Abstract
Tamoxifen continues to be a standard endocrine therapy for the prevention and treatment of estrogen receptor (ER)-positive breast cancer. Tamoxifen can be considered a classic “pro-drug,” requiring metabolic activation to elicit pharmacological activity. CYP2D6 is the rate-limiting enzyme catalyzing the conversion of tamoxifen into metabolites with significantly greater affinity for the ER and greater ability to inhibit cell proliferation. Both genetic and environmental (drug-induced) factors that alter CYP2D6 enzyme activity directly affect the concentrations of the active tamoxifen metabolites and the outcomes of patients receiving adjuvant tamoxifen. The a priori knowledge of the pharmacogenetic variation known to abrogate CYP2D6 enzyme activity may provide a means by which the hormonal therapy of breast cancer can be individualized.
Breast cancer is the most common malignancy among women. Its lifetime risk amounts to a total of 10%,1 and approximately 15–20% of all breast cancers are associated with the occurrence of familial breast and/or ovarian cancer.2 During the past two decades, various high- and low-risk cancer susceptibility genes have been detected, including high-risk susceptibility genes such as breast cancer gene 1 (BRCA1) and breast cancer gene 2 (BRCA2). Although the role of inheritance in breast cancer carcinogenesis is well established, an emerging area of research is pharmacogenetics, a field that studies the role of genetic inheritance in individual variation in drug response and toxicity. Recently, genetic and drug-induced variation in the phase I drug-metabolizing enzyme cytochrome P450 2D6 (CYP2D6) has emerged as an important contributor to the interindividual variability in response after the administration of tamoxifen.
For three decades, tamoxifen has been a standard endocrine therapy for the treatment of ER-positive breast cancer. When administered to women with ER-positive breast cancer for 5 years after surgery, tamoxifen almost halves the annual recurrence rate and reduces the breast cancer mortality rate by one-third in both pre- and post-menopausal women.3 Tamoxifen’s continued importance is reflected by its status as the only hormonal agent approved by the US Food and Drug Administration (FDA) for the prevention of breast cancer,4 the treatment of ductal carcinoma in situ,5 and the treatment of pre-menopausal breast cancer.6
Although tamoxifen is still the accepted standard of care for pre-menopausal breast cancer, aromatase inhibitors (AIs) are now an accepted therapy for post-menopausal breast cancer. In this setting, there are two accepted treatment algorithms: an AI for 5 years7,8 or tamoxifen for 2–3 years followed by an AI for 2–3 years.9–11 Compared with 5 years of tamoxifen, administration of an AI for 5 years reduces the risk of a disease event by 13%, but does not prolong survival.7,12 In contrast, sequencing of hormonal therapy with tamoxifen for 2–3 years followed by an AI reduces the risk of a disease event by 40%9,10 and prolongs survival13,14 compared with 5 years of tamoxifen. For this reason, tamoxifen is still commonly employed for treatment of post-menopausal breast cancer.15
When more than one effective option exists for treating a given disease, there is much interest in identifying biomarkers which can reproducibly identify patients that should preferentially receive or be excluded from a given therapy. Notably, tumors or somatic markers (e.g., ER, HER-2) are not predictive of a preferential response to breast cancer endocrine therapy (e.g., tamoxifen vs. aromatase inhibitor). In this study, we review the pharmacology of tamoxifen biotransformation, the importance of CYP2D6 pharmacogenetics accounting for the variability in exposure to the active tamoxifen metabolites, and evidence to suggest that CYP2D6 can be used as a predictive marker for the individualization of endocrine therapy.
TAMOXIFEN PHARMACOLOGY
The classic understanding of tamoxifen pharmacology has been that the parent drug and its metabolites interact with the ER in both breast and non-breast tissues to produce a complex phenotype of both agonist and antagonist effects. Although investigators observed wide interindividual variability in the concentrations of tamoxifen and its metabolites,16 there was no evidence linking variability in tamoxifen concentrations (or its metabolites) with response or side effects.17,18 However, recent in vitro and in vivo human studies have characterized an important tamoxifen metabolic pathway leading to the formation of active metabolites, which exhibit significantly greater affinity for the ER,19 and greater potency in suppressing cell proliferation (compared to tamoxifen).20,21 Knowledge of the genetic and environmental factors, which contribute to the wide variability in the formation of these active metabolites, has led to a new understanding of tamoxifen pharmacology.
