Summary
Background
A common feature of memory and its underlying synaptic plasticity is that each can be dissected into short-lived forms involving modification or trafficking of existing proteins and long-term forms that require new gene expression. An underlying assumption of this cellular view of memory consolidation is that these different mechanisms occur within a single neuron. At the neuro-anatomical level, however, different temporal stages of memory can engage distinct neural circuits, a notion that has not been conceptually integrated with the cellular view.
Results
We have investigated this issue in the context of aversive Pavlovian olfactory memory in Drosophila. Previous studies have demonstrated a central role for cAMP signaling in the mushroom body (MB). The Ca++ responsive adenylyl cyclase rutabaga is believed to be a coincidence detector in γ neurons, one of the three principle classes of MB Kenyon cells. We are able to separately restore short-term or long-term memory to a rutabaga mutant with expression of rutabaga in different subsets of MB neurons.
Conclusions
Our findings suggest a model in which the learning experience initiates two parallel associations: a short-lived trace in MB γ neurons, and a long-lived trace in α/β neurons.
Introduction
Memory consolidation and the underlying synaptic plasticity each have been dissected into short, intermediate and long-term forms [1-3]. Short-term plasticity generally involves modification of pre-existing proteins whereas long-lasting plasticity and memory involve recruitment of a cascade of new gene expression [4-6]. This cellular view is consistent with the idea that both short- and long-lived modifications occur sequentially in the same set of neurons. In contrast, anatomical lesions suggest a dissection of temporal phases of memory into different circuits [7-11].
We have investigated the relationship between biochemical signaling and circuit function in memory consolidation using a Pavlovian olfactory task in Drosophila. We used cell type restricted expression of the rutabaga adenylyl cyclase, which is believed to be a major coincidence detector for this task, to map the spatial requirements for each temporal phase of memory.
The cAMP signaling cascade has been shown to play a key and conserved role in memory formation [12-14]. In Drosophila, this has been investigated in most detail in the context of an aversive Pavlovian olfactory conditioning assay [15]. Genetic experiments have revealed a clear role in memory for many components of this pathway, from G-protein coupled signaling at the membrane to CREB activation in the nucleus [16, 17]. A wide variety of experiments indicate that cAMP signaling in a neural structure called the mushroom bodies (MBs) plays a central role and may be sufficient at least for short-term memory (STM).
MBs are paired neuropils located in the dorsal protocerebrum of many insect brains [18]. The MB Kenyon cell axons form a bundle that bifurcates into several distinct lobes that contain most of the axon terminals. Importantly, the Drosophila MBs consist of at least three major classes of Kenyon cells whose axonal branches occupy distinct subsets of lobes [19], the α/β neurons, α’/β’ neurons, and the γ lobe neurons.
Multiple components of the cAMP signaling pathway have been shown to be expressed at elevated levels in MBs [16, 17, 20, 21]. In the case of the rutabaga adenylyl cyclase, expression is elevated in MBs [22] and transgenic expression in MBs of a rutabaga+ cDNA is sufficient to rescue the learning defect of the rutabaga mutant [23-25]. Moreover, expression just in the γ lobe subset of Kenyon cells is sufficient to restore short-term memory [23, 26]. In contrast, expression in the α/β subset of MB neurons has been reported to have no effect or only a modest effect on short-term memory (depending on odor combinations used during training) [26]. Together, the data support the hypothesis that odor-shock associations occur largely in MB γ lobe neurons. The cellular notion of memory consolidation would therefore suggest that long-term memory (LTM) might involve cAMP signaling onto CREB, within γ lobe neurons. But this notion is at odds with two recent findings. First, LTM has been reported to require NMDA receptor function in a different neural structure, the R4m neurons of the ellipsoid body [27]. Second, spatially restricted expression of a CREB repressor [28] specifically in α/β MB neurons inhibits an associative increase in calcium influx and blocks memory [29].
We have investigated the process of memory consolidation at the circuit level by expressing a rutabaga+ cDNA in each of the three major subsets of mushroom body neurons in animals that were otherwise mutant for the rutabaga gene. We then assayed the ability of spatially restricted expression of rutabaga to support each of the temporal stages of memory consolidation. Using this approach, we were able to independently restore either STM or LTM performance to a rutabaga mutant animal depending on the sub-type of MB neurons in which we express the transgene.
Our findings suggest that the learning experience initiates a rapidly formed but short-lived memory trace in the MB γ neurons and also causes a long-lived memory to form more slowly in the MB α/β neurons. We propose that the γ lobe and α/β lobe neurons support independent memory traces with different kinetics of formation and decay.
Results
rutabaga adenylyl cyclase is required for short and long-term memory
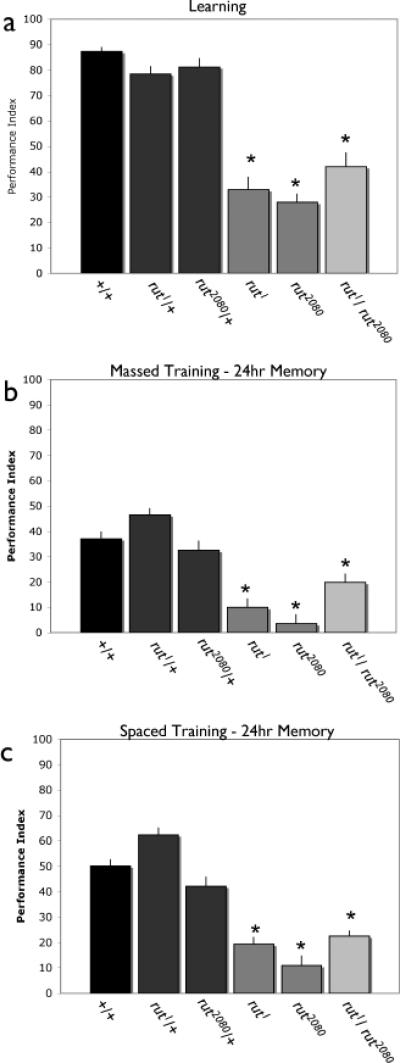
As previously reported [23, 30, 31], each of two rutabaga mutant alleles exhibit severely reduced performance 2 minutes as well as 3 hours after a single aversive Pavlovian training session (Figure 1a, 3b). We next examined two different forms of consolidated memory in the rutabaga mutants. We used 10 repetitive training sessions, either massed together (massed training) or with a 15 minute rest interval between training sessions (spaced training) [32]. Massed training yields a memory that is stable for over 24 hours, but is believed to be independent of CREB-mediated gene expression. In contrast, spaced training induces an additional consolidated LTM that is sensitive to cycloheximide and requires CREB mediated transcription [28, 32]. The rutabaga mutant animals exhibit severely reduced performance 24 hours after either 10 massed (Fig. 1b) or 10 spaced sessions of training (Fig. 1c; see also [33]). The fact that rutabaga mutants exhibit severely reduced STM and LTM provided an opportunity to map the spatial requirements for rutabaga signaling for each.
Figure 1. The rutabaga gene is required to support all memory phases.
Female flies that were wild type (W1118 isoCJ1), heterozygous for rut1 or rut2080, homozygous rut1 or rut2080, or trans-heterozygous for both alleles were tested for immediate memory after a single training session a), or 24 hour memory after massed b) or spaced c) training. Both homozygous rut1 and rut2080 as well as the transheterozygous animals exhibited significantly lower performance indices from wild type controls. P<0.05, a)N=6 for all groups, b) N=12 for all groups, c) N=13 for all groups.
