Abstract
Material remains of ancestor nucleotides and proteins are largely unavailable, thus sequence comparison among homologous genes in present-day organisms forms the core of current knowledge of molecular evolution. Variation in protein three-dimensional structure is a basis for functional diversity. To study the evolution of three-dimensional structures in related proteins would significantly improve our understanding of protein evolution and function. A protein may contain ancestor conformations that have been allosterically suppressed by evolutionarily additive structures. Using monoclonal antibody probes to detect such conformation in proteins after removing the suppressor structure, our study demonstrated three-dimensional structure evidence for the evolutionary relationship between troponin I and troponin T, two subunits of the troponin complex in the Ca2+-regulatory system of striated muscle, and among their muscle type-specific isoforms. The experimental data showed the feasibility to detect evolutionarily suppressed history-telling structural states in proteins by removing conformational modulator segments added during evolution. In addition to identifying structural modifications that were critical to the emerging of diverged proteins, investigating this novel mode of evolution will help to understand the origin and functional potential of protein structures.
Keywords: Molecular evolution, protein structure, conformational modulation, atavism, epitope analysis, troponin
Introduction
The study of evolution depends crucially on fossil materials. However, material remains of ancestor nucleotides and proteins are largely unavailable, thus the evolutionary relationship between proteins is evaluated by sequence comparisons. Sequence alignment among homologous genes in present-day species is the primary method in current molecular evolution studies and has provided powerful information for how evolution has worked (Benner et al., 2002). Nonetheless, the functional evolution of a protein is determined by changes in three-dimensional structures. To further investigate how the three-dimensional structure of a protein has evolved would significantly strengthen the evolutionary relationships indicated by nucleotide and amino acid sequence comparisons. In addition, protein three-dimensional structures may contain allosteric information that reflects the evolutionary past. The small size of current database of high-resolution three-dimensional structures, especially allosteric states, determined by X-ray crystallography and nuclear magnetic resonance spectroscopy (NMR) is insufficient to lead the study of molecular evolution. Higher throughput approaches to detect homologous three-dimensional structures for their evolutionary relationships are needed to compensate for the limitations of sequence analysis and structural biology data (Blundell et al., 2006).
The binding affinity of an antibody to an antigen depends on three-dimensional structure fits between the antibody variable region (the paratope) and the epitope on the antigen protein (Fig. 1A). Immunogenic epitopes are often variable structures and it is established that the degree of cross-reactivity of an antibody to homologous proteins reflects their structural similarity and diversity (Miller and Cohen, 1991). This “immunological distance” has been used to evaluate the phylogeny of homologous proteins (Li et al., 1987; Kaminogawa et al., 1989; Prager, 1993). While the cross-reactivity of polyclonal antibodies reflects the overall structural similarity of related proteins, monoclonal antibodies (mAbs1) (Köhler and Milstein, 1975) detect similarities and differences in specific epitopic structures. Selected mAbs against conserved or evolving epitopes may be used as three-dimensional structure probes to investigate evolutionary conservation and variation. In addition to detecting a static structure, antibodies against allosteric epitopes provide probes for measuring conformational changes in a protein (Wang and Jin, 1998; Jin et al., 2000a; 2000b). Protein epitopic structures can be quantitatively studied by microtiter plate enzyme-linked immunosorbant assay (ELISA) that effectively detects conformational differences and changes under native conditions (Wang and Jin, 1998; Jin et al., 2007).
Figure 1. Background information.
(A) Antigen-antibody binding depends on structural fit between the antigenic epitope and the antibody paratope. A change in the conformation of an antigen protein may alter the affinity of an antibody. (B) TnI and TnT are two subunits of the troponin complex and play interrelated functions in the thin filament Ca2+ regulatory system of striated muscle. (C) Three pairs of linked TnI and TnT genes are present in vertebrates. Supporting the functional pairing of slow TnI and cardiac TnT genes, the Western blots showed that slow TnI was expressed in embryonic and neonatal hearts together with cardiac TnT. (D) The Western blots using polyclonal anti-TnI and anti-TnT antibodies RATnI and RATnT detected multiple TnI and TnT bands in the skeletal and cardiac muscles of Atlantic hagfish (Myxine glutinosa) indicating early emergence and divergence of the TnI-TnT gene super family. The results were confirmed by Western blots using pan-TnI mAb 4D12 and pan-TnT mAb 2C8 (data not shown).
Gene duplication and sequence divergence produce genetic variation and molecular diversity that fuel evolution (Raes and Van de Peer, 2003). Troponin I (TnI) and troponin T (TnT) are two subunits of the troponin complex that regulates the contraction of striated (skeletal and cardiac) muscles (Perry, 1998; Perry, 1999; Jin et al., 2008). The two proteins function together but distinctively in the troponin complex (Fig. 1B) and genes encoding TnI and TnT are closely linked in the vertebrate genome (Huang and Jin, 1999), suggesting their origin from the duplication of a single ancestral gene followed by neofunctionalization. As linked pairs of genes, TnI and TnT have each evolved into three isoforms with specific expressions in cardiac, slow and fast skeletal muscles (Barton et al., 1997; Tiso et al., 1997; Huang and Jin, 1999; Perry, 1998; 1999) (Fig. 1C), implying a series of duplication events from an ancestral TnI-TnT gene pair followed by subfunctionization and neofunctionalization. The fast TnI-fast TnT gene pair shows linked expression. Although the cardiac TnI-slow TnT and slow TnI-cardiac TnT gene pairs are apparently scrambled, the slow TnI-cardiac TnT gene pair co-expresses in embryonic cardiac muscle as a functional pair (Fig. 1C). These data suggest that the evolution of the three linked TnI-TnT gene pairs has a functional basis.
Distinct TnI and TnT isoforms are found in the cardiac and skeletal muscles of hagfish (Atlantic hagfish, Myxine glutinosa), an elementary vertebrate that shared with true vertebrates a common ancestor lived some half a billion years ago (Fock and Hinssen, 2002) (Fig. 1D), indicating an early emergence of TnI and TnT and their isoform genes. This long existing TnI-TnT gene family encoding genetically and functionally close-linked allosteric protein isoforms provides an excellent system to experimentally study the evolution of protein three-dimensional structures. Using a collection of antibody probes to compare epitope structures and detect evolutionarily suppressed molecular conformation after removing a suppressor structure, the present study demonstrated three-dimensional structure evidence for the evolutionary relationships between TnI and TnT and among their muscle type-specific isoforms. The experimental data showed the feasibility to detect suppressed history-telling structural states in proteins by removing conformational modulator segments added during evolution, suggesting a novel mode of protein evolution.