PRIMARY TAMOXIFEN METABOLISM
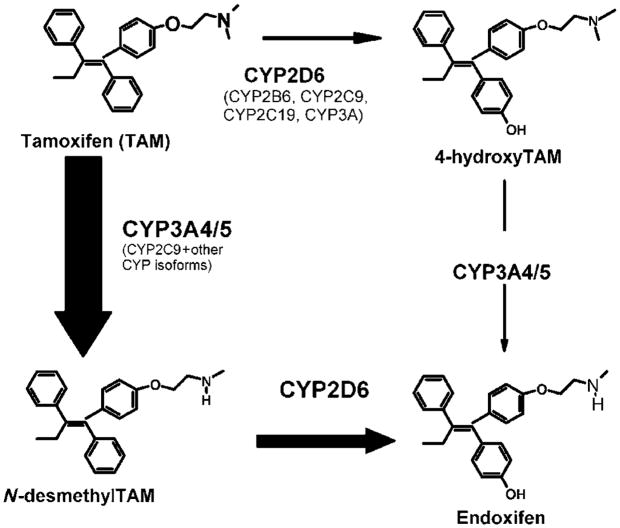
Tamoxifen undergoes extensive hepatic oxidation by the cytochrome P450 (P450) isoforms to several primary and secondary metabolites22–24 with variable potencies toward the ER (see Figure 1). Major tamoxifen metabolites include N-desmethyltamoxifen (NDM), 4-hydroxytamoxifen (4-OH-Tam), tamoxifen-N-oxide, α-hydroxytamoxifen, and N-didesmethyltamoxifen.19,24–27
Figure 1.
Selected transformation pathways of tamoxifen and the main CYP enzymes involved. The relative contribution of each pathway to the overall oxidation of tamoxifen is shown by the thickness of the arrow, and the principal P450 isoforms responsible are highlighted in larger fonts. Modified in part from Jin, Y. et al. J. Natl. Cancer Inst. 97, 30–39 (2005).
NDM, resulting from the CYP3A4/5-mediated catalysis of tamoxifen, is quantitatively the major primary metabolite of tamoxifen and accounts for approximately 92% of primary tamoxifen oxidation.24 In women receiving tamoxifen at a dose of 20 mg/day, plasma steady-state concentrations of tamoxifen and NDM are 362.5 and 654.9 nM, respectively, whereas the steady-state concentrations of 4-OH-Tam are extremely low (9 nM).28
SECONDARY TAMOXIFEN METABOLISM
Using NDM as an intermediary substrate, the secondary metabolism of tamoxifen has recently been more fully elucidated.24,28,29 NDM is predominantly biotransformed to α-hydroxy N-desmethyl-, N-didesmethyl-, and 4-hydroxy-N-desmethyl-tamoxifen (endoxifen). Whereas the biotransformation of NDM to endoxifen is exclusively catalyzed by CYP2D6 (see Figure 1), all other routes of N-desmethyl biotransformation are catalyzed predominantly by the CYP3A subfamily24 (data not shown in Figure 1). Recent clinical studies have demonstrated that common CYP2D6 genetic variation (leading to low or absent CYP2D6 activity), or the inhibition of CYP2D6 enzyme activity significantly lowers the plasma concentrations of endoxifen.19,28 Patients homozygous for a CYP2D6 null allele had significantly lower endoxifen concentrations (mean, 21.9 nM) than patients with one (mean, 64.2 nM) or two (mean, 88.6 nM) CYP2D6 functional alleles.30
ANTIPROLIFERATIVE EFFECTS OF TAMOXIFEN AND ITS METABOLITES
It has been known for many years that 4-OH-Tam31,32 possesses a much higher affinity for ERs and is 30- to 100-fold more potent than tamoxifen in suppressing estrogen-dependent cell proliferation.33,34 For this reason, many considered 4-OH-Tam to be the active metabolite of tamoxifen and it is frequently used to characterize tamoxifen activity in vitro. However, new data suggest that endoxifen has identical properties and potency as 4-OH-Tam, but is present in concentrations up to 10-fold higher than 4-OH-Tam.30
Lien et al.35,36 first identified “metabolite Bx”, or endoxifen, in 1989. However, only recently did Stearns et al.29 publish that endoxifen exhibits potency similar to 4-OH-Tam with respect to ER-binding affinity and suppression of estrogen-dependent cell growth. Subsequent to this work, Flockhart’s group performed a series of studies to further characterize endoxifen pharmacology. These studies confirmed that endoxifen has equivalent potency in vitro similar to 4-OH-Tam in ER-α and ER-β binding,20 in suppression of ER-dependent breast cancer proliferation,20,37 and in global ER-responsive gene expression.38
Recently, Buck et al.21 confirmed these findings through the characterization of the effects of tamoxifen and its metabolites on the antiproliferative transforming growth factor-β signal-transduction pathway in human breast cancer cells. They analyzed the growth inhibitory effect of tamoxifen and its major metabolites, and demonstrated that only 4-OH-Tam and endoxifen had significant antiproliferative activity and were able to induce transforming growth factor-β2 and transforming growth factor-β type II receptor.