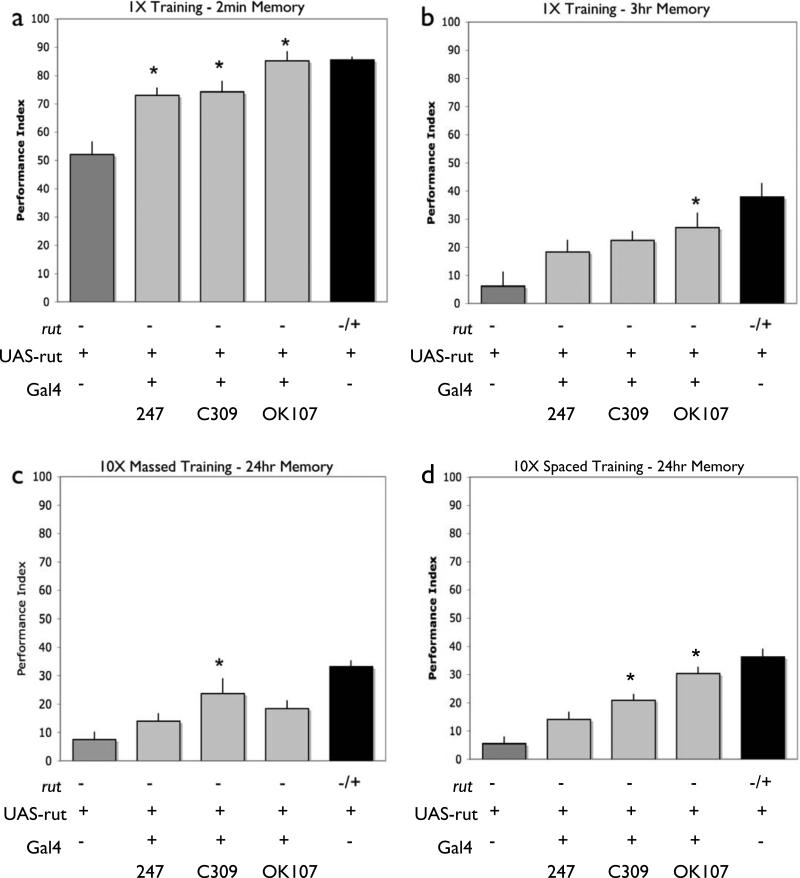
Figure 3. Broad MB expression of rutabaga can support all memory phases.
Memory retention was tested 2 minutes (a) and 3 hours (b) after a single training session as well as 24 hours after either massed (c) or spaced (d) training. In each case, performance was compared among the following groups: rut2080 mutant males with a UAS-rut+ transgene but no Gal4 driver (rut2080/Y; UAS-rut), rut2080 heterozygous females with a UAS-rut+ transgene but no Gal4 driver (rut2080/+; UAS-rut), rut2080 mutant males with a UAS-rut+ transgene and one of three MB Gal4 lines (247, C309, or OK107) or, rut2080 heterozygous females with a UAS-rut+ transgene and one of the three Gal4 lines (Because rut is X linked, we used hemizygous males for these experiments. In this and all figures that follow, the males are shown in white bars and the heterozygous female siblings that do not contain a Gal4 are shown black, and the heterozygous females containing Gal4 lines are shown in an associated supplemental figure. The control females for this figure are shown in Fig. S1). In contrast with the rut2080/Y; UAS-rut mutant males, rut2080mutant males with both a UAS-rut+ transgene and each of the MB Gal4 drivers (247, C309, or OK107), exhibit significantly improved levels of performance measured 2-min after training [P<0.05, N=6 for all groups] (a), improved performance either three hours a single training session, with OK107 showing significant improvement [P<0.05, N=7 for all groups] (b) or 24 hours after either massed, with C309 showing significant improvement [P<0.05, N=15 for all groups] (c) or spaced training with both C309 and OK107 showing significant improvement [P<0.05, N=23 for all groups] (d). In all cases, no significant improvements were observed in control females that were rut2080/+; UAS-rut and contained a Gal4 line (Figure S1).
Mushroom body expression of rutabaga is sufficient to support both short and long-term memory
We tested whether rutabaga expression in MB neurons could rescue LTM as has already been shown to be the case for STM and for intermediate memory during the first 3 hours after a single training session [23, 26, 31]. We used the established rutabaga+ cDNA construct under control of the Gal4 trans-activator system to spatially restrict rutabaga+ expression in an otherwise rutabaga mutant animal. Because there is some evidence that choice of odor combinations can impact conclusions [26], we consistently used one combination, OCT and MCH for all experiments (see methods). We used three different Gal4 lines (247, C309 and OK107) to drive the expression of the rutabaga transgene in MB neurons. The C309 and 247 Gal4 drivers each yield expression in the in α/β and γ lobes (Fig. 2b, c; [34]) with little or no expression in α’/β’ lobes. The OK107 Gal4 line yields expression in α/β, γ and α’/β’ neurons, which includes all three of the major classes of Kenyon cells (Fig. 2d). Of these three pan-MB drivers, 247 shows the most restricted expression, labeling only approximately one third of MB neurons[31]. With MB expression of rutabaga, we are able to improve memory of the rutabaga mutant both 2 minutes and 3 hours after one training session (Figure 3a,b; see [23, 26, 31]) as well as 24 hours after massed training (Fig 3c) . We also are able to restore the long-term memory of rutabaga animals measured 24 hours after spaced training (Fig. 3d). In contrast, expression in olfactory projection neurons (PNs) under control of Gal4 line GH146 (Fig. 2h) does not improve memory performance (Figs. 4b and d, 5b and d). This is consistent with the prior observation that expression of rutabaga in PNs supports only an appetitive memory trace, but not an aversive one [35]. Together, the above findings are consistent with previous results [23, 24, 26] and the broadly accepted hypothesis that rutabaga-dependent cAMP signaling in MBs is sufficient to restore the aversive STM defect of the rutabaga mutant. Our findings further demonstrate that MB expression can restore LTM. Thus rutabaga signaling in MBs appears to be largely sufficient to support each of the temporal phases of memory that have been observed with this task.
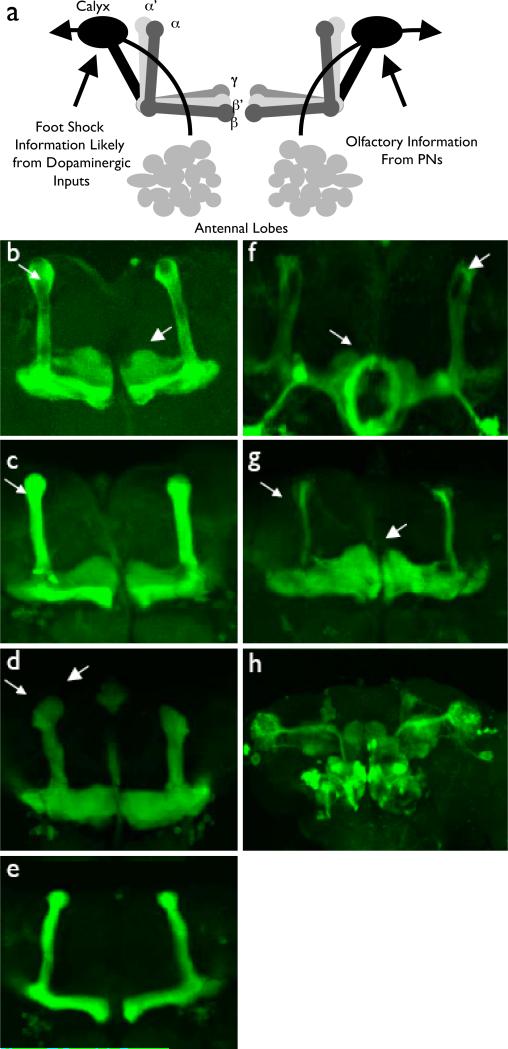
Figure 2.
