Abstract
The fungal neurotoxin α-cyclopiazonic acid (CPA), a nanomolar inhibitor of Ca2+-ATPase, has a pentacyclic indole tetramic acid scaffold that arises from one molecule of tryptophan, acetyl-CoA, malonyl-CoA and dimethylallyl pyrophosphate by consecutive action of three enzymes CpaS, D, O. CpaS is a hybrid, two module polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS) that makes and releases cyclo-acetoacetyl-L-tryptophan (cAATrp), the tetramic acid that serves as substrate for subsequent prenylation and oxidative cyclization to the five ring CPA scaffold. The NRPS module in CpaS has a predicted four domain organization of Condensation, Adenylation, Thiolation, Reductase* (C-A-T-R*) where R* lacks the critical Ser-Tyr-Lys catalytic triad of the short chain dehydrogenase/reductase (SDR) super family. By heterologous overproduction in E. coli of the 56 kDa Aspergillus flavus CpaS TR* didomain, and the single T and R* domains, we demonstrate that CpaS catalyzes a Dieckmann type cyclization on the N-acetoacetyl-Trp intermediate bound in thioester linkage to the phosphopantetheinyl arm of the T domain to form and release cAATrp. This occurs without any participation of NAD(P)H, so R* does not function as a canonical SDR family member. Use of the T and R* domains in in trans assays enabled multiple turnovers and evaluation of specific mutants. Mutation of the D3803 residue in the R* domain, conserved in other fungal tetramate synthetases, abolished activity both in in trans and in cis (TR*) activity assays. It is likely that cyclization of β-ketoacyl-aminoacyl-S-pantetheinyl intermediates to released tetramates represents a default cyclization/release route for redox-incompetent R* domains embedded in NRPS assembly lines.
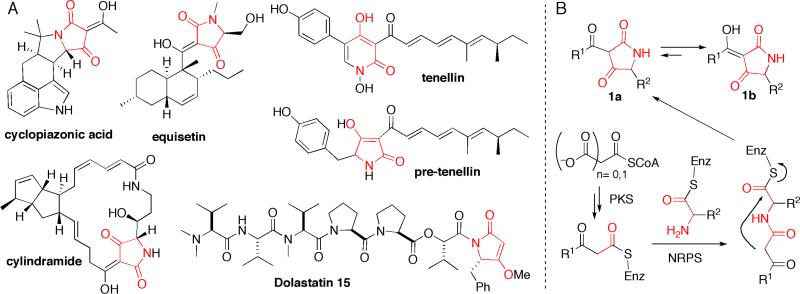
The heterocyclic pyrrolidine-2,4-dione ring systems commonly known as tetramic acids are found in many natural product scaffolds originating from a variety of terrestrial and marine species such as actinobacteria, fungi, sponges and cyanobacteria (for selected examples, see Figure 1A). (1) Due to both the conformational constraint of the five membered ring and the dicarbonyl functionality, tetramic acid moieties are specific pharmacophore elements for recognition by a range of biological targets. Such natural products thus exhibit a wide range of activities, including antibacterial, antiviral and antitumor. The great majority of tetramic acids are 3-acyl substituted (1a) with the enol isomer (1b) as the predominating tautomer (Figure 1B), providing an additional group decorating the tetramic acid scaffold and interacting with cellular targets. Inspection of 3-acyl tetramate natural products suggests a polyketide origin for the 3-acyl chain, with a single amino acid moiety providing the nitrogen atom and the C5 side chain (Figure 1B). Several biosynthetic gene clusters have been recently identified and sequenced that validate the polyketide synthase (PKS)-nonribosomal peptide synthetase (NRPS) hybrid assembly prediction. (2-9) In general these assembly lines contain a series of polyketide synthase modules sufficient to build the full length 3-ketoacyl chain and then a single nonribosomal peptide synthetase module as the chain-terminating catalyst. Therefore, the 3-acyl-2,4-pyrrolidinedione (tetramate) rings are thought to be formed in the release step of the PK-NRP hybrid chain from its enzymatic assembly line (Figure 1B).
Figure 1.
(A) Selected tetramic acid natural products. (B) Proposed biosynthetic route to 3-acyltetramic acid.
This mode of NRPS-mediated release of thioester-linked chains, constituting pyrrolidine-2,4-dione formation, would join a variety of other known NRPS release mechanisms, including hydrolysis, macrolactonization, macrolactamization, and reduction. (10-12) A typical chain termination NRPS module, both in pure NRPS assembly lines and in hybrid PKS-NRPS assembly lines, has a minimal domain composition either C-A-T-TE, for hydrolysis or macrocyclization fates, or C-A-T-R, for reductive outcomes, (C = condensation, A= adenylation, T = thiolation). The chain terminating TE and R domains mediate the different product outcomes. The TE (thioesterase) domains are 35 kDa αβ hydrolase protein folds, while the R (reductase) domains belong to the short chain dehydrogenase/reductase (SDR) superfamily with Rossmann-fold nucleotide binding motifs, and use NAD(P)H to effect reductive release of the acyl chain as an aldehyde.
Identification of several fungal tetramate natural product biosynthetic gene clusters, including fusarin C, (2) equisetin, (3) tenellin, (4) pseutotin A, (5) aspyridone (6) and chaetoglobosin, (7) highlighted conserved hybrid PKS-NRPS genes that encode a N-terminal iterative PKS module and a C-terminal NRPS module. The apparent function of the NRPS module would be two fold: (a) selection and, installation of the chain-terminating amino acid moiety into the polyketide scaffold generated by the iterative PKS and (b) carrying out the tetramic acid formation in the release step. Bioinformatic analysis of the chain-terminating NRPS modules in fungal tetramate synthetase indicates a C-A-T-R four domain composition rather than a typical C-A-T-TE. An initial hypothesis was formulated (2-7) that the tetramate ring formation occurred via a reductive release to give an aldehyde intermediate that undergoes non-enzymatic ring closure to give a pyrrolidine-2-one, which was further oxidatively tailored by one or more P450 enzymes. However, a recent study by Cox and coworkers revealed that the heterologous overexpression of tenellin synthetase in Aspergillus oryzae led to in vivo production of a mature tetramic acid containing pre-tenellin, suggesting for the first time the R domain in tenellin synthetase might function as a simple condensation catalyst, which we would label R* to indicate a nonredox function. (13) In addition, Schmidt and colleagues have examined the excised, purified TR* didomain and R* domain of equisetin synthetase with a model substrate N-acetocetyl-l-alanyl-S-N-acetyl-cysteamine (N-acac-l-Ala-SNAC) as a soluble mimic of the complex acylpeptidyl chain that would normally be covalently tethered on the T domain. (14) They observed the redox-independent generation of the corresponding tetramate moiety by EqiS_TR* and EqiS_R*, consistent with a Dieckmann type formation of the 3-acyltetramate ring, although they could not obtain saturation kinetics with the model SNAC substrate. (14) These results raise the general question whether the reductase-like (R*) domains in fungal tetramate synthetases are indeed not aldehyde generators, but instead act in a default nonredox mode to carry out different carbonyl chemistry.
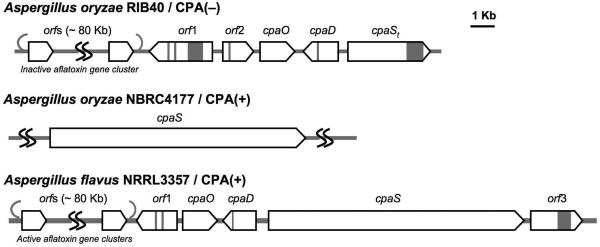
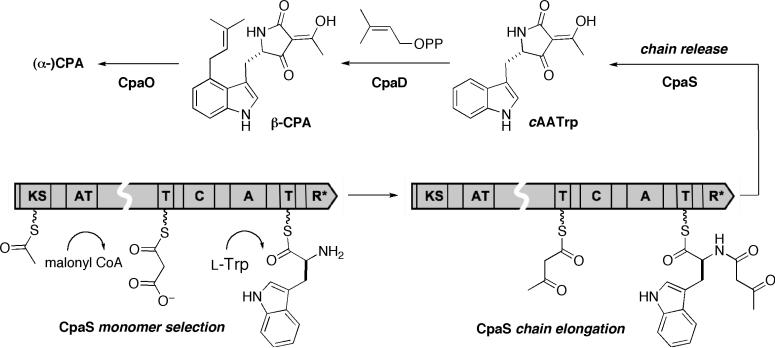
The latest addition to the family of fungal tetramate biosynthetic gene clusters is for cyclopiazonic acid (CPA or α-CPA, for structure see, Figure 1A), a potent neurotoxin with a rigid pentacyclic indole tetramate scaffold that acts as a nanomolar inhibitor of the sarcoplasmic Ca2+ ATPase. (15) Identification of CPA-related biosynthetic genes (16, 17) from two CPA-producing and one CPA-non-producing Aspergillus species correlates (Figure 2) with the previous biosynthetic predictions that the cyclopiazonate scaffold is derived from two biosynthetic intermediates (Scheme 1), cyclo-acetoacetyl L-tryptophan (cAATrp) and β-CPA, which are assembled from tryptophan, two molecules of acetate, and dimethylallyl diphosphate in a three-enzyme pathway. (18) The immediate precursor of α-CPA is the tricyclic β-CPA, converted by the flavoprotein oxidocyclase (CpaO) in a redox step forming two rings, (19-21); in turn β-CPA should be derived from the regioselective prenylation of the indole moiety of cAATrp by a dimethylallyltransferase (CpaD), (22) the second enzyme in this short metabolic pathway (Scheme 1). The first enzyme is a hybrid two module PKS-NRPS (CpaS), homologous to other sequenced fungal tetramate synthetases, that should be responsible for elaborating one molecule of acetyl-CoA, malonyl-CoA and tryptophan, to a tethered N-acetoacetyl-Trp-S-T domain intermediate on the way to the released tetramate cAATrp (Scheme 1). The occurrence of a truncated, rather than full length, CpaS in A. oryzae RIB40 would thus account for its CPA non-producing status (Figure 2). (17)
Figure 2.
