Abstract
Pattern recognition receptors (PRRs) including Toll-like receptors (TLRs) and RIG like helicase (RLH) receptors are involved in innate immune antiviral responses. Here we show that nucleotide-binding oligomerization domain 2 (NOD2) can also function as a cytoplasmic viral PRR by triggering activation of interferon regulatory factor-3 (IRF3) and production of interferon-β (IFN). Following recognition of viral ssRNA genome, NOD2 utilized the adaptor protein MAVS (mitochondrial antiviral signaling) to activate IRF3. NOD2-deficient mice failed to produce IFN efficiently and exhibited enhanced susceptibility to virus-induced pathogenesis. Thus, the function of NOD2 as a viral PRR highlights the important role of NOD2 in host antiviral defense mechanisms.
INTRODUCTION
Innate immune antiviral responses are the first line of defense against virus infection 1, 2. Interferon-α/β (IFN) plays an important role during innate antiviral responses by activating the JAK-STAT signaling pathway 3. Virus-infected cells utilize pattern recognition receptors (PRRs) to recognize pathogen (virus) associated molecular patterns (PAMPs) and to trigger phosphorylation of the transcription factor interferon regulatory factor 3 (IRF3), which then translocates to the nucleus to transactivate IFN genes 4. So far two classes of viral PRRs have been identified: the Toll-like receptors (TLRs) 5 and the RLH (RIG like helicases) receptors such as RIGI and Mda5 6.
A third class of PRR includes members of the nucleotide binding domain (NBD) and leucine-rich-region (LRR) containing family (known as NLRs) of cytoplasmic proteins; these proteins respond to bacterial PAMPs to activate NF-κB and MAPK pathways 7, 8, 9. For example, the NLR NOD2 detects bacterial PAMPs including muramyl dipeptide 10. However, to date, no NLRs were reported to respond to virus-specific PAMPs and activate an antiviral response.
Recently it was demonstrated that the NLR family member NLRX1 interacts with IPS-1 to negatively regulate the IFN pathway 11, and induces reactive oxygen species (ROS) formation 12. Thus, members of NLR family of proteins may modulate (either positively or negatively) the host antiviral apparatus. In addition, NOD2 facilitates production of human β-defensin-2 (HBD2) after MDP stimulation, and HBD2 is also upregulated in human respiratory syncytial virus (RSV) infected cells 13, 14.
Here, to investigate the involvement of NLRs in innate antiviral response we examined the ability of various NLRs to activate IRF3 and IFN production following infection with RSV, a negative-sense single-stranded RNA (ssRNA) genome containing paramyxovirus that causes severe lung disease in infants, children, elderly and immuno-compromised individuals 15, 16. Among the various NLRs, NOD2 activated IRF3 and IFN production in RSV-infected cells. Activation of NOD2 by ssRNA resulted in signaling dependent on the mitochondrial antiviral signaling (MAVS) protein (also known as IPS-1, VISA, CARDIF). Both synthetic and viral ssRNA genome were capable of activating the MAVS-IRF3-IFN pathway. The important physiological role of NOD2 in antiviral defense was evident from the enhanced RSV pathogenesis, lung disease and greater viral susceptibility in NOD2-deficient mice. Similar roles for NOD2 were observed in responses to influenza A and parainfluenza viruses. Thus, these studies revealed a previously unknown function of NOD2 as a viral PRR important for host defense against virus infection.
RESULTS
ssRNA induces IFN production via NOD2
To study the involvement of NLR proteins in antiviral responses, we expressed various HA-tagged human NLR proteins (e.g. NOD1, NOD2, IPAF, NAIP, NOD3) in 293 cells, which do not endogenously express most NLRs. We treated these cells with synthetic ssRNA and analyzed IFN production and IRF3 activation. We utilized ssRNA because several highly pathogenic viruses including paramyxoviruses (RSV, Sendai virus, human parainfluenza viruses, measles virus), rhabdoviruses (rabies virus, vesicular stomatitis virus) and orthomyxoviruses (influenza viruses) contain ssRNA genomes, and because ssRNA activates PRRs including TLR7, TLR8 and RIGI 5, 6. NOD2 but not NOD1 facilitated ssRNA-induced IRF3 activation (Fig. 1a) and IFN-β production (Supplementary Fig. 1a, 1b). Such activation was lacking in cells treated with CpG DNA (Fig. 1a, Supplementary Fig. 1a, 1b). HA-tagged NOD constructs were expressed in high amounts in transfected 293 cells (Supplementary Fig. 1c); in addition, HA-NOD2 and HA-NOD1 proteins were functional as they facilitated activation of NF-κB in 293 cells treated with MDP or iE-DAP, respectively (Supplementary Fig. 1d).
Figure 1.
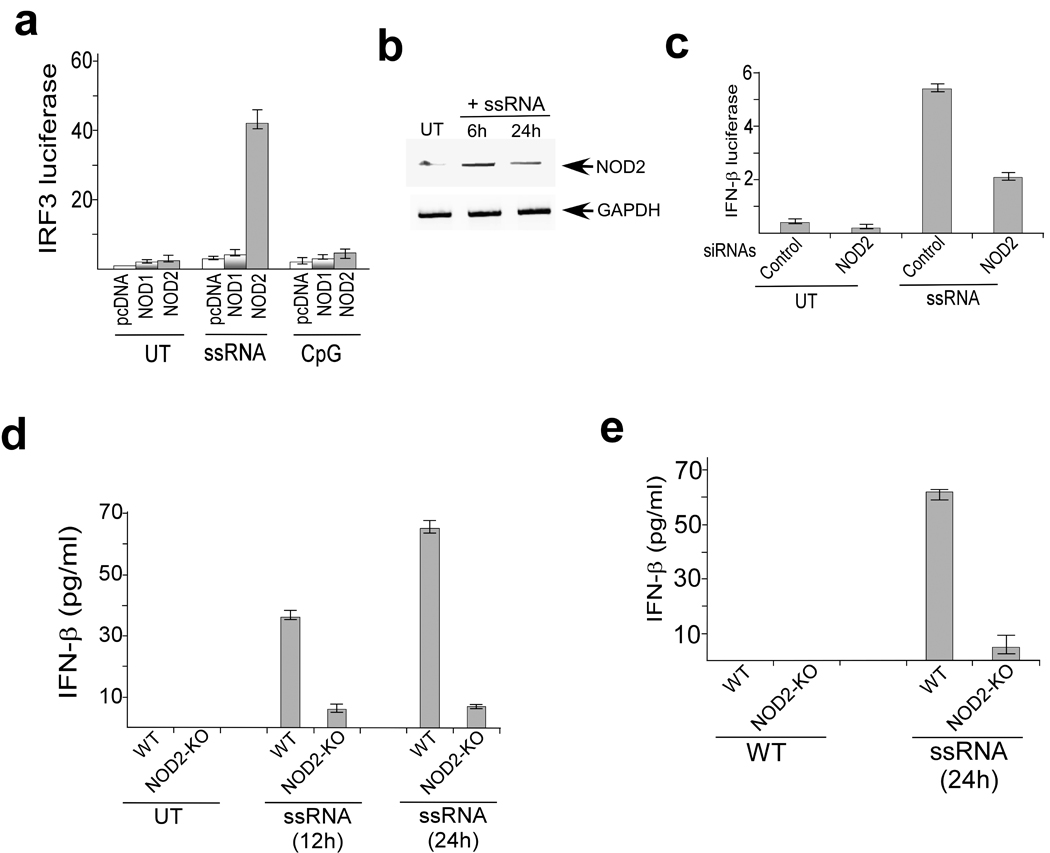
ssRNA activates NOD2. (a) Activation of an IRF3 luciferase reporter in untreated (UT), ssRNA, and CpG DNA treated (6h) 293 cells transfected with pcDNA, human HA-NOD1 or human HA-NOD2. (b) RT-PCR analysis of NOD2 expression in A549 cells left untreated or stimulated for the indicated time periods with ssRNA. (c) Activation of IFN-β luciferase reporter in A549 cells transfected with either control siRNA or NOD2 siRNA and left untreated or stimulated (6h) with ssRNA. The luciferase assay results are presented as mean ± s.d. from three independent experiments. (d,e) Bone marrow-derived macrophages (BMM) or mouse embryonic fibroblasts (MEFs) were isolated from wild-type and NOD2-KO mice and left untreated or stimulated with ssRNA for the indicated time periods. IFN-β production was measured by ELISA. Values represent the mean ± s.d. of three independent experiments.
To establish the physiological relevance of NOD2-mediated ssRNA-induced activation of IRF3, we next evaluated the role of endogenous NOD2 in inducing IRF3-IFN following ssRNA treatment. For these studies we utilized human lung epithelial A549 cells, since these cells are permissive to the majority of viruses that contain ssRNA genomes and endogenously express various PRRs. Treatment of these cells with ssRNA resulted in increased NOD2 expression (Fig. 1b); NOD1 expression remained unchanged (data not shown). NOD2-specific siRNA diminished NOD2 expression in ssRNA-treated A549 cells (Supplementary Fig. 2a), and impaired ssRNA-induced activation of IRF3 and IFN-β production (Fig. 1c, Supplementary Fig. 2b).
Similarly, ssRNA-induced IRF3 activation and IFN-β production was reduced in bone marrow derived macrophages (BMM) and mouse embryo fibroblast (MEF) isolated from NOD2-KO compared to wild-type mice (Fig. 1d,e). In contrast, wild-type and NOD2-KO BMM and MEFs produced similar amounts of IFN-β after treatment with poly-IC (dsRNA that activates TLR3) (Supplementary Fig. 2c). The ability of poly-IC to induce IFN (via TLR3) in both wild-type and NOD2-KO MEFs showed that IRF3-IFN pathway is intact in NOD2-KO animals. Furthermore, poly-IC administration in vivo resulted in production of similar concentrations of IFN-β in wild-type and NOD2-KO animals (Supplementary Fig. 2d). Thus our results obtained with cell lines and primary cells demonstrated that activation of NOD2 by ssRNA results in IFN-β production.