CYP2D6 GENOTYPIC HETEROGENEITY
The CYP2D6 enzyme is an important phase I drug enzyme involved in the metabolism of up to 25% of all drugs. More than 48 different drug substrates for this enzyme have been identified, include drugs from the following classes: β-blockers, antidepressants, antiarrhythmics, and antipsychotics.39
The CYP2D6 gene is highly polymorphic, currently with 63 different major alleles known, many of which are associated with increased, decreased, or abolished function of the final gene product. The CYP2D6 phenotypes associated with these different alleles include poor (PM), intermediate (IM), extensive (EM), and ultrarapid (UM) metabolizers. Some of the most common and important variant alleles distributed in different ethnic groups are listed in Table 1, and all variant alleles are presented on the homepage of the human CYP allele nomenclature committee (http://www.imm.ki.se/cypalleles/cyp2d6.htm). Carriers of any two of approximately 20 known null alleles are phenotypic poor metabolizers, representing 7–10% of the European and North American Caucasian population.39 One of the most important functionally altered null variants, among others includes CYP2D6*4 (15–21% in Caucasians). Important alleles associated with reduced enzyme activity include CYP2D6*10 (up to 57% in Asians40) and CYP2D6*17 (20–34% in African and African Americans).41 Individuals at the high end of the activity spectrum (UM) carry gene duplications and multiduplications of functional alleles, which lead to higher CYP2D6 expression and enzyme activity, with relatively low frequency observed in Caucasians and Asians,39 but commonly observed in Ethiopians (up to 30%42).
Table 1.
Major CYP2D6 alleles, effect on enzyme metabolism, and allele frequencies in selected populations
| Allele frequencies (%) |
||||
|---|---|---|---|---|
| Major variant alleles | Consequence | Caucasians | Asians | Black Africans |
| CYP2D6*2xn | Increased enzyme activity | 1–5 | 0–2 | 2 |
| CYP2D6*4 | Inactive enzyme | 12–21 | 1 | 2 |
| CYP2D6*5 | No enzyme | 27 | 6 | 4 |
| CYP2D6*10 | Decreased activity | 1–2 | 51 | 6 |
| CYP2D6*17 | Decreased activity | 0 | 0 | 20–35 |
| CYP2D6*41 | Decreased activity | 8–10 | 0–2 | 11–14 |
Modified from Ingelman-Sundberg, M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J. 5, 6–13 (2005).
Borges et al.30 recently reported an updated analysis of their prospective tamoxifen pharmacology cohort by assessing the combined effect of 33 different CYP2D6 alleles on the plasma concentrations of tamoxifen and its metabolites. Notable in this analysis was that patients heterozygous for a reduced (e.g., *10) and null (e.g., *4) allele (i.e., individuals typically classified as IM) had similar endoxifen concentrations compared to PM, indicating the importance of comprehensive CYP2D6 genotyping to account for the variability in endoxifen plasma concentrations. In this same study, patients with multiple copies of any functional allele (UM) had higher mean endoxifen concentrations compared to patients who did not carry a UM allele.30
TRANSLATION OF CYP2D6 PHARMACOGENETICS IN TAMOXIFEN-TREATED BREAST CANCER
We were the first to show evidence that genetic variability in CYP2D6 may affect the treatment outcomes of patients receiving tamoxifen.43 We performed a retrospective analysis of a prospective adjuvant tamoxifen trial (NCCTG 89-30-52) in post-menopausal women with surgically resected ER-positive breast cancer (stages I–III) to determine the role of genetic variation in CYP2D6.43 Because of the difficulty in amplifying DNA from formalin-fixed paraffin-embedded tissue, only the CYP2D6*4 (the most common null allele contributing to the PM state44) and the CYP2D6*6 (an infrequent null allele) were studied. No *6 variants were detected. Of the 256 women enrolled in the tamoxifen-only arm, 223 paraffin-embedded tissue blocks were available for DNA extraction and CYP2D6*4 was amplified in 190. Women with the CYP2D6*4/*4 genotype had shorter relapse-free time and worse disease-free survival compared to women with either one or no *4 alleles (log rank P = 0.030 and P = 0.020, respectively). In a multivariate analysis, *4/*4 patients still tended to have worse relapse-free survival (RFS) (hazard ratio (HR) = 1.86, P = 0.089).43 Additionally, we observed differences in the incidence of moderate or severe hot flashes in patients homozygous for the *4 allele (0%) versus those with one or no *4 alleles (20%; P = 0.06). These data supported the hypothesis that the CYP2D6-mediated formation of the potent antiestrogens may lead to differences not only in response but also in side effects.
CYP2D6 INHIBITORS AND THEIR EFFECT ON TAMOXIFEN ACTIVATION
The newer antidepressant drugs, such as the selective serotonin reuptake inhibitors and the serotonin and norepinephrine reuptake inhibitors, are commonly administered with tamoxifen in breast cancer patients to treat depression or alleviate hot flashes.19,45–48 Notably, Jin et al.28 demonstrated that the co-administration of fluoxetine and/or paroxetine can convert a CYP2D6 extensive metabolizer to a phenotypic PM, as demonstrated by a reduction in plasma endoxifen concentrations to a similar level as genotypic PM. Many other clinically important drugs have been reported to inhibit the CYP2D6 enzyme system, but their effects on tamoxifen metabolism and treatment outcome have not been carefully studied.
CYP2D6 METABOLISM AND TAMOXIFEN TREATMENT OUTCOME
We recently updated our analysis of the NCCTG 89-30-52 tamoxifen trial by assessing the combined effect of CYP2D6 genetic variation and drug-induced inhibition of the enzyme system on breast cancer outcomes.49 In this analysis, we reviewed the medical records at each randomizing site to determine whether potent (fluoxetine and paroxetine) or weak/moderate (sertraline, cimetidine, amiodarone, doxepin, ticlopidine, and haloperidol) CYP2D6 inhibitors were co-prescribed with tamoxifen.