Summary of CPA biosynthetic genes identified in three Aspergillus sp. including two CPA producing/CPA(+) strains and one CPA non-producing/CPA(−) strain
Scheme 1.
CPA biosynthetic enzymes (CpaS, CpaD and CpaO) and their roles to generate and tailor intermediates cyclo-acetoacetyl l-tryptophan (cAATrp) and β-CPA to afford (α-)CPA.
Our close inspection of the CpaS C-terminal R domain disclosed a mutation (Tyr→Leu) within the SDR Ser-Tyr-Lys catalytic triad that is of paramount importance for reductase activities. (23-25) The absence of any redox ORFs such a cytochrome P450s in the vicinities of CpaS, D, O, further suggests that the tetramate moiety of cAATrp is likely derived from the direct action of CpaS in a non-redox fashion.
In this study we have overproduced the C-terminal T-R* didomain of CpaS and establish that it does carry out a redox-independent Dieckmann cyclization, but only when the N-acac-tryptophanyl moiety is presented to the CpaS R* domain on its normal pantetheinyl covalent tether on the immediately upstream T domain. The development of an in trans assay between the T and R* domain of CpaS allowed us to carry out mutagenesis studies on CpaS_R* and identify unique amino acid residues within this fungal tetramate synthetase that are important for its Dieckmann cyclization activity.
Experimental Section
General methods and materials
Standard molecular biology procedures were performed as described(47). Oligonucleotide primers were synthesized by Integrated DNA Technologies (Coralville, IA). PCR was performed either using KAPA HiFi DNA polymerase (KAPA Biosystems) or Fusion high fidelity DNA polymerase (New England Biolab) under suggested conditions on a BioRAD MyCycler™ thermocycler. For general cloning, E. coli NovaBlue(DE3) cells (Novagen) were used. For protein overproduction, BL21 Star (DE3) cells (Invitrogen) were used. pET24b plasmid was obtained from Novagen. Coventional cloning was performed using DNA ligation kit Ver 2.1 (Takara). pET151/D-Topo expression kit was obtained from Invitrogen. DNA sequencing was performed at the Molecular Biology Core Facilities of the Dana Farber Cancer Institute (Boston, MA). Aspergillus oryzae RIB40 strain (ATCC 42149) and genomic DNA were obtained from American Type Culture Collection (ATCC). Aspergillus flavus NRRL3357 strain and genomic DNA were obtained from Dr. Jiujiang Yu (USGA). Dimethylallylpyrophosphate (DMAPP) was obtained from Isoprenoids LC. [1-14C]Acetyl-CoA (56 Ci/mol) was obtained from GE Healthcare. Phosphopantetheinyl transferase Sfp was heterologously overproduced and purified to apparent homogeinity as previously described (44). All other chemicals were purchased from Sigma-Aldrich unless noted otherwise. HPLC was performed on a Beckman System Gold (Beckman Coulter) instrument on Phenomenex Luna C18 columns (250 × 4.5 mm, for analytical purpose; 250 × 21.6 mm, for preparative purpose). The eluent was monitored for absorption at 280 nm for all LC analysis of tryptophan derivatives. LC-MS analysis was carried out on Shimadzu LCMS-QP8000a (for low resolution mass) and Agilent series 1100 LC/MSD-TOF (for high resolution mass) machines. 1H NMR spectra were recorded on a Varian 600 MHz spectrometer. Chemical shifts are reported in ppm with the solvent resonance resulting from incomplete deuteration as the internal standard (CDCl3 δ7.26, CD3OD δ3.31). Data are reported as follows: chemical shift, multiplicity (s = singlet, d = doublet, t = triplet, q = quartet, p = pentet, br = broad, m = multiplet), coupling constants (Hz), and integration.
Cloning of cpaS_TR*, cpaS_T, cpaS_R*
Domain boundary of CpaS (3906 aa) was analyzed using Pfam database (http://pfam.sanger.ac.uk/). cpaS_TR*(3433-3906), cpaS_T(3460-3543) and cpaS_R*(3544-3906) were amplified directly from A. flavus NRRL 3357 genomic DNA using the following primers: cpaS_TR* forward, CACCATGCGCCCTTCCACAATCATC; cpaS_TR* reverse, CTATTAATTACCCAAGTAGGGATATCG; cpaS_T forward, CACCATGCCATGGACGGCGCC ; cpaS_T reverse, CTATTATGATGGTGATTCAGC ; cpaS_R* forward, GAGTCATATGGGCAATGCGCCGTACGATATC ; cpaS_R* reverse, CTTAGCGGCCGCATTACCCAAGTAGGGATATCG. Amplified genes were purified by agarose gel. cpaS_TR* and cpaS_T genes were cloned into pET151/D-Topo vector according to the manufacture instruction to yield N-terminal His6-tagged expression plasmids. cpaS_R* was ligated to the NdeI and NotI sites of pET24b vector to give C-terminal His6-tagged expression plasmid. Correct clones were obtained by the subsequent restrictive digestions and sequencing of the isolated plasmids.
Overproduction and purification of CpaS_TR*, CpaS_T, CpaS_R*
Plasmids containing the verified cpaS_TR*, cpaS_T and cpaS_R* genes were transferred into E. coli BL21 Star (DE3) competent cells for protein expression. Cells were cultured in Luria Bertani medium containing appropriate antibiotics (100 μg/mL of ampicillin for pET151 vector and 50 μL/mL kanamycin for pET24b vector). A fresh overnight culture from a single colony was used to inoculate 2 × 1L medium. Cells were grown at 37 °C in a shaker at 200 rpm till mid-log phase (A600 ≈ 0.5–0.7). The cultures were then cooled to 16 °C and induced by isopropyl β-D-1-thiogalactopyranoside with a final concentration of 0.1 mM. The cultures were allowed to further grow at 16 °C for 12–16 h. The cells were harvested by centrifugation and cell pellets from the 2L culture were resuspended in a buffer (30 mL) containing 20 mM Tris-HCl pH 8, 500 mM NaCl, 20 mM imidazole, 10 mM MgCl2, 1mM DTT and homogenized. The homogenized cells were lysed by passing through a cell disruptor (Avestin EmulsiFlex-C5) three times at 5000-10,000 psi, and the lysate was clarified by ultracentrifugation (35,000 g × 35 min). The supernatant was incubated with 2 mL of Ni-NTA-agarose resin (Qiagen) at 4 °C for 2 h and loaded to a column. The resin was then washed with a buffer (20 mL) containing 20 mM Tris-HCl pH 8, 500 mM NaCl, 20 mM imidazole. His6-tagged proteins was eluted with 200 mM imidazole in 20 mM Tris-HCl pH 8, 500 mM NaCl. The protein was further buffer-exchanged and purified by gel filtration chromatography using a HiLoad 26/60 Superdex 200 column (GE Biosciences) with a buffer containing 25 mM HEPES pH 7.0, 50 mM NaCl, 1mM DTT, 5 mM MgCl2, 5% glycerol. Fractions containing desired proteins were pooled and concentrated using Amicon Ultra centrifugal filter device (Millipore). The protein concentration was determined by Bradford assay and the protein's absorbance at 280 nm with the predicted molar extinction coefficient. Proteins were aliquot-flash-frozen in liquid nitrogen and stored at −80 °C for further usages. The overproduction yield was ca. 12 mg/mL for CpaS_TR*, 7 mg/L for CpaS_T and 15 mg/L for CpaS_R*.
Phosphopantetheinylation assays of CpaS_TR*, CpaS_T
Phosphopantetheinylation assay was performed by measuring the incorporation of radiolabeled [1-14C]acetyl-SCoA onto heterologously overproduced CpaS_TR* and CpaS_T and the conditions are as the following. 75mM Tris•HCl (pH 7.5), 10mM MgCl2, 0.5mM TCEP, 100 μM [1-14C]acetyl-CoA (12.19 Ci/mol, final), and 10 μM CpaS_TR* (wt and mutants) or CpaS_T. Reactions were started by adding 3 μM Sfp. Aliquotes of the reaction mixtures were quenched at certain time intervals with of 10% (wt/vol) TCA, followed by the addition of BSA (100 mg). The protein pellet was washed with 10% (wt/vol) TCA and resuspended in formic acid, and the amount of radioactive label was measured by liquid scintillation counting. Mass spectrometry analysis of overproduced CpaS_TR* and CpaS_R* were performed according to the described procedure (lit) at the Dana Farber Cancer Institute Core Facility.
Preparation of cAATrp
To a suspension of 4Å molecular sieves (0.5 g) in THF (10 mL) was added L-Trp-OMe•HCl (254 mg, 1.0 mmol) and Et3N (280 μL, 2.2 mmol) at room temperature. After stirred for 15 min, S-tert-butyl acetothioacetate (360 μL, 2 mmol) was added, followed by the dropwise addition of a solution of silver trifluoroacetate (660 mg, 3 mmol) in THF (5 mL). The mixture was stirred vigorously overnight. The precipitates and molecular sieves were filtered off and the filtrate was evaporated to give the crude N-Acac-l-Trp-OMe. Further purification by silica gel chromatography to give N-Acac-l-Trp-OMe (156 mg, 52%). 1H NMR (600 MHz, CDCl3) δ 8.36 (s, 1H), 7.52 (d, 1H, J = 7.8 Hz), 7.32 (d, 1H, J = 8.4 Hz), 7.18-7.14 (m, 1H), 7.11-7.07 (m, 1H), 7.03 (s, 1H), 4.93-4.89 (m, 1H), 3.68 (s, 3H), 3.33-3.30 (m, 1H), 3.27 (s, 2H), 2.11 (s, 3H). HRMS (ESI): Calcd for C16H18N2O4 [M + Na]+: 325.1164, Found: 325.1163. To a solution of N-Acac-l-Trp-OMe (61 mg, 0.2 mmol) in THF (5 mL) was added methanolic NaOMe (0.8 mL, 0.4 mmol, 0.5 M in MeOH) at 0 °C. The mixture was warmed to room temperature and further stirred for 1h before stopped by adding drops of acetic acid. The solvents were evaporated and the residue was subjected to silica gel chromatography to give cAAtrp (47 mg, 87%) as a pale yellow solid. [α]D24 = −117.8 (c = 0.62, MeOH). 1H NMR (600 MHz, CD3OD) δ 7.56 (d, 1H, J = 8.4 Hz), 7.31 (d, 1H, J = 8.4 Hz), 7.11 (s, 1H), 7.08-7.06 (m, 1H), 7.01-6.98 (m, 1H), 4.76 (dd, 1H, J = 7.8, 5.4 Hz), 3.36 (dd, 1H, J = 14.4, 5.4 Hz), 3.19 (dd, 1H, J = 14.4, 7.8 Hz), 2.05 (s, 3H). HRMS (ESI): Calcd for C15H14N2O3 [M + Na]+ : 293.0902, Found: 293.0902.