NOD2 facilitates virus-induced IFN production
To further validate the ability of NOD2 to launch an antiviral response, we infected 293 cells with RSV. RSV induced activation of IRF3 and IFN-β in 293 cells expressing HA-NOD2 but not in 293 cells expressing HA-NOD1 (Fig. 2a, 2b). Inactivation of virion particles with ultra-violet (UV) light abolished the ability of RSV to activate IRF3 in NOD2 expressing cells (Fig. 2a). The inability of UV inactivated RSV to activate IRF3 indicated that an intact viral RNA genome is essential for NOD2 activation. The direct role of viral components in NOD2 activation was further confirmed by the loss of IRF3 activation following inhibition of RSV cellular entry with an RSV neutralizing antibody (specific for the RSV fusion or F protein) (Supplementary Fig. 2e).
Figure 2.
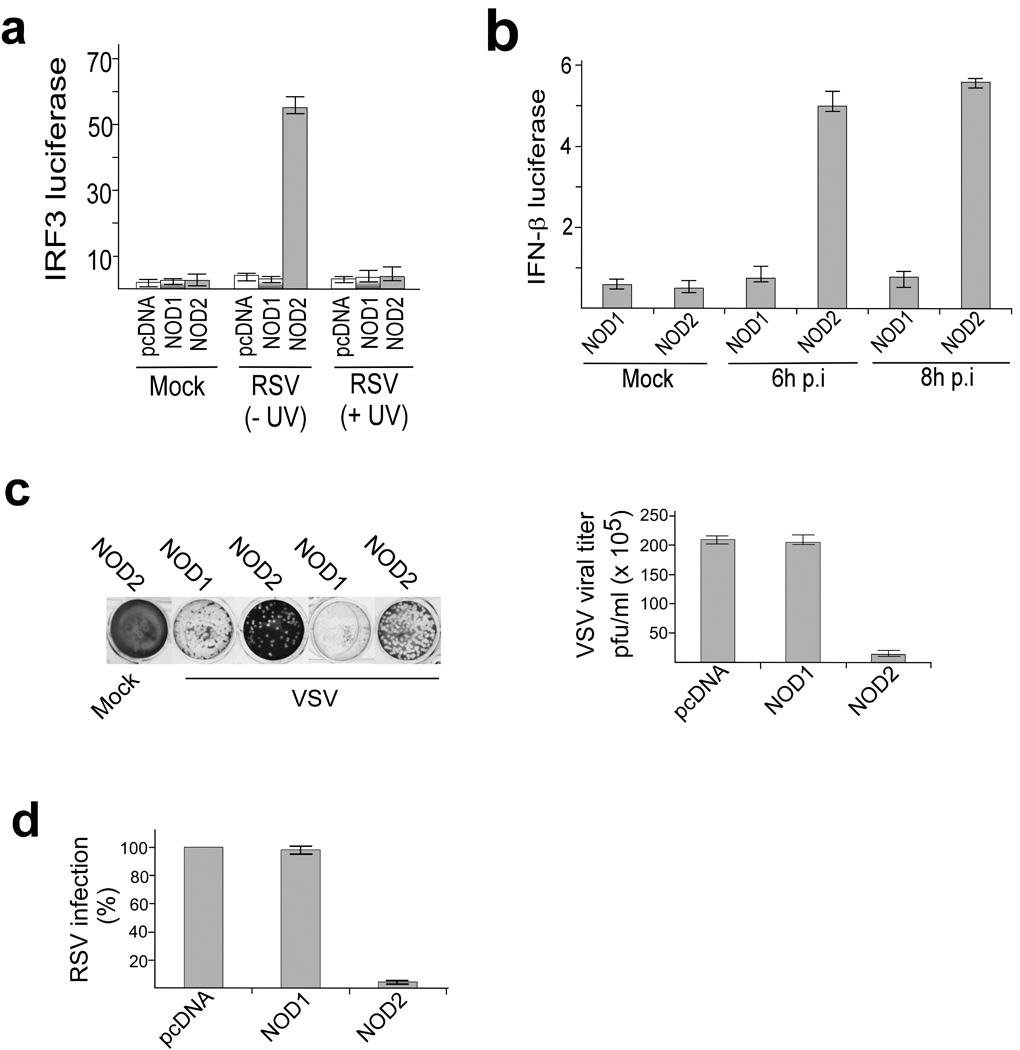
Activation of antiviral response by NOD2 in virus infected cells. (a,b) Activation of IRF3 and IFN-β luciferase reporter genes in mock infected and RSV infected 293 cells expressing pcDNA, HA-NOD1, or HA-NOD2. In (a), luciferase was measured 6 h post-infection (p.i.) and cells were infected with ultraviolet radiation (UV) treated or UV untreated RSV as indicated. (c) Plaque assay of VSV infectivity in 293 cells expressing pcDNA, HA-NOD1 or HA-NOD2. Crystal violet staining and VSV titer expressed as pfu per ml are shown. (d) RSV infectivity in 293 cells expressing pcDNA, HA-NOD1 or HA-NOD2. 100% infectivity represents the viral titer from cells expressing pcDNA. The plaque assay values represent the mean ± s.d. from three independent experiments. The luciferase assay results are presented as mean ± s.d. from three independent experiments.
The functional significance of NOD2 in antiviral responses was established using the IFN -sensitive vesicular stomatitis virus (VSV). Plaque assay analysis of VSV titers obtained from 293 cells expressing HA-NOD2 or HA-NOD1 revealed a marked reduction in viral titer in cells expressing HA-NOD2 (Fig. 2c). Like VSV, RSV titers were lower in NOD2 expressing cells compared to NOD1 expressing cells (Fig. 2d). Human parainfluenza virus type 3 17 and VSV 18 also activated IRF3 in NOD2 expressing 293 cells (Supplementary Fig. 3a). In contrast, vaccinia virus—a DNA virus—failed to activate IRF3 in NOD2 expressing cells (Supplementary Fig. 3b). These results demonstrated that, like the RLH receptors like RIGI and Mda5, NOD2 can function as a cytoplasmic PRR for viral ssRNA.
We next evaluated the role of endogenous NOD2 in inducing IRF3-IFN in response to RSV infection. Previous studies described activation of IRF3 and production of IFN-β from RSV infected A549 cells as early as 2h post-infection 19, 20. Although uninfected A549 cells did not express detectable amounts of NOD2, RSV infection resulted in increased NOD2 expression within 2h post-infection (Fig. 3a). RSV infection failed to induce NOD1 expression (data not shown). NOD2 siRNA markedly diminished NOD2 expression after RSV infection (Supplementary Fig. 4a) and resulted in reduced activation of IRF3 and production of IFN-β after RSV infection (Fig. 3b, Supplementary Fig. 4b). The effect was more pronounced during early (4h and 6h) compared to late (10h) time points after infection (Fig. 3b). This result suggests that NOD2 is critical for the early antiviral response, whereas during late infection time periods other PRRs (e.g. RIGI) may activate the IRF3-IFN pathway.
Figure 3.
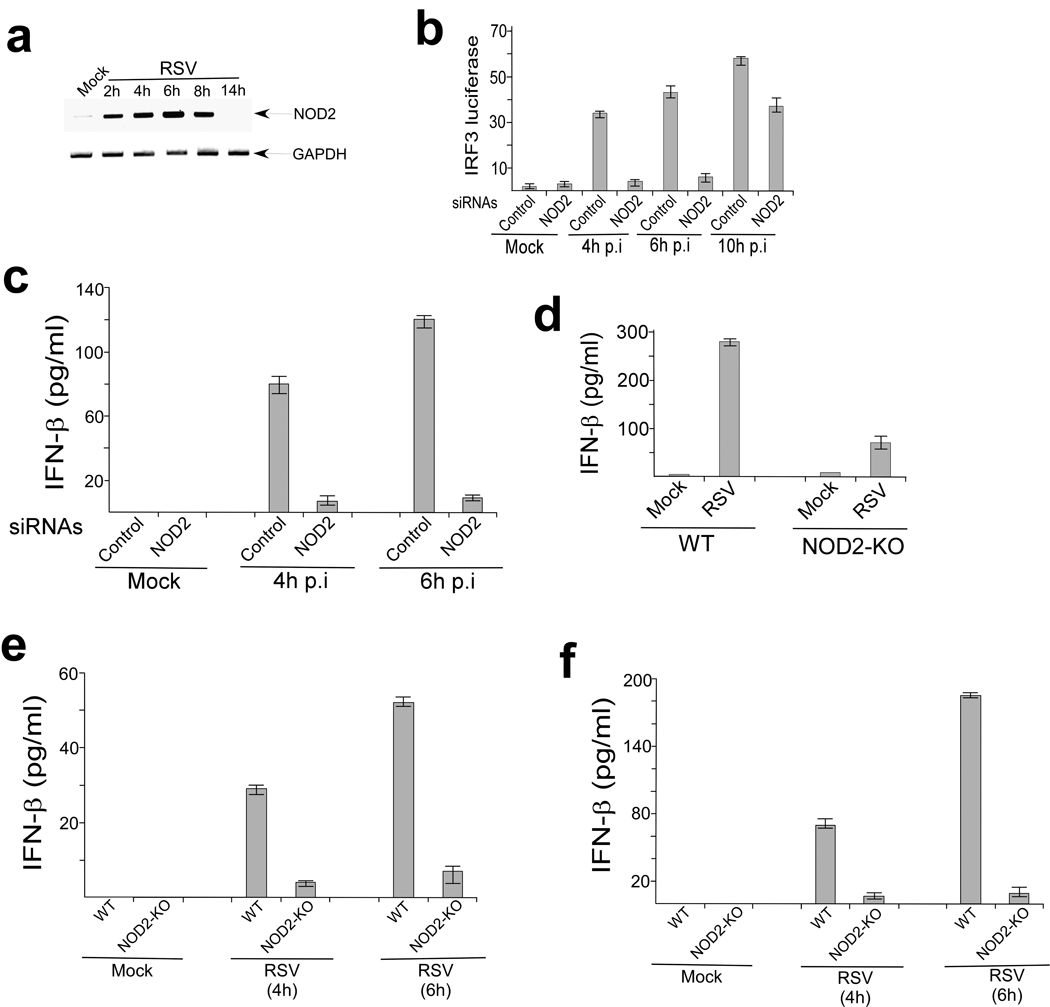
NOD2 is required for IFN production. (a) RT-PCR analysis of NOD2 expression in mock and RSV infected A549 cells. (b) Activation of IRF3 luciferase reporter in mock and RSV infected (hours post-infection. p.i) A549 cells transfected with either control siRNA or NOD2 siRNA. The luciferase assay results are presented as mean ± s.d. from three independent experiments. (c) IFN-β production from mock and RSV infected primary normal human bronchial epithelial (NHBE) cells transfected with either control siRNA or NOD2 siRNA. (d–f) IFN-β production from mock and RSV infected alveolar macrophages (d), BMM (e) and MEFs (f) isolated from wild-type (WT) or NOD2-KO mice. IFN-β was measured by ELISA and each value represents the mean ± s.d. from three independent experiments.
