome of C. parvum infected cells [13, 20, 21] have found genes associated with the inflammatory response, cytoskeleton function, cell proliferation and apoptosis to be differentially expressed in response to C. parvum infection. Here we describe changes in host cell gene expression in C. parvum infected cell monolayers using microarrays, and present evidence that one over-expressed membrane protein localizes to the cellular membrane overlaying the developing parasite. These observations demonstrate for the first time that the composition of the host cell membrane is affected by this infection.
2. Materials and Methods
2.1. Cell culture
Human ileocecal epithelial cells (HCT-8) (American Type Culture Collection #CCL-244) were cultured in RPMI 1640 (GIBCO/Invitrogen, Parsley, United Kingdom) supplemented with 10% heat-inactivated horse serum (GIBCO), 1% penicillin/streptomycin, 2 mM L-glutamine, and 1 mM sodium pyruvate (Sigma, St. Louis, MO). Cells were maintained in a 5% CO2 atmosphere at 37°C and 85% humidity. Monolayers were seeded in 25 or 75 cm2 flasks at 1×106 cells/25 cm2 or 3×106/75 cm2 and grown to 80% confluence.
2.2. Parasites
C. parvum oocysts were obtained from experimentally infected calves or immunosuppressed rodents. Oocysts were purified from stool as described [22]. Oocysts for inoculation of cell monolayers were surface-sterilized with 10% bleach for 7 min on ice and washed twice with phosphate buffered saline (PBS). Oocysts for mock infections were heat-inactivated at 80°C for 20 min.
2.3. Cell monolayer infection, immunofluorescent labeling and flow cytometry
HCT-8 cells were infected with C. parvum oocysts at a 1/1 of oocyst/host cell ratio and incubated for 24 h. Taurocholic acid at a concentration of 0.05% was added with the oocysts to promote infection [23]. After incubation, cell monolayers were recovered by treating with Accutase (Innovation Cell Technologies, Inc) and processed for immunofluorescence labelling with a Cryptosporidium-specific antiserum as described [24]. Labelled cells were re-suspended at a concentration of 106/ml in PBS and were analyzed by flow cytometry (FACSCalibur; BD Biosciences, San Jose, California) equipped with a 488 nm argon laser. Alexa 488 fluorescence emitted by the secondary anti-rabbit IgG antibody conjugate was acquired in the FL1 channel. FL1 and scatter signal amplifiers were set in log mode. Cell populations were displayed on scatter plots using CellQuest software (BD Biosciences).
2.4. RNA extraction for microarrays
RNA was extracted from about 3×106 cells using 1 ml TRI reagent (Sigma, St. Louis, Missouri). RNA was precipitated in isopropanol, and converted to double-stranded cRNA. cRNA was transcribed with T7 RNA polymerase in the presence of biotinilitated UTP analogue and purified to remove unincorporated nucleotides. Fragmented cRNA was added to hybridization cocktail and heated to 99°C. Oligonucleotide microarrays containing 14,500 human genes (Human Genome U133A 2.0; Affymetrix, Santa Clara, CA) were hybridized at 45°C for 16 h while being rotated at 60 rpm. Arrays were then washed, stained with streptavidin-phycoerythrin (Molecular Probes, Eugene, OR), and scanned.
2.5. Microarrays and data analysis
Background subtraction and normalization of Affymetrix U133A 2.0 microarray data were carried out using the Robust Multichip Average (RMA; [25]) module in BioConductor. A two-class paired Significance Analysis of Microarrays (SAM) module in TIGR Microarray Data Analysis System (MIDAS, version 2.19) [26], which utilizes a modified t-test statistic and sample-label permutations to evaluate statistical significance, was used to identify differentially regulated genes in C. parvum infected vs. mock infected monolayers. The false-discovery rate [27], an estimate of the fraction of falsely called infection-responsive genes, was set at 4.9%.
The list of differentially regulated genes identified in Affymetrix U133A 2.0 was further subdivided into functional categories using the bioinformatics analysis resource DAVID (Database for Annotation, Visualization and Integrated Discovery) of the National Institute of Allergy and Infectious Diseases [28, 29]. The Modified Fisher Exact test was used to measure the probability of observing a number of differentially regulated genes for a specific category given a category’s representation on the array.
2.6. Reverse-transcriptase PCR
Fourteen (of 147) significantly up- or down-regulated genes were randomly selected for reverse-transcriptase (RT) PCR analysis based on them belonging to different biological processes. Primers were designed to amplify fragments of 200-350 basepair from the selected transcripts (Table 1). A portion of 10 ng of the same RNA as used for array hybridization was reverse transcribed using M-MLV RT (Promega, Madison, Wisconsin). Control amplification reactions lacking reverse transcriptase were included to control for any DNA contamination. The cDNA product was added to 10 μl Fast Start DNA Master SYBR Green I master mix (Roche Applied Science, Indianapolis, Indiana) and amplified using a LightCycler® 2.0 thermal cycler. Triplicate samples of a ten-fold dilution series of each cDNA were amplified. Amplification conditions were as follows; initial denaturation at 95°C for 10 min; 40-45 cycles of 95°C for 1 sec, specific annealing temperature for 15 sec, and 1 sec/25 basepair extension at 72°C ; A melting cycle from 65°C to 95°C was applied to acquire melting curves. In all cases HCT-8 DNA was included as a positive PCR control and HCT-8 cDNA was used to draw standard curves.