Preparation of acacTrp-SCoA and acacTrp-SNAC
acac-l-Trp was prepared according to the literature procedure.(48)To a solution of acac-l-Trp (6.8 mg) in CH3CN (0.3 mL) and THF (0.3 mL) was added PyBOP ( 24.6 mg). After stirred for 15 min at room temperature, coenzyme A tri-lithium salt (18.5 mg) in H2O (0.1 mL) was added. The resulting mixture was stirred vigorously for 1h and K2CO3 (6.5 mg) was added. Reaction was stirred further for 1 h before quenched by the addition of AcOH (30 μL). The mixture was diluted with THF/H2O mixture to provide a homogeneous solution and directly subjected to preparative HPLC purification. Fractions containing the target acacTrp-SCoA was identified by LC-MS, combined and lyophilized to give a white powder (15.6 mg, 62%). 1H NMR (600 MHz, D2O) δ 8.45 (s, 1H), 8.02 (s, 1H), 7.32 (d, 1H, J = 7.8 Hz), 7.22 (d, 1H, J = 7.8 Hz), 7.00 (app t, 1H, J = 7.8 Hz), 6.98 (s, 1H), 6.90 (app t, 1H, J = 7.8 Hz), 5.99 (d, 1H, J = 6.0 Hz), 4.80-4.74 (m, 1H), 4.73-4.71 (m, 1H), 4.70-4.68 (m, 1H), 4.51-4.49 (m, 1H), 4.23-4.17 (m, 2H), 3.95 (s, 1H), 3.81 (dd, 1H, J = 9.6, 4.8 Hz), 3.56 (dd, 1H, J = 9.6, 4.8 Hz), 3.38-3.31 (m, 3H), 3.00-2.96 (dd, 1H, J = 15.0, 8.4 Hz), 2.88-2.84 (m, 2H), 2.29 (t, 1H, J = 6.0 Hz), 1.97 (s, 3H), 0.86 (s, 3H), 0.73 (s, 3H). HRMS (ESI): Calcd for C36H50N9O19P3S [M+H]+ : 1038.2235, Found: 1038.2240. acacTrp-SNAC was prepared in an identical procedure except replacing coenzyme A tri-lithium salt with N-Ac cysteamine.
Mutagenesis of CpaS_TR* and CpaS_R* and mutant protein overproduction
The mutants of cpaS_TR* and cpaS_R* were generated according to the protocol described in Stratagene QuikChange® Multi Site-Directed Mutagenesis Kit, using plasmids pET151::cpaS_TR* and pET24b::cpaS_R* as the templates and primers listed in Table 1. Primer oligonucleotides were in situ 5’-phosphorylated using T4 polynucleotide kinase (New England Biolab). Correct mutant plasmids were verified by sequencing. The mutant proteins were overproduced and purified in the same manner as those described for the wild types.
Table 1.
Oligonucleotide primers used to generate CpaS_TR* and CpaS_R* mutants
| CpaS_TR* CpaS_R* mutant | Primer sequencea |
|---|---|
| S3508A | 5'-CTCCTCGGGGGAAATGCGACGCTGCTGGTCAGG |
| C3619A | 5'-GTGGAAAGAGTGCATGCGATCGCCGTGGAAAAGG |
| D3663A | 5'-CTGCAAGACCGGATCGCGGTGGTGATCCACAAC |
| H3667A | 5'-ATCGACGTGGTGATCGCGAACGGCTCGAATGGC |
| S3688A | 5'-TCCTCCGCTCGACGGGGCGGACGGGCTCACAGCAAC |
| S3707A | 5'-CCCCTGCACTATATCGCGTCCGGAAGGGTCATC |
| K3742A | 5'-GGGCTCACAGCAACCGCGTGGGCCAGCGAAGTC |
| D3777A | 5'-CAGGCTCCAGCACAGGCGGCACTGAACTCGCTC |
| D3803A | 5'-ATGGAAGGATACTTGGCGTTTCAGAAGGTAGAG |
| H3843A | 5'-GGTTTCCTTTTTCCACGCGTCGAGCAATGTCAAG |
bold: modified sequences
CpaS_TR* activity assay
The single time point assay (Figure 4A) was carried out in a 100 μL scale at room temperature with CpaS_TR* (70 μM), acacTrp-SCoA (50 μM) and buffer containing HEPES (50 mM, pH 7.0), MgCl2 (5 mM). The reaction was initiated by the addition of Sfp (30 μM) and terminated after 10 min with 0.2 mM TCA in MeOH (100 μL). Proteins were pelleted centrifugally (×13,000g) for 10 min at 4 °C and the supernatants were subjected to HPLC and LC-MS analyses. To examine if NADH and/or NADPH is required for the activity of CpaS_TR*, NADH (10 mM) or NADPH (10 mM) was included in the reaction buffer prior to the addition of Sfp. Control reactions that exclude CpaS_TR* and/or Sfp from the assay components were also carried out. Products were examined in the same fashion by HPLC.
Figure 4.
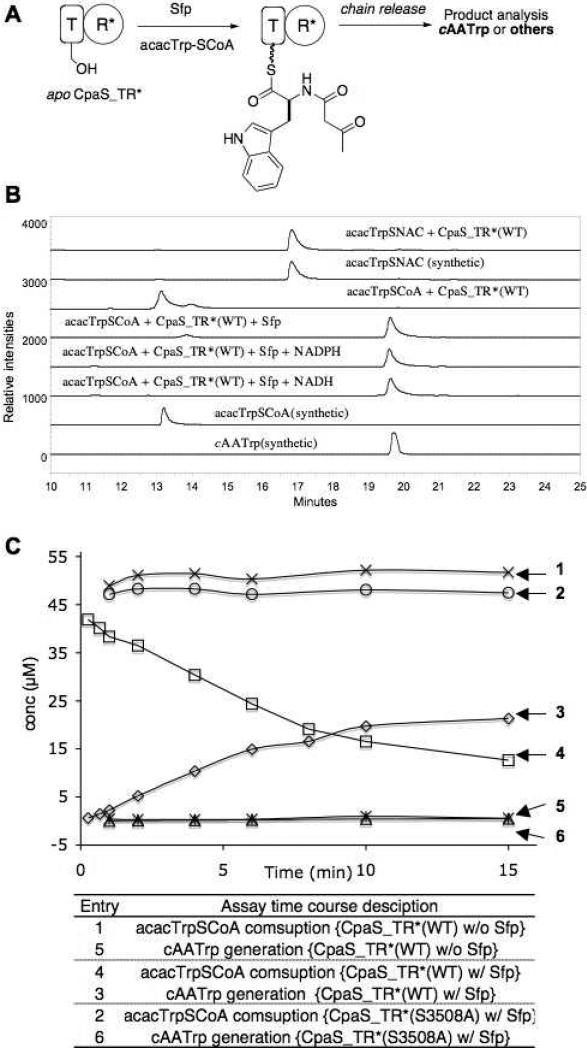
CpaS_TR* catalyzed formation of cAATrp requires the modification of apo CpaS_TR* by phosphopantetheinyltransferase Sfp with acacTrp-SCoA, independent of NADH and NADPH. (A) Schematized CpaS_TR* assay procedure. (B) HPLC traces of cAATrp formation from acacTrp-SCoA primed CpaS_TR* with or without NADH and NADPH (line 3-5), in comparison with synthetic standards of acacTrp-SCoA and cAATrp (line 1,2). CpaS_TR* did not take acacTrp-SCoA and acacTrp-SNAC as substrates (line 2 vs 6 and line 7 vs 8). (B) Time course analysis of cAATrp generation and acacTrp-SCoA consumptions, where CpaS_TR* was primed with acacTrp-SCoA with Sfp (entries 3, 4); CpaS_TR* was incubated with acacTrp-SCoA only (entries 1, 5); CpaS_TR* (S3508A) mutant was incubated with acacTrp-SCoA and Sfp (entries 2, 6).
The time course analysis of CpaS_TR* (WT or mutants) mediated cAATrp generation and acacTrp-SCoA consumption (Figure 4B&7) was carried out in a similar fashion as the single point assay except in a 400-500 μL scale. At each selected time point, 50 μL of the reaction mixture was quenched with 0.2 mM TCA in MeOH (50 μL), protein pelleted and supernatant was subjected to HPLC analysis. The areas of HPLC peaks corresponding to cAATrp and acacTrp-SCoA were obtained after manually adjusting HPLC chromatograph baselines. The area was fitted into a standard curve (concentration v.s. area) of cAATrp and acacTrp-SCoA generated using synthetic standards, to calculate the corresponding concentration.
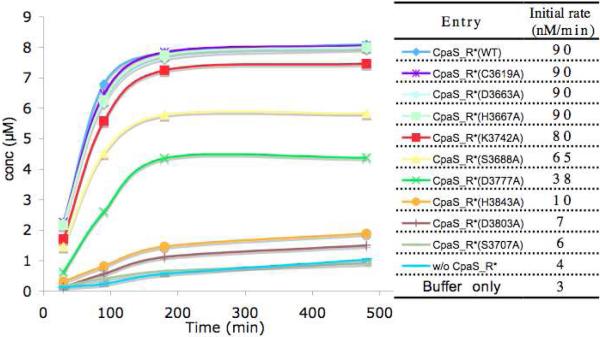
Figure 7.