Materials and Methods
Analysis of nucleotide and amino acid sequences
Amino acid sequences of TnI and TnT were aligned with Clustal W 2.0.10 computer program (Larkin et al., 2007) and a phylogenetic tree was built with Geneious 4.5.3 program using Jukes-Cantor genetic distance model and neighbor-joining tree building method (Drummond et al., 2008). The arbitrary random-seed was 562,379 at 100 samples, creating a consensus tree with a support threshold percentage of 50%.
The Genbank accession numbers for the protein and nucleic acid sequences are: Human fast TnI (NM_003282), human slow TnI (BC012600), human cardiac TnI (NM_000363); mouse fast TnI (NM_009405), mouse slow TnI (NM_021467), mouse cardiac TnI (NM_009406); chicken fast TnI (NM_205417), chicken slow TnI (XM_419242), chicken cardiac TnI (NM_213570); Xenopus laevis fast TnI (AF480427), Xenopus tropicalis slow TnI (AAH61268.1), Xenopus tropicalis cardiac TnI (NM_001011410); zebrafish fast TnI (AF425744), zebrafish slow TnI (CAD59124.1), zebrafish cardiac TnI (XM_686456); human fast TnT (NM_001042782), human cardiac TnT (NM_001001430), human slow TnT (S69208); mouse fast TnT (L49467), mouse cardiac TnT (L47549), mouse slow TnT (AF020946); chicken fast TnT (M22158), chicken cardiac TnT (NM_205449), chicken slow TnT (NM_205114); Xenopus laevis fast TnT (AY114144), Xenopus laevis cardiac TnT (AF467920), toad slow TnT (AY773671); Salmo salar fast TnT (AF072687), zebrafish cardiac TnT (XM_685694), and zebrafish slow TnT (NM_181499).
Protein engineering, expression and purification
cDNAs encoding TnI and TnT isoforms were cloned into the T7 RNA polymerase-based pAED4 plasmid vector (Jin, 1995) for protein expression in E. coli. Modified cDNAs encoding TnI and TnT fragments (Fig. 2) were constructed using polymerase chain reaction (PCR) as previously described, in which forward and reverse primers were designed to contain an ATG codon prior to the N-terminal deletion site or a stop codon at the C-terminal deletion site (Biesiadecki et al., 2007). The PCR amplified cDNA fragments were restriction enzyme modified at the ends and cloned into the pAED4 expression vector. The recombinant plasmid was verified by DNA sequencing. The deletion constructs as well as intact TnI and TnT were expressed in BL21(DE3)pLysS E. coli as non-fusion proteins and purified as described (Jin et al., 2001; Biesiadecki et al., 2004; Biesiadecki et al., 2007).
Figure 2. TnI and TnT constructs and site-specific mAbs.
Intact TnI and TnT and their fragments were prepared to map the location of epitopes recognized by the mAbs used in our study and for conformational analysis. (A) Mouse cardiac TnI fragments were constructed with a deletion of the cardiac TnI-specific N-terminal 28 amino acids (cTnI-ND) or deletions of the N-terminal 28 amino acids and the C-terminal 19 amino acids (cTnI-ND-CD). The Western blots showed that mAb 4H6 recognizes both fragments as well as intact cardiac TnI, indicating specificity to a middle epitope. In contrast, mAbs TnI-1 and 4D12 recognize intact cardiac TnI and cTnI-ND but not cTnI-ND-CD, indicating specificity to the C-terminus. (B) Mouse cardiac, fast and slow TnT fragments with N-terminal deletions were constructed by selectively deletion of the N-terminal variable region (TnT-ND), or extended deletion into the conserved middle region (cardiac TnT-ND92 and cardiac TnT-ND130), or deletion of both N-terminal and middle regions (TnT-T2, Heeley et al., 1987). The Western blot analysis localized the epitopes of mAbs 2C8, T12 and 1G9 in the middle or C-terminal regions of the TnT polypeptide.
Anti-TnI and anti-TnT antibodies
An anti-TnI polyclonal antiserum RATnI was raised by immunization of a New Zealand White rabbit with purified chicken breast muscle fast TnI (Jin, 1996).
An anti-TnT polyclonal antiserum RATnT was raised by immunization of a New Zealand White rabbit with purified chicken adult breast muscle fast TnT (Wang and Jin, 1998).
A mouse mAb TnI-1 was developed by immunization with purified chicken breast muscle fast TnI (Jin et al., 2001). A mouse mAb 4D12 was developed by immunization with purified N-terminal truncated mouse cardiac TnI (Barbato et al., 2005). On TnI fragments expressed in E. coli, the Western blots in Fig. 2A demonstrate the specificity of mAbs TnI-1 and 4D12 against C-terminal epitopes.
A mouse mAb T12 (provided by Prof. Jim Lin, University of Iowa) was generated by immunization with purified rabbit fast skeletal muscle TnT (Lin et al., 1984). Western blots on TnT fragments revealed its specificity against an epitope in the middle region of the TnT polypeptide (Fig. 2B).
A mouse mAb 1G9 was developed by immunization with purified human cardiac TnT as described previously (Wang and Jin, 1998). The Western blots on TnT fragments in Fig. 2B showed its specificity against an epitope in the C-terminal T2 region.
A mouse mAb 4H6 was developed by immunization with a purified middle region fragment of mouse cardiac TnI (Du et al., 2008). A mouse mAb CT3 recognizing cardiac TnT and slow TnT was generated by immunization with purified bovine cardiac TnT (Jin et al., 2000). A mouse mAb 2C8 recognizing a stable epitope with similar affinities among all three TnT isoforms in the vertebrates was developed by immunization with purified human cardiac TnT (Jin et al., 2003; Zhang et al., 2006).
Despite the notion that antisera produced in vivo are adapted to a non-self-recognizing rule, numerous studies including our development of multiple mAbs against troponin subunits including 4D12 and 4H6 shown in the present study, have demonstrated that mAbs generated by hybridoma technology can strongly cross-react to homologous antigens from the host species, i.e., the mouse, in which the immunization was carried out. This epitopic level cross reactivity allows an inclusion of the host species in the study of phylogenetic relationship of a protein using mAbs as three-dimensional structure probes.