Table 1.
PCR primers
| ATAD4a | FORb | 5′- GAG CAG TCG GGA TTA CAG -3′ |
| REV | 5′- CTT CTC CAG GCG GGT GTT A -3′ | |
| BTG2 | FOR | 5′- CAT TCG CAT CAA CCA CAA -3′ |
| REV | 5′- CTG GAG ACT GCC ATC A -3′ | |
| CCL5 | FOR | 5′- CCT CGG ACA CCA CAC CCT -3′ |
| REV | 5′- GGA CAA GAG CAA GCA GAA AC -3′ | |
| EGFR | FOR | 5′- CCA AGG CAC GAG TAA CAA G -3′ |
| REV | 5′- GAC TGC TAA GGC ATA GGA A -3′ | |
| EMP1 | FOR | 5′- GTA TCA CCA CGG CTA TTC -3′ |
| REV | 5′- GGT TTG GGA TTT GAC CTC -3′ | |
| F2RL1 | FOR | 5′- ATG GCA ACA ACT GGA TTT AT -3′ |
| REV | 5′- TCA CGA CAT ACA AAG GG -3′ | |
| FGF18 | FOR | 5′- CTG CTG CTG TGC TTC CA -3′ |
| REV | 5′- GGC ATA CTT GTC CCC AT -3′ | |
| FOS | FOR | 5′- CTT CCT TCG TCT TCA CCT A -3′ |
| REV | 5′- ATG CGT TTT GCT ACA TCT CC -3′ | |
| GAPDH | FOR | 5′- AAT GAC CCC TTC ATT GAC C -3′ |
| REV | 5′- TGG TGC AGG AGG CAT TGC -3′ | |
| KLK10 | FOR | 5′- CCA TAA AGT CAT ACG CTC C -3′ |
| REV | 5′- GCC CAA AGT CAC ACA G -3′ | |
| KRT4 | FOR | 5′- GCA GTT CTT AGA GCA ACA -3′ |
| REV | 5′- CTC CGC ATC ATA GAG G -3′ | |
| MALL | FOR | 5′- ATG AAC TTG CTC TTC TAG -3′ |
| REV | 5′- GCT AAT ACG GTG AAA CC -3′ | |
| PHLDA1 | FOR | 5′- GTT TGG ACA TCA CCC TAC T -3′ |
| REV | 5′- GCT GGC ACA ACA ATG AAA G -3′ | |
| PLAUR | FOR | 5′- GTG TAA GAC CAA CGG GGA TTG -3′ |
| REV | 5′- GCT TCG GGA ATA GGT GAC AG -3′ | |
| SDHA | FOR | 5′- CAT CCA CTA CAT GAC GGA GCA -3′ |
| REV | 5′- ATC TTC CCA TCT TCA GTT CTG CTA -3′ | |
| TNFRSF12A | FOR | 5′- GGA CCT GGA CAA GTG C -3′ |
| REV | 5′- GAA TGG ATG AAT GAA TGA TGA GT -3′ |
For gene titles see Table 2.
FOR, forward primer; REV, reverse primer
2.7. Immunofluorescence of Protease Activated Receptor-2
HCT-8 cells were infected with C. parvum oocysts (ratio 1/1) and processed 24 h post-infection for immunofluorescent labeling as described above for C. parvum labelling with the following modifications: methanol fixed cells were incubated with a 1:200 dilution of Cryptosporidium specific 2E5 mouse monoclonal primary antibody and a 1:200 dilution of anti-Protease Activated Receptor-2 (PAR2) rabbit antibody (abcam, Inc.). This antibody recognizes a peptide located in the PAR2 cytoplasmic carbox-terminal region. The sequence of the peptide is not disclosed by the company. Subsequently, the cells were incubated with a 1:1000 dilution of secondary Alexa Fluor 488 labeled rabbit anti-mouse IgG antibody pre-adsorbed against human IgG and a 1:1000 dilution of Alexa Fluor 568 conjugated goat anti-rabbit IgG antibody pre-adsorbed against goat, mouse, and human IgG. Labelled cells were examined by fluorescence microscopy. Uninfected cells labeled with anti-PAR2 or anti-Cryptosporidium antibody were used to assess background fluorescence. The lack of cross-reactivity of the anti-PAR2 antibody with parasite antigen was investigated by Western blotting. Briefly, lysates of 106 oocysts or of 3 × 105 infected, mock-infected, and uninfected HCT-8 cells were resolved by SDS-PAGE under reducing conditions on a 4-12% PAGE Bis-Tris gel and protein electrophoretically transferred to nitrocellulose. The blot was incubated with the anti-PAR2 antibody or control rabbit serum as negative control. Bound antibody was visualized with a horseradish peroxidase secondary anti-rabbit antibody and a luminol based enhanced chemiluminescence peroxidase substrate.