We defined “extensive” metabolizers as patients without a CYP2D6*4 allele who were not prescribed a CYP2D6 inhibitor. “Decreased” CYP2D6 metabolism was defined as patients with one or two *4 alleles, or the confirmation that a CYP2D6 inhibitor was co-administered with tamoxifen (regardless of genotype).
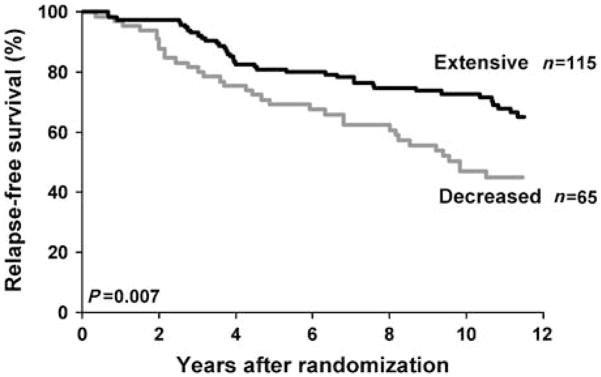
Medication history was available in 225 out of 256 eligible patients, and we confirmed that 13 patients (6%) were co-prescribed a CYP2D6 inhibitor with a median duration of 2–3 years. Knowledge of both medication history and CYP2D6 genotype (available in 180 patients) allowed us to classify patients as having “extensive” (n = 115) or “decreased” (n = 65) CYP2D6 metabolism. In a multivariate analysis, patients with “decreased” metabolism had significantly shorter time to recurrence (P = 0.034; HR = 1.91) and worse RFS (P = 0.017; HR = 1.74) relative to patients with “extensive” metabolism (Figure 2).
Figure 2.
Kaplan–Meier estimates of RFS based on CYP2D6 metabolism (extensive vs. decreased). Reprinted with permission from Goetz, M.P. et al. Breast Cancer Res. Treat. 101, 113–121 (2007).
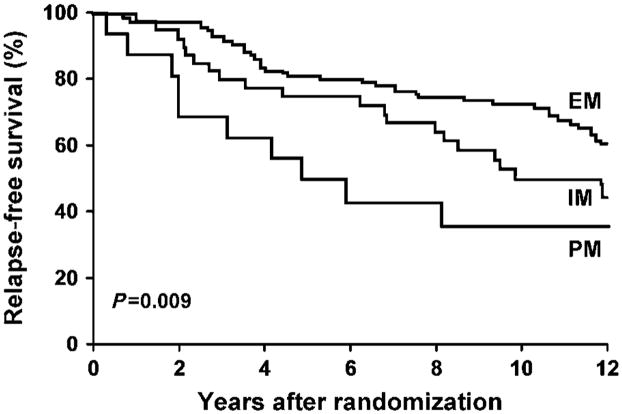
Because the cohort of patients with “decreased” metabolism comprised both phenotypic PM and IM, we used the in vivo data regarding the combined effect of CYP2D6 genotype and CYP2D6 inhibitors on endoxifen plasma concentrations28,30 to classify patients with “decreased” CYP2D6 metabolism into “intermediate” or “poor” metabolizer categories. Intermediate metabolizers were defined as patients (1) heterozygous for the *4 allele (*4/Wt) without co-prescription of a CYP2D6 inhibitor, or (2) no *4 alleles (Wt/Wt) with co-administration of a weak/moderate inhibitor. Poor metabolizers were defined as (1) patients homozygous for the *4 allele (*4/*4), (2) *4/Wt and co-administration of any CYP2D6 inhibitor, or (3) Wt/Wt and co-administration of a potent inhibitor. As expected, Cox’s modeling demonstrated a stepwise decrement in breast cancer outcomes based on the extent of impairment of CYP2D6 metabolism. Whereas women classified as “intermediate” metabolizers tended to exhibit worse RFS (HR = 1.63, P = 0.075), patients classified as “poor metabolizers” had the worst outcome, with significantly worse RFS (HR = 2.69, P = 0.005)49 (Figure 3).
Figure 3.
Kaplan–Meier estimates of RFS based on metabolizer status (extensive, intermediate, or poor). Reprinted with permission from Goetz, M.P. et al. Breast Cancer Res. Treat. 101, 113–121 (2007).
OTHER DATA IN SUPPORT OF CYP2D6 AS A MARKER OF TREATMENT RESPONSE
Recently, our findings were validated by Schroth et al.,50 who demonstrated the significance of CYP2D6 and CYP2C19 genetic variation in a retrospective cohort of 206 tamoxifen-treated and 280 tamoxifen-untreated breast cancer patients. Tamoxifen-treated women with CYP2D6 alleles associated with absent or reduced enzyme function (*4, *5, *10, *41) had significantly more recurrences, shorter relapse-free time (HR = 2.24; 95% CI (confidence interval) = 1.16–4.33; P = 0.02), and worse event-free survival (HR = 1.89; 95% CI = 1.10–3.25; P = 0.02) compared to carriers of functional alleles.50 As expected, there was no association between treatment outcome and CYP2D6 genotype in the cohort of patients that did not receive tamoxifen.