Indeed, we (Supplementary Fig. 4c) and others 20 have shown that RIGI expression in A549 cells is detectable only at late time points post-infection with RSV. In addition, the early antiviral response was independent of RIGI because silencing of RIGI had no effect in IFN-β expression during early RSV infection (Supplementary Fig. 4d,e). Like A549 cells, MEFs did not express abundant NOD2 until early time points after RSV infection, and RIGI was undetectable until late time points after RSV infection (Supplementary Fig. 4f,g). Thus temporal expression of NOD2 and RIGI during early and late infection, respectively, may facilitate optimal sustained IFN production from virus-infected cells.
Next we examined the role of NOD2 in primary normal human bronchial epithelial cells (NHBE), as these cells constitute the major cell type infected by RSV in humans. RSV rapidly induced NOD2 expression in NHBE cells (Supplementary Fig. 5). NOD2 was essential for IFN production, as NOD2 siRNA markedly reduced IFN-β production from RSV infected NHBE cells (Fig. 3c). The critical role of NOD2 was further established by demonstrating that BMM, MEFs and alveolar macrophages derived from NOD2-KO mice produced less IFN-β than wild-type counterparts after RSV infection (Fig. 3d–f). NOD2-KO MEFs and BMM also exhibited defective IFN-β production following influenza A/PR/8/34 (H1N1) virus infection (Supplementary Fig. 6). Collectively these data reveal an important role for endogenous NOD2 in the induction of antiviral immune responses.
NOD2 interacts with viral ssRNA
We next investigated the role of the viral ssRNA genome (viral-ssRNA) in NOD2 activation. Viral-ssRNA isolated from purified RSV virion particles activated IRF3 only in NOD2 expressing cells (Fig. 4a). An intact ssRNA genome was required for NOD2 activation since treatment of the viral-ssRNA with RNAse abolished IRF3 activation (Fig. 4a). Viral-ssRNA utilizes NOD2 for IFN production, as diminished IFN-β production following viral-ssRNA treatment was observed in MEFs and BMM isolated from NOD2-KO compared to wild-type mice (Fig. 4b,c).
Figure 4.
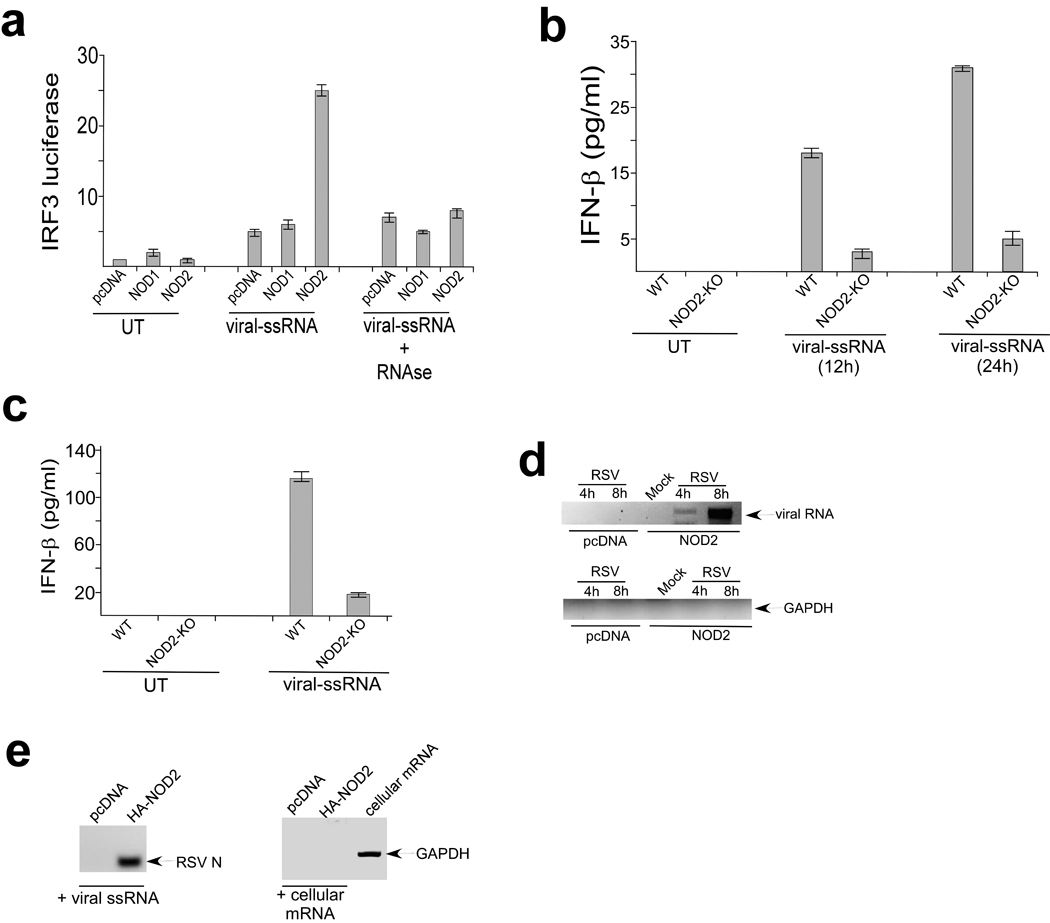
Activation of NOD2 by viral ssRNA genome. (a) Activation of IRF3 luciferase reporter in 293 cells expressing pcDNA, HA-NOD1, HA-NOD2 that were left untreated (UT) or stimulated with RSV ssRNA genome (viral-ssRNA). Where indicated viral-ssRNA was treated with RNAse. The luciferase assay results are presented as mean ± s.d. from three independent experiments. (b,c) IFN-β production from BMM (b) and MEFs (c) isolated from wild-type (WT) or NOD2-KO mice that were left untreated or stimulated with viral-ssRNA. IFN-β was measured by ELISA and each value represents the mean ± s.d. from three independent experiments. (d) 293 cells were transfected with pcDNA or HA-NOD2 and were mock infected or infected with RSV. At 4h or 8h post-infection, NOD2 was immunoprecipitated with HA-agarose and bound RNA was amplified using primers specific for GAPDH or RSV nucleocapsid (N) protein. The amplified products were analyzed on the agarose gel. (e) RSV ssRNA genome and total cellular mRNA was incubated with HA-NOD2 bound to HA-agarose beads. Bound RNA was amplified using the primers in (d). The amplified products were analyzed on the agarose gel. Total cellular mRNA amplified with GAPDH specific primers served as a positive control.
The role of viral-ssRNA as an activator of NOD2 was further demonstrated by observing interaction of viral ssRNA with NOD2 within a cellular milieu. We immunoprecipitated HA-NOD2 from RSV-infected 293 cells that were transfected with HA-NOD2, and amplified bound RNA with either GAPDH (control) or RSV nucleocapsid (N) protein specific primers. These experiments revealed association of NOD2 with viral but not control RNA (Fig. 4d). In a cell-free assay, HA-NOD2 bound to HA-agarose beads was incubated with RSV ssRNA genome or mRNA isolated from cells. After incubation, bound RNA was amplified with either GAPDH (control) or RSV nucleocapsid (N) protein specific primers; we detected interaction of NOD2 with RSV ssRNA (Fig. 4e). In contrast, GAPDH mRNA (which is enriched in total cellular mRNA) did not associate with NOD2 (Fig. 4e). We also observed failure of NOD1 to interact with viral ssRNA genome (data not shown). These results demonstrate that interaction of viral ssRNA genome with NOD2 results in its activation and subsequent induction of IFN production.
MAVS is required for NOD2-mediated responses
We next focused on the mechanism utilized by NOD2 to activate IRF3-IFN. As both RIGI and NOD2 posses CARD (caspase recruitment domain) domains 6, 7, 8, 9, we speculated that similar to RIGI, NOD2 may also interact with MAVS. In addition, a recent study showed that the NLR family member NLRX1 interacts with mitochondrial localized MAVS via its nucleotide binding domain (NBD), a domain also found in NOD2 11. Interaction of NOD2 with MAVS was essential for NOD2- mediated activation of antiviral responses, as MAVS siRNA (Supplementary Fig. 7) diminished RSV-induced IFN-β production from infected NHBE cells to an extent similar as NOD2 siRNA (Fig. 5a). Similar observations were noted in MEFs derived from MAVS-KO mice that were infected with RSV or transfected with viral or synthetic ssRNA (Fig. 5b,c). Influenza A virus also required MAVS for IFN production, as IFN-β production from NOD2-KO and MAVS-KO MEFs was reduced to a similar extent following influenza A infection (Supplementary Fig. 8). These results demonstrate that MAVS is critical for virus-induced NOD2-mediated IFN production.