Time course and rates comparison of cAATrp generation from N-acac-Trp-S-TCpaS with CpaS_R* (WT and mutants) in trans.
CpaS_T assay with CpaS_R* in trans
The assay (Figure 5, entries 1&2) was carried out at room temperature with CpaS_T (50 μM), acacTrp-SCoA (25 μM), Sfp (5 μM) and buffer containing HEPES (50 mM, pH 7.0), MgCl2 (5 mM). The reaction was initiated by the addition of CpaS_R* (2.5 μM, 5mol%). Analogous to CpaS_TR* time point assays, aliquots (50 μL) of the reaction mixture was quenched at each selected time point with 0.2 mM TCA in MeOH (50 μL). Protein was pelleted and supernatant was subjected to HPLC analysis to determine the conversion rate based on HPLC area of cAATrp and acacTrp-SCoA and the established standard curves. As control (Figure 5, entries 3&4), the assay was carried out without the addition of CpaS_R* and the rate of cAATrp generation and acacTrp-SCoA consumption were monitored in the same fashion. The in trans activities of nine CpaS_R* mutants with acacTrp-SCoA primed CpaS_T (Figure 6) were examined in the same fashion as CpaS_R (WT) and the time courses of cAATrp generation were analyzed.
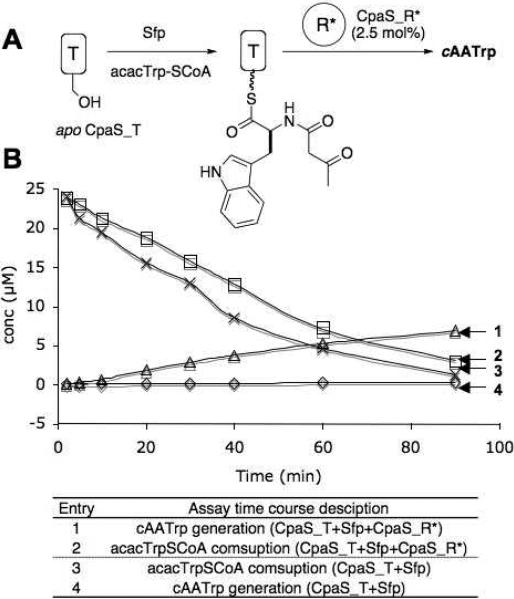
Figure 5.
acacTrp-S-CpaS_T and CpaS_R* activities in trans. (A) Schematized CpaS_T and CpaS_R* in trans assay procedure. (B) Time course analysis of cAATrp generation and acacTrp-SCoA consumption from acacTrp-SCoA primed CpaS_T with or without CpaS_R* present in trans.
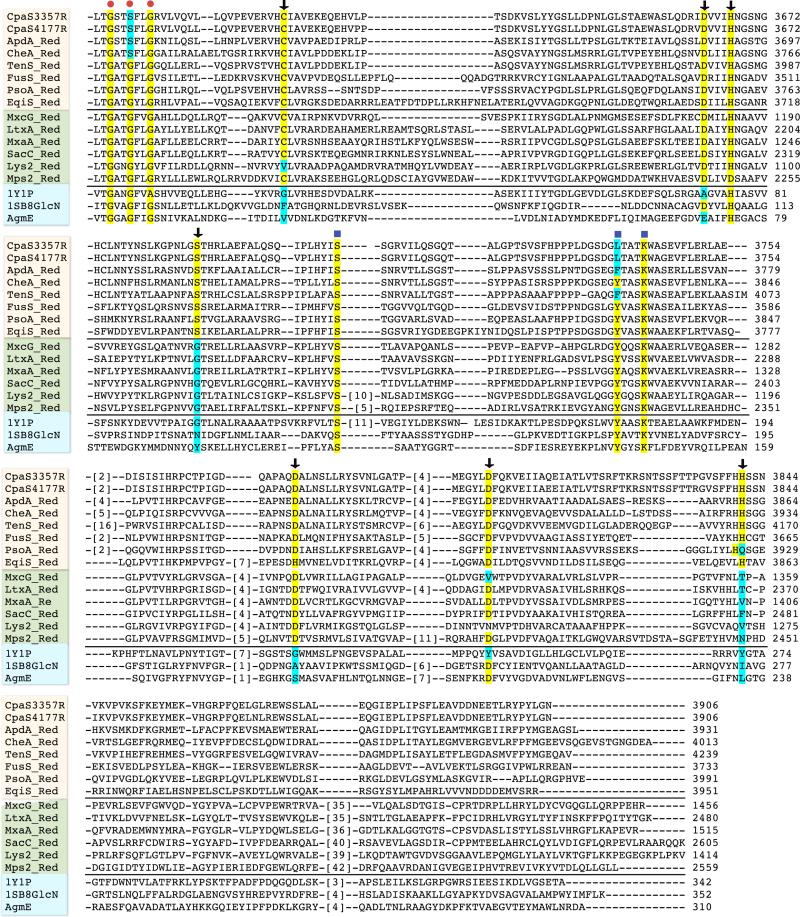
Figure 6.
Sequence alignment of the C-terminal reductase domains of CpaS from A. flavus NRRL3357 and A. oryzae NBRC4177 with 1) C-terminal reductase domains of all other known fungal tetramate synthases; 2) known redox-active C-terminal reductase domains from selected nonribosomal peptide synthetases; 3) selected structurally characterized short-chain reductase/dehydrogenases. Sequences were aligned using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2), starting from the conserved nucleotide binding domain. Sequence numbers represent the actual position number of the amino acid in the full-length synthetase. Red dot indicates the conserved NADH/NADPH binding domain; blue square highlights the positions of the SDR Ser-Tyr-Lys catalytic triad; black arrows points the conserved polar amino acid residues in the R* domain of fungal tetramate synthase that distinguish from either other NRPS redox-active R domains and/or structural characterized SDRs. Protein name abbreviations: aspyridone synthetase (ApdA), chaetoglobosin synthetase (CheA), tenellin synthetase (TenS), fusarin synthetase (FusS), pseurotin synthetase (PsoA), equisetin synthetase (EqiS), myxochelin NRPS module (MxcG), lyngbyatoxin synthetase (LtxA), myxalamid NRPS module (MxaA), safracin NRPS (SacC), α-aminoadipate reductase (Lys2), mycobacterial glycopeptidolipid synthetase (Mps2), aldehyde reductase (1Y1P), UDP-N-acetyl glucosamine 4-epimase (1SB8), ADP-L-glycero-mannoheptose 6-epimerase (AgmE).
Results
Cloning and overexpression of CpaS_TR*, CpaS_R* and CpaS_T
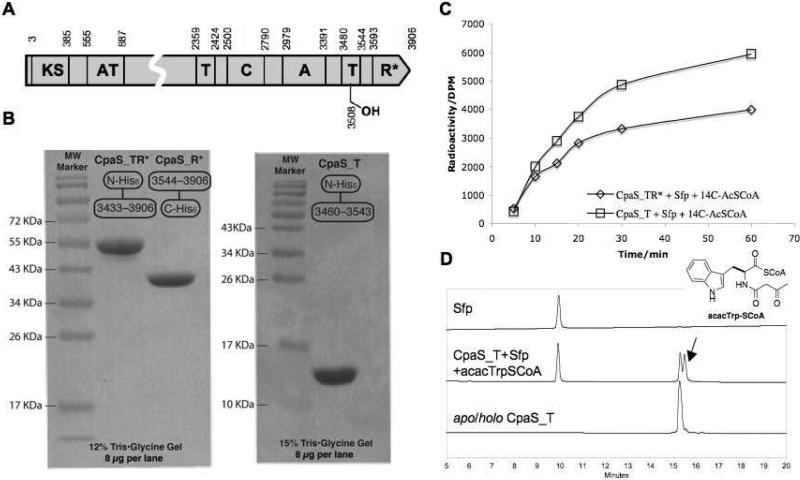
CpaS is a 431KDa hybrid PKS-NRPS that contains 3906 amino acids (Figure 3A). Two complete cpaS genes have been disclosed from A. flavus NRRL3357 and A. oryzae NBRC4177 (Figure 2), which are 93% identical. (16, 17) A. flavus NRRL3357 cpaS was used in this study. To study the tetramate ring formation and chain release mechanism of CpaS, we undertook to clone and overproduce truncated forms of CpaS protein in E. coli, including the 56 kDa C-terminal TR* didomain (CpaS_TR*), the 40 kDa R* domain (CpaS_R*) and a 14 kDa stand-alone carrier protein/thiolation domain construct (CpaS_T). The prediction of CpaS domain boundaries was based on bioinformatic comparisons with characterized NRPS modular enzymes. The genes were amplified directly from A. flavus NRRL3357 genomic DNA and cloned into p151ET/Topo and pET24b vectors to afford N-terminal His6-tagged CpaS_TR*, CpaS_T and C-terminal His6-tagged CpaS_R* expression constructs respectively (see methods). Overproductions in E. coli gave soluble CpaS_TR*, CpaS_T and CpaS_R* proteins in good yields, which were further purified to apparent homogeneities (Figure 3B) by successive nickel-affinity and gel filtration chromatographies. All truncated CpaS proteins appeared monomeric in solution, as estimated by size exclusion chromatography.
Figure 3.
