Abstract
Nitrification, a key process in the global nitrogen cycle that generates nitrate through microbial activity, may enhance losses of fertilizer nitrogen by leaching and denitrification. Certain plants can suppress soil-nitrification by releasing inhibitors from roots, a phenomenon termed biological nitrification inhibition (BNI). Here, we report the discovery of an effective nitrification inhibitor in the root-exudates of the tropical forage grass Brachiaria humidicola (Rendle) Schweick. Named “brachialactone,” this inhibitor is a recently discovered cyclic diterpene with a unique 5-8-5-membered ring system and a γ-lactone ring. It contributed 60–90% of the inhibitory activity released from the roots of this tropical grass. Unlike nitrapyrin (a synthetic nitrification inhibitor), which affects only the ammonia monooxygenase (AMO) pathway, brachialactone appears to block both AMO and hydroxylamine oxidoreductase enzymatic pathways in Nitrosomonas. Release of this inhibitor is a regulated plant function, triggered and sustained by the availability of ammonium (NH4+) in the root environment. Brachialactone release is restricted to those roots that are directly exposed to NH4+. Within 3 years of establishment, Brachiaria pastures have suppressed soil nitrifier populations (determined as amoA genes; ammonia-oxidizing bacteria and ammonia-oxidizing archaea), along with nitrification and nitrous oxide emissions. These findings provide direct evidence for the existence and active regulation of a nitrification inhibitor (or inhibitors) release from tropical pasture root systems. Exploiting the BNI function could become a powerful strategy toward the development of low-nitrifying agronomic systems, benefiting both agriculture and the environment.
Keywords: global warming, nitrogen pollution, nitrous oxide emissions, root exudation, climate change
Most modern agricultural systems are based on large inputs of inorganic nitrogen (N), with ammonium (NH4+) being the primary N source (1, 2). Also, current crop management practices result in the development of highly nitrifying soil environments (3, 4). Nitrification results in the transformation of the relatively immobile NH4+ to highly mobile nitrate (NO3−), making inorganic N susceptible to losses through leaching of NO3− and/or gaseous N emissions, potentially initiating a cascade of environmental and health problems (1, 2, 5, 6). Nitrous oxide (N2O) is one of the three major biogenic greenhouse gases contributing to global warming, produced primarily from denitrification processes in agricultural systems (5, 7). Also, assimilation of NO3− by plants can result in further N2O emissions directly from plant canopies (8). The low agronomic N-use efficiency (NUE) found in many agricultural systems is largely the result of N losses associated with nitrification (i.e., N losses from NO3− leaching and denitrification) (9–11). Most plants have the ability to assimilate both NH4+ and NO3− (12); therefore, nitrification does not need to be a dominant process in the N cycle for efficient N use.
Nitrification is low in some forest and grassland soils (13–17). Since the early 1960s, some tropical grasses have been suspected of having the capacity to inhibit nitrification (18–21). However, this concept remained controversial due to the lack of direct evidence showing such inhibitory effects or the identification of specific inhibitors (22).
We adopted a very sensitive bioassay using a recombinant luminescent Nitrosomonas europaea to detect biological nitrification inhibition (BNI) in plant–soil systems with the inhibitory activity of roots expressed in allylthiourea units (ATU) (23). Using this methodology, we were able to show that certain plants release nitrification inhibitors from their roots (23–26). Such BNI capacity appears to be relatively widespread among tropical pasture plants, with Brachiaria spp. showing the highest capacity among the pasture grasses tested (24). The potential for high BNI capacity also exists in wild wheat (26). A pasture grass, Brachiaria humidicola (Rendle) Schweick, native to tropical Africa and grown extensively in tropical South American grasslands, releases substantial amounts of BNIs from its roots, ranging from 17 to 50 ATU per gram of root dry weight per day (23, 24). Here, we report the identity of the major nitrification inhibitor (which is a recently discovered cyclic diterpene), its most likely mode of inhibitory action, the regulatory nature of its release from roots, and the effectiveness of this BNI function in suppressing soil nitrification and N2O emissions from Brachiaria pastures. The potential implications of the BNI function limiting N losses from agricultural systems and in reducing the ecological footprint of food production are indicated.
Results and Discussion
Isolation, Characterization, and Structure Determination of Brachialactone.
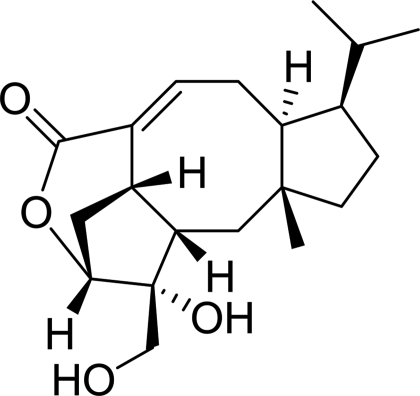
Bioassay-guided fractionation of the root exudates achieved the isolation of a cyclic diterpene, which we named “brachialactone” (Fig. 1 or its enantiomer). It has a unique dicyclopenta[a,d]cyclooctane skeleton (5-8-5 ring system) with a γ-lactone ring bridging one of the five-membered rings and the eight-membered ring. Similar 5-8-5 tricyclic terpenoids (ophiobolanes and fusicoccanes) are found in fungi and plants (27). However, to our knowledge, any derivative that has a lactone ring is novel. Fusicoccane-type cyclic diterpenes are biologically synthesized from geranylgeranyl diphosphate by two-step cyclization catalyzed by terpene cyclases (27, 28). Fusicoccins can act as activators of H+-ATPases in plants by modifying the function of 14-3-3 proteins and also exhibit unique biological activity (i.e., anticancer activity) in animal cells (28).