To examine the time course PAR2 expression, HCT-8 cells were infected with C. parvum oocysts (oocysts/host cell ratio 1/1) and processed 2, 6, 10, 15, 20, 24, 30, 35, 40, and 48 h post-infection for immunofluorescence labelling with C. parvum anti-2E5 antibody or anti-PAR2 antibody as described above. Labelled cells were examined by microscopy and flow cytometry. CellQuest Pro Software was used to analyze the flow cytometry data.
3. Results
3.1. Analysis of global gene expression
In triplicate experiments with Affymetrix HG-U133A 2.0 arrays, and based on a false-discovery rate of 4.95%, a total of 147 genes were found to be differentially regulated 24 h after infection with C. parvum, including 73 up-regulated and 74 down-regulated genes. Fig. 1 shows the relative expression of 14 randomly selected genes. At 24 h post-infection the following gene ontology (GO) biological process terms were found to be over-represented among up-regulated genes (p-value ≤ 5.2E-4, EASE Score): Cell Communication, Signal Transduction, Organ Development, and Response to External Stimulus. Biological process terms found to be over-represented among down-regulated genes (p-value ≤ 8.6E-3, EASE Score) include Morphogenesis of an Epithelium and Amino Acid Metabolic Processes.
Fig. 1. Gene expression assessed by microarrays and quantitative RT PCR.
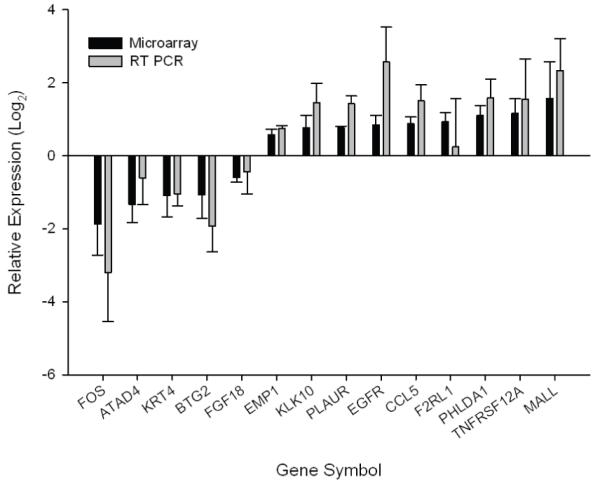
Fourteen mRNA species were RT PCR amplified using the primers shown in Table 1. For each sample, expression values were normalized against SDHA mRNA and relative expression values obtained by comparing RNA extracted from corresponding infected and uninfected monolayer. RT PCR (grey bars) was compared with microarray data (black bar) (n = 3 experiments). For gene symbols see Table 2.
3.2. Confirmatory RT PCR analysis
Portions of the same RNA samples analyzed in triplicate Affymetrix microarrays were used as substrate for RT PCR to compare the expression of the 14 selected genes (Table 2) in infected and mock infected monolayers. These genes were randomly selected among the 30 most up-regulated and 17 most down-regulated genes. The concentration of each mRNA transcript was estimated using a standard curve and normalized against the concentration of GAPDH (not shown) or succinate dehydrogenase complex subunit A (SDHA) mRNA. The RT PCR results were in good agreement with the microarray data (Fig. 1). Normalization against GAPDH or SDHA did not significantly change the results.
Table 2.
Gene symbol and title of 14 genes selected for RT PCR analysis
| Gene Symbol | Gene Title | fold change (log2 ) | GenBank accession |
|---|---|---|---|
| ATAD4 | ATPase family, AAA domain containing 4 | -1.32 | NM_024320 |
| BTG2 | BTG family, member 2 | -1.21 | NM_006763 |
| CCL5 | chemokine (C-C motif) ligand 5 | 0.87 | NM_002985 |
| EGFR | epidermal growth factor receptor | 0.84 | NM_005228 |
| EMP1 | epithelial membrane protein 1 | 0.58 | NM_001423 |
| F2RL1 | coagulation factor II (thrombin) receptor-like 1 | 0.94 | NM_005242 |
| FGF18 | fibroblast growth factor 18 | -0.60 | NM_003862 |
| FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | -1.87 | NM_005252 |
| KLK10a | kallikrein-related peptidase 10 | 0.76 | NM_002776 |
| KRT4 | keratin 4 | -1.09 | NM_002272 |
| MALLa | mal, T-cell differentiation protein-like | 1.56 | NM_005434 |
| PHLDA1 | pleckstrin homology-like domain, family A, member 1 | 1.42 | NM_007350 |
| PLAUR | plasminogen activator, urokinase receptor | 0.77 | NM_002659 |
| TNFRSF12A | tumor necrosis factor receptor superfamily, member 12A | 1.16 | NM_016639 |
These two genes are not among the 147 most differentially regulated genes. They were included in the RT PCR validation based on a preliminary set of microarray data.
