Abstract
Gene conversion -- the substitution of genetic material from another gene -- is recognized as the underlying cause of a growing number of genetic diseases. While in most cases conversion takes place between a normal gene and its pseudogene, here we report an occurrence of disease-associated gene conversion between two functional genes. Chronic pancreatitis in childhood is frequently associated with mutations of the cationic trypsinogen gene (serine protease 1; PRSS1). We have analyzed PRSS1 in 1106 patients with chronic pancreatitis, and identified a novel conversion event affecting exon 2 and the subsequent intron. The recombination replaced at least 289 nucleotides with the paralogous sequence from the anionic trypsinogen gene (serine protease 2; PRSS2), and resulted in the PRSS1 mutations c.86A>T and c.161A>G, causing the amino acid substitutions N29I and N54S, respectively. Analysis of the recombinant N29I-N54S double mutant cationic trypsinogen revealed increased autocatalytic activation, which was solely due to the N29I mutation. In conclusion, we have demonstrated that gene conversion between two functional paralogous trypsinogen genes can occur and cause genetically determined chronic pancreatitis.
Keywords: chronic pancreatitis, conversion mutation, anionic trypsinogen, PRSS2, cationic trypsinogen, PRSS1, SPINK1
INTRODUCTION
The term “gene conversion” describes a non-reciprocal exchange of genetic information between homologous DNA sequences. The donor gene(s) remain unmodified while an acceptor gene acquires the DNA sequence from the first gene within the recombined segment (Flajnik, 2002). Gene conversion events are essential for the evolution of gene families and have been implicated in a growing number of genetic diseases which include congenital adrenal hyperplasia (OMIM #201910), Gaucher disease (OMIM #230800), von Willebrand disease (OMIM #193400), polycystic kidney disease (OMIM #601313) and Shwachman-Diamond syndrome (OMIM #260400). In the majority of cases, the donor gene is a duplicated pseudogene, which accumulated mutations over time and recombination of the mutated sequences with the normal gene results in the pathogenic genotype.
Mutations of the PRSS1 gene (protease, serine, 1; OMIM #276000) encoding human cationic trypsinogen are associated with chronic pancreatitis. The R122H and N29I mutations are frequently found in hereditary pancreatitis (OMIM #167800) families, whereas other mutations associated with sporadic chronic pancreatitis cases are rare but diverse (Whitcomb et al., 1996; Gorry et al., 1997; Witt et al., 1999; Teich, 2001). Trypsinogen genes are tandemly repeated within the T-cell-receptor beta (TCR-beta) locus (Rowen et al., 1996). This is a hot spot for gene conversion events to generate a broad variety of TCR-beta genes (Flajnik, 2002). Therefore, conversion mutations within the interpolated trypsinogen gene family are very likely. This mechanism, which was first suggested for the N29I mutation (Chen and Férec, 2000b), is evident in case of the R122H mutation associated with the c.365 366 GC>AT di-nucleotide substitution, but remains speculative for the single-nucleotide substitutions causing mutations R122H (c.365G>A), N29I (c.86A>T) and A16V (c.47C>T) (Chen and Férec, 2000a; Chen et al., 2000). In this report, we prove this concept by describing the clinical, genetic and biochemical properties of a large gene conversion event between the anionic trypsinogen gene (PRSS2, donor gene) and the cationic trypsinogen gene (PRSS1, acceptor gene), which resulted in a novel mutant cationic trypsinogen protein and chronic pancreatitis.
METHODS AND MATERIALS
Case report
We report on a girl who presented at 6 years of age with chronic abdominal pain. Initially, a pancreatic pseudocyst was diagnosed. The cyst was drained operatively by retrocolic cystojejunostomy. Extensive anamnestic and metabolic work-up for drugs, trauma, parathormone, serum triglycerides, pilocarpine iontophoresis as well as detailed viral serology did not reveal any abnormality. The patient has no relatives with chronic pancreatitis or pancreatic cancer. Within two years of presentation, the patient had two severe episodes of acute pancreatitis. Endoscopic retrograde pancreaticography at 8 years of age demonstrated an irregular pancreatic duct, which led into another large pseudocyst in the pancreatic tail. The pancreatic tail was resected. At 11 years of age pancreaticolithiasis and portal vein thrombosis were diagnosed. The latter caused splenomegaly and mild thrombocytopenia. At present, the 16 year-old patient is well with pancreatic enzyme supplementation. She complains of occasional abdominal pain, but has not required hospitalisation during the last years. Glucose tolerance is not impaired. So far, the clinical course of chronic pancreatitis in the index patient was severe, but self-limiting. Her German mother is well, whereas her father is said to be Italian, but remains unknown.