Fig. 1.
Chemical structure of brachialactone, the major nitrification inhibitor isolated from root exudates of B. humidicola.
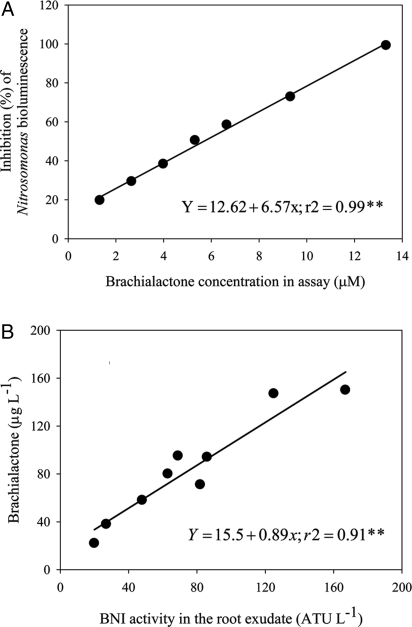
Inhibition of nitrification in our in vitro assay system of N. europaea was linearly related to brachialactone concentration over the range of 1.3–13.3 μM (r2 = 0.99, P < 0.01; Fig. 2A). Brachialactone, with an ED80 of 10.6 μM, should be considered a potent nitrification inhibitor when compared with nitrapyrin or dicyandiamide, two of the most widely used synthetic nitrification inhibitors (ED80 of 5.8 μM for nitrapyrin and 2,200 μM for dicyandiamide). Contribution of brachialactone to the total inhibitory activity in these root exudates ranged from 60% to 96% (r2 = 0.91, P < 0.01; Fig. 2B and Table S1).
Fig. 2.
Inhibition of nitrification by brachialactone and contribution of brachialactone to BNI activity released from roots. (A) Inhibitory effect of brachialactone on N. europaea in an in vitro assay. (B) Contribution of brachialactone to the BNI activity released from roots (i.e., in root exudates) of B. humidicola. Root exudates were collected from intact plants using 1 L of aerated solution of 1 mM NH4Cl with 200 μM CaCl2 over 24 h. Each data point represents root exudates collected from hydroponically grown plants in a glasshouse during March to May of 2007 and 2008.
Mode of Inhibitory Action of Brachialactone on Nitrosomonas.
Brachialactone inhibited Nitrosomonas function, possibly by blocking both enzymatic pathways, ammonia monooxygenase (AMO) and hydroxylamino oxidoreductase (HAO), that are involved in ammonia oxidation; however, the inhibitory effect on the HAO pathway is less than its effect on the AMO pathway (Table 1; for details, see Table S2). Crude extract of root exudates containing BNI activity showed an inhibitory effect of similar strength on both enzymatic pathways (Table 1; Table S2), indicating that other BNIs released from roots have a mode of action different from that of brachialactone. Recently, linolenic acid, a major BNI compound present in the leaf tissue of B. humidicola, was shown to block both AMO and HAO enzymatic pathways in a manner similar to BNI activity of crude root exudates, indicating the possibility of a single inhibitor affecting both the enzymatic pathways in Nitrosomonas (29). When a fatty acid binding protein, BSA, was added (after the addition of linolenic acid) to the Nitrosomonas pure cultures, a major portion of the inhibitory effect was removed, indicating the reversible nature of the inhibitory effect from linolenic acid (29). The reducing power generated from the oxidation of hydroxylamine by HAO is thought to pass through cytochrome c-554 to both cytochrome aa3 oxidase and ubiquinone (30), which is subsequently used for the reduction of NAD(P)+, as well as for the maintenance of the AMO reaction (31). We cannot as yet rule out the possibility of brachialactone disrupting the generation of reductive power, NAD(P)H2, by interfering directly with the electron transfer pathways of the cytochrome chain in the inner membrane of Nitrosomonas; thus, loss of light emission, which is independent of the direct effect on enzymatic pathways (i.e., AMO and HAO). In contrast, the inhibitory effect of nitrapyrin, a synthetic nitrification inhibitor, was nearly eliminated when hydroxylamine was added to the assay, indicating that only the AMO enzymatic pathway was affected (Table 1), which is in agreement with its known mode of action (32). High concentrations of monoterpenes found in conifer forest systems are reported to suppress nitrifier activity by blocking the AMO pathway (33). Only a few compounds—phenyl, methyl, or hydroxyethyl hydrazine and hydrogen peroxide—are known to inhibit the HAO enzymatic pathway in Nitrosomonas (34). Most commercial nitrification inhibitors (such as dicyandiamide or nitrapyrin) suppress nitrifier activity by targeting primarily the AMO pathway; thus, they could be vulnerable to genetic changes in nitrifier populations or to natural genetic diversity in ammonia-oxidizers (AOs) (35, 36). Given the inherent genetic variability in nitrifier populations (35), it is likely that BNIs released from Brachiaria spp. will be less vulnerable to genetic changes due to their more diverse modes of action on Nitrosomonas.