3.3. Immunofluorescence analysis of F2RL1 gene products
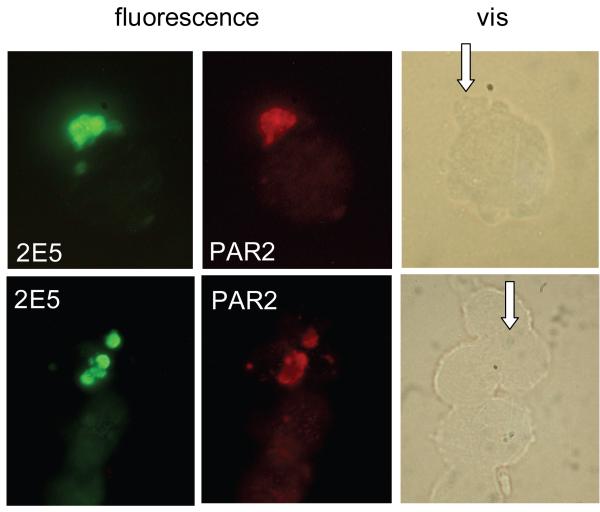
Based on the microarray and RT PCR quantification described above, immunofluorescence was used to examine the expression of PAR2, the protein encoded by gene F2RL1. This choice was motivated by the fact that PAR2 may play an important role in the innate immune response, as further discussed below. From a practical point of view, this analysis was facilitated by the availability of antibody specific for human PAR2. Infected HCT-8 cells were processed for immunofluorescence 24 h post-infection, and reacted with fluorescent anti-PAR2 antibody and anti-C. parvum antibody as described in Materials and Methods. Labelling with secondary anti-rabbit Alexa 568 conjugate antibody and anti-mouse Alexa 488 conjugate revealed that PAR2 fluorescence was localized to the cell membrane which surrounds the intracellular stages of the parasite (Fig. 2). Interestingly, PAR2 fluorescence was strongly concentrated within this well-defined region of host cell membrane. The absence of cross-reaction between anti-PAR2 rabbit antibody and secondary anti-mouse IgG antibody, or between the anti-Cryptosporidium monoclonal mouse antibody and secondary anti-rabbit IgG antibody was confirmed in control monolayers (not shown). Uninfected cells were used as the negative control and labeled with 2E5 or PAR2 antibodies, together with their fluorescent secondary antibody. No fluorescence from antibodies against 2E5 or PAR2 was observed on the uninfected cells (not shown). Infected cells labelled separately with each antibody combination were also examined to confirm that the green and red immunofluorescent signals were specific and that no bleed-over from red to green and vice versa took place. Moreover, a Western blot of lysed C. parvum oocysts, and of infected, uninfected and mock-infected HCT-8 cells were probed with anti-PAR2 antibody. The three cell lysates displayed an identical profile indicating that the antibody does not recognize any antigen produced by intracelluar parasite stages.
Fig. 2. Immunofluorescence analysis of PAR2 expression on infected cells.
Cells recovered from a HCT-8 monolayer 24 h post-infection were double-labelled for C. parvum (2E5 monoclonal antidoby) and PAR2 as indicated. In unlabelled cells the parasites are difficult to discern and their position are indicated with arrows.
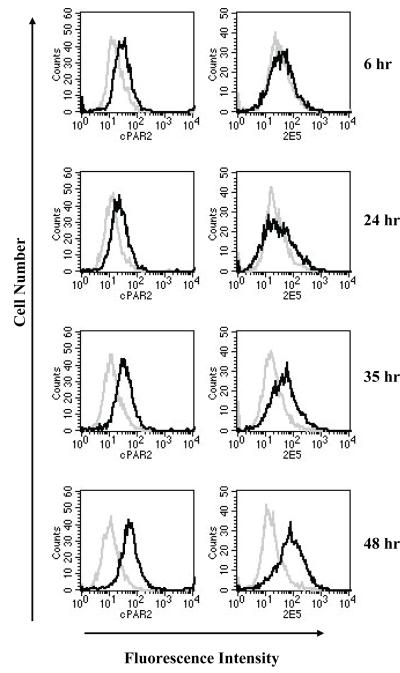
The change in PAR2 fluorescent signal in C. parvum infected cell monolayers was investigated over a 48-h period by flow cytometry using infected and uninfected cells labelled separately with anti-PAR2 or anti-Cryptosporidium antibody. Each cell population was then reacted separately with Alexa 488 conjugated anti-rabbit and anti-mouse IgG, respectively, and green fluorescence emitted by each cell population displayed on histograms (Fig. 3). The parasite immunofluorescence signal increased during the 48-h period, which is consistent with previous reports indicating parasite proliferation in the initial 48-72-h [11, 30]. At 48 h post-infection, the magnitude of parasite immunofluorescence as compared to that emitted by uninfected cells indicates that a majority of host cells was infected. PAR2 fluorescence also increased during this period, and the PAR2 signal paralleled parasite fluorescence, reaching a maximum 48 h post-infection. These observations are consistent with the microarray and microscopic observations in showing PAR2 over-expression in response to the infection.
Fig. 3. Time course of PAR2 and C. parvum immunofluorescence.
Cells from an infected monolayer were fixed at different time points post-infection and two portions of the cell suspensions immunolabelled for C. parvum antigen or PAR2, respectively. C. parvum and PAR2 immunofluorescence was acquired by flow cytometry in the FL1 channel. Uninfected cells labeled with C. parvum or PAR2 specific antibody were used as negative control. Histogram overlay shows fluorescence of infected (black) and uninfected cells (grey).