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting
Muscle tissues were homogenized in Laemmli SDS-PAGE sample buffer (containing 2% SDS that prevents protein degradation) using a high-speed mechanical homogenizer. The muscle homogenates and SDS-PAGE samples of purified TnI, TnT and the engineered protein fragments were resolved by Laemmli or Tris-Tricine gel electrophoresis (Jin, 1995). The gels were stained with Coomassie Brilliant Blue R250 to reveal the protein bands. Duplicate gels were electrically blotted to nitrocellulose or PVDF membranes. After blocking in Tris-buffered saline (TBS) containing 0.5% Triton X-100, 0.05% SDS and 1% bovine serum albumin (BSA), the membranes were incubated with an anti-TnI or anti-TnT antibody in TBS containing 0.1% BSA, washed using TBS containing 0.5% Triton X-100, 0.05% SDS, incubated with alkaline phosphatase-conjugated anti-rabbit IgG or anti-mouse IgG second antibody (Sigma Co, St. Louis, MO), washed again, and processed for colorimetric development in 5-bromo-4-chloro-3-indolylphosphate-nitro blue tetrazolium substrate solution, as described previously (Wang and Jin, 1998).
ELISA epitope analysis
ELISA epitope analysis (Wang and Jin, 1998; Jin et al., 2007) was employed to compare the relative binding affinity of the antibody epitope probes to the isoforms and fragments of TnI and TnT for their relationships in three dimensional structure. The purified TnI or TnT protein was dissolved in Buffer A (0.1 M KCl, 3 mM MgCl2, 10 mM PIPES, pH 7.0) at 2 μg/mL and used at 100 μL/well to non-covalently coat 96-well microtiter plates by incubation at 4 °C overnight. Bovine serum albumin coated wells were set as background controls. After removing unbound proteins and blocking any remaining free plastic surface by washing three times with Buffer A containing 0.05% Tween-20 (Buffer T), the immobilized TnI or TnT was incubated with 100 μL/well serial dilutions of anti-TnI or anti-TnT first antibody in Buffer T containing 0.1% BSA at room temperature for 2 h. Following three washes with Buffer T to remove the unbound first antibody, the plates were incubated with 100 μL/well horseradish peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin second antibody (Sigma, Co, St. Louis, MO) in Buffer T containing 0.1% BSA at room temperature for 1 h. The plates were washed three times as above to remove unbound second antibody before the addition of H2O2-2,2′-azinobis-(3-ethylbenzthiazolinesulfonic acid) substrate solution (100 μL/well). The enzymatic color reaction in each assay well was monitored for A405nm vs. A655nm at a series of time points using an automated microplate reader (BioRad Benchmark, Hercules, CA). The values in the linear course of the color development were used to plot titration curves for comparing the relative binding affinity of the antibodies to various TnI or TnT constructs. Antibody dilutions at 50% maximum binding were calculated from the titration curves to compare the relative binding affinity. The experiments were done in triplet and repeated one or more times. Statistical analysis was performed using Student’s t test. All values are presented as mean ± SD.
Results
Phylogeny of TnI and TnT isoforms deduced from sequence analysis
Although TnI and TnT are significantly diverged proteins, the close genomic linkage of TnI and TnT genes and their shared feature of variable N-terminal region (Jin et al., 2008) suggest the origination of TnI and TnT by duplication of an ancestor gene. Supporting this hypothesis, phylogenetic analysis of amino acid sequences showed a distant homology between the TnI and TnT gene families (Fig. 3), consistent with their placement in the same sequence family (PF00992) in the Pfam database (http://pfam.janelia.org/; Mistry and Finn, 2007; Finn et al., 2008).
Figure 3. Sequence divergence among TnI and TnT isoforms.
The amino acid sequence divergence is shown in the phylogenetic tree. The evolutionary distances are indicated with a ruler bar for the rate of amino acid substitution.
Sequence analysis of the muscle fiber type isoforms of TnI and TnT further demonstrated that the divergence of the three muscle type-specific isoforms is greater than the divergence of each isoform in different species (Fig. 3), consistent with the previously data from analyzing mammalian and avian TnT isoforms (Jin et al., 1998). Although a [fast skeletal (slow skeletal, cardiac)] evolutionary relationship was indicated by a previous sequence-based study of other muscle proteins (Oota and Saitou, 1999), this order of divergence was unclear from the sequence analysis of the muscle type-specific TnI and TnT isoforms.
Unidirectional immunological cross reactivity of polyclonal anti-TnI and anti-TnT antibodies
To investigate whether the distant relationship between TnI and TnT genes suggested by sequence analysis can be detected in similarities of folded protein structure, we first tested cross-reactivity of an anti-TnI polyclonal antiserum with TnT and that of an anti-TnT polyclonal antiserum to TnI for overall similarity between epitopic structures of the two proteins. The Western blots in Fig. 4A showed that antiserum RATnI raised by chicken fast TnI immunization cross-reacted with mouse fast TnT but not cardiac TnT or slow TnT. RATnI did not show detectable cross-reaction to chicken fast TnT in the Western blots. This could be due to the non-self-recognizing mechanism discussed above. On the other hand, the non-reactivity of RATnI to chicken fast TnT excludes the possibility of TnT contamination in the chicken fast TnI immunogen used in generating the RATnI antiserum. Therefore, the cross reactivity of RATnI to TnT indicates the presence of TnI-like epitope structures in TnT inherited from a common ancestor. This results also supports the notion that TnI and TnT genes were evolved by duplication, other than splitting, of an ancestor gene followed by neofunctionalization.
Figure 4. Detection of TnI-like epitope structures in TnT.
(A) The Western blots showed that polyclonal antibody RATnI raised by immunization with chicken fast skeletal muscle TnI cross reacts with mouse fast TnT but not slow TnT and cardiac TnT that are identified by the anti-cardiac TnT/slow TnT mAb CT3. In contrast, polyclonal antibody RATnT raised using chicken fast skeletal muscle TnT immunization does not have detectable cross-reaction to TnI. The presence of TnI-like epitopic structures in TnT but not vice versa indicates inheritances from a TnI-like ancestor. (B) The Western blot and ELISA affinity titration curves showed that while intact cardiac TnT was not recognized by RATnI, removal of the N-terminal domain significantly increased the affinity to RATnI as shown by the significantly lowered antibody concentration (the higher fold of dilution) to reach 50% maximum binding (from >2×10-3 to 6.40±0.36×10-4, **P<0.001). The detection of TnI-like epitope conformation in cardiac TnT after deletion of the N-terminal region was further demonstrated by Western blotting and ELISA using mAb TnI-1 raised against the conserved C-terminal domain of TnI (antibody dilutions for 50% maximum binding increased from >1×10-3 to 9.93±4.41×10-6, **P<0.001). A cardiac TnT fragment slightly smaller than cTnT-ND produced as a side product during bacterial expression and purification (the very light band in the SDS-gel) had even stronger binding to mAb TnI-1 (indicated in the Western blot by an *), suggesting a further unsuppressed molecular conformation.