Table 1.
Inhibitory strength of brachialactone on AMO or HAO enzymatic pathways of N. europaea
| Compound | Concentration in the in vitro assay, μM | Inhibition, % |
|
|---|---|---|---|
| AMO pathway | HAO pathway | ||
| BNI-root exudate (crude methanol extract) | — | 63.4 ± 0.8 | 63.8 ± 0.8 |
| Brachialactone | 5.0 | 59.7 ± 0.9 | 37.7 ± 0.9 |
| Nitrapyrin | 3.0 | 82.3 ± 1.5 | 8.1 ± 1.2 |
Root exudate was collected from intact BH (CIAT 679) plants (root fresh weight of ≈20 g) using aerated solutions of 1 mM NH4Cl, evaporated to dryness, extracted with methanol, and evaporated to dryness and dissolved in 200 μL of dimethyl sulfoxide; 1 μL of the crude extract was used for the determination of BNI activity. Values are means ± SE from four replications.
Influence of NH4+ on the Release of Brachialactone.
Assuming that the BNI function evolved as a mechanism to conserve N by limiting nitrification and the associated N losses, we hypothesized that the inhibitory activity would respond to the presence of NH4+ in soil, because its availability determines the extent of soil nitrifier activity (37). We showed earlier that BNIs are released from roots of B. humidicola plants when the sole N source was NH4+, but not when they were grown with NO3− (38). Results from the present study demonstrate that N form (NH4+ vs. NO3−) in the root environment has a major influence on the release of brachialactone, which is accelerated only in the presence of NH4+ (Table 2).
Table 2.
Influence of NH4+ in the root environment on the release of BNI activity and brachialactone from roots of Brachiaria humidicola grown with NH4+ as the N source
| Exudate-collection solution | BNI activity released from the roots, ATU per g root dry wt per day | Brachialactone released from the roots, μg per g root dry wt per day |
|---|---|---|
| Distilled water | 6.6 ± 1.2 | 1.3 ± 0.1 |
| Acidified water (pH 3.0) | 4.8 ± 0.4 | 1.6 ± 0.6 |
| KNO3 (1 mM) | ND | ND |
| NH4Cl (1 mM) | 13.9 ± 1.6 | 8.3 ± 0.2 |
All collection solutions contained 200 μM CaCl2. Values are means ± SE from three replications. ND, not detected.
For inhibitors to be most effective, their release should be concentrated in the area where the root system is exposed to NH4+. To test this hypothesis, we used a split-root system in which plants were initially grown with (NH4)2SO4 as the sole N source. Then one half of the root system was exposed to NH4+ and the other half to NO3− in separate tanks. Release of brachialactone was triggered only in the part of the root system exposed to NH4+, and not in the entire root system (Table 3), indicating a localized release response.
Table 3.
Influence of nitrogen form (NH4+ vs. NO3−) in the exudate collection solutions on the release of BNI activity and brachialactone from the roots of B. humidicola in a split-root system
| Split-root system treatment | BNI activity released from the roots, ATU per g root dry wt per day | Brachialactone released from the roots, μg per g root dry wt per day |
|---|---|---|
| The half of the root system exposed to NH4+ | 9.5 ± 1.4 | 9.4 ± 0.2 |
| The half of the root system exposed to NO3− | ND | ND |
Values are means ± SE from three replications.
Validation of Effectiveness of BNI Function in Suppressing Soil Nitrification and N2O Emissions.
Based on conservative estimates of the live root biomass from a long-term grass pasture being ≈1.5 Mg·ha−1 (39) with a BNI capacity of 17–50 ATU per g of root dry wt per day (24), we estimate that BNI activity of 2.6 × 106 to 7.5 × 106 ATU·ha−1·day−1 can potentially be released from B. humidicola roots. This estimate amounts to an inhibitory potential equivalent to the application of ≈6.2–18 kg of nitrapyrin per ha per year (based on 1 ATU being equal to 0.6 μg of nitrapyrin), which is large enough to have a significant influence on the function of soil nitrifier populations and nitrification rates.