Gal4 Driven MB Driver GFP Expression. For each Gal4 Driver, unless otherwise noted, a projection of the MB lobe region of male flies heterozygous for Gal4 and UAS-mcd8:GFP is shown. a) Schematic of olfactory system in Drosophila. Olfactory information from Antennal Lobes is conveyed to the MB calyx via projection neurons (PNs). Foot-shock (Unconditioned Stimulus, US) is thought to be conveyed by dopaminergic inputs to MBs (not shown). MB Kenyon cells are made up of three principle neuron types: α’/β’ and α/β neurons have both a vertical and horizontal branch whereas γ lobes neurons consist of a single, horizontal projection. b) 247 driven GFP expression. Expression is restricted to alpha/beta (small arrowhead) and gamma neurons (large arrowhead). c) C309 driven GFP expression. Again, expression is restricted to alpha/beta (small arrowhead) and gamma neurons (large arrowhead). d) OK107 driven GFP expression. OK107 expression pattern labels alpha’/beta’ (small arrowhead), alpha/beta (large arrowhead), and gamma neurons. e) C739 driven GFP expression. Expression is restricted to alpha/beta type neurons in the MB. f) C305a driven GFP expression. The C305a expression pattern labels approximately half of alpha’/beta’ (large arrowhead) MB neurons, ellipsoid body neurons (small arrowhead), as well as antennal lobes. g) 201Y driven GFP expression. The 201Y expression pattern labels gamma (large arrowhead) and a small number of core alpha/beta neurons (small arrowhead). h) GH146 driven GFP expression. A whole brain projection of male flies heterozygous for GH146 Gal4 and UAS-mcd8:GFP is shown. GH146 labels olfactory projection neurons (PNs).
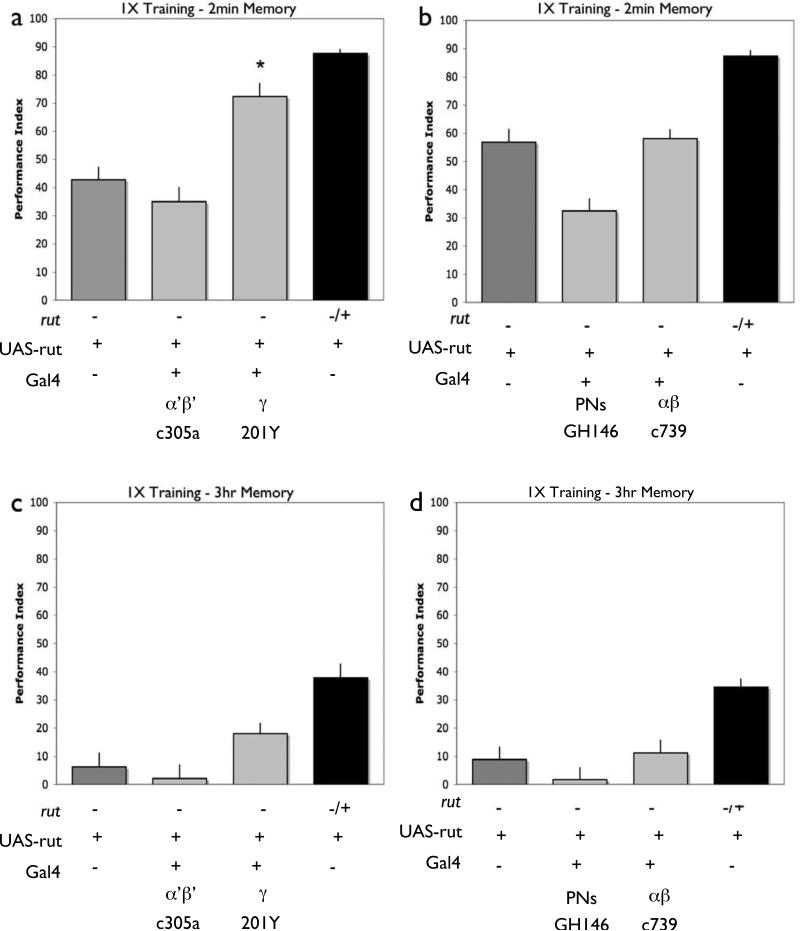
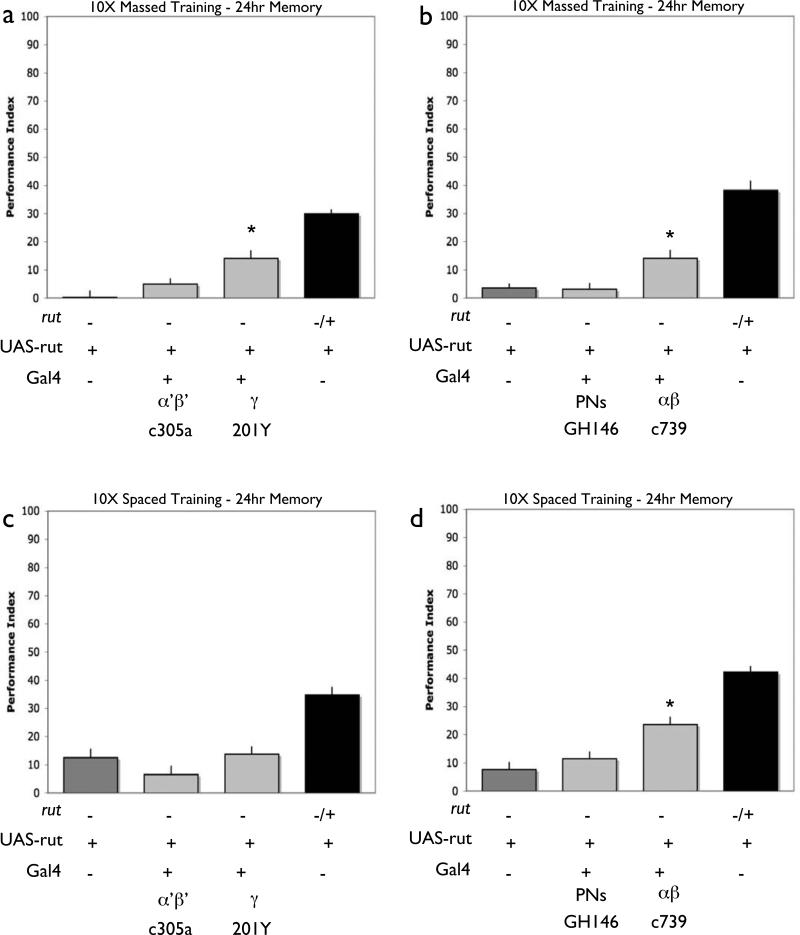
Figure 4. MB γ Lobe expression of rut supports early memory.
Memory retention was tested 2 minutes (a,b) and 3 hours (c,d) after a single training session rut2080males with a UAS-rut+ transgene but no Gal4 driver (rut2080/Y; UAS-rut) exhibit reduced performance relative to heterozygous sisters (rut2080/+; UAS-rut). In contrast, rut2080mutant males with both a UAS-rut+ transgene and the 201Y gamma lobe Gal4 driver exhibit significantly improved levels of performance and significantly improved performance relative to mutant levels (a). However, rut2080mutant males with both a UAS-rut+ transgene and an alpha’/beta’ Gal4 driver (C305a) (a), alpha/beta Gal4 driver (C739) (b) or PN driver (GH146) (b), were not significantly improved from mutant controls. P<0.05, (a)N=8 for all groups, (b)N=12 for all groups. Flies of the same genotypes were also tested for three hour memory after a single training session. In this case, expression with the lobe specific Gal4 drivers 201Y, C305a, (c), C739 and GH146 (d) was not sufficient to significantly improve performance above mutant levels. P<0.05, (c)N=7 for all groups, (d)N=8 for all groups. In all cases, no significant improvements were observed in control females that were rut2080/+; UAS-rut and contained a Gal4 line (Figure S2).
Figure 5. rutabaga expression in both γ and α/β neurons supports 24 hour memory.