Interestingly, the polyclonal antiserum RATnT raised by fast TnT immunization did not show detectable cross-reactivity to TnI (Fig. 4A). This unidirectional immunological cross reactivity between TnI and TnT indicates that the dominant immunogenic epitopes in the TnT immunogen are not present in TnI, suggesting evolutionarily diverged structures that were absent in the common ancestor of TnI and TnT. The RATnI and RATnT antibodies produced with identical immunization procedures exhibited similar overall avidities, indicating comparable immunogenecities of TnT and TnI. Therefore, it is less likely that the dominant immunogenic epitopes in present day TnT had existed in the common ancestor but could have been lost in present day TnI to allow weaker immunogenic epitopes to be effective.
Troponin I is the inhibitory subunit of troponin complex whose primary function is an inhibitor of actin-myosin interactions (Perry, 1999). Consistent with this conserved function, the presence of TnI-like epitope structure in present day TnI and TnT further suggested a hypothesis that the ancestor of TnI and TnT was likely a TnI-like inhibitory protein.
The result that RATnI discriminately cross-reacts to fast TnT than cardiac or slow TnT (Fig. 4A) indicates a shorter immunological distance to fast TnI from fast TnT than that from cardiac or slow TnT, suggesting further hypothesis that the original TnI-TnT gene pair evolved by duplication of the ancestral troponin gene was a fast TnI-TnT-like pair and the present-day fast TnI and fast TnT genes had less neofunctionalization from the original gene pair as compared to that of the other two TnI-TnT gene pairs.
Detection of evolutionarily suppressed TnI-like epitope structures in TnT
To further investigate the above hypotheses beyond static immunological distance, we tested the feasibility of detecting ancestor-like three-dimensional structure states that are suppressed in the evolutionarily diverged present-day protein. The N-terminal variable region of TnT is an evolutionarily additive structure (Jin and Samanez, 2001) that conveys conformational and functional modulations (Wang and Jin, 1998; Jin and Root, 2000; Biesiadecki et al., 2007). Therefore, we made N-terminal truncated TnT (Fig. 2B) to examine the effect of removing this evolutionary addition on the three dimensional structure of the conserved regions of TnT.
The Western blots in Fig. 4B showed no detectable cross-reaction between polyclonal antiserum RATnI with intact cardiac TnT. However, deletion of the N-terminal region of cardiac TnT resulted in significant binding of RATnI. Epitope structures detected by Western blotting have gone through denaturation-renaturation that may affect the folding states. Therefore, we verified the detection of evolutionarily suppressed TnI-like three-dimensional structures in cardiac TnT by ELISA affinity titration under native conditions. The results in Fig. 4B confirmed that deletion of the N-terminal domain of cardiac TnT significantly increased the binding affinity of RATnI.
The C-terminal region of TnI is evolutionarily conserved among isoforms and across species (Jin et al., 2001). We employed mAb TnI-1 against a highly conserved C-terminal epitope of TnI as a probe for ancestor-like three-dimensional structure to further demonstrate the detection of evolutionarily suppressed TnI-like three-dimensional structure inherited in TnT. Consistent with the result from using polyclonal antibody RATnI, the Western blot and ELISA titration curves in Fig. 4B showed a significant cross-reaction of TnI-1 to N-terminal truncated, but not intact, cardiac TnT.
These experiments demonstrated that after the three-dimensional structure of TnT evolved to become distinct from that of TnI as the lack of cross-reactivity of RATnI and TnI-1 with intact cardiac TnT, removal of the evolutionarily additive and conformationally modulating N-terminal region resulted in an ancestor-like conformation as detected by the anti-TnI antibodies. The experimental data indicate the presence of evolutionarily suppressed ancestor-like three-dimensional structures in present-day TnT, which can be detected by removing an evolutionary determinant.
Detection of evolutionarily suppressed fast TnT-like epitope structure in cardiac TnT
Since the sequence phylogeny shown in Fig. 3 did not provide clear information, we further exmined the evolutionary relationship among the muscle type TnI and TnT isoforms using folded structure information, we investigated epitope similarities among the three TnT isoforms. The immunological distance between fast TnT and cardiac or slow TnT was examined using ELISA affinity titration. The results showed that the polyclonal antiserum RATnT raised with fast TnT immunization cross-reacted more strongly to cardiac TnT than that to slow TnT (Fig. 5A). The Western blot in Fig. 5B further showed that mAb T12 raised with fast TnT immunization cross-reacted with cardiac TnT but not slow TnT. The result that fast TnT is related more closely to cardiac TnT than slow TnT suggests that the fast TnI-fast TnT and slow TnI-cardiac TnT gene pairs shared a common ancestor (a fast TnI-fast TnT-like gene pair as suggested above).
Figure 5. Epitope mapping of the evolutionary lineage of TnT isoforms.
(A) The ELISA affinity titration curves showed that polyclonal antiserum RATnT raised against fast TnT cross-reacts to cardiac TnT stronger than that to slow TnT (the antibody dilutions for 50% maximum binding were 2.90±0.10×10-4 and 3.00±0.69×10-3, respectively, *P<0.01), indicating that fast TnT is related closer to cardiac TnT than to slow TnT. (B) The Western blot reactivity and ELISA affinity titration curves demonstrate that mAb T12 raised against fast TnT cross-reacts with cardiac TnT (cTnT) but not slow TnT (sTnT). The affinity of T12 to cardiac TnT was significantly increased after deletion of the N-terminal variable region (the antibody dilutions for 50% maximum binding increased from 2.90±2.76×10-4 to 1.03±0.32×10×10-5, **P<0.001), demonstrating an enhanced fast TnT-like epitope structure. (C) The Western blots and ELISA affinity titration curves demonstrate that mAb 1G9 raised against cardiac TnT cross-reacts with slow TnT but not fast TnT (fTnT). The affinity of 1G9 to slow TnT is significantly increased after deletion of the N-terminal variable region (the antibody dilutions for 50% maximum binding increased from >1×10-2 to 1.02±0.07×10-4, **P<0.001) and further increased in isolated C-terminal T2 domain (50% maximum binding dilution = 5.47±0.06×10-5, **P<0.001), demonstrating an enhanced cardiac TnT-like epitope structure.
This evolutionary lineage was further demonstrated by detecting suppressed fast TnT-like epitope structure in cardiac TnT after removing the evolutionarily additive N-terminal domain. The Western blot in Fig. 5B detected a much increased binding of mAb T12 to N-terminal truncated cardiac TnT in comparison to that to intact cardiac TnT. In contrast, N-terminal truncation of slow TnT did not produce such effect (Fig. 5B). The detection of evolutionarily suppressed fast TnT-like conformational state in cardiac TnT after removing the evolutionary effect of the N-terminal domain was confirmed by ELISA affinity titration under native conditions (Fig. 5B). It is worth noting that TnT is an elongated protein in which the N-terminal region is an extended structure (Cabral-Lilly et al., 1997; Wendt et al., 1997). Therefore, removal of the N-terminal domain enhances antibody affinity to the distant T12 epitope structure by modulating conformation other than unmasking a static structure.