Figure 5.
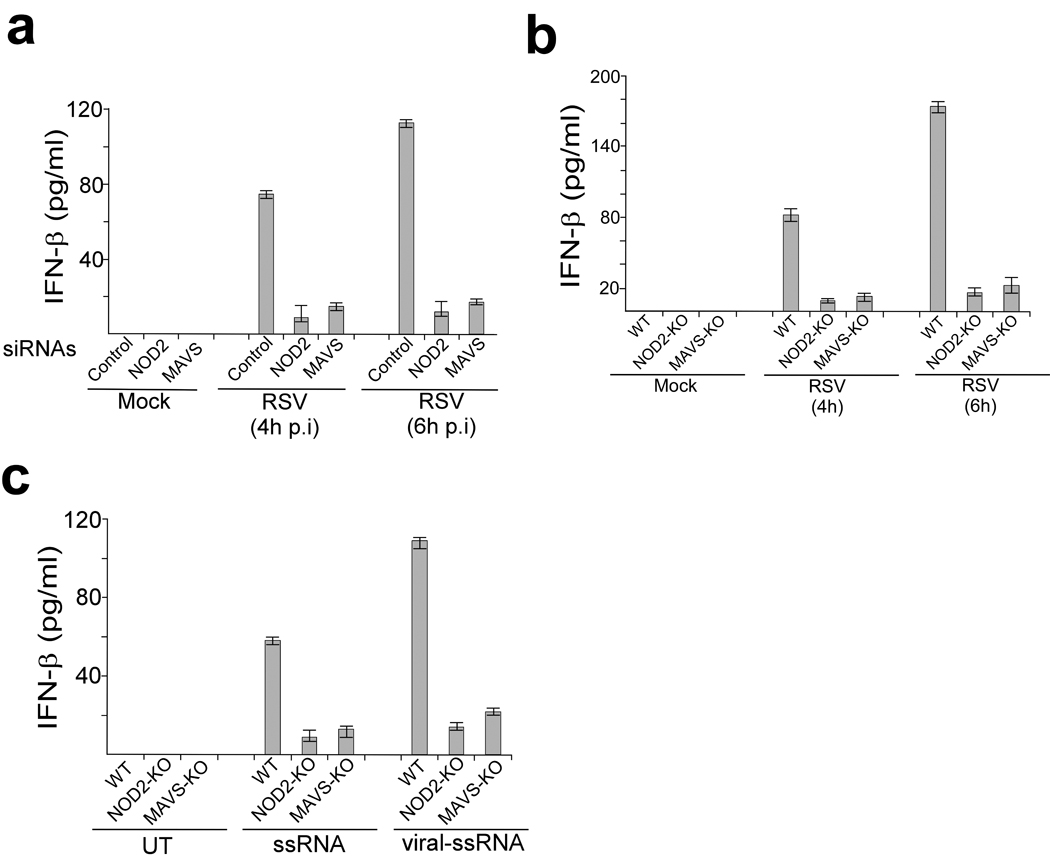
Role of MAVS during NOD2-mediated activation of the antiviral pathway. (a) IFN-β production from mock and RSV infected primary normal human bronchial epithelial (NHBE) cells transfected with either control siRNA, NOD2 siRNA or MAVS siRNA. (b) IFN-β production from mock and RSV infected MEFs isolated from wild-type (WT), NOD2-KO or MAVS-KO mice. (c) IFN-β production from MEFs left untreated (UT) or stimulated with synthetic or viral (RSV) ssRNA. IFN-β was measured by ELISA and each value represents the mean ± s.d. from three independent experiments.
We next examined interaction of NOD2 with MAVS. Initially we investigated the ability of activated NOD2 to translocate to the mitochondria. Immunoblot analysis of mitochondrial extract from RSV-infected NOD2 expressing cells revealed that although approximately 6%–7% of NOD2 is localized in mitochondria in uninfected cells, RSV infection resulted in enrichment (40%–45% of total cellular NOD2) of NOD2 in mitochondria (Supplementary Fig. 9a,b). Immunofluorescence analysis also revealed co-localization of endogenous NOD2 with mitochondria in RSV-infected A549 cells (Supplementary Fig. 9c). To study the interaction of NOD2 with MAVS, 293 cells were transfected with HA-NOD2 and GFP tagged MAVS. Co-immunoprecipitation analysis revealed interaction of NOD2 with MAVS (Fig. 6a), and this interaction was enhanced following RSV infection. Please note that high levels of GFP-IPS-1 was expressed in cells (as deduced by Western blot analysis of cell lysate with anti-GFP antibody) (Fig. 6a, lower panel) and substantial amount of HA-NOD2 was bound to the anti-HA-agarose beads that were used to pull-down HA-NOD2/GFP-IPS-1 complex (Fig. 6b). In contrast to NOD2, NOD1 failed to interact with MAVS (Supplementary Fig. 9d). Double labeled immunofluorescence studies with RSV-infected 293 cells expressing GFP-MAVS and HA-NOD2 confirmed co-localization of NOD2 and MAVS (Fig. 6c). Similarly, RSV infection of A549 (Fig. 6d) and NHBE (Fig. 6e) cells enhanced colocalization of endogenous NOD2 with endogenous MAVS. These results demonstrated that NOD2 interacts with MAVS during virus infection.
Figure 6.
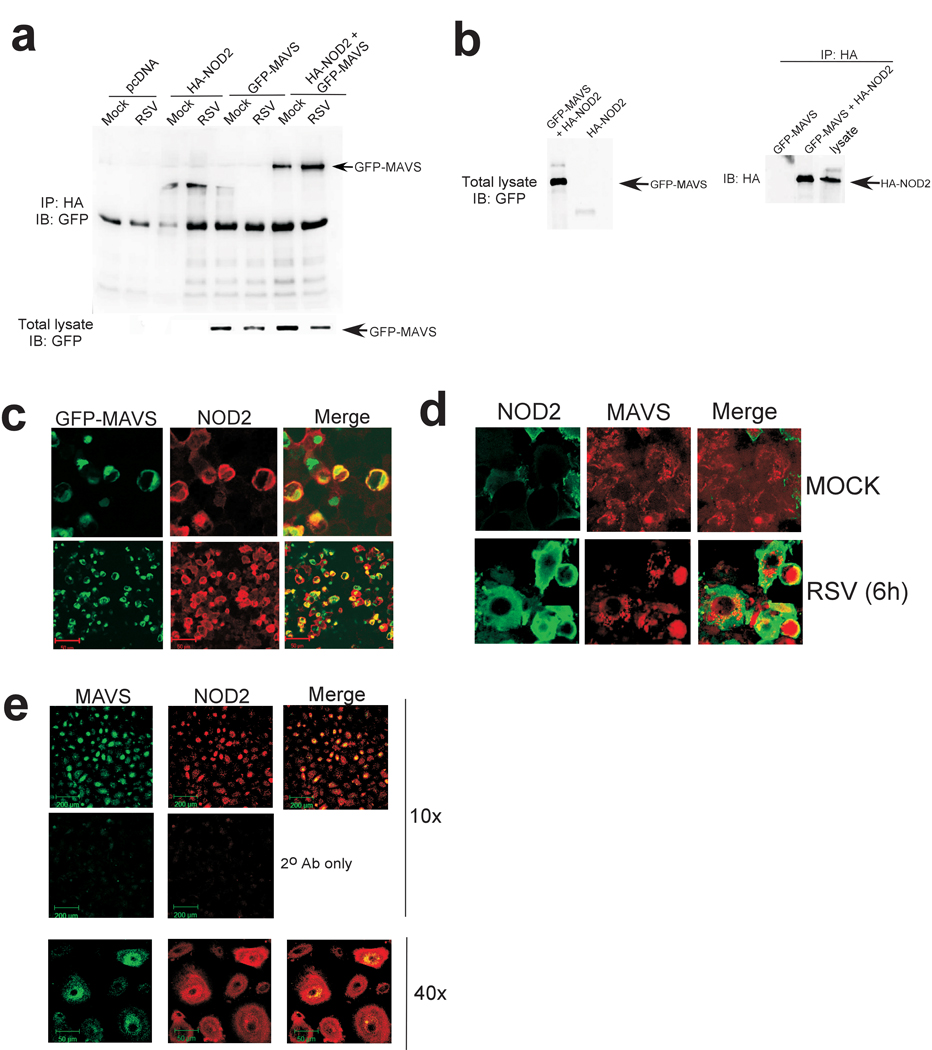
Interaction of MAVS with NOD2. (a) 293 cells were transfected with pcDNA, HA-NOD2 and-or GFP-MAVS and mock infected or infected with RSV. Lysates were immunoprecipitated with anti-HA agarose beads and bound proteins were immunoblotted with anti-GFP. Expression of GFP-IPS-1 in the cell lysate (by immunoblotting 25 µg of total cellular lysate with anti-GFP antibody) is also shown in the lower panel. (b) Expression of GFP-IPS-1 and HA-NOD2 in the cell lysate (by immunoblotting with anti-GFP and anti-HA antibodies) and amount of HA-NOD2 bound to anti-HA-agarose beads is also shown. For immunoblotting with cell lysates, 25 µg of total cellular lysate protein was used to detect GFP-IPS-1 and HA-NOD2. (c) RSV-infected (4h) 293 cells co-expressing GFP-MAVS (green) and HA-NOD2 (red) were imaged using confocal microscopy. (d,e) Mock or RSV-infected (6h) A549 cells (d) or RSV-infected (4h) NHBE cells (e) were stained with anti-NOD2 and anti-MAVS and imaged by confocal microscopy to detect endogenous NOD2 and MAVS.