Genotyping of the cationic trypsinogen gene
Local ethics committee approval and informed consent of the patient and her mother was obtained before analysis. Leukocyte DNA was extracted from anti-coagulated blood. The coding sequences of PRSS1 (RefSeq: NM_002769.2) and PRSS2 (protease, serine 2, OMIM #601564; RefSeq NM_002770.2) were amplified and subsequently sequenced as described previously (Teich et al., 2002). The N34S mutation of the SPINK1 gene (serine protease inhibitor, Kazal type 1, OMIM #167790), which is frequently associated with sporadic chronic pancreatitis, has been investigated as described recently (Witt et al., 2000; Teich et al., 2002). For PCR amplification of exon 2 of the PRSS1 gene, we have used reverse primers (PCR primers B and D), which bind to both the PRSS1 and PRSS2 sequences, and completely PRSS1-specific forward primers (PCR primers A and C), ensuring that only PRSS1 is amplified (Table 1, Fig 2; Supplementary Fig S1). Primer pair A–B was used for routine amplifications, whereas primer pair C–D was used to evaluate the 3’ border of the gene conversion. PCR products were sequenced using sequencing primers I and II, which can anneal to both PRSS1 and PRSS2 sequences (Table 1, Fig 2, Supplementary Fig S1). PCR and sequencing reactions were carried out at 57° and 62°C annealing temperature, respectively.
Table 1.
PCR and sequencing primers. Numbering based on Rowen et al. (1996).
| orientation | Sequence | PRSS1 nt number | PRSS1 homology | PRSS2 nt number | PRSS2 homology | |
|---|---|---|---|---|---|---|
| PCR primerA | forward | 5′-ccc aaa atc ttg cat ttg ac-3′ | 130287-306 | 100% | 172162-82 | 85% |
| PCR primer B | reverse | 5′-aca gtt agc aga ggt aga gtg-3′ | 132488-68 | 100% | 174339-59 | 95% |
| Sequencing primer I | forward | 5′-tga tca cca ggg gtg gca gag 3′ | 131752-72 | 100% | 173624-44 | 90% |
| PCR primer C | forward | 5′-gca ctc agt ggg aga gac aa-3′ | 131585-604 | 100% | 173457-76 | 65% |
| PCR primer D | reverse | 5′-gtt ctg atg cat act cag gtt-3′ | 132538-58 | 100% | 174409-29 | 100% |
| Sequencing primer II | forward | 5′-aca aga tcg ttg ggg gct ac-3′ | 131924-43 | 100% | 173796-815 | 100% |
Figure 2.
Alignment of the PRSS1 gene between c.41–148 (IVS1-148) and c.200+499 (IVS2+499) with the corresponding PRSS2 sequence. Exon 2 is shown in capital letters. Positions of the forward sequencing primers and reverse PCR primers B and D are underlined. Forward PCR primers A and C are not shown, as these anneal upstream of the sequence region displayed here. The maximal converted region is highlighted in gray; the minimal converted region is framed.
Recombinant expression of trypsinogen mutants
Construction of the expression plasmid harboring the human cationic trypsinogen gene (PRSS1) was described previously (Sahin-Tóth, 2000; Sahin-Tóth and Tóth, 2000). Mutations N29I and N54S were introduced into the cationic trypsinogen gene by oligonucleotide-directed site-specific mutagenesis using the overlap-extension two-step PCR mutagenesis method. Small scale expression, in vitro refolding and affinity-purification of human trypsinogens was carried out as reported previously (Sahin-Tóth, 2000; Sahin-Tóth and Tóth, 2000; Kukor et al., 2003). Properties of wild-type and mutant trypsinogens were compared in activation experiments using the physiological activator enterokinase; or the potentially pathological activators trypsin (autoactivation) and cathepsin B. Active trypsins were further characterized with respect to catalytic activity, autocatalytic degradation (autolysis) and inhibition by the physiological inhibitor SPINK1. For experimental details see (Kukor et al., 2003; Teich et al., 2004).