Detection of evolutionarily suppressed cardiac TnT-like epitope structure in slow TnT
To investigate whether the cardiac TnI-slow TnT gene pair had evolved from duplication of the original fast TnI-fast TnT-like gene pair or from a slow TnI-cardiac TnT-like gene pair, the fact that mAb T12 raised by fast TnT immunization cross-reacts to cardiac TnT but not slow TnT supports the later hypothesis. To further test this hypothesis, we experimentally examined the similarity of folded structures of cardiac TnT and slow TnT. mAb 1G9 raised with cardiac TnT immunization exhibited a weak cross-reaction to slow TnT in Western blot but did not react to fast TnT (Fig. 5C), indicating a presence of cardiac TnT-like epitope in slow but not fast TnT.
Showing the effects of serial N-terminal truncations of slow TnT on the conformation of 1G9 epitope in the C-terminal region (Fig. 2B), the Western blots in Fig. 5C detected progressive increases in the affinity of 1G9 when the N-terminal and middle regions were removed. These effects were further demonstrated by ELISA affinity titrations (Fig. 5C). The results indicate that the cardiac TnT-like 1G9 epitope structure has significantly diverged in slow TnT, but the evolutionarily suppressed three-dimensional structure can be more clearly detected when the conformational modulating effect of the N-terminal region was removed. The restoration of cardiac TnT-like three-dimensional structure in slow TnT was further demonstrated by the even higher affinity of 1G9 to the isolated C-terminal domain of slow TnT, indicating its nature as a “default” structure (Fig. 5C).
In contrast, removing the N-terminal and middle regions of fast TnT did not produce detectable binding of mAb 1G9 (data not shown). The absence of 1G9 epitope structure in fast TnT suggest its emergence after the divergence of the ancestors of slow TnI-cardiac TnT and fast TnI-fast TnT gene pairs. Our unpublished observation that the majority of a large of mAbs raised by immunization using cardiac TnT cross-react with slow TnT but not fast TnT also supports this evolutionary lineage.
Discussion
In the present study, we attempted to experimentally determine the evolutionary lineage of proteins through the analysis of folded structures. By using representative antibody epitope probes (summarized in Table 1) to dissect protein structure and conformational states, we experimentally determined the evolutionary lineages in the TnI-TnT gene family (Fig. 6A). In addition to confirming that TnI and TnT are distantly related proteins as suggested by sequence analysis, the results demonstrated that TnI and TnT arose from a TnI-like ancestor protein.
Table 1. Antibody epitope probes.
| Antibody | Immunogen | Polyclonal/Monoclonal | Host Species | Epitope Mapping | Cross-Reaction to TnI | Cross-Reaction to TnT | Affected by TnT N-terminal Deletion |
|---|---|---|---|---|---|---|---|
| RATnI | Chicken Fast TnI | Polyclonal | Rabbit | Multiple | Fast+Slow+Cardiac | Fast | Yes |
| RATnT | Chicken Fast TnT | Polyclonal | Rabbit | Multiple | No | Fast+Slow+Cardiac | N/A |
| TnI-1 | Chicken Fast TnI | Monoclonal | Mouse | C-Terminal | Fast+Slow+Cardiac | Fast | Yes |
| 4D12 | Mouse Cardiac TnI | Monoclonal | Mouse | C-Terminal | Fast+Slow+Cardiac | Fast | N/A |
| T12 | Rabbit Fast TnT | Monoclonal | Mouse | Central | No | Fast+Cardiac | Yes |
| 1G9 | Human Cardiac TnT | Monoclonal | Mouse | C-Terminal | No | Cardiac+Slow | Yes |
| 4H6 | Mouse Cardiac TnI | Monoclonal | Mouse | N/A | Cardiac | N/A | N/A |
| 2C8 | Human Cardiac TnT | Monoclonal | Mouse | N/A | No | Cardiac+Slow+Fast | N/A |
| CT3 | Bovine Cardiac TnT | Monoclonal | Mouse | N/A | No | Cardiac+Slow | N/A |
N/A, not analyzed in the present study.
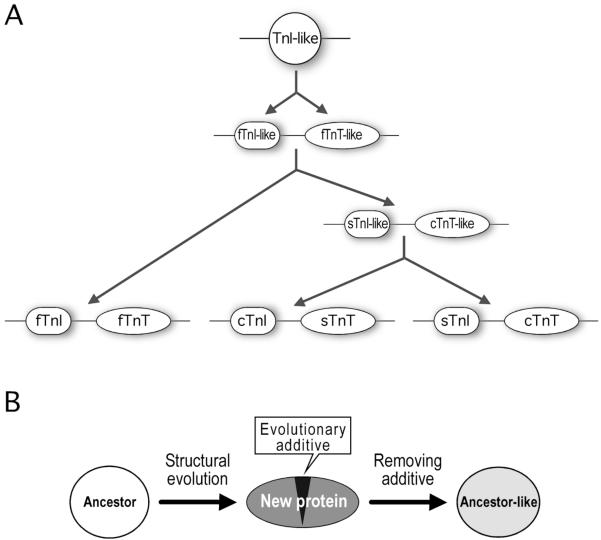
Figure 6. Summary and hypothesis.
(A) The evolution of TnI, TnT and the isoform gene pairs is summarized from data of sequence analysis, immunological distance, and experimental determination of evolutionarily suppressed conformational states. This evolutionary lineage began from a TnI-like ancestor that was duplicated to form a fast TnI-fast TnT-like gene pair. A later duplication event resulted in the emergences of a slow TnI-cardiac TnT-like gene pair that was further duplicated to form the present-day slow TnI-cardiac TnT and cardiac TnI-slow TnT gene pairs. (B) A hypothesis is proposed to suggest that the evolution of a protein three-dimensional structure could result from addition and/or alteration of a critical structure that modulates the global conformation and function of the protein. Ancestor-like three-dimensional structures may be suppressed in the new protein and become detectable after removing the modulatory effect.