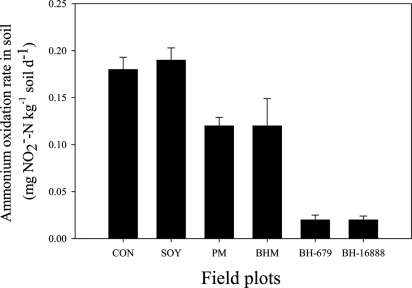
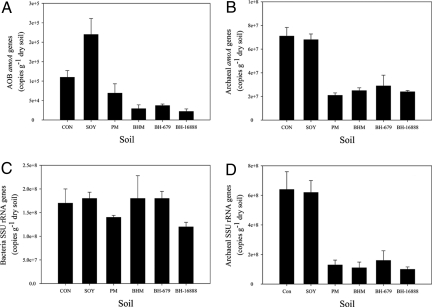
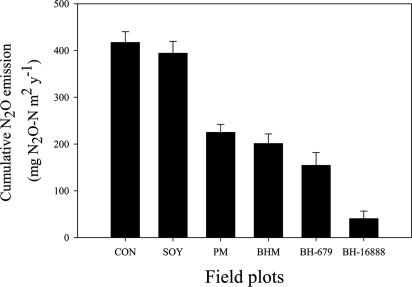
Field studies at Centro Internacional de Agricultura Tropical (CIAT; Palmira, Colombia) indicated a 90% decline in soil ammonium oxidation rates (Fig. 3) due to extremely small populations of nitrifiers [AO bacteria (Fig. 4B) and AO archaea (Fig. 4A); determined as amoA genes] in B. humidicola plots within 3 years of establishment (Fig. 4 A and B). Ammonium availability was relatively high (≥10 mg of NH4 N per kg of soil) in the field plots during the experimental period of 2005 to 2007 (Fig. S1). This observation suggests that the extremely low nitrifier populations observed in B. humidicola plots were not due to lack of soil ammonium nitrogen. However, there was little effect on the total soil bacterial population (expressed as gene copy number) (Fig. 4C), indicating the highly specific nature of the inhibitory effect toward the AO bacterial populations. Nevertheless, the archaea population in general is suppressed by Brachiaria sp. and Panicum sp. pastures (Fig. 4D), indicating that inhibitors produced by the root systems of these pastures may not be entirely specific to AO archaea. N2O emissions were also suppressed >90% in field plots of B. humidicola (CIAT 16888) compared with plots of soybean, which lacks BNI capacity, or control plots (plant-free field-plots) (Fig. 5). Two other pasture grasses, Panicum maximum and Brachiaria spp. hybrid cv. Mulato that have a low to moderate level of BNI capacity (3 to 10 ATU g−1 root dwt d−1), showed only an intermediate level of inhibitory effect on soil ammonium oxidation rates (Fig. 3).
Fig. 3.
Soil ammonium oxidation rates (mg of NO2− N per kg of soil per day) in field plots planted with tropical pasture grasses (differing in BNI capacity) and soybean (lacking BNI capacity in roots) [over 3 years from establishment of pastures (September 2004 to November 2007); for soybean, two planting seasons every year and after six seasons of cultivation]. CON, control (plant-free) plots; SOY, soybean; PM, P. maximum; BHM, Brachiaria hybrid cv. Mulato; BH-679, B. humidicola CIAT 679 (standard cultivar); BH-16888, B. humidicola accession CIAT 16888 (a germ plasm accession). Values are means ± SE from three replications.
Fig. 4.
Influence of tropical pasture grass cultivation (in field plots, over 3 years: September 2004 to November 2007) on soil microorganism populations at 1 day after ammonium sulfate fertilization by estimating copy number of AOB amoA genes (A); AOA amoA genes (B); bacterial small-subunit (SSU) rRNA genes (C); and archaea SSU 16S rRNA genes (D). Plots are identified in Fig. 3 legend. Gene copy number was expressed as copy number per gram of dried soil and obtained through absolute quantification by using real-time PCR. Values are means ± SE from three replications.
Fig. 5.
Cumulative N2O emissions (mg of N2O N per m2 per year) from field plots of tropical pasture grasses (monitored monthly over a 3-year period, from September 2004 to November 2007). Plots are identified in Fig. 3 legend. Values are means ± SE from three replications.
Conclusions and Perspectives
These results provide evidence for the existence of a plant-controlled mechanism by which nitrification inhibitors are produced and delivered by roots to soil-nitrifier sites. Here, a biological molecule providing a major portion of the BNI activity from Brachiaria root systems has been identified and characterized, and its release has been shown to be a tightly controlled physiological function. This report solves nearly 3 decades of mystery surrounding the low nitrification rates found in Brachiaria-dominated tropical pastures.
Fertilizer-N consumption is expected to reach 200 Tg·y−1 by 2025 from the present 150 Tg·y−1 (2, 40, 41). Environmental damage could result, given the pervasive inefficiencies in N use (<40% of applied N is recovered by most field crops) (2, 42), due largely to nitrification and its associated processes (1, 13). The economic implications of this “wasted N” can be enormous; they are currently estimated as US$ 17 billion from cereal production systems alone (13, 43). N2O emissions from agricultural systems are expected to reach 25.7 Tg of N per year by 2025, contributing significantly to global warming (1, 7, 10, 44).
Our results suggest potential differences in N2O emissions among plant species (Fig. 5) linked to their differential BNI capacities (Table S4) (24); however, such differences are not presently considered by the Intergovernmental Panel on Climate Change in their estimations of projected N2O emissions from agricultural systems (45). For example, South American savannas occupy ≈250 million (M)ha, largely under native grass or pastures of introduced Brachiaria spp. (39). These pastures could be of low nitrifying (24) and low N2O emitting systems. If these grasslands were converted to soybean and maize, crops that lack BNI capacity (24, 46), there could be major implications for N2O emissions. Approximately 11 Mha of pastoral land in the Cerrados region of Brazil has already been converted to soybean and maize (37, 47), and an additional 35–40 Mha could suffer such conversion. Such land-use changes could have major consequences on N2O emissions from this region.