RESULTS
In our index patient, direct DNA sequencing of exon 2 of the PRSS1 gene revealed that the wild type sequence changes at nucleotide c.86 (based on cDNA sequence) to a mixed sequence, which contains 22 heterozygous sequence differences (Supplementary Fig S1). This phenomenon stops at nucleotide 175 after the end of exon 2 (IVS2+175). Subtraction of the overlaid sequence from the PRSS1 (RefSeq NM_002769.2) sequence revealed the corresponding PRSS2 (RefSeq NM_002770.2) gene sequence (Fig 1; GenBank accession number AY605657). This indicates a conversion mutation affecting exon 2 and intron 2 between the PRSS2 gene (donor) and the PRSS1 gene (acceptor). Due to the high degree of similarity between the two genes, the 5’ and 3’ conversion break points could not be located to two distinct nucleotides. The PRSS1 and PRSS2 sequences are identical between IVS1-34 and c.85 (5‘ conversion border) and IVS2+175 and IVS2+263 (3‘ conversion border), respectively. The region between the conversion borders, which is the minimal converted sequence, is 289 nucleotides in length. It exhibits 92 percent identity between the PRSS1 and PRSS2 genes. Figure 2 shows a schematic alignment of the PRSS1 gene with the corresponding PRSS2 sequence in the converted region.
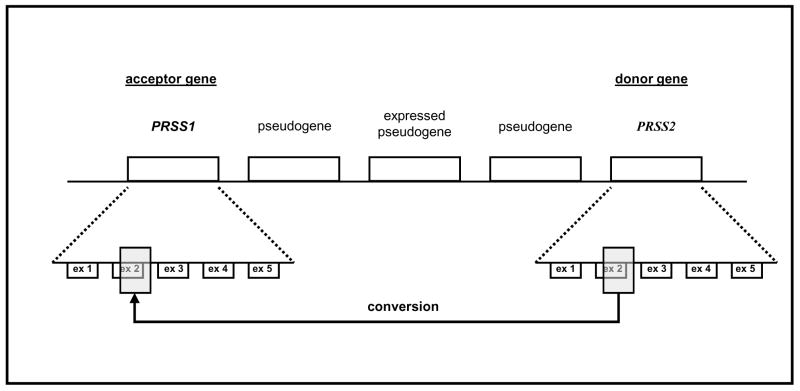
Figure 1.
Schematic representation of the conversion mutation within the human trypsinogen gene family at the TCR-beta locus on chromosome 7q35. Each gene represents a tandem 10-kb repeat (Rowen et al., 1996). A region comprising parts of exon 2 and intron 2 of the PRSS1 acceptor gene is converted by the PRSS2 donor gene. The PRSS2 gene remains unaltered (adapted from Chen et al., 2001).
The conversion results in six exonic and sixteen intronic nucleotide substitutions. Four of the six exonic substitutions are “silent”, as they do not alter the PRSS1 amino acid sequence. At nucleotide position 86, an A>T transversion substitutes asparagine by isoleucine (N29I) and at position 161, an A>G transition substitutes asparagine by serine (N54S). Thus, the amino acid sequence of PRSS1 exon 2 is converted completely to the PRSS2 sequence. The remainder of the PRSS1 gene is without further abnormalities, the PRSS2 gene is wild type. The presence of the novel mutation has been excluded in 1105 further patients with chronic pancreatitis and the mother of the index patient by direct DNA sequencing of exon 2 and flanking intronic sequences. The SPINK1-N34S mutation was not present in the index patient and her mother.