NOD2 can activate NF-κB and MAPK pathways via the kinase RICK (also known as Rip2, CARDIAK, CCK and Ripk2) 21, 22. Bacterial products like MDP specifically stimulate NOD2 and result in NF-κB activation via RICK. However, treatment of NOD2 expressing 293 cells with MDP did not activate IRF3 (Supplementary Fig. 10a). Likewise, MDP treatment of NHBE and BMM did not result in IFN-β production (Supplementary Fig. 10b,c). In addition, RICK may not play a major role in IFN induction by NOD2 since silencing endogenous RICK expression did not alter NOD2 mediated IFN-β production in RSV infected cells (Supplementary Fig. 11). We also examined the efficiency of interaction between NOD2 and MAVS compared to RIGI and MAVS. Although both NOD2 and RIGI associated with MAVS, the RIGI-MAVS interaction was slightly more efficient than NOD2-MAVS interaction (Supplementary Fig. 12a). In addition, we also observed that in RSV-infected cells, NOD2 more efficiently interacted with MAVS than with RICK (Supplementary Fig. 12b). However, NOD2 efficiently interacted with RICK in cells stimulated with the well-established NOD2 stimulator MDP (Supplementary Fig. 12b). Thus, we speculate that NOD2 utilizes either RICK or MAVS depending on the stimulus (e.g. MDP vs. ssRNA) to activate either IRF3 or NF-κB.
In addition to IRF3, NF-κB activation is required for IFN gene expression. Although IRF3 alone is capable of inducing IFN gene transcription, the transactivating function of NF-κB synergistically acts with IRF3 to promote optimal IFN expression 23. This is also true for NOD2 mediated IFN expression, as suppressing NF-κB activity in RSV infected cells diminished IFN expression via activated NOD2 by 30%–35%; as expected, expression of the NF-κB-dependent TNF gene in RSV infected cells was reduced by 80% (Supplementary Fig. 13). Based on these results, we speculate that NOD2 activated by stimulation by viral ssRNA interacts with MAVS to induce activation of both IRF3 and NF-κB in a manner similar to that of RLHs 6. In contrast, NOD2 activated by bacterial products (e.g. MDP) activates NF-κB via RICK. Further detailed studies are required to investigate the role of NOD2 in activating NF-κB pathway during virus infection and the role of MAVS and RICK during these events.
MAVS interacts with LRR-NBD domains of NOD2
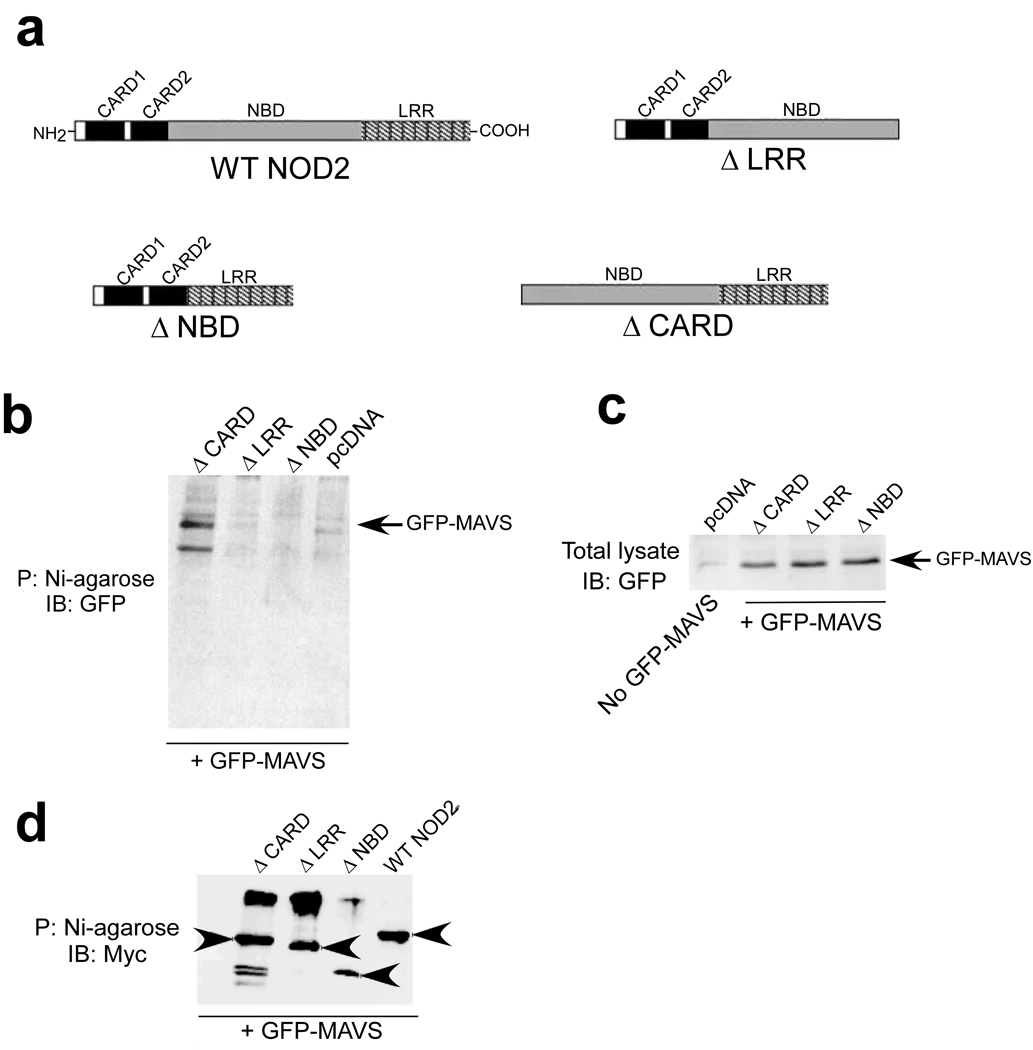
The CARD domain of RIGI promotes its association with MAVS, whereas NLRX1 utilizes its NBD domain to interact with MAVS. Thus, we next investigated the role of NBD, CARD and LRR domains of NOD2 in MAVS association. For these studies, we generated various His-Myc tagged versions of NOD2 deletion mutants − ΔCARD (NOD2 mutant lacking both CARD domains), ΔNBD (NOD2 mutant lacking the NBD domain) and ΔLRR (NOD2 mutant lacking the LRR domain) (Fig. 7a). These mutants were expressed in 293 cells along with GFP-MAVS. Lysates obtained from these cells were precipitated with nickel-agarose beads and, after washing, the proteins bound to the beads were subjected to immunoblotting with anti-GFP. While ΔCARD was capable of interacting with MAVS, both ΔNBD and ΔLRR failed to associate with MAVS (Fig. 7b). Comparable amounts of GFP- MAVS protein were expressed in the various NOD2 mutant expressing cells (Fig. 7c). In addition, similar amounts of His-Myc tagged NOD2 mutants were bound to the Nickel-agarose beads during the experimental condition used to study interaction of MAVS with NOD2 mutants (Fig. 7d). These results indicate that unlike RIGI, the CARD domains of NOD2 are not important for its interaction with MAVS. However, the NBD and LRR domains of NOD2 are required for MAVS association.
Figure 7.
NBD and LRR domains of NOD2 are essential for interaction with MAVS. (a) Schematic showing the various NOD2 constructs with deletion in specific domains. WT, wild-type; ΔCARD (NOD2 mutant lacking both CARD domains), ΔNBD (NOD2 mutant lacking the NBD domain) and ΔLRR (NOD2 mutant lacking the LRR domain). (b) 293 cells expressing various His-Myc tagged NOD2 constructs or pcDNA along with GFP- MAVS were lysed and lysates were incubated with Nickel-agarose (Ni-agarose). Following washing of the beads, the bound proteins were subjected to immunoblot analysis with anti-GFP. P; precipitation. (c) Lysates from cells in (b) were immunoblotted with anti-GFP to examine expression of GFP- MAVS. The first lane shows cells transfected with pcDNA only. (d) Cell lysates obtained from 293 cells co-expressing WT and NOD2 deletion mutants (his-myc-NOD2 constructs) along with GFP-IPS-1 was incubated with Ni-agarose, followed by immunoblotting with myc antibody (to detect his-myc tagged NOD2 protein constructs bound to the Ni-agarose beads). Please note that the WT and deleted version of NOD2 proteins are indicated with arrowheads.
This conclusion was further validated by examining the functionality of the NOD2 mutants in activating IRF3. Infection of 293 cells transfected with either wild-type or mutant (ΔCARD, ΔNBD, ΔLRR) NOD2 constructs revealed that wild-type NOD2 and ΔCARD, but neither ΔNBD nor ΔLRR, induced IRF3 activation after infection with RSV (Supplementary Fig. 14).
Role of NOD2 in host antiviral defense
Finally, we assessed the physiological role of NOD2 by infecting wild-type and NOD2-KO mice with RSV. The mouse model of RSV infection mimics virus infection in humans, as infected mice can develop disease states resembling pneumonia 24, 25; in addition, mice induce a robust antiviral response characterized by production of IFN-β and expression of IFN-dependent genes like Mx during early RSV infection (within 12h post-infection) 24, 25, 26, 27, 28, 29, 30. Moreover, RSV is sensitive to IFN in infected mice because as low as 200 units per ml of IFN inhibits RSV infection in mice by 100 fold 26. It is important to mention that during RSV infection of mouse respiratory tract, IFN is induced early during infection (at 12h-2d post-infection) but its production is lost at 3d post-infection 24, 25, 26, 27, 28, 29, 30. This observation suggests that IFN is important to restrict RSV spread during early infection and the production of IFN dictates the clinical outcome of the disease (for e.g. lung inflammation and apoptosis of airway cells).
Wild-type and NOD2-KO mice were infected with sublethal dose of RSV (5×106 pfu per animal delivered by intra-nasal inoculation) followed by collection of lungs and bronchoalveolar lavage (BAL) fluid at different time periods. We observed expression of murine NOD2 in RSV-infected lungs at 1d post-infection, and such expression was lost at 4d post-infection (Supplementary Fig. 15a). This result suggested an important role for NOD2 in IFN expression, as the IFN induction kinetics correlated with the NOD2 expression kinetics 24, 25, 26, 27, 28, 29, 30. Compared to wild-type mice, NOD2-KO mice showed diminished IFN-β production in the respiratory tract and increased viral titer (Fig. 8a,b)
Figure 8.