4. Discussion
Cell monolayers infected with Cryptosporidium parasites are complex systems in which two eukaryotic species interact. The system is difficult to standardize, as Cryptosporidium oocysts cannot be produced in vitro, but are purified from the feces of experimentally infected animals. Even when conventional laboratory rodent models, such as inbred, immuno-suppressed [31] or -deficient mice [32] are used to propagate C. parvum, the oocysts recovered from the feces are a heterogeneous population which can greatly vary in infectivity [33]. C. parvum isolates are also genetically heterogeneous and readily generate new recombinants with different infectivity phenotypes [34]. Oocyst age has also been shown to affect infectivity [30]. As far as the host cells are concerned, the age of the monolayer can modulate the infection [35]. These variables may have negatively affected the correlation between replicate microarray experiments.
The comparison of infected and control monolayers using microarrays and RT PCR detects host transcripts which are up-regulated in infected cells, as well as transcripts which are up-regulated as a result of changes in the monolayer induced by the infection. Widespread apoptosis, extrusion of infected cells and induction of mitosis to restore monolayer integrity have been documented [9, 36-38]. Consistent with these changes, two of the three published microarray analyses have found that genes involved in cell proliferation and apoptosis are particularly abundant among up-regulated genes in C. parvum infected monolayers [20, 21]. Liu et al. [21] reported that pro-apoptotic genes were induced and anti-apoptotic genes were down-regulated at 24 h post-infection. In contrast, we observe an over-representation of apoptosis-related genes among up- and down-regulated genes at the same time point, which is similar to the observations of Deng et al. [20]. Castellanos-Gonzalez et al. reported the over-expression of five genes in C. parvum or C. hominis infected cell monolayers [13]. Among these genes they found the osteoprotegerin gene, which in our data shows only a modest level of up-regulation (0.31 log2) and does therefore not feature in the supplemental table. Among the four other up-regulated genes found by Castellanos-Gonzalez and co-workers, only CD24 is found among the up-regulated genes in our study (Supplemental Table). There are thus significant differences among sets of microarray data from C. parvum infected cells, which is not surprising given the complexity of the experimental system and the lack of standardized conditions. More studies performed under similar conditions are needed to discriminate between genes which are modulated as a result of the infection and experimental noise.
Since in C. parvum infected monolayers many cells remain uninfected, we were interested in investigating whether up-regulation of F2RL1 transcription was specific to infected cells, and whether this process resulted in increased PAR2 expression. PAR2 was of particular interest in this context as it is widely distributed in the gastrointestinal tract and is involved in regulating mucus production [39-41], a process which could be relevant to the innate host defense against intestinal pathogens. The PAR2 immunofluorescence analysis confirms a direct link between infection and up-regulation and is significant in showing that the host cell membrane is modified during infection. PAR2, a G-protein-coupled receptor, is activated by proteolytic cleavage of the N-terminal domain. PAR2 is expressed in many tissues, including the intestinal epithelium [42-44]. Its involvement in the regulation of salivary gland secretion, pancreatic juice secretion and small ion transport [39, 45, 46] make it potentially relevant for understanding the host’s innate defense against intestinal pathogens. Expression of PAR2 has also been reported to increase cytosolic Ca2+ concentration [43, 46-50] but preliminary observations in our system using Calcium Green-1, AM (Molecular Probes, Eugene, Oregon) failed to demonstrate changes in intracellular Ca2+ (data not shown). The observation that agonists of PAR2 stimulate the secretion of pro-inflammatory cytokines [51, 52], induce NFκB pathway activation, IL-8 production, and activate p-38, MAPK and ERK1/2 signaling [53, 54] may be relevant to understanding the mechanisms involved in C. parvum induced release of cytokines and chemokines from infected cells [55, 56]. Whether these changes are regulated by PAR2 signaling remains to be investigated. Infections with Helicobacter pylori and Pseudomonas aeruginosa pneumonia can also activate PAR2 signaling [57, 58]. However, the functional significance of PAR2 activation in response to Cryptosporidium infection, or how many proteins respond in this manner, is unknown.
The over-expression and localization of host membrane proteins in C. parvum infected cells may have practical implications as a better understanding of the physiology of the cell membrane may shed light on the lack of anti-cryptosporidial activity of many compounds which are effective against other intracellular pathogens [59]. If resistance to such compounds is caused by the inability of certain molecules to reach the trophozoite and meront stages of the parasite, understanding the membrane composition may facilitate the targeted modification of existing anti-protozoal compounds into a form which may better penetrate the cell membrane.
Supplementary Material
Acknowledgments
This study was supported by grant R01AI055347 from the National Institutes of Allergy and Infectious Diseases. Y-L. Y. was supported by an Agnes Varis Graduate Fellowship. Our thanks to Yue Shao, Chris Parkin and Lax Iyer from the Tufts Center for Neuroscience Research core facility, Linghui Zhang and Chuck Shoemaker from TCSVM, and John Leong and Brian Akerley from the University of Massachusetts Medical Center for feedback and suggestions.