Memory retention was 24 hours after either massed (a,c) or spaced (b,d) training. rut2080males with a UAS-rut+ transgene but no Gal4 driver (rut2080/Y; UAS-rut) exhibit reduced performance relative to heterozygous sisters (rut2080/+; UAS-rut) 24 hours after massed training. rut2080mutant males heterozygous for both a UAS-rut+ transgene and the 201Y gamma lobe Gal4 driver (a) or the C739 alpha/beta driver (b) exhibit significantly improved levels of performance compared to mutant controls. Performance levels were not significantly improved with flies carrying the C305a (a), or GH146 (b) Gal4 drivers. P<0.05, a)N=16 for all groups, b) N=18 for all groups. Flies of the same genotypes were also tested 24 hours after spaced training. In this case, only flies with carrying the C739 alpha/beta driver showed performance significantly above mutant levels (d). Flies carrying the 201Y, C305a, or GH146 drivers were not improved compared to mutant levels (c, d). P<0.05, c)N=16 for all groups, d) N=18 for all groups. In all cases, no significant improvements were observed in control females that were rut2080/+; UAS-rut and contained a Gal4 line (Figure S3).
rutabaga expression with Gal4 201Y γ lobe but not Gal4 C739 α/β neurons is sufficient for STM
We tested the effects on both STM and LTM of rutabaga expression in each of the three major classes of MB Kenyon cells. For each of these experiments, we used a collection of well characterized Gal4 lines [23, 31, 36] that together dissect the MB neuron subtypes. Gal4 line C739 yields expression in all or most α/β lobe neurons but not in the other classes of MB neurons (Fig. 2e). For expression in α’/β’ lobes, we used the C305a Gal4 driver that expresses in approximately 50% of the prime lobes (Fig. 2f) [36] as well as expressing outside the MB, notably in antennal lobes and ellipsoid body. Gal4 line 201Y drives expression in all or most γ lobe neurons as well as a small number of core α/β lobe neurons (Fig. 2g). A recently published report provides detailed characterization of each of the Gal4 drivers utilized in this study [37]. While no Gal4 driver is entirely specific to a given cell type, the ones we have chosen are the most specific for each MB cell type among those available.
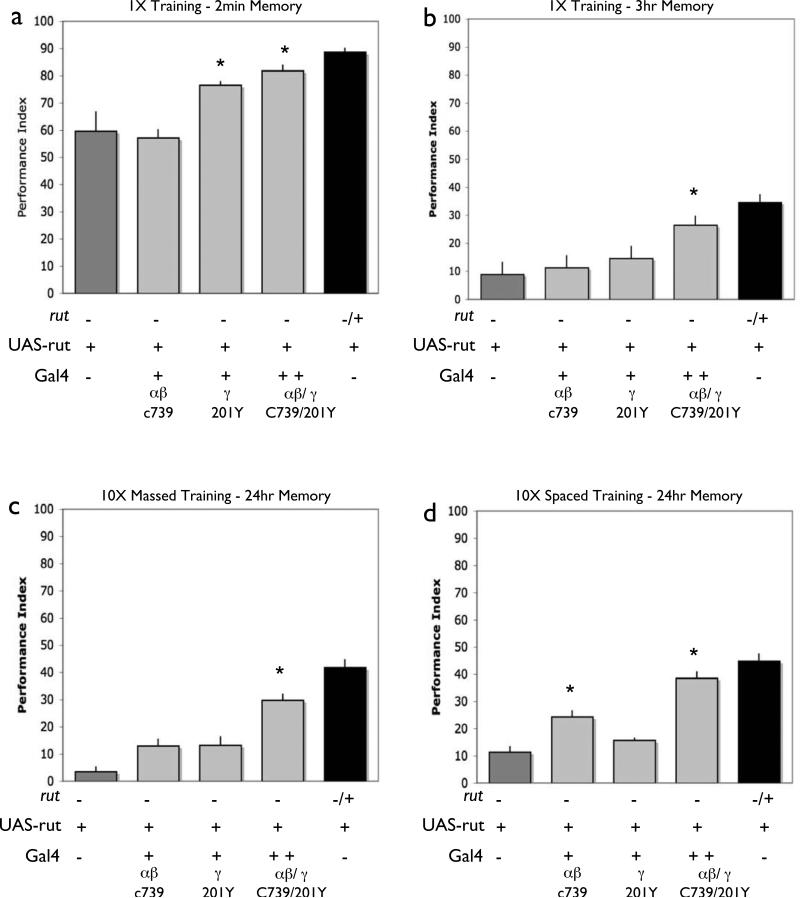
As previously reported [23, 26, 31], we find that expression primarily in γ neurons, with the 201Y driver, is sufficient to restore 2-min memory performance (Fig. 4a). In contrast, expression in α/β or α’/β’ lobes do not improve STM performance of the rutabaga mutant (also previously observed [23, 26, 31](Fig. 4a and b). Interestingly, memory at the 3-hour time-point cannot be significantly improved with expression in 201Y γ lobe neurons alone. In fact, we do not significantly restore 3-hour memory with expression in any single MB neuron sub-type (Fig. 4c,d). This is in contrast with the performance increase seen with broad MB expression with OK107 (compare with Fig. 3b). This demonstrates that for memory 3 hours after training, additional expression in combination with γ lobes is needed to fully support memory performance (see below).
rutabaga expression with Gal4 C739 α/β but not Gal4 201Y γ lobe neurons supports long-term memory
We next tested the effects of expression in each of these three neuron sub-types on memory measured 24 hours after massed or spaced training. Massed training yields a memory trace that is long-lived, but genetically and pharmacologically distinct from CREB dependent LTM. Spaced training induces this CREB-dependent long-term memory [32]. Our experiments with spaced training yielded two unexpected results. First, in contrast with STM after one training session, we see no improvement with 201Y γ lobe expression (Fig. 5c). Instead, after spaced training we only obtain significant restoration of memory performance with expression in C739 α/β lobe neurons (Fig. 5d). These two findings are surprising because C739 α/β expression does not improve STM performance of the rutabaga mutants. Thus rutabaga signaling in α/β lobes is sufficient to support a long-lived memory but not STM (Figs. 4b,d). In contrast, γ lobe expression supports STM but not LTM. As was the case with single training sessions and massed training, we again see no improvement in memory performance with expression in C305a α’/β’ lobe neurons or with expression in olfactory PNs (Fig. 5c,d). Together, these findings support a specific requirement of rutabaga in C739 α/β neurons to support memory 24 hours after spaced training. It also is of interest that memory after massed training can be partially supported with either 201Y γ or C739 α/β lobe neurons (Fig. 4c).
Combined expression of rutabaga in 201Y γ and C739 α/β neurons restores both short and long-term memory
Expression of the rutabaga+ transgene in α/β and γ lobes with Gal4 line C309 improves memory measured out to 24 hours after either massed or spaced training (Fig 3). In contrast, we observed only partial rescue of memory 24 hours after massed training with 201Y γ lobe or C739 α/β expression and a partial rescue of memory after spaced training with C739 α/β lobe expression (Fig. 5a,b). Together, these results suggest that for these memory retention intervals, some combination of α/β and γ lobe expression is needed. As an independent test of the effects of combined expression in α/β and γ lobes, we generated rutabaga mutant animals that contained both the 201Y and C739 Gal4 lines, as well as the rutabaga+ transgene. When combined in the same fly, these two Gal4 lines yield expression that includes both the α/β and γ lobes (Fig. 7a). Immediately or 3 hours after a single training session, combined expression in both γ lobes and α/β lobes resulted in nearly normal levels of performance [24](Fig. 6a, b, see also Fig. 4a). On its own, α/β lobe expression with C739 was not sufficient to improve short-term memory (Figs. 6a,4b). Moreover, at the 3-hour time point, expression in either one of these cell types on its own cannot significantly improve memory but the combination does (Figs. 4c, d and 6b).
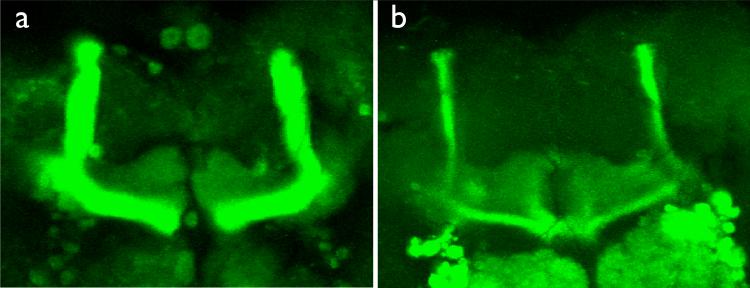
Figure 7. Gal4 expression pattern of double Gal4 lines.
A projection of the MB lobe region of male flies heterozygous for each of two Gal4 drivers, and UAS-mcd8:GFP is shown. (a) combined 201Y and C739 driven GFP expression. 201Y expression in γ lobes, and C739 expression in α/β lobes are each visible. (b) combined 201Y and C305a driven GFP expression. 201Y expression in γ lobes, and C305a expression in α’/β’, as well in ellipsoid body and antennal lobe are visible.