We further detected inheritances in the slow TnI-cardiac TnT gene pair from the fast TnI-fast TnT-like ancestor pair and in cardiac TnI-slow TnT pair from the slow TnI-cardiac TnT-like ancestor gene pair. This pattern is consistent with the [fast skeletal (slow skeletal, cardiac)] evolutionary relationship indicated by a previous sequence analysis of other muscle proteins (Oota and Saitou, 1999). Among the three subunits of troponin (Fig. 1B), troponin C (TnC) is a member of the calmodulin family of Ca2+ binding proteins. Different from TnI and TnT that have evolved into three isoforms for the three muscle types of vertebrates, TnC is present in only two isoforms: fast TnC (Gahlmann and Kedes, 1990) and slow-cardiac TnC (Parmaceck and Leiden, 1989). The undifferentiated utilization of the same TnC isoform in cardiac and slow skeletal muscles supports a hypothesis that the duplication of the cardiac and slow TnI-TnT gene pairs was a relatively recent event.
The retention and evolution of TnI and TnT from duplication of a common ancestor provide an excellent example for the initial role of neofunctionalization since the linked expression of the duplicated genes precludes the role of subfunctionalization. On the other hand, the later duplication and retention of the muscle type-specific TnI-TnT gene pairs represent typical subfunctionalizations (Li et al., 2005).
Beyond static state immunological distance, the effect of removing an evolutionarily additive structure from TnT on the detection of evolutionarily suppressed TnI-like epitopic structures inherited from the TnI-like ancestor is consistent with the role of the N-terminal variable region of TnT as a conformational modulator (Wang and Jin, 1998; Jin and Root, 2000; Biesiadecki et al., 2007). In contrast to the commonly seen destructive effects in which deleting a portion of a protein destroys the integrity of antibody epitopes, the dissection of TnT proteins based on their structural and functional domains demonstrated three-dimensional structures showing ancestor-like conformational states. This reversal effect indicates that ancestor-like structures may be retained in a suppressed state in present-day proteins and be exhibited when the evolutionary modulating determinant(s) is removed. The N-terminal region of TnT is a conformational modulator and, in a broad sense, acts as a molecular switch. However, the N-terminal region of TnT does not simply act in activating and inactivating TnT function. Both intact and N-terminal truncated TnTs are active in regulating muscle contraction and TnI does not represent a different functional state of TnT. Therefore, the detection of TnI-like epitopes in TnT after removing the evolutionarily additive N-terminal modulator indicates structural changes reflecting a reversal of structural evolution to an ancestor-like state.
In addition to providing experimental evidence for the evolution of TnI, TnT and their isoforms, the detection of ancestor-like epitopic structures in present-day proteins by removing evolutionarily additive structures has a two-fold significance. First, it suggests a hypothesis that while genetic variations produce gradual changes in the amino acid sequence of proteins to provide the raw material for natural selection and molecular evolution, functional divergence and fitness value are mainly based on three-dimensional structural changes that could be a global conformational change produced by critical sequence changes in a modulator domain. This model may also explain quantum functional changes and leaps in an evolutionary lineage. Our data suggest a critical role of “evolutionary determinants” that are new structures capable of tuning the global conformation and function during protein evolution.
Second, our findings suggest that the evolution of protein three-dimensional structure is under a selection in which a trait would be fixed and maintained depending on the final functional outcome. Therefore, present-day proteins could retain potentials of conferring ancestor-like three-dimensional structures that are suppressed by conformational modulations occurred during evolution. When the stabilizing effect of the conformational modulator is removed, the suppressed ancestor-like three-dimensional structures will be exhibited (Fig. 6B). This hypothesis may explain the molecular basis of atavism, in which a mutation that causes the loss of evolutionary modulation in a protein would result in the expression of an ancestral trait in contrast to a radical new trait.
Although the post genome era databases allow extensive sequence analyses for detecting evolutionary relationship of proteins or their functional domains, protein three-dimensional structure analysis has a special value. Ancestral information may be retained in a present-day protein as an ancestor-like alternative conformation that might be more common for one protein than another derived from the same ancestor. Our approach to experimentally detecting evolutionarily suppressed ancestor-like alternative conformations in present-day proteins makes it feasible to dig “molecular fossils” to study the origin of present-day protein structures. While the protein three-dimensional structure database is rapidly growing, the precise correlation between sequence and folded structure remains to be established. At the present, an effective and practical method to compare protein three-dimensional structure differences is extremely valuable for the study of protein evolution. Our data have demonstrated that ancestor-like conformational states can be detected with antibodies against epitopes that represent the ancestor-like alternative conformation. In addition, knowing that an ancestor-like alternative conformation is retained in a protein structure will guide the search for the same information “hidden” in the amino acid sequences.
The sensitive and rapid throughput ELISA epitope analysis (Wang and Jin, 1998; Jin et al., 2007) can readily detect similarities, differences and especially changes in protein conformation. Although the binary comparisons of epitope affinity titration curves were not able to obtain exact evolutionary distance between protein isoforms, our data demonstrated that this approach is powerful not only in detecting protein allosteric changes during a switch between two active states (Jin et al., 2007) but also in evaluating three-dimensional structure changes in geological time scale to identify fossils of molecular evolution. Since many mAbs as well as polyclonal antibodies can detect protein conformational changes (Ogut and Jin, 1996; Wang and Jin, 1998; Jin et al., 2000a; 2000b; Jin and Root, 2000; Biesiadecki et al, 2007; Jin et al., 2007), application of this approach can be readily explored on many proteins of interest using existing antibodies.
In addition to strengthening the sequence evidence that TnI and TnT arose from gene duplication, our study provided data for the presence of alternative conformational components, or conformers, shared by TnI and TnT, which might have been retained for a functional benefit. It would be interesting to investigate where these components are and the role that they play. For example, the C-terminal domain of TnI is the most conserved region in the TnI polypeptide chain, which undergoes conformational changes in the troponin complex when Ca2+ binds to TnC (Jin et al., 2001). Three-dimensional structure reconstruction studies suggested a possible role for the C-terminal domain of TnI in trapping tropomyosin during myofilament regulation (Pirani et al., 2006). Since the N-terminal region of TnT may be removed by restricted proteolysis during acute functional adaptation (Zhang et al., 2006; Feng et al., 2008), the presence and exhibition of TnI C-terminus-like structure inherited in TnT intrigues future investigations of the structural basis of TnI and TnT for their role in the regulation of muscle contraction. To explore the functional significance of ancestor-like conformational states inherited in present-day proteins will test a novel structure-function relationship of proteins.
In perspective, our study reports an experimental approach to demonstrate that Evolution is Data. Investigation of the evolution of protein three-dimensional structures opens a door to the study of molecular paleontology despite the absence of fossil materials. In addition to identifying structural modifications that were critical to the emerging of present-day proteins, to understand this novel mode of evolution and the role of conformational modulator sites that determined evolutionary divergence of a protein and the consequence of their manipulation will help to predict the structural and functional potentials of the protein and guide the design of structural modifications and the search for effective drug targets. Mutations in such evolutionary determining structure of a protein might have high potency to modify phenotypes and cause diseases. The potential of a protein to exhibit suppressed ancestor-like structures suggests a hypothesis that the diverged molecular conformation would be more likely to fall back an ancestor-like state than to produce a radical new conformation, which calls for further experimental investigation.