List of abbreviations
- RT
reverse-transcriptase
- PAR2
Protease Activated Receptor-2
- PAGE
polyacrylamide gel electrophoresis
- SDS
sodium dodecyl sulfate
- PBS
phosphate buffered saline
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Griffiths JK. Human cryptosporidiosis: epidemiology, transmission, clinical disease, treatment, and diagnosis. Adv Parasitol. 1998;40:37–85. doi: 10.1016/s0065-308x(08)60117-7. [DOI] [PubMed] [Google Scholar]
- [2].Colford JM, Jr., Tager IB, Hirozawa AM, et al. Cryptosporidiosis among patients infected with human immunodeficiency virus. Factors related to symptomatic infection and survival. Am J Epidemiol. 1996;144:807–816. doi: 10.1093/oxfordjournals.aje.a009015. [DOI] [PubMed] [Google Scholar]
- [3].Tzipori S. Cryptosporidiosis in perspective. Adv Parasitol. 1988;27:63–129. doi: 10.1016/S0065-308X(08)60353-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Huang BQ, Chen XM, LaRusso NF. Cryptosporidium parvum attachment to and internalization by human biliary epithelia in vitro: a morphologic study. J Parasitol. 2004;90:212–221. doi: 10.1645/GE-3204. [DOI] [PubMed] [Google Scholar]
- [5].Aji T, Flanigan T, Marshall R, et al. Ultrastructural study of asexual development of Cryptosporidium parvum in a human intestinal cell line. J Protozool. 1991;38:82S–84S. [PubMed] [Google Scholar]
- [6].Lefkowitch JH, Krumholz S, Feng-Chen KC, et al. Cryptosporidiosis of the human small intestine: a light and electron microscopic study. Hum Pathol. 1984;15:746–752. doi: 10.1016/s0046-8177(84)80165-3. [DOI] [PubMed] [Google Scholar]
- [7].Elliott DA, Clark DP. Cryptosporidium parvum induces host cell actin accumulation at the host-parasite interface. Infect Immun. 2000;68:2315–2322. doi: 10.1128/iai.68.4.2315-2322.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].O’Hara SP, Small AJ, Chen XM, et al. Host cell actin remodeling in response to Cryptosporidium. Subcell Biochem. 2008;47:92–100. doi: 10.1007/978-0-387-78267-6_7. [DOI] [PubMed] [Google Scholar]
- [9].Chen XM, Levine SA, Tietz P, et al. Cryptosporidium parvum is cytopathic for cultured human biliary epithelia via an apoptotic mechanism. Hepatology. 1998;28:906–913. doi: 10.1002/hep.510280402. [DOI] [PubMed] [Google Scholar]
- [10].Ojcius DM, Perfettini JL, Bonnin A, et al. Caspase-dependent apoptosis during infection with Cryptosporidium parvum. Microbes Infect. 1999;1:1163–1168. doi: 10.1016/s1286-4579(99)00246-4. [DOI] [PubMed] [Google Scholar]
- [11].Widmer G, Corey EA, Stein B, et al. Host cell apoptosis impairs Cryptosporidium parvum development in vitro. J Parasitol. 2000;86:922–928. doi: 10.1645/0022-3395(2000)086[0922:HCAICP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- [12].McCole DF, Eckmann L, Laurent F, et al. Intestinal epithelial cell apoptosis following Cryptosporidium parvum infection. Infect Immun. 2000;68:1710–1713. doi: 10.1128/iai.68.3.1710-1713.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Castellanos-Gonzalez A, Yancey LS, Wang HC, et al. Cryptosporidium infection of human intestinal epithelial cells increases expression of osteoprotegerin: a novel mechanism for evasion of host defenses. J Infect Dis. 2008;197:916–923. doi: 10.1086/528374. [DOI] [PubMed] [Google Scholar]
- [14].Chen XM, Gores GJ, Paya CV, et al. Cryptosporidium parvum induces apoptosis in biliary epithelia by a Fas/Fas ligand-dependent mechanism. Am J Physiol. 1999;277:G599–608. doi: 10.1152/ajpgi.1999.277.3.G599. [DOI] [PubMed] [Google Scholar]
- [15].Blader IJ, Manger ID, Boothroyd JC. Microarray analysis reveals previously unknown changes in Toxoplasma gondii-infected human cells. J Biol Chem. 2001;276:24223–24231. doi: 10.1074/jbc.M100951200. [DOI] [PubMed] [Google Scholar]
- [16].Gail M, Gross U, Bohne W. Transcriptional profile of Toxoplasma gondii-infected human fibroblasts as revealed by gene-array hybridization. Mol Genet Genomics. 2001;265:905–912. doi: 10.1007/s004380100487. [DOI] [PubMed] [Google Scholar]
- [17].Min W, Lillehoj HS, Kim S, et al. Profiling local gene expression changes associated with Eimeria maxima and Eimeria acervulina using cDNA microarray. Appl Microbiol Biotechnol. 2003;62:392–399. doi: 10.1007/s00253-003-1303-x. [DOI] [PubMed] [Google Scholar]
- [18].Imai K, Mimori T, Kawai M, et al. Microarray analysis of host gene-expression during intracellular nests formation of Trypanosoma cruzi amastigotes. Microbiol Immunol. 2005;49:623–631. doi: 10.1111/j.1348-0421.2005.tb03654.x. [DOI] [PubMed] [Google Scholar]