Given the current environmental concerns, it is desirable to develop new technologies and approaches for combating the rampant and rapid nitrification in agricultural systems to reduce N pollution and improve NUE (1). Development of improved forage grasses for low nitrifying pasture-based production systems is possible given the significant genetic variability found for the BNI function within the Brachiaria spp. (24). Also, introducing high BNI capacity from wild wheat (Leymus racemosus) into cultivated wheat could be an option in the foreseeable future (26, 48). A fundamental shift toward NH4+-dominated crop nutrition can be achieved by using crops and pastures that have high BNI capacity or integrating annual crop production with a high BNI-capacity forage component, resulting in low-nitrifying agronomic production systems, benefiting both agriculture and the environment.
Materials and Methods
Bioassay-Guided Fractionation and Structure Elucidation.
Isolation of brachialactone from root exudates.
B. humidicola (Rendle) Schweick (CIAT 679) plants were raised hydroponically in the greenhouse (23). Root exudates were collected by using aerated solutions of NH4Cl (1 mM), the water was evaporated, and the residue was extracted with dichloromethane (23). The dichloromethane was evaporated, and the residue was dissolved in methanol and separated by using an HPLC system (Tosoh 8020 photodiode array system) equipped with a C18 reverse-phase column (Tosoh TSKgel SuperODS). BNI activity of HPLC effluents was determined (23). The major BNI-active compound was eluted in the 20–50% acetonitrile gradient in water. The active fractions were pooled and further purified by repeated HPLC to yield 0.7 mg (purity, >95% on HPLC) as an amorphous powder.
Characterization and structure determination of brachialactone.
The UV and CD spectra of brachialactone in methanol at room temperature were recorded on a UV-1600 spectrophotometer (Shimadzu) and a J-820 spectropolarimeter (JASCO), respectively. As shown in Fig. S2, the UV absorption spectrum showed the absorption maximum at a wavelength of 230 nm (ε = 2.4 × 103 M−1·cm−1). CD spectrum gave a positive band at 232 nm ([θ] = +6.5°·cm2·dmol−1, 0.027 mmol/100 mL).
Brachialactone was dissolved in methanol and applied to EI mass spectrometry on a QP2010 (Shimadzu) at 70 keV by direct inlet. The peaks observed (m/z, %) were 334 (M+, 17), 291 (9), 245 (5), 227 (5), 199 (11), 149 (21), 137 (100), 136 (75), 121 (47), and 107 (22) (Fig. S3). For the accurate mass analysis, the sample was diluted using 50% aqueous methanol including 1% acetic acid and measured in the positive-ion mode on an ESI-FTICR mass spectrometer (ApexII 70e; Bruker Daltonics). The measurement gave the protonated molecular ion at m/z 335.2213 corresponding to C20H31O4 (335.2217) with 1.2 × 10−6 error [M + H]+. These results contributed to determine the molecular formula of brachialactone to be C20H30O4.
The 1H, 13C and 2D NMR spectra were measured in CD3OD on Avance 800 and Avance 500 spectrometers (Bruker Biospin). Assignment of NMR signals is listed in Table S3. Chemical bond connections in the molecule were determined based on DQF-COSY, TOCSY, HSQC, and HMBC correlations. The presence of a lactone ring was confirmed by HMBC from carbonyl to H4. Relative configuration was determined by using NOE information, and the absolute configuration in Fig. 1 and Fig. S4 was drawn according to that of ophiobolin D (27).
Determining the Mode of Brachialactone Inhibitory Action on Nitrosomonas.
Mode of inhibitory action on Nitrosomonas was determined by incubating the pure cultures of luminescent N. europaea with brachialactone in the presence of hydroxylamine (i.e., inhibition of the HAO enzymatic pathway) or in the absence of hydroxylamine (i.e., inhibition of the AMO enzymatic pathway) using a previously reported protocol (23, 29); 1 μL of purified brachialactone (dissolved in dimethyl sulfoxide) to give 5 μM in the assay medium was added to 250 μL of bacterial culture and 199 μL of distilled water; the contents were incubated for 10 min before the addition of 200 μL of 1 mM hydroxylamine (to give 307 μM) to give an assay volume of 650 μL. The mean of the five bioluminescence measurements made during the subsequent 10-min incubation at 15 οC was taken as the activity level. Every measurement was repeated four times, and they were considered to be replications for the calculation of the SE (Table S2).
Influence of NH4+ on the Release of Brachialactone.
Plants were grown with NH4+ or NO3− as sole N sources (38). Root exudates were collected using aerated treatment solutions and BNI activity was extracted (23); brachialactone levels were determined by using HPLC.
Split-Root System Studies.
Plants were raised hydroponically with (NH4)2SO4 as the sole N source. After 6 months of growth, the root system of each plant was divided in half and each half was grown in a separate nutrient tank. After 2 weeks of separate growth, one half of the root system was used for collecting root exudates in a 1 mM N solution from NH4Cl; and the other half was used for collecting root exudates using 1 mM KNO3; root exudate solutions were evaporated, and BNI activity was determined (23); brachialactone levels were determined by using HPLC.