Figure 6. rutabaga expression in both γ and α/β lobes combined supports all memory phases.
Memory retention was tested 2 minutes (a) and 3 hours (b) after a single training session as well as 24 hours after either massed (c) or spaced (d) training. In each case, performance was compared among the following groups: rut2080 mutant males with a UAS-rut+ transgene but no Gal4 driver (rut2080/Y; UAS-rut), rut2080 heterozygous females with a UAS-rut+ transgene but no Gal4 driver (rut2080/+; UAS-rut), rut2080 mutant males with a UAS-rut+ transgene and either the 201Y or C739 drivers alone, or the 201Y and C739 Gal4 drivers combined, and rut2080 heterozygous females with a UAS-rut+ transgene and these Gal4 lines (these control females are shown in Fig. S4). In contrast with the rut2080/Y; UAS-rut mutant males, rut2080mutant males with both a UAS-rut+ transgene and either the 201Y, or 201Y combined with C739 Gal4 drivers exhibit nearly normal levels of performance measured 2-min after training, while C739 expression caused no improvement [P<0.05, N=6 for all groups] (a), and only expression combined with both the 201Y and C739 drivers significantly improved performance three hours after a single training session [P<0.05, N=8 for all groups] (b) Only expression combined with both the 201Y and C739 drivers significantly improved performance 24 hours after massed training [P<0.05, N=18 for all groups] (c). For 24 hours after spaced training, expression with the C739 driver alone resulted in significant improvement of performance, however, this effect was augmented by combining both C739 and 201Y expression. [P<0.05, N=23 for all groups]. In all cases, no significant improvements were observed in control females that were rut2080/+; UAS-rut and contained a Gal4 line with the exception of flies carrying both the 201Y and C739 drivers combined after spaced training (Figure S4).
We next examined the effects of combined 201Y γ lobe and C739 α/β expression on memory measured 24 hours after massed and spaced training. With spaced training, we once again observe no improvement with 201Y γ lobe expression alone and a partial but significant effect with C739 α/β lobe expression (Compare Fig 6d with Fig. 5c,d). The partial rescue with α/β lobe expression appears, however, to be significantly bolstered with the addition of γ lobe expression (Fig. 6d). A potential caveat to this interpretation is that in this particular experiment we also observed modestly increased performance in the female control siblings that are heterozygous for rutabaga and contain both of these two gal4 lines. Nevertheless, we strongly favor the interpretation that 201Y γ lobe expression bolsters the effects of C739 α/β lobe expression for two reasons. First, the robust rescue observed with Gal4 line C309 (Fig. 3d), which also expresses in both of these MB subtypes but not in α’/β’. Second, in an independent series of experiments (Fig. 8), we again observe the increased performance in males that contain C739 ad 201Y, but did not observe the non-specific increase in females (Fig. S6).
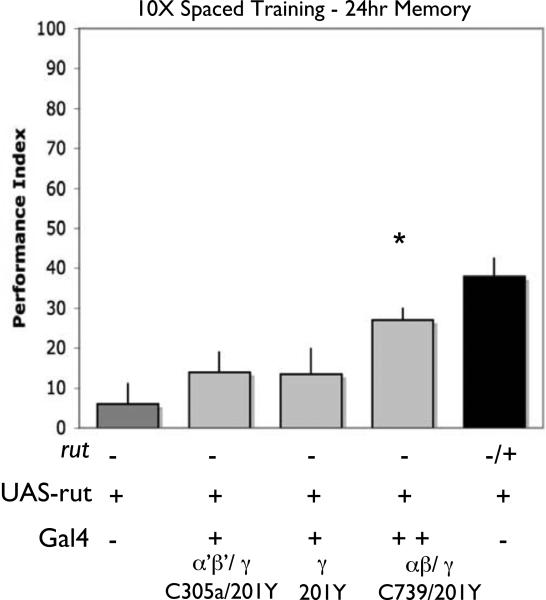
Figure 8. rutabaga expression in both γ and α’/β’ lobes combined does not restore memory 24 hours after spaced training.
Memory retention was tested 24 hours after spaced training. In each case, performance was compared among the following groups: rut2080 mutant males with a UAS-rut+ transgene but no Gal4 driver (rut2080/Y; UAS-rut), rut2080 heterozygous females with a UAS-rut+ transgene but no Gal4 driver (rut2080/+; UAS-rut), rut2080 mutant males with a UAS-rut+ transgene and the 201Y driver alone, or the 201Y and c305a, or 201y and C739 Gal4 drivers combined. For 24 hours after spaced training, expression with the either the 201Y driver alone, or with both the c305a and 201y drivers combined did not significantly improve performance compared to rut2080 mutant males with a UAS-rut+ transgene but no Gal4 line. As first observed in Fig. 6, we see significant improvement when we combine C739 and 201Y. [P<0.05, N=8 for all groups]. In all cases, no significant differences were observed among control females that were rut2080/+; UAS-rut and contained a Gal4 line (Fig. S6).
It is of note that the 247 driver also expresses in both α/β and γ neurons, but expression with this driver does not significantly improve performance 3 hours after a single training session, or 24 hours after massed or spaced training (Fig3b,c,d). This likely is due to the fact that the 247 driver expresses in a smaller subset of cells than either the C309 driver, or the combination of 201Y and C739 drivers [37].
Nevertheless, this prompted us to do an additional control experiment to rule out the possibility that combined expression from C739/201Y improved performance after spaced training because of an increase in the number of MB neurons expressing rutabaga rather than the types of MB neurons. We created animals that were mutant for rutabaga, contained the UAS-rutabaga+ transgene, and also contained both the 201Y and C305a drivers. In this way, we increased the cell number of MB neurons as before, but now included the γ and α’/β’ neurons (Fig 7b) instead of γ and α/β neurons (Fig. 7a). In contrast with the C739/201Y combination (Figs. 6d and 8), we observe no restoration of memory with the C305a/201Y combination (Fig. 8). Taken together, these findings strongly support they hypothesis that γ lobe expression of rutabaga bolsters the impact of expression in α/β.
Discussion
Pavlovian olfactory learning in Drosophila is believed to involve rutabaga-dependent coincidence detection of conditioned stimulus (CS; odor) and unconditioned stimulus (US; shock) pathways primarily in MB γ lobe neurons [23, 26, 31]. The CS olfactory information is carried by PNs from the antennal lobe and the US is thought to be mediated by dopaminergic or octopaminergic neurons for aversive and appetitive learning respectively [38, 39]. In this model, the rutabaga adenylyl cyclase is synergistically activated by concurrent elevation in intracellular calcium, driven by the CS stimulus, and by G-protein coupled protein receptor activation driven by the US. rutabaga stimulation results in elevated levels of cAMP/activation of PKA, which in turn is assumed to drive synaptic plasticity underlying memory [40]. STM is thought to involve transient elevations in PKA activity with impacts on trafficking and post-translational modifications of synaptic proteins. In contrast, LTM is believed to involve more stable elevation in PKA levels that are induced by repetitive spaced training. Activated PKA that is translocated to the nucleus is thought to cause phosphorylation of CREB and the activation of a cascade of gene-expression [2, 6, 41]. This explanation of olfactory learning in flies derives in part from genetic manipulations of cAMP pathway in MB neurons and in part from a parallel dissection of synaptic plasticity underlying learning in Aplysia [3].