Two additional retrospective studies have reported no association between CYP2D6 genotype and the outcomes of tamoxifen-treated patients.51,52 Nowell et al.53 demonstrated no association between CYP2D6*4 genotype and overall survival in a retrospective cohort of 162 patients treated with tamoxifen for ER-positive and negative disease. Additionally, Wegman et al.52,54 demonstrated no association between the CYP2D6*4 allele with distant disease-free survival or overall survival.
The reasons for the conflicting outcomes of these studies are likely due to one or more of the following: (1) the lack of central testing of ER, resulting in the probable inclusion of ER-negative patients; (2) use of retrospectively assembled cohorts biased toward patients with available tumor specimens; (3) the inclusion of patients receiving varying lengths (2–5 years) and varying doses (20–40 mg) of tamoxifen; (4) the potential confounder of not accounting for additional treatments (e.g., chemotherapy), known to impact outcome; and (5) not accounting for CYP2D6 inhibitors.
TAMOXIFEN AND FDA RELABELING
On 18 October 2006, an FDA Advisory Committee met to discuss the tamoxifen research findings to date and to make a recommendation regarding a label change. The FDA committee considered the following question: “Does the clinical evidence demonstrate that post-menopausal women with ER-positive breast cancer who are CYP2D6-poor metabolizers (by genotype or drug interaction) are at increased risk for breast cancer recurrence?” The consensus of the Subcommittee was that the label should be updated to reflect the increased risk for breast cancer along with the mechanistic data presented. However, regarding the recommendation to test women, the committee did not reach a consensus. Some members believed that the genetic test should be recommended, whereas others believed that it should be mentioned in the label as an option for discussion between the health-care provider and patient. However, the majority indicated that it should be included in an appropriate section of the package insert.
INTEGRATION OF CYP2D6 PHARMACOGENETICS INTO THE CLINIC
Our data suggest that CYP2D6 may provide a means by which the hormonal therapy of breast cancer can be individualized. In addition to recommending the discontinuation of potent CYP2D6 inhibitors, it is our practice to fully inform patients of the importance of CYP2D6 genotype as a predictor of tamoxifen treatment outcome, and to consider CYP2D6 genotyping in settings wherein alternative therapies are known to be equivalent or superior to tamoxifen monotherapy (e.g., post-menopausal adjuvant breast cancer). In this setting, based on our data43,49 and the data by Schroth et al.,50 genotyping should encompass at least the common PM (e.g., *3–6) and IM (*9, *10, *17, *41) alleles, and PM and IM should be considered for alternative therapy. At this time, the clinical utility of measuring plasma endoxifen levels as a surrogate for CYP2D6 phenotype is unknown, but is an important area of ongoing research. An additional further area of research is needed to determine the optimal sequence of hormonal therapy (tamoxifen for some duration followed by an AI) for patients considered to be CYP2D6 extensive metabolizer or UM. For node-positive, ER-positive patients who are at high risk of recurrence, CYP2D6 extensive metabolizer and UM might be optimally treated with tamoxifen for upwards of 4–5 years followed by 5 years of an AI, as was established in the MA17 trial.11 This strategy would allow for the administration of therapy for a longer duration of time, which is currently not available if AIs are administered upfront for 5 years.7
For pre-menopausal breast cancer, there are no published data regarding CYP2D6 genotype and treatment outcomes. In the setting of chemoprevention, a small case–control study from the Italian chemoprevention group demonstrated a higher likelihood that tamoxifen-treated women homozygous for the *4 allele would develop breast cancer compared with those who did not carry a *4 allele.55 These data are considered preliminary, and further research is needed in pre-menopausal women before using CYP2D6 genotype to exclude patients from tamoxifen.
SUMMARY OF TAMOXIFEN PHARMACOLOGY
Tamoxifen can be considered a classic “pro-drug,” requiring metabolic activation to elicit pharmacological activity. Importantly, CYP2D6 appears to be the rate-limiting enzyme converting the pharmacologically inactive metabolites (tamoxifen and NDM) to endoxifen, and additionally contributes to the formation of 4-OH-Tam (from tamoxifen). Our findings suggest that both genetic and environmental (drug-induced) factors that alter CYP2D6 enzyme activity affect tamoxifen treatment outcomes. In the era of third-generation AI, CYP2D6 may provide a means by which the hormonal therapy of breast cancer can be individualized.
Acknowledgments
Supported in part by Paul Calabresi Program in Clinical-Translational Research at Mayo Clinic (MPG), CA 90628; Career Development Award from Mayo Cancer Center Breast Cancer SPORE (MPG), CA 116201; and Mayo Comprehensive Cancer Center Grant (MMA), CA 15083.
Footnotes
CONFLICT OF INTEREST
Drs Goetz and Ames are co-inventors on a pending patent application, which is owned by the Mayo Foundation for Medical Education and Research and is related to CYP2D6 and tamoxifen. If Mayo is successful in commercializing the technology, Mayo and the inventors will stand to receive a royalty.