Acknowledgments
We sincerely thank Dr. M. Moazzem Hossain for the preparation of mAbs 4D12 and 1G9, Dr. Jim Lin, University of Iowa, for providing mAb T12, and Dr. Stacia Sower, University of New Hampshire, for sharing the hagfish muscle samples. This study was supported by grants from the National Institutes of Health AR048816 and HL078773 and from the National Aeronautic and Space Administration NNA04Ck26G to J-PJ.
Footnotes
- mAb
- monoclonal antibody
- BSA
- bovine serum albumin
- ELISA
- enzyme-linked immunosorbant assay
- PAGE
- polyacrylamide gel electrophoresis
- PCR
- polymerase chain reaction
- TBS
- Tris-buffered saline
- TnC
- troponin C
- TnI
- troponin I
- TnT
- troponin T
References
- Barbato JC, Huang QQ, Hossain MM, Bond M, Jin JP. Proteolytic N-terminal truncation of cardiac troponin I enhances ventricular diastolic function. J. Biol. Chem. 2005;280:6602–6609. doi: 10.1074/jbc.M408525200. [DOI] [PubMed] [Google Scholar]
- Barton PJ, Townsend PJ, Brand NJ, Yacoub MH. Localization of the fast skeletal muscle troponin I gene (TNNI2) to 11p15.5: genes for troponin I and T are organized in pairs. Ann. Human. Genet. 1997;61:519–523. doi: 10.1046/j.1469-1809.1997.6160519.x. [DOI] [PubMed] [Google Scholar]
- Bautista JA, Zhang Z, Jin JP. Proteolytic sensitivity of cardiac troponin I regulated by conformational modification. Biophys. J. 2007:474a. Abstract: 2263-Pos. [Google Scholar]
- Benner SA, Caraco MD, Thomson JM, Gaucher EA. Planetary biology--paleontological, geological, and molecular histories of life. Science. 2002;296:864–868. doi: 10.1126/science.1069863. [DOI] [PubMed] [Google Scholar]
- Biesiadecki BJ, Schneider KL, Yu ZB, Chong SM, Jin JP. An R111C polymorphism in wild turkey cardiac troponin I accompanying the dilated cardiomyopathy-related abnormal splicing variant of cardiac troponin T with potentially compensatory effects. J. Biol. Chem. 2004;279:13825–13832. doi: 10.1074/jbc.M314225200. [DOI] [PubMed] [Google Scholar]
- Biesiadecki BJ, Chong SM, Nosek TM, Jin JP. Troponin T core structure and the regulatory NH2-terminal variable region. Biochemistry. 2007;46:1368–1379. doi: 10.1021/bi061949m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell TL, Sibanda BL, Montalvao RW, Brewerton S, Chelliah V, Worth CL, Harmer NJ, Davies O, Burke D. Structural biology and bioinformatics in drug design: opportunities and challenges for target identification and lead discovery. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006;361:413–423. doi: 10.1098/rstb.2005.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral-Lilly D, Tobacman LS, Mehegan JP, Cohen C. Molecular polarity in tropomyosin-troponin T co-crystals. Biophys. J. 1997;73:1763–1770. doi: 10.1016/S0006-3495(97)78206-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ashton B, Cheung M, Heled J, Kearse M, Moir R, Stones-Havas S, Thierer T, Wilson A. Geneious v4.0. 2008 Available from http://www.geneious.com/
- Du J, Liu J, Feng HZ, Hossain MM, Gobara N, Zhang C, Li Y, Jean-Charles PY, Jin JP, Huang XP. Impaired relaxation is the main manifestation in transgenic mice expressing a restrictive cardiomyopathy mutation, R193H, in cardiac TnI. Am J Physiol Heart Circ Physiol. 2008;294:H2604–2613. doi: 10.1152/ajpheart.91506.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng HZ, Biesiadecki BJ, Yu ZB, Hossain MM, Jin JP. Restricted N-terminal truncation of cardiac troponin T: A novel mechanism for functional adaptation to energetic crisis. J. Physiol. (London) 2008;586:3537–3550. doi: 10.1113/jphysiol.2008.153577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fock U, Hinssen H. Nebulin is a thin filament protein of the cardiac muscle of the agnathans. J. Muscle Res. Cell Motil. 2002;23:205–213. doi: 10.1023/a:1020909902462. [DOI] [PubMed] [Google Scholar]
- Gahlmann R, Kedes L. Cloning, structural analysis, and expression of the human fast twitch skeletal muscle troponin C gene. J. Biol. Chem. 1990;265:12520–12528. [PubMed] [Google Scholar]
- Heeley DH, Golosinska K, Smillie LB. The effects of troponin T fragments T1 and T2 on the binding of nonpolymerizable tropomyosin to F-actin in the presence and absence of troponin I and troponin C. J. Biol. Chem. 1987;262:9971–9978. [PubMed] [Google Scholar]
- Huang QQ, Jin JP. Preserved close linkage between the genes encoding troponin I and troponin T, reflecting an evolution of adapter proteins coupling the Ca(2+) signaling of contractility. J. Mol. Evol. 1999;49:780–788. doi: 10.1007/pl00006600. [DOI] [PubMed] [Google Scholar]
- Jin JP. Cloned rat cardiac titin class I and class II motifs. Expression, purification, characterization, and interaction with F-actin. J. Biol. Chem. 1995;270:6908–6916. [PubMed] [Google Scholar]
- Jin JP. Alternative RNA splicing-generated cardiac troponin T isoform switching: a non-heart-restricted genetic programming synchronized in developing cardiac and skeletal muscles. Biochem. Biophys. Res. Commun. 1996;225:883–889. doi: 10.1006/bbrc.1996.1267. [DOI] [PubMed] [Google Scholar]
- Jin JP, Chen A, Huang QQ. Three alternatively spliced mouse slow skeletal muscle troponin T isoforms: Conserved primary structure and regulated expression during postnatal development. Gene. 1998;214:121–129. doi: 10.1016/s0378-1119(98)00214-5. [DOI] [PubMed] [Google Scholar]
- Jin JP, Root DD. Modulation of troponin T molecular conformation and flexibility by metal ion binding to the NH2-terminal variable region. Biochemistry. 2000;39:11702–11713. doi: 10.1021/bi9927437. [DOI] [PubMed] [Google Scholar]