In flies, convergent data from several different types of experiment support this model. First, mutant strains with disrupted MB structure or pharmacological disruption of MB development [42-44], each block Pavlovian olfactory memory without impacting the ability to sense and respond to the US (shock) and CS (odor) stimuli. Second, several canonical members of the cAMP signaling pathway exhibit elevated levels of expression in MB [17]. Third, a wide variety of experiments using spatially restricted transgenic manipulations support the hypothesis that cAMP signaling in MB is sufficient to support memory performance [17]. In particular, rutabaga expression in the γ lobe subset of MB neurons is sufficient to restore STM to a rutabaga mutant animal [23, 26] and (Fig. 4a,6a). Fourth, functional imaging studies reveal an associative increase in calcium influx in MBs following training [29]. Finally, reversible disruptions of dynamin-dependent neurotransmission in MB supports the conclusion that output from α/β and/or γ neurons is required during memory retrieval (Although it is worth note that blocking 201y γ neurons alone did not appear to inhibit 3-minute retention [47]), but not for acquisition or storage [26, 45, 46]. The findings are largely consistent with the hypothesis that the synaptic plasticity that underlies acquisition is caused by inputs to α/β and/or γ neurons. Output from these neurons is only required to drive the behavioral responses during retrieval. In contrast, the neurotransmission in α’/β’ neurons is required during acquisition and storage, but not during retrieval [34, 47]. Given our finding that expression of rutabaga expression is sufficient in γ and α/β (but not α’/β’ lobe) neurons to support STM and LTM respectively, we propose that odor driven α’/β’ lobe activity is required for plasticity in α/β and γ neurons (see also [34]).
But several key aspects remain poorly understood. First, although this model explicitly proposes rutabaga as the coincidence detector in γ lobes, approximately 50% of memory performance remains intact in rutabaga null mutant animals. Thus rutabaga independent mechanisms are capable of supporting olfactory associations, but we do not know where this occurs or what mechanisms are involved. In addition, the few investigations of circuits involved with LTM are hard to interpret in the context of the simple model outlined above.
Genetic disruptions of MB development prevent LTM [44] and in several cases including that of Notch, spatially restricted gene manipulations support a role for MB [48, 49] [33, 50-53]. More recently, functional imaging studies have revealed an elevated odor driven calcium influx in α/β neurons after spaced training. Both this cellular correlate and LTM performance can be blocked by expression in α/β neurons of a CREB-blocker isoform [29]. Together these findings indicate a role for α/β lobe neurons in LTM.
The findings reported here impact our understanding in several ways. First, we provide strong evidence that rutabaga signaling in mushroom bodies can support both STM and LTM. Viewed in the context of the literature discussed above, this suggests that the NMDA receptor requirement observed in ellipsoid body neurons [27] represents a separate signaling pathway from that of rutabaga in MB. Second, our data strongly support the conclusion that STM and LTM involve distinct and functionally independent rutabaga signaling in γ and α/β lobes respectively. Our findings are consistent with a model in which two different coincidence detection mechanisms are at play in MB. One likely occurs in γ lobes, and requires rutabaga for its formation. A second coincidence detection mechanism appears to be rutabaga independent, but requires rutabaga signaling in α/β lobes for it's stabilization.
Several of our key findings support the above model. First, broad MB expression of a UAS-rut+ transgene is sufficient to improve performance in rut mutant animals at each of the time points after either one, 10 massed, or 10 spaced training sessions (Fig. 3). Thus the need for rutabaga expression appears to be largely or solely in MB. It is worth note that for the 3-hour and 24 hour massed training time-points, the findings are not entirely consistent. With 3-hour retention, we can improve memory with OK107, but with 247 and C309, we observe only a trend of improvement that is not significant. This may be due to differences in levels of expression, but given this discrepancy, we cannot rule out a role for neurons outside MB for this retention interval. Our results with massed training are similar. Here, we are able to restore memory performance with C309 and partially with C739 or 201Y, but observe only a trend that is not significant with 247 and OK107. Here again, we cannot distinguish if this reflects subtle differences in expression levels or cell type within MBs, or an additional requirement for expression outside mushroom bodies. If the latter notion were true, however, it would imply a common expression outside mushroom bodies for the Gal4 lines C739, 201Y and C309. In the case of STM and LTM after spaced training, however, the data clearly indicate that the primary rutabaga dependent contribution to this form of olfactory memory is in MB.
Our findings support the established hypothesis that rutabaga expression in γ lobes is sufficient to support rutabaga dependent STM and further indicate that expression in α/β or α’/β’ cannot on their own support STM. In contrast, the most striking set of findings are that expression in γ lobe neurons with the 201Y driver yields no significant improvement in long-term memory performance after spaced training, while expression in C739 α/β neurons supports LTM but does not impact STM (Figs. 5c,6d, 8). The reciprocal outcomes seen with 201Y γ lobe and C739 α/β lobe Gal4 drivers supports the hypothesis that rutabaga plays at least two roles: In γ lobe neurons to support STM and in α/β neurons for consolidated memory. The rutabaga function in α/β lobes appears to be required to consolidate an association whose formation is rutabaga independent.
While we cannot formally rule out a contribution of rutabaga expression in the few ellipsoid body neurons labeled by the C739 Gal4 line, we view this possibility as unlikely for three reasons. First, the ellipsoid body neurons labeled by C739 are not of the R4m type which require NMDA receptor function for LTM (A.S. Chiang, personal communication). Second, the C305a α’/β’ line also broadly labels ellipsoid body, but expression of rutabaga in this pattern does not improve memory. Finally, we also observe a significant improvement of memory after spaced training with the C309 and OK107 Gal4 lines, which on their own give expression in both γ and α/β neurons, but not ellipsoid body. Given the known role for NMDA receptors in ellipsoid body [27], our results suggest the interesting hypothesis that there is a dynamic circuit level interaction rather than just a biochemical consolidation within mushroom bodies.
A common feature of memory across phyla is an apparent dissection of the neuro-anatomical requirements for different memory phases. In mammalian systems, the notion of memory transfer has been invoked, but whether this involves an actual transfer of information or reflects an evolving circuit requirement for some other reason is not understood. Our experiments provide evidence that this anatomical dissection of STM and LTM also occurs in Drosophila and offers circuit level and mechanistic insight into this process.
Experimental Procedures
Fly Strains
The wild type flies utilized in behavior experiments were Canton-S w1118 (iso1CJ). Mutant strains used were rut1 and rut2080. The X linked rutabaga alleles were crossed into a background containing the iso1CJ autosomes. Behavioral rescue experiments were conducted by crossing rut2080;UAS-rut females (from Martin Heisenberg) with males from each of the Gal4 lines c309, OK107, 247, 201Y, C305a, (from Ann-Shyn Chiang), C739, GH146. A control cross to iso1CJ also was used. The experimental groups utilized in the rescue experiments were male progeny of the above cross that were rut2080 hemizygous; UAS-rut heterozygous, and Gal4 heterozygous (or no Gal4 for control cross). Female progeny from the same cross that were heterozygous for rut2080 were used as controls (See supplementary figures S1, S2, S3, and S4). GFP imaging was performed by crossing each Gal4 line with a UAS-mCD8::GFP reporter.
Behavior
All behavioral experiments were performed in a genotype balanced manner, with the experimentor blind to genotype. Data in each figure represent independent sets of experiments even in cases where genotypes and training paradigms are identical. In each case, experiments within a figure panel were performed in parallel. Olfactory associative learning was tested by training 2−3 day old flies in a t-maze apparatus using a Pavlovian conditioning paradigm [15]. Odors used were 3-octanol and 4-methyl-cyclohexanol. Each individual N consisted of two groups of 125 flies, each of which was shocked in the presence of one of these two odors. Thus a single N consisted of approximately 250 flies, with half of the flies trained to one odor, and half to the other. A half performance index in calculated by dividing the number of flies that chose correctly, minus the flies that chose incorrectly by the total number of flies in the experiment. A final performance index is calculated by averaging both reciprocal half-performance indexes for the two odors.
For 24 hour memory experiments, animals were subjected to ten such training sessions either massed together or spaced out with a 15 minute rest interval [32, 54]. For these multiple training protocols, robotic trainers were used and in all cases the animals were manually tested in the t-maze apparatus 24 hours after training. All genotypes are trained and tested in parallel, and rotated between all the robotic trainers to ensure a balanced experiment.
Statistics
The behavioral data from this paradigm is normally distributed, and thus can be analyzed with ANOVA. JMP software was utilized to perform ANOVA and Tukey-Kramer Honestly Significant Difference test, with comparisons made between all genotypes. Statistical significance in the figures represents a significant increase in performance in comparison to mutant male control levels with P<0.05, except in supplementary figures where statistical signficance represents a significant increase compared to heterozygous female controls. Error bars in behavioral data graphs represent the standard error of the mean.