References
- 1.Collaborative Group on Hormonal Factors in Breast Cancer. Familial breast cancer: collaborative reanalysis of individual data from 52 epidemiological studies including 58,209 women with breast cancer and 101,986 women without the disease. Lancet. 2001;358:1389–1399. doi: 10.1016/S0140-6736(01)06524-2. [DOI] [PubMed] [Google Scholar]
- 2.Easton DF, Bishop DT, Ford D, Crockford GP. Genetic linkage analysis in familial breast and ovarian cancer: results from 214 families. The breast cancer linkage consortium. Am J Hum Genet. 1993;52:678–701. [PMC free article] [PubMed] [Google Scholar]
- 3.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) Effects of chemotherapy hormonal therapy for early breast cancer on recurrence 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 4.Fisher B, et al. Tamoxifen for the prevention of breast cancer: current status of the National Surgical Adjuvant Breast and Bowel Project P-1 study. J Natl Cancer Inst. 2005;97:1652–1662. doi: 10.1093/jnci/dji372. [DOI] [PubMed] [Google Scholar]
- 5.Fisher B, et al. Tamoxifen, radiation therapy, or both for prevention of ipsilateral breast tumor recurrence after lumpectomy in women with invasive breast cancers of one centimeter or less. J Clin Oncol. 2002;20:4141–4149. doi: 10.1200/JCO.2002.11.101. [DOI] [PubMed] [Google Scholar]
- 6.Colleoni M, et al. Tamoxifen after adjuvant chemotherapy for premenopausal women with lymph node-positive breast cancer: International Breast Cancer Study Group Trial 13–93. J Clin Oncol. 2006;24:1332–1341. doi: 10.1200/JCO.2005.03.0783. [DOI] [PubMed] [Google Scholar]
- 7.Howell A, et al. Results of the ATAC (Arimidex, Tamoxifen, Alone or in Combination) trial after completion of 5 years’ adjuvant treatment for breast cancer. Lancet. 2005;365:60–62. doi: 10.1016/S0140-6736(04)17666-6. [DOI] [PubMed] [Google Scholar]
- 8.Thurlimann B, et al. A comparison of letrozole and tamoxifen in postmenopausal women with early breast cancer. N Engl J Med. 2005;353:2747–2757. doi: 10.1056/NEJMoa052258. [DOI] [PubMed] [Google Scholar]
- 9.Coombes RC, et al. A randomized trial of exemestane after two to three years of tamoxifen therapy in postmenopausal women with primary breast cancer. N Engl J Med. 2004;350:1081–1092. doi: 10.1056/NEJMoa040331. [DOI] [PubMed] [Google Scholar]
- 10.Jakesz R, et al. Switching of postmenopausal women with endocrine-responsive early breast cancer to anastrozole after 2 years’ adjuvant tamoxifen: combined results of ABCSG trial 8 and ARNO 95 trial. Lancet. 2005;366:455–462. doi: 10.1016/S0140-6736(05)67059-6. [DOI] [PubMed] [Google Scholar]
- 11.Goss PE, et al. Randomized trial of letrozole following tamoxifen as extended adjuvant therapy in receptor-positive breast cancer: updated findings from NCIC CTG MA.17. J Natl Cancer Inst. 2005;97:1262–1271. doi: 10.1093/jnci/dji250. [DOI] [PubMed] [Google Scholar]
- 12.Coates AS, et al. Five years of letrozole compared with tamoxifen as initial adjuvant therapy for postmenopausal women with endocrine-responsive early breast cancer: update of study BIG 1–98. J Clin Oncol. 2007;25:486–492. doi: 10.1200/JCO.2006.08.8617. [DOI] [PubMed] [Google Scholar]
- 13.Jonat W, et al. Effectiveness of switching from adjuvant tamoxifen to anastrozole in postmenopausal women with hormone-sensitive early-stage breast cancer: a meta-analysis. Lancet Oncol. 2006;7:991–996. doi: 10.1016/S1470-2045(06)70948-2. [DOI] [PubMed] [Google Scholar]
- 14.Coombes RC, et al. Survival and safety of exemestane versus tamoxifen after 2–3 years’ tamoxifen treatment (Intergroup Exemestane Study): a randomized controlled trial. Lancet. 2007;369:559–570. doi: 10.1016/S0140-6736(07)60200-1. [DOI] [PubMed] [Google Scholar]
- 15.Winer EP, et al. American Society of Clinical Oncology technology assessment on the use of aromatase inhibitors as adjuvant therapy for postmenopausal women with hormone receptor-positive breast cancer: status report 2004. J Clin Oncol. 2005;23:619–629. doi: 10.1200/JCO.2005.09.121. [DOI] [PubMed] [Google Scholar]
- 16.Ingle JN, et al. Evaluation of tamoxifen plus letrozole with assessment of pharmacokinetic interaction in postmenopausal women with metastatic breast cancer. Clin Cancer Res. 1999;5:1642–1649. [PubMed] [Google Scholar]