Overproduction of truncated CpaS proteins and their phosphopantentienylation assays. (a) Domain architecture and boundaries of CpaS (NRRL3357). (B) SDS-PAGE purity of overproduced CpaS_TR*, CpaR* and CpaS_T. (C) Phosphopantenthenylation of CpaS_TR* and CpaS_T by Sfp. Time course was shown by analyzing the incorporation of [1-14C]acetyl-S-phosphopantetheinyl group from [1-14C]acetyl-CoA. (D) HPLC analysis of overproduced apo/holo CpaS_T and acacTrp-SCoA modified CpaS_T (arrow indicates N-acacTrp-S-T intermediate, predicted mass: 14625.4 Da; observed mass, 14625.0 Da)
Phosphopantetheinylation assay of CpaS_TR* and CpaS_T
With ample amounts of protein in hand, we first sought to assess the apo/holo states of overproduced CpaS_TR* and CpaS_T arising during heterologous expression. The time-dependent incorporation of [14C] acetyl-pantetheinyl groups onto the apo forms of the T domains in either construct from radiolabeled [1-14C]acetyl-SCoA, catalyzed by the promiscuous phosphopantentheinyl transferase Sfp (5 mol%), were observed within 60 min (Figure 3C). By increasing the amount of Sfp to 30 mol%, the posttranslational phosphopantetheinylation process can be expedited to reach saturation within 10 min. The measured total incorporation of 14C-radiolabel corresponds to ca. 50-60% of the calculated value, suggesting the presence of 40-50% holo forms of the T domains in the overproduced CpaS_TR* and CpaS_T that are posttranslationally primed by endogenous phosphopantetheinyl transferase activity of E. coli cells. This was further confirmed by mass spectrometry analysis of the intact proteins (data not shown). We further demonstrated the apo CpaS_T can be primed with synthetic acacTrp-S-CoA (see methods for preparation) using Sfp, as shown by the generation of N-acac-l-Trp-S-T covalent adduct (Figure 3D). HPLC analysis showed baseline separation between apo/holo forms of CpaS_T and N-acac-l-Trp-S-T species, whose identity was confirmed by Q-TOF MS analysis (Figure 3D)
Stoichiometric Generation of cAATrp by priming apo-CpaS_TR* with acacTrp-S-CoA using Sfp
To address the role of C-terminal R* domain of CpaS in cAATrp generation, we first sought to prime the apo form of the T domain in CpaS-TR* with synthetic acacTrp-S-CoA using Sfp to generate a transient N-acac-l-Trp-S-T tethered thioester (Figure 4A). This covalent intermediate, should be presentable in cis to the downstream R* domain. If the R* domain acts as a Dieckmann cyclization catalyst, the cAATrp product should be released and be detectable. As shown in line 5 of Figure 4B, incubation of an equimolar amount of apo CpaS_TR* with acacTrp-SCoA in the presence of Sfp at pH 7, room temperature for 10 min, led to the generation of a new peak that corresponds to cAATrp, as verified by high resolution MS analysis ([M-H]− cal for C15H13N2O3: 269.0926, obs: 269.0924) and HPLC co-elution with synthetic cAATrp. The generation of cAATrp from acacTrp-SCoA at pH 7 was independent of and did not require NAD(P)H cofactor (Figure 4B lines 3 and 4), reminiscent of the recent observation with EqiS_TR* didomain from equisetin synthetase. (14) However, in distinction to the Eqi-TR*, which accepts soluble aminoacyl SNACs, the production of cAATrp by CpaS-TR* strictly depends on the presence of both CpaS_TR* and the priming phosphopantetheinyltransferase Sfp, as schematized in Figure 4A. We did not observe the conversions of acacTrp-SCoA or acacTrp-SNAC to cAATrp by either CpaS_TR* or CpaS_R* alone (for CpaS_TR* assay, see Figure 4B lines 1 and 6–8; CpaS_R* assay data not shown), indicating the R* domain in CpaS recognized the N-acac-L-tryptophanyl group only when it is tethered to the phosphopantetheinyl arm of the upstream CpaS_T domain and presented in its native context.
We next carried out a time course analysis of cAATrp generation versus acacTrp-SCoA consumption in the presence of CpaS_TR* and Sfp. As shown in Figure 4C (entries 3 & 4), the rate of cAATrp generation correlates closely with the consumption of acacTrp-SCoA, indicating the priming of the carrier protein/thiolation domain of CpaS_TR* with Sfp is likely a rate limiting step; any generated N-acac-l-Trp-S-T intermediate is rapidly acted on by the intramolecular R* domain to release the cAATrp. To validate the requirement for display of the N-acac-Trp moiety on the pantetheinyl arm of the T domain, the CH2OH side chain of Ser3508 was mutated to an alanine, blocking posttranslational priming. In this CpaS_TR*(S3508A) T domain active site mutant, the generation of cAATrp dropped to the baseline level (Figure 4C, entry 6), identical to what is observed for the assay using wild type CpaS_TR* without Sfp (Figure 4C, entry 5). In both cases, there is minimal consumption of acacTrp-SCoA (Figure 4C, entries 1&2). These studies thus consolidate the evidence that CpaS R* domain is a nicotinamide-independent condensation catalyst that releases the N-acac-L-tryptophanyl group tethered to the upstream T domain. In the didomain TR* construct, where the N-acacTrp-pantetheinyl arm is delivered by Sfp, only single turnovers occur and the kinetics of interdomain chain transfer and cyclization cannot be readily measured.
acacTrp-S-CpaS_T and CpaS_R* activities in trans
Given the above findings that the CpaS_R* domain was active in the T_R* didomain construct, the question arises why the CpaS R* domain and the EqiS R* domain, and by extension the reductase-like domains of other PKS-NRPS hybrid tetramate synthetases, are different from the traditional redox-active SDR enzyme family members. Our approach was to make mutants in the R* domain of CpaS and analyze effects on activity. We have noted above the major obstacle that when the T and R* domains are in cis within the same protein, as they are in native CpaS, and the substrate is introduced onto the apo form of the T domain by the phosphopantetheinyltransferase Sfp, only single turnovers can occur. Further, since the apo/holo levels of T domains in T-R* didomains varied from prep to prep for both wild type and mutant forms of CpaS fragments, quantitation of differences in sub-stoichiometric yields of product was not viable. Another limitation to kinetics is that the R* domain would not accept soluble thioester substrates (N-acacTrp-SNAC or –SCoA). To overcome this limitation, we sought to excise CpaS_T and R* as separate, folded domains and establish a CpaS_T/CpaS_R* assay in trans.
To this end, an N-His tagged CpaS_T and a C-His tagged CpaS*_R were generated (Figure 3B) and used for assays. Now the acacTrp-pantetheinyl-primed CpaS_T domain could be used as substrate in excess over the CpaS_R* domain as catalyst and multiple turnovers assessed between the in trans pair of domains (Figure 5A). CpaS_R* did mediate release of cAATrp. Using the same HPLC conditions as for CpaS_TR*, the in trans generation of cAATrp and the consumption of acacTrp-S-CoA were monitored over a 90-min time course (Figure 5B). While the incorporation rate of acacTrp-S-pantetheinyl from acacTrp-SCoA into CpaS_T appeared similar with or without the presence of CpaS_R* (2.5 mol%) (Figure 5B, entry 1&3), the generation of cAATrp from acacTrp-SCoA-primed CpaS_T was clearly accelerated (Figure 5B, entry 2&4). The relatively low turnover rate (0.03 ± 0.004 min−1) is likely due to the disrupted CpaS_T and CpaS_R* interaction (now in trans rather than in the native in cis array), and constitutes three turnovers in the 90 minute assay. This gave 30 fold signal to noise which allowed us to proceed with initial evaluation of mutants in the R* domain.
Identification of residues of CpaS R* domain related to its unusual nonredox Dieckmann cyclase activity
To identify residues within CpaS R* that contribute to its unusual redox-independent cyclase activity, a sequence comparison of the C-terminal R* domains of known fungal tetramate synthetases were mapped onto two comparator lists. One involved selected NRPS C-terminal reductase domains that are known to be bona fide redox active, enzyme domains including those involved in myxochelin, (26, 27) lyngbyatoxin, (28) myxalamid, (29) safracin, (30) lysine (yeast) (31) and mycobacterial glycopeptidolipid biosynthesis. (32) The second comparator set was three well characterized SDR enzymes, an aldehyde reductase, (33) UDP-N-acetyl glucosamine 4-epimerase (34) and ADP-L-glycero-mannoheptose 6-epimerase. (35) Multiple sequence alignments using ClustalW2 (Figure 6) showed the presence of the well conserved nucleotide binding domain GXX(G/S)XXG within all 17 sequences. However, the SDR catalytic triad Ser-Tyr-Lys (24) is not well conserved in the R* domains. Noteably, Tyr→Phe, Tyr→Leu mutations are present in the R* domains of tenellin, aspyridone and cyclopiazonic acid synthetases. The critical roles of tyrosine residues in the SDR Ser-Tyr-Lys catalytic triad have been examined and discussed in many cases, where the mutation Tyr→Phe would abolish the reductase activities. (23-25)
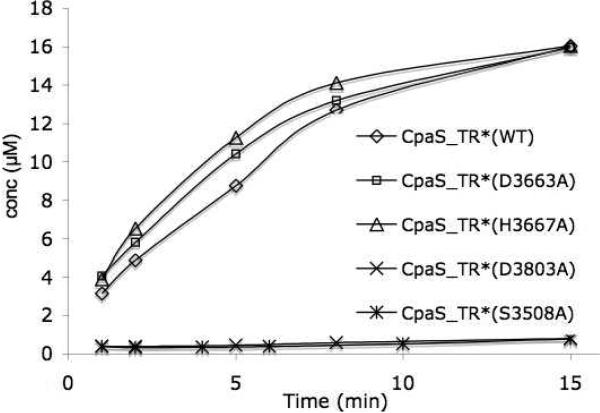
Further detailed examination of the aligned sequences highlighted seven other polar amino acid residues (C3619, D3663, H3667, S3688, D3772, D3803, H3843) that are exclusively conserved within all R* domains of known fungal tetramate synthetases, but not for redox-active NRPS R domains or the selected SDRs. We therefore chose these residues, along with S3707 and K3742 that are present in the Ser-Tyr-Lys triad, to carry out focused mutagenesis studies. CpaS_R* carrying the alanine mutations of these targeted nine polar residues were generated and the single mutant proteins were overproduced as for the wild type. Most of the CpaS_R* mutant proteins were obtained in good yields (5-15 mg/L), comparable to the wild type, except for CpaS_R*(S3707A) and CpaS_R*(H3843A) that overexpressed poorly in E. coli with yields ca. 0.1 mg/L. All CpaS_R* mutant proteins were purified to homogeneity by successive nickel affinity and gel filtration chromatographies. They were subjected to in trans assay with acacTrp-SCoA-primed CpaS_T, in an identical manner to the wild type CpaS_R* and the generation of cAATrp was monitored by HPLC over a time course of 6 hours. The results are summarized in Figure 7 and clearly indicate the activities of three CpaS_R mutants H3843A, D3803A, S3707A dropped dramatically to the background level, close to the background rate of non-enzymatic generation of cAATrp by N-acac-Trp-CpaS_T alone or by buffer (pH 7.0) from acacTrp-S-CoA.