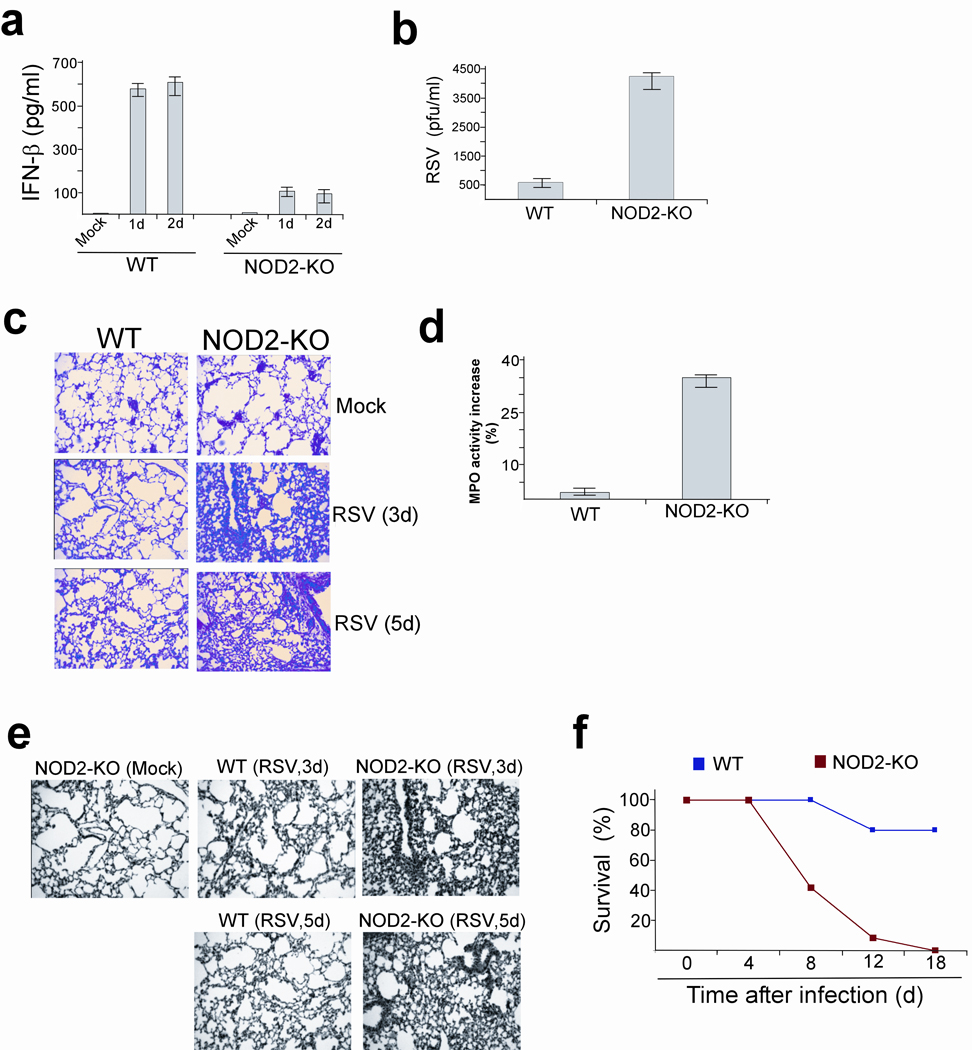
NOD2 is essential for host defense against virus infection. (a) IFN-β concentrations in the bronchoalveolar lavage (BAL) of RSV infected wild-type (WT) and NOD2-KO mice. Values (n=four mice per group) are mean ± s.e.m. P < .05, by t-test when data were normally distributed, or by Mann-Whitney Rank sum test when data were not normally distributed. (b) RSV titer in the BAL (3d post-infection) of WT and NOD2-KO mice. Values are mean ± s.e.m. P < .05, by t-test when data normally distributed, or by Mann- Whitney Rank sum test when data were not normally distributed. (c) H&E staining of lung sections obtained from RSV-infected WT and NOD2-KO mice. (d) Neutrophil sequestration in lungs of RSV infected (2d post-infection) WT and NOD2-KO mice was assessed by myeloperoxidase (MPO) activity assay with total lung homogenate. MPO activity is shown as percentage increase over enzyme activity in mock infected mice. Results are mean ± s.e., n=5, P < 0.05. (e) TUNEL staining of lung sections obtained from RSV infected WT and NOD2-KO mice. (f) Survival of WT and NOD2-KO mice infected with RSV (5×108 pfu per animal). P › 0.02 between NOD2-KO and WT mice as deduced by Wilcoxon test.
It is well known that RSV causes lung disease by inducing pneumonia, a massive inflammation of the lungs 31. Higher virus burden ultimately results in enhanced inflammation and exaggerated lung disease due to flooding of alveolar spaces with edema fluid. This occurs as a result of enhanced permeability of the epithelial barrier due to apoptosis of airway epithelial cells. RSV infection resulted in more severe lung pathology in NOD2-KO mice (as deduced by H&E staining of lung sections 3 and 5 days post-infection) (Fig. 8c). We noted massive peribronchial lymphocytic inflammation and filling of the lumen with exudates of infiltrating neutrophils and mucus. Neutrophils constitute the major immune cells infiltrating the lung of RSV-infected mice and humans and high number of these cells in the airway causes severe immunopathology associated with RSV clinical disease 32, 33, 34. To examine neutrophil accumulation in the lungs, we performed a myeloperoxidase (MPO) activity assay 35, 36 with lung homogenates of RSV infected wild-type and NOD2-KO mice. Higher RSV-induced enhancement of neutrophil activity was visible in the lung tissue of NOD2-KO (~35%) compared to wild-type (~4%) mice (Fig. 8d). The enhanced inflammation in the respiratory tract of NOD2-KO mice was also confirmed by higher concentrations of pro-inflammatory cytokines and chemokines (e.g. tumor necrosis factor, IL-10, RANTES) in the BAL of infected NOD2-KO compared to wild-type animals (Supplementary Fig. 15b–d). High RSV load has been associated with enhanced apoptosis of airway epithelial cells and infiltrating neutrophil granulocytes, which contributes to the development of lung lesions and injury 37. In deed, in situ apoptosis analysis of lung sections by TUNEL assay revealed enhanced apoptosis in the lungs of NOD2-KO compared to wild-type animals infected with RSV (Fig. 8e).
Notably, RSV-infected NOD2-KO mice lost significantly more body weight and exhibited reduced survival than wild-type counterparts (Supplementary Fig. 16, Fig. 8f). We also observed diminished IFN-β production in the BAL of NOD2-KO compared to wild-type mice infected with influenza A virus (Supplementary Fig. 17). These results demonstrated that NOD2 is a critical component of host antiviral defense mechanisms.
DISCUSSION
In the current study we identified NOD2 as a viral PRR that can sense viral ssRNA genomes to activate IFN production and antiviral defense. Like RLH receptors NOD2 associated with MAVS to activate IRF3 and promote IFN production. The importance of NOD2 in host defense was evident from the ability of both immune (e.g. macrophages) and non-immune (e.g. epithelial cells, MEFs) cells to utilize NOD2 for IFN production. The in vivo importance of NOD2 in antiviral responses was evident from the enhanced RSV-induced pathogenesis in infected NOD2-KO mice.
It is important to mention that other PRRs including RIGI may also be involved in activating an antiviral response against paramyxoviruses like RSV 20. For quite some time, the in vivo relevance of RIGI in antiviral function was not documented, due to the embryonic lethal phenotype of majority of RIGI-KO mice 38. However, one strain of RIGI-KO mice generated by crossing RIGI heterozygous mice with ICR outbred mice, followed by intercrossing of the resulting RIGI heterozygous mice 38, survived to adulthood. These RIGI-KO mice exhibited impaired IFN production and enhanced susceptibility to two positive sense ssRNA viruses: Encephalomyocarditis virus (EMCV) and Japanese encephalitis virus (JEV) 38. However no studies were conducted to demonstrate the importance of RIGI in activating the antiviral host defense apparatus against negative sense ssRNA viruses (e.g. paramyxoviruses).
NOD2 is expressed in low amounts in uninfected mice 39, and we showed that its expression increased after viral infection. Our observation is similar to previous studies demonstrating that majority of PRRs (for e.g. RIGI) are expressed at low abundance, but their expression is elevated following pathogen invasion 40, 41. Similarly, we observed viruses mediated induction of NOD2 in various cells. Although in our studies we noted induction of NOD2 in virus infected MEFs; one study reported an inability of MDP to induce NOD2 expression in wild-type MEFs, this observation may have been due to a defect in MDP transport to the cytoplasm 42. Several other studies showed that NOD2 expression is stimulated by various bacteria and bacterial components 40, 41. Similarly, expression of RIGI 43 and Mda-5 38 in uninfected, unstimulated wild-type MEFs is negligible, but treatment of cells with PAMPs results in up-regulation of RIGI expression 43. This mechanism of restricting expression of PRRs in un-stimulated cells may be critical to prevent uncontrolled inflammation.
Although NOD2 can be activated by MDP 7, 8, 9, 10, so far no studies have demonstrated direct binding of MDP to NOD2; it is only known that lack of NOD2 expression results in loss of MDP responsiveness. Thus, MDP could directly interact with NOD2, or associate with protein(s) that form a complex with NOD2. In that context, the interaction of viral-ssRNA with NOD2 demonstrated here could also be mediated indirectly via “bridging” protein(s). Interaction of ssRNA genomes of viruses with RIGI has also been noted previously 44, 45. In addition to NOD2, cryopyrin (also called Nalp3), another NLR protein, activates the inflammasome and leads to IL-1 production upon stimulation with bacterial and viral (influenza A) RNA 46, 47, 48. Although NLRs like cryopyrin 46,47, 48 and NLRX1 11 play an important role in innate immunity by activating the inflammasome and inhibiting IFN production, respectively, no studies have determined whether other NLRs, like NOD2, can directly contribute to antiviral responses by inducing IFN production.