Field Validation of the Effectiveness of B. humidicola in Suppressing Soil Nitrification and N2O Emissions.
A field experiment was established on August 30, 2004, and continued until November 2007 at CIAT headquarters (3°30′N, 76°21′W) on a Vertisol (Typic Pellustert), pH 7.4, with an annual mean rainfall of ≈1,000 mm, annual mean temperature of 26 °C, and an elevation of 965 m above sea level. The six treatments were: (i) B. humidicola (CIAT 679, a standard cultivar with a BNI capacity of 17.3 ATU per g of root dry wt per day) (Table S4); (ii) B. humidicola (CIAT 16888, a germ plasm accession with a BNI capacity of 53.8 ATU pr g of root dry wt per day) (Table S4); (iii) Brachiaria hybrid cv. Mulato (an improved pasture with a BNI capacity of 10.2 ATU g of root dry wt per day) (Table S4); (iv) P. maximum cv. Common (with a BNI capacity of 3.3 ATU g of root dry wt per day) (24); (v) soybean cv. ICAP 34 (lacks BNI capacity in roots) (24); and (vi) control (bare soil, no plants). The experimental unit was a 10 × 10 m plot. The treatments were replicated three times in randomized complete blocks. For each soybean crop cycle, fertilizer was applied twice: for the first application (4 weeks after planting), plots were fertilized (kg·ha−1) with: 48 N, 48 K, 16 P, 0.4 Zn, and 0.4 B, and for the second application (8 weeks after planting) only N was applied at 48 kg·ha−1. The pasture grass plots and the bare soil plots also received the same amount of fertilizers at the same time. Pastures were cut twice a year, coinciding with the harvesting of the soybean crop. Soil inorganic N levels were monitored (30 days after ammonium sulfate fertilization) twice a year.
Soil Sampling, Determination of Ammonium Oxidation Rates, and AOB and AOA Gene Quantification.
Two 1-m2 quadrants within each experimental unit were marked as permanently treated sampling subplots. N fertilizer was applied as ammonium sulfate solution in water to the sampling subplots. Three years after establishing the pastures (November 2007), soil samples (five samples from the top 10-cm soil layer and pooled for each experimental unit) were collected from subplots 1 day after fertilizer application and ammonium oxidation rates in the soil were determined (49). Quantification of AOB and AOA functional genes was carried out on these soil samples by real-time PCR using the primer combinations amoA-1F/amoA-2R (50), amoA19F (6)/amoA643R (51), BACT1369F/PROK1541R (52), and Arch20F-/-Arch958R (53) for AOB amoA gene, AOA amoA gene, bacterial SSU rRNA gene, and archaeal SSU rRNA gene, respectively (for details, see SI Methods).
N2O Emission Measurements in the Field.
N2O emissions from the sampling subplots were measured monthly from September 2004 to November 2007. When fertilizer was applied, gas sampling was done 1 day after application of fertilizer to the subplots by using the static chamber technique and analyzed for N2O on a gas chromatograph equipped with an electron capture detector (ECD) (54). N2O flux was determined from concentration plotted against time. Results presented in Fig. 5 are the cumulative amounts of N2O expressed in mg of N2O N per m2 per year from September 2004 to November, 2007 (for detailed methods, see SI Methods).
Acknowledgments.
We thank Dr. Naoyoshi Kawano (JIRCAS, Japan), Ms. Akane Notazawa (JIRCAS, Japan), and Dr. Monrawee Fukuda (JIRCAS, Japan) for setting up of experiments, collection of root exudates, and HPLC determination of brachialactone; Dr. Ikuko Maeda (NFRI, Japan) for NMR measurement; Ms. Tomoko Sato (NFRI, Japan) for high-resolution MS measurement; Dr. Rika Iwaura (NFRI, Japan) for CD spectrum measurement; Ms. Alba Lucia Chavez (CIAT, Colombia) and Mr. Michael Matiasek (CIAT, Colombia) for research assistance related to molecular analysis of AOB and AOA in soil samples; Ms. Myriam Duque (CIAT, Colombia) for biometric analysis of molecular data from soil; Mr. Hernan Mezu' (CIAT, Colombia) for field and laboratory technical assistance; and Dr. J. Miles (CIAT, Colombia), for his intellectual support to this work. This research was partially supported by the restricted funding to the CIAT from the Ministry of Foreign Affairs of the Government of Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903694106/DCSupplemental.