- 17.Bratherton DG, et al. A comparison of two doses of tamoxifen (Nolvadex) in postmenopausal women with advanced breast cancer: 10 versus 20 mg bd. Br J Cancer. 1984;50:199–205. doi: 10.1038/bjc.1984.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacCallum J, et al. Concentrations of tamoxifen and its major metabolites in hormone responsive and resistant breast tumours. Br J Cancer. 2000;82:1629–1635. doi: 10.1054/bjoc.2000.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stearns V, et al. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst. 2003;95:1758–1764. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 20.Johnson MD, et al. Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat. 2004;85:151–159. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]
- 21.Buck MR, Coller JK, Murdter TE, Eichelbaum M, Knabbe C. TGFb2 and TbRII are valid molecular biomarkers for the antiproliferative effects of tamoxifen and tamoxifen metabolites in breast cancer cells. Breast Cancer Res Treat. 2007 doi: 10.1007/s10549-007-9526-7. in press. [DOI] [PubMed] [Google Scholar]
- 22.Lonning PE, Lien EA, Lundgren S, Kvinnsland S. Clinical pharmacokinetics of endocrine agents used in advanced breast cancer. Clin Pharmacokinet. 1992;22:327–358. doi: 10.2165/00003088-199222050-00002. [DOI] [PubMed] [Google Scholar]
- 23.Crewe HK, Ellis SW, Lennard MS, Tucker GT. Variable contribution of cytochromes P450 2D6, 2C9 and 3A4 to the 4-hydroxylation of tamoxifen by human liver microsomes. Biochem Pharmacol. 1997;53:171–178. doi: 10.1016/s0006-2952(96)00650-8. [DOI] [PubMed] [Google Scholar]
- 24.Desta Z, Ward BA, Soukhova NV, Flockhart DA. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther. 2004 doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- 25.Crewe HK, Notley LM, Wunsch RM, Lennard MS, Gillam EM. Metabolism of tamoxifen by recombinant human cytochrome P450 enzymes: formation of the 4-hydroxy, 4′-hydroxy and N-desmethyl metabolites and isomerization of trans-4-hydroxytamoxifen. Drug Metab Dispos. 2002;30:869–874. doi: 10.1124/dmd.30.8.869. [DOI] [PubMed] [Google Scholar]
- 26.Lee KH, Ward BA, Desta Z, Flockhart DA, Jones DR. Quantification of tamoxifen and three metabolites in plasma by high-performance liquid chromatography with fluorescence detection: application to a clinical trial. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;791:245–253. doi: 10.1016/s1570-0232(03)00218-6. [DOI] [PubMed] [Google Scholar]
- 27.Lien EA, Solheim E, Ueland PM. Distribution of tamoxifen and its metabolites in rat and human tissues during steady-state treatment. Cancer Res. 1991;51:4837–4844. [PubMed] [Google Scholar]
- 28.Jin Y, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97:30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 29.Stearns V, Beebe KL, Iyengar M, Dube E. Paroxetine controlled release in the treatment of menopausal hot flashes: a randomized controlled trial. JAMA. 2003;289:2827–2834. doi: 10.1001/jama.289.21.2827. [DOI] [PubMed] [Google Scholar]
- 30.Borges S, et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin Pharmacol Ther. 2006;80:61–74. doi: 10.1016/j.clpt.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 31.Jordan VC, Collins MM, Rowsby L, Prestwich G. A monohydroxylated metabolite of tamoxifen with potent antioestrogenic activity. J Endocrinol. 1977;75:305–316. doi: 10.1677/joe.0.0750305. [DOI] [PubMed] [Google Scholar]
- 32.Jordan VC. Metabolites of tamoxifen in animals and man: identification, pharmacology and significance. Breast Cancer Res Treat. 1982;2:123–138. doi: 10.1007/BF01806449. [DOI] [PubMed] [Google Scholar]
- 33.Borgna JL, Rochefort H. Hydroxylated metabolites of tamoxifen are formed in vivo and bound to estrogen receptor in target tissues. J Biol Chem. 1981;256:859–868. [PubMed] [Google Scholar]
- 34.Robertson DW, Katzenellenbogen JA, Long DJ, Rorke EA, Katzenellenbogen BS. Tamoxifen antiestrogens. A comparison of the activity, pharmacokinetics, and metabolic activation of the cis and trans isomers of tamoxifen. J Steroid Biochem. 1982;16:1–13. doi: 10.1016/0022-4731(82)90137-6. [DOI] [PubMed] [Google Scholar]
- 35.Lien EA, Solheim E, Lea OA, Lundgren S, Kvinnsland S, Ueland PM. Distribution of 4-hydroxy-N-desmethyltamoxifen and other tamoxifen metabolites in human biological fluids during tamoxifen treatment. Cancer Res. 1989;49:2175–2183. [PubMed] [Google Scholar]
- 36.Lien EA, Anker G, Lonning PE, Solheim E, Ueland PM. Decreased serum concentrations of tamoxifen and its metabolites induced by aminoglutethimide. Cancer Res. 1990;50:5851–5857. [PubMed] [Google Scholar]