Previous studies 49 have shown that transfection of NOD2 alone (in the absence of any stimulant) in wild-type MEFs resulted in substantial NF-κB activation. However, we found that overexpression of NOD2 in 293 cells did not induce marked activation of IRF3 in the absence of external stimuli (e.g. RSV, synthetic or viral ssRNA). Thus, it appears that NOD2-mediated activation of IRF3 is stimulus dependent, whereas RICK activation is stimulus independent.
In summary, our findings demonstrated that in addition to RIGI, Mda5 and TLRs, NOD2 can also function as a viral PRR and participate in inducing antiviral signaling. Distinct temporal activation of various PRRs may be required to generate optimal antiviral responses, and various viruses may trigger induction of different classes of PRRs.
METHODS
Virus and cell culture
RSV (A2 strain) and VSV were propagated in Hela and BHK cells, respectively 13, 18. Influenza A [A/PR/8/34 (H1N1)] virus was grown in the allantoic cavities of 10-day-old embryonated eggs. All viruses were purified by centrifugation (two times) on discontinuous sucrose gradients. Human lung epithelial A549 cells and 293 cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS), penicillin, streptomycin, and glutamine. Primary normal human bronchial epithelial (NHBE) cells (from Lonza) were maintained in bronchial epithelial growth medium (BEGM) according to the supplier’s instruction.
Luciferase assay
293 cells were transfected (Lipofectamine 2000 from Invitrogen) with 1µg of various plasmids (HA-NOD2, HA-NOD1, pcDNA6.1, IRF3-luciferase, IFN-β-luciferase) and 100 ng of pRL-null-renilla luciferase. 293 cells were then infected or treated with RSV, ssRNA, or CpG DNA. A549 cells were transfected (Lipofectamine 2000 from Invitrogen) with 80nM of NOD2 siRNA or control siRNA. 24 h post-siRNA transfection, cells were co-transfected with pRL-null-renilla luciferase (100 ng), IRF3-luciferase (1µg) or IFN-β-luciferase (1µg). After 24h, cells were either infected with 0.5 MOI of RSV or treated with 1µg per ml ssRNA40-LyoVec (Invivogen) for different time periods. Luciferase activity was measured using Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol. Transfection efficiency was normalized by measuring expression of renilla luciferase. Luciferase units were measured by standard methodology.
RT-PCR
The primers used to detect the various genes by RT-PCR are provided in a supplementary table.
siRNA
All the siRNAs were ordered from Qiagen. The sequences of siRNA oligonucleotide used in the current study are provided in a supplementary table. As a negative control, AllStars Negative Control siRNA from Qiagen (catalog number 1027281, proprietary sequence) was used. A549 or 293 cells were transfected with siRNAs using Lipofectamine 2000 (Invitrogen) and NHBE cells were transfected with siRNAs with PrimeFect Primary Cell siRNA Transfection Reagent (Lonza) according to the manufacturer’s protocol.
Viral infection
293 or A549 cells were infected with purified RSV at 0.5 multiplicity of infection (MOI) in serum free antibiotic free OPTI-MEM medium (GIBCO). Following adsorption for 1.5h at 37°C, cells were washed twice with serum containing DMEM and the infection was continued in the presence of serum containing DMEM for the specified time points. MEFs were infected with purified RSV or influenza A (A/PR/8/34 virus) at 1 MOI in serum free antibiotic free OPTI-MEM medium.
Co-immunoprecipitation
293 cells were transfected with indicated tagged constructs and were then infected with RSV. Cell pellets were lysed (in TBS containing 1% Triton-X100) and sonicated. All lysates were incubated for 12h (at 4°C) with anti-HA-agarose beads (Sigma-Aldrich). Proteins bound to washed anti HA-agarose were eluted at pH 2.8. Eluted proteins were subjected to immunoblot analysis with anti-GFP (Santa Cruz) or anti-HA (Sigma, clone HA-7)
Immunofluorescence analysis
Cells plated on four well glass chamber slides were transfected with indicated tagged constructs. Cells were then infected with RSV (1 MOI) for 4h or 6h. Following infection, cells were fixed with 3.7% formaldehyde and permeabilized and blocked in the permeabilizing buffer containing Triton X-100 (0.2%) and BSA (3%), and then incubated with either anti-HA (Sigma), anti-NOD2 (Cayman Chemical company) or anti-MAVS (Cell Signaling Technology) antibodies for 1 hr at 37°C. The washed cells were then incubated with the secondary antibody (Vector laboratories). Finally, the washed cells were mounted and the imaging of the cells was carried out using Zeiss LSM510 META laser scanning confocal microscopy.
Interaction of NOD2 with viral ssRNA
293 cells were transfected with HA-NOD2 and then infected with RSV. Lysates were immunoprecipitated with anti-HA agarose for 4h at 4°C. After washing the beads with TBS, Tri-reagent was added to isolate bound RNA. RT-PCR was performed using RSV nucleocapsid (N protein) protein and GAPDH specific primers. For the cell-free interaction assay, lysate obtained from 293 cells expressing HA-NOD2 was incubated with HA-agarose beads. The HA-NOD2 bound to the beads were incubated with RSV ssRNA genome or total cellular mRNA (cellular mRNA was isolated by using the RNeasy minikit) for 45min incubation at 4°C. Beads were washed and RNA isolated from the washed beads was amplified using primers described above.
Virus infection of mice
6–8-week old pathogen-free C57BL/6 and NOD2-KO (C57BL/6J background) mice were obtained from Jackson laboratory. We further back-crossed these NOD2-KO mice to the C57BL/6 genetic background for a total of eight generations. Genome wide SNP analysis on these animals (Harlan Laboratories, Inc.), revealed that wild-type and NOD2-KO mice are genetically identical, with the exception of the NOD2 deletion (data not shown). Mice were anesthetized using inhaled methoxyfluorane and intranasally inoculated with RSV (5×106 pfu per animal) in 100 µl of low serum Opti-MEM medium (Invitrogen). Uninfected control animals were sham-inoculated with 100 µl of Opti-MEM. For another set of studies, mice were infected intranasally with RSV at 5×108 pfu per animal and the survival of infected mice was followed for 18 d.
TUNEL assay and MPO assay
Formalin fixed lungs were stained using the in situ TUNEL assay kit from Promega. Lung neutrophil content was assessed by measuring myeloperoxidase (MPO) activity 35, 36.
Generation of NOD2 mutants
The NOD2 cDNA was cloned into pcDNA6-Myc-His vector (Invitrogen) and deletion mutants of NOD2 were constructed by PCR.
Treatment with synthetic and viral ssRNA
Cells were treated 1µg per ml of synthetic ssRNA that is already conjugated with trasfection reagent (ssRNA40-LyoVec from Invivogen). For isolation of viral ssRNA, purified RSV virion particles were centrifuged for 4h at 28,000 rpm using SW32Ti rotor. The ssRNA genome was isolated from the viral pellet by using the RNeasy minikit. Cells were transfected with viral ssRNA using Lipofectamine 2000 from Invitrogen.
Isolation of MEFs and macrophages
Alveolar macrophages were collected by centrifuging bronchoalveolar lavage fluid at 2500 rpm for 10 min at 4°C. After washing the cell pellet was seeded in a 24-well plate. MEFs were prepared as described previously 50. Bone marrow-derived macrophages were obtained from femurs and tibias of wild-type and NOD2-KO mice and were cultured for 6–8 days.
ELISA
ELISA was performed using human or mouse IFN-β specific ELISA kits (PBL interferon source).
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health grants AI069062 (to S.B.), CA129246 (to S.B.), American Lung Association National Biomedical Research Grant-RG-49629-N (to S.B.), and AI067716 (to P.H.D). A.S. is supported by National Institutes of Health training grant T32-DE14318. We would like to thanks K. Li (University of Tennessee Health Science Center) for providing several reagents used in the study. We would also like to thank A. Garcia-Sastre (Mt. Sinai School of Medicine, New York, NY) and Z. J. Chen (Howard Hughes Medical Institute and Department of Molecular Biology, University of Texas Southwestern Medical Center, Dallas, TX) for providing influenza A virus and MAVS-KO MEFs, respectively. Confocal images were generated in the University Core Optical Imaging Facility and the FACS analysis was performed by K. Moncada Gorena and C. Thomas in the Flow Cytometry Core Facility (supported by NIH grants P30 CA54174-San Antonio Cancer Institute, P30 AG013319-Nathan Shock Center and P01AG19316).