References
- 1.Galloway JN, et al. Transformation of the nitrogen cycle: Recent trends, questions, and potential solutions. Science. 2008;320:889–892. doi: 10.1126/science.1136674. [DOI] [PubMed] [Google Scholar]
- 2.Schlesinger WH. On the fate of anthropogenic nitrogen. Proc Natl Acad Sci USA. 2009;106:203–208. doi: 10.1073/pnas.0810193105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dinnes DL, et al. Nitrogen management strategies to reduce nitrate leaching in tile drained Midwestern soils. Agron J. 2002;94:153–171. [Google Scholar]
- 4.Bellamy PH, Loveland PJ, Ian Bradley R, Murray Lark R, Kirk GJD. Carbon losses from all soils across England and Wales 1978–2003. Nature. 2005;437:245–248. doi: 10.1038/nature04038. [DOI] [PubMed] [Google Scholar]
- 5.Bremner JM, Blackmer AM. Nitrous oxide: Emission from soils during nitrification of fertilizer nitrogen. Science. 1978;199:295–296. doi: 10.1126/science.199.4326.295. [DOI] [PubMed] [Google Scholar]
- 6.Leninger S, et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- 7.Kroeze C. Nitrous oxide and global warming. Sci Total Environ. 1994;143:193–209. doi: 10.1016/0048-9697(94)90310-7. [DOI] [PubMed] [Google Scholar]
- 8.Smart DR, Bloom AJ. Wheat leaves emit nitrous oxide during nitrate assimilation. Proc Nat Acad Sci USA. 2001;98:7875–7878. doi: 10.1073/pnas.131572798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryden JC, Ball PR, Garwood EA. Nitrate leaching from grassland. Nature. 1984;311:50–53. [Google Scholar]
- 10.Smith KA, McTaggart IP, Tsuruta H. Emissions of N2O and NO associated with nitrogen fertilization in intensive agriculture, and the potential for mitigation. Soil Use Manage. 1997;13:296–304. [Google Scholar]
- 11.Robertson GP. Nitrogen use efficiency in row crop agriculture: Crop nitrogen use and soil nitrogen loss. In: Jackson L, editor. Ecology in Agriculture. New York: Academic; 1997. pp. 347–365. [Google Scholar]
- 12.Salsac L, Chaillou S, Morot-Gaudry J, Lesaint C. Nitrate and ammonium nutrition in plants. Plant Physiol Biochem. 1987;25:805–812. [Google Scholar]
- 13.Subbarao GV, et al. Scope and strategies for regulation of nitrification in agricultural systems–challenges and opportunities. Crit Rev Plant Sci. 2006;25:303–335. [Google Scholar]
- 14.Cooper AB. Suppression of nitrate formation with an exotic conifer plantation. Plant Soil. 1986;93:383–394. [Google Scholar]
- 15.Wedin DA, Tilman D. Species effects on nitrogen cycling: A test with perennial grasses. Oecologia. 1990;84:433–441. doi: 10.1007/BF00328157. [DOI] [PubMed] [Google Scholar]
- 16.Northup RR, Yu Z, Dahigren RA, Vogt KA. Polyphenol control of nitrogen release from pine litter. Nature. 1995;377:227–229. [Google Scholar]
- 17.Robertson GP. Nitrification in forested ecosystems. Philos Trans R Soc London Ser B. 1982;296:445–457. [Google Scholar]
- 18.Munro PE. Inhibition of nitrite-oxidizers by roots of grass. J Appl Ecol. 1966;3:227–229. [Google Scholar]
- 19.Sylvester-Bradley R, Mosquera D, Mendez JE. Inhibition of nitrate accumulation in tropical grass-land soils: Effect of nitrogen fertilization and soil disturbance. J Soil Sci. 1988;39:407–416. [Google Scholar]
- 20.Lata JC, et al. Grass populations control nitrification in savanna soils. Funct Ecol. 2004;18:605–611. [Google Scholar]
- 21.Boudsocq S, Lata JC, Mathieu J, Abbadie L, Barot S. Modelling approach to analyse the effects of nitrification inhibition on primary production. Funct Ecol. 2009;23:220–230. [Google Scholar]
- 22.Fillery IRP. Plant-based manipulation of nitrification in soil: A new approach to managing N loss? Plant Soil. 2007;294:1–4. [Google Scholar]
- 23.Subbarao GV, et al. A bioluminescence assay to detect nitrification inhibitors released from plant roots: A case study with Brachiaria humidicola. Plant Soil. 2006;288:101–112. [Google Scholar]
- 24.Subbarao GV, et al. Biological nitrification inhibition (BNI)–is it a widespread phenomenon? Plant Soil. 2007;294:5–18. [Google Scholar]
- 25.Zakir HAKM, et al. Detection, isolation and characterization of a root-exuded compound, methyl 3-(4-hydroxyphenyl)propionate, responsible for biological nitrification inhibition by sorghum (Sorghum bicolor) New Phytol. 2008;180:442–451. doi: 10.1111/j.1469-8137.2008.02576.x. [DOI] [PubMed] [Google Scholar]
- 26.Subbarao GV, et al. Can biological nitrification inhibition (BNI) genes from perennial Leymus racemosus (Triticeae) combat nitrification in wheat farming? Plant Soil. 2007;299:55–64. [Google Scholar]
- 27.Muromtsev GS, et al. Occurrence of fusicoccanes in plants and fungi. J Plant Growth Regul. 1994;13:39–49. [Google Scholar]