- [19].Lovegrove FE, Gharib SA, Patel SN, et al. Expression microarray analysis implicates apoptosis and interferon-responsive mechanisms in susceptibility to experimental cerebral malaria. Am J Pathol. 2007;171:1894–1903. doi: 10.2353/ajpath.2007.070630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Deng M, Lancto CA, Abrahamsen MS. Cryptosporidium parvum regulation of human epithelial cell gene expression. Int J Parasitol. 2004;34:73–82. doi: 10.1016/j.ijpara.2003.10.001. [DOI] [PubMed] [Google Scholar]
- [21].Liu J, Deng M, Lancto CA, et al. Biphasic modulation of apoptotic pathways in Cryptosporidium parvum-infected human intestinal epithelial cells. Infect Immun. 2009;77:837–849. doi: 10.1128/IAI.00955-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Widmer G, Tzipori S, Fichtenbaum CJ, et al. Genotypic and phenotypic characterization of Cryptosporidium parvum isolates from people with AIDS. J Infect Dis. 1998;178:834–840. doi: 10.1086/515373. [DOI] [PubMed] [Google Scholar]
- [23].Feng H, Nie W, Sheoran A, et al. Bile acids enhance invasiveness of Cryptosporidium spp. into cultured cells. Infect Immun. 2006;74:3342–3346. doi: 10.1128/IAI.00169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Theodos CM, Griffiths JK, D’Onfro J, et al. Efficacy of nitazoxanide against Cryptosporidium parvum in cell culture and in animal models. Antimicrob Agents Chemother. 1998;42:1959–1965. doi: 10.1128/aac.42.8.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Irizarry RA, Bolstad BM, Collin F, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Saeed AI, Sharov V, White J, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- [27].Benjamini Y, Drai D, Elmer G, et al. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- [28].Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- [29].Dennis G, Jr., Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- [30].Rochelle PA, Ferguson DM, Johnson AM, et al. Quantitation of Cryptosporidium parvum infection in cell culture using a colorimetric in situ hybridization assay. J Eukaryot Microbiol. 2001;48:565–574. doi: 10.1111/j.1550-7408.2001.tb00192.x. [DOI] [PubMed] [Google Scholar]
- [31].Yang S, Healey MC, Du C. Infectivity of preserved Cryptosporidium parvum oocysts for immunosuppressed adult mice. FEMS Immunol Med Microbiol. 1996;13:141–145. doi: 10.1016/0928-8244(95)00096-8. [DOI] [PubMed] [Google Scholar]
- [32].Tzipori S. Cryptosporidiosis: laboratory investigations and chemotherapy. Adv Parasitol. 1998;40:187–221. doi: 10.1016/s0065-308x(08)60121-9. [DOI] [PubMed] [Google Scholar]
- [33].Bukhari Z, Smith HV. Cryptosporidium parvum: oocyst excretion and viability patterns in experimentally infected lambs. Epidemiol Infect. 1997;119:105–108. doi: 10.1017/s0950268897007590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Feng X, Rich SM, Tzipori S, et al. Experimental evidence for genetic recombination in the opportunistic pathogen Cryptosporidium parvum. Mol Biochem Parasitol. 2002;119:55–62. doi: 10.1016/s0166-6851(01)00393-0. [DOI] [PubMed] [Google Scholar]
- [35].Sifuentes LY, Di Giovanni GD. Aged HCT-8 cell monolayers support Cryptosporidium parvum infection. Appl Environ Microbiol. 2007;73:7548–7551. doi: 10.1128/AEM.01579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Griffiths JK, Moore R, Dooley S, et al. Cryptosporidium parvum infection of Caco-2 cell monolayers induces an apical monolayer defect, selectively increases transmonolayer permeability, and causes epithelial cell death. Infect Immun. 1994;62:4506–4514. doi: 10.1128/iai.62.10.4506-4514.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Widmer G, Yang YL, Bonilla R, et al. Preferential infection of dividing cells by Cryptosporidium parvum. Parasitology. 2006;133:131–138. doi: 10.1017/S0031182006000151. [DOI] [PubMed] [Google Scholar]
- [38].Savidge TC, Shmakov AN, Walker-Smith JA, et al. Epithelial cell proliferation in childhood enteropathies. Gut. 1996;39:185–193. doi: 10.1136/gut.39.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kawabata A, Kinoshita M, Nishikawa H, et al. The protease-activated receptor-2 agonist induces gastric mucus secretion and mucosal cytoprotection. J Clin Invest. 2001;107:1443–1450. doi: 10.1172/JCI10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lin KW, Park J, Crews AL, et al. Protease-activated receptor-2 (PAR-2) is a weak enhancer of mucin secretion by human bronchial epithelial cells in vitro. Int J Biochem Cell Biol. 2008;40:1379–1388. doi: 10.1016/j.biocel.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Nishikawa H, Kawai K, Nishimura S, et al. Suppression by protease-activated receptor-2 activation of gastric acid secretion in rats. Eur J Pharmacol. 2002;447:87–90. doi: 10.1016/s0014-2999(02)01892-7. [DOI] [PubMed] [Google Scholar]