- Jin JP, Chen A, Ogut O, Huang QQ. Conformational modulation of slow skeletal muscle troponin T by an NH(2)-terminal metal-binding extension. Am J. Physiol.:Cell Physiol. 2000a;279:1067–1077. doi: 10.1152/ajpcell.2000.279.4.C1067. [DOI] [PubMed] [Google Scholar]
- Jin JP, Walsh MP, Sutherland C, Chen W. A role for serine-175 in modulating the molecular conformation of calponin. Biochem. J. 2000b;350:579–588. [PMC free article] [PubMed] [Google Scholar]
- Jin JP, Samanez R. Evolution of a metal-binding cluster in the NH(2)-terminal variable region of avian fast skeletal muscle troponin T: functional divergence on the basis of tolerance to structural drifting. J. Mol. Evol. 2001;52:103–116. doi: 10.1007/s002390010139. [DOI] [PubMed] [Google Scholar]
- Jin JP, Yang F, Yu Z, Ruse C, Bond M, Chen A. The highly conserved COOH terminus of troponin I forms a Ca2+-modulated allosteric domain in the troponin complex. Biochemistry. 2001;40:2623–2631. doi: 10.1021/bi002423j. [DOI] [PubMed] [Google Scholar]
- Jin JP, Brotto MA, Hossain MM, Huang QQ, Brotto LS, Nosek TM, Morton DH, Crawford TO. Truncation by Glu180 nonsense mutation results in complete loss of slow skeletal muscle troponin T in a lethal nemaline myopathy. J. Biol. Chem. 2003;278:26159–26165. doi: 10.1074/jbc.M303469200. [DOI] [PubMed] [Google Scholar]
- Jin JP, Chong SM, Hossain MM. Microtiter plate monoclonal antibody epitope analysis of Ca2+- and Mg2+-induced conformational changes in troponin C. Arch. Biochem. Biophys. 2007;466:1–7. doi: 10.1016/j.abb.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminogawa S, Shimoda M, Kurisaki J, Yamauchi K. Application of a monoclonal antibody to a comparative study of alpha-lactalbumins from various species. J. Dairy Sci. 1989;72:1124–1129. doi: 10.3168/jds.s0022-0302(89)79214-6. [DOI] [PubMed] [Google Scholar]
- Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li WH, Tanimura M. The molecular clock runs more slowly in man than in apes and monkeys. Nature. 1987;326:93–96. doi: 10.1038/326093a0. [DOI] [PubMed] [Google Scholar]
- Li WH, Yang J, Gu X. Expression divergence between duplicate genes. Trends Genet. 2005;21:602–607. doi: 10.1016/j.tig.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Lin JJ, Matsumura F, Yamashiro-Matsumura S. Tropomyosin-enriched and alpha-actinin-enriched microfilaments isolated from chicken embryo fibroblasts by monoclonal antibodies. J. Cell Biol. 1984;98:116–127. doi: 10.1083/jcb.98.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EJ, Cohen AB. Use of antibodies in the study of protein structure and function in lung diseases. Am. J. Physiol. 1991;260:L1–12. doi: 10.1152/ajplung.1991.260.2.L1. [DOI] [PubMed] [Google Scholar]
- Mistry J, Finn R. Pfam: a domain-centric method for analyzing proteins and proteomes. Methods Mol Biol. 2007;396:43–58. doi: 10.1007/978-1-59745-515-2_4. [DOI] [PubMed] [Google Scholar]
- Ogut O, Jin JP. Expression, zinc-affinity purification, and characterization of a novel metal-binding cluster in troponin T: metal-stabilized alpha-helical structure and effects of the NH2-terminal variable region on the conformation of intact troponin T and its association with tropomyosin. Biochemistry. 1996;35:16581–16590. doi: 10.1021/bi961712y. [DOI] [PubMed] [Google Scholar]
- OOta S, Saitou N. Phylogenetic relationship of muscle tissues deduced from superimposition of gene trees. Mol Biol Evol. 1999;16:856–867. doi: 10.1093/oxfordjournals.molbev.a026170. [DOI] [PubMed] [Google Scholar]
- Parmaceck MS, Leiden JM. Structure and expression of the murine slow/cardiac troponin C gene. J. Biol. Chem. 1989;264:13217–13225. [PubMed] [Google Scholar]
- Perry SV. Troponin T: genetics, properties and function. J. Muscle Res. Cell Molti. 1998;19:575–602. doi: 10.1023/a:1005397501968. [DOI] [PubMed] [Google Scholar]
- Perry SV. Troponin I: inhibitor or facilitator. Mol. Cell. Biochem. 1999;190:9–32. [PubMed] [Google Scholar]
- Pirani A, Vinogradova MV, Curmi PM, King WA, Fletterick RJ, Craig R, Tobacman LS, Xu C, Hatch V, Lehman W. An atomic model of the thin filament in the relaxed and Ca2+-activated states. J. Mol. Biol. 2006;357:707–717. doi: 10.1016/j.jmb.2005.12.050. [DOI] [PubMed] [Google Scholar]
- Prager EM. The sequence-immunology correlation revisited: data for cetacean myoglobins and mammalian lysozymes. J. Mol. Evol. 1993;37:408–416. doi: 10.1007/BF00178870. [DOI] [PubMed] [Google Scholar]
- Raes J, Van de Peer Y. Gene duplication, the evolution of novel gene functions, and detecting functional divergence of duplicates in silico. Appl. Bioinformatics. 2003;2:91–101. [PubMed] [Google Scholar]
- Tiso N, Rampoldi L, Pallavicini A, Zimbello R, Pandolfo D, Valle G, Lanfranchi G, Danieli A. Fine mapping of five human skeletal muscle genes: alpha-tropomyosin, beta-tropomyosin, troponin-I slow-twitch, troponin-I fast-twitch, and troponin-C fast. Biochem. Biophys. Res. Commun. 1997;230:347–350. doi: 10.1006/bbrc.1996.5958. [DOI] [PubMed] [Google Scholar]
- Wang J, Jin JP. Conformational modulation of troponin T by configuration of the NH2-terminal variable region and functional effects. Biochemistry. 1998;37:14519–14528. doi: 10.1021/bi9812322. [DOI] [PubMed] [Google Scholar]
- Wendt T, Guenebaut V, Leonard KR. Structure of the Lethocerus troponin-tropomyosin complex as determined by electron microscopy. J. Struct. Biol. 1997;118:1–8. doi: 10.1006/jsbi.1996.3834. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Biesiadecki BJ, Jin JP. Selective deletion of the NH2-terminal variable region of cardiac troponin T in ischemia reperfusion by myofibril-associated mu-calpain cleavage. Biochemistry. 2006;45:11681–11694. doi: 10.1021/bi060273s. [DOI] [PMC free article] [PubMed] [Google Scholar]