- 37.Lim YC, Desta Z, Flockhart DA, Skaar TC. Endoxifen (4-hydroxy-N-desmethyl-tamoxifen) has anti-estrogenic effects in breast cancer cells with potency similar to 4-hydroxytamoxifen. Cancer Chemother Pharmacol. 2005;55:471–478. doi: 10.1007/s00280-004-0926-7. [DOI] [PubMed] [Google Scholar]
- 38.Lim YC, et al. Endoxifen, a secondary metabolite of tamoxifen, and 4-OH-tamoxifen induce similar changes in global gene expression patterns in MCF-7 breast cancer cells. J Pharmacol Exp Ther. 2006;318:503–512. doi: 10.1124/jpet.105.100511. [DOI] [PubMed] [Google Scholar]
- 39.Ingelman-Sundberg M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J. 2005;5:6–13. doi: 10.1038/sj.tpj.6500285. [DOI] [PubMed] [Google Scholar]
- 40.Sistonen J, Sajantila A, Lao O, Corander J, Barbujani G, Fuselli S. CYP2D6 worldwide genetic variation shows high frequency of altered activity variants and no continental structure. Pharmacogenet Genomics. 2007;17:93–101. doi: 10.1097/01.fpc.0000239974.69464.f2. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez-Antona C, Ingelman-Sundberg M. Cytochrome P450 pharmacogenetics and cancer. Oncogene. 2006;25:1679–1691. doi: 10.1038/sj.onc.1209377. [DOI] [PubMed] [Google Scholar]
- 42.Aklillu E, Persson I, Bertilsson L, Johansson I, Rodrigues F, Ingelman-Sundberg M. Frequent distribution of ultrarapid metabolizers of debrisoquine in an Ethiopian population carrying duplicated and multiduplicated functional CYP2D6 alleles. J Pharmacol Exp Ther. 1996;278:441–446. [PubMed] [Google Scholar]
- 43.Goetz MP, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol. 2005;23:9312–9318. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 44.Sachse C, Brockmoller J, Bauer S, Roots I. Cytochrome P450 2D6 variants in a Caucasian population: allele frequencies and phenotypic consequences. Am J Hum Genet. 1997;60:284–295. [PMC free article] [PubMed] [Google Scholar]
- 45.Loprinzi CL, et al. Venlafaxine in management of hot flashes in survivors of breast cancer: a randomised controlled trial. Lancet. 2000;356:2059–2063. doi: 10.1016/S0140-6736(00)03403-6. [DOI] [PubMed] [Google Scholar]
- 46.Loprinzi CL, et al. Phase III evaluation of fluoxetine for treatment of hot flashes. J Clin Oncol. 2002;20:1578–1583. doi: 10.1200/JCO.2002.20.6.1578. [DOI] [PubMed] [Google Scholar]
- 47.Kimmick GG, Lovato J, McQuellon R, Robinson E, Muss HB. Randomized, double-blind, placebo-controlled, crossover study of sertraline (Zoloft) for the treatment of hot flashes in women with early stage breast cancer taking tamoxifen. Breast J. 2006;12:114–122. doi: 10.1111/j.1075-122X.2006.00218.x. [DOI] [PubMed] [Google Scholar]
- 48.Barton DL, et al. Pilot evaluation of citalopram for the relief of hot flashes. J Support Oncol. 2003;1:47–51. [PubMed] [Google Scholar]
- 49.Goetz MP, et al. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat. 2007;101:113–121. doi: 10.1007/s10549-006-9428-0. [DOI] [PubMed] [Google Scholar]
- 50.Schroth W, et al. Breast cancer treatment outcome with adjuvant tamoxifen in relation to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol. doi: 10.1200/JCO.2007.12.2705. in press. [DOI] [PubMed] [Google Scholar]
- 51.Nowell S, et al. Association between sulfotransferase 1A1 genotype and survival of breast cancer patients receiving tamoxifen therapy. J Natl Cancer Inst. 2002;94:1635–1640. doi: 10.1093/jnci/94.21.1635. [DOI] [PubMed] [Google Scholar]
- 52.Wegman P, et al. Genotype of metabolic enzymes and the benefit of tamoxifen in postmenopausal breast cancer patients. Breast Cancer Res. 2005;7:R284–R290. doi: 10.1186/bcr993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nowell SA, et al. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res Treat. 2005;91:249–258. doi: 10.1007/s10549-004-7751-x. [DOI] [PubMed] [Google Scholar]
- 54.Wegman P, Elingarami S, Carstensen J, Stal O, Nordenskjold B, Wingren S. Genetic variants of CYP3A5, CYP2D6, SULT1A1, UGT2B15 and tamoxifen response in postmenopausal patients with breast cancer. Breast Cancer Res. 2007;9:R7. doi: 10.1186/bcr1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonanni B, et al. Polymorphism in the CYP2D6 tamoxifen-metabolizing gene influences clinical effect but not hot flashes: data from the Italian Tamoxifen Trial. J Clin Oncol. 2006;24:3708–3709. doi: 10.1200/JCO.2006.06.8072. author reply 3709. [DOI] [PubMed] [Google Scholar]