We further attempted to correlate the observed CpaS_R* mutant activities in trans to N-acac-Trp-CpaS_T to the native context in cis by generating three corresponding CpaS_TR* didomain mutants and assaying cAATrp release. One mutant, D3803A had markedly diminished activity to generate cAATrp from N-acac-Trp-CpaS_T in trans, while two mutants, H3667A and D3663A, had activity with comparable to wild type, and could serve as positive controls. The three TR* didomain mutant proteins were overproduced, purified, and activities compared both to wild type and the anticipated catalytically dead mutant (S3508A) carrying the Ser-Ala mutation in the T active site. As observed in Figure 8, CpaS_TR* (H3667A and D3663A) still had comparable activities as the wild type; however CpaS_TR (D3803A) was as inactive for cAATrp generation as that of CpaS_TR(S3508A). This result further validated that the D3803 residue in the R* domain of CpaS is catalytically consequential.
Figure 8.
Time course comparison of cAATrp generation from acacTrp-SCoA primed CpaS_TR* (WT) with selected CpaS_TR* mutants (S3508A, D3663A, H3667A, D3803A).
Discussion
Pyrrolinedione ring systems are found in a variety of polyketide and nonribosomal natural products and are pharmacophoric elements in medicinally active members of these natural product classes. The 2,5–pyrrolinedione ring system is a succinamide ring and this regiosiomer is found in andrimid and moiramide, (36-38) but the 2,4-pyrrolinedione, known commonly as the tetramate ring system, is more widespread. (1) The tetramate ring can be a terminal structural element as in the anti HIV molecule equisetin and the related trichosetin or in the antitumor peptide dolatstain 15 or it can be an embedded ring system as found in the antitumor metabolite cylindramide and in the fused pentacyclic scaffold of α-cyclopiazonate (for structures see, Figure 1A). The nitrogen atom of the tetramate scaffolds arise from an amino acid monomer that is incorporated as part of the chain termination process, and the tetramate side chain at C5 derives from the side chain of that amino acid building block. Most of the naturally occurring tetramate rings have a polyketide-derived substituent at C3, reflecting the hybrid PKS-NRPS assembly lines that generate them. The 2,5-dicarbonyl groups offer H-bond interactions with protein targets, as validated by the x-ray structures of streptolydigin complexed with RNA polymerase (39) and of α-cyclopiazonate with sarcoplasmic reticulum Ca++-ATPase. (40, 41)
Because of the combined structural constraints and hydrogen bonding potential that 2,4-pyrrolinedione rings impart to the hybrid PK-NRP natural products, there is interest to decipher the molecular logic by which these ring systems are installed during biosynthesis. Fungal genomics and bioinformatic analyses for the biosynthetic assembly lines for the hybrid PK-NRP molecules show one or more PKS modules with an NRPS module that acts as the most downstream, chain-terminating module. It is therefore likely that tetramate ring formation is the last step in all such PKS-NRPS hybrid assembly lines, raising the question of how cyclization is coupled to disconnection of the full length acyl/peptidyl chain from the enzymatic assembly lines.
The cyclopiazonate biosynthetic pathway in the Aspergillus strains has the virtue of a short three enzyme pathway. The first enzyme is the single two module protein, CpaS, with a PKS module upstream of the NRPS module. The single PKS module converts an acetyl CoA and a malonyl CoA to an acetoacetyl-S-pantetheinyl intermediate (Scheme 1). The NRPS module selects and activates Trp, installing it on its thiolation domain. Action by the NRPS condensation domain makes the amide linkage and presumably generates the N-acetoacteyl-Trp-S-pantetheinyl intermediate in the NRPS module (Scheme 1) that is the last tethered intermediate. Action of the R* domain converts the acacTrp-S-pantetheinyl-T into cyclo-acetoacetyl-Trp product. The indole side chain of Trp becomes the C5 substituent of the released tetramate ring and the acetoacetyl moiety has functioned as intramolecular nucleophile, leaving an acetyl group as the C3 substituent, which exists predominantly in the enol form. The next two enzymes in the pathway CpaD and CpaO are post-assembly line tailoring catalysts, with CpaD effecting a regiospecific prenylation at the indole C4 poisition of cAATrp to yield β-CPA (Scheme 1). CpaO then carries out the deceptively simple dehydrogenation of β-CPA to yield an unstable ene-imine product, which is captured by intramolecular cyclization to create the pentacyclic fused scaffold of α-cyclopiazonate. (19) In this double cyclization step the nitrogen of the tetramate moiety can be viewed as the initiating nucleophile for the cascade.
Inspection of the NRPS tetramate-forming modules shows that the typical four domain termination module (C-A-T-TE) is replaced with a C-A-T-R* module, in which the terminal thioesterase domain is replaced with a domain of the short chain reductase/dehydrogenase superfamily. The thioesterase (TE) domains of NRPS termination modules have been shown to be the catalysts for hydrolytic and /or macolactamizing/macrolactonizing release of full length peptidyl chains from the covalent thioester linkage to the pantetheinyl arm of the neighboring T (thiolation) domain. It therefore followed that replacement of a TE domain by an R domain in a module would indicate reductive release by delivery of a hydride from NAD(P)H to the thioester to yield the tethered thiohemiacetal which unravels nonenzymatically to the free aldehyde and the HS-pantetheinyl arm on the T domain. (10-12) Indeed, in earlier efforts we proved this NAD(P)H-dependent reductive release mechanism by purified yeast Lys 5 in conversion of aminoadipate to the cyclic aldimine in lysine biosynthesis. (31) We have also described a tandem reduction of N-Me-Val-Trp-S-LtxA by two molecules of NADPH to yield first the dipeptidyl aldehyde and then the four electron-reduced alcohol in the lyngbyatoxin biosynthetic pathway. (28)
While a reduction/reoxidation pathway is conceivable for action of terminal R* domains in tetramate assembly, (2-7) studies on the T and R* domains from the fungal equisetin synthetase showed conclusively that the Eqi_R* domain was redox incompetent. (14) Instead, it acted on soluble N-acac-lAla-SNAC substrate mimics to produce the corresponding tetramate by an intramolecular Dieckmann cyclization.
In this study we made a similar T_R* didomain construct excised from the Aspergillus flavus CpaS bimodular enzyme and confirmed that this R* did not carry out reductive release of the N-acac-Trp thioester intermediate. Unlike EqiS_R*, CpaS R* domain did not accept low molecular weight N-acac-Trp thioester mimics (N-acac-Trp-SNAC and –SCoA), but only recognized the authentic N-acac-Trp thioester intermediate present natively on the pantetheinyl prosthetic group of the upstream thiolation domain. The tethered N-acac-Trp thioester intermediate was generated using the methodology we have discovered and developed in previous NRPS assembly line studies, (42-44) by combining the partially apo form of the CpaS_TR* construct heterologously overproduced in E. coli, synthetic acacTrp-S-CoA and the promiscuous phosphopantetheinyltransferase Sfp. The acacTrp chain was thus installed regiospecifically on the T domain of CpaS_TR* and then rapidly acted on in situ by the R* domain to release authentic cAATrp.
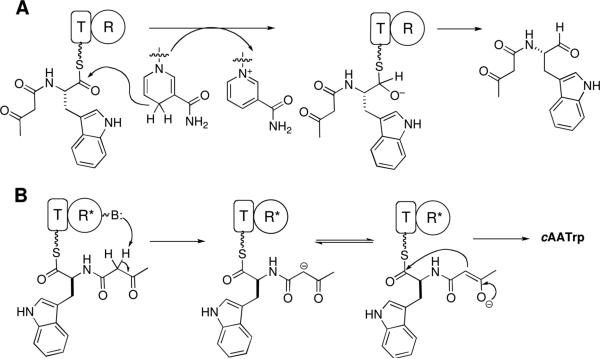
We likewise formulate this tetramate-forming chain release step as a Dieckmann cyclization with the R* domain recognizing the N-acac-Trp-S-pantetheinyl arm for capture of the thioester carbonyl by the C2 carbanion of the acac moiety (Scheme 2B). As in the equisetin case, this transformation was NAD(P)H independent. (14) While it is on the one hand satisfying to study the native tethered acac-Trp-S-pantetheinyl thioester as Dieckman cyclization substrate for the action of the adjacent downstream R* in the purified CpaS T-R* didomain, it is very difficult to study kinetics and mechanism when the substrate (N-acac-Trp-S-T domain) and enzyme (R* domain) are present in stoichiometric amounts and coupled intramolecularly. To that end we were able to overexpress, purify and assay separated CpaS T and R* domain protein fragments, allowing initiation of kinetic studies with substrate concentrations in large excess over enzyme. While rates were sluggish, they sufficed for some initial examinations of the residues of R* domains that are different from canonical R domains.
Scheme 2.
Comparison of NRPS C-terminal R vs R* domain using CpaS as an example. (A) Genuine R domain generates aldehyde product via hydride transfer from NAD(P)H cofactor and a thioacetal intermediate. (B) Redox-incompetent R* domain subtracts C2 hydrogen of β-ketoamide to form a resonance-stabilized carbanion intermediate that undergoes intermolecular Claisen condensation to give cAATrp.