Confocal Imaging
Fly brains of 2−3 day old adult male flies that were heterozygous for a Gal4 driver and UAS-mCD8::GFP were dissected in PBS. The fly brains were then transferred into 4% Paraformaldehyde in PBS and fixed overnight at 4° C. Brains were placed in a vacuum for 40 minutes to remove air from trachae prior to mounting. Brains were then cleared and mounted in FocusClear solution, and imaged immediately.
The confocal images of brains were acquired using a ZEISS LSM 510 confocal microscope. The following confocal settings were used: 20× lens, 1 μm spacing in the z-axis with 1024 × 1024 resolution in x- and y-axes. The GFP signal was captured with an Argon/2 488 nm laser. The raw data were processed by LSM Image Browser Rel.4.2 (ZEISS) and later manipulated as figures in Powerpoint.
Supplementary Material
Acknowledgements
We are grateful to G. Turner and Y. Zhong for careful reading of the manuscript. We thank members of the Dubnau and Turner Labs for helpful discussions. We thank Dan Valente for assistance with statistical analysis. The UAS-rutabaga flies were a gift from Martin Heisenberg. We also thank the Beckman Young Investigator, Human Frontiers Science Program, DART neurosciences inc. and NIMH Grant #5R01MH069644 for financial support to JD. AB was a Barbara McClintock Fellow at the Watson School of Biological Sciences and was supported by Grant 5T32GM065094 from the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- 1.Dubnau J, Tully T. Gene discovery in Drosophila: new insights for learning and memory. Annu Rev Neurosci. 1998;21:407–444. doi: 10.1146/annurev.neuro.21.1.407. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9530502. [DOI] [PubMed]
- 2.Hawkins RD, Kandel ER, Bailey CH. Molecular mechanisms of memory storage in Aplysia. Biol Bull. 2006;210:174–191. doi: 10.2307/4134556. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16801493. [DOI] [PubMed]
- 3.Stough S, Shobe JL, Carew TJ. Intermediate-term processes in memory formation. Curr Opin Neurobiol. 2006;16:672–678. doi: 10.1016/j.conb.2006.10.009. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17097872. [DOI] [PubMed]
- 4.Martin KC, Casadio A, Zhu H, Yaping E, Rose JC, Chen M, Bailey CH, Kandel ER. Synapse-specific, long-term facilitation of aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91:927–938. doi: 10.1016/s0092-8674(00)80484-5. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9428516. [DOI] [PubMed]
- 5.Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12194863. [DOI] [PubMed]
- 6.Silva AJ, Kogan JH, Frankland PW, Kida S. CREB and memory. Annu Rev Neurosci. 1998;21:127–148. doi: 10.1146/annurev.neuro.21.1.127. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9530494. [DOI] [PubMed]
- 7.Chen C, Kim JJ, Thompson RF, Tonegawa S. Hippocampal lesions impair contextual fear conditioning in two strains of mice. Behav Neurosci. 1996;110:1177–1180. doi: 10.1037//0735-7044.110.5.1177. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8919020. [DOI] [PubMed]
- 8.Frankland PW, Cestari V, Filipkowski RK, McDonald RJ, Silva AJ. The dorsal hippocampus is essential for context discrimination but not for contextual conditioning. Behav Neurosci. 1998;112:863–874. doi: 10.1037//0735-7044.112.4.863. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9733192. [DOI] [PubMed]
- 9.Squire LR. Memory and the hippocampus: a synthesis from findings with rats, monkeys, and humans. Psychol Rev. 1992;99:195–231. doi: 10.1037/0033-295x.99.2.195. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=1594723. [DOI] [PubMed]
- 10.Scoville WB, Milner B. Loss of recent memory after bilateral hippocampal lesions. 1957. J Neuropsychiatry Clin Neurosci. 2000;12:103–113. doi: 10.1176/jnp.12.1.103. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10678523. [DOI] [PubMed]
- 11.Milner B. Disorders of learning and memory after temporal lobe lesions in man. Clin Neurosurg. 1972;19:421–446. doi: 10.1093/neurosurgery/19.cn_suppl_1.421. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=4637561. [DOI] [PubMed]
- 12.Davis RL, Cherry J, Dauwalder B, Han PL, Skoulakis E. The cyclic AMP system and Drosophila learning. Mol Cell Biochem. 1995;149−150:271–278. doi: 10.1007/978-1-4615-2015-3_31. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8569740. [DOI] [PubMed]
- 13.Nguyen PV, Woo NH. Regulation of hippocampal synaptic plasticity by cyclic AMP-dependent protein kinases. Prog Neurobiol. 2003;71:401–437. doi: 10.1016/j.pneurobio.2003.12.003. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15013227. [DOI] [PubMed]
- 14.Mayford M, Kandel ER. Genetic approaches to memory storage. Trends Genet. 1999;15:463–470. doi: 10.1016/s0168-9525(99)01846-6. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10529810. [DOI] [PubMed]
- 15.Tully T, Quinn WG. Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol [A] 1985;157:263–277. doi: 10.1007/BF01350033. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=3939242. [DOI] [PubMed]
- 16.Margulies C, Tully T, Dubnau J. Deconstructing memory in Drosophila. Curr Biol. 2005;15:R700–713. doi: 10.1016/j.cub.2005.08.024. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16139203. [DOI] [PMC free article] [PubMed]
- 17.McGuire SE, Deshazer M, Davis RL. Thirty years of olfactory learning and memory research in Drosophila melanogaster. Prog Neurobiol. 2005;76:328–347. doi: 10.1016/j.pneurobio.2005.09.003. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16266778. [DOI] [PubMed]
- 18.Strausfeld NJ, Hansen L, Li Y, Gomez RS, Ito K. Evolution, discovery, and interpretations of arthropod mushroom bodies. Learn Mem. 1998;5:11–37. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10454370. [PMC free article] [PubMed]
- 19.Jefferis GS, Marin EC, Watts RJ, Luo L. Development of neuronal connectivity in Drosophila antennal lobes and mushroom bodies. Curr Opin Neurobiol. 2002;12:80–86. doi: 10.1016/s0959-4388(02)00293-3. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11861168. [DOI] [PubMed]
- 20.Heisenberg M. Mushroom body memoir: from maps to models. Nat Rev Neurosci. 2003;4:266–275. doi: 10.1038/nrn1074. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12671643. [DOI] [PubMed]
- 21.Keene AC, Waddell S. Drosophila olfactory memory: single genes to complex neural circuits. Nat Rev Neurosci. 2007;8:341–354. doi: 10.1038/nrn2098. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17453015. [DOI] [PubMed]
- 22.Han PL, Levin LR, Reed RR, Davis RL. Preferential expression of the Drosophila rutabaga gene in mushroom bodies, neural centers for learning in insects. Neuron. 1992;9:619–627. doi: 10.1016/0896-6273(92)90026-a. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=1382471. [DOI] [PubMed]
- 23.Zars T, Fischer M, Schulz R, Heisenberg M. Localization of a short-term memory in Drosophila. Science. 2000;288:672–675. doi: 10.1126/science.288.5466.672. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10784450. [DOI] [PubMed]
- 24.McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14657498. [DOI] [PubMed]
- 25.Mao Z, Roman G, Zong L, Davis RL. Pharmacogenetic rescue in time and space of the rutabaga memory impairment by using Gene-Switch. Proc Natl Acad Sci U S A. 2004;101:198–203. doi: 10.1073/pnas.0306128101. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14684832. [DOI] [PMC free article] [PubMed]
- 26.Akalal DB, Wilson CF, Zong L, Tanaka NK, Ito K, Davis RL. Roles for Drosophila mushroom body neurons in olfactory learning and memory. Learn Mem. 2006;13:659–668. doi: 10.1101/lm.221206. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16980542. [DOI] [PMC free article] [PubMed]