Recombinant cationic trypsinogen double-mutant N29I-N54S showed increased susceptibility for spontaneous activation to trypsin (Figure 3). When compared to the common hereditary-pancreatitis associated N29I mutant, the degree of increased autoactivation was identical, indicating that the N29I mutation is solely responsible for the phenotypic change in the N29I-N54S double mutant. In agreement with this notion, the N54S mutation alone had no effect on the autocatalytic activation of cationic trypsinogen (not shown). Activation of the double-mutant N29I-N54S by enterokinase or cathepsin B was normal. Catalytic parameters of wild-type cationic trypsin (kcat 67 ± 1.2 s−1; KM 26 ± 1.2 μM; kcat/KM 2.6*106 s−1 M−1) and double-mutant N29I-N54S trypsin (kcat 64.5 ± 1.7 s−1; KM 21.3 ± 1.5 μM; kcat/KM 3.0*106 s−1 M−1) were essentially identical, as determined on the N-CBZ-Gly-Pro-Arg-pNA chromogenic peptide substrate. Similarly, inhibition by SPINK1 and autocatalytic degradation of the active N29I-N54S trypsin were indistinguishable from wild type cationic trypsin (not shown).
Figure 3.
Autocatalytic activation of wild type (PRSS1), single-mutant (N29I) and double-mutant (N29I-N54S) cationic trypsinogen. 2 μM trypsinogen (final concentration, in a final volume of 100 μL) was incubated at 37 oC, in 0.1 M Tris-HCl (pH 8.0, upper panel) or 0.1 M Na-acetate buffer (pH 5.0, lower panel), in the presence of 2 mg/mL bovine serum albumin. Aliquots of 2 μL were withdrawn from reaction mixtures at indicated times and trypsin activity was determined with 0.14 mM N-CBZ-Gly-Pro-Arg-p-nitroanilide (final concentration). Activity was expressed as percentage of the potential total activity, which was determined on similar zymogen samples activated with bovine enterokinase at 22 oC in 0.1 M Tris-HCl (pH 8.0), 2 mg/mL bovine serum albumin and 1 mM CaCl2.
DISCUSSION
We report a unique gene-conversion mutation, which effectively leads to the complete replacement of exon 2 of the PRSS1 gene (acceptor gene) by the corresponding PRSS2 sequence (donor gene). This observation proves that gene conversion is a relevant mechanism for the development of pancreatitis-associated trypsinogen mutations (Chen and Férec, 2000a, b). The generally accepted minimum homology requirement for a conversion mutation to occur is about 200 nt homologous sequence both at the start and at the end of the converted gene fragment. The conversion rate is directly proportional to the extent of homology and even a single-nucleotide difference between the donor and acceptor sequences may reduce the probability of a conversion significantly (Waldman and Liskay, 1988; Liskay et al., 1987). In our case, the 5’ and 3’ conversion borders count 79 and 89 nucleotides only, respectively. The fact that recombination occurred despite the shortness of both of these homologous regions may be explained by the “conversion-provoking” genetic environment within the T-cell-receptor beta gene complex (Chen and Férec, 2003).
At the protein level, the gene-conversion event introduces two mutations into cationic trypsinogen. One of these, N29I, is also frequently found in hereditary pancreatitis, and previous biochemical analysis indicated that the mutation causes increased autocatalytic activation of cationic trypsinogen (Sahin-Tóth, 2000; Sahin-Tóth and Tóth, 2000; Szilágyi et al., 2001). Similarly, a higher propensity for autoactivation was observed with a number of other pancreatitis-associated mutants (N29T, R122H, D19A, D22G, K23R), indicating that autoactivation represents a risk factor for premature, intra-pancreatic trypsin-generation, which is believed to be the triggering event of hereditary pancreatitis (Sahin-Tóth, 2000; Sahin-Tóth and Tóth, 2000; Teich et al., 2000; Chen et al., 2003). Biochemical characterization of the N29I-N54S double mutant revealed no difference from the previously investigated N29I mutant, indicating that the N54S mutation has no additional deleterious effect.
During the last years, the impact of gene conversion has been established in many human diseases (see introduction). In most of these, the disease associated mutations in the functional genes have been copied from their highly homologous pseudogenes (Collier et al., 1993; Eikenboom et al, 1994, Watnick et al., 1998). A recent and relevant example is the Shwachman-Diamond syndrome, an autosomal recessive syndrome which includes exocrine pancreatic insufficiency. This condition is associated with a germline conversion in the Shwachman-Bodian-Diamond Syndrome gene with faulty sequences from the similar, but non-functional, pseudogene (Boocock et al., 2003). In contrast, the donor gene of the conversion mutation reported here is the anionic trypsinogen gene, a functionally important gene of the exocrine pancreas. Thus, our results highlight a unique example of how gene conversions between two normal genes can result in a novel, disease-associated allele.