Given that both EqiS R* and CpaS R* domains are not redox active but still are members of the nicotinamide-dependent short chain reductase (SDR) superfamily, we assume that R* domains may have evolved from R domains that were functional redox catalysts as in Lys5 and LtxA and other bona fide NRPS R domain reductases. The proposed default pathway of redox-dead R* domains on N-acyl-aminoacyl-S-pantetheinyl-T domain substrates to be a Dieckmann cyclization/chain release requires the N-acyl substituents to be 3-ketoacyl groups. These can yield the resonance-stabilized C2 carbanions, from dissociation of an acidic C2-H, as kinetically accessible carbon nucleophiles (Scheme 2B).
We looked for clues as to how R domains might evolve/decay to R* domains by sequence comparisons, first around the conserved R domain catalytic triad of Ser-Tyr-Lys and then for residues conserved in R but not R*, or vice-versa. In canonical SDR enzymes, such as androsterone 17-keto reductase, mutation of either the active site Ser to Ala or Tyr to Phe residue completely abolishes measurable activity. (25) This catalytic Tyr is indeed mutated to Leu in the CpaS R* domain and to Phe in the corresponding tenellin synthetase R*. However, this cannot be the only mechanism for loss of redox activity in R* domains since other tetramate synthetases, including the equisetin R* that maintains the catalytic triad. As noted in the results and Figure 8, we then mutagenized a series of polar/charged residues that are conserved in the R* domains but not in the known redox active R domains. One aspartate residue, D3803, when mutated to Ala brought Dieckman condensation levels back to baseline. It remains to be seen if this result is generalizable across R* domains and will probably require an x-ray structure with bound substrates or substrate surrogates to determine the role of this aspartate carboxylate side chain; for example it could function as a base to generate the C2 carbanion of the acac moiety of tethered acacTrp substrate to initiate the tetramate ring cyclization. Also, we will need to achieve heterologous expression and isolation of the full length two module eight domain 3906 residue (431 kDa) CpaS to study the mechanism of chain release in the intact PKS-NRPS assembly line in vitro and any residual role of NAD(P)H in modulating the nonredox tetramate ring formation pathways; it is not yet clear if this will be achievable in bacterial or fungal systems. (45)
One distinction between NRPS chain termination modules with composition C-A-T-TE vs C-A-T-R/R* involves the distinction between thioester chemistry and oxoester chemistry in the release step. For all modules with a thioesterase domain, the full length NRP or PK-NRP chain is transferred covalently from the pantetheinyl arm of the most downstream T domain to the CH2OH side chain of the active site Ser of the TE, prior to product release by hydrolysis or macrocyclization. Thus, the assembly line switches from thioester chemistry to oxoester chemistry at the chain dismount step in the TE active site. In contrast it is more likely that the R domains do not transfer the acyl/peptidyl chains from those pantetheinyl thiol tethers but deliver a hydride ion from NAD(P)H to yield the thiohemiacetal still tethered to the pantetheinyl arm of the immediately upstream T domain. The hemithioacetal unravels to the released peptide aldehyde and regenerates the HS-pantetheinyl arm on the T domain so the assembly line can run through another cycle (Scheme 2A).
As the R domains may act on thioester not oxoester substrates, the thioester linkage may be consequential for the R* domains to effect tetramate ring cyclization. As a non-redox default pathway the R* domain performs a Dieckmann reaction on thioester not oxoester substrate. The R* domains may not need to be very efficient since background cyclization of acac-aminoacyl thioesters can proceed nonenzymatically because the S-pantetheinyl arm is a good leaving group. This may be of relevance in consideration of how α-lipomycin (8) and kirromycin (46) assembly lines form tetramate and pyridone rings in their chain release steps, as those NRPS modules have only C-A-T domains: they lack both TE and R or R* release catalytic domains. Finally, it is likely that Nature's strategies for disconnection of assembled covalent chains by Dieckmann cyclization routes include additional variants to explain tetramate ring formation in the fungal signaling molecule HSAF and cyclindramide where the tetramate lies between polyketide regions. (9)
ACKNOWLEDGMENT
We thank Dr. Kevin McCluskey (FGSC) and Dr. Jiujiang Yu (USGA) for providing the NRRL3357 strain and its genomic DNA.
ABBREVIATIONS
- CPA
cyclopiazonic acid
- cAATrp
cyclo-acetoacetyl-l-tryptophan
- PKS
polyketide synthase
- NRPS
nonribosomal peptide synthetase
- SDR
short chain dehydrogenase/reductase
- C
condensation
- A
adenylation
- T
thiolation
- R
reductase
- TE
thioesterase
- SNAC
S-N-acetyl cystamine
- acac
acetoacetyl
Footnotes
This work was supported in part by National Institute of Health Grant GM20011 (to C.T.W.) and Swiss National Science Foundation and Ernst Schering Foundation (postdoctoral fellowships to X.L.).
REFERENCES
- 1.Schobert R, Schlenk A. Tetramic and tetronic acids: An update on new derivatives and biological aspects. Bioorg. Med. Chem. 2008;16:4203–4221. doi: 10.1016/j.bmc.2008.02.069. [DOI] [PubMed] [Google Scholar]
- 2.Song ZS, Cox RJ, Lazarus CM, Simpson TJ. Fusarin C biosynthesis in Fusarium moniliforme and Fusarium venenatum. Chembiochem. 2004;5:1196–1203. doi: 10.1002/cbic.200400138. [DOI] [PubMed] [Google Scholar]
- 3.Sims JW, Fillmore JP, Warner DD, Schmidt EW. Equisetin biosynthesis in Fusarium heterosporum. Chem. Comm. 2005:186–188. doi: 10.1039/b413523g. [DOI] [PubMed] [Google Scholar]
- 4.Eley KL, Halo LM, Song ZS, Powles H, Cox RJ, Bailey AM, Lazarus CM, Simpson TJ. Biosynthesis of the 2-pyridone tenellin in the insect pathogenic fungus Beauveria bassiana. Chembiochem. 2007;8:289–297. doi: 10.1002/cbic.200600398. [DOI] [PubMed] [Google Scholar]
- 5.Maiya S, Grundmann A, Li X, Li SM, Turner G. Identification of a hybrid PKS/NRPS required for pseurotin A biosynthesis in the human pathogen Aspergillus fumigatus. Chembiochem. 2007;8:1736–1743. doi: 10.1002/cbic.200700202. [DOI] [PubMed] [Google Scholar]
- 6.Bergmann S, Schumann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nature Chemical Biology. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- 7.Schumann J, Hertweck C. Molecular basis of cytochalasan biosynthesis in fungi: Gene cluster analysis and evidence for the involvement of a PKS-NRPS hybrid synthase by RNA silencing. J. Am. Chem. Soc. 2007;129:9564–9565. doi: 10.1021/ja072884t. [DOI] [PubMed] [Google Scholar]
- 8.Bihlmaier C, Welle E, Hofmann C, Welzel K, Vente A, Breitling E, Muller M, Glaser S, Bechthold A. Biosynthetic gene cluster for the polyenoyltetramic acid alpha-lipomycin. Antimicrob. Agents Chemother. 2006;50:2113–2121. doi: 10.1128/AAC.00007-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu F, Zaleta-Rivera K, Zhu X, Huffman J, Millet JC, Harris SD, Yuen G, Li X-C, Du L. Structure and biosynthesis of heat-stable antifungal factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob. Agents Chemother. 2007;51:64–72. doi: 10.1128/AAC.00931-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic, machinery, and mechanisms. Chem. Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 11.Kopp F, Marahiel MA. Macrocyclization strategies in polyketide and nonribosomal peptide biosynthesis. Nat. Prod. Rep. 2007;24:735–749. doi: 10.1039/b613652b. [DOI] [PubMed] [Google Scholar]
- 12.Sattely ES, Fischbach MA, Walsh CT. Total biosynthesis: in vitro reconstitution of polyketide and nonribosomal peptide pathways. Nat. Prod. Rep. 2008;25:757–793. doi: 10.1039/b801747f. [DOI] [PubMed] [Google Scholar]
- 13.Halo LM, Marshall JW, Yakasai AA, Song Z, Butts CP, Crump MP, Heneghan M, Bailey AM, Simpson TJ, Lazarus CM, Cox RJ. Authentic heterologous expression of the tenellin iterative polyketide synthase nonribosomal peptide synthetase requires coexpression with an enoyl reductase. Chembiochem. 2008;9:585–594. doi: 10.1002/cbic.200700390. [DOI] [PubMed] [Google Scholar]
- 14.Sims JW, Schmidt EW. Thioesterase-like role for fungal PKS-NRPS hybrid reductive domains. J. Am. Chem. Soc. 2008;130:11149–11155. doi: 10.1021/ja803078z. [DOI] [PubMed] [Google Scholar]
- 15.Seidler NW, Jona I, Vegh M, Martonosi A. Cyclopiazonic acid is a specific inhibitor of the Ca2+-ATPase of sarcoplasmic reticulum. J. Biol. Chem. 1989;264:17816–17823. [PubMed] [Google Scholar]