- [42].Nystedt S, Emilsson K, Wahlestedt C, et al. Molecular cloning of a potential proteinase activated receptor. Proc Natl Acad Sci U S A. 1994;91:9208–9212. doi: 10.1073/pnas.91.20.9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bohm SK, Kong W, Bromme D, et al. Molecular cloning, expression and potential functions of the human proteinase-activated receptor-2. Biochem J. 1996;314( Pt 3):1009–1016. doi: 10.1042/bj3141009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].D’Andrea MR, Derian CK, Leturcq D, et al. Characterization of protease-activated receptor-2 immunoreactivity in normal human tissues. J Histochem Cytochem. 1998;46:157–164. doi: 10.1177/002215549804600204. [DOI] [PubMed] [Google Scholar]
- [45].Kawabata A. PAR-2: structure, function and relevance to human diseases of the gastric mucosa. Expert Rev Mol Med. 2002;4:1–17. doi: 10.1017/S1462399402004799. [DOI] [PubMed] [Google Scholar]
- [46].Nguyen TD, Moody MW, Steinhoff M, et al. Trypsin activates pancreatic duct epithelial cell ion channels through proteinase-activated receptor-2. J Clin Invest. 1999;103:261–269. doi: 10.1172/JCI2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kim MH, Choi BH, Jung SR, et al. Protease-activated receptor-2 increases exocytosis via multiple signal transduction pathways in pancreatic duct epithelial cells. J Biol Chem. 2008;283:18711–18720. doi: 10.1074/jbc.M801655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Sato S, Ito Y, Kondo M, et al. Ion transport regulated by protease-activated receptor 2 in human airway Calu-3 epithelia. Br J Pharmacol. 2005;146:397–407. doi: 10.1038/sj.bjp.0706330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Sun G, Stacey MA, Schmidt M, et al. Interaction of mite allergens Der p3 and Der p9 with protease-activated receptor-2 expressed by lung epithelial cells. J Immunol. 2001;167:1014–1021. doi: 10.4049/jimmunol.167.2.1014. [DOI] [PubMed] [Google Scholar]
- [50].Mirza H, Yatsula V, Bahou WF. The proteinase activated receptor-2 (PAR-2) mediates mitogenic responses in human vascular endothelial cells. J Clin Invest. 1996;97:1705–1714. doi: 10.1172/JCI118597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cenac N, Garcia-Villar R, Ferrier L, et al. Proteinase-activated receptor-2-induced colonic inflammation in mice: possible involvement of afferent neurons, nitric oxide, and paracellular permeability. J Immunol. 2003;170:4296–4300. doi: 10.4049/jimmunol.170.8.4296. [DOI] [PubMed] [Google Scholar]
- [52].Uehara A, Muramoto K, Takada H, et al. Neutrophil serine proteinases activate human nonepithelial cells to produce inflammatory cytokines through protease-activated receptor 2. J Immunol. 2003;170:5690–5696. doi: 10.4049/jimmunol.170.11.5690. [DOI] [PubMed] [Google Scholar]
- [53].Macfarlane SR, Sloss CM, Cameron P, et al. The role of intracellular Ca2+ in the regulation of proteinase-activated receptor-2 mediated nuclear factor kappa B signalling in keratinocytes. Br J Pharmacol. 2005;145:535–544. doi: 10.1038/sj.bjp.0706204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Yoshida N, Katada K, Handa O, et al. Interleukin-8 production via protease-activated receptor 2 in human esophageal epithelial cells. Int J Mol Med. 2007;19:335–340. doi: 10.3892/ijmm.19.2.335. [DOI] [PubMed] [Google Scholar]
- [55].Pantenburg B, Dann SM, Wang HC, et al. Intestinal immune response to human Cryptosporidium sp. infection. Infect Immun. 2008;76:23–29. doi: 10.1128/IAI.00960-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Lean IS, McDonald V, Pollok RC. The role of cytokines in the pathogenesis of Cryptosporidium infection. Curr Opin Infect Dis. 2002;15:229–234. [Google Scholar]
- [57].Kajikawa H, Yoshida N, Katada K, et al. Helicobacter pylori activates gastric epithelial cells to produce interleukin-8 via protease-activated receptor 2. Digestion. 2007;76:248–255. doi: 10.1159/000113041. [DOI] [PubMed] [Google Scholar]
- [58].Moraes TJ, Martin R, Plumb JD, et al. Role of PAR2 in murine pulmonary pseudomonal infection. Am J Physiol Lung Cell Mol Physiol. 2008;294:L368–377. doi: 10.1152/ajplung.00036.2007. [DOI] [PubMed] [Google Scholar]
- [59].Woods KM, Nesterenko MV, Upton SJ. Efficacy of 101 antimicrobials and other agents on the development of Cryptosporidium parvum in vitro. Ann Trop Med Parasitol. 1996;90:603–615. doi: 10.1080/00034983.1996.11813090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.