Supplementary Material
Abbreviations
- PRSS1
protease, serine, 1, cationic trypsinogen
- PRSS2
protease, serine, 2, anionic trypsinogen
- SPINK1
serine protease inhibitor, Kazal type 1
Footnotes
Databases: PRSS1, OMIM #276000; PRSS2, OMIM #601564; the mutation in this report has been submitted to GenBank, under accession number AY605657.
References
- Boocock GR, Morrison JA, Popovic M, Richards N, Ellis L, Durie PR, Rommens JM. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet. 2003;33:97–101. doi: 10.1038/ng1062. [DOI] [PubMed] [Google Scholar]
- Chen JM, Férec C. Gene conversion-like missense mutations in the human cationic trypsinogen gene and insights into the molecular evolution of the human trypsinogen family. Mol Genet Metab. 2000a;71:463–9. doi: 10.1006/mgme.2000.3086. [DOI] [PubMed] [Google Scholar]
- Chen JM, Férec C. Origin and implication of the hereditary pancreatitis-associated N21I mutation in the cationic trypsinogen gene. Hum Genet. 2000b;106:125–6. doi: 10.1007/s004390051019. [DOI] [PubMed] [Google Scholar]
- Chen JM, Férec C. Trypsinogen genes: evolution. In: Cooper DN, editor. Nature encyclopedia of the human genome. Macmillan publishers, nature publishing group; 2003. pp. 645–50. [Google Scholar]
- Chen JM, Kukor Z, Le Maréchal C, Tóth M, Tsakiris L, Raguénès O, Férec C, Sahin-Tóth M. Evolution of trypsinogen activation peptides. Mol Biol Evol. 2003;20:1767–77. doi: 10.1093/molbev/msg183. [DOI] [PubMed] [Google Scholar]
- Chen JM, Montier T, Férec C. Molecular pathology and evolutionary and physiological implications of pancreatitis-associated cationic trypsinogen mutations. Hum Genet. 2001;109:245–52. doi: 10.1007/s004390100580. [DOI] [PubMed] [Google Scholar]
- Chen JM, Raguénès O, Férec C, Deprez PH, Verellen-Dumoulin C. A CGC>CAT gene conversion-like event resulting in the R122H mutation in the cationic trypsinogen gene and its implication in the genotyping of pancreatitis. J Med Genet. 2000;37:E36. doi: 10.1136/jmg.37.11.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier S, Tassabehji M, Sinnott P, Strachan T. A de novo pathological point mutation at the 21-hydroxylase locus: implications for gene conversion in the human genome. Nat Genet. 1993;3:260–5. doi: 10.1038/ng0393-260. [DOI] [PubMed] [Google Scholar]
- Eikenboom JC, Vink T, Briet E, Sixma JJ, Reitsma PH. Multiple substitutions in the von Willebrand factor gene that mimic the pseudogene sequence. Proc Natl Acad Sci U S A. 1994;91:2221–4. doi: 10.1073/pnas.91.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajnik MF. Comparative analyses of immunoglobulin genes: surprises and portents. Nat Rev Immunol. 2002;2:688–98. doi: 10.1038/nri889. [DOI] [PubMed] [Google Scholar]
- Gorry MC, Gabbaizedeh D, Furey W, Gates LK, Jr, Preston RA, Aston CE, Zhang Y, Ulrich C, Ehrlich GD, Whitcomb DC. Mutations in the cationic trypsinogen gene are associated with recurrent acute and chronic pancreatitis. Gastroenterology. 1997;113:1063–8. doi: 10.1053/gast.1997.v113.pm9322498. [DOI] [PubMed] [Google Scholar]
- Kukor Z, Tóth M, Sahin-Tóth M. Human anionic trypsinogen: properties of autocatalytic activation and degradation and implications in pancreatic diseases. Eur J Biochem. 2003;270:2047–58. doi: 10.1046/j.1432-1033.2003.03581.x. [DOI] [PubMed] [Google Scholar]