- 16.Chang PK, Horn BW, Dorner JW. Clustered genes involved in cyclopiazonic acid production are next to the aflatoxin biosynthesis gene cluster in Aspergillus flavus. Fungal Genet. Biol. 2009;46:176–182. doi: 10.1016/j.fgb.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Tokuoka M, Seshime Y, Fujii I, Kitamoto K, Takahashi T, Koyama Y. Identification of a novel polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS) gene required for the biosynthesis of cyclopiazonic acid in Aspergillus oryzae. Fungal Genet. Biol. 2008;45:1608–1615. doi: 10.1016/j.fgb.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 18.Holzapfel CW, Wilkins DC. On the Biosynthesis of Cyclopiazonic Acid. Phytochemistry. 1971;10:351–358. [Google Scholar]
- 19.Steenkamp DJ, Schabort JC, Ferreira NP. Beta-Cyclopiazonate Oxidocyclase from Penicillium-Cyclopium .3. Preliminary Studies on Mechanism of Action. Biochim. Biophys. Acta. 1973;309:440–456. doi: 10.1016/0005-2744(73)90042-9. [DOI] [PubMed] [Google Scholar]
- 20.Schabort JC, Wilkens DC, Holzapfe CW, Potgieter DJJ, Neitz AW. Beta-Cyclopiazonate Oxidocyclase from Penicillium-Cyclopium .1. Assay Methods, Isolation and Purification. Biochim. Biophys. Acta. 1971;250:311–328. doi: 10.1016/0005-2744(71)90188-4. [DOI] [PubMed] [Google Scholar]
- 21.Schabort JC, Potgieter DJJ. Beta Cyclopiazonate Oxidocyclase from Penicillium-Cyclopium .2. Studies on Electron Acceptors, Inhibitors, Enzyme Kinetics, Amino Acid Composition, Flavin Prosthetic Group and Other Properties. Biochim. Biophys. Acta. 1971;250:329–345. doi: 10.1016/0005-2744(71)90189-6. [DOI] [PubMed] [Google Scholar]
- 22.McGrath RM, Steyn PS, Ferreira NP, Neethling DC. Biosynthesis of Cyclopiazonic Acids in Penicillium-Cyclopium - Isolation of Dimethylallylpyrophosphate - Cyclo-Acetoacetyltryptophanyl Dimethylallyltransferase. Bioorg. Chem. 1976;5:11–23. [Google Scholar]
- 23.Thorn A, Egerer-Sieber C, Jager CM, Herl V, Muller-Uri F, Kreis W, Muller YA. The crystal structure of progesterone 5 beta-reductase from Digitalis lanata defines a novel class of short chain dehydrogenases/reductases. J. Biol. Chem. 2008;283:17260–17269. doi: 10.1074/jbc.M706185200. [DOI] [PubMed] [Google Scholar]
- 24.Ladenstein R, Winberg JO, Benach J. Structure-function relationships in short-chain alcohol dehydrogenases. Cell. Mol. Life Sci. 2008;65:3918–3935. doi: 10.1007/s00018-008-8590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Filling C, Berndt KD, Benach J, Knapp S, Prozorovski T, Nordling E, Ladenstein R, Jornvall H, Oppermann U. Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J. Biol. Chem. 2002;277:25677–25684. doi: 10.1074/jbc.M202160200. [DOI] [PubMed] [Google Scholar]
- 26.Li YY, Weissman KJ, Muller R. Myxochelin biosynthesis: Direct evidence for two- and four-electron reduction of a carrier protein-bound thioester. J. Am. Chem. Soc. 2008;130:7554–+. doi: 10.1021/ja8025278. [DOI] [PubMed] [Google Scholar]
- 27.Gaitatzis N, Kunze B, Muller R. In vitro reconstitution of the myxochelin biosynthetic machinery of Stigmatella aurantiaca Sg a15: Biochemical characterization of a reductive release mechanism from nonribosomal peptide synthetases. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11136–11141. doi: 10.1073/pnas.201167098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Read JA, Walsh CT. The lyngbyatoxin biosynthetic assembly line: chain release by four-electron reduction of a dipeptidyl thioester to the corresponding alcohol. J. Am. Chem. Soc. 2007;129:15762–15763. doi: 10.1021/ja077374d. [DOI] [PubMed] [Google Scholar]
- 29.Silakowski B, Nordsiek G, Kunze B, Blocker H, Muller R. Novel features in a combined polyketide synthase/non-ribosomal peptide synthetase: the myxalamid biosynthetic gene cluster of the myxobacterium Stigmatella aurantiaca Sga15. Chem. Biol. 2001;8:59–69. doi: 10.1016/s1074-5521(00)00056-9. [DOI] [PubMed] [Google Scholar]
- 30.Velasco A, Acebo P, Gomez A, Schleissner C, Rodriguez P, Aparicio T, Conde S, Munoz R, de la Calle F, Garcia JL, Sanchez-Puelles JM. Molecular characterization of the safracin biosynthetic pathway from Pseudomonas fluorescens A2-2: designing new cytotoxic compounds. Mol. Microbiol. 2005;56:144–154. doi: 10.1111/j.1365-2958.2004.04433.x. [DOI] [PubMed] [Google Scholar]
- 31.Ehmann DE, Gehring AM, Walsh CT. Lysine biosynthesis in Saccharomyces cerevisiae: Mechanism of alpha-aminoadipate reductase (Lys2) involves posttranslational phosphopantetheinylation by Lys5. Biochemistry. 1999;38:6171–6177. doi: 10.1021/bi9829940. [DOI] [PubMed] [Google Scholar]
- 32.Sonden B, Kocincova D, Deshayes C, Euphrasie D, Rhayat L, Laval F, Frehel C, Daffe M, Etienne G, Reyrat JM. Gap, a mycobacterial specific integral membrane protein, is required for glycolipid transport to the cell surface. Mol. Microbiol. 2005;58:426–440. doi: 10.1111/j.1365-2958.2005.04847.x. [DOI] [PubMed] [Google Scholar]
- 33.Kamitori S, Iguchi A, Ohtaki A, Yamada M, Kita K. X-ray structures of NADPH-dependent carbonyl reductase from Sporobolomyces salmonicolor provide insights into stereoselective reductions of carbonyl compounds. J. Mol. Biol. 2005;352:551–558. doi: 10.1016/j.jmb.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 34.Ishiyama N, Creuzenet C, Lam JS, Berghuis AM. Crystal structure of WbpP, a genuine UDP-N-acetylglucosamine 4-epimerase from Pseudomonas aeruginosa - Substrate specificity in UDP-hexose 4-epimerases. J. Biol. Chem. 2004;279:22635–22642. doi: 10.1074/jbc.M401642200. [DOI] [PubMed] [Google Scholar]
- 35.Deacon AM, Ni YS, Coleman WG, Ealick SE. The crystal structure of ADP-L-glycero-D-mannoheptose 6-epimerase: catalysis with a twist. Structure. 2000;8:453–462. doi: 10.1016/s0969-2126(00)00128-3. [DOI] [PubMed] [Google Scholar]
- 36.Liu XY, Fortin PD, Walsh CT. Andrimid producers encode an acetyl-CoA carboxyltransferase subunit resistant to the action of the antibiotic. Proc. Natl. Acad. Sci. U.S.A. 2008;105:13321–13326. doi: 10.1073/pnas.0806873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin M, Fischbach MA, Clardy J. A Biosynthetic Gene Cluster for the Acetyl-CoA Carboxylase Inhibitor Andrimid. J. Am. Chem. Soc. 2006;128:10660–10661. doi: 10.1021/ja063194c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Freiberg C, Brunner NA, Schiffer G, Lampe T, Pohlmann J, Brands M, Raabe M, Haebich D, Ziegelbauer K. Identification and Characterization of the First Class of Potent Bacterial Acetyl-CoA Carboxylase Inhibitors with Antibacterial Activity. J. Biol. Chem. 2004;279:26066–26073. doi: 10.1074/jbc.M402989200. [DOI] [PubMed] [Google Scholar]
- 39.Tuske S, Sarafianos SG, Wang XY, Hudson B, Sineva E, Mukhopadhyay J, Birktoft JJ, Leroy O, Ismail S, Clark AD, Dharia C, Napoli A, Laptenko O, Lee J, Borukhov S, Ebright RH, Arnold E. Inhibition of bacterial RNA polymerase by streptolydigin: Stabilization of a straight-bridge-helix active-center conformation. Cell. 2005;122:541–552. doi: 10.1016/j.cell.2005.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laursen M, Bublitz M, Moncoq K, Olesen C, Moller JV, Young HS, Nissen P, Morth JP. Cyclopiazonic Acid Is Complexed to a Divalent Metal Ion When Bound to the Sarcoplasmic Reticulum Ca2+-ATPase. J. Biol. Chem. 2009;284:13513–13518. doi: 10.1074/jbc.C900031200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moncoq K, Trieber CA, Young HS. The molecular basis for cyclopiazonic acid inhibition of the sarcoplasmic reticulum calcium pump. J. Biol. Chem. 2007;282:9748–9757. doi: 10.1074/jbc.M611653200. [DOI] [PubMed] [Google Scholar]
- 42.Sieber SA, Walsh CT, Marahiel MA. Loading Peptidyl-Coenzyme A onto Peptidyl Carrier Proteins: A Novel Approach in Characterizing Macrocyclization by Thioesterase Domains. J. Am. Chem. Soc. 2003;125:10862–10866. doi: 10.1021/ja0361852. [DOI] [PubMed] [Google Scholar]
- 43.Belshaw PJ, Walsh CT, Stachelhaus T. Aminoacyl-CoAs as probes of condensation domain selectivity in nonribosomal peptide synthesis. Science. 1999;284:486–489. doi: 10.1126/science.284.5413.486. [DOI] [PubMed] [Google Scholar]
- 44.Quadri LE, Weinreb PH, Lei M, Nakano MM, Zuber P, Walsh CT. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- 45.Seshime Y, Juvvadi PR, Tokuoka M, Koyama Y, Kitamoto K, Ebizuka Y, Fujii I. Functional expression of the Aspergillus flavus PKS-NRPS hybrid CpaA involved in the biosynthesis of cyclopiazonic acid. Bioorg. Med. Chem. Lett. 2009;19:3288–3292. doi: 10.1016/j.bmcl.2009.04.073. [DOI] [PubMed] [Google Scholar]
- 46.Weber T, Laiple KJ, Pross EK, Textor A, Grond S, Welzel K, Pelzer S, Vente A, Wohlleben W. Molecular analysis of the kirromycin biosynthetic gene cluster revealed beta-alanine as precursor of the pyridone moiety. Chem. Biol. 2008;15:175–188. doi: 10.1016/j.chembiol.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor Laboratory Press; Painview, NY: 2001. [Google Scholar]
- 48.D'Angeli F, Filira F, Giormani V, Di Bello C. Acetoacetyl derivatives of amino acids, as protected units for peptide synthesis. Ricerca Scientifica. 1966;36:11–16. [Google Scholar]