- 27.Wu CL, Xia S, Fu TF, Wang H, Chen YH, Leong D, Chiang AS, Tully T. Specific requirement of NMDA receptors for long-term memory consolidation in Drosophila ellipsoid body. Nat Neurosci. 2007;10:1578–1586. doi: 10.1038/nn2005. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17982450. [DOI] [PMC free article] [PubMed]
- 28.Yin JC, Wallach JS, Del Vecchio M, Wilder EL, Zhou H, Quinn WG, Tully T. Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila. Cell. 1994;79:49–58. doi: 10.1016/0092-8674(94)90399-9. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=7923376. [DOI] [PubMed]
- 29.Yu D, Akalal DB, Davis RL. Drosophila alpha/beta mushroom body neurons form a branch-specific, long-term cellular memory trace after spaced olfactory conditioning. Neuron. 2006;52:845–855. doi: 10.1016/j.neuron.2006.10.030. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17145505. [DOI] [PMC free article] [PubMed]
- 30.Livingstone MS, Sziber PP, Quinn WG. Loss of calcium/calmodulin responsiveness in adenylate cyclase of rutabaga, a Drosophila learning mutant. Cell. 1984;37:205–215. doi: 10.1016/0092-8674(84)90316-7. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=6327051. [DOI] [PubMed]
- 31.Schwaerzel M, Heisenberg M, Zars T. Extinction antagonizes olfactory memory at the subcellular level. Neuron. 2002;35:951–960. doi: 10.1016/s0896-6273(02)00832-2. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12372288. [DOI] [PubMed]
- 32.Tully T, Preat T, Boynton SC, Del Vecchio M. Genetic dissection of consolidated memory in Drosophila. Cell. 1994;79:35–47. doi: 10.1016/0092-8674(94)90398-0. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=7923375. [DOI] [PubMed]
- 33.Lu Y, Lu YS, Shuai Y, Feng C, Tully T, Xie Z, Zhong Y, Zhou HM. The AKAP Yu is required for olfactory long-term memory formation in Drosophila. Proc Natl Acad Sci U S A. 2007;104:13792–13797. doi: 10.1073/pnas.0700439104. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17690248. [DOI] [PMC free article] [PubMed]
- 34.Krashes MJ, Keene AC, Leung B, Armstrong JD, Waddell S. Sequential use of mushroom body neuron subsets during drosophila odor memory processing. Neuron. 2007;53:103–115. doi: 10.1016/j.neuron.2006.11.021. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17196534. [DOI] [PMC free article] [PubMed]
- 35.Thum AS, Jenett A, Ito K, Heisenberg M, Tanimoto H. Multiple memory traces for olfactory reward learning in Drosophila. J Neurosci. 2007;27:11132–11138. doi: 10.1523/JNEUROSCI.2712-07.2007. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17928455. [DOI] [PMC free article] [PubMed]
- 36.Lin HH, Lai JS, Chin AL, Chen YC, Chiang AS. A map of olfactory representation in the Drosophila mushroom body. Cell. 2007;128:1205–1217. doi: 10.1016/j.cell.2007.03.006. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17382887. [DOI] [PubMed]
- 37.Aso Y, Grubel K, Busch S, Friedrich AB, Siwanowicz I, Tanimoto H. The mushroom body of adult Drosophila characterized by GAL4 drivers. J Neurogenet. 2009;23:156–172. doi: 10.1080/01677060802471718. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19140035. [DOI] [PubMed]
- 38.Schwaerzel M, Monastirioti M, Scholz H, Friggi-Grelin F, Birman S, Heisenberg M. Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila. J Neurosci. 2003;23:10495–10502. doi: 10.1523/JNEUROSCI.23-33-10495.2003. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14627633. [DOI] [PMC free article] [PubMed]
- 39.Wang Y, Chiang AS, Xia S, Kitamoto T, Tully T, Zhong Y. Blockade of neurotransmission in Drosophila mushroom bodies impairs odor attraction, but not repulsion. Curr Biol. 2003;13:1900–1904. doi: 10.1016/j.cub.2003.10.003. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14588247. [DOI] [PubMed]
- 40.Mons N, Guillou JL, Jaffard R. The role of Ca2+/calmodulinstimulable adenylyl cyclases as molecular coincidence detectors in memory formation. Cell Mol Life Sci. 1999;55:525–533. doi: 10.1007/s000180050311. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10357223. [DOI] [PMC free article] [PubMed]
- 41.Yin JC, Tully T. CREB and the formation of long-term memory. Curr Opin Neurobiol. 1996;6:264–268. doi: 10.1016/s0959-4388(96)80082-1. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8725970. [DOI] [PubMed]
- 42.Heisenberg M, Borst A, Wagner S, Byers D. Drosophila mushroom body mutants are deficient in olfactory learning. J Neurogenet. 1985;2:1–30. doi: 10.3109/01677068509100140. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=4020527. [DOI] [PubMed]
- 43.de Belle JS, Heisenberg M. Associative odor learning in Drosophila abolished by chemical ablation of mushroom bodies. Science. 1994;263:692–695. doi: 10.1126/science.8303280. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8303280. [DOI] [PubMed]
- 44.Pascual A, Preat T. Localization of long-term memory within the Drosophila mushroom body. Science. 2001;294:1115–1117. doi: 10.1126/science.1064200. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11691997. [DOI] [PubMed]
- 45.Dubnau J, Grady L, Kitamoto T, Tully T. Disruption of neurotransmission in Drosophila mushroom body blocks retrieval but not acquisition of memory. Nature. 2001;411:476–480. doi: 10.1038/35078077. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11373680. [DOI] [PubMed]
- 46.McGuire SE, Le PT, Davis RL. The role of Drosophila mushroom body signaling in olfactory memory. Science. 2001;293:1330–1333. doi: 10.1126/science.1062622. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11397912. [DOI] [PubMed]
- 47.Wang Y, Mamiya A, Chiang AS, Zhong Y. Imaging of an early memory trace in the Drosophila mushroom body. J Neurosci. 2008;28:4368–4376. doi: 10.1523/JNEUROSCI.2958-07.2008. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18434515. [DOI] [PMC free article] [PubMed]
- 48.Didelot G, Molinari F, Tchenio P, Comas D, Milhiet E, Munnich A, Colleaux L, Preat T. Tequila, a neurotrypsin ortholog, regulates long-term memory formation in Drosophila. Science. 2006;313:851–853. doi: 10.1126/science.1127215. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16902143. [DOI] [PubMed]
- 49.Pavlopoulos E, Anezaki M, Skoulakis EM. Neuralized is expressed in the alpha/beta lobes of adult Drosophila mushroom bodies and facilitates olfactory long-term memory formation. Proc Natl Acad Sci U S A. 2008;105:14674–14679. doi: 10.1073/pnas.0801605105. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18794519. [DOI] [PMC free article] [PubMed]
- 50.Presente A, Boyles RS, Serway CN, de Belle JS, Andres AJ. Notch is required for long-term memory in Drosophila. Proc Natl Acad Sci U S A. 2004;101:1764–1768. doi: 10.1073/pnas.0308259100. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14752200. [DOI] [PMC free article] [PubMed]
- 51.Ge X, Hannan F, Xie Z, Feng C, Tully T, Zhou H, Xie Z, Zhong Y. Notch signaling in Drosophila long-term memory formation. Proc Natl Acad Sci U S A. 2004;101:10172–10176. doi: 10.1073/pnas.0403497101. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15220476. [DOI] [PMC free article] [PubMed]
- 52.Qian M, Pan G, Sun L, Feng C, Xie Z, Tully T, Zhong Y. Receptor-like tyrosine phosphatase PTP10D is required for long-term memory in Drosophila. J Neurosci. 2007;27:4396–4402. doi: 10.1523/JNEUROSCI.4054-06.2007. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17442824. [DOI] [PMC free article] [PubMed]
- 53.Ho IS, Hannan F, Guo HF, Hakker I, Zhong Y. Distinct functional domains of neurofibromatosis type 1 regulate immediate versus long-term memory formation. J Neurosci. 2007;27:6852–6857. doi: 10.1523/JNEUROSCI.0933-07.2007. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17581973. [DOI] [PMC free article] [PubMed]
- 54.Dubnau J, Chiang AS, Grady L, Barditch J, Gossweiler S, McNeil J, Smith P, Buldoc F, Scott R, Certa U, Broger C, Tully T. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12593794. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.