REFERENCES
- 1.Kawai T, Akira S. Innate immune recognition of viral infection. Nat. Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 2.Bose S, Banerjee AK. Innate immune response against nonsegmented negative strand RNA viruses. J. Interferon. Cytokine. Res. 2003;23:401–412. doi: 10.1089/107999003322277810. [DOI] [PubMed] [Google Scholar]
- 3.Stark GR, Kerr IM, Williams BRG, Silverman RH, Schreiber RD. How cells respond to interferons. Annu. Rev. Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 4.Uematsu S, Akira S. Toll-like receptors and Type I interferons. J. Biol. Chem. 2007;282:15319–15323. doi: 10.1074/jbc.R700009200. [DOI] [PubMed] [Google Scholar]
- 5.O'Neill LA. How Toll-like receptors signal: what we know and what we don't know. Curr. Opin. Immunol. 2006;18:3–9. doi: 10.1016/j.coi.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Basler CF, Garcia-Sastre A. Sensing RNA virus infections. Nat. Chem. Biol. 2007;3:20–21. doi: 10.1038/nchembio0107-20. [DOI] [PubMed] [Google Scholar]
- 7.Martinon F, Tschopp J. NLRs join TLRs as innate sensors of pathogens. Trends. Immunol. 2005;26:447–454. doi: 10.1016/j.it.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Fritz JH, Ferrero RL, Philpott DJ, Girardin SE. Nod-like proteins in immunity, inflammation and disease. Nat. Immunol. 2006;7:1250–1257. doi: 10.1038/ni1412. [DOI] [PubMed] [Google Scholar]
- 9.Kanneganti TD, Lamkanfi M, Núñez G. Intracellular NOD-like receptors in host defense and disease. Immunity. 2007;27:548–559. doi: 10.1016/j.immuni.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Franchi L, Warner N, Viani K, Nuñez G. Function of Nod-like receptors in microbial recognition and host defense. Immunol. Rev. 2009;227:106–128. doi: 10.1111/j.1600-065X.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore CB, et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- 12.Tattoli I, et al. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-kappaB and JNK pathways by inducing reactive oxygen species production. EMBO. Rep. 2008;9:293–300. doi: 10.1038/sj.embor.7401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kota S, et al. Role of human beta defensin-2 during tumor necrosis factor-alpha/NF-kappa B mediated innate anti-viral response against human respiratory syncytial virus. J. Biol. Chem. 2008;283:22417–22429. doi: 10.1074/jbc.M710415200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voss E, et al. NOD2/CARD15 mediates induction of the antimicrobial peptide human beta-defensin-2. J. Biol. Chem. 2006;281:2005–2011. doi: 10.1074/jbc.M511044200. [DOI] [PubMed] [Google Scholar]
- 15.Hall CB. Respiratory syncytial virus and parainfluenza virus. N. Engl. J. Med. 2001;344:1917–1928. doi: 10.1056/NEJM200106213442507. [DOI] [PubMed] [Google Scholar]
- 16.Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. Respiratory syncytial virus infection in elderly and high-risk adults. N. Engl. J. Med. 2005;352:1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- 17.Bose S, Malur A, Banerjee AK. Polarity of human parainfluenza virus type 3 infection in polarized human lung epithelial A549 cells : Role of microfilament and microtubule. J. Virol. 2001;75:1984–1989. doi: 10.1128/JVI.75.4.1984-1989.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bose S, Kar N, Maitra R, DiDonato JA, Banerjee AK. Temporal activation of NF-κB regulates an interferon-independent innate antiviral response against cytoplasmic RNA viruses. Proc. Natl. Acad. Sci. USA. 2003;100:10890–10895. doi: 10.1073/pnas.1832775100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jamaluddin M, et al. IFN-β mediates coordinate expression of antigen-processing genes in RSV-infected pulmonary epithelial cells. Am. J. Physiol. Lung. Cell. Mol. Physiol. 2001;280:L248–L257. doi: 10.1152/ajplung.2001.280.2.L248. [DOI] [PubMed] [Google Scholar]
- 20.Liu P, et al. Retinoic acid-inducible gene I mediates early antiviral response and Toll-like receptor 3 expression in respiratory syncytial virus-infected airway epithelial cells. J. Virol. 2007;81:1401–1411. doi: 10.1128/JVI.01740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inohara N, Ogura Y, Nuñez G. Nods: a family of cytosolic proteins that regulate the host response to pathogens. Curr. Opin. Microbiol. 2002;5:76–80. doi: 10.1016/s1369-5274(02)00289-8. [DOI] [PubMed] [Google Scholar]
- 22.Franchi L, et al. Intracellular NOD-like receptors in innate immunity, infection and disease. Cell. Microbiol. 2008;10:1–8. doi: 10.1111/j.1462-5822.2007.01059.x. [DOI] [PubMed] [Google Scholar]
- 23.Wathelet MG, Lin CH, Parekh BS, Ronco LV, Howley PM, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol. Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 24.Jafri HS, et al. Respiratory syncytial virus induces pneumonia, cytokine response, airway obstruction, and chronic inflammatory infiltrates associated with long-term airway hyperresponsiveness in mice. J. Infect. Dis. 2004;189:1856–1865. doi: 10.1086/386372. [DOI] [PubMed] [Google Scholar]
- 25.Bolger G, et al. Primary infection of mice with high titer inoculum respiratory syncytial virus: characterization and response to antiviral therapy. Can. J. Physiol. Pharmacol. 2005;83:198–213. doi: 10.1139/y05-007. [DOI] [PubMed] [Google Scholar]
- 26.Guerrero-Plata A, Baron S, Poast JS, Adegboyega PA, Casola A, Garofalo RP. Activity and regulation of alpha interferon in respiratory syncytial virus and human metapneumovirus experimental infections. J. Virol. 2005;79:10190–10199. doi: 10.1128/JVI.79.16.10190-10199.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guerrero-Plata A, Casola A, Garofalo RP. Human metapneumovirus induces a profile of lung cytokines distinct from that of respiratory syncytial virus. J. Virol. 2005;79:14992–14997. doi: 10.1128/JVI.79.23.14992-14997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pletneva LM, Haller O, Porter DD, Prince GA, Blanco JC. Induction of type I interferons and interferon-inducible Mx genes during respiratory syncytial virus infection and reinfection in cotton rats. J. Gen. Virol. 2008;89:261–270. doi: 10.1099/vir.0.83294-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chávez-Bueno S, et al. Respiratory syncytial virus-induced acute and chronic airway disease is independent of genetic background: an experimental murine model. Virol. J. 2005;2:46. doi: 10.1186/1743-422X-2-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castro SM, et al. Antioxidant treatment ameliorates respiratory syncytial virus-induced disease and lung inflammation. Am. J. Respir. Crit. Care Med. 2006;174:1361–1369. doi: 10.1164/rccm.200603-319OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hippenstiel S, Opitz B, Schmeck B, Suttorp N. Lung epithelium as a sentinel and effector system in pneumonia--molecular mechanisms of pathogen recognition and signal transduction. Respir. Res. 2006;7:97. doi: 10.1186/1465-9921-7-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yasui K, et al. Neutrophil-mediated inflammation in respiratory syncytial viral bronchiolitis. Pediatr. Int. 2005;47:190–195. doi: 10.1111/j.1442-200x.2005.02039.x. [DOI] [PubMed] [Google Scholar]
- 33.Wang SZ, Forsyth KD. The interaction of neutrophils with respiratory epithelial cells in viral infection. Respirology. 2000;5:1–10. doi: 10.1046/j.1440-1843.2000.00219.x. [DOI] [PubMed] [Google Scholar]
- 34.Wang SZ, Xu H, Wraith A, Bowden JJ, Alpers JH, Forsyth K. Neutrophils induce damage to respiratory epithelial cells infected with respiratory syncytial virus. Eur. Respir. J. 1998;12:612–618. doi: 10.1183/09031936.98.12030612. [DOI] [PubMed] [Google Scholar]
- 35.Bubeck SS, Cantwell AM, Dube PH. Delayed inflammatory response to primary pneumonic plague occurs in both outbred and inbred mice. Infect. Immun. 2007;75:697–705. doi: 10.1128/IAI.00403-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilmott RW, Kitzmiller JA, Fiedler MA, Stark JM. Generation of a transgenic mouse with lung-specific overexpression of the human interleukin-1 receptor antagonist protein. Am. J. Respir. Cell. Mol. Biol. 1998;18:429–434. doi: 10.1165/ajrcmb.18.3.2983. [DOI] [PubMed] [Google Scholar]
- 37.Welliver TP, et al. Severe human lower respiratory tract illness caused by respiratory syncytial virus and influenza virus is characterized by the absence of pulmonary cytotoxic lymphocyte responses. J. Infect. Dis. 2007;195:1126–1136. doi: 10.1086/512615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 39.Opitz B, et al. Nucleotide-binding Oligomerization Domain Proteins Are Innate Immune Receptors for Internalized Streptococcus pneumoniae. J. Biol. Chem. 2004;279:36426–36432. doi: 10.1074/jbc.M403861200. [DOI] [PubMed] [Google Scholar]
- 40.Matikainen S, et al. Tumor necrosis factor alpha enhances influenza A virus-induced expression of antiviral cytokines by activating RIG-I gene expression. J. Virol. 2006;80:3515–3522. doi: 10.1128/JVI.80.7.3515-3522.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le Goffic R, Pothlichet J, Vitour D, Fujita T, Meurs E, Chignard M, Si-Tahar M. Influenza A virus activates TLR3-dependent inflammatory and RIG-I-dependent antiviral responses in human lung epithelial cells. J. Immunol. 2007;178:3368–3372. doi: 10.4049/jimmunol.178.6.3368. [DOI] [PubMed] [Google Scholar]
- 42.Abbott DW, Yang Y, Hutti JE, Madhavarapu S, Kelliher MA, Cantley LC. Coordinated regulation of Toll-like receptor and NOD2 signaling by K63-linked polyubiquitin chains. Mol. Cell. Biol. 2007;27:6012–6025. doi: 10.1128/MCB.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, et al. Retinoic Acid-Inducible Gene-I Mediates Late Phase Induction of TNF-alpha by Lipopolysaccharide. J. Immunol. 2008;180:8011–8019. doi: 10.4049/jimmunol.180.12.8011. [DOI] [PubMed] [Google Scholar]
- 44.Hornung V, et al. 5'-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 45.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 46.Kanneganti TD, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 47.Kanneganti TD, et al. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J. Biol. Chem. 2006;281:36560–36568. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- 48.Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J. Exp. Med. 2009;206:79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kobayashi K, et al. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature. 2002;416:194–199. doi: 10.1038/416194a. [DOI] [PubMed] [Google Scholar]
- 50.Tominaga K, et al. MRG15 regulates embryonic development and cell proliferation. Mol. Cell. Biol. 2005;25:2924–2937. doi: 10.1128/MCB.25.8.2924-2937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.