- 28.Toyomasu T, et al. Fusicoccins are biosynthesized by an unusual chimera diterpene synthase in fungi. Proc Natl Acad Sci USA. 2007;104:3084–3088. doi: 10.1073/pnas.0608426104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subbarao GV, et al. Free fatty acids from the pasture grass Brachiaria humidicola and one of their methyl esters as inhibitors of nitrification. Plant Soil. 2008;313:89–99. [Google Scholar]
- 30.McTavish H, Arciero DM, Hooper AB. Interaction with membranes of cytochrome c554 from Nitrosomonas europaea. Arch Biochem Biophys. 1995;324:53–58. doi: 10.1006/abbi.1995.9930. [DOI] [PubMed] [Google Scholar]
- 31.Hooper AB, Vannelli T, Bergmann DJ, Arciero DM. Enzymology of the oxidation of ammonia to nitrite by bacteria. Antonie Leeuwenhoek. 1997;71:59–67. doi: 10.1023/a:1000133919203. [DOI] [PubMed] [Google Scholar]
- 32.McCarty GW. Modes of action of nitrification inhibitors. Biol Fertil Soils. 1999;29:1–9. [Google Scholar]
- 33.Ward BB, Courtney KJ, Langerheim JH. Inhibition of Nitrosomonas europaea by monoterpenes from coastal redwood (Sequoia sempervirens) in whole-cell studies. J Chem Ecol. 1997;23:2583–2598. [Google Scholar]
- 34.Logan MSP, Hooper AB. Suicide inactivation of hydroxylamine oxidoreductase of Nitrosomonas europaea by organohydrazines. Biochemistry. 1995;34:9257–9264. doi: 10.1021/bi00028a039. [DOI] [PubMed] [Google Scholar]
- 35.Norton JM, Alzerreca JJ, Suwa Y, Klotz MG. Diversity of ammonia monooxygenase operon in autotrophic ammonia oxidizing bacteria. Arch Microbiol. 2002;177:139–149. doi: 10.1007/s00203-001-0369-z. [DOI] [PubMed] [Google Scholar]
- 36.Belser LW. Population ecology of nitrifying bacteria. Annu Rev Microbiol. 1979;33:309–333. doi: 10.1146/annurev.mi.33.100179.001521. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki I, Dular U, Kwok SC. Ammonium and ammonium ion as substrate for oxidation by Nitrosomonas cells and extracts. J Bacteriol. 1974;176:6623–6630. doi: 10.1128/jb.120.1.556-558.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subbarao GV, Wang HY, Ito O, Nakahara K, Berry WL. NH4+ triggers the synthesis and release of biological nitrification inhibition compounds in Brachiaria humidicola roots. Plant Soil. 2007;290:245–257. [Google Scholar]
- 39.Fisher MJ, et al. Carbon storage by introduced deep-rooted grasses in the South American savannas. Nature. 1994;371:236–238. [Google Scholar]
- 40.Vitousek PM, et al. ) Human alteration of the global nitrogen cycle: Sources and consequences. Ecol Applic. 1997;7:737–750. [Google Scholar]
- 41.IFA. World Fertilizer Use. Paris: International Fertilizer Association; 2005. p. 124. [Google Scholar]
- 42.Ju X-T, et al. Reducing environmental risk by improving N management in intensive Chinese agricultural systems. Proc Nat Acad Sci USA. 2009;106:3041–3046. doi: 10.1073/pnas.0813417106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raun WR, Johnson GV. Improving nitrogen use efficiency for cereal production. Agron J. 1999;91:357–363. [Google Scholar]
- 44.Intergovernmental Panel on Climate Change. Climate Change 2007: The Physical Science Basis–Summary for Policy Makers. Paris: World Meteorological Organization/United Nations Environ Programme; 2007. [Google Scholar]
- 45.Stehfest E, Bouwman L. N2O and NO emission from agricultural fields and soils under natural vegetation: Summarizing available measurement data and modeling of global annual emissions. Nutr Cycl Agroecosys. 2006;74:207–228. [Google Scholar]
- 46.Brown JC, Koeppe M, Coles B, Price KP. Soybean production and conversion of tropical forest in the Brazilian Amazon: The case of Vilhena, Rondonia. AMBIO. 2005;34:462–469. [PubMed] [Google Scholar]
- 47.Zimmer AH, et al. Integrated agropastoral production systems. In: Guiamaraes EP, et al., editors. Agropastoral Systems for the Tropical Savannas of Latin America. Cali, Colombia: CIAT; 2004. pp. 253–290. EMBRAPA, Brasilia, Brazil. [Google Scholar]
- 48.Zahn LM. A boost from wild wheat. Science. 2007;318:171. [Google Scholar]
- 49.Belser LW, Mays EL. Specific inhibition of nitrite oxidation by chlorate and its use in assessing nitrification in soils and sediments. Applied Environ Microbiol. 1980;39:505–510. doi: 10.1128/aem.39.3.505-510.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rotthauwe JH, Witzel KP, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: Molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Treusch AH, et al. Novel gene for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ Microbiol. 2005;7:1985–1995. doi: 10.1111/j.1462-2920.2005.00906.x. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki M, Lance T, DeLong E. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl Environ Microbiol. 2000;66:4605–4614. doi: 10.1128/aem.66.11.4605-4614.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeLong EF. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holland E, et al. Standard Soil Methods for Long-term Ecological Research. Oxford: Oxford Univ Press; 1999. pp. 185–201. [Google Scholar]