- Liskay RM, Letsou A, Stachelek JL. Homology requirement for efficient gene conversion between duplicated chromosomal sequences in mammalian cells. Genetics. 1987;115:161–7. doi: 10.1093/genetics/115.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowen L, Koop BF, Hood L. The complete 685-kilobase DNA sequence of the human beta T cell receptor locus. Science. 1996;272:1755–62. doi: 10.1126/science.272.5269.1755. [DOI] [PubMed] [Google Scholar]
- Sahin-Tóth M, Tóth M. Gain-of-function mutations associated with hereditary pancreatitis enhance autoactivation of human cationic trypsinogen. Biochem Biophys Res Commun. 2000;278:286–9. doi: 10.1006/bbrc.2000.3797. [DOI] [PubMed] [Google Scholar]
- Sahin-Tóth M. Human cationic trypsinogen. Role of Asn-21 in zymogen activation and implications in hereditary pancreatitis. J Biol Chem. 2000;275:22750–5. doi: 10.1074/jbc.M002943200. [DOI] [PubMed] [Google Scholar]
- Szilágyi L, Kénesi E, Katona G, Kaslik G, Juhász G, Gráf L. Comparative in vitro studies on native and recombinant human cationic trypsins. Cathepsin B is a possible pathological activator of trypsinogen in pancreatitis. J Biol Chem. 2001;276:24574–80. doi: 10.1074/jbc.M011374200. [DOI] [PubMed] [Google Scholar]
- Teich N, Bauer N, Mössner J, Keim V. Mutational screening of patients with nonalcoholic chronic pancreatitis: identification of further trypsinogen variants. Am J Gastroenterol. 2002;97:341–6. doi: 10.1111/j.1572-0241.2002.05467.x. [DOI] [PubMed] [Google Scholar]
- Teich N, Le Maréchal C, Kukor Z, Caca K, Witzigmann H, Chen JM, Tóth M, Mössner J, Keim V, Férec C, Sahin- Tóth M. Interaction between trypsinogen isoforms in genetically determined pancreatitis. Mutation E79K in cationic trypsin (PRSS1) causes increased transactivation of anionic trypsinogen (PRSS2) Hum Mutat. 2004;23:22–31. doi: 10.1002/humu.10285. [DOI] [PubMed] [Google Scholar]
- Teich N, Ockenga J, Hoffmeister A, Manns M, Mössner J, Keim V. Chronic pancreatitis associated with an activation peptide mutation that facilitates trypsin activation. Gastroenterology. 2000;119:461–5. doi: 10.1053/gast.2000.9312. [DOI] [PubMed] [Google Scholar]
- Teich N. [accessed 2001];Database of genetic variants in patients with chronic pancreatitis. Available from: URL: http://www.uni-leipzig.de/pancreasmutation.
- Waldman AS, Liskay RM. Dependence of intrachromosomal recombination in mammalian cells on uninterrupted homology. Mol Cell Biol. 1988;8:5350–7. doi: 10.1128/mcb.8.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watnick TJ, Gandolph MA, Weber H, Neumann HP, Germino GG. Gene conversion is a likely cause of mutation in PKD1. Hum Mol Genet. 1998;7:1239–43. doi: 10.1093/hmg/7.8.1239. [DOI] [PubMed] [Google Scholar]
- Whitcomb DC, Gorry MC, Preston RA, Furey W, Sossenheimer MJ, Ulrich CD, Martin SP, Gates LK, Jr, Amann ST, Toskes PP, Liddle R, McGrath K, Uomo G, Post JC, Ehrlich GD. Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene. Nat Genet. 1996;14:141–5. doi: 10.1038/ng1096-141. [DOI] [PubMed] [Google Scholar]
- Witt H, Luck W, Becker M. A signal peptide cleavage site mutation in the cationic trypsinogen gene is strongly associated with chronic pancreatitis. Gastroenterology. 1999;117:7–10. doi: 10.1016/s0016-5085(99)70543-3. [DOI] [PubMed] [Google Scholar]
- Witt H, Luck W, Hennies HC, Classen M, Kage A, Lass U, Landt O, Becker M. Mutations in the gene encoding the serine protease inhibitor, Kazal type 1 are associated with chronic pancreatitis. Nat Genet. 2000;25:213–6. doi: 10.